Abstract
The post-translational modification of proteins expands the regulatory scope of the proteome far beyond what is achievable through genome regulation. The field of protein citrullination has seen significant progress in the last two decades. The small family of peptidylarginine deiminase (PADI or PAD) enzymes, which catalyse citrullination, have been implicated in virtually all facets of molecular and cell biology, from gene transcription and epigenetics to cell signalling and metabolism. We have learned about their association with a remarkable array of disease states and we are beginning to understand how they mediate normal physiological functions. However, while the biochemistry of PADI activation has been worked out in exquisite detail in vitro, we still lack a clear mechanistic understanding of the processes that regulate PADIs within cells, under physiological and pathophysiological conditions. This review summarizes and discusses the current knowledge, highlights some of the unanswered questions of immediate importance and gives a perspective on the outlook of the citrullination field.
Keywords: protein, citrullination, peptidylarginine deiminase, disease
1. Introduction
Protein expression is subject to several, super-imposing layers of regulation. Transcriptional, post-transcriptional and translational control mechanisms determine whether a protein is expressed within a certain cell, at a certain time. Once a protein is translated, it can be chemically modified through enzymatic and non-enzymatic reactions. These post-translational modifications (PTMs) can impact the structure, stability, sub-cellular localization and activity of a protein and modulate its binding affinity to other proteins, metabolites and nucleic acids. Protein modifications can be spatially and temporally controlled, allowing cells to respond to environmental changes such as stress signals, developmental cues, changes in nutrient or oxygen availability and oncogenic insults. PTMs, therefore, expand the functional proteome far beyond the complexity of the genome, and add an enormous degree of sophistication to biological systems, making them responsive and adaptable. Over 200 types of PTMs have been described to date [1].
Citrullination, or peptidylarginine deimination, is the post-translational conversion of an arginine residue to citrulline and involves the hydrolysis of the arginine and concomitant release of ammonia. Citrullination converts the guanidinium group of arginine to a ureido group, resulting in the loss of positive charge and two potential hydrogen bond donors [2]. Depending on the location of the modification within the protein, this may have profound consequences for protein function by altering local electrostatic interactions and hydrogen bonding ability. Indeed, like other PTMs, citrullination has been shown to impact several aspects of protein biology, such as structure, stability, localization, protein and nucleic acid binding and catalytic activity, as well as affect the subsequent deposition of other PTMs. As citrulline is a non-coded amino acid, its presence within a protein can only result through modification, implying a change in the cell's state or environment and the initiation of a relevant response.
Citrullination is catalysed by a small family of enzymes, the peptidylarginine deiminases (PADIs or PADs). The five PADI family members, PADI1, 2, 3, 4 and 6, are structurally similar and likely to operate via common regulatory mechanisms, but they show varying tissue distributions and sub-cellular localizations, suggesting that they have specific and non-overlapping organismal roles [3,4]. A large number of citrullinated proteins have been identified in different biological and disease systems [5–10] and this is likely to be a reflection of the wide regulatory capacity of citrullination. It is noteworthy however that, unlike kinases, ubiquitinases, methyltransferases or acetyltransferases, PADIs comprise a very small family of enzymes, with only four proteins having catalytic activity. This, coupled with their remarkably strong association with the development of pathology, makes it worth considering how PADIs are regulated and which molecular and cellular functions they modulate in health and disease.
Although the presence of citrulline within proteins has been known for nearly 50 years [11], having been suggested as early as the 1930s [12], and the first PADI was isolated 30 years ago [13], citrullination has been a rather obscure PTM for a long time. Historically, PADIs have been best known for their role in disease development. Research over many years has established strong and, in some cases, causal associations between aberrantly high levels of citrullination and the development of autoimmunity, neurodegeneration and cancer [14–16]. Conversely, lack of PADIs is associated with defects in embryo development, neurodevelopment and infertility [10,17,18]. The involvement of deregulated citrullination in disease has necessitated the development of biochemical, proteomic and computational methods for its detection [5,8,19–22], as well as pharmacological approaches for its inhibition [23,24], and important progress has been achieved in these areas.
A lot remains to be understood about the organismal functions of PADIs, the molecular and cellular mechanisms that underlie their physiological roles and how these relate to the deregulation of the modification in disease. Central to this is understanding the precise mechanisms of PADI activation within cells in different physiological and pathophysiological contexts. This review will summarize our current understanding, discuss some of the immediate open questions and provide a perspective on the future outlook of the field.
2. Distribution of citrullination across the tree of life and evolution in animals—an evolutionary accident?
The distribution of PADIs across the tree of life is highly unusual and the PADI sequence has been subject to extensive losses, modifications and duplications across evolution [25]. PADIs are present in some bacteria and fungi and, although their functions in these organisms are completely unknown, they have been shown to be catalytically active [25–27]. Paradoxically, PADIs are absent from yeast, worms and flies, but are ubiquitous across vertebrates. Ray-finned fish have a single PADI gene, but duplications down the vertebrate lineage have resulted in five paralogues in mammals. At least two different PADI types can be found across life [25], the animal-type three-domain PADI and the fungal-type, two-domain PADI. The animal-type PADI emerged during cyanobacterial evolution, while the fungal-type PADI can be found in actinobacteria and other bacteria. This and other complementary pieces of evidence, including phylogeny and sequence evolution rate analyses, indicate that the ancestral cyanobacterial gene was introduced into animals by horizontal gene transfer [25]. The cyanobacterial PADI is catalytically active and can citrullinate mammalian proteins including histones, which are absent from bacteria, suggesting that the horizontal transfer event introduced citrullination as a new catalytic activity in animals.
Two other types of enzymes can catalyse citrullination: pPAD, an extended agmatine deiminase found in the human pathogen Porphyromonas gingivalis [28] and gADI, an extended free L-arginine deiminase found in the human parasite Giardia Lamblia [29]. These have evolved independently from PADIs and are found in bacteria and some eukaryotic organisms [25]. Furthermore, a recent study identified a protein with peptidylarginine deiminase activity in the plant Arabidopsis thaliana [30]. Protein At5g08170 was identified after searching the Arabidopsis thaliana genome for a protein motif generated by aligning the catalytic core sequences of bacterial deiminases, after the authors identified citrullinated proteins within the Arabidopsis proteome. It is unclear whether At5g08170 is a PADI homologue, since a comprehensive search for PADI orthologues across the tree of life identified no PADI homologues in plants [25]. Regardless of this, however, At5g08170 was shown to act in a calcium-dependent manner, similarly to PADIs, to citrullinate proteins with nucleic acid functions, upon cold stress. The authors suggest that citrullination in plants is responsive to stress and may be associated with cell reprogramming, although more evidence is required to support this suggestion.
It is therefore intriguing that citrullination, as a catalytic function, has emerged in a variety of ways across evolution (via PADIs, pPAD, gADI and At5g08170). Further research into the function of the enzymes described above, in their corresponding host organisms, will ascertain whether they operate via similar underlying molecular principles and have similar cellular functions across different species (e.g. modulation of histone function or cellular reprogramming). This is a fascinating area of research and an exciting next frontier for the citrullination field.
3. Structure, sub-cellular localization, tissue distribution and substrate specificities of PADIs—a PADI for nearly every cell
The PADI family enzymes exhibit high protein sequence homology, both between the five paralogues (greater than 50%), as well as between orthologues from different mammalian species [4,25]. Human PADIs consist of three structural domains, the N-terminal (PAD_N, Pfam annotation: PF08526), middle (PAD_M, Pfam annotation: PF08527) and catalytic C-terminal domain (PAD_C, Pfam annotation: PF03068). PADI2, 3 and 4 are active as head-to-tail homodimers, where the N-terminal domain of one monomer is in contact with the C-terminal domain of the other [3,4,31,32]. PADI1-4 are calcium-dependent and have highly conserved calcium binding and catalytic residues, while PADI6 lacks some of the calcium-binding residues and the catalytic cysteine [3,4]. PADI6 is therefore considered to be enzymatically dead and, to the author's knowledge, no PADI6 protein substrates have been identified to date.
PADIs differ in their sub-cellular localization and tissue distribution. PADI1, 3 and 6 localize in the cytoplasm, PADI2 can shuttle between the cytoplasm and the nucleus and PADI4, the only PADI that possesses a bona fide nuclear localization signal (NLS), is found predominantly in the nucleus [33,34]. Intriguingly, PADI4 has also been shown to be exposed on the cell surface of resting human neutrophils and PADI2 to be released to the extracellular space by the same cells [35]. Although it remains to be determined whether these findings translate across different cell types, they open new possibilities regarding the regulatory scope of PADI2 and PADI4.
Under physiological conditions, PADIs show highly variable tissue-specific distribution. PADI1 is expressed in the skin and the endometrium [36,37], while expression of the protein in the esophagus, testis, kidney and cervix is also suggested in the Human Protein Atlas [38]. PADI2 expression is most widespread and is found in the brain, uterus, spleen, pancreas, skeletal muscle and secretory glands [39,40], while Human Protein Atlas additionally suggests expression in the human digestive and gastrointestinal tract, kidney, bladder, testis and bone marrow [38]. In fact, Human Protein Atlas suggests some level of PADI2 mRNA expression in virtually all tissues, making it possible that all tissues have the potential for PADI2 expression under certain conditions, or within certain cell sub-populations. The fact that PADI2, which is the ancestral vertebrate PADI paralogue [25], is also the most widely expressed, may suggest that the remaining PADIs, which arose by duplication, were selected during evolution to carry out specific functions. Support for this idea comes from the finding that, while PADI2 expression in the pituitary is not indicated by the Human Protein Atlas, it has been shown to increase in this tissue during the estrous cycle [41]. PADI3 expression is restricted mainly to the skin and hair follicles [42–44]. PADI4 (also known as PAD V) is only strongly expressed in the bone marrow, spleen and blood, with highest expression in monocytes and granulocytes and some expression in haematopoietic progenitor cells [33,45,46], while emerging evidence demonstrates that it is also expressed in the uterus, sperm, oocytes and mammalian embryos [10,47]. Like PADI2, however, data from Human Protein Altas suggests that PADI4 mRNA is detectable at low levels in cell sub-populations of most tissues [38], suggesting that its expression and, possibly activation, is possible upon certain stimuli. PADI6 (also known as ePAD), is expressed in oocytes and mammalian embryos [48]. While PADIs show a well-defined tissue distribution under physiological conditions, their tissue expression can be highly deregulated in disease [49].
Although PADIs share high structural homology, they have different substrate specificities in vivo [50,51]. The structural basis for substrate recognition specificities is not yet understood. The differing substrate preferences may be a consequence of their differing sub-cellular localizations and expression patterns, but may also be determined by whether conditions are permissive for their activation in certain cellular contexts, for example, the presence of cofactors or inhibitory interactions. We don't have sufficient information at present to deduce general principles about PADI target specificities, such as consensus amino acid sequence motifs, however our ever-increasing understanding and advancements in proteomic and analytical methodologies, along with better biochemical understanding of their regulation in vivo, is likely to yield these insights in the years to come.
4. Molecular and cellular functions regulated by citrullination—a small family with a big reach
We are gaining an ever-increasing understanding of the molecular and cellular processes impacted by citrullination. This knowledge has come both from large-scale proteomic studies, which have aimed to deduce general principles through identifying the ‘citrullinome’ [5,6,8], as well as targeted studies that have examined the role of PADIs and citrullination of specific substrates.
