Abstract
Purpose
In women under the age of 40, primary ovarian insufficiency (POI) is a devastating diagnosis with significant prevalence of 1–4% (Rajkovic and Pangas, Semin Reprod Med. 35(3):231–40, 2017). POI is characterized by amenorrhea with elevated levels of follicle stimulating hormone (FSH) and reduced estrogen levels, mimicking the menopausal state. Genetic determinants account for just over 10% of POI cases, yet determining whether particular single nucleotide polymorphisms (SNPs) are pathogenic is challenging.
Methods
We performed exome sequencing on a cohort of women with POI. CRISPR mutagenesis was employed to create a mutation in a conserved amino acid in the nematode protein. Functional relevance was assessed by analysis of bivalents and aberrant DNA morphologies in diakinesis nuclei.
Results
We identified a nonsynonymous c.C1051G; p.R351G variant, in a conserved region of the MSH5 protein. Mutation of this conserved amino acid in the C. elegans homolog, msh-5, revealed defective crossover outcomes in the homozygous and hemizygous states.
Conclusions
These studies further implicate MSH5 as a POI gene and c.C1051G; p.R351G variant as likely playing a functional role in mammalian meiosis. This approach also highlights the ability of model organisms, such as C. elegans, to rapidly and inexpensively identify alleles of interest for further studies in mammalian models.
Keywords: POI, MSH5, Crossover, Infertility, C. elegans, Meiosis
Background
Over the last several decades, as women have delayed childbirth into their thirties, the diagnosis of primary ovarian insufficiency (POI) has increased. POI is defined as loss of ovarian follicles function with amenorrhea prior to the age of 40 [1]. A clinically heterogenous disorder, POI can be caused by infection, chronic disease, X-linked chromosomal abnormalities, and environment. Single gene defects are associated with > 10% of POI cases. More than forty genes (reviewed in [2, 3]), functioning in various oogenic processes, have been implicated through both traditional cytogenetic and mapping studies and more recently through genome-wide associate studies (GWAS).
The age of menopause is directly related to the size of ovarian reserve: the lower the size of the initial oocyte pool, the earlier the onset of menopause. Accordingly, genes involved in early steps of oogenesis, such as Nanos 3, Solh1, and Sohlh2, have been implicated in POI [4–8]. During embryogenesis, after migration into the genital ridge, female germ cells divide to produce upwards of a million progenitor cells that enter meiosis, undergoing the events of double-strand break formation, pairing, synapsis, and homologous recombination-mediate repair to form crossovers between homologous chromosomes. Errors in meiotic processes lead to checkpoint activation and oocyte loss [9–11], decreasing the oocyte anlage. This is in addition to the process of atresia which eliminates over 90% oocytes prior to birth, leaving a pool of oocytes that will be continuously lost throughout childhood and through reproductive life. While meiotic crossover genes are prime candidates for POI-associated loci [2], functional data supporting this role is only available for a handful of these genes. These include the crossover commitment factors MSH4 and MSH5 [12, 13], the cohesin STAG3 [14], the synaptonemal complex protein SYCE1 [15, 16], and, more recently, the nuclease EXO1 and the recombinase RAD51 [17].
While exome sequencing studies have increased the number of putative POI genes over the last decade, determining whether specific variants are pathogenic is particularly challenging when rare variants cause missense mutations rather than the premature stop or frameshift mutations which would be easily classified as pathogenic. In the current study, we identified the c.C1051G;p.R351G variant in a conserved position of MSH5. To test the pathogenicity of this allele, we took advantage of the genetic model system, the nematode C. elegans, to create this variant and test its function in vivo. Importantly, C. elegans shares many of the features of meiosis with humans, including a critical role for the MSH4/MSH5 dimer in specifying crossover sites [18, 19]. We show here that hemizygosity for the MSH5 mutation causes crossover defects implicating arginine 351 as critical for MSH5 function.
Materials and methods
Patient recruitment
We investigated pathogenic variants in a cohort of 173 participants diagnosed with POI. The study was approved by the Institutional Review Board of the University of Pittsburgh (PRO09080427).
