Abstract
Atezolizumab plus bevacizumab (ATZ/BV) treatment is a combined immunotherapy consisting of immune checkpoint inhibitor (ICI) and anti-vascular endothelial growth factor monoclonal antibody, which has brought a major paradigm shift in the treatment of unresectable hepatocellular carcinoma (HCC). Gain-of-function mutation of CTNNB1 contributes to resistance of ICI monotherapy through the framework of non-T-cell-inflamed tumor microenvironment. However, whether CTNNB1 mutation renders resistance to ATZ/BV similar to ICI monotherapy remains to be elucidated. In this study, a liquid biopsy sample in plasma of 33 patients with HCC treated with ATZ/BV was subjected to droplet digital PCR for detecting hotspot mutations at the exon 3 of CTNNB1 locus. A total of eight patients (24.2%) exhibited at least one CTNNB1 mutation. The objective response rate (ORR) in patients with wild-type (WT) and mutant (MT) CTNNB1 was 8.0% and 12.5%, respectively, and the disease control rate (DCR) was 68.0% and 87.5%, respectively. No significant difference in both ORR and DCR has been observed between the two groups. The median progression-free survival in patients with WT and MT CTNNB1 was 6.6 and 7.6 months, respectively (not statistically significant). Similarly, no significant difference in overall survival has been observed between patients with WT and MT CTNNB1 (13.6 vs. 12.3 months). In conclusion, the treatment effect of ATZ/BV in patients with HCC with MT CTNNB1 was comparable to those patients with WT CTNNB1. These results implicate that BV added to ATZ might improve immunosuppressive tumor microenvironment caused by CTNNB1 mutation.
Keywords: Liquid biopsy, CTNNB1, atezolizumab plus bevacizumab, HCC
Introduction
Recent advances in next-generation sequencer (NGS) and analysis solutions have made it possible to analyze the genomes in a variety of cancers. Although a number of driver gene mutations was detected in hepatocellular carcinoma (HCC), among which TERT promoter, CTNNB1, and TP53, are found to be most frequent 1, 2. CTNNB1 mutations are usually detected in exon 3 which encodes serine-threonine phosphorylation sites for GSK-3β that activates β-catenin degradation. Subsequently, CTNNB1 mutation at the site leads to constitutive activation of Wnt/β-catenin signaling 3. Recent omics analyses have successfully shown that HCC with CTNNB1 mutation is characterized by small-sized and well-differentiated tumor and are considered to be a group with a favorable prognosis 4, 5. Additionally, CTNNB1 mutation is also enriched in non-T-cell-inflamed tumors 6, which render poor clinical response to immune checkpoint inhibitor (ICI) in HCC 7.
Recently, atezolizumab plus bevacizumab (ATZ/BV) has been approved as a first-line treatment for advanced HCC 8. This treatment consists of ICI and anti-vascular endothelial growth factor (VEGF) antibody and is positioned as an immune complex therapy. The association between CTNNB1 mutation and the therapeutic effect of ATZ/BV is of interest but remains to be elucidated. In this study, hot spot mutations of CTNNB1 have been detected at Ser33, Ser37, Thr41 and Ser45 by droplet digital polymerase chain reaction (ddPCR) using circulating tumor DNA (ctDNA) of patients with HCC, and their relationship with ATZ/BV treatment response and prognosis has been investigated.
Materials and Methods
Among the patients treated with ATZ/BV for HCC in our hospital between October 2020 and June 2021, this study included a total of 33 patients of whom blood samples were collected before treatment initiation. After obtaining the informed consent, ctDNA was extracted from the plasma samples using the MagMAX Cell-Free DNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA) with King Fisher Duo Prime (Thermo Fisher Scientific). The cell-free DNA concentration was measured using Qubit 4 Fluorometer (Thermo Fisher Scientific). The ddPCR Mutation Detection Assays and the QX200 droplet digital PCR system (Bio-Rad, Hercules, CA) have been used for detecting mutant CTNNB1 in exon 3 at Ser33 (A95G/A95T), Ser37 (C98G), Thr41 (A121G), and Ser45 (T133C/C134T) 9. A95G/A95T and T133C/C134T have been detected using cocktail mutant primer/probe mixture. Mutation allele frequency (MAF) value greater than 0.1% was considered to be positive for the mutation. Patients received ATZ/BV intravenously every 3 weeks. Radiological assessments have been evaluated using contrast-enhanced CT or MRI at every two cycles according to the Response Evaluation Criteria in Solid Tumors, version 1.1 (RECIST 1.1) 10. Adverse events (AEs) were evaluated according to the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE), version 4.0 11. Statistical comparisons of clinical variables between two groups (CTNNB1 wild-type (WT) and mutant (MT)) have been performed using unpaired t-test, Fisher's exact test or Pearson's chi-square test. The Kaplan-Meier method and the log-rank test have been used to determine progression-free survival (PFS) and overall survival (OS). P-values <0.05 were considered statistically significant. All statistical analyses were performed using the SPSS statistical software version 24 (IBM, Chicago, IL). This study was approved by the research ethics committees of the Graduate School of Medicine, Chiba University (approval number: 1090, 3416, and 3950).
