Abstract
The incidence of hepatocellular carcinoma (HCC) is increasing worldwide. Extracellular vesicles (EVs) contain sufficient bioactive substances and are carriers of intercellular information exchange, as well as delivery vehicles for nucleic acids, proteins and drugs. Although EVs show great potential for the treatment of HCC and their role in HCC progression has been extensively studied, there are still many challenges such as time-consuming extraction, difficult storage, easy contamination, and low drug loading rate. We focus on the biogenesis, morphological characteristics, isolation and extraction of EVs and their significance in the progression of HCC, tumor invasion, immune escape and cancer therapy for a review. EVs may be effective biomarkers for molecular diagnosis of HCC and new targets for tumor-targeted therapy.
Keywords: hepatocellular carcinoma, tumor microenvironment, hypoxia, vesicle drug delivery, extracellular vesicles
1 Introduction
China is the world’s top liver cancer country, and the 2020 Global Oncology Report showed that 906,000 patients of liver cancer occurred worldwide, of which 410,000 new cases occurred in China, accounting for >45% (1, 2). HCC is a common and fatal cancer, accounting for approximately 90% of all liver cancer cases (3). Although much progress has been made in diagnostic and treatment of HCC, such as liver excision, chemotherapy embolism and Sorafenib, it remains a health problem worldwide, with the incidence expected to exceed one million cases in a few years, due to its metastatic nature, high recurrence rate and low long-term survival (4, 5). EVs exist in tissues, various body fluids and supernatant, such as saliva (6), pleural effusion (7, 8), plasma (9, 10), urine (11), breast milk (12, 13), cerebrospinal fluid (14) and ascites (15, 16), which are greatly released by a variety of cells in a constitutive or inducible manner. EVs can regulate many biological processes, such as migration and extracellular matrix remodeling (17). Recently, some studies have shown that EVs play an important part in regulating cell signaling. Particularly, HCC cell-derived EVs may lead to local spread, distant metastasis and multifocal growth (18). HCC cell can secrete more EVs and promote tumor metastasis. After exposure to anti-tumor drugs, the release of EVs from hepatoma cell also increased, which activate natural killer cells and induce anti-tumor immunity. Besides, tumor cell-derived EVs can produce direct immune effects to stimulate target cells. It has been reported that EVs-mediated intercellular transfer may promote the invasion of HCC by affecting the tumor microenvironment (TME) (18, 19). EVs-mediated signaling in liver disease makes them a unique therapeutic tool that can provide targeted delivery of tissue siRNAs, miRNAs and circRNAs to affect gene expression (20). Notably, EVs are natural nanomaterials. Compared with drugs, modified EVs have many advantages, which significantly improve the specificity, efficacy, and safety of EVs-based cancer therapies and become ideal candidates for drug development and delivery (20). Nowadays, the use of biogenic EVs as drug delivery has become a research hotspot, and its complex phospholipid membrane structure may be conducive to immune escape, site-specific transmission, cell uptake and intracellular transport (21). In addition, some microRNAs in EVs have also been introduced as potential biomarkers, and their expression level is related to the invasiveness of HCC (22). It has been reported that EVs play a key role in biological functions, including intercellular transfer, angiogenesis, immune response, tumor growth and metastasis of HCC (23–25).
2 Introduction of EVs
2.1 Biogenesis and Morphological Characteristics of EVs
It is known that EVs can be a key role in human physiological and pathological diseases with various subtypes of cell-released membrane structures. EVs of particle diameters <200 nm are referred to as small EVs (sEVs) and medium-to-larger particles of diameters >200 nm are referred to as m/lEVs (26). Depending on the description of conditions or cell of origin, EVs can also be classified as apoptotic body, large oncosome, hypoxic EV, podocyte EV, etc, which are showed as follows (22, 26–28) ( Table 1 ).
Table 1.
The types of EVs.
| Classification | Subtypes | Diameter | Source | Marker | Ref |
|---|---|---|---|---|---|
| Physical characteristics/Size | sEV | < 200nm | Originates from the inward outgrowth of multivesicular bodies (MVB), endosomal system | Transmembrane proteins CD9, CD63 and CD81; ALIX; TSG101 | (5, 22, 29) |
| Derived from hepatocytes, macrophages, NK cells, T cells, B cells | |||||
| m/lEV | > 200nm | Plasma membrane outward budding production | Integrin; Selectin; CD40; Most membrane-associated proteins in source cells | (30, 31) | |
| Derived from almost all healthy living cells. | |||||
| Descriptions of conditions/Cell of origin | Apoptotic body | 1-5μm | generated from cell fragments undergoing apoptosis | Phosphatidylserine; Genomic DNA; It is similar to the surface markers of its derived cells and rich in caspases-3 and caspases-7 | (30, 32) |
| Large oncosome | 1-10μm | originates from the shedding of the membrane bubbles | CK18 | (33–36) | |
| released by Invasive prostate cancer cells, urinary bladder, and glioblastoma | |||||
| Hypoxic EV | – | Hypoxic cell | include mRNA and proteins (MMPs, IL-8, PDGFs, caveolin 1, and lysyl oxidase, etc) | (37, 38) | |
| Podocyte EV | – | from the tip of the microvilli of the podocytes | — | (39) |
The sEVs (<200nm) originate from the inward outgrowth of endosomal membranes, are one of subpopulations of EVs (30), which can be produced from different cells such as hepatocytes (40), NK cells (41), T cells (42), and B cells (43), and surface markers of sEVs include CD9, CD63, CD81, and CD82 (44). sEVs are formed by the endonuclear body system and transmit information to the recipient cell through three main processes:First, the cytoplasmic membrane is initially invaginated by lipid raft-mediated endocytosis to form endocytic vesicles, which fuse with each other to form early endosomes (Endocytosis) (45);Second, early intranuclear bodies regenerate and invaginate, and intracellular material forms multiple intraluminal vesicles (ILVs), which are further transformed into late intranuclear bodies and multivesicular bodies (MVBs). This process also involves the inversion of cytoplasmic contents, transmembrane proteins, and peripheral proteins (Receptor-ligand Interaction) (46). Finally, MVBs fuse with the cytoplasmic membrane to form sEVs (Fusion With the Plasma Membrane) (5, 23, 47). In addition, MVBs have also been reported to fuse with lysosomes and promote the degradation of vesicle contents (27, 44, 48). The formation, release and sorting of sEVs are a series of regulated processes, which mainly require the endosomal sorting complex required for transport (ESCRT), members of the ESCRT family [apoptosis contiguous gene 2-interacting protein X (ALIX), also called PDCD6IP (49), tumor susceptibility gene 101 (TSG101)] (50, 51), four transmembrane proteins family (49, 52) and lipid raft-associated proteins (53, 54) and many substances are involved. As we all know, ESCRT is composed of ESCRT-0, ESCRT-I, ESCRT-II and ESCRT-III (55), and is associated with delivery of ubiquitinated proteins, degradation of lysosomes and recycling of proteins (20). Moreover, ESCRT plays an important part in luminal vesicle biogenesis and cargo aggregation (49). ESCRT-independent processes also seem to be involved in the formation and secretion of sEVs in an intertwined manner (56). Intracellular transport of sEVs involves many molecular switches, such as RAB GTpase proteins, membrane linked proteins, actin and microtubulin (23). Besides, Rab family proteins,including Rab7, Rab11, Rab27, and Rab35, also play a crucial role in the process of sEVs secretion (25). The secretion of sEVs also requires the involvement of the SNARE complex and the synaptic binding protein family (30). Furthermore, the involvement of sphingomyelinase in vesicle release was confirmed by the elevated ceramide levels in sEVs and less release of sEVs after sphingomyelinase inhibition (20, 56). Overall, sEVs regulate signaling pathways in receptor cells, coordinate TME and communication between different cells.
The m/lEVs (>200nm) are released by the plasma membrane to the outgoing buds, so the membrane composition of the m/lEVs is extremely close to the plasma membrane. The cell membrane surface is full of phosphatidylserine and most of the membrane-associated proteins, which can regulate the intercellular information exchange and affect the functions of target cells (30). The mechanism of m/lEVs formation is related to intracellular calcium signaling stimulation (23, 57), membrane bending proteins and the asymmetric distribution of phospholipids. The inward flow of calcium ions in the cytoplasm activates phospholipid crawling enzymes to disrupt phospholipid asymmetry, leading to redistribution of phospholipids in the cell membrane bilayer (58). The junctional protein ARRDC1 recruits ESCRT proteins and VPS4 (an ATPase) to the cell membrane (59); ESCRT-1 protein interacts directly with inhibitory proteins; pro-caspase3 stimulates Rho-related protein kinase 1 to promote apoptogenesis and induces myocardin contraction, contributing to the release of m/lEVs.
Apoptotic body (50-2000 nm), also known as apoptotic vesicles, are produced by debris cells that undergo apoptosis (60, 61). When cells undergo apoptosis, the cell membrane folds inward and wraps around the cytoplasm, organelles and nuclear fragments to form vesicles, which are the largest subpopulation of EVs. Apoptotic vesicles have surface markers and are enriched in caspases-3 and caspases-7, caspases-3 and Rho/Rock pathway taking part in membrane blistering (30, 32, 62). Moreover, apoptotic vesicles play a key role in attracting phagocytes, promoting the clearance of apoptotic cell debris, and regulating antigen presentation and immune cell responses (30). Apoptotic cells have been reported that can facilitate the encapsulation of chemotherapeutic drugs or nanoparticles into EVs (22). In addition, apoptotic vesicles from apoptotic cells can be preferentially taken up by macrophages and produce antitumor effects (22). Thus, apoptotic vesicles may also be an ideal delivery system, but the use of apoptotic vesicles as therapeutic nanovesicles (NVs) has been less studied, which may be related to their large cell size and uneven distribution.