Whole exome sequencing
We conducted exome sequencing on the proband from a cohort of women with POI recruited at the University of Pittsburgh. Exons and splice sites were captured with the Agilent SureSelectXT Human Exon V4 + UTRs Kit, and 2 × 100 bp paired-end exome sequencing was performed on an Illumina HiSeq 2500. We prepared reads for analysis with Cutadapt version 1.2.1 to remove the adapters and with the FASTX-Toolkit version 0.0.13.2 to trim the first 5 bp at the 5’ end of reads. We aligned data to UCSC Genome Browser hg19 by using Burrows-Wheeler Aligner version 0.7.3a MEM (maximal exact match). Local realignment around insertions and deletions, recalibration of read base quality, and variant calling were conducted with Genome Analysis Toolkit (GATK) version 2.6–5. GATK Haplotype Caller was used for calling variants.
CRISPR editing of C. elegans
Alignments between the C. elegans, mouse, and human MSH5 homologs were performed using ClustalW [20] and the relevant region of homology is shown in Fig. 1. R351 corresponds to R404 in the worm protein. Nearby Cas9 recognition sites were identified using the CRISPOR tool at mit.crispor.edu. Two cuts were made to remove the wild-type nucleotides and allow repair with a ssDNA oligonucleotide (IDT). Sequences of CRISPR reagents:
Fig. 1.
Identification of putative pathogenic allele in POI patient. A Pedigree of POI patient. B CLUSTAL multi-sequence alignment of the C. elegans, human, and mouse MSH5 proteins showing the conserved arginine that is mutated in the human patient and the CRISPR-edited worms (grey highlight)
msh-5 sgRNA 1: rGrArArUrArUrUrArUrCuAuGrGrCrArGrArCrArCrCrGrG.
msh-5 sgRNA 2: rGrArArCrArGrCrUrCrArGrCrUrCrArUrUrCrArUrUrGrG.
Repair template: 5’-TTC ATT TTT GGC CGA TTT CAG AGT GTA TTC CAA AAA TTT CAA AGT GGA ACC GCC CAG CTT ATC CAC TGG GAG TGC TTC GTC TCG ACA GTC AAC GCG CTT GTT GAA ATC TTG AAT ATT ATC GGA CAG ACA CCA GTA AAT TCA AAT AAT TAA TTT TTA TAA AAA AGC GAG GTT TCA AAT AAT TT-3’.
Analysis of diakinesis chromosomes
Whole mount fixation of 1-day-old adults was performed using Carnoy’s Fixation (six parts acetic acid, 3 parts 100% ethanol, 1 part chloroform). Worms were collected in 5 µl of 1 × M9, rinsed once, and 10 µl Carnoy’s fixative was added. Just prior to drying, 50 µl of 1 × phosphate-buffered saline (PBS) with DAPI (4’,6-diamidino-2-phenylindole) was added for 15 min in a humidity chamber. After removal of the PBS + DAPI solution, samples were mounted in Prolong Gold with DAPI (Invitrogen) and cured overnight prior to imaging. Imaging was performed on a Nikon A1r confocal microscope as 0.2 µm Z-stacks using a 60 × oil immersion objective. Reconstruction of images for quantifying diakinesis figures was performed using Volocity 3D imaging software (Quorum Technologies).
C. elegans strains and growth
C. elegans were grown on NGM media [21] seeded with OP50 bacteria. Strains utilized in this study were CB128: dpy-10(e128) II, AV115: msh-5(me23) IV/nT1 [unc?9n745) let-? qIs50] (IV;V), QP989: msh-5(ea36), QP1790: dpy-1(e1) II; msh-5(ea36) IV. To generate transheterozygotes, msh-5(ea23)/nT1 was heat-shocked to generate males which were backcrossed to maintain a male-producing stock. dpy-1; msh-5(ea36) hermaphrodites were crossed to msh-5(ea36)/nT1 males and non-Dpy, non-GFP cross-progeny were collected for analysis. These were compared to control non-Dpy, non-GFP heterozygotes from dpy-10 hermaphrodites crossed to msh-5(me23)/nT1 males and to dpy-1; msh-5(ea36) × N2 males.
