Abstract
Feruloyl esterases can remove aromatic residues (e.g., ferulic acid) from plant cell wall polysaccharides (xylan, pectin) and are essential for complete degradation of these polysaccharides. Expression of the feruloyl esterase-encoding gene (faeA) from Aspergillus niger depends on d-xylose (expression is mediated by XlnR, the xylanolytic transcriptional activator) and on a second system that responds to aromatic compounds with a defined ring structure, such as ferulic acid and vanillic acid. Several compounds were tested, and all of the inducing compounds contained a benzene ring which had a methoxy group at C-3 and a hydroxy group at C-4 but was not substituted at C-5. Various aliphatic groups occurred at C-1. faeA expression in the presence of xylose or ferulic acid was repressed by glucose. faeA expression in the presence of ferulic acid and xylose was greater than faeA expression in the presence of either compound alone. The various inducing systems allow A. niger to produce feruloyl esterase not only during growth on xylan but also during growth on other ferulic acid-containing cell wall polysaccharides, such as pectin.
Feruloyl esterases are enzymes that release aromatic residues, such as ferulic acid, from plant cell wall polymers (5). Ferulic acid residues in plant cell walls are linked mainly to xylan and pectin and may cross-link cell wall polymers, which increases rigidity, inhibits cell elongation, and reduces biodegradability by microorganisms (15). In xylans, ferulic acid is attached to arabinose residues, which are linked to the xylan backbone (26, 32). In pectins, ferulic acid can be attached to galactose or arabinose residues in the side chains (15). Ferulic acid can be enzymatically converted into vanillin, a major flavor compound (10, 13, 18).
Feruloyl esterases can be isolated from a wide range of microorganisms (3, 5, 8, 11, 16) when they are grown on complex substrates, such as xylan, pectin, wheat bran, or sugar beet pulp. Most of these enzymes are active with feruloylated oligomers obtained from xylan or pectin but exhibit little or no activity with the polymeric substrates. The feruloyl esterase A (FaeA)-encoding gene (faeA) from Aspergillus niger has been cloned, and induction of this gene has been studied at the protein level (5). A high level of FaeA activity in the culture filtrate was detected when an overproducing transformant was grown on medium containing wheat arabinoxylan. Low levels of FaeA activity were detected when A. niger was grown on sugar beet pectin (31). Addition of ferulic acid to media containing xylan increased the levels of FaeA activity in culture filtrates (12).
In A. niger, two regulatory systems affect the expression of genes encoding enzymes involved in the degradation of cell wall polymers. The carbon catabolite repressor protein, CreA, prevents transcription of these genes in the presence of substrates that are easy to metabolize, such as glucose or fructose (9, 23). The xylanolytic transcriptional activator, XlnR (28), is required for expression of all of the xylanolytic genes (including faeA) and some cellulolytic genes when A. niger is grown on xylose or xylan (29). These two systems are not sufficient to account for the induction pattern observed for faeA (5, 12). The fact that induction on pectin and enhanced induction on xylan in the presence of ferulic acid occur suggests that there is a more complex regulatory system. Our objectives in this study were (i) to determine the involvement of XlnR and CreA in the regulation of faeA expression, (ii) to demonstrate the presence of a third factor regulating faeA expression, (iii) to identify the nature of the inducing compounds for this new factor, and (iv) to analyze the relationships among the three regulatory factors.
MATERIALS AND METHODS
Strains.
All of the strains used were derived from A. niger N400 (= CBS120.49). A low-sporulating mutant, N402 (cspA1), was used as a wild-type strain in all experiments. Mutations were obtained by UV mutagenesis. The A. niger creA mutant NW200 (bioA1 cspA1 creAd4 pyrA13::pGW635 areA1::pAREG1), which had a derepressed phenotype, was selected in an areA1 background (24) and was cotransformed with pAREG1 (containing the A. niger areA gene [20]) and pGW635 (containing the pyrA selection marker). A. niger NXA1-4 [argB13 cspA1 nicA1 pyrA6 UAS(xlnA)-pyrA xlnR1] has been described previously (28). No expression of xylanolytic genes on xylose was observed with this loss-of-function mutant (29). Escherichia coli DH5αF′ (BRL Life Technologies Inc., Gaithersburg, Md.) was used for routine plasmid propagation.