Results
Plasma samples from 33 patients treated with ATZ/BV, including 26 men and 7 women whose average age was 72 years (range, 48-89 years), have been analyzed (Table 1). Chronic liver damage was due to HBV (n = 3), HCV (n = 8), alcohol (n = 3), and others (n = 19). According to the Child-Pugh classification, they were classified as class A5 (n = 18), and class A6 (n = 15). The number of patients with BCLC stage B and C was 8 and 25, respectively. Eighteen patients were treated with ATZ/BV as a first-line systemic chemotherapy. Fourteen and eighteen patients were accompanied by macrovascular invasion (MVI) and extrahepatic spread (EHS), respectively.
Table 1.
Patients' characteristics at baseline
| Characteristics | All patients (n = 33) |
CTNNB1 WT (n = 25) |
CTNNB1 MT (n = 8) |
p-value |
|---|---|---|---|---|
| Age (years, median) | 72 | 70 | 76 | 0.110 |
| Gender (male/female) | 26/7 | 20/5 | 6/2 | >0.999 |
| Etiology (Viral/Non-viral)※ | 11/22 | 10/15 | 1/7 | 0.218 |
| Child-Pugh grade (A5/A6) | 18/15 | 12/13 | 6/2 | 0.242 |
| BCLC stage (B/C) | 8/25 | 4/21 | 4/4 | 0.366 |
| Prior systemic therapy (yes/no) | 15/18 | 13/12 | 2/6 | 0.242 |
| AFP (ng/mL, median) | 13326.3 | 16826.9 | 2387.0 | 0.555 |
| Tumor number (≧4/<4) | 15/18 | 10/15 | 5/3 | 0.418 |
| Macrovascular invasion (yes/no) | 14/19 | 13/12 | 1/7 | 0.098 |
| Extrahepatic spread (yes/no) | 18/15 | 14/11 | 4/4 | >0.999 |
Abbreviations: WT, wild-type; MT, mutant; AFP, alpha-fetoprotein; BCLC, Barcelona clinic liver cancer
※Viral group was defined as patients who were HBs antigen-positive and/or HCV antibody-positive, while the non-viral group was defined as all others.
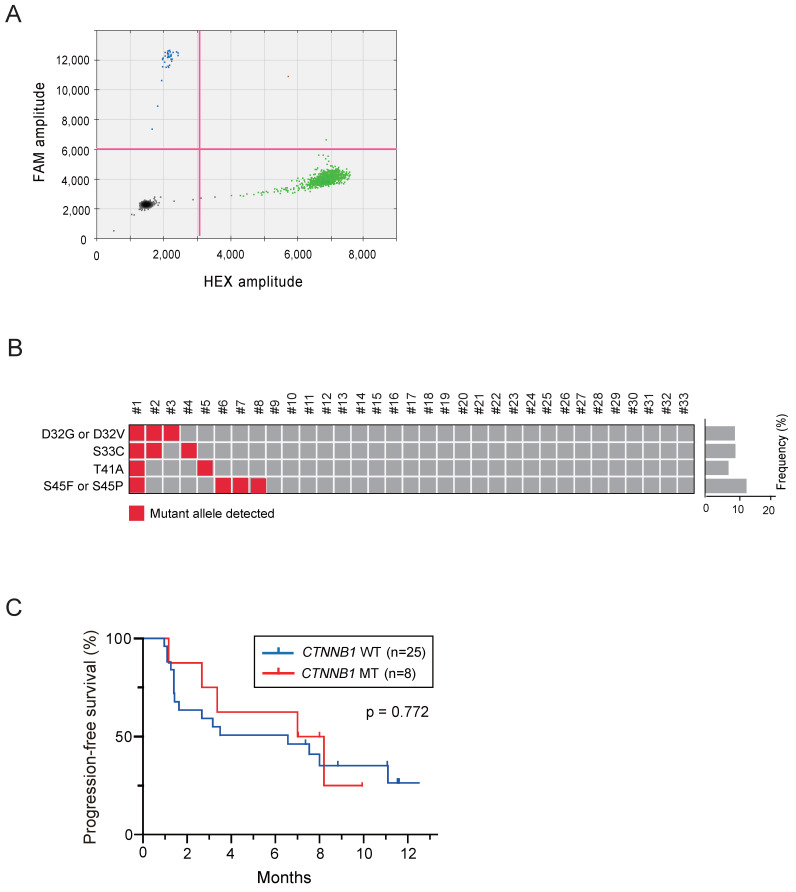
The ctDNA of these patients was successfully subjected to ddPCR assays (Figure 1A). At least one missense mutation in the CTNNB1 in exon 3 was found in 8 of 33 patients (24.2%), and the median MAF was 0.47% (range: 0.1%-26.8%) (Figure 1B). No significant difference in clinical background variables has been observed between the WT and MT groups (Table 1). In addition, there was no significant difference in the size, vascularity, and forms in nodules with the largest diameters among two groups 12.