Results
Novel missense variant in MSH5 identified in POI patient
We conducted exome sequencing on a cohort of women with POI recruited at the University of Pittsburgh. PPOF22 is a white American who was diagnosed at 18 years of age with primary amenorrhea and experienced normal puberty. There were no significant structural abnormalities noted on her physical exam. She did not have a history of cancer, previous chemo or radio therapy, and no history of pelvic or ovarian surgeries. Ultrasound examination showed small ovaries and absent follicles. She had elevated FSH level (85 mIU/ml), LH level (72.8 mIU/ml), undetectable AMH, 46, XX, negative FMR1, negative chromosomal microarray, and low estradiol level (15 pg/ml).
There was no evidence of autoimmune disease: both anti-thyroid and anti-adrenal gland antibodies were negative in the affected individual. She was diagnosed with nonsyndromic POI. Her mother had menarche at 13 years of age and menopause at 48 years of age. ES was performed on her and her parents to identify nucleotide variants that may account for the idiopathic and nonsyndromic hypergonadotropic hypogonadism. Variants were filtered for quality and significance as previously reported [22]. We filtered for nonsynonymous variants (in exons or splice sites) that are presumed to be damaging, with a minor allele frequency of < 5% and assumed recessive inheritance given the lack of other family members with POI. A homozygous variant in MSH5, c.C1051G; p.R351G, was identified, and inherited from each parent (Fig. 1A). Alignments of the MSH5 protein identified R351 as a highly conserved amino acid across from worms to humans (Fig. 1B). R351G variant was previously deposited as pathogenic in ClinVar by the Center for Reproductive Medicine, Shandong Provincial Hospital. However, no functional evidence for the pathogenicity of this variant was provided. Moreover, this particular variant was present at a relatively high allele frequency of 1.4% in the gnomAD database [23]. Given the high degree of conservation of the R351 amino acid among multiple species, as well as PPOF22’s and Shandong Center’s association with POI, we decided to study its functional significance in C. elegans.
MSH-5(R404G) displays meiotic defects in C. elegans
Using CRISPR gene editing, we created mutant worms containing the R404 mutation, which is the amino acid corresponding to human R351, and designated the allele msh-5(ea36). Unlike the canonical null allele, msh-5(me23) which produces only rare viable offspring and therefore needs to be maintained as a heterozygous, balanced strain [18], msh-5(ea36) could be maintained as a homozygote and was highly fertile at both 20 °C and the elevated growth temperature of 25 °C. Furthermore, the strain did not segregate males or a large fraction of dead eggs (not shown), both of which result from nondisjunction of the X chromosome and autosomes, respectively. Therefore, we inferred that ea36 does not substantially abrogate crossover formation.
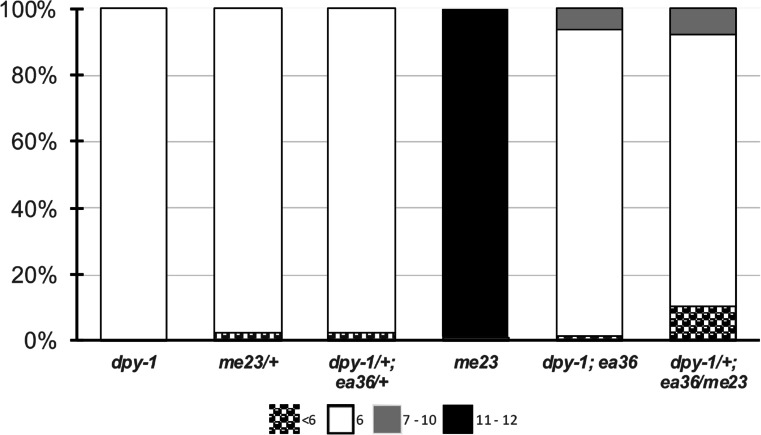
To directly assess the integrity of crossover formation in these animals, we fixed 1-day-old adults and assessed crossover formation by counting the number of DAPI staining figures in diakinesis-arrested (equivalent to dictyate) oocytes. In wild type, 6 DAPI bodies are observed in diakinesis since the 6 pairs of chromosomes achieve crossovers and form bivalents. In msh-5(me23), the impairment of crossover designation prevents bivalent formation and 12 univalent chromosomes are instead observed in almost all nuclei (Fig. 2). In a small subset of nuclei, fewer than 12 DAPI are seen (10 or 11), either because 2 univalents abut one another or because 2 chromosomes have fused as a result of non-homologous recombination (HR)-mediated repair. By contrast, in msh-5(ea36) mutant animals, 6 bivalents were observed in the vast majority of diakinesis oocytes (Fig. 2). However, unlike in wild type, 6% of oocytes contained 7 or 8 DAPI bodies, suggesting that one or two homolog pairs did not receive a crossover. The presence of univalent in msh-5(ea36) suggests that the R404G mutation mildly impairs MSH-5 function.