Chemicals.
Caffeic acid, cinnamic acid, p-coumaric acid, ferulic acid, 4-hydroxybenzoic acid, 3,4-dimethoxycinnamic acid, protocatechuic acid, sinapic acid, syringic acid, vanillic acid, veratric acid, veratryl alcohol, vanillyl alcohol, and anisyl alcohol were obtained from Acros (Oxon, United Kingdom). d-Glucose, d-fructose, and d-xylose were obtained from Merck (Darmstadt, Germany). l-Arabinose was obtained from Sigma (Zwijndrecht, The Netherlands). All other standard chemicals were obtained from either Sigma or Merck. 3-Methoxy-4-hydroxyphenyl propionic acid was a gift from Gary Williamson, Institute of Food Research (Norwich, United Kingdom).
Media and culture conditions.
Minimal medium contained (per liter) 6 g of NaNO3, 1.5 g of KH2PO4, 0.5 g of KCl, 0.5 g of MgSO4 · 7H2O, trace elements (30), and 1% (mass/vol) glucose as a carbon source unless otherwise indicated. For complete medium, minimal medium was supplemented with 0.2% (mass/vol) tryptone, 0.1% (mass/vol) yeast extract, 0.1% (mass/vol) Casamino Acids, and 0.05% (mass/vol) yeast RNA. Liquid cultures were inoculated with 106 spores/ml and incubated at 30°C in an orbital shaker at 250 rpm. Bacto-Agar (Difco) (1.5%, mass/vol) was used to solidify the media. To grow auxotrophic strains, the necessary supplements were added to the media at the following concentrations: biotin, 4 μg/liter; uridine, 200 mg/liter; arginine, 200 mg/liter; and nicotinamide, 1 mg/liter.
For all transfer experiments, the strains were grown first in complete medium (containing 2% fructose) for 16 h at 30°C in an orbital shaker at 250 rpm. The mycelium was harvested by suction over a nylon membrane (100-μm mesh) and washed with minimal medium or complete medium without a carbon source. Aliquots (1.5 g, wet weight) of the mycelium were transferred to minimal medium or complete medium containing different carbon sources and incubated at 30°C. The mycelium was again harvested by suction over a nylon membrane, frozen in liquid nitrogen, and stored at −70°C. Aromatic compounds were used at a concentration of 0.03% (mass/vol).
DNA manipulation.
Standard methods, such as subcloning, DNA digestion, and plasmid DNA isolation (25), were used for DNA manipulation. A sequence analysis was performed for both strands of DNA by using either a Cy 5 AutoCycle sequencing kit or a Cy 5 Thermo Sequenase dye terminator kit (Pharmacia Biotech, Uppsala, Sweden). The reaction mixtures were analyzed with an ALFred DNA sequencer (Pharmacia Biotech). Nucleotide sequences were analyzed with computer programs based on the programs of Devereux et al. (4).
Northern analysis.
Mycelium was powdered with a Micro-dismembrator (Braun, Melsungen, Germany). Total RNA was isolated from the powdered mycelium by using TRIzol reagent (Life Technologies) as recommended by the supplier. For Northern analysis, 5 μg of total RNA was incubated with 3.3 μl of 6 M glyoxal, 10 μl of dimethyl sulfoxide, and 2 μl of 0.1 M phosphate buffer (pH 7) in a 20-μl (total volume) mixture for 1 h at 50°C in order to denature the RNA. RNA samples were separated on a 1.5% agarose gel by using 0.01 M phosphate buffer (pH 5) and were transferred to Hybond-N filters (Amersham, Little Chalfont, United Kingdom) by capillary blotting. The filters were hybridized at 42°C in a solution containing 50% (vol/vol) formamide, 10% (mass/vol) dextran sulfate, 0.9 M NaCl, 90 mM trisodium citrate, 0.2% (mass/vol) Ficoll, 0.2% (mass/vol) polyvinylpyrrolidone, 0.2% (mass/vol) bovine serum albumin, 0.1% (mass/vol) sodium dodecyl sulfate, and 100 μg of single-stranded herring sperm DNA per ml. Washing was performed under similar stringency conditions (30 mM NaCl, 3 mM trisodium citrate, 0.5% [mass/vol] sodium dodecyl sulfate; 68°C). A 0.6-kb cDNA fragment of faeA, a 0.8-kb EcoRV-KpnI fragment of aguA encoding an α-glucuronidase (6), and a 0.7-kb EcoRI fragment of the 18S rRNA subunit (21) were used as probes. The aguA gene was used to compare the expression of faeA to the expression of another gene regulated by XlnR, and the 18S rRNA probe was used as an RNA loading control.