Large oncosomes(1-10 μm) are released by cancer cells and may play a role in the tumor microenvironment. It has been shown that CK18 is a marker of large oncosomes and can be identified in circulation and tissues (63).
The mechanism of production of hypoxic EVs may depend on hypoxia-inducible factors and RAB22A, which in a hypoxic environment relies on the mediating action of the small GTPase RAB22A to dislodge hypoxic EVs from the cells (38). Hypoxic EVs are influenced by the environment and containing biomarkers such as mRNA and proteins, among which proteins include MMPs, IL-8, PDGFs, caveolin 1, and lysyl oxidase (37).
Podocyte EVs (100-200 nm) derived from the tip vesicles of podocyte microvilli (39). It can be expressed before other markers of nephropathy and therefore may serve as a new marker of glomerular and tubular injury.Medeiros et al. have shown that EVs can be produced by podocyte cells after exposure to high glucose and expressed before proteinuria (64). It remains to be proven about the biomarkers contained in EVs produced by podocytes.
2.2 Contents of EVs
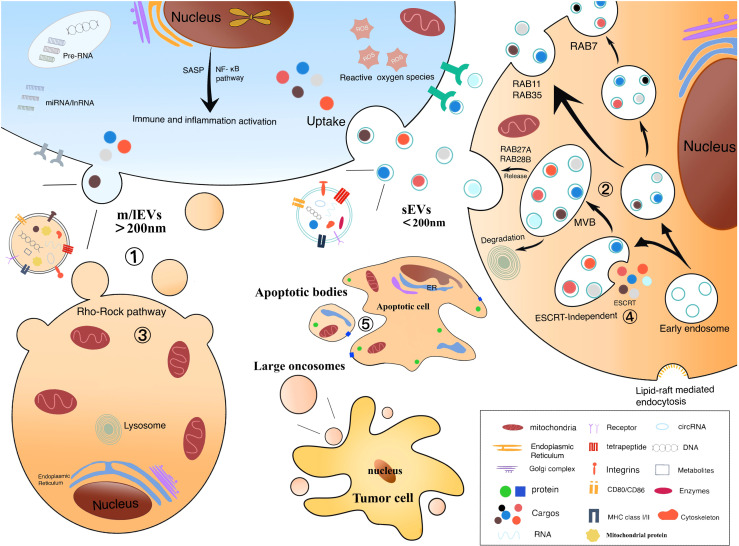
EVs are usually secreted under physiological conditions and rich in nucleic acids, proteins, lipids, and metabolites (31) (Figure 1). In response to stimuli such as differentiation, neuronal signaling or immune response, the secretory content varies depending on the cells of EVs origin and their function. Surface proteins were abundant, with high enrichment of tetraspanins (CD9, CD63) and lysosome-associated membrane protein 2b (Lamp2b) (20). Besides, RNA is presented in EVs, including miRNA, long non-coding RNA (lncRNA), transfer RNA (tRNA), etc, which range from approximately 25 to 700 nucleotides in length and vary in content depending on the different origin of EVs (5). To be interest, EVs from tumor cells are particularly rich in RNA. According to the Vesiclepedia database, 213 unique proteins were identified in HCC cell-derived EVs. The sEVs proteins include cargo proteins and membrane proteins, the latter being associated with exocytosis of recipient cells and target organ selection (65, 66). The composition of cargo proteins in sEVs varies across tumor cells (5). Studies have found that the ultraconserved lncRNA (ucRNA) expression is dramatically altered within EVs as compared to donor cells. For example, HCC cell-derived EVs transfer ultraconserved lncRNA TUC339 enrichment to neighboring cells in the microenvironment, which is transcribed in host cells and promotes HCC proliferation and diffusion (66). In addition, Yang, B et al. suggested that EVs promote hepatocellular carcinoma metastasis because some substances in EVs are involved in epithelial mesenchymal transition (EMT) (40).
Figure 1.
Biological origin of electric vehicles: ① m/lEV formation is the result of mass membrane foaming. Calcium relies on the cellular scale of protein hydrolysis degrading membrane binding, which can help cell membranes germinate and promote their secretion. ② Formation of sEV includes endocytosis, the formation of nucleosomes and MVBs, and the release of sEVs. The vesicles contained in MVBs fuse with the plasma membrane, causing their release. ③ Refactoring is related to the Rho/Rock pathway. ④ Composition of ESCRT is related to the biological occurrence of sEV and MVB. Rab protein facilitates the transport and docking of MVBs over the plasma membrane, leading to cytoplasmic vomiting and the release of sEVs. ⑤ Extensive membrane vesicles occur on the membrane of apoptotic cells to form apoptotic body.
Moreover, many studies have reported that mitochondrial proteins are also cargoes of EVs (67–70). EVs can carry mitochondria, mitochondrial proteins, or mitochondrial DNA to travel between organelles (67, 71). Kiran Todka et al. found that mitochondrial proteins are selectively enriched in EVs and that delivery of mitochondrial proteins to EVs requires sorting nexin 9(SNX9)-dependent mitochondria-derived vesicles (MDVs). MDVs are responsible for carrying mitochondrial proteins between mitochondria and other organelles (72). Intercellular transfer of mitochondria (including mtDNA) results in altered mitochondrial function. If mitochondria are localized within the mitochondrial network of the recipient cell, it may elevate the intracellular ATP levels, further generate metabolic stress and ROS to regulate innate immunity, which may have a significant impact on the tumor microenvironment (73–75). For example, it has been found that mitochondrial DNA (12S rRNA (RNR1) G709A) play an important role in the development of HCC (76). However, whether the process of mitochondrial can influence the hepatocellular carcinoma progression associated with EVs needs to be further explored.
2.3 Specific Mechanisms of Uptake and Internalization Between EVs and the Target Cells
Since our current knowledge about the physiology, diversity, internalization, and cargo delivery of EVs is still somewhat limited, it remains impossible to derive a clear mechanism about how EVs interact with and modify receptor cells. However, determining the intracellular pathways and mechanisms of their cargo delivery could help us to utilize EVs as therapeutic agents appropriately (77).
The uptake pathways of EVs are known to be greatly diverse by cells and EVs type, which may be more dependent on the receptor cell type than EVs itself (22, 78, 79). EVs can translocate their contents to recipient cells by different mechanisms such as direct fusion, direct binding, endocytosis or phagocytosis (22). Although the mechanism of EVs uptake and cargo translocation into the cytoplasm of the receptor cell is still not fully defined, it mainly occurs in three steps: targeting the receptor cell, entering point into the receptor cell, and delivering the contents to the receptor cell. However, the end point of EVs internalization is still uncertain, and the function of EVs-mediated cargo transfer cannot being well defined (78).
The pathway of EVs internalization determines the functional response and efficiency of cargo delivery, while the internalization of EVs is mediated by a variety of mechanisms (80), including grid protein dependence and endocytosis of grid protein non-dependent pathways (78). In general, endocytosis is usually divided into two main subgroups: phagocytosis and cytokinesis. Phagocytosis is a type of endocytosis of relatively large (>1µm) particles and is usually restricted to specialized professional phagocytes. In contrast, all cells are capable of cytokinesis (81–83). Grid protein-mediated endocytosis is a recognized pathway for extracellular substance uptake (84). Meanwhile, studies have shown that EVs enter cells mainly through grid protein-independent endocytosis and macrocytosis (83). Non-dependent endocytosis of grid proteins, including the formation of inverted influxes of vesicle-coated cells on cell membranes (77, 84, 85). Alternatively, fusing with the exoplasmic membrane, EVs can enter cell directly, thereby release their contents into the cytoplasm (80).
3 Separation Methods of EVs
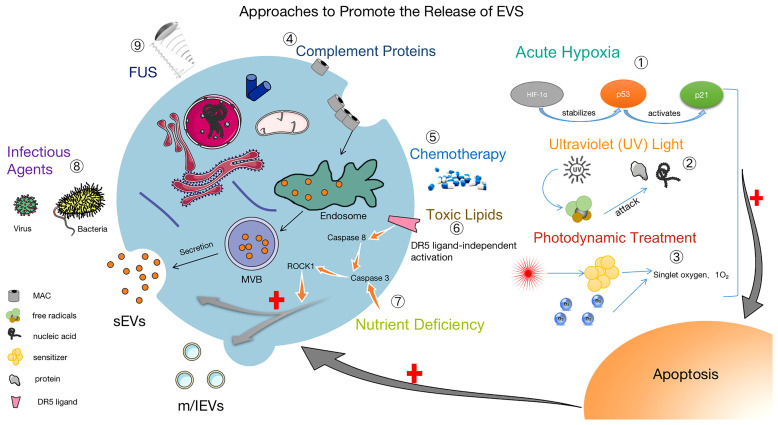
The isolation and collection of EVs is a necessary condition for biomedical research and clinical transformation. Researchers have developed many methods to separate EVs, and it is particularly significant to use the proper isolation method under different conditions. For better clinical applications, improving existing technologies for the isolation and storage of EVs are facing great challenges (20). Efficient access to EVs is extremely important for research, and in addition to the use of suitable isolation techniques, promoting the production and release of EVs is also of great value. Upon increased release of EVs, cargo and surface marker proteins may cause altered biological functions (86, 87). Notably, EVs induced by tapping membrane complexes have been reported to play important physiological roles in enhancing immunity, promoting coagulation, wound healing and growth (88, 89). Hirsova, P found that toxic lipids induce the release of EVs from hepatocytes and can activate the pro-inflammatory response of macrophages, which also suggests that inhibiting the release of EVs could be a therapeutic strategy for patients with NASH (90). Based on the therapeutic potential of EVs, we believe that it is of great interest to select suitable methods to facilitate or inhibit the release of EVs depending on the purpose. Thus, some approaches to promote the release of EVs are summarized in Figure 2 .
Figure 2.