Fig. 2.
msh-5(ea36) is impaired in crossover formation. Shown are the numbers of DAPI+ bodies in diakinesis oocytes for the respective genotypes. Control dpy-1 animals had only the expected 6 DAPI+ bodies. msh-5 heterozygous worms similarly had predominantly 6 DAPI bodies, whereas the msh-5(me23) null worms had 12 univalents in almost all nuclei. msh-5(ea36) mutant animals have increased numbers of univalents and chromosome fusions.
To more rigorously test ea36 functionality, we wanted to determine how much of wild-type function that it confers in the hemizygous state (mutation/null). To this end, we first confirmed that msh-5 mutation is not haploinsufficient by observing diakinesis figures in dpy-10/ + ; + /msh-5(me23) hermaphrodites. As shown in Fig. 2, no univalent chromosomes were observed in animals heterozygous for the msh-5 null allele. Therefore, we infer that a single wild-type copy of the msh-5 locus is able to confer full meiotic functions at the normal growth temperature of 20 °C.
We next generated animals transheterozygous for the null and ea36 alleles (complete genotype: dpy-1/ + ; msh-5(me23)/msh-5(ea36)). These hemizygous animals showed worse meiotic crossover outcomes, with 18% of diakinesis oocytes exhibiting defects. Nearly 9% of diakinesis nuclei contained univalent chromosomes, the vast majority of these with a single pair of univalents, although 2 and 3 pairs were also observed. We also noted a substantial increase in nuclei with fewer than 6 DAPI bodies (8/117 with 5 and 3/117 with 4 DAPI bodies). Five or fewer DAPI bodies may arise from the close juxtaposition of bivalents or from the fusion of two or more homolog pairs. While 5 DAPI figures were seen rarely in wild type or in heterozygous controls (Fig. 2), the prevalence in the msh-5(ea36)/msh-5(me23) animals suggests that partial abrogation of MSH-5 function may lead to chromosome fusions. No adverse phenotypes were observed in dpy-1/ + ; msh-5(ea36)/ + heterozygotes, confirming that the dpy-1 mutation in the strain did not contribute to the phenotype. The diakinesis nuclei with univalents and/or fusion chromosomes in the msh-5(ea36)/msh-5(me23) hemizygous animals strongly supports the conclusion that msh-5(ea36) is impaired in function.
Discussion
POI affects 1–4% of women in the USA and has significant consequences for fertility as well as other adult disorders, including increased morbidity and mortality [1].
Using exome sequencing analysis of POI patients, we were able to identify a variant in the MSH-5 gene that has been previously assigned as pathogenic without functional evidence. The mutated arginine lies is a highly conserved region of the protein between mouse and humans and a well-conserved domain between worms and humans. Arginine 351 is conserved between worms, mice, and humans suggesting possible conservation in function. The variant is present in gnomad database at 1.4% allele frequency. Although not as rare as other pathogenic variants identified with POI, common variants, such as nonsynonymous MCM8 rs16991615 variant, have significant impact on reproductive life span and age of menopause (PMID: 24,493,794). To determine if this variant of interest might be pathogenic, we mutated it in the worm and analyzed meiotic crossover outcomes. The appearance of non-bivalent chromosomes in the arrested diakinesis nuclei in both the homozygous and hemizygous ea36 mutant animals indicated that R404 is required for full MSH-5 function. By analogy, these data also support a mutagenic role of R351G in the human protein.