In a recent review, Genander et al. analysed published human citrullinomes of cells under resting and inflammatory conditions and identified the gene ontology (GO) categories represented most highly [52]. They demonstrate that, in resting cells, citrullinomes are enriched for proteins involved in RNA metabolic processes and gene expression, while under inflammatory conditions they are overwhelmingly enriched for GO categories relating to immune responses [52]. This may be a manifestation of the magnitude of PADI activation achieved in the two different scenarios, while it is also worth considering the conditions under which the citrullinome was measured. For example, in studies that probed the citrullinome under conditions of aberrant activation (for example in autoimmune disease or upon activation with broad stimuli, such as calcium ionophores), many proteins that become citrullinated are not PADI targets under physiological conditions. In this respect, we note that the GO categories found to be enriched within the citrullinomes of resting cells generally comprise highly abundant proteins, such as cytoskeletal and ribonuclear proteins [52]. Advances in mass spectrometric methods and associated computational analysis tools will undoubtedly lead to the identification of low abundance substrates, or citrullination events that may be present in sub-stoichiometric concentrations but nonetheless be functionally relevant, in the not too distant future. A better understanding of the mechanisms that govern PADI activation in vivo, beyond inflammation, will also allow researchers to tailor experiments so they reveal classes of PADI substrates that are relevant in new contexts.
For the purposes of this review, the molecular and cellular processes impacted by PADIs, as have been studied through specific protein substrates, are discussed.
4.1. Gene regulation
PADIs modulate gene expression in a number of ways: (i) via histone citrullination; (ii) via citrullination of transcription factors and epigenetic regulators; and (iii) via citrullination of signalling mediators that impact on gene expression. In addition, citrullination can modulate the likelihood and functional consequences of other PTMs on neighbouring amino acids.
4.1.1. Histone citrullination
Core histones represent one of the prototypical PADI substrate classes [53] and citrullination of the N-terminal tails of histones is intimately linked to transcriptional regulation. PADI4-mediated citrullination of histone H3 at residues Arg2, Arg8 and Arg17 (H3R2/8/17) and histone H4 at Arg3 (H4R3) were initially shown to repress transcription through the pS2 promoter by antagonizing CARM-1-mediated arginine methylation [54,55]. A subsequent study, which probed PADI4 localization and activity more globally, found that it is primarily associated with gene activation, as PADI4 localizes near the transcriptional start sites of active genes and its binding correlates with the binding of activating transcription factors, while it anti-correlates with repressive chromatin modifications [16]. In addition, PADI2-mediated citrullination of histone H3 Arg26 (H3R26) has also been shown to promote the transcriptional activation of estrogen receptor α (ERα) target genes in breast cancer [56,57] and of interleukin 6 (IL-6) in multiple myeloma [58]. Further support for the role of histone citrullination as a gene regulation mechanism comes from work in leukaemia cells, which demonstrated that PADI4 acts as a co-activator of translocated in leukaemia 1 (Tal1) through citrullination of H3R2 [59]. Beyond mediating gene regulation, histone citrullination was also recently shown to regulate non-coding RNA expression in pituitary cancers [60].
Although the above studies demonstrated disease-associated, aberrant PADI-mediated transcriptional regulation, this has also been demonstrated in physiological contexts. A study of normal canine mammary epithelial tissue suggested that PADI2-mediated H3 citrullination may regulate lactation-related genes during diestrus [61]. Additionally, PADI2-mediated citrullination of H3R26 mediates the expression of genes involved in oligodendrocyte differentiation [18], while PADI4-mediated H3 citrullination regulates the expression of pluripotency genes in embryonic stem cells [10].
Histone modifications act as a platform for the binding of transcriptional regulators. It is, therefore, possible that citrullination ‘readers’ bind histone citrulline marks and direct transcriptional protein complexes to specific genomic sites. Support for this mechanism comes from a study that showed that citrullinated H3R26 (H3Cit26) is bound by the chromatin remodelling factor Smarcad1 in pluripotent stem cells [62]. On the other hand, citrullination can modulate the binding of transcription factors and other chromatin-associated proteins to nearby histone marks and has been shown to work in conjunction with lysine methylation and lysine acetylation. Citrullination of H3R8 reduces the binding of heterochromatin protein 1 (HP1) to sites carrying trimethylation of H3 lysine 9 (H3K9me3), a prototypical heterochromatic mark, resulting in de-repression of cytokine genes and human endogenous retroviruses [63]. In addition, histone H3 citrullination was shown to be coordinated with H3 lysine deacetylation, through association between PADI4 and histone deacetylase 1 (HDAC1) during repression of the pS2 promoter in cancer cells [64] and in haematopoietic progenitor cells [65], suggesting that the crosstalk between citrullination and deacetylation may operate more broadly. It is likely that many more instances of citrullinated-histone-associated factors and interplay between citrullination and other PTMs will be uncovered in the future. Provided that PADI2 and PADI4 have clear regulatory roles in development and disease, systematic approaches for the identification of citrullination readers are warranted. In addition, it is exciting to consider how citrullination may play into the ‘histone code’ [66].
Citrullination of linker histones impacts chromatin structure. PADI4-mediated citrullination of linker H1 at Arg54 (H1R54), which is positioned within the H1 globular domain, results in reduced association with nucleosomal DNA and chromatin decondensation in mammalian embryonic stem cells and neutrophils [10,67], while similar results have been obtained with the avian H1 orthologue, histone H5 [68]. Citrullination of linker histones mediates both the local decondensation associated with a transcriptionally permissive chromatin state, and the global and dramatic chromatin decompaction associated with Neutrophil Extracellular Trap (NET) formation (discussed in detail below). The factors that determine the degree of linker histone citrullination and the subsequent degree of chromatin decondensation are unclear, however, it is likely that the nature or magnitude of PADI activation differs between the two scenarios. As discussed above, understanding the mechanistic details of PADI activation within the nucleus will not only allow us to understand this dichotomy but also potentially allow us to manipulate the process to achieve a desired degree of chromatin decondensation.
4.1.2. Citrullination of transcriptional and epigenetic regulators
In addition to modifying histones, PADIs can regulate transcription via citrullination of transcription factors and epigenetic regulators and by acting as transcriptional cofactors. PADI4-mediated citrullination of Elk-1, a transcription factor and downstream target of MAPK-ERK signalling, facilitates its phosphorylation by ERK2, which in turn increases its association with the histone methyltransferase p300, ultimately leading to transcriptional activation of target genes [16]. An independent study reported that, in 293T cells, PADI4 can target p300 directly, resulting in its enhanced association with co-activator GRIP1 [69].
Additionally, citrullination can impact epigenetic state through modulation of the DNA methyltransferases (DNMTs). In cancer cells, PADI4-mediated citrullination of the de novo methyltransferase DNMT3a results in its stabilization and increased DNA methylation at certain promoters [70]. Citrullination of DNMT3b has also been reported in mouse embryonic stem cells [10], suggesting this may be a more widely applicable mechanism. Notably, citrullination of RNA polymerase II (RNApolII) has been shown to impact on gene transcription [71]. In breast cancer cells, PADI2 citrullinates the C-terminal domain (CTD) of RNApolII, which acts as a regulatory hub where PTMs mediate the binding of auxiliary factors, determining polymerase progression [72]. Citrullination of the RNApolII mediates its association with the positive transcription elongation factor b (P-TEFb) kinase complex and its recruitment to chromatin, regulating RNApolII processivity.
It is unclear whether the above mechanisms operate in normal cells and are co-opted by cancer cells upon positive selection of PADI upregulating mutations. An alternative scenario is that such mechanisms only operate within transformed cells, where deregulated PADIs have the capacity to acquire new protein targets (due to a breakdown in the regulation of their levels or activity) and ‘hijack’ additional molecular pathways. The above studies have generated sufficient knowledge and novel reagents, such as citrullination-specific antibodies, to render possible comparative studies between normal and cancer cells, allowing deeper understanding of how PADIs impact on cancer initiation and progression.
Beyond studies in cancer cells, a significant amount of our current knowledge on citrullination-mediated transcriptional regulation comes from studies of cytokine gene regulation in inflammatory cells. PADI4-mediated citrullination of the transcription factor E2F1, cooperates with its acetylation to enhance its binding to the BET family bromodomain reader BRD4 (bromodomain-containing protein 4) [73]. This facilitates binding of E2F1 specifically to cytokine genes within granulocytes during inflammation. Additionally, PADI4-mediated citrullination of the nuclear factor κB (NFκB) p65 promotes its nuclear localization and transcriptional activity by enhancing its interaction with importin α3 [74]. The PADI4-mediated and enhanced transactivation activity of NFκB is specifically targeted to inflammatory cytokine genes interleukin 1β (IL1β) and tumor necrosis factor α (TNFα), promoting inflammation. Additionally, PADI2 was shown to directly citrullinate transcription factors GATA3 and RORγt and modulate their DNA binding activity, ultimately skewing T-cell differentiation outcomes [75].
4.1.3. PADI recruitment to chromatin
Although the effects of histone citrullination on transcription and chromatin compaction are well documented and we are beginning to map the mechanisms underlying this type of regulation, the mechanisms that govern the association of PADIs with chromatin remain unknown. PADIs do not possess a bona fide DNA binding domain and we currently lack data that will allow us to ascertain whether they preferentially associate with specific DNA sequences. Binding to specific genomic sites may be mediated by the DNA sequence itself, by other histone modifications, or by association with other transcriptional regulators or protein complexes. An example of the latter comes from studies on the regulation of ERα-mediated transcription, where it was found that stimulation of breast cancer cells with 17β-estradiol leads to association of PADI2 with ERα and recruitment of PADI2 to ERα target promoters [56]. Notably, in this system PADI2-mediated histone citrullination is observed mere minutes after stimulation with 17β-estradiol and facilitates ERα binding to chromatin, suggesting that it is one of the initiating events in ERα-mediated transcriptional regulation [57]. It is therefore unclear at this point whether additional factors may mediate association of PADI2 with DNA, or whether this is mediated exclusively by ERα. The complexity of these scenarios, coupled with the fact that chromatin-associated PADIs may, in principle, be catalytically active or inactive under different conditions, necessitates detailed studies that will allow us to precisely map PADIs and their activation across the genome under different cellular stimuli. This will undoubtedly be facilitated by the generation of new and specific reagents, such as anti-PADI and citrullination-specific antibodies.
In addition to modifying chromatin and associated proteins, PADIs may modulate the transcriptional and epigenetic state of cells indirectly, through modulation of signalling pathways that affect DNA-based events. The emerging role of citrullination in signal transduction is discussed next.
4.2. Cell signalling
A number of studies have started to integrate citrullination within cell signalling cascades. PADIs are activated by inflammatory signalling and impact upon it. In addition to PADI-mediated transcriptional regulation of cytokine genes, which is described above, citrullination of cytokines has also been shown to biochemically regulate their function. PADI2 citrullinates chemokines CXCL10 and CXCL11 and reduces their chemoattracting capacity, as well as their ability to signal through the chemokine receptor CXCR3, through a mechanism that's independent of receptor binding impairment [76]. Similarly, citrullination of CXCL8/IL-8 results in reduced CXCR2-mediated signaling [77], while citrullination of CXCL12 at different sites differentially modulates its binding to receptors CXCR4 and CXCR7 [78]. Furthermore, TNFα citrullination leads to reduced production of chemokines CXCL8 and CXCL10 [79]. The above findings seem paradoxical when considered against the strong association between PADI activation, inflammation and inflammatory disorders, as well as the evidence for PADI-mediated transcriptional activation of cytokine genes [58,63,73,74].