Methods to facilitate the release of EVs. ① Acute Hypoxia: Catabolism of HIF-1α is inhibited by acute hypoxia, which stabilizes the P53 gene and activates the P21 gene, leading to apoptosis and promoting the release of EVs (91–93). ② UV: After UV irradiation, a large number of free radicals are generated to attack nucleic acids and proteins, causing apoptosis and increasing the release of EVs (94). ③ Photodynamic Treatment: Laser irradiation at a specific wavelength excites the tissue-absorbing photosensitizer, and the excited state of the photosensitizer transmits energy to the surrounding oxygen, generating strongly reactive monomorphic oxygen, which may reacts oxidatively with the surrounding neighboring biomolecules, resulting in a cytotoxic effect that causes apoptosis and also promoting the release of EVs (95). ④ Complement Proteins: The membrane attack complex (MAC) is composed of complement proteins (C5b, C6, C7, C8 and C9). MAC is cleared from the cell surface by cytosolic or cytocytic action to help release EVs (96, 97). ⑤ Chemotherapy: The use of chemotherapeutic agents (e.g., doxorubicin, methotrexate, and cisplatin) causes cellular damage and EVs release (95, 98). ⑥ Toxic Lipids: Toxic lipids activates the DR5 pro-apoptotic signaling cascade, which in turn activates ROCK1 and promotes the release of EVs from hepatocytes (90). ⑦ Nutritional Deficiency: Activation of Caspase 3, ROCK1 signaling pathway and promotion the release of EVs (99). ⑧ Infection factors (100) and ⑨ focused ultrasound (101) can also promote the release of EVs.
3.1 Traditional Methods
3.1.1 Ultracentrifugation
Ultracentrifugation is considered as the “gold standard” for the separation of EVs (102). Due to the different particle size and density, its settling speed is also different, using gradually increasing centrifugal speed or low speed and high speed alternate centrifugation, can be separated in batches at different separation speeds and centrifugal time (30). Cellular impurities were removed with a low speed of 300 g, and high centrifugal force of 16,000 g can be used to separate apoptotic bodies, 20,000 g to separate m/lEVs, and 100,000 g to precipitate and concentrate sEVs (103, 104). This method is widely used, but the purity of sample is not satisfied for the supernatant will contain 40% EVs, which leads to protein contamination and lower yield. There is an overlap in the size of sEVs and m/lEVs, and slightly larger sEVs and smaller microvesicles are difficult to isolate (105). In addition, it generally requires multiple centrifugation processes to achieve better separation, but it is prone to vesicle destruction and also has many disadvantages such as the large size of the instrument, high cost, lengthy and laborious processing, and few samples (106).
3.1.2 Gradient Ultracentrifugation
The requirements of gradient ultracentrifugation are more stringent, when there is a small difference in settling velocity between different particles, they are placed on the top of a medium with different density gradient. Under the action of a certain centrifugal force, the particles are separated by aggregating into the layer of the medium with a similar density to theirs, and the commonly used medium is sucrose (107). Sucrose gradient centrifugation can be used to isolate sEVs (108, 109). This method is popular because of good separation effect, high purity, no extrusion and deformation of the particles, and the ability to maintain the activity of the particles. However, it needs to prepare inert gradient media solution, be complicated to operate, not easy to master, time-consuming and labor-intensive (20-24 h), and high cost. What’s more, the density of EVs and high-density lipoprotein particles (HDL) is similar and they can be separated out together, so the samples are prone to contamination (110). Besides, the use of newer isotonic gradients contribute to better maintenance of the physical properties of the vesicles (111).
3.1.3 Precipitation Method
The precipitation method mainly includes polymer precipitation and organic solvent precipitation. Commercial kits that rely on polymer co-precipitation have been reported being used for the isolation and purification of EVs, decreasing solubility and promoting precipitation. The precipitated EVs can be easily and reproducibly separated and avoid prolonged ultracentrifugation (112, 113). Unfortunately, the main problems with this method are that co-precipitation is susceptible to contamination by non-EVs substances and that mechanical forces or chemical additives can damage EVs (114). In addition, the method relies more on manual manipulation with low throughput and recovery, and purification of polymers from EVs may interfere with downstream analysis. Therefore, co-precipitation is not suitable for most research and clinical applications.
3.1.4 Molecular Exclusion Chromatography
The principle of molecular exclusion chromatography is that different solute molecules, such as EVs and protein impurities, are separated from each other as they pass through porous packings due to differences in size resulting in different rates of passing through the pores (115, 116). This approach yields purified EVs from complex biological media (117–119), removed soluble plasma proteins and HDLs effectively, preserved the biological activity and integrity of EVs and also reduced aggregation (115). A variety of influencing factors such as media type, pore size, column size, and flow rate should be considered for EVs separation (20, 116). This method is efficient and inexpensive, and it is more suitable for small volumes of blood samples because of the upper sample volume limitation.
3.1.5 Asymmetric Flow Field Flow Classification Method
Asymmetric flow field flow fractionation (AF4) is a technique in which a force field is applied to achieve the separation of EVs with different sizes and molecular weights (120). AF4 contains permeable plates and when a vertical force field is applied, the analytes in the sample will be moved to the boundary by the force and smaller particles will undergo Brownian motion to reach a new equilibrium position (121). The advantages of this method are rapid (<1 h), high resolution, gentle, label-free, and reproducible. It can be applied to a variety of eluates, contributing to the successful separation of different subpopulations of EVs.
3.2 New Methods
3.2.1 Immunoaffinity Capture
Obviously, EVs are rich in proteins. Immunoaffinity capture is the specific binding of antibodies to the corresponding antigens on the surface of EVs such as adhesion proteins, tetra-transmembrane proteins and integrins, achieving the separation of EVs by immune reactions (122, 123). Magnetic beads provide a large surface area to capture EVs, targeting antigens on the surface of EVs to select specific subgroups, improving separation efficiency, specificity and purity, making it more suitable for marker detection of EVs and clinical diagnostic studies (124). However, the expensive antibody reagents, stringent reaction conditions, reduction of isolation yields, and the vulnerability of the biological activity of the EVs contents to PH and salt concentration have made it inappropriate to isolate large volume samples (125).
3.2.2 Microfluidics
Based on different molecular size, microfluidics can isolate EVs from large cellular debris (126). Compared to conventional separation methods, with smaller sample volumes (50µL - 500µL), microfluidic techniques are faster (30 min-2 h), portable, cost effective and automated, resulting in high purity of EVs. However, some microfluidic technologies allow only small sample input, lack method validation and standardization, which may influence the application of downstream analysis.
3.2.3 Contactless Classification
The use of acoustic waves for contactless separation of EVs has recently been proposed by some researchers. This separation method applies forces based on the size and density of vesicles (127). Particles in the acoustic region migrate toward the pressure nodes after the force is applied. Acoustic interaction forces are proportional to vesicle volume, with larger vesicles moving more rapidly. This method can separate EVs very quickly and without contact.
4 Quantification Methods of EVs
Currently, the quantification of EVs has been challengig. It is suggested that for conditioned medium, the number of cells at the time of initiation and collection should be clearly indicated. In addition, proper characterization of EVs at the time of separation helps to understand their properties. Several techniques for measuring the size of EVs are being investigated, including lateral-flow immunochromatographic assay (LFIA), nanoparticle tracking analysis (NTA), and nanopore tunable resistive pulse sensing techniques(TRPS), high resolution flow cytometry, multi-angle light scattering coupled to asymmetric flow field-flow fractionation (AF4), fluorescence correlation spectroscopy (FCS), enzyme linked immunosorbent assay (ELISA) and Raman spectroscopy, etc. Here, we talk about some advantages and disadvantages of some techniques. LFIA, with its high degree of flexibility, is a good tool for cost-effective field detection, but the assay lacks sensitivity (128). The AF4 system is highly repeatable (120), however, it requires skilled operators. NTA and TRPS can be used for particle size analysis of EVs, and their detection sensitivity is 70-90 nm and 70-100 nm, respectively. NTA technology allows one-time measurement and quantification of EVs, but the equipment is expensive and difficult to operate (129, 130). The ELISA technique is greatly flexible and can be modified appropriately for the analyte, but it is also time-consuming.
In addition, EVs are rich in proteins, lipids, nucleic acids and other biomolecules, and it can be quantified by quantifying these specific molecules. For example, total protein amounts were determined by using Bradford, micro-bicinchonic acid (BCA), fluorimetric assays, global protein stainon sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), etc. However, due to the possible presence of protein contaminants, the measurements are on the high side. The amount of total lipids can be measured by sulfofphosphovanilin assay (131) and total reflection fourier-transform infraredspectroscopy (132). RNA can be quantified by global RNA assays (133). In conclusion, the quantification of EVs is a critical topic that still lacks consensus and standardization both domestically and internationally, and we expect more studies to be reported in the future.
5 Interactions Between HCC and HCC Cell-Derived EVs
In the microenvironment where tumor cells and normal cells are located, HCC cell-derived EVs build a bridge to communicate with each other and promote HCC proliferation, invasion and distant metastasis, etc. EVs origined from HCC often regulate tumor progression through autocrine and/or paracrine cellular communication. HCC cell-derived EVs stimulate recipient cells to produce cytokines and promote the migration of HCC, such as matrix metalloproteinase 2 (MMP2) and matrix metalloproteinase 9 (MMP9) (134). Meanwhile, HCC is a typical hyperangiogenic tumor. HCC cells secrete EVs loaded with different miRNAs, LncRNAs, circRNAs that can activate signaling pathways in the recipient cells, thus causing the recipient cells to respond, promoting HCC migration or inhibiting HCC proliferation, which have an impact on tumor angiogenesis (47). For example, HCC cell-derived EVs carry oncogenic RNAs and proteins, which allows EVs to activate the PI3K/AKT and MAPK signaling pathways and promote distant tumor metastasis (46).