R351 is a surface residue situated in an alpha helical bundle above the ATP binding pocket of the MSH4-MSH5 heterodimer. While the helices in this region are conserved from across phyla, no function-blocking mutations have been identified in these domains. However, the human MSH5 protein has at least 13 additional putative interaction partners (https://thebiogrid.org/110576), a number of which, e.g., MLH1 and MLH3, FANCA, and BRCA1, contribute to crossover recombination [22–27]. Thus, it is possible that R351G interferes with these binding interfaces, conferring a partial recombination defect.
With the advent of CRISPR, the use of model organisms to test variants of unknown interest from exome and genome sequencing studies has gained traction (e.g., [28–30]) due to the rapidity, ease, and low cost of creating mutations and analyzing mutant phenotypes. We show herein the ease of testing conserved meiotic functions in the nematode C. elegans, where changes in the morphology and number of chromosome masses in diakinesis nuclei serve as a readout for proper crossover formation [31]. Given the growing list of meiotic genes that are implicated in premature ovarian failure and the high conservation between the repair machinery involved in homologous recombination repair, the worm provides a tractable and affordable option for assaying conserved functions. While the absence of a phenotype may not rule out pathogenicity in humans, the appearance of mutant phenotypes, we believe is strongly suggestive of a deleterious role and would warrant additional studies in mammalian systems. In the case of this particular family, the studied variant, MSH5, c.C1051G; p.R351G, may contribute to the overall phenotype. Additional studies and additional patients with MSH5 c.C1051G homozygous variant will be needed to determine if observed variant is truly pathogenic or a significant modifier for the loss of ovarian function.
Author contribution
JLY and AR contributed to the study conception and design. MST, NM, and JLY performed data collection. All authors contributed to analysis. JLY wrote the manuscript which was edited and approved by all authors.
Funding
This work was funded by R01GM104007 (JLY and NM) and R01HD070647 and R21HD074278 (AR).
Declarations
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of University of Pittsburgh (PRO09080427). Informed consent was obtained from all individual participants in the study.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rajkovic A, Pangas S. Ovary as a biomarker of health and longevity: insights from genetics. Semin Reprod Med. 2017;35(3):231–240. doi: 10.1055/s-0037-1603571. [DOI] [PubMed] [Google Scholar]
- 2.Chon SJ, Umair Z, Yoon MS. Premature ovarian insufficiency: past, present, and future. Front Cell Dev Biol. 2021;9:672890. doi: 10.3389/fcell.2021.672890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venturella R, De Vivo V, Carlea A, D’Alessandro P, Saccone G, Arduino B, et al. The genetics of non-syndromic primary ovarian insufficiency: a systematic review. Int J Fertil Steril. 2019;13(3):161–168. doi: 10.22074/ijfs.2019.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jolly A, Bayram Y, Turan S, Aycan Z, Tos T, Abali ZY, et al. Exome sequencing of a primary ovarian insufficiency cohort reveals common molecular etiologies for a spectrum of disease. J Clin Endocrinol Metab. 2019;104(8):3049–3067. doi: 10.1210/jc.2019-00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin Y, Jiao X, Dalgleish R, Vujovic S, Li J, Simpson JL, et al. Novel variants in the SOHLH2 gene are implicated in human premature ovarian failure. Fertil Steril. 2014;101(4):1104–9 e6. doi: 10.1016/j.fertnstert.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Santos MG, Machado AZ, Martins CN, Domenice S, Costa EM, Nishi MY, et al. Homozygous inactivating mutation in NANOS3 in two sisters with primary ovarian insufficiency. Biomed Res Int. 2014;2014:787465. doi: 10.1155/2014/787465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Wang B, Dong Z, Zhou S, Liu Z, Shi G, et al. A NANOS3 mutation linked to protein degradation causes premature ovarian insufficiency. Cell Death Dis. 2013;4:e825. doi: 10.1038/cddis.2013.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao S, Li G, Dalgleish R, Vujovic S, Jiao X, Li J, et al. Transcription factor SOHLH1 potentially associated with primary ovarian insufficiency. Fertil Steril. 