Figure 1.
Droplet digital PCR assays using circulating tumor DNA extracted from plasma of patients with hepatocellular carcinoma treated with atezolizumab plus bevacizumab. (A) Representative droplet digital PCR assay for detecting CTNNB1 mutation at Thr41 (A121G). Black, green, blue, and orange dots indicate empty droplets, wild-type DNA HEX-positive droplets, mutant DNA FAM-positive droplets, and wild-type and mutant double-positive droplets, respectively. (B) Summary of results for the detection of CTNNB1 mutations in all patients. (C) Progression-free survival of patients based on CTNNB1 mutations.
AEs of any grades were observed in 6 (75.0%) and 19 patients (76.0%) with WT and MT CTNNB1, respectively. The most frequent adverse events in WT group were hypertension (28.0%), fatigue (20.0%), and proteinuria (16.0%). Similarly, those in MT group were hypertension, proteinuria, and rush (25.0%). Immune-related adverse events (irAEs) were suspected in 5 patients (hypothyroidism, hypopituitarism, and rush) in WT group and 3 patients (hyperthyroidism, rush, and type 1 diabetes) in MT group.
Subsequently, treatment response and prognosis have been investigated based on the presence or absence of CTNNB1 mutation. The objective response rate (ORR) in patients with WT and MT CTNNB1 was 8.0% (2/25) and 12.5% (1/8), respectively (Table 2). The disease control rate (DCR) in patients with WT and MT CTNNB1 was 68.0% (17/25) and 87.5% (7/8), respectively. Collectively, no significant difference in both ORR and DCR has been observed. Even if limited to patients who have not undergone prior systemic chemotherapy, there was no significant difference in ORR and DCR among the patients with or without CTNNB1 mutation. During the follow-up period (median: 8.2 months), no statistically significant difference in PFS has been observed between the two groups (median: 6.6 vs. 7.6 months, p = 0.772) (Figure 1C). Similarly, no statistically significant difference in OS has been also observed between the two groups (median: 13.6 vs. 12.3 months). Together, although the study was based on a limited number of cases, both treatment response and prognosis in patients with CTNNB1 mutations were comparable to those in patients without CTNNB1 mutations.
Table 2.
Response to treatment
| CTNNB1 WT (n = 25) | CTNNB1 MT (n = 8) | p-value | |
|---|---|---|---|
| Best response | |||
| CR | 0 | 0 | |
| PR | 2 | 1 | |
| SD | 15 | 6 | |
| PD | 8 | 1 | |
| ORR (%) | 8.0 | 12.5 | >0.999 |
| DCR (%) | 68.0 | 87.5 | 0.394 |
Abbreviations: WT, wild-type; MT, mutant; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; ORR, objective response rate; DCR, disease control rate
Discussion
To eradicate cancer cells, it is important that the series of stepwise events, so called “cancer-immunity cycle,” functions properly in tumors, lymph nodes, and vessels 13. However, tumor cells are known to have immune escape mechanisms including PD-1/PD-L1 and CTLA4 pathways 14. They were called immune checkpoint pathways, which make it difficult to eliminate tumor cells by cytotoxic T cells (CTLs) 15. ICIs including ATZ are inhibitory antibodies against checkpoint molecules and reactivate suppressed immune responses 16. However, tumors with CTNNB1 mutations are known as non-T-cell-inflamed tumors, which do not respond well to ICI 17. Conversely, VEGF, the target of bevacizumab, binds to VEGF receptor 2 specifically expressed on vascular endothelial cells 18. VEGF not only promotes the proliferation and migration of vascular endothelial cells, but also suppresses tumor immunity by acting on various immune cells 19. Therefore, this study sought to evaluate the therapeutic efficacy of ATZ/BV combination therapy in patients with HCC with CTNNB1 mutation who would not respond to ICI alone.