EVs secreted by HCC cells containing some specific miRNAs will play a specific role in HCC. For example, hypomethylation causes increased expression of mir -429 in HCC cells, and these large EVs mediated by mir -429 are shed and bind to Rb-binding protein 4 (RBBP4) in surrounding target cells, promoting the transcriptional activity of E2F1 and ultimately upregulating the expression of POU class 5 homeobox 1 (POU5F1) in target cells, thereby promoting HCC development (46). Meanwhile, EVs-loaded miR-221 binds to the 3’-UTR target site of the p27/Kip1 oncogene and promotes HCC proliferation and migration (135). EVs containing protein CD147 released by HCC cells activate the NF-κB pathway of surrounding fibroblasts, induce MMP-9 expression, and stimulate the ERK1/2 and p38 MAPK pathways, leading to extracellular matrix degradation and tumor invasion (136, 137). In addition, EVs containing miR-25 released from HCC cells inhibited p53 expression in surrounding HCC cells, thereby restoring FOXM1 (a key regulator of cell cycle progression) expression, activating the HGF/Ras pathway, reversing the expression of sorafenib-induced apoptotic markers BCL2 and BAX, making HCC cells resistant to sorafenib (138). miR-34a is reduced in the large EVs released by CHB or HCC cells, resulting in increased levels of mRNA and protein in c-Mets in surrounding cells, promoting phosphorylation of c-Met-induced extracellular signal- regulated kinases 1 and 2 (ERK1/2), thereby facilitating CHB conversion to HCC (139, 140). Intracellular TLR4 signaling in HCC cells is transduced to the actin cytoskeleton via the MyD88 pathway, leading to the release of large EVs. Peripheral tumor macrophages take up large EVs containing microRNA let-7b, which attenuates tumor inflammation by targeting the pro-inflammatory cytokine IL-6 (141). Upregulation of ANXA2 expression in HCC cells promotes the shedding of CD147-containing large EVs and the production of MMP-2 in surrounding fibroblasts, thereby promoting HCC development (142). Thus, HCC cell-derived EVs can also act as a bridge between surrounding tumor cells or other cells, and their loaded cargo can have an impact on HCC progression when taken up by target cells.
5.1 HCC Cell-Derived EVs Promote HCC Migration by Directly Activating or Inhibiting Signaling Pathways
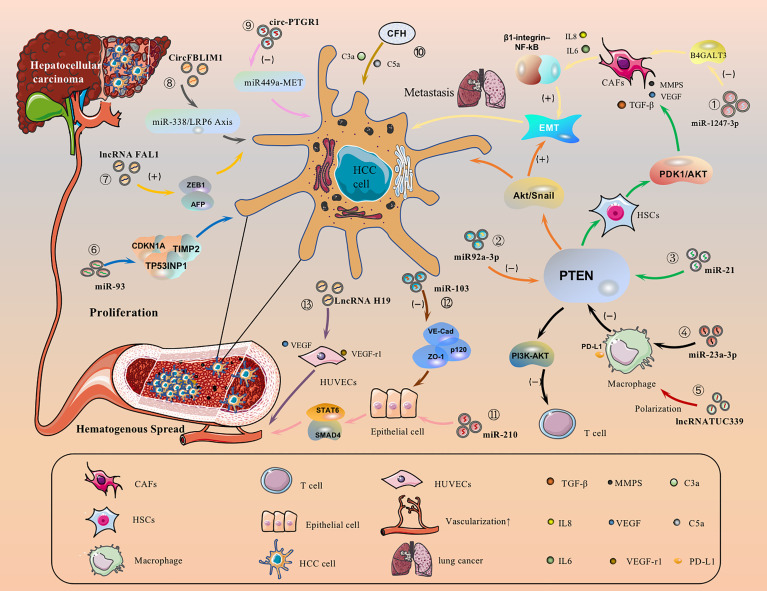
HCC cell-derived EVs-loaded cargoes can promote cancer cell migration by directly activating or inhibiting signaling pathways. For example, EVs-miR-1247-3p secreted by HCC cells directly transferred to lung pre-metastasisniche fibroblasts, decreased the expression of β-1,4-galactosyltransferases III (B4GALT3, a protein mediating glycosylation), thereby converting them into CAFs, and then activated the β1-integrin-NF-κB signaling pathway to promote EMT, thereby promoting the metastasis of hepatocellular carcinoma to the lung, and IL-6 and IL-8 secreted by CAFs to promote the development of HCC ( Figure 3 .①) (143). Meanwhile, EVs-miR92a-3p can promote HCC metastasis and EMT by inhibiting PTEN activation of the Akt/Snail signaling pathway ( Figure 3 .②) (40). Besides, HCC cells can also secrete EVs-miRNA-21 that directly targets PTEN and activates the PDK1/AKT signaling pathway. Moreover, it transforms hepatic stellate cells (HSC) into activated cancer-associated fibroblasts(CAF), which can further promote HCC growth by secreting vascular growth factors(VEGF, MMP2, MMP9 and TGF-β) ( Figure 3 .③) (144). Under endoplasmic reticulum stress, HCC cells inhibit PTEN and activate the PI3K-AKT pathway by delivering EVs-miR-23a-3p to macrophages, increasing macrophage PD-L1 expression and inhibiting T-cell function, promoting immune escape ( Figure 3 .④) (145). In addition, EVs-lncRNA TUC339 can be taken up by THP-1 cells, resulting in reduced production of pro-inflammatory cytokines, reduced expression of costimulatory molecules, impaired phagocytosis, and promotion of macrophage M (IL-4) polarization ( Figure 3 .⑤) (146). EVs-miR-93 promotes HCC tumorigenesis by affecting CDKN1A, TP53INP1, and TIMP2, and sEVs-miR-93 overexpression predicts poor prognosis ( Figure 3 .⑥) (147). It has been reported that lncRNA FAL1 are taken up by surrounding HCC cells and promote HCC cell proliferation and migration by competitively binding miR-1236 in recipient cells, which in turn upregulates the expression of their target genes AFP and ZEB1 ( Figure 3 .⑦) (148). sEVs-CircFBLIM1 can promote HCC progression through the miR-338/LRP6 axis ( Figure 3 .⑧) (149). The sEVs-circ-PTGR1 downregulates miR449a-MET expression, disrupts tumor microenvironment homeostasis, and promotes HCC migration and invasion ( Figure 3 .⑨) (150). EVs complement factor H (CFH) elevates C3a and C5a levels, exacerbating inflammatory responses and tumor growth ( Figure 3 .⑩) (151).
Figure 3.
HCC cell-derived EVs carry cargo and regulate different receptor cells.
5.2 The Role of HCC Cell-Derived EVs on Angiogenesis
HCC is typically a highly angiogenic tumor and therefore angiogenesis is closely related to the prognosis. We have known that EVs-loaded cargo is able to promote angiogenesis and increase vascular permeability. Altered vascular permeability implies altered endothelial continuity, allowing cancer cells to infiltrate and attach to the microvascular endothelial lining and form tumor metastases. For example, Lin, XJ et al. found that delivery of EVs-miR-210 to endothelial cells to target SMAD4 and STAT6 for pro-angiogenesis ( Figure 3 .⑪) (152). Besides, EVs-miR-103 inhibits the expression of VE-Cad, p120 and ZO-1 and reduces endothelial integrity to promote tumor invasion ( Figure 3 . ⑫) (153). EVs-LncRNA H19 induces the production of the pro-angiogenic cytokine (VEGF) and its receptor VEGF-r1 in HUVECs and stimulates angiogenesis ( Figure 3 . ⑬) (154). Interestingly, Y Zhou et al. found that ovarian cancer-derived EVs carry NID1 through ERK/MAPK to promote EMT, accelerate angiogenesis, and promote tumor invasion (155), but the role of NID1 in HCC is still unclear (156). In addition, HCC cell-derived EVs can promote angiogenesis in HUVECs, and the amount of HepG2-derived EVs determines the amount of angiogenesis, lumen formation. The sEVs may influence human umbilical vein lumen formation via the VEGF receptor and the angiogenesis-associated heat shock protein HSP70 (157).
HCC cells-derived EVs carrying proteins were found to inhibit angiogenesis by reducing VEGF through activation of AMPK signaling iynamic network microenvironment consisting of hepatocytes and their surroundings, suchn HCC (158). At the genetic level, CLEC3B-related genes are closely associated with angiogenic genes. In experiments, cells with high levels of CLEC3B formed fewer vessels than those with low levels. Likewise, in animal studies, immunohistochemical detection of tumor tissue from in situ tumor-implanted mice showed a significant reduction in CD31-positive and CD34-positive endothelium (EC) in CLEC3B high-isogenic grafts. Thus, high levels of CLEC3B EVs significantly reduce the expression of endothelial growth factor (EGF) in HCC, thereby reducing angiogenesis.
5.3 Inhibition of HCC Growth by EVs-Loaded Cargo of Different Cellular Origin
When certain signaling pathways are blocked by EVs-loaded cargo, the growth and distant metastasis of HCC may also be inhibited. For example, when Vps4A is overexpressed in HCC cell-derived EVs, it inhibits the PI3K-Akt pathway and thereby inhibits the metastasis of HCC (159). When normal cells secrete sEVs containing SENP3-EIF4A1, SENP3-EIF4A1 inhibits HCC cell proliferation by suppressing miR-9-5p in HCC cells and activating the expression of ZFP36 (160). In contrast, EVs-circ-0051443 promotes HCC cell apoptosis and inhibits tumor growth by competing with miR-331-3p in HCC cells and upregulating BAK1 expression (161). Interestingly, Huang, X et al. proposed that IncRNA 85 regulates the invasion of cancer cell by targeting miR-324-5p and through ceRNA mechanisms, and more importantly, miR-324-5p overexpressed can reducing migration by regulating the expression of MMPs, ETS1 and SP1 genes in HCC (162, 163). When tumor-associated fibroblasts (CAFs) secrete EVs containing miR-320a, miR-320a inhibits HCC growth by suppressing the PBX3/ERK1/2/CDK2 pathway in HCC cells (164). For example, EVs enriched in LncRNA H19 were secreted by CD90+ cancer cells to promote angiogenesis, inducing the production and secretion of the pro-angiogenic cytokine VEGF and its receptor in HUVECs (154). What’s more, it has been shown that co-culture of Huh7 cells with HepG2 cells, where Huh7 secretes EVs containing miR-122, has an inhibitory effect on tumor growth, when co-cultured HepG2 cells attenuate this inhibitory effect by secreting IGF1 (165).