2015;103(2):548–53 e5. doi: 10.1016/j.fertnstert.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M. Distinct DNA-damage-dependent and -independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc Natl Acad Sci U S A. 2005;102(3):737–742. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt PA, Hassold TJ. Sex matters in meiosis. Science. 2002;296(5576):2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- 11.Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K, Hoog C. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science. 2002;296(5570):1115–1118. doi: 10.1126/science.1070594. [DOI] [PubMed] [Google Scholar]
- 12.Carlosama C, Elzaiat M, Patino LC, Mateus HE, Veitia RA, Laissue P. A homozygous donor splice-site mutation in the meiotic gene MSH4 causes primary ovarian insufficiency. Hum Mol Genet. 2017;26(16):3161–3166. doi: 10.1093/hmg/ddx199. [DOI] [PubMed] [Google Scholar]
- 13.Guo T, Zhao S, Zhao S, Chen M, Li G, Jiao X, et al. Mutations in MSH5 in primary ovarian insufficiency. Hum Mol Genet. 2017;26(8):1452–1457. doi: 10.1093/hmg/ddx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franca MM, Nishi MY, Funari MFA, Lerario AM, Baracat EC, Hayashida SAY, et al. Two rare loss-of-function variants in the STAG3 gene leading to primary ovarian insufficiency. Eur J Med Genet. 2019;62(3):186–189. doi: 10.1016/j.ejmg.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 15.de Vries L, Behar DM, Smirin-Yosef P, Lagovsky I, Tzur S, Basel-Vanagaite L. Exome sequencing reveals SYCE1 mutation associated with autosomal recessive primary ovarian insufficiency. J Clin Endocrinol Metab. 2014;99(10):E2129–E2132. doi: 10.1210/jc.2014-1268. [DOI] [PubMed] [Google Scholar]
- 16.McGuire MM, Bowden W, Engel NJ, Ahn HW, Kovanci E, Rajkovic A. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril. 2011;95(5):1595–1600. doi: 10.1016/j.fertnstert.2010.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo W, Guo T, Li G, Liu R, Zhao S, Song M, et al. Variants in homologous recombination genes EXO1 and RAD51 related with premature ovarian insufficiency. J Clin Endocrinol Metab. 2020;105(10). 10.1210/clinem/dgaa505. [DOI] [PubMed]
- 18.Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156(2):617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153(3):1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13(3):336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 23.Cressman VL, Backlund DC, Avrutskaya AV, Leadon SA, Godfrey V, Koller BH. Growth retardation, DNA repair defects, and lack of spermatogenesis in BRCA1-deficient mice. Mol Cell Biol. 1999;19(10):7061–7075. doi: 10.1128/MCB.19.10.7061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 24.Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85(7):1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 26.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31(4):385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 27.Santucci-Darmanin S, Neyton S, Lespinasse F, Saunieres A, Gaudray P, Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11(15):1697–1706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- 28.Cui M, Ying R, Jiang X, Li G, Zhang X, Zheng J, et al. A model of hereditary sensory and autonomic neuropathy type 1 reveals a role of glycosphingolipids in neuronal polarity. J Neurosci. 2019;39(29):5816–5834. doi: 10.1523/JNEUROSCI.2541-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDiarmid TA, Au V, Loewen AD, Liang J, Mizumoto K, Moerman DG, et al. CRISPR-Cas9 human gene replacement and phenomic characterization in Caenorhabditis elegans to understand the functional conservation of human genes and decipher variants of uncertain significance. Dis Model Mech. 2018;11(12). 10.1242/dmm.036517. [DOI] [PMC free article] [PubMed]
- 30.Wong WR, Brugman KI, Maher S, Oh JY, Howe K, Kato M, et al. Autism-associated missense genetic variants impact locomotion and neurodevelopment in Caenorhabditis elegans. Hum Mol Genet. 2019;28(13):2271–2281. doi: 10.1093/hmg/ddz051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillers KJ, Jantsch V, Martinez-Perez E, Yanowitz JL. Meiosis WormBook. 2017;2017:1–43. doi: 10.1895/wormbook.1.178.1. [DOI] [PMC free article] [PubMed] [Google Scholar]