RESULTS
Analysis of the promoter region of faeA.
We sequenced the promoter region of faeA (EMBL accession no. Y09330) and identified a consensus sequence for the CreA repressor protein, SYGGRG (S, Y, and R represent C or G, C or T, and A or G, respectively [17]) at position −376 upstream of the ATG in the 3′→5′ orientation, and for the xylanolytic transcriptional activator, XlnR (GGCTAA [29]) at position −265 upstream of the ATG in the 5′→3′ orientation. An XlnR-like sequence (GGCTAG) was detected at position −225 upstream of the ATG in the 5′→3′ orientation.
Expression of faeA on aromatic compounds.
High levels of faeA expression were observed when the A. niger wild-type strain was grown on ferulic acid, vanillic acid, vanillyl alcohol, coniferyl alcohol, and 3-methoxy-4-hydroxyphenyl propionic acid (Fig. 1). Lower levels of expression were observed when this strain was grown on vanillin, veratric acid, and veratryl alcohol. No expression was detected with any of the other aromatic compounds.
FIG. 1.
Expression of faeA in the presence of aromatic compounds. A. niger N402 mycelium was incubated for 4 h in complete medium (blank), in complete medium containing 0.03% (mass/vol) fructose, and in complete medium containing different aromatic compounds at a concentration of 0.03% (mass/vol). The blots are Northern blots prepared as described in the text. 18S rRNA was used as an RNA loading control. Abbreviations: hydr., hydroxy; meth., methoxy; dimeth., dimethoxy; ppa, phenylpropionic acid.
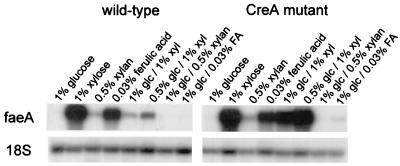
CreA represses expression of faeA.
A. niger N402 and NW200 mycelia were transferred to minimal medium containing different carbon sources and were harvested after 4 h. The faeA gene was expressed in the presence of xylose, xylan, and ferulic acid at similar levels in the two strains (Fig. 2). Glucose strongly reduced the expression of faeA by the wild-type strain on xylose, but only a small decrease was observed with the creA mutant. The presence of glucose eliminated expression of faeA in the presence of ferulic acid and xylan with the wild-type strain, but a low level of expression in the presence of ferulic acid was detected with the creA mutant.
FIG. 2.
CreA repression of faeA expression. To determine the influence of the carbon catabolite repressor protein CreA on faeA expression, A. niger N402 and NW200 (CreA mutant) mycelia were incubated for 4 h in minimal medium containing different carbon sources. The blots are Northern blots prepared as described in the text. 18S rRNA was used as an RNA loading control. Abbreviations: glc, glucose; xyl, xylose; FA, ferulic acid.
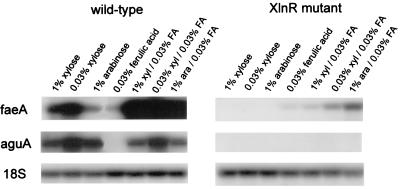
Interaction between the different systems for induction of faeA expression.
We compared expression of faeA to expression of aguA, which encodes an α-glucuronidase and also is regulated by XlnR (29), in the wild type and an XlnR-deficient mutant 2 h after induction. Expression of faeA and expression of aguA were both greater in the presence of 0.03% xylose than in the presence of 1% xylose in the wild-type strain (Fig. 3). Addition of ferulic acid to the carbon sources strongly increased faeA expression but did not have a significant effect on aguA expression. aguA expression was not observed in the presence of any carbon source in the XlnR mutant, but faeA expression was detected in this mutant in the presence of ferulic acid, especially when arabinose was also present.