Alteration of original physiological functions between HCC cells through the delivery of cargo molecules in EVs. Some goods are markers to diagnose HCC from other liver diseases; Some can determine the effectiveness of HCC treatment and predict the recurrence rate of HCC; Some can be used as vehicles for delivering drugs for the treatment of HCC. In conclusion, EVs loaded with cargo play different roles in the migration of HCC, regulating the talks between HCC and cells ( Table 2 ).
Table 2.
The cargos and functions of EVs related with HCC.
| Name of the Cargo in EVs | Cargo Type | Mechanism of the Cargo | Function of the Cargo | Vivo or vitro | Cell lines | Refs |
|---|---|---|---|---|---|---|
| miR-429 | miRNA | Targeting the RBBP4/E2F1/OCT4 axis in recipient cells, promote liver T-ICs properties | Facilitate HCC | Vitro | T-ICs | (46) |
| miR-142-3p | miRNA | Down-regulation of RAC1 | Suppressed migration of HCC | Vivo | Hepa1-6 | (166) |
| miR-221 | miRNA | Binding to the target sites in the 3’-UTR of p27/Kip1 tumor suppressor gene | Promote proliferation of HCC | Vitro | SMMC-7721 | (135) |
| miR-25 | miRNA | Attenuating p53 and enhancing FOXM1 expression | Mediate sorafenib resistance in HCC | Vitro | HepG2 | (138) |
| miRNA let7b | miRNA | Targeting proinflammatory cytokine IL-6 | Attenuates tumor inflammation | Vivo,Vitro | H22 | (136, 141) |
| miR-34a | miRNA | miR-34a was down-expressed in HCC, promoted the translation of antiapoptotic factors | Promote the conversion of CHB to HCC | Vitro | – | (139) |
| CD147 | protein | Induce upregulation of MMPs in fibroblasts, leading to extracellular matrix degradation | Promote tumoral invasion | Vitro | – | (136) |
| miR-1247-3p | miRNA | Targets B4GALT3, activate β1-integrin–NF-κB signaling, activated CAFs secrete pro-inflammatory cytokines | Promote lung migration of liver cancer | Vivo,Vitro | CSQT-2 | (143) |
| miR-103 | miRNA | Inhibiting the expression of VE-Cad, p120 and ZO-1, attenuated the endothelial junction integrity | Promote vascular permeability and metastasis | Vivo | MHCC97H | (153, 167) |
| miR-638 | miRNA | Attenuate endothelial junction integrity | Promote vascular permeability and metastasis | Vivo | HuH-7M | (168) |
| miR-93 | miRNA | Directly inhibiting the expression of TIMP2/TP53INP1/CDKN1A | Promote proliferation and metastasis of HCC | Vitro | SKHEP1 | (147) |
| miR-23a-3p | miRNA | Promotes PD-L1 expression in macrophages and inhibits T-cell function through miR-23a–PTEN–AKT signaling pathway | Promote proliferation and metastasis of HCC | Vivo,Vitro | HepG2 | (145) |
| lncRNAFAL1 | lncRNA | Competitively binding to miR-1236, indirectly up-regulated the expression of AFP and ZEB1 | Promote proliferation of HCC | Vitro | Huh7 | (5, 148) |
| IncRNA 85 | lncRNA | Targeted miR-324-5p and regulated its expression through a ceRNA mechanism | Promote proliferation and metastasis of HCC | Vitro | HepG2 | (163) |
| lncRNATUC339 | lncRNA | Excess lncTUC339 expression in macrophages promoted M(IL-4) polarization | Suppress the immune response to tumor cells | Vitro | HL-7702 | (147) |
| circUHRF1 | circRNA | Upregulate TIM-3 expression and suppress the production of IFN-γ and TNF-α | Inhibit NK cell function | Vivo | SMMC-7721 | (169) |
| Vps4A | protein | PI3K/Akt pathway was inactivated by Vps4A-overexpression | Inhibit the growth and metastasis of HCC | Vivo | Hep3B | (159) |
| CFH | protein | Increase the production of C3a and C5a | Promote proliferation and metastasis of HCC | Vivo,Vitro | Huh7 | (151) |
6 Regulation of HCC by Different Cell-Derived EVs in the Microenvironment
There is growing evidence that the dynamic network microenvironment consisting ofhepatocytes and their surroundings, such as cancer cells, immune cells, cytokines andextracellular matrix is also a key factor in tumor metastasis. Liver is rich in immune cells, which can greatly produce EVs, and has a unique immune-tolerant microenvironment, which is a huge challenge for HCC immunotherapy (170). Among them, various immunosuppressive cell subsets and signaling pathway-mediated pre-tumor immune responses play a key role in “tumor immune escape”. EVs are not restricted by space and material and can interact with cancer cells anywhere in the body. EVs produced by cancer cells can also interact with nearby immune cells (171, 172). The interaction between tumor and the immune system determines the progression of the tumor at the early stage. In conclusion, HCC occurs not only because hepatocytes contain sufficient genetic mutations, but multiple interrelated factors in the hepatic microenvironment influence the progression of HCC, and the mechanistic features of these new factors have prompted the search for new therapeutic approaches to treat not only the tumor itself but also the hepatic microenvironment to prevent recurrence and treatment resistance, some of which have yet to be fully elucidated.
6.1 Mesenchymal Stem Cells-Derived EVs
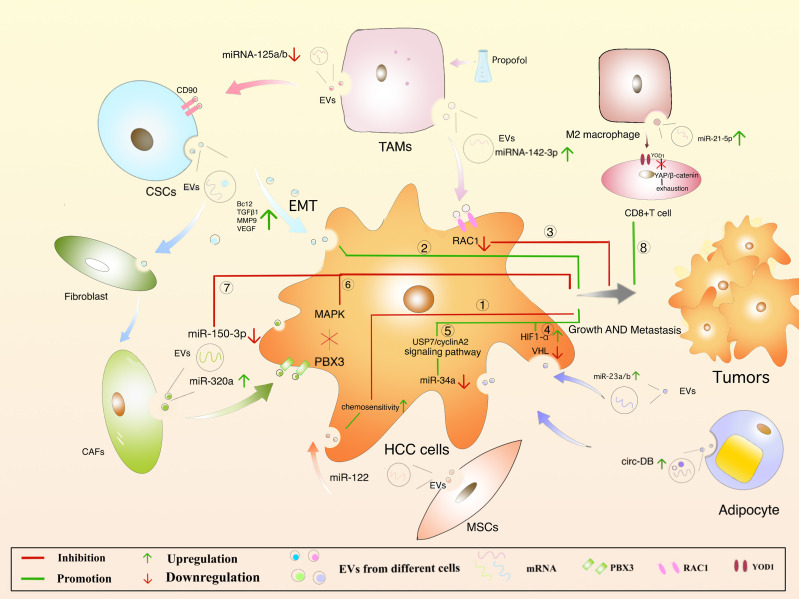
MSCs are present in bone marrow, umbilical cord blood and adipose tissue and are adult stem cells with multidirectional differentiation potential (173). MSCs attenuate fibrosis by upregulating hepatocyte growth factor (HGF) (174, 175), insulin growth factor (176), and MSCs-derived EVs improve hepatocyte regeneration and modulate immune activity, demonstrating therapeutic benefits in various liver diseases (173). Meanwhile, the role of MSCs-derived EVs cannot be ignored. Experiments have shown that ADMSC (adipose-derived mesenchymal stem cells)-derived EVs promote anti-tumor responses of NKT cells, leading to early ADC increase and low-grade tumor differentiation (177). In addition, an anti-tumorigenic effect of MSC-EVs was also observed in a CCl4-induced mouse liver tumor model. After treatment with EVs, the growth of liver tumor was significantly inhibited by inhibiting oxidative stress (178). Bruno, S et al. have demonstrated that EVs in human BM-MSCs can induce HepG2 cell cycle blockers and apoptosis necrosis in vitro, which inhibit tumor growth in the body. However, EVs secreted by fibroblasts formed by differentiation of human derived MSCs lack antitumor effects (179). In addition, miR-122 delivered via AMSC-derived EVs may provide new therapeutic options for HCC ( Figure 4 . ①) (180). It remains unclear that whether MSCs-derived EVs can inhibit HCC progression by carrying cargo, and it provides a new direction for the possibility of using MSCs-derived EVs as carriers to exert anti-tumor effects.
Figure 4.
Regulation of HCC by different cell-derived EVs.
6.2 Cancer Stem Cells-Derived EVs
Cancer Stem Cells (CSCs), with proliferative and differentiation potential, is more easily contributing to tumor recurrence (181–184). It is reported that EVs derived from CSCs can induce tumor growth, metastasis, participating in angiogenesis and maintaining the stem cell phenotypes (185–188). EVs released from CSCs containing multiple cargoes, including proteins and multiple RNA (189). EVs can make the microenvironment to change in the direction of promoting tumor occurrence and metastasis. For example, Domenis, R et al. found that CSC- derived EVs inhibits T cells through monocyte-specific secretion of IL-10 (190). In addition, fibroblasts can be converted into cancer-associated fibroblasts (CAF) through the uptake of CSC-derived EVs, promoting tumor progression and metastasis (191). It was also found that CSCs-like CD90+ hepatocytes regulate the endothelial phenotype by releasing EVs containing H19 lncRNA, significantly increase VEGF expression, and promote intercellular adhesion, induce angiogenesis, and affect the tumor microenvironment (154). What’s more, Alzahrani FA et al. showed that hepatic CSCs-derived EVs were able to increase the expression of Bcl2, TGFβ1, NFκB, MMP9, VEGF, 13K, ERK and decrease the levels of Bax, p53, TIMP1 mRNA in the liver of mice, suggesting that CSCs-derived EVs promote hepatocellular carcinoma cell invasion while upregulating TGFβ1-induced EMT ( Figure 4 . ②) (192). However, it is of interest that CSCs-derived EVs and MSCs derived EVs had opposite effects on HCC growth and progression in vivo, and neither involved promotion or inhibition of HCC-induced oxidative stress or antioxidant activity. As can be seen, these studies have showed new insights into the treatment of HCC, and more research is needed to clarify the mechanisms involved.