PADIs have also been implicated in signal transduction pathways that regulate cell growth, invasiveness and the transition between the epithelial and mesenchymal states. PADI4 citrullinates glycogen synthase kinase-3β (GSK3β) and mediates its translocation to the nucleus, thereby dampening TGFβ (transforming growth factor beta) signalling, while knock-down of PADI4 in this system promotes epithelial-to-mesenchymal transition and enhances the metastatic potential of cancer cells [80]. In addition, PADI2 was shown to inhibit Wnt signalling through citrullination of β-catenin, which enhances its degradation in a manner that's independent of its GSK3β-mediated regulation [81]. These two studies therefore describe anti-tumorigenic functions of PADI2 and PADI4, through dampening of cancer promoting signal transduction pathways. On the other hand, it was shown that PADI1 is over-expressed and may promote development of triple-negative breast cancer (TNBC) through direct citrullination of MEK1 kinase and modulation of its potential to phosphorylate ERK1/2 [82]. A study by the same laboratory showed that MEK1 is targeted by PADI2 in endometrial cancer and the citrullination event promotes MEK1-mediated phosphorylation of ERK1/2, facilitating tumour progression [83].
The implication of citrullination in classical signal transduction pathways is both fascinating and daunting and the fact that these pathways are highly pleiotropic may complicate, rather than clarify, our understanding of the regulation and roles of PADIs. As discussed above, future efforts in the field may build on these studies to assess whether PADIs impact on growth promoting signaling pathways under conditions of homeostasis.
4.3. Cell and tissue structure
PADIs impact on cell and tissue robustness through citrullination of structural proteins. PADI1- and PADI3-mediated citrullination of structural and intermediate filament-associated proteins such as keratins, trichohyalin and filaggrin provide mechanical strength and contribute to the maintenance of healthy skin and hair follicles, while their deregulation is associated with skin diseases such as psoriasis [84–88]. PADI2-mediated citrullination of the ECM protein Fibulin-5 protects it from proteolytic degradation and maintains elastogenic tissue function in the lungs [89]. Furthermore, citrullination of collagen type I is thought to mediate mesenchymal-to-epithelial transition [90], promoting cancer metastasis, while citrullination of fibronectin was also shown to promote cell invasion through altering integrin-mediated signalling [91]. Similarly, citrullination of collagen type II modulates its binding to integrins and decreases cell adhesion [92]. Finally, PADI6 localizes to the intermediate filament structures of oocytes and mediates cytoskeletal integrity [48,93].
Another type of structural protein whose function is regulated by citrullination is Myelin Basic Protein (MBP), a major constituent of the nerve myelin sheath and essential in the transmission of action potentials and motor function [94]. A significant body of work has determined that the correct level of citrullination is essential for proper MBP function. Aberrantly high levels of MBP citrullination are associated with the development of multiple sclerosis (MS) and have been shown to lead to destabilization of the myelin sheath [15,95].
The above studies demonstrate that precise regulation of PADI activity is essential for the maintenance of healthy tissue function. PADI inhibition has been suggested as a therapeutic strategy for autoimmune and neurodegenerative disorders, as well as cancer, due to the fact that these disease states are typically associated with aberrantly high levels of citrullination. However, caution is warranted to ensure therapeutic interventions that perturb the activity of PADIs are developed on solid understanding of their roles in tissue physiology.
4.4. Emerging functions
Recent work has implicated citrullination in a number of new biological processes. These are discussed briefly below.
4.4.1. Cell metabolism
A study published last year showed that PADI1- and PADI3-mediated citrullination of the glycolytic enzyme pyruvate kinase M2 (PKM2) alters its binding to amino acids and promotes glycolysis in cancer cells [96]. A more recent study showed that citrullination of glukokinase leads to reduction of its catalytic activity and results in reduced insulin secretion in response to glucose stimulation [97]. These are, to the author's knowledge, the first reports that mechanistically link protein citrullination to the regulation of metabolic pathways. However, the first study identified several citrullinated proteins within the glycolytic pathway, opening up the possibility that PADIs can impinge on cellular metabolism more broadly.
4.4.2. RNA biology
Another recently reported function of citrullination is in RNA metabolic processes. As mentioned above, proteins within the GO categories relating to RNA biology are highly represented within citrullinome datasets [52], but a functional role for PADIs in such processes has not been studied widely. Citrullination of the splicing factor PSF was shown to regulate its binding to mRNA [98]. In this respect, it is notable that a significant subset of citrullinated proteins identified in one study contain the RG/RGG RNA binding motif [9]. This particular study showed that PADI4-mediated citrullination inhibits arginine methylation of those proteins and prevents their aggregation. However, these findings open the possibility that citrullination of RNA binding motifs may be a more general mechanism of regulating the function of protein-RNA binding.
The fact that conversion of arginine to citrulline removes a positive change, which would be predicted to modulate interactions with charged molecules such as nucleic acids or certain charged metabolites, suggests that wider roles of citrullination in the regulation of metabolism or RNA-mediated processes are plausible. Formal experimentation will determine whether this is indeed the case and whether such mechanisms operate under physiological conditions or only become relevant under conditions of PADI deregulation.
4.4.3. Autophagy
Functional links are also emerging between citrullination and autophagy. Induction of autophagy has been shown to lead to activation of PADI4 and generation of citrullinated neo-epitopes that are associated with autoimmunity [99], while processing of proteins via autophagic vesicles was shown to lead to the generation and presentation of citrullinated peptides specifically, by antigen presenting cells, synoviocytes and fibroblasts [99,100]. It is not understood how activation of autophagy leads to citrullination. PADI activation within autophagic vesicles may be a result of increased local calcium availability, however further investigation is required to ascertain whether specific autophagy-associated signalling can modulate PADI activity. On the other hand, PADIs have been implicated in the regulation of autophagy [101–103], suggesting that PADI activation may happen independently of autophagy signalling. Understanding the mechanistic details of the interplay between autophagy and citrullination may provide new ways of modulating autophagy in cancer therapy and new avenues of alleviating tissue destruction in autoimmunity.
5. Citrullination in health and disease—the virtue and the vice
The last decade has seen significant progress in our understanding of the physiological functions of PADIs in health and disease. Depending on their tissue distribution, different PADIs have been shown to modulate the innate immune response, skin homeostasis, nerve myelination, stem cell biology, fertility and reproductive functions (figure 1). Conversely, deregulation of PADI levels or PADI activating pathways may lead to a breakdown in the tight regulation of PADI enzymatic activity and result in the non-physiological citrullination of additional proteins and disruption of their normal functions. This is illustrated in the fact that aberrant PADI activity is associated with an increasingly broad array of diseases, from autoimmunity and neurodegeneration to atherosclerosis and cancer (figure 1).
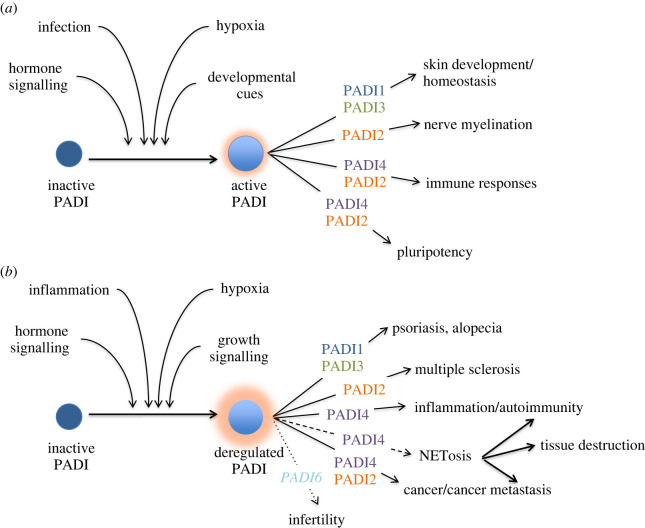
Figure 1.
Organismal functions of citrullination in physiology and disease. (a) Under physiological conditions, PADIs are activated in response to stimuli such as infection, hormone stimulation, developmental signals and, possibly, hypoxia. Depending on their tissue expression, PADIs can regulate skin homeostasis, nerve myelination, immune responses and embryo development. PADI6, which is considered to be catalytically inactive, is not included in this schematic. (b) When one of the activating signals or the level of PADI activity are deregulated, aberrant levels of citrullination can underlie the development of skin disorders, multiple sclerosis, cancer progression, metastasis and autoimmunity. Deregulation of NETosis can exacerbate autoimmunity, cancer metastasis and tissue destruction associated with a number of disease states. Lack of PADI6 leads to compromised female fertility.
5.1. Neurobiology and neurodegeneration
Citrullination has been studied both in the context of central nervous system (CNS) physiology and neurodegeneration [94]. PADI2 is the most highly expressed PADI in the brain and CNS [18,104]. Its function as a transcriptional regulator is required for oligodendrocyte differentiation, while a tightly regulated level of citrullination of MBP maintains myelin sheath stability, as demonstrated by the fact that PADI2-KO mice display motor dysfunction [18]. On the other hand, aberrantly high levels of MBP citrullination also destabilize myelin and are thought to underlie the development of MS, as described above. This is supported by in vivo studies, which demonstrated that mice that over-express PADI2 in oligodendrocytes exhibit myelin loss [15]. In addition, PADI-mediated inflammation in the brain and PADI4-mediated transcriptional deregulation in oligodendrocytes have also been suggested as mechanisms underlying the development of MS [105]. In this respect, PADI inhibition has been suggested as a therapeutic strategy against MS [106]. Aberrantly high levels of citrullination have also been associated with neurodegenerative disorders such as Alzheimer's and prion diseases [107,108], however, a clear mechanistic understanding of how PADI deregulation may mediate disease progression in these cases still needs to be developed.
5.2. Skin homeostasis and skin diseases
A detailed review of the roles of PADIs in skin homeostasis and discussion of PADI-mediated mechanisms that may underlie skin diseases has recently been published elsewhere [109]. Briefly, PADI1 and PADI3 are expressed in the epidermis and hair follicles, where they mediate skin cell differentiation and maintain epidermal barrier function through citrullination of skin structural proteins [86]. It is not yet clear whether histone citrullination-mediated transcriptional regulation has a role in skin differentiation and homeostasis [52]. Genetic Polymorphisms of PADI3 are associated with skin disorders such as uncombable hair syndrome and certain forms of alopecia [110,111], while reduced levels of keratin K1 citrullination are thought to compromise skin structure and are associated with psoriasis and atopic dermatitis [87].
5.3. Reproductive biology, fertility and embryo development
PADIs have a regulatory role in the female reproductive system [112]. As described above, PADI6 is specific to the oocyte and is essential for female fertility through maintaining the stability of the oocyte cytoskeleton [48,93]. PADI2 and PADI4 have been shown to regulate the expression of insulin-like growth factor-binding protein 1 (IGFBP1) in response to progesterone, in the uterine cells of pregnant sheep [113,114]. As IGRBP1 is important for embryo implantation, it is suggested that PADI activity is important for the establishment of pregnancy. Further indication that PADIs may regulate female fertility comes from the finding that PADI2 expression in the mouse pituitary shows cyclic expression according to the estrous cycle [41].
Citrullination also plays a role in embryo development. PADI4 and citrullinated H3 are detectable in the mouse and pig pre-implantation embryo from the two-cell-stage onwards and PADI inhibition leads to a reduction in the number of pluripotent cells of the Inner Cell Mass (ICM), while PADI4 also regulates the expression of pluripotency genes in embryonic stem cells [10,115–117]. PADI4-KO mice are viable, but are born with skewed Mendelian ratios [118], suggesting that loss of PADI4 compromises embryonic development. It is possible that the loss of PADI4 is compensated by one of the other PADI members, for example PADI2 which can also mediate some of the same transcriptional mechanisms [18], however mouse models that are null for more than one PADI are required to ascertain this.