FIG. 3.
Interaction of different systems for induction of faeA expression. Relationships between XlnR and ferulic acid induction of faeA expression were studied by incubating A. niger N402 and NXA1-4 (XlnR mutant) mycelia for 2 h in minimal medium containing different carbon sources. The blots are Northern blots prepared as described in the text. 18S rRNA was used as an RNA loading control. Abbreviations: xyl, xylose; ara, arabinose; FA, ferulic acid.
DISCUSSION
The data described in this paper revealed that regulation of faeA transcription is complex. faeA is subject to carbon catabolite repression, and its expression is subject to the carbon catabolite repressor CreA, the xylanolytic transcriptional activator XlnR, and a factor that responds to the presence of certain aromatic compounds.
Expression of faeA on xylose in the presence of glucose is reduced strongly in the A. niger wild-type strain but only slightly in the creAd4 mutant, which indicates that CreA regulates the expression of faeA. The creAd4 mutant is not a complete loss-of-function mutant, and the degree of derepression is target and allele specific (31). This phenotype may explain in part the reduced faeA expression on ferulic acid by the creA mutant when glucose is present in the medium. Additional repression by glucose, independent of CreA, also may be important. The residual glucose-mediated repression of faeA expression in the creA mutant indicates that the ferulic acid-dependent regulatory system is more sensitive to glucose repression than XlnR. The xylose concentration influences XlnR-induced expression of xylanolytic genes, as revealed by the expression of faeA and aguA in the presence of 1 and 0.03% xylose. This phenomenon has been explained by modulation of gene expression via CreA (7). Expression of faeA and aguA on arabinose may be due to xylose contamination of the arabinose (6). Adding ferulic acid to xylose or arabinose resulted in increased levels of expression of faeA but not aguA, xlnB, and xlnD (data not shown), indicating that ferulic acid does not influence the overall regulation of the xylanolytic enzyme spectrum. The difference in auxotrophic markers might have some effect on the levels of expression observed in the strains used in this study, as demonstrated previously for Saccharomyces cerevisiae (3). However, a comparison of strains with different auxotrophic markers did not reveal significant differences in the expression of the genes examined in this study (data not shown).
faeA is expressed in the presence of aromatic compounds with the following substituents on the aromatic ring: a methoxy group at C-3, a hydroxy group at C-4, and an unsubstituted C-5. The C-1 substituent seems to be less important for the level of expression. When we examined a group of aromatic compounds with the substitutions on the aromatic ring described above, only vanillin resulted in a low level of faeA expression. Perhaps an aldehyde linkage at C-1 results in a lower level of induction of faeA. Alternatively, the low level of expression may be caused by the toxicity of vanillin to A. niger. All of the aromatic compounds which we tested are toxic to A. niger to some extent (14, 19). Vanillin is the most toxic of these compounds, as determined by plate growth experiments (data not shown). The regulatory system described here differs from other systems in A. niger that respond to the presence of aromatic compounds in that benzoic acid and 4-hydroxybenzoic acid, which induce the production of enzymes involved in the catabolism of benzoic acid via 4-hydroxybenzoic acid and 3,4-dihydroxybenzoic acid to protocatechuic acid (1, 2, 27), do not induce expression of faeA. Thus, at least two independent regulatory systems respond to induction by aromatic compounds in A. niger. Milstein et al. (22) suggested a model for the degradation of certain aromatic compounds by Aspergillus japonicus. They suggested that there are four groups of aromatic compounds with individual catabolic pathways that converge at protocatechuic acid, followed by ring opening via a common pathway. Ferulic acid and 4-hydroxybenzoic acid are in different groups, and this model also could explain the data presented here.
faeA expression in A. niger is affected by at least three regulatory systems. faeA is coexpressed with other genes involved in xylan degradation and is under the control of the transcriptional activator XlnR. Xylose simultaneously induces this system and causes carbon catabolite repression, which is mediated by CreA. The response of an additional regulatory system to the release of ferulic acid from xylan and pectin might make these plant cell wall polymers more accessible for degradation even at higher xylose concentrations. The data presented here provide more insight into the complex regulation of A. niger genes that encode cell wall-degrading enzymes and a better understanding of the changes in the fungal enzyme spectrum that occur during degradation of plant cell wall components.