6.3 Macrophages-Derived EVs
Depending on the state and functional status of macrophages after activation, they can be divided into M1 and M2 macrophages, with M1 macrophages playing a tumoricidal role and M2 macrophages promoting tumorigenesis (193). M1 macrophages are involved in the polarization of Th1 and high expression of IL-6, IL-12, TNF-α, iNOS, ROS to promote the occurrence of inflammation (194). EVs from M1 macrophages induce stronger antigen-specific cytotoxic T-cell responses in lymph nodes, enhance immune responses to cancer vaccines, and are used as effective vaccine adjuvants (195). In the TME, tumor-associated macrophage (TAM)-derived EVs significantly downregulate miRNA-125a and miRNA-125b (miRNA-125a/b targets CD90, a stem cell marker for HCC) and promote the progression of HCC (196). The macrophages were treated with propofol to help secrete more EVs with miRNA-142-3p, which can be absorbed by HCC cells, and furtherly, RAC1 inhibited the migration and tumor growth in mice ( Figure 4 . ③) (166). M2 macrophages are involved in Th2 polarization and highly express IL-4, IL-10, TGF-β, CD206, CD163, CCL22, etc., while reduce the expressing of IL-12,downregulate the immune response and promote tumor progression (197). In an experiment by Jian Pu et al. in which EVs were injected into a mouse model of liver cancer, M2 macrophage-derived EVs were found to promote CD8+ T cell failure via the miR-21-5p/YOD1/YAP/β-catenin axis ( Figure 4 . ⑧) (198). Thus, it seems that M2 macrophages are closely associated with the malignant development of HCC (199).
6.4 Adipocytes-Derived EVs
Adipocytes mainly play a role in providing metabolic substrates for tumor cells. There is evidence that adipose-derived EVs can promote tumor growth in HCC by downregulating VHL, delivery of miR-23a/b. Studies in vivo have shown that increasing levels of EVs-miR-23a/b, VEGF, GLUT1 and HIF1α accelerated tumor growth and rate in high fat diet mice ( Figure 4 . ④) (200). Visceral adipocyte exocytosis induces dysregulation of the TGF-b pathway in HepG2 cells in high body fat individuals, but not in low body fat individuals (201). Zhang, H et al. suggested that EVs-circ-DB was upregulated in HCC patients with high body fat and its positively correlated USP7 was also increased (202). Mature adipocyte-derived EVs and HCC cellular effects lead to a decrease in miRNA-34a (tumor suppressor), while an increase in the USP7/Cyclin A2 signaling pathway (pro-cancer), a promotion of HCC cell growth, and a reduction in DNA damage ( Figure 4 . ⑤). Nevertheless, once circ-DB is knocked out, these effects will disappear. Furthermore, adiponectin is an abnormally abundant adipocytokine that regulates sEVs biogenesis by binding to T-cadherin and reduces cytosolic ceramide levels by releasing EVs (203, 204). sEVs are formed through the non-dependent mechanism of ESCRT, a process in which ceramide is essential and accordingly lipocalin is crucial in regulating their exocytosis. sEVs as a biological delivery vehicle for cancer treatment has been a hot research topic recently, but the role of adipocyte-derived EVs in HCC still requires further investigation.
6.5 Fibroblasts-Derived EVs
The connective tissue is rich in fibroblasts. Understanding the regulation of CAF in HCC is critical. CAFs-derived EVs are low in miR-320a, which binds to its direct downstream target PBX3 and inhibits HCC by suppressing MAPK pathway activation ( Figure 4 . ⑥) (164). The expression of CAFs-derived EVs-MiR-150-3p is reduced, which can inhibit the migration and invasion of hepatocellular carcinoma cells ( Figure 4 .⑦) (205), suggesting it may be a new therapeutic option. Meanwhile, studies have reported that miR-195 in HCC has been downgraded to VEGF, CDC42, CDK1, CDK4, CDK6, and CDC25 (206, 207). As described, understanding the mechanism of fibroblasts-derived EVs on HCC can help design new therapeutic approaches.
7 Hypoxia-Induced Microenvironment Affects the Regulation of HCC by EVs
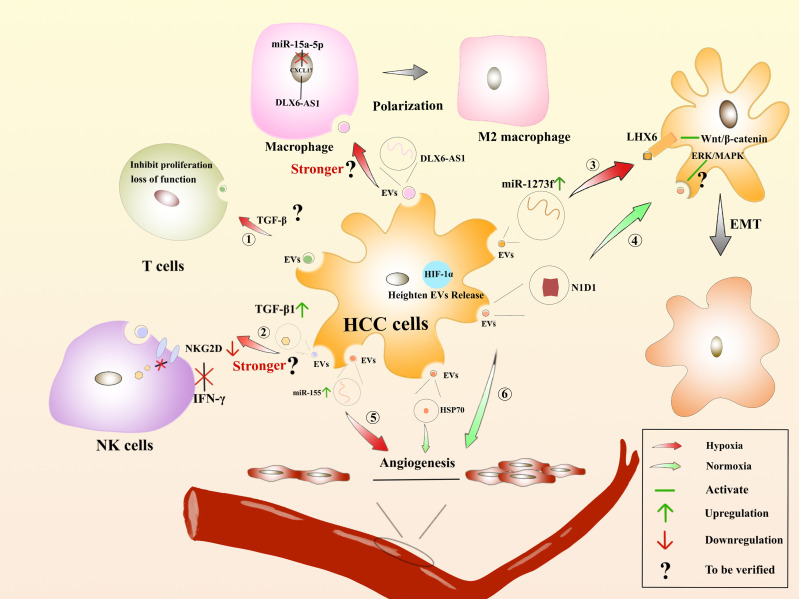
Many solid tumors live in the hypoxic microenvironment. Hypoxia promotes the production and release of EVs from cancer cells. Studies have showed that the number of sEVs in breast cancer cells and oral squamous carcinoma cells was significantly increased under hypoxic conditions (208). Hypoxia-inducible factor-α1 is a regulator of cells under hypoxic conditions and can facilitate the release of EVs (209). The proteins and nucleic acids of sEVs are also altered in the hypoxic environment (210). Under hypoxic conditions, miR-1273f carried by sEVs could accelerate the progression of HCC, targeting LHX6, which further inhibits HCC tumorigenesis or malignant transformation by targeting the Wnt/β-catenin signaling pathway (211). Hypoxia-generated sEVs can inhibit the expression of E-cadherin, thereby promoting EMT (212). EVs derived from HCC cells could affect angiogenic endothelial cells under the hypoxic conditions through upregulation of miR-155, thereby affecting tumor angiogenesis (213). Furthermore, EVs released from epithelial ovarian cancer (EOC) cells can express more miR-21-3p, miR-125b-5p and miR-181d-5p under the hypoxic conditions, thus facilitating M2 macrophage polarization (214). Additionally, DLX6-AS1 carried by HCC is in competition with miR-155 to regulate CXCL17. M2 macrophage polarization is induced, and migration, invasion, and EMT of HCC will be accelerated (215). Unfortunately, the authors did not investigate whether hypoxia accelerates this process. Rong, L et al. saying that hypoxia enhanced the secretion of sEVs in breast cancer cells, thereby inhibiting the proliferation of T cells (216). Moreover, hypoxia induced a significant increase in TGF-β1 content in cancer cell-derived EVs, decreased the expression of the activation receptor NKG2D, and inhibited the cytotoxicity of NK cells and also reduced the production of IFN-γ (217). Therefore, the tumor hypoxic microenvironment is closely related to tumor development, treatment and prognosis, which has become a research hotspot to find new treatments for HCC (Figure 5).
Figure 5.
Hypoxia-induced microenvironment affects the regulation of HCC by EVs : The role of EVs derived from HCC on immune cells in the hypoxic environment. ① Suppressing the proliferation of T cells or rendering them incompetent. ② Whether the inhibitory effect on IFN-β production by NK cells and the process of inducing macrophage polarization are enhanced remains to be verified. The role of HCC-derived EVs facilitates EMT. ③ In the hypoxic environments, miR-1273f is upregulated in HCC-derived EVs, acting on LHX6 to activate Wnt/β-catenin to promote EMT. ④ In the normoxic environment, HCC-derived EVs contain N1D1, which may activate the ERK/MAPK pathway in recipient HCC cells to promote EMT. Regulation of angiogenesis by HCC-derived EVs. ⑤ In the hypoxic environments, miR-155 is upregulated in HCC-derived EVs and promotes angiogenesis. ⑥ In the normoxic environment, HCC-derived EVs are enriched in N1D1 and HSP1, which promote angiogenesis.
8 Biomarkers
EVs are providing important links for intercellular information transfer (218), and specific proteins and nucleic acids in EVs are important biomarkers for clinical diagnosis of various liver diseases.At present, the clinical assessment of liver damage is mainly based on liver enzyme profiles, such as aspartate aminotransferase (AST), alanine aminotransferase (ALT) (219–221). However, these enzyme markers lack specificity for liver diseases. Traditional tumor markers such as AFP, AFP-L3 are susceptible to other liver diseases and cannot analyze HCC for etiology, which has certain limitations. Therefore, to find new specific markers for patients with liver disease is significant. Much research mentioned that the proteins and nucleic acids carried by EVs can serve as markers to predict the prognosis of patients with liver disease (222–224).