5.4. Immunity, inflammation and autoimmunity
One of the best-established functions of protein citrullination is in immune responses and sterile inflammation (figure 2). PADI4 is most prominently expressed in neutrophils and other leucocytes and can mediate immune signalling via transcriptional and biochemical regulation of cytokines [74]. Furthermore, PADI4 is strongly activated during the innate immune response to infection and mediates NETosis, a process of inflammation-induced cell death, which involves profound decondensation and release of chromatin from neutrophils [119]. PADIs also impact adaptive immune responses through the regulation of T cell differentiation. As mentioned above, PADI2-mediated citrullination of transcription factors GATA3 and RORγt affects T cell responses by modulating the relative numbers of helper T cell sub-populations and, consequently, tissue inflammation [75]. Recent work suggested that differentiation of helper T cells is also affected by NET-associated extracellular histones [120], a process likely mediated by citrullination.
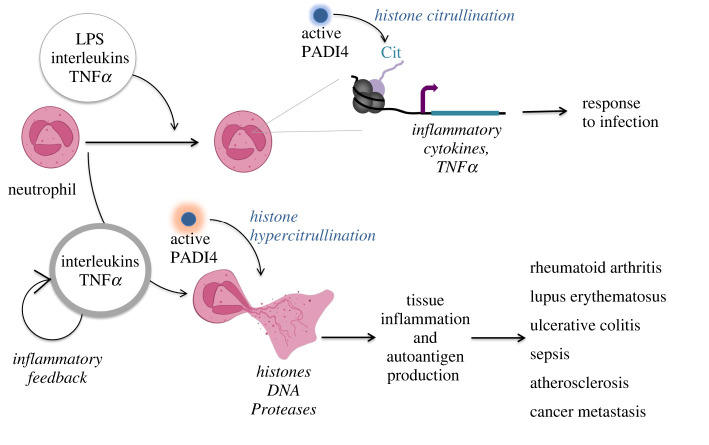
Figure 2.
Modes and outcomes of PADI4 activation in the immune system. Schematic representation of the outcomes of PADI4 activation in neutrophils. Upon infection, active PADI4 can citrullinate histones to mediate transcriptional activation of inflammatory cytokines. Increased inflammatory signalling can lead to histone hypercitrullination and NETosis, which can cause tissue destruction and exacerbate a number of pathologies.
Citrullination is also inextricably associated with autoimmunity. The Padi4 gene falls within a susceptibility locus for Rheumatoid Arthritis (RA), and certain single nucleotide polymorphisms are associated with the disease [14]. The adaptive immune system reacts specifically to citrullinated peptides of endogenous proteins and presentation of these modified peptides by Major Histocompatibility Complex (MHC) molecules on antigen-presenting cells elicit highly specific T cell responses [121], which are thought to be associated with autoimmunity. Citrullinated peptides have also been shown to bind with higher affinity to certain HLA alleles, which are associated with susceptibility to RA [122], providing a mechanistic basis for the strong links between citrullination and autoimmunity, however more recent work has demonstrated that this may not a universally applicable explanation [123]. Autoantibodies against citrullinated versions of such proteins (anti-citrullinated protein autoantibodies, or ACPA) serve as diagnostic and prognostic RA biomarkers and their presence is associated with faster progressing and more destructive disease [124], while similar autoantigens have also been associated with lupus erythematosus and other autoimmune disorders [67].
5.5. NETs and NET-associated pathologies
NETosis constitutes a fast and effective response to infection, however, the deregulation of this process and associated release of inflammatory cytokines, proteases and active PADIs to the extracellular space, are highly destructive for the surrounding tissue and have been associated with the exacerbation of a remarkable array of pathologies, including RA, lupus erythematosus, ulcerative colitis, inflammatory bowel disease, atherosclerosis, sepsis, type I diabetes and severe Covid-19 (reviewed recently in detail in [125]) (figure 2).
A large body of literature suggests that PADI4 activation is an essential step and operates in the early stages of NETosis [68,118,126,127]. However, detailed imaging methods developed recently have mapped the sequence of events that take place during NETosis and suggest that histone citrullination is a late, rather than initiating event [128,129]. More recently, it was suggested that citrullination does not mediate NETosis in neutrophils, but modulates the binding of histones to Toll-like receptors, thereby mediating downstream signalling [130]. Additionally, a significant number of studies have shown that NETosis can progress in a PADI4-independent manner [131,132]. While some of these studies may be complicated by the stimuli used to induce NETosis in the different studies [133,134], in vivo experiments that examined the role of PADI4 in NETosis associated with bacterial infection, diabetes, sepsis and cancer, among others, show that PADI4 inhibition interferes with the ability of neutrophils to release NETs [118,135,136]. As a result, PADI4 inhibition has been studied extensively as an approach to alleviate NET-associated pathologies that manifest in diabetes, sepsis, infection-induced lung injury, age-related cardiac fibrosis, deep-vein thrombosis and myocardial ischemia [137–142].
5.6. Cancer
A substantial body of work draws a strong association between PADI deregulation and cancer. Evidence suggests that PADIs can impact on tumour development through modulating cell signalling, transcription and the extracellular matrix (ECM), thereby regulating growth, apoptosis and the epithelial-to-mesenchymal transition. PADI2-mediated transcriptional regulation is associated with the development and metastasis of breast, gynecological and prostate cancers [60,143,144]. PADI4 is detected in the blood of cancer patients and in a large array of malignant tumours whose normal tissue and benign tumour counterparts lack PADI4 expression, while metastatic tumours exhibit significantly higher PADI4 expression than the corresponding primary tumours [49,90,145]. This suggests that cells that aberrantly upregulate or activate PADI4, either through genetic mutation or deregulation of PADI4-activating signalling pathways, have a growth advantage and are selected during cancer development and metastasis. This has been mechanistically attributed to PADI4-mediated remodelling of the ECM and initiation of metastatic colony formation [90]. In addition, PADI4-mediated regulation of gene transcription has been shown to interfere with apoptosis and growth arrest in cancer cell lines [146–148]. An expanding body of literature also suggests that PADI4 may have an indirect role in supporting cancer progression, through the promotion of NETs [149]. Importantly, these studies showed that loss of PADI4 in mice reduced both NETs formation and cancer metastasis [150,151]. The above studies suggest that when PADIs act to promote cancer their activation is co-opted by existing and emerging tumors rather than acting as a tumour-initiating event. However, some evidence also exists to suggest that PADI2 over-expression can act as a tumour initiating mechanism [152].
On the other hand, PADIs have also been suggested to have anti-tumour effects. PADI2 has been shown to mediate the action of an anti-cancer compound that acts through the dampening Wnt signalling [81], while PADI4 was shown to inhibit epithelial-to-mesenchymal transition [80]. Additionally, PADI4 has been suggested to act as a tumour suppressor as Padi4-null mice showed resistance to DNA damage-induced apoptosis in the thymus and H4R3 citrullination was associated with smaller tumour size in a cohort of non-small cell lung carcinoma patients [153]. However, this study did not detect a significant association between levels of H4R3 citrullination in the tumours and increase in patient survival. While PADIs cannot be broadly and unequivocally summarized as pro- or anti-tumourigenic factors, the field has amassed enough evidence to warrant the serious consideration of PADI inhibitors in cancer therapy, especially for late stage and metastatic cancers.
Beyond modulation of PADI activity as an approach to cancer therapy, an emerging body of work suggests that citrullination may be exploited in the development of anti-cancer vaccines [154]. Citrullinated epitopes of endogenous proteins such as vimentin have been found to be presented by cancer cells, in addition to antigen-presenting cells [155]. The authors demonstrated that immunization with a citrullinated vimentin peptide resulted in a specific T cell response and increased the survival of tumour bearing mice [156].
The studies discussed above collectively demonstrate that inhibition of PADIs, and PADI4 more specifically, holds significant promise as a therapy for a large array of clinical conditions. Indeed, significant effort has been devoted toward generating potent and specific PADI inhibitors [24,101,127]. The demonstration that loss of PADI4 function in the haematopoietic system, where it is most highly expressed, has no adverse effects for normal haematopoiesis in mice [46], suggests that PADI4 inhibition may be well tolerated and increases the urgency for the development of inhibitors suitable for clinical use.
6. Understanding regulation of PADI activity in vivo—how to exalt the virtues while keeping the vices at bay
Given the strong association between aberrantly high levels of citrullination and disease states such as autoimmunity, neurodegeneration and cancer [24], it is important to consider the mechanisms that govern PADI activation, how they may operate in different cellular contexts and how they may be deregulated in disease. An important open question in citrullination biology is how PADIs are regulated in vivo.
PADI1-4 are calcium-dependent enzymes and their activation mechanisms have been mapped in great detail in vitro, showing that calcium binding is an obligatory step in generating the active cleft [3]. PADI4 is bound by five calcium ions, which induce the activity of the enzyme by greater than 105-fold [3]. Elegant structural and biochemical studies of PADI2 by Slade et al. have shown that calcium binding sites are bound by si calcium ions in a stepwise fashion and identified a calcium switch that exposes the enzyme active site and positions the catalytic cysteine, rendering the enzyme greater than 7 × 105-fold more active [4]. The putative calcium switch residues are highly conserved among PADI1-4, suggesting that the calcium switch is a universal activation mechanism.
Within a cellular context, PADI activation is tightly regulated and it is possible for the enzyme to be expressed but catalytically inactive. For example, neutrophils express robust amounts of PADI4 but citrullination is undetectable until the cells are stimulated with a calcium ionophore or inflammatory stimuli such as TNFα and lipopolysaccharide (LPS) [157]. The calcium concentration necessary to activate PADIs in vitro is at least 10-fold higher than reported intracellular calcium concentrations, which reach up to 10 µM [158]. While activation is likely to be temporally and spatially controlled, it is as yet unclear whether PADIs are always activated as a result of local calcium influx or whether their activation is subject to allosteric regulation. Opening of calcium channels can lead to local calcium concentrations in the micromolar range [159,160] and this would be sufficient to activate catalysis. Indeed, it is known that some of the inflammatory stimuli that lead to PADI4 activation in neutrophils, such as chemotactic peptides, incite a calcium influx and a similar mechanism has been attributed to progesterone-dependent PADI2 activation in breast cancer cells [114,161]. However, it is unclear how PADIs may be recruited to the proximity of high calcium areas, especially when considering PADI activation within the nucleus, as in the case of PADI4- or PADI2-mediated citrullination of histone proteins [54,55].
An alternative mechanism involves allosteric regulation. Binding to a cofactor, such as an interacting protein or complex, may elicit a conformational change that mimics the calcium switch by exposing the catalytic cysteine, thus rendering catalysis possible in intracellular calcium concentrations. Support for this mechanistic scenario comes from studies that showed that RA-specific autoantibodies against PADI4 bind near the calcium switch site and lower the calcium concentration required for activation [162,163]. Similarly, it is plausible that the cell signalling pathways that are engaged under conditions of PADI activation result in PTMs that confer a similar structural change. It has been suggested that autocitrullination of PADI4 occurs during neutrophil activation and alters its catalytic activity [164], while the heavy involvement of kinases within the inflammatory signalling pathways that are known to lead to PADI activation makes it likely that phosphorylation events and other PTMs have a regulatory role. In breast cancer cells, PADI4-mediated citrullination of the transcription factor Elk-1 happens upon stimulation with Epidermal Growth Factor (EGF), suggesting that PADI4 may be activated by a component of EGF signalling [16], although it is yet unclear from this study whether it is the catalytic activity of PADI4, or its association with Elk-1 that is regulated in this manner. Similarly, ATP-dependent PADI2 activation in mast cells is mediated by the kinases p38 MAPK and PKC, supporting the possibility that PADI2 activation is modulated by phosphorylation [165].