Liquid biopsy is a minimally invasive approach, mainly by collecting blood, and can be performed repeatedly 20. Unlike tissue biopsy samples, liquid samples are less susceptible to the bias of tumor heterogeneity 21. Firstly, ctDNA was extracted from plasma by liquid biopsy, and subsequently ddPCR assays were conducted. As a result, CTNNB1 mutation was detected in 8 of 33 patients (24.2%). Considering that the frequency of mutations has been reported to be ranging from 23% to 36% in NGS analyses of HCC tissue samples 22, 23, although the detection rate was slightly lower in this study, it was considered acceptable for ddPCR to detect specific mutations. Given that tumor-derived DNA is a small fraction of cell-free DNA in plasma, it is often difficult to detect mutations by liquid biopsy in cases with low tumor burden. Consistent with this finding, it has been reported that the concordance rate of mutation detection between tissue and liquid biopsies is up to 70% 24. COSMIC database shows that CTNNB1 mutation rate in HCC is higher in Europe and Americas than in Asia 25. In addition, CTNNB1 mutation is reported to be associated with HCV-related HCC 26. The current analysis population was a relatively small number of Japanese patients with 24.2% of HCV-positive rate. This might be also one of the reasons for the lower CTNNB1 mutation detection rate compared to previous reports.
Importantly, no significant difference in the percentage of RECIST-based treatment response has been observed between WT and MT CTNNB1 groups. Concordant with these results, no significant difference was observed in both PFS and OS. It is well known that OS of patients with advanced cancer is also affected by later-line treatment. Our current study was conducted over a relatively short observation period. Taking into consideration that clinical applications of Wnt inhibitors are being attempted for advanced HCC 27, the impact of CTNNB1 mutation on survival in patients treated with ATZ/BV should be further investigated.
Blockade of VEGF is known to increase CTL infiltration and decrease immunosuppressive cells, including regulatory T cells and myeloid-derived suppressor cells, thereby promoting tumor cell recognition and cancer cell death 28, 29. Additionally, it is known that anti-VEGF promotes dendritic cell maturation and accelerates T-cell priming 30. Altogether, BV added to ATZ might change the “non-inflamed” pathological features of HCC with mutant CTNNB1 to the “inflamed” thorough the modification of “cancer-immunity cycle.” This should be confirmed by pathological examination of paired tumor tissues before and after ATZ/BV administration.
In conclusion, our results suggest the possibilities that the therapeutic effect of ATZ/BV is not attributable to the presence or absence of CTNNB1 mutation. Further analysis with a large number of patients and for a longer observation period would be necessary to build solid evidences.
Acknowledgments
The authors thank Risa Kakiuchi and Anna Sakaue for technical assistance.
Funding
This study was partially supported by grants from the Japan Society for the Promotion of Science (JSPS) and the Program for Basic and Clinical Research on Hepatitis from Japan Agency for Medical Research and Development (AMED, #JP21fk0210054s0103).
Availability of data
Data can be made available upon reasonable request.
Author Contributions
KO, HK, and TC designed the study, performed statistical analyses and prepared the manuscript. KO, HK, TC, HU, SY, TI (Takamasa Ishino), MK, TI (Terunao Iwanaga), MN (Miyuki Nakagawa), SF, NF, TS (Takafumi Sakuma), KK (Keisuke Koroki), YK, KK (Kazufumi Kobayashi), NK, SK, MN (Masato Nakamura), TK (Takayuki Kondo), TS (Tomoko Saito), SO, ES, and SN collected the samples. JA, NQ, YM, JZ, RN, and RM. analyzed and interpreted the data. HK and TC performed statistical analyses and prepared the manuscript. TK (Tatsuo Kanda), HM, NM, JK, SM, and NK edited and reviewed the manuscript. Informed consent was obtained from all patients.
Ethical Committee Approval and Patient Consent
This study was approved by the research ethics committees of the Graduate School of Medicine, Chiba University (approval number: 1090, 3416, and 3950).