8.1 EVs-Associated Nucleic Acids as Biomarkers for HCC Diagnosis
8.1.1 miRNAs
miRNAs in serum EVs hold great potential as novel diagnostic biomarkers, and some of which have been reported worldwide ( Table 3 ). Elevated levels of miRNA-21 and lncRNA-ATB expression were found to have higher specificity and sensitivity for HCC (232, 239). Patients with postoperative recurrence of HCC have significantly reduced the expression of miRNA-718, which was associated with the highly aggressive nature of HCC (233). Interestingly, Wang, Y et al. proposed that EVs-miR-122, EVs-miR-148a and EVs-miR-1246 in HCC patients serum were apparently higher than those in the LC and the NC group, and that these miRNAs combined with AFP could effectively reduce the rate of misdiagnosis (227). However, for HCC patients with low AFP expression, whether or not with hepatitis virus infection, sEVs’ miRNAs are more indicative of being markers of HCC when they are expressed as miR-10b-5p+ miR-221-3p+ miR-223-3p and miR-10b-5p+ miR-221-3p+ miR-223-3p+ miR-21-5p (234). Tian X et al. indicated that an acidic environment triggers HIF-1α and HIF-2α activation and facilitates the expression of EVs-miR-21 and EVs-miR-10b, significantly promoting the progression of HCC both in vivo and vitro (235, 249). We also find that several miRNAs are studied at high frequency, such as miR-21 and miR-122, and the results may differ in different study contexts. Besides, we read that some serum miRNAs are biomarkers of HCC (250–256), but it is not explicitly stated that these miRNAs are associated with EVs, and their roles in the progression and recurrence of HCC need to be further explored.
Table 3.
EVs as biomarkers for the diagnosis of HCC.
| Classification | Biomarkers | Expression | Species | Type of biological fluid | AUROC | Clinical significance | Refs | |
|---|---|---|---|---|---|---|---|---|
| m/lEvs | AnnexinV+EpCAM+ASPGR1+CD133+taMPs | ↑ | Human | serum | 0.7439 | Diagnosis of HCC/CCA from LC | (225) | |
| EpCAM+AnnexinV +ASGPR1+taMPs | ↑ | Human | serum | 0.7322 | Diagnosis of HCC/CCA from LC | (225) | ||
| Total m/lEVs of peripheral blood | ↑ | Human | serum | 0.83 | Diagnosis of E-HCC from LC (TNM stage I) | (226) | ||
| sEVs | microRNA | miR-148a | ↑ | Human | serum | 0.871 | Diagnosis of HCC from NC Diagnosis of E-HCC from LC | (227) |
| 0.860 | ||||||||
| miR-122 | ↑ | Human | serum | 0.990 | Diagnosis of HCC from NC Diagnosis of E-HCC from LC | (227) | ||
| 0.795 | ||||||||
| miR-1246 | ↑ | Human | serum | 0.825 | Diagnosis of HCC from NC Diagnosis of E-HCC from LC | (227) | ||
| 0.761 | ||||||||
| miR-638 | ↑ | Human | serum | —— | Associated with tumor recurrence, As a prognostic marker | (228) | ||
| miR-125b | ↑ | Human | serum | 0.739 | Prediction of recurrence and survival | (229) | ||
| miR-93 | ↑ | Human | serum | 0.825 | The prognosis and diagnosis of HCC | (147) | ||
| miR-665 | ↑ | Human, | serum | —— | Diagnosis and prognosis of HCC | (230) | ||
| Mice | ||||||||
| miR-92b | ↑ | Human, | serum | 0.702 | Prediction of E-HCC relapse after LDLT | (231) | ||
| Rats | ||||||||
| miR-21 | ↑ | Human | serum | —— | Detection of E-HCC, Prognostic marker | (232) | ||
| miR-718 | ↑ | Human | serum | —— | Prediction of HCC relapse after LDLT | (233) | ||
| miR-21-5p | ↑ | Human | serum | 0.71 | Diagnosis of HCC from LC | (234) | ||
| miR-21, miR-10b | ↑ | Human, | serum | —— | Prognostic markers of E-HCC | (235) | ||
| Mice | ||||||||
| miR-18a, miR221, miR-222, miR224 | ↑ | Human | serum | —— | Diagnosis of HCC from LC/CHB | (223) | ||
| miR-101, miR106b, miR-122, miR-195 | ↑ | Human | serum | —— | Diagnosis of HCC from CHB | (223) | ||
| miR-122, miR148a, miR-1246 | ↑ | Human | serum | —— | Diagnosis of HCC from LC | (227) | ||
| miRNA-519d, miR-595, miR-939 | ↑ | Human | serum | —— | Diagnosis of HCC from LC | (222) | ||
| miR-10b-5p, miR-221-3p, miR-223-3p, miR-21-5p | ↑ | Human | plasma | 0.86 | Diagnosis of HCC from CH or LC | (234) | ||
| lncRNA | lncRNA-HEIH | ↑ | Human | serum | —— | Diagnosis of HCV-associated HCC from CHC | (236) | |
| LINC02394 | ↑ | Human | serum | 0.719 | Diagnosis of HCC from CHB | (237) | ||
| LINC00635 | ↑ | Human | serum | 0.750 | Diagnosis of HCC from CHB | (237) | ||
| LINC00161 | ↑ | Human | serum | 0.794 | Prediction of HCC growth and metastasis | (238) | ||
| IncRNA-ATB | ↑ | Human | serum | —— | The prognosis of HCC | (239) | ||
| Lnc85 | ↑ | Human | plasma | 0.869 | Diagnosis of AFP-negative HCC from healthy controls and LC | (163) | ||
| SENP3-EIF4A1 | ↑ | Human,Mice | plasma | 0.8028 | The diagnosis of HCC | (160) | ||
| circRNA | circFBLIM1 | ↑ | Human,Mice | serum | —— | The therapeutic target of HCC | (149) | |
| circ-0051443 | ↑ | Human,Mice | plasma | 0.8089 | The diagnosis and therapeutic target of HCC | (161) | ||
| circRNA-100338 | ↑ | Human,Mice | serum | —— | The diagnosis and therapeutic target of HCC | (240) | ||
| circUHRF1 | ↑ | Human,Mice | plasma | —— | The therapeutic target of HCC | (169) | ||
| circ-DB | ↑ | Human,Mice | adipocyte | —— | The prognosis of HCC | (202) | ||
| proteins | LAPTM4B-35 | ↑ | Human | serum | —— | Prediction of recurrence and diagnosis of HCC | (241) | |
| SMAD3 | ↑ | Human,Mice | peripheral blood | 0.70 | The diagnosis of HCC | (242) | ||
| RAB5A | ↑ | Human | serum | —— | The diagnosis and therapeutic target of HCC | (243) | ||
| ENO1 | ↑ | Human,Mice | serum | —— | The prognosis of HCC | (244) | ||
| Other combinations | miR-122, miR-148a, AFP | ↑ | Human | serum | 0.931 | Diagnosis of HCC from LC | (227) | |
| SMAD3+ATP | ↑ | Human,Mice | peripheral blood | 0.90 | The diagnosis of HCC | (242) | ||
| lncRNA-RP11-513I15.6, miR-1262/RAB11A | ↑ | Human | serum | —— | Diagnosis of E-HCC from CHB | (245) | ||
| miRNA-21, lncRNA-ATB | ↑ | Human | serum | —— | The prognosis of HCC, overall survival | (239) | ||
| ENSG00000258332.1, LINC00635, AFP | ↑ | Human | serum | 0.894 | The diagnosis and prognosis of HCC | (237) | ||
| AFP、ENST00000248932.1, ENST00000440688.1, ENST00000457302.2 | ↑ | Human | plasma | 0.905 0.879 |
Predict the probability of HCC in the cancer‐free groups Predict the probability of HCC in the CH groups |
(246) | ||
| Total EVs | Total EV | ↑ | Human | serum | 0.83 | Detection of HCC | (225) | |
| AFP, GPC3, ALB, APOH, FABP1, FGB, FGG, AHSG, RBP4, TF | ↑ | Human | plasma | 0.93 | Diagnosis of E-HCC from LC | (247) | ||
| LINC00853 | ↑ | Human | serum | 0.956 | Diagnosis of E-HCC from CH、LC | (248) | ||
8.1.2 lncRNAs
In recent years, the potential of EVs-derived lncRNAs in the prognosis of HCC has also attracted growing research interest. lncRNAs alter lncRNA expression can contribute to the cancer phenotype by stimulating cell proliferation, angiogenesis, immune evasion, and inhibition of apoptosis. Among them, linc-VLDLR was identified as a lncRNA enriched in EVs that contributes to the cellular stress response (257). ENSG00000248932.1, ENST00000440688.1 and ENST00000457302.2 were significantly increased in HCC patients, suggesting that lncRNAs may predict tumorigenesis and can be used to dynamically monitor HCC metastases (246). The expression of lncRNA-HEIH was higher in patients with HCV (hepatitis C virus)-associated HCC than that of CHC (chronic hepatitis C) patients (236, 258). Some indicated that sEVs levels of ENSG00000258332.1 and LINC00635 in serum were significantly high and it would be more specific and sensitive when they combined with serum AFP to detect HCC (237). Huang X and Kim S et al. suggested that EVs-derived Lnc85 and LINC00853 showed high positivity in AFP-negative patients with early HCC and were significantly better than AFP, respectively, which is particularly relevant to patients with AFP-negative tumors (163, 248). The potential of EVs containing lncRNAs as biomarkers in the process of HCC diagnosis cannot be ignored, and to find more specific markers for HCC is the next research direction.