Understanding the biochemical mechanisms that lead to PADI activation requires the identification of the precise conditions and cell stimuli that elicit a citrullination response. In this respect, the fields of neutrophil activation and inflammatory signalling offer fertile ground for investigation, although significant progress in this regard has been impeded by the seeming heterogeneity and differing outcomes of neutrophil responses depending on the experimental context, as well as variability in the antibody reagents typically used to access PADI4 activation [166].
A recently reported context of PADI4 activation is likely to provide additional research avenues in this area. Wang et al. discovered that PADI4 is activated under hypoxic conditions and mediates hypoxia-inducible factor (HIF) dependent transcriptional regulation in cancer cells [167]. Although the activation is closely associated with hypoxia-mediated upregulation of PADI4, kinetic experiments in this study also report conditions under which PADI4 activation is observed within a few hours of the hypoxic stimulus and before any appreciable upregulation of the protein. This suggests that hypoxia leads to the enzymatic activation of PADI4, as well as its upregulation. Disentangling the catalytic and transcriptional/translational regulation of PADIs will be a key step toward understanding mechanisms of allosteric regulation and hypoxic stimuli may provide a cleaner system than growth promoting or inflammatory signalling.
A study published earlier this year identified that cytomegalovirus infection elicits upregulation and activation of PADI2 and PADI4 and consequent citrullination of a wide range proteins [168]. The timing of PADI activation in this context makes it difficult to ascertain whether viral infection regulates the catalytic activity of PADIs, or whether the citrullination observed is due to increased levels of the enzymes. However, more detailed temporal studies in this context may provide an additional useful system in which to study PADI activation.
An alternative, but not mutually exclusive, possibility is that PADIs are subject to inhibitory PTMs, which are removed or reversed under activating conditions. A study by Chang et al. reported that association with the protein tyrosine phosphatase PTPN22 suppresses the catalytic activity of PADI4 and subsequent NETosis [169]. Although in this context PADI4 inhibition is not mediated via the phosphatase activity of PTPN22, it is possible that association with other phosphatases or other PTM-reversing regulates PADI activity.
An exciting possibility is that detailed understanding of the mechanistic principles of PADI activation and how they differ between physiological and disease conditions may enable the design of a next generation of therapeutics based on modulators that will achieve precise perturbation or activation of citrullination. It will therefore be a valuable endeavour for the field to pursue studies that will delineate the cellular conditions, signalling pathways and biochemical mechanisms that underlie PADI regulation in physiology and disease.
7. Discussion
PADI enzymes are ubiquitous across vertebrates and are becoming increasingly appreciated as key regulators in many aspects of mammalian physiology. They are absent from classical model organisms such as yeast, worms and flies [25] and, arguably, this has impeded progress into understanding the physiological functions of citrullination, as the field wasn't afforded the rich information that can be derived from systematic genetic screens. The discovery that PADIs arose in animals by horizontal gene transfer leads us to think about citrullination and its roles in animal biology in a new light. The fact that the PADI sequence was not only retained throughout vertebrate evolution, but duplicated multiple times, suggests that citrullination confers a fitness or survival advantage to animals. While there is still a lot to understand regarding the physiological functions of PADIs, an explanation for their retention may be the role of citrullination in the defense against infection. Indeed, it has been suggested that genes that are horizontally transferred from bacteria may augment the innate immune functions of eukaryotes [170]. Alternatively, the emerging role of citrullination in mammalian embryonic development [10,109,110] may explain the selective pressure for retaining the sequence. A notable insight into this question is offered by a recent study in fish. Golenberg et al. deleted the single PADI in zebrafish and found that this protein regulates wound healing and fin regeneration [171]. It is not yet known how the PADI-null zebrafish respond to infection and it is possible that PADIs have acquired different roles, even throughout animal evolution. However, we can safely assume, and indeed we are starting to appreciate, that some of their functions are of fundamental importance in animal physiology.
The future is undoubtedly bright for citrullination research and we can predict that the field will see yet greater progress in the short- and mid-term, as researchers are building ever more specific and powerful tools for the manipulation of PADIs and the detection of their activation, as well as animal models where their function is perturbed. It has been pleasing to see that citrullination was highlighted as one of the PTMs that are likely to have a significant impact in the field of signal transduction, in an editorial on ‘The Future of Signaling’ a few years back [172].
8. Outlook
Some questions are largely unresolved and it is the author's view that the following are of immediate importance: Firstly, is citrullination a bona fide signal transduction mechanism, one that mediates the translation of environmental cues into transcriptional and epigenetic changes in normal physiology? Secondly, what determines ‘normal’ versus ‘pathogenic’ levels of citrullination in a tissue? Is it the magnitude, or the nature of the activating signal that is altered in the various pathologies exacerbated by aberrant citrullination? If it is the mechanism, can we exploit this in disease therapy? Thirdly, the long-standing question on the elusive reversing mechanism: after a stimulus that induces citrullination, for example of histones, the citrulline mark disappears with time [54]. Is this due to the existence of a citrullination ‘eraser’, akin to a phosphatase or deacetylase? Or is citrullination only lost through degradation and recycling of the modified protein? And lastly, can we map specific residues or regions within PADIs that may be perturbed to achieve certain outcomes, for example a constitutively active PADI, or one that only responds to certain stimuli? We can dare to speculate that, by continuing to build upon current knowledge and moving towards precise biochemical understanding of PADI activation and substrate engagement mechanisms, it will be possible to answer these outstanding questions.
Acknowledgements
I am grateful to A. Finch, whose support made the writing of this article possible. I would like to thank P. Bhagwatwar for help with figure generation and the members of the Christophorou laboratory for valuable discussions and proofreading of the manuscript. I dedicate this article to the memory of Kerri Mowen—a strong Citrulline-phile, generous collaborator and fearless science advocate.
Data accessibility
This article has no additional data.
Authors' contributions
M.A.C.: conceptualization, data curation, funding acquisition, investigation, visualization, writing—original draft, writing—review and editing.
Conflict of interest declaration
The author declares no competing interests.
Funding
M.A.C. is supported by a Sir Henry Dale Fellowship, jointly funded by the Wellcome Trust and the Royal Society (grant no. 105642/A/14/Z) and by funding from the UK Biotechnology and Biological Sciences Research Council (BBS/E/B/000C0421).
References
- 1.Beltrao P, Bork P, Krogan NJ, Van Noort V. 2013. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 9, 714. ( 10.1002/msb.201304521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slade DJ, Subramanian V, Fuhrmann J, Thompson PR. 2014. Chemical and biological methods to detect post-translational modifications of arginine. Biopolymers 101, 133-143. ( 10.1002/bip.22256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. 2004. Structural basis for Ca2+-induced activation of human PAD4. Nat. Struct. Mol. Biol. 11, 777-783. ( 10.1038/nsmb799) [DOI] [PubMed] [Google Scholar]
- 4.Slade DJ, et al. 2015. Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design. ACS Chem. Biol. 10, 1043-1053. ( 10.1021/cb500933j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilvawala R, Nguyen SH, Maurais AJ, Nemmara VV, Nagar M, Salinger AJ, Nagpal S, Weerapana E, Thompson PR. 2018. The Rheumatoid Arthritis-Associated Citrullinome. Cell Chem. Biol. 25, 691-704.e6. ( 10.1016/j.chembiol.2018.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurais AJ, Salinger AJ, Tobin M, Shaffer SA, Weerapana E, Thompson PR. 2021. A streamlined data analysis pipeline for the identification of sites of citrullination. Biochemistry 60, 2902-2914. ( 10.1021/acs.biochem.1c00369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewallen DM, et al. 2015. Chemical proteomic platform to identify citrullinated proteins. ACS Chem. Biol. 10, 2520-2528. ( 10.1021/acschembio.5b00438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CY, et al. 2018. Mining the human tissue proteome for protein citrullination. Mol. Cell. Proteomics 17, 1378-1391. ( 10.1074/mcp.RA118.000696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanikawa C, et al. 2018. Citrullination of RGG motifs in FET Proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 22, 1473-1483. ( 10.1016/j.celrep.2018.01.031) [DOI] [PubMed] [Google Scholar]
- 10.Christophorou MA, et al. 2014. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507, 104-108. ( 10.1038/nature12942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers GE. 1962. Occurrence of citrulline in proteins. Nature 194, 1149-1151. ( 10.1038/1941149a0) [DOI] [PubMed] [Google Scholar]
- 12.Fearon WR, Boggust WA. 1952. Isolation of a carbamido acid from autolysed yeast. Nature 170, 372-373. ( 10.1038/170372b0) [DOI] [PubMed] [Google Scholar]
- 13.Fujisaki M, Sugawara K. 1981. Properties of peptidylarginine deiminase from the epidermis of newborn rats. J. Biochem. 89, 257-263. ( 10.1093/oxfordjournals.jbchem.a133189) [DOI] [PubMed] [Google Scholar]
- 14.Suzuki A, et al. 2003. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34, 395-402. ( 10.1038/ng1206) [DOI] [PubMed] [Google Scholar]
- 15.Musse AA, Li Z, Ackerley CA, Bienzle D, Lei H, Poma R, Harauz G, Moscarello MA, Mastronardi FG. 2008. Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. DMM Dis. Model. Mech. 1, 229-240. ( 10.1242/dmm.000729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR, Roberson MS, Kraus WL, Coonrod SA. 2011. Genome-Wide analysis reveals PADI4 cooperates with Elk-1 to activate C-Fos expression in breast cancer cells. PLoS Genet. 7, e1002112. ( 10.1371/journal.pgen.1002112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, et al. 2016. Mutations in PADI6 Cause Female Infertility Characterized by Early Embryonic Arrest. Am. J. Hum. Genet. 99, 744-752. ( 10.1016/j.ajhg.2016.06.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcão AM, et al. 2019. PAD2-Mediated Citrullination Contributes to Efficient Oligodendrocyte Differentiation and Myelination. Cell Rep. 27, 1090-1102.e10. ( 10.1016/j.celrep.2019.03.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensen SMM, Pruijn GJM. 2014. Methods for the detection of peptidylarginine deiminase (pad) activity and protein citrullination. Mol. Cell. Proteom. 13, 388-396. ( 10.1074/mcp.R113.033746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clancy KW, Weerapana E, Thompson PR. 2016. Detection and identification of protein citrullination in complex biological systems. Curr. Opin Chem. Biol. 30, 1-6. ( 10.1016/j.cbpa.2015.10.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudmann NS, Hansen NUB, Jensen ACB, Karsdal MA, Siebuhr AS. 2015. Biological relevance of citrullinations: diagnostic, prognostic and therapeutic options. Autoimmunity 48, 73-79. ( 10.3109/08916934.2014.962024) [DOI] [PubMed] [Google Scholar]