Abbreviations
- AE
adverse event
- ATZ
atezolizumab
- ATZ/BV
atezolizumab plus bevacizumab
- BV
bevacizumab
- ctDNA
circulating tumor DNA
- CTL
cytotoxic T cell
- DCR
disease control rate
- ddPCR
droplet digital polymerase chain reaction
- EHS
extrahepatic spread
- ICI
immune checkpoint inhibitor
- irAE
immune-related adverse event
- HCC
hepatocellular carcinoma
- MAF
mutation allele frequency
- MT
mutant
- MVI
macrovascular invasion
- NGS
next-generation sequencer
- ORR
objective response rate
- OS
overall survival
- PCR
polymerase chain reaction
- PFS
progression-free survival
- WT
wild-type
References
- 1.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M. et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–73. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 2.Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S. et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–11. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–92. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M. et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology. 2017;153:812–26. doi: 10.1053/j.gastro.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin Cancer Res. 2019;25:3074–83. doi: 10.1158/1078-0432.CCR-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M. et al. Prospective Genotyping of Hepatocellular Carcinoma: Clinical Implications of Next-Generation Sequencing for Matching Patients to Targeted and Immune Therapies. Clin Cancer Res. 2019;25:2116–26. doi: 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY. et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med. 2020;382:1894–905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 9.Asano K, Mikata R, Chiba T, Kan M, Maruta S, Yamada T. et al. Analysis of circulating cell-free DNA after endoscopic ultrasound-guided fine needle aspiration in pancreatic ductal adenocarcinoma. Pancreatology. 2021;21:1030–7. doi: 10.1016/j.pan.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz LH, Seymour L, Litière S, Ford R, Gwyther S, Mandrekar S. et al. RECIST 1.1 - Standardisation and disease-specific adaptations: Perspectives from the RECIST Working Group. Eur J Cancer. 2016;62:138–45. doi: 10.1016/j.ejca.2016.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brahmer JR, Lacchetti C, Schneider BJ, Atkins MB, Brassil KJ, Caterino JM. et al. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2018;36:1714–68. doi: 10.1200/JCO.2017.77.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai T, Hirohashi S, Upton MP, Noguchi M, Kishi K, Makuuchi M. et al. Pathology of small hepatocellular carcinoma. A proposal for a new gross classification. Cancer. 1987;60:810–9. doi: 10.1002/1097-0142(19870815)60:4<810::aid-cncr2820600417>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 13.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotte A, Jin JY, Lemaire V. Mechanistic overview of immune checkpoints to support the rational design of their combinations in cancer immunotherapy. Ann Oncol. 2018;29:71–83. doi: 10.1093/annonc/mdx686. [DOI] [PubMed] [Google Scholar]
- 16.Akinleye A, Rasool Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J Hematol Oncol. 2019;12:92. doi: 10.1186/s13045-019-0779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pai SG, Carneiro BA, Mota JM, Costa R, Leite CA, Barroso-Sousa R. et al. Wnt/beta-catenin pathway: modulating anticancer immune response. J Hematol Oncol. 2017;10:101. doi: 10.1186/s13045-017-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibuya M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer. 2011;2:1097–105. doi: 10.1177/1947601911423031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukumura D, Kloepper J, Amoozgar Z, Duda DG, Jain RK. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15:325–40. doi: 10.1038/nrclinonc.2018.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet) Cancer Cell. 2017;31:172–9. doi: 10.1016/j.ccell.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415–21. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenetrol Hepatol. 2014;11:340–9. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 23.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139–52. doi: 10.1038/s41575-019-0229-4. [DOI] [PubMed] [Google Scholar]
- 25.Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74–83. doi: 10.1016/j.ygeno.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Javanmard D, Najafi M, Babaei MR, Karbalaie Niya MH, Esghaei M. et al. Investigation of CTNNB1 gene mutations and expression in hepatocellular carcinoma and cirrhosis in association with hepatitis B virus infection. Infect Agent Cancer. 2020;15:37. doi: 10.1186/s13027-020-00297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada K, Hori Y, Inoue S, Yamamoto Y, Iso K, Kamiyama H, Yamaguchi A. et al. E7386, a Selective Inhibitor of the Interaction between β-Catenin and CBP, Exerts Antitumor Activity in Tumor Models with Activated Canonical Wnt Signaling. Cancer Res. 2021;81:1052–62. doi: 10.1158/0008-5472.CAN-20-0782. [DOI] [PubMed] [Google Scholar]
- 28.Wallin JJ, Bendell JC, Funke R, Sznol M, Korski K, Jones S. et al. Atezolizumab in combination with bevacizumab enhances antigen-specific T-cell migration in metastatic renal cell carcinoma. Nat Commun. 2016;7:12624. doi: 10.1038/ncomms12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegde PS, Wallin JJ, Mancao C. Predictive markers of anti-VEGF and emerging role of angiogenesis inhibitors as immunotherapeutics. Semin Cancer Biol. 2018;52(Pt 2):117–24. doi: 10.1016/j.semcancer.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Gabrilovich DI, Chen HL, Girgis KR, Cunningham HT, Meny GM, Nadaf S. et al. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat Med. 1996;2:1096–103. doi: 10.1038/nm1096-1096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data can be made available upon reasonable request.