ACKNOWLEDGMENT
We thank Danisco Ingredients, Brabrand, Denmark, for financial support.
REFERENCES
- 1.Boschloo J G, Paffen A, Koot T, van den Tweel W J J, van Gorcom R F M, Cordewener J H G, Bos C J. Genetic analysis of benzoate metabolism in Aspergillus niger. Appl Microbiol Biotechnol. 1990;34:225–228. [Google Scholar]
- 2.Chopra R, Sharma V M, Ganesan K. Elevated growth of Saccharomyces cerevisiae ATH1 null mutants on glucose is an artifact of nonmatching auxotrophies of mutant and reference strains. Appl Environ Microbiol. 1999;65:2267–2268. doi: 10.1128/aem.65.5.2267-2268.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christov L P, Prior B A. Esterases of xylan-degrading microorganisms: production, properties, and significance. Enzyme Microbiol Technol. 1993;15:460–475. doi: 10.1016/0141-0229(93)90078-g. [DOI] [PubMed] [Google Scholar]
- 4.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries R P, Michelsen B, Poulsen C H, Kroon P A, van den Heuvel R H H, Faulds C B, Williamson G, van den Hombergh J P T W, Visser J. The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in the degradation of complex cell wall polysaccharides. Appl Environ Microbiol. 1997;63:4638–4644. doi: 10.1128/aem.63.12.4638-4644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries R P, Poulsen C H, Madrid S, Visser J. aguA, the gene encoding an extracellular α-glucuronidase from Aspergillus tubingensis, is specifically induced on xylose and not on glucuronic acid. J Bacteriol. 1998;180:243–249. doi: 10.1128/jb.180.2.243-249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries R P, de Graaff L H, Visser J. CreA modulates the XlnR induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol. 1999;150:281–285. doi: 10.1016/s0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- 8.Donaghy J, McKay A M. Purification and characterisation of a feruloyl esterase from the fungus Penicillium expansum. J Appl Microbiol. 1997;83:718–726. doi: 10.1046/j.1365-2672.1997.00307.x. [DOI] [PubMed] [Google Scholar]
- 9.Drysdale M R, Kolze S E, Kelly J M. The Aspergillus niger carbon catabolite repressor encoding gene, creA. Gene. 1993;130:241–245. doi: 10.1016/0378-1119(93)90425-3. [DOI] [PubMed] [Google Scholar]
- 10.Falconnier B, Lapierre C, Lesage-Meessen L, Yonnet G, Brunerie P, Colonna-Ceccaldi B, Corrieu G, Asther M. Vanillin as a product of ferulic acid biotransformation by the white-rot fungus Pycnoporus cinnabarinus I-937: identification of metabolic pathways. J Biotechnol. 1994;37:123–132. [Google Scholar]
- 11.Faulds C B, Williamson G. Purification and characterisation of a ferulic acid esterase (FAE-III) from Aspergillus niger: specificity for the phenolic moiety and binding to microcrystalline cellulose. Microbiology. 1994;140:779–787. [Google Scholar]
- 12.Faulds C B, de Vries R P, Kroon P A, Visser J, Williamson G. Influence of ferulic acid on the production of feruloyl esterases by Aspergillus niger. FEMS Microbiol Lett. 1997;157:239–244. doi: 10.1111/j.1574-6968.1997.tb12779.x. [DOI] [PubMed] [Google Scholar]
- 13.Gasson M J, Kitamura Y, McLauchlin W R, Narbad A, Parr A J, Lindsay E, Parsons H, Payne J, Rhodes M J C, Walton N J. Metabolism of ferulic acid to vanillin. J Biol Chem. 1998;273:4163–4170. doi: 10.1074/jbc.273.7.4163. [DOI] [PubMed] [Google Scholar]
- 14.Guiraud P, Steiman R, Seigle-Murandi F, Benoit-Guyod J L. Comparison of the toxicity of various lignin-related aromatic compounds towards selected fungi perfecti and fungi imperfecti. Ecotoxicol Environ Safety. 1995;32:29–33. doi: 10.1006/eesa.1995.1081. [DOI] [PubMed] [Google Scholar]