- 22.Chaerkady R, et al. 2021. Characterization of citrullination sites in neutrophils and mast cells activated by ionomycin via integration of mass spectrometry and machine learning. J. Proteome Res. 20, 3150-3164. ( 10.1021/acs.jproteome.1c00028) [DOI] [PubMed] [Google Scholar]
- 23.Bicker KL, Thompson PR. 2013. The protein arginine deiminases: structure, function, inhibition, and disease. Biopolymers 99, 155-163. ( 10.1002/bip.22127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis HD, Nacht M. 2016. IPAD or PADi - ‘tablets’ with therapeutic disease potential? Curr. Opin Chem. Biol. 33, 169-178. ( 10.1016/j.cbpa.2016.06.020) [DOI] [PubMed] [Google Scholar]
- 25.Cummings TFM, et al. 2021. Citrullination was introduced into animals by horizontal gene transfer from cyanobacteria. Mol. Biol. Evol. 39, msab317. ( 10.1093/molbev/msab317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crisp A, Boschetti C, Perry M, Tunnacliffe A, Micklem G. 2015. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 16, 1-3. ( 10.1186/s13059-015-0607-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Sayed ASA, Shindia AA, Abouzaid AA, Yassin AM, Ali GS, Sitohy MZ. 2019. Biochemical characterization of peptidylarginine deiminase-like orthologs from thermotolerant Emericella dentata and Aspergillus nidulans. Enzyme Microb. Technol. 124, 41-53. ( 10.1016/j.enzmictec.2019.02.004) [DOI] [PubMed] [Google Scholar]
- 28.Goulas T, et al. 2015. Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci. Rep. 5, 1-17. ( 10.1038/srep11969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touz MC, Rópolo AS, Rivero MR, Vranych CV, Conrad JT, Svard SG, Nash TE. 2008. Arginine deiminase has multiple regulatory roles in the biology of Giardia lamblia. J. Cell Sci. 121, 2930-2938. ( 10.1242/jcs.026963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marondedze C, Elia G, Thomas L, Wong A, Gehring C. 2021. Citrullination of proteins as a specific response mechanism in plants. Front. Plant Sci. 12, 377. ( 10.3389/fpls.2021.638392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CY, Lin C-C, Liu Y-L, Liu G-Y, Liu J-H, Hung H-C. 2017. Molecular interplay between the dimer interface and the substrate-binding site of human peptidylarginine deiminase 4. Sci. Rep. 7, 1-4. ( 10.1038/srep42662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saijo S, et al. 2016. Monomeric form of peptidylarginine deiminase type I revealed by X-ray crystallography and small-angle X-ray scattering. J. Mol. Biol. 428, 3058-3073. ( 10.1016/j.jmb.2016.06.018) [DOI] [PubMed] [Google Scholar]
- 33.Asaga H, Nakashima K, Senshu T, Ishigami A, Yamada M. 2001. Immunocytochemical localization of peptidylarginine deiminase in human eosinophils and neutrophils. J. Leukoc. Biol. 70, 46-51. ( 10.1189/jlb.70.1.46) [DOI] [PubMed] [Google Scholar]
- 34.Zheng L, Nagar M, Maurais AJ, Slade DJ, Parelkar SS, Coonrod SA, Weerapana E, Thompson PR. 2019. Calcium regulates the nuclear localization of protein arginine deiminase 2. Biochemistry 58, 3042-3056. ( 10.1021/acs.biochem.9b00225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, et al. 2017. Spontaneous secretion of the citrullination enzyme PAD2 and cell surface exposure of PAD4 by neutrophils. Front. Immunol. 8, 1200. ( 10.3389/fimmu.2017.01200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rus'd AA, Ikejiri Y, Ono H, Yonekawa T, Shiraiwa M, Kawada A, Takahara H. 1999. Molecular cloning of cDNAs of mouse peptidylarginine deiminase type I, type III and type IV, and the expression pattern of type I in mouse. Eur. J. Biochem. 259, 660-669. ( 10.1046/j.1432-1327.1999.00083.x) [DOI] [PubMed] [Google Scholar]
- 37.Guerrin M, Ishigami A, Méchin MC, Nachat R, Valmary S, Sebbag M, Simon M, Senshu T, Serre G. 2003. cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem. J. 370, 167-174. ( 10.1042/BJ20020870) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlén M, et al. 2015. Tissue-based map of the human proteome. Science 347, 1260419. ( 10.1126/science.1260419) [DOI] [PubMed] [Google Scholar]
- 39.Watanabe K, Akiyama K, Hikichi K, Ohtsuka R, Okuyama A, Senshu T. 1988. Combined biochemical and immunochemical comparison of peptidylarginine deiminases present in various tissues. BBA - Gen. Subj. 966, 375-383. ( 10.1016/0304-4165(88)90088-8) [DOI] [PubMed] [Google Scholar]
- 40.Watanabe K, Senshu T. 1989. Isolation and characterization of cDNA clones encoding rat skeletal muscle peptidylarginine deiminase. J. Biol. Chem. 264, 15 255-15 260. ( 10.1016/s0021-9258(19)84818-4) [DOI] [PubMed] [Google Scholar]
- 41.Khan SA, Edwards BS, Muth A, Thompson PR, Cherrington BD, Navratil AM. 2016. GnRH stimulates peptidylarginine deiminase catalyzed histone citrullination in gonadotrope cells. Mol. Endocrinol. 30, 1081-1091. ( 10.1210/me.2016-1085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanno T, et al. 2000. Human peptidylarginine deiminase type III: molecular cloning and nucleotide sequence of the cDNA, properties of the recombinant enzyme, and immunohistochemical localization in human skin. J. Invest. Dermatol. 115, 813-823. ( 10.1046/j.1523-1747.2000.00131.x) [DOI] [PubMed] [Google Scholar]
- 43.Nachat R, Méchin MC, Charveron M, Serre G, Constans J, Simon M. 2005. Peptidylarginine deiminase isoforms are differentially expressed in the anagen hair follicles and other human skin appendages. J. Invest. Dermatol. 125, 34-41. ( 10.1111/j.0022-202X.2005.23763.x) [DOI] [PubMed] [Google Scholar]
- 44.Chavanas S, Méchin MC, Takahara H, Kawada A, Nachat R, Serre G, Simon M. et al. 2004. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 330, 19-27. ( 10.1016/j.gene.2003.12.038) [DOI] [PubMed] [Google Scholar]
- 45.Vossenaar ER, Radstake TR, van der Heijden A, van Mansum MA, Dieteren C, de Rooij DJ, Barrera P, Zendman AJ, van Venrooij WJ. 2004. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann. Rheum. Dis. 63, 373-381. ( 10.1136/ard.2003.012211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young C, Russell JR, Lawson H, Mapperley C, Kranc KR, Christophorou MA. 2021. Peptidylarginine deiminase IV (PADI4) is not essential for cell-autonomous HSC maintenance and normal haematopoiesis. bioRxiv. [DOI] [PMC free article] [PubMed]
- 47.Brahmajosyula M, Miyake M. 2013. Localization and expression of peptidylarginine deiminase 4 (PAD4) in mammalian oocytes and preimplantation embryos. Zygote 21, 315-324. ( 10.1017/S0967199411000633) [DOI] [PubMed] [Google Scholar]
- 48.Wright PW, et al. 2003. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev. Biol. 256, 74-89. ( 10.1016/S0012-1606(02)00126-4) [DOI] [PubMed] [Google Scholar]
- 49.Chang X, Han J. 2006. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol. Carcinog. 45, 183-196. ( 10.1002/mc.20169) [DOI] [PubMed] [Google Scholar]
- 50.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. 2010. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry 49, 4852-4863. ( 10.1021/bi100363t) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darrah E, Rosen A, Giles JT, Andrade F. 2012. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann. Rheum. Dis. 71, 92-98. ( 10.1136/ard.2011.151712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vikhe Patil K, Mak KHM, Genander M. 2021. A Hairy Cituation – PADIs in Regeneration and Alopecia. Front. Cell Dev. Biol. 9, 789676. ( 10.3389/fcell.2021.789676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. 2002. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem. Biophys. Res. Commun. 290, 979-983. ( 10.1006/bbrc.2001.6303) [DOI] [PubMed] [Google Scholar]
- 54.Cuthbert GL, et al. 2004. Histone deimination antagonizes arginine methylation. Cell 118, 545-553. ( 10.1016/j.cell.2004.08.020) [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, et al. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279-283. ( 10.1126/science.1101400) [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, et al. 2012. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor α target gene activation. Proc. Natl Acad. Sci. USA 109, 13 331-13 336. ( 10.1073/pnas.1203280109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guertin MJ, Zhang X, Anguish L, Kim S, Varticovski L, Lis JT, Hager GL, Coonrod SA. 2014. Targeted H3R26 deimination specifically facilitates estrogen receptor binding by modifying nucleosome structure. PLoS Genet. 10, e1004613. ( 10.1371/journal.pgen.1004613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McNee G, et al. 2017. Citrullination of histone H3 drives IL-6 production by bone marrow mesenchymal stem cells in MGUS and multiple myeloma. Leukemia 31, 373-381. ( 10.1038/leu.2016.187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolodziej S, et al. 2014. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat. Commun. 5, 1-2. ( 10.1038/ncomms4995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeVore SB, et al. 2018. Histone citrullination represses MicroRNA expression, resulting in increased oncogene mRNAs in somatolactotrope cells. Mol. Cell. Biol. 38, e00084-18. ( 10.1128/mcb.00084-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cherrington BD, Morency E, Struble AM, Coonrod SA, Wakshlag JJ. 2010. Potential role for peptidylarginine deiminase 2 (PAD2) in citrullination of canine mammary epithelial cell histones. PLoS ONE 5, e11768. ( 10.1371/journal.pone.0011768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao S, et al. 2017. SMARCAD1 contributes to the regulation of naive pluripotency by interacting with histone citrullination. Cell Rep. 18, 3117-3128. ( 10.1016/j.celrep.2017.02.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma P, Azebi S, England P, Christensen T, Møller-Larsen A, Petersen T, Batsche E, Muchardt C. et al. 2012. Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLoS Genet. 9, e1002934. ( 10.1371/journal.pgen.1002934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denis H, Deplus R, Putmans P, Yamada M, Métivier R, Fuks F. 2009. Functional connection between deimination and deacetylation of histones. Mol. Cell. Biol. 29, 4982-4993. ( 10.1128/mcb.00285-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakashima K, et al. 2013. PAD4 regulates proliferation of multipotent haematopoietic cells by controlling c-myc expression. Nat. Commun. 4, 1-8. ( 10.1038/ncomms2862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293, 1074-1080. ( 10.1126/science.1063127) [DOI] [PubMed] [Google Scholar]
- 67.Dwivedi N, et al. 2014. Deimination of linker histones links neutrophil extracellular trap release with autoantibodies in systemic autoimmunity. FASEB J. 28, 2840-2851. ( 10.1096/fj.13-247254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, et al. 2009. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184, 205-213. ( 10.1083/jcb.200806072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl Acad. Sci. USA 102, 3611-3618. ( 10.1073/pnas.0407159102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deplus R, Denis H, Putmans P, Calonne E, Fourrez M, Yamamoto K, Suzuki A, Fuks F. 2014. Citrullination of DNMT3A by PADI4 regulates its stability and controls DNA methylation. Nucleic Acids Res. 42, 8285-8296. ( 10.1093/nar/gku522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma P, et al. 2019. Citrullination at the C-terminal domain controls RNA polymerase II transcription. Mol. Cell 73, 84– 96.e7. ( 10.1016/j.molcel.2018.10.016) [DOI] [PubMed] [Google Scholar]
- 72.Eick D, Geyer M. 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 113, 8456-8490. ( 10.1021/cr400071f) [DOI] [PubMed] [Google Scholar]
- 73.Ghari F, et al. 2016. Immunology: Citrullination-acetylation interplay guides E2F-1 activity during the inflammatory response. Sci. Adv. 2, e1501257. ( 10.1126/sciadv.1501257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun B, et al. 2017. Citrullination of NF-B p65 promotes its nuclear localization and TLR-induced expression of IL-1 and TNF. Sci. Immunol. 2, eaal3062. ( 10.1126/sciimmunol.aal3062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun B, et al. 2019. Reciprocal regulation of Th2 and Th17 cells by PAD2-mediated citrullination. JCI Insight. 4, e129687. ( 10.1172/jci.insight.129687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loos T, Mortier A, Gouwy M, Ronsse I, Put W, Lenaerts J-P, Van Damme J, Proost P. 2008. Citrullination of CXCL10 and CXCL11 by peptidylarginine deiminase: a naturally occurring posttranslational modification of chemokines and new dimension of immunoregulation. Blood 112, 2648-2656. ( 10.1182/blood-2008-04-149039) [DOI] [PubMed] [Google Scholar]
- 77.Proost P, et al. 2008. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J. Exp. Med. 205, 2085-2097. ( 10.1084/jem.20080305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Struyf S, et al. 2009. Citrullination of CXCL12 Differentially Reduces CXCR4 and CXCR7 Binding with Loss of Inflammatory and Anti-HIV-1 Activity via CXCR4. J. Immunol. 182, 666-674. ( 10.4049/jimmunol.182.1.666) [DOI] [PubMed] [Google Scholar]
- 79.Moelants EAV, Mortier A, Grauwen K, Ronsse I, Van Damme J, Proost P. 2013. Citrullination of TNF-α by peptidylarginine deiminases reduces its capacity to stimulate the production of inflammatory chemokines. Cytokine. 61, 161-167. ( 10.1016/j.cyto.2012.09.011) [DOI] [PubMed] [Google Scholar]
- 80.Stadler SC, et al. 2013. Dysregulation of PAD4-mediated citrullination of nuclear GSK3β activates TGF-β signaling and induces epithelialto-mesenchymal transition in breast cancer cells. Proc. Natl Acad. Sci. USA 110, 11 851-11 856. ( 10.1073/pnas.1308362110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qu Y, et al. 2018. Small molecule promotes β-catenin citrullination and inhibits Wnt signaling in cancer. Nat. Chem. Biol. 14, 94-101. ( 10.1038/nchembio.2510) [DOI] [PubMed] [Google Scholar]
- 82.Qin H, et al. 2017. PAD1 promotes epithelial-mesenchymal transition and metastasis in triple-negative breast cancer cells by regulating MEK1-ERK1/2-MMP2 signaling. Cancer Lett. 409, 30-41. ( 10.1016/j.canlet.2017.08.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xue T, et al. 2021. PADI2-Catalyzed MEK1 Citrullination Activates ERK1/2 and Promotes IGF2BP1-Mediated SOX2 mRNA Stability in Endometrial Cancer. Adv. Sci. 8, 2002831. ( 10.1002/advs.202002831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rogers GE, Harding HWJ, Llewellyn-Smith IJ. 1977. The origin of citrulline-containing proteins in the hair follicle and the chemical nature of trichohyalin, an intracellular precursor. BBA - Protein Struct. 495, 159-175. ( 10.1016/0005-2795(77)90250-1) [DOI] [PubMed] [Google Scholar]
- 85.Senshu T, Akiyama K, Kan S, Asaga H, Ishigami A, Manabe M. 1995. Detection of deiminated proteins in rat skin: probing with a monospecific antibody after modification of citrulline residues. J. Invest. Dermatol. 105, 163-169. ( 10.1111/1523-1747.ep12317070) [DOI] [PubMed] [Google Scholar]
- 86.Senshu T, Kan S, Ogawa H, Manabe M, Asaga H. 1996. Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem. Biophys. Res. Commun. 225, 712-719. ( 10.1006/bbrc.1996.1240) [DOI] [PubMed] [Google Scholar]
- 87.Ishida-Yamamoto A, Takahashi H, Iizuka H, Senshu T, Akiyama K, Nomura K. 2000. Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J. Invest. Dermatol. 114, 701-705. ( 10.1046/j.1523-1747.2000.00936.x) [DOI] [PubMed] [Google Scholar]
- 88.Steinert PM, Parry DAD, Marekov LN. 2003. Trichohyalin mechanically strengthens the hair follicle. J. Biol. Chem. 278, 41 409-41 419. ( 10.1074/jbc.m302037200) [DOI] [PubMed] [Google Scholar]
- 89.Sun B, et al. 2021. PAD2-mediated citrullination of Fibulin-5 promotes elastogenesis. Matrix. Biol. 102, 70-84. ( 10.1016/j.matbio.2021.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuzhalin AE, et al. 2018. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat. Commun. 9, 4783. ( 10.1038/s41467-018-07306-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stefanelli VL, et al. 2019. Citrullination of fibronectin alters integrin clustering and focal adhesion stability promoting stromal cell invasion. Matrix. Biol. 82, 86-104. ( 10.1016/j.matbio.2019.04.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sipilä K, et al. 2014. Citrullination of collagen II affects integrin-mediated cell adhesion in a receptor-specific manner. FASEB J. 28, 3758-3766. ( 10.1096/fj.13-247767) [DOI] [PubMed] [Google Scholar]
- 93.Esposito G, Vitale AM, Leijten FPJ, Strik AM, Koonen-Reemst AMCB, Yurttas P, Robben TJAA, Coonrod S, Gossen JA. 2007. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol. Cell. Endocrinol. 273, 25-31. ( 10.1016/j.mce.2007.05.005) [DOI] [PubMed] [Google Scholar]
- 94.Harauz G, Musse AA. 2007. A tale of two citrullines - Structural and functional aspects of myelin basic protein deimination in health and disease. Neurochem. Res. 32, 137-158. ( 10.1007/s11064-006-9108-9) [DOI] [PubMed] [Google Scholar]
- 95.Moscarello MA, et al. 2013. Inhibition of peptidyl-arginine deiminases reverses protein- hypercitrullination and disease in mouse models of multiple sclerosis. DMM Dis. Model. Mech. 6, 467-478. ( 10.1242/dmm.010520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coassolo S, Davidson G, Negroni L, Gambi G, Daujat S, Romier C, Davidson I. 2021. Citrullination of pyruvate kinase M2 by PADI1 and PADI3 regulates glycolysis and cancer cell proliferation. Nat. Commun. 12, 1718. ( 10.1038/s41467-021-21960-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang M, et al. 2021. Citrullination of glucokinase linked to autoimmune diabetes. Nat.Comms. 13, 1870. ( 10.1038/s41467-022-29512-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Snijders AP, et al. 2015. Arginine methylation and citrullination of splicing factor proline- and glutamine-rich (SFPQ/PSF) regulates its association with mRNA. RNA 21, 347-359. ( 10.1261/rna.045138.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sorice M, et al. 2016. Autophagy generates citrullinated peptides in human synoviocytes: a possible trigger for anti-citrullinated peptide antibodies. Rheumatol. (United Kingdom). 55, 1374-1385. ( 10.1093/rheumatology/kew178) [DOI] [PubMed] [Google Scholar]
- 100.Ireland JM, Unanue ER. 2011. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J. Exp. Med. 208, 2625-2632. ( 10.1084/jem.20110640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, et al. 2012. Anticancer peptidylarginine deiminase (PAD) inhibitors regulate the autophagy flux and the mammalian target of rapamycin complex 1 activity. J. Biol. Chem. 287, 25 941-25 953. ( 10.1074/jbc.M112.375725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fan T, et al. 2014. Peptidylarginine deiminase IV promotes the development of chemoresistance through inducing autophagy in hepatocellular carcinoma. Cell Biosci. 4, 49. ( 10.1186/2045-3701-4-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cau L, Takahara H, Thompson PR, Serre G, Méchin MC, Simon M. 2019. Peptidylarginine deiminase inhibitor Cl-amidine attenuates cornification and interferes with the regulation of autophagy in reconstructed human epidermis. J. Invest. Dermatol. 139, 1889-1997. ( 10.1016/j.jid.2019.02.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeisel A, et al. 2015. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138-1142. ( 10.1126/science.aaa1934) [DOI] [PubMed] [Google Scholar]
- 105.Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch H-M, Probert L, Casaccia-Bonnefil P, Moscarello MA. 2006. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J. Neurosci. 26, 11 387-11 396. ( 10.1523/JNEUROSCI.3349-06.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang L, Tan D, Piao H. 2016. Myelin basic protein citrullination in multiple sclerosis: a potential therapeutic target for the pathology. Neurochem. Res. 41, 1845-1856. ( 10.1007/s11064-016-1920-2) [DOI] [PubMed] [Google Scholar]
- 107.Jang B, Ishigami A, Maruyama N, Carp RI, Kim YS, Choi EK. 2013. Peptidylarginine deiminase and protein citrullination in prion diseases Strong evidence of neurodegeneration. Prion 7, 42-46. ( 10.4161/pri.22380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mukherjee S, et al. 2021. Citrullination of amyloid-β peptides in Alzheimer's Disease. ACS Chem. Neurosci. 12, 3719-3732. ( 10.1021/acschemneuro.1c00474) [DOI] [PubMed] [Google Scholar]
- 109.Méchin MC, Takahara H, Simon M. 2020. Deimination and peptidylarginine deiminases in skin physiology and diseases. Int. J. Mol. Sci. 21, 566. ( 10.3390/ijms21020566) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Basmanav FBÜ, et al. 2016. Mutations in three genes encoding proteins involved in hair shaft formation cause uncombable hair syndrome. Am. J. Hum. Genet. 99, 1291-1304. ( 10.1016/j.ajhg.2016.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Malki L, et al. 2019. Variant PADI3 in central centrifugal cicatricial alopecia. N. Engl. J. Med. 380, 833-841. ( 10.1056/nejmoa1816614) [DOI] [PubMed] [Google Scholar]
- 112.Horibata S, Coonrod SA, Cherrington BD. 2012. Role for peptidylarginine deiminase enzymes in disease and female reproduction. J. Reprod. Develop. 58, 274-282. ( 10.1262/jrd.2011-040) [DOI] [PubMed] [Google Scholar]
- 113.Young CH, Rothfuss HM, Gard PF, Muth A, Thompson PR, Ashley RL, Cherrington BD. 2017. Citrullination regulates the expression of insulin-like growth factor-binding protein 1 (IGFBP1) in ovine uterine luminal epithelial cells. Reproduction 153, 1-10. ( 10.1530/REP-16-0494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Young CH, Snow B, Devore SB, Mohandass A, Nemmara VV, Thompson PR, Thyagarajan B, Navratil AM, Cherrington BD. 2021. Progesterone stimulates histone citrullination to increase IGFBP1 expression in uterine cells. Reproduction 162, 117-127. ( 10.1530/REP-21-0132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brahmajosyula M, Wakayama T, Miyake M. 2010. Role of peptidylarginine deiminase 4 in mammalian preimplantation embryonic development. Biol. Reprod. 83, 253. ( 10.1093/biolreprod/83.s1.253) [DOI] [Google Scholar]
- 116.Kan R, Jin M, Subramanian V, Causey CP, Thompson PR, Coonrod SA. 2012. Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Dev. Biol. 12, 19. ( 10.1186/1471-213X-12-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brahmajosyula M, Miyake M. 2013. Role of peptidylarginine deiminase 4 (PAD4) in pig parthenogenetic preimplantation embryonic development. Zygote 21, 385-393. ( 10.1017/S0967199412000160) [DOI] [PubMed] [Google Scholar]
- 118.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. 2010. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207, 1853-1862. ( 10.1084/jem.20100239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303, 1532-1535. ( 10.1126/science.1092385) [DOI] [PubMed] [Google Scholar]
- 120.Wilson AS, et al. 2022. Neutrophil extracellular traps and their histones promote Th17 cell differentiation directly via TLR2. Nat. Commun. 13, 528. ( 10.1038/s41467-022-28172-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ireland J, Herzog J, Unanue ER. 2006. Cutting edge: unique t cells that recognize citrullinated peptides are a feature of protein immunization. J. Immunol. 177, 1421-1425. ( 10.4049/jimmunol.177.3.1421) [DOI] [PubMed] [Google Scholar]
- 122.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. 2003. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC Class II Molecule. J. Immunol. 171, 538-541. ( 10.4049/jimmunol.171.2.538) [DOI] [PubMed] [Google Scholar]