- 15.Ishii T. Structure and function of feruloylated polysaccharides. Plant Sci. 1997;127:111–127. [Google Scholar]
- 16.Kroon P A, Faulds C B, Williamson G. Purification and characterization of a novel esterase induced by growth of Aspergillus niger on sugarbeet pulp. Biotechnol Appl Biochem. 1996;23:255–262. [PubMed] [Google Scholar]
- 17.Kulmburg P, Mathieu M, Dowzer C, Kelly J M, Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CreA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 18.Lesage-Meessen L, Delattre M, Haon M, Thibault J-F, Colonna-Ceccaldi B, Brunerie P, Asther M. A two-step conversion process for vanillin production from ferulic acid combining Aspergillus niger and Pycnoporus cinnabarinus. J Biotechnol. 1996;50:107–113. doi: 10.1016/0168-1656(96)01552-0. [DOI] [PubMed] [Google Scholar]
- 19.López-Malo A, Alzamora S M, Argaiz A. Effect of vanillin concentration, pH and incubation temperature on Aspergillus flavus, Aspergillus niger, Aspergillus ochraceus and Aspergillus parasiticus growth. Food Microbiol. 1997;14:117–124. [Google Scholar]
- 20.McCabe A P, Vanhanen S, Sollewijn Gelpke M D, van de Vondervoort P J I, Arst H N, Jr, Visser J. Identification, cloning and sequence of the Aspergillus niger areA wide domain regulatory gene controling nitrogen utilisation. Biochim Biophys Acta. 1998;1396:163–168. doi: 10.1016/s0167-4781(97)00212-1. [DOI] [PubMed] [Google Scholar]
- 21.Melchers W J G, Verweij P E, van den Hurk P, van Belkum A, de Pauw B E, Hoogkamp-Korstanje A A, Meis J F G M. General primer-mediated PCR for detection of Aspergillus species. J Clin Microbiol. 1994;32:1710–1717. doi: 10.1128/jcm.32.7.1710-1717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milstein O, Trojanowski J, Hüttermann A, Gressel J. Catabolism of single ring aromatic acids by four Aspergillus species. Microbios. 1988;55:7–16. [PubMed] [Google Scholar]
- 23.Ruijter G J G, Visser J. Carbon repression in aspergilli. FEMS Microbiol Lett. 1997;151:103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x. [DOI] [PubMed] [Google Scholar]
- 24.Ruijter G J G, Vanhanen S, Gielkens M M C, van de Vondervoort P J I, Visser J. Isolation of Aspergillus niger creA mutants and effects of the mutations on expression of arabinases and l-arabinose catabolic enzymes. Microbiology. 1997;143:2991–2998. doi: 10.1099/00221287-143-9-2991. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Saulnier L, Vigouroux J, Thibault J-F. Isolation and partial characterization of feruloylated oligosaccharides from maize bran. Carbohydr Res. 1995;272:241–253. doi: 10.1016/0008-6215(95)00053-v. [DOI] [PubMed] [Google Scholar]
- 27.van Gorcom R F M, Boschloo J G, Kuijvenhoven A, Lange J, van Vark A J, Bos C J, van Balken J A M, Pouwels P, van den Hondel C A M J J. Isolation and molecular characterisation of the benzoate-para-hydroxylase gene (bphA) of Aspergillus niger: a member of a new gene family of the cytochrome P450 superfamily. Mol Gen Genet. 1990;233:192–197. doi: 10.1007/BF00265053. [DOI] [PubMed] [Google Scholar]
- 28.van Peij N N M E, Visser J, de Graaff L H. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 29.van Peij N N M E, Gielkens M M C, de Vries R P, Visser J, de Graaff L H. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visser, J. Unpublished data.
- 32.Wende G, Fry S C. O-feruloylated, O-acetylated oligosaccharides as side-chains of grass xylans. Phytochemistry. 1997;44:1011–1018. doi: 10.1016/s0031-9422(96)00648-6. [DOI] [PubMed] [Google Scholar]