- 123.Becart S, Whittington KB, Prislovsky A, Rao NL, Rosloniec EF. 2021. The role of posttranslational modifications in generating neo-epitopes that bind to rheumatoid arthritis-associated HLA-DR alleles and promote autoimmune T cell responses. PLoS ONE 16, e0245541. ( 10.1371/journal.pone.0245541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Van Venrooij WJ, Van Beers JJBC, Pruijn GJM. 2011. Anti-CCP antibodies: the past, the present and the future. Nat. Rev. Rheumatol. 7, 391-398. ( 10.1038/nrrheum.2011.76) [DOI] [PubMed] [Google Scholar]
- 125.Ciesielski O, Biesiekierska M, Panthu B, Soszyński M, Pirola L, Balcerczyk A. 2022. Citrullination in the pathology of inflammatory and autoimmune disorders: recent advances and future perspectives. Cell. Mol. Life Sci. 79, 94. ( 10.1007/s00018-022-04126-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leshner M, Wang S, Lewis C, Zheng H, Chen XA, Santy L, Wang Y. 2012. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front. Immunol. 3, 307. ( 10.3389/fimmu.2012.00307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lewis HD, et al. 2015. Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation. Nat. Chem. Biol. 11, 189-191. ( 10.1038/nchembio.1735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen X, et al. 2016. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods. 13, 1013-1020. ( 10.1038/nmeth.4031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thiama HR, Wong SL, Qiu R, Kittisopikul M, Vahabikashi A, Goldman AE, Goldman RD, Wagner DD, Waterman CM. 2020. NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture. Proc. Natl Acad. Sci. USA 117, 7326-7337. ( 10.1073/pnas.1909546117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsourouktsoglou TD, Warnatsch A, Ioannou M, Hoving D, Wang Q, Papayannopoulos V. 2020. Histones, DNA, and citrullination promote neutrophil extracellular trap inflammation by regulating the localization and activation of TLR4. Cell Rep. 31, 107602. ( 10.1016/j.celrep.2020.107602) [DOI] [PubMed] [Google Scholar]
- 131.Konig MF, Andrade F. 2016. A critical reappraisal of neutrophil extracellular traps and NETosis mimics based on differential requirements for protein citrullination. Front. Immunol. 7, 461. ( 10.3389/fimmu.2016.00461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tatsiy O, de Carvalho Oliveira V, Mosha HT, McDonald PP. 2021. Early and late processes driving NET formation, and the autocrine/paracrine role of endogenous RAGE ligands. Front. Immunol. 12, 675315. ( 10.3389/fimmu.2021.675315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boeltz S, et al. 2019. To NET or not to NET:current opinions and state of the science regarding the formation of neutrophil extracellular traps. Cell Death Differ. 26, 395-408. ( 10.1038/s41418-018-0261-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rosazza T, Warner J, Sollberger G. 2021. NET formation – mechanisms and how they relate to other cell death pathways. FEBS J. 288, 3334-3350. ( 10.1111/febs.15589) [DOI] [PubMed] [Google Scholar]
- 135.Cools-Lartigue J, Spicer J, Mcdonald B, Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P, Ferri L. 2013. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J. Clin. Invest. 123, 3448-3458. ( 10.1172/JCI67484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kolaczkowska E, Jenne CN, Surewaard BGJ, Thanabalasuriar A, Lee W-Y, Sanz M-J, Mowen K, Opdenakker G, Kubes P. 2015. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat. Commun. 6, 6673. ( 10.1038/ncomms7673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, Kahn CR, Wagner DD. 2015. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 21, 815-819. ( 10.1038/nm.3887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun S, et al. 2021. Neutrophil extracellular traps impair intestinal barrier functions in sepsis by regulating TLR9-mediated endoplasmic reticulum stress pathway. Cell Death Dis. 12, 1-2. ( 10.1038/s41419-021-03896-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alflen A, et al. 2020. Neutrophil extracellular traps impair fungal clearance in a mouse model of invasive pulmonary aspergillosis. Immunobiology 225, 151867. ( 10.1016/j.imbio.2019.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martinod K, et al. 2017. Peptidylarginine deiminase 4 promotes age-related organ fibrosis. J. Exp. Med. 214, 439-458. ( 10.1084/jem.20160530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Martinod K, Demers M, Fuchs TA, Wong SL, Brill A, Gallant M, Hu J, Wang Y, Wagner DD. 2013. Neutrophil histone modification by peptidylarginine deiminase 4 is critical for deep vein thrombosis in mice. Proc. Natl Acad. Sci. USA 110, 8674-8679. ( 10.1073/pnas.1301059110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Savchenko AS, Borissoff JI, Martinod K, De Meyer SF, Gallant M, Erpenbeck L, Brill A, Wang Y, Wagner DD. 2014. VWF-mediated leukocyte recruitment with chromatin decondensation by PAD4 increases myocardial ischemia/reperfusion injury in mice. Blood 123, 141-148. ( 10.1182/blood-2013-07-514992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang L, et al. 2017. PADI2-mediated citrullination promotes prostate cancer progression. Cancer Res. 77, 5755-5768. ( 10.1158/0008-5472.CAN-17-0150) [DOI] [PubMed] [Google Scholar]
- 144.Horibata S, Rogers KE, Sadegh D, Anguish LJ, Mcelwee JL, Shah P, Thompson PR, Coonrod SA. 2017. Role of peptidylarginine deiminase 2 (PAD2) in mammary carcinoma cell migration. BMC Cancer 17, 378. ( 10.1186/s12885-017-3354-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. 2009. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 9, 40. ( 10.1186/1471-2407-9-40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. 2008. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J. Biol. Chem. 283, 20 060-20 068. ( 10.1074/jbc.M802940200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo Q, Fast W. 2011. Citrullination of Inhibitor of Growth 4 (ING4) by Peptidylarginine Deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J. Biol. Chem. 286, 17 069-17 078. ( 10.1074/jbc.M111.230961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li P, et al. 2010. Coordination of PAD4 and HDAC2 in the regulation of p53-target gene expression. Oncogene 29, 3153-3162. ( 10.1038/onc.2010.51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Masucci MT, Minopoli M, Del Vecchio S, Carriero MV. 2020. The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front. Immunol. 11, 1749. ( 10.3389/fimmu.2020.01749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tohme S, et al. 2016. Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Res. 76, 1367-1380. ( 10.1158/0008-5472.CAN-15-1591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang L, et al. 2020. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 583, 133-138. ( 10.1038/s41586-020-2394-6) [DOI] [PubMed] [Google Scholar]
- 152.McElwee JL, Mohanan S, Horibata S, Sams KL, Anguish LJ, Mclean D, Cvitaå¡ I, Wakshlag JJ, Coonrod SA. 2014. PAD2 overexpression in transgenic mice promotes spontaneous skin neoplasia. Cancer Res. 74, 6306-6317. ( 10.1158/0008-5472.CAN-14-0749) [DOI] [PubMed] [Google Scholar]
- 153.Tanikawa C, et al. 2012. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat. Commun. 3, 676. ( 10.1038/ncomms1676) [DOI] [PubMed] [Google Scholar]
- 154.Brentville VA, Vankemmelbeke M, Metheringham RL, Durrant LG. 2020. Post-translational modifications such as citrullination are excellent targets for cancer therapy. Semin. Immunol. 47, 101393. ( 10.1016/j.smim.2020.101393) [DOI] [PubMed] [Google Scholar]
- 155.Durrant LG, Metheringham RL, Brentville VA. 2016. Autophagy, citrullination and cancer. Autophagy 12, 1055-1056. ( 10.1080/15548627.2016.1166326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M, Cook K, Xue W, Durrant LG. 2016. Citrullinated vimentin presented on MHC-II in tumor cells is a target for CD4+ T-Cell-mediated antitumor immunity. Cancer Res. 76, 548-560. ( 10.1158/0008-5472.CAN-15-1085) [DOI] [PubMed] [Google Scholar]
- 157.Neeli I, Khan SN, Radic M. 2008. Histone Deimination As a Response to Inflammatory Stimuli in Neutrophils. J. Immunol. 180, 1895-1902. ( 10.4049/jimmunol.180.3.1895) [DOI] [PubMed] [Google Scholar]
- 158.Youn HD, Sun L, Prywes R, Liu JO. 1999. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286, 790-793. ( 10.1126/science.286.5440.790) [DOI] [PubMed] [Google Scholar]
- 159.Llinás R, Sugimori M, Silver RB. 1992. Microdomains of high calcium concentration in a presynaptic terminal. Science 256, 677-679. ( 10.1126/science.1350109) [DOI] [PubMed] [Google Scholar]
- 160.Tour O, Adams SR, Kerr RA, Meijer RM, Sejnowski TJ, Tsien RW, Tsien RY. 2007. Calcium Green FlAsH as a genetically targeted small-molecule calcium indicator. Nat. Chem. Biol. 3, 423-431. ( 10.1038/nchembio.2007.4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Demaurex N, Monod A, Lew DP, Krause KH. 1994. Characterization of receptor-mediated and store-regulated Ca2+ influx in human neutrophils. Biochem. J. 297, 595-601. ( 10.1042/bj2970595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Darrah E, Giles JT, Ols ML, Bull HG, Andrade F, Rosen A. 2013. Erosive rheumatoid arthritis is associated with antibodies that activate PAD4 by increasing calcium sensitivity. Sci. Transl. Med. 5, 186ra65. ( 10.1126/scitranslmed.3005370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Demoruelle MK, Wang H, Davis RL, Visser A, Hoang J, Norris JM, Holers VM, Deane KD, Darrah E. 2021. Anti-peptidylarginine deiminase-4 antibodies at mucosal sites can activate peptidylarginine deiminase-4 enzyme activity in rheumatoid arthritis. Arthritis Res. Ther. 23, 163. ( 10.1186/s13075-021-02528-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X. 2010. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 62, 1630-1640. ( 10.1002/art.27439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arandjelovic S, McKenney KR, Leming SS, Mowen KA. 2012. ATP induces protein arginine deiminase 2-dependent citrullination in mast cells through the P2X7 purinergic receptor. J. Immunol. 189, 4112-4122. ( 10.4049/jimmunol.1201098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Neeli I, Radic M. 2016. Current challenges and limitations in antibody-based detection of citrullinated histones. Front. Immunol. 7, 528. ( 10.3389/fimmu.2016.00528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y, et al. 2021. Histone citrullination by PADI4 is required for HIF-dependent transcriptional responses to hypoxia and tumor vascularization. Sci. Adv. 7, eabe3771. ( 10.1126/sciadv.abe3771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Griffante G, et al. 2021. Human cytomegalovirus-induced host protein citrullination is crucial for viral replication. Nat. Commun. 12, 3910. ( 10.1038/s41467-021-24178-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chang HH, Dwivedi N, Nicholas AP, Ho IC. 2015. The W620 polymorphism in PTPN22 disrupts its interaction with peptidylarginine deiminase type 4 and enhances citrullination and netosis. Nat.Comms 13, 1870. ( 10.1002/art.39215) [DOI] [PubMed] [Google Scholar]
- 170.Chou S, et al. 2015. Transferred interbacterial antagonism genes augment eukaryotic innate immune function. Nature 518, 98-101. ( 10.1038/nature13965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Golenberg N, Squirrell JM, Bennin DA, Rindy J, Pistono PE, Eliceiri KW, Shelef MA, Kang J, Huttenlocher A. et al. 2020. Citrullination regulates wound responses and tissue regeneration in zebrafish. J. Cell Biol. 219, e201908164. ( 10.1083/jcb.201908164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yaffe MB. 2018. Predicting the future of signaling for 2018. Sci. Signal. 11, eaar7429. ( 10.1126/scisignal.aar7429) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.