8.1.3 CircRNA
There is growing evidence that circRNA in EVs has certain advantages in terms of abundance and stability, indicating that they are promising therapeutic targets for HCC. Similar to miRNA and lncRNA, changes in circRNA expression can also affect the occurrence and progression of HCC (259). In addition, circFBLIM1 was significantly expressed in HCC serum sEVs and promoted HCC progression by affecting the miR-338/LRP6 axis (149). Similarly, Bai N et al. found that circFBLIM1 acts as ceRNA to facilitate HCC by sponging miR-346 (260). In contrast, sEVs-circ-0051443 inhibits HCC progression by regulating miR-331-3p/BAK1 (161). Moreover, Huang XY et al. indicated that HUVECs receiving the circRNA-100,338 could boost the metastatic capacity of HCC cells, which may be related to the regulation of angiogenesis (209). Furthermore, serum EVs-circrna-100, 338 in patients with radical hepatic resection HCC are persistently hyperexpressed, dedicating lung metastases and low survival (240). Ultimately, circMTO1 (261), circSETD3 (262), cSMARCA5 (263), and hsa_circ_0068669 (264) also play key roles in HCC and are potential therapeutic targets, but it remains unclear whether these circRNAs and EVs are related.
8.2 EVs-Associated Proteins as Biomarkers of Liver Disease
EVs proteins change with the environment and state of liver cells, it can be used directly or indirectly as a biomarker in different liver diseases (265, 266) to predict the progression of the corresponding liver disease ( Table 4 ). CYP450-2E1 (227) and protein tyrosine phosphatase receptor (sPTPRG) isoforms associated with EVs are biomarkers of liver injury, and sPTPRG in plasma reflects the extent of liver injury (274, 278). If CD8, CD14, and connective tissue growth factor (CCN2) are highly expressed in EVs, they can be used to assess the degree of liver fibrosis (272, 273, 279). High expression of Apolipoprotein A-1 by EVs elevates liver-specific proteins such as FGB, causing toxic acute liver injury (269). Studies have shown that EVs containing Carboxylesterase-1 and Carboxylesterase-3 can be evaluated for hepatotoxicity (269, 270). JH H et al. indicated that EVs highly express AnnexinV+EpCAM+ASGPR1+CD133+ taMPs, which can be a novel biomarker for HCC and CCA liquid biopsies (225). If MMP-7 is highly expressed in EVs, it could be a marker for the differential diagnosis of CCA (271). Hepatocytes secrete EVs if ASGPR1+, which can be an alternative non-invasive biomarker of portal hypertension in NASH patients (267).
Table 4.
EVs-associated proteins as biomarkers of liver disease.
| Liver disease | Biomarkers | Types | Function | References |
|---|---|---|---|---|
| Non-alcoholic steatohepatitis(NASH) | ASGPR1+ | Protein | A surrogate noninvasive biomarker of portal hypertension in patients with cirrhotic NASH. | (267) |
| CD4+ | Protein | Biomarkers of nonalcoholic fatty liver(NAFL)and CHC | (268) | |
| Toxic acute liver injury | Apolipoprotein A-1 | Protein | Tentative hepatotoxic markers during hepatic damage | (269) |
| Carboxylesterase-1 | Protein | Hepatotoxic markers during hepatic damage | (269) | |
| Carboxylesterase-3 | Protein | Non-invasive indicator of drug toxicity | (270) | |
| CCA | AnnexinV+EpCAM+ASGPR1+CD133+ taMPs | – | A novel biomarker of HCC and CCA liquid biopsy | (225) |
| MMP-7 | Protein | Biomarkers for the diagnosis of CCA | (271) | |
| Liver fibrosis | CD8+ | Protein | A biomarker for liver fibrosis | (272) |
| CD14+ | Protein | A tamps biomarker for liver fibrosis | (273) | |
| Alcoholic steatohepatitis(ASH) | CYP450-2E1 | Cytochrome | A potential biomarker for liver injury | (274) |
| CD40L | Protein | A potential biomarker for ASH | (275) | |
| Alcoholic hepatitis | CD34+ ASGPR | Protein | Biomarkers of alcoholic hepatitis | (276) |
| CK18 | Protein | Biomarkers of alcoholic hepatitis | (277) |
High CD4+ expression in EVs can be a biomarker to diagnosis nonalcoholic fatty liver (NASH) from chronic hepatitis C (CHC) (268). Positive CD34+ with ASGPR (heavy alcoholic hepatitis) or CK18 (alcoholic hepatitis) in EVs can be used as biomarkers (276, 277), among them, CD34 can also be used as a biomarker to determine heavy alcoholic hepatitis (276). ENO1 upregulates the expression of integrin α6β4 and activates the FAK/Src-p38MAPK pathway (244). Gorji-Bahri G et al. suggested that RAB5A knockdown could be used as a therapeutic target to control the progression of HCC (243). Pang Y et al. saying that LAPTM4B-35 is associated with the HCC relapse, drug resistance, and it is expected to be a new diagnostic marker for HCC (241).
Many studies have shown that EVs affect the progression of various liver diseases by regulating cellular functions and activating key signaling pathways in receptor cells, obviously, which are newly discovered potential biomarkers, to open up new ways to clinically distinguish different kinds of liver disease. Unfortunately, the role of EVs in the diagnosis, prognosis determination and predictive value of liver diseases is still lacking sufficient clinical evidence. Studies on the sensitivity and specificity of these markers in liver disease have also been reported relatively rarely, and relevant applications remain to be further investigated.
9 Vesicle-Loaded Drugs
9.1 EVs are Natural Nanocarriers
EVs are endogenous cell-derived membranous structures, natural nanocarriers with very low cytotoxicity and immunogenicity, protecting the transported RNA from disassembly and phagocytosis by ribonucleases, with inherent activity targeting and ability to cross biological barriers (30). EVs can transport a wide variety of bioactive molecules, thus altering the physiological functions of the recipient cells and reducing the accumulation of chemotherapeutic drugs in non-target organs, thereby reducing off-target toxicity. Additionally, EVs can bind to each other through various ligand receptors, especially cytokinesis (280). EVs are efficient as synthetic nanocarriers. EVs as nucleic acid and drug delivery vehicles has been extensively studied (281, 282). Notably, EVs as drug carriers need to find an efficient method as cargo loading. Different techniques such as electroporation (283), incubation (284), sonication (285), and freeze-thawing have been applied for the EVs loading (286). What’s more, EVs can also be loaded with specific cargoes with endogenous mechanisms such as direct transfection or co-incubation to deliver the cargo to the cytoplasm (287, 288). However, these loading techniques may lead to some changes in the morphological characteristics and physicochemical properties of EVs, as well as aggregation of themselves or of the cargo they carry (289, 290). A more accurate understanding of the proteomic profile of EVs and the factors influencing protein composition will facilitate the development of protein-based therapeutic strategies for EVs in the future (291, 292).
9.2 Application of Drug-Carrying EVs in HCC
We review emerging strategies for targeted delivery using EVs and explore the use of them for the treatment of hepatocellular carcinoma. Treatment of H22 cells with the chemotherapeutic drug methotrexate (MTX) and irradiation with UV light, which could secrete Microparticles (MPs) when co-incubate with the remaining H22 cells, effectively kill tumor cells and reduce adverse effects, while impeding drug efflux (98). We read that RBC-EVs loaded with doxorubicin or sorafenib showed enhanced therapeutic effects in mouse models of in situ HCC through a macrophage-dependent mechanism compared with conventional doses of doxorubicin and sorafenib (293). More importantly, drug-loaded RBC-EVs did not show systemic toxicity, whereas conventional doses of doxorubicin and sorafenib did. The main challenges in the current clinical application of EVs are the limited yield and the susceptibility to contamination of EVs with various centrifugation methods (105, 114), which affects the purity and biological properties of EVs. In addition, although EVs are good natural carriers, how to load substances efficiently such as antitumor drugs or genes into EVs is still an urgent technical problem to be solved. Drug-carrying EVs are promising for clinical applications in the treatment of liver diseases, and careful selection of cells of origin for EVs, the creation of appropriate methods for loading the molecules they carry, overcoming low yields, etc. are current research hotspots.
10 Discussion
EVs are sensory molecules for information exchange between tumor cells in the microenvironment, activating different signaling pathways and influencing the development, progression and metastasis of tumors (294–297). In recent years, EVs have become promising vehicles in liver disease for their low toxicity, high stability and preferential absorption (298). Today, the application of EVs is still in its early stages. Although there have been clinical trials choosing miRNAs for liver disease, they are still not available for clinical use (298), lacking a number of clinical trials to demonstrate the effectiveness of EVs. The mechanisms and clinical applications of EVs in liver disease need to be studied in more depth. EVs may be an effective intervention in the future, showing a new light for oncology patients. What’s more, EVs can also alter the function of recipient cells and is crucial in the genesis, development and pathogenesis of HCC. Circulating EVs, as a novel signaling modality, which are involved in multiple processes including tumor development and metastatic drug resistance, are promising biomarkers for diagnosing liver disease and monitoring treatment response (46).
Notably, our current understanding of EVs is still inadequate and standard methods for isolating and tracking EVs are lacking. EVs are nearly released by all cells in the body, and many mechanisms involved in their production, transport, uptake and involvement in cancer development have not been fully explored (299), and challenges remain in the extraction, identification and processing of EVs biomarkers for analysis. In addition, the complexity of the immune response and microenvironment in the liver poses a significant challenge to the routine treatment of patients with HCC (300). Therefore, it is important to improve isolation techniques, tracking methods, screening for tissue-specific markers of EVs or the identifying EVs of tissue-specific origin in lesions. Making full use of the different extraction techniques available and optimising them is an important next step in research. In addition, experiments in vitro and in vivo on EVs still have many limitations, so there is an urgent need to establish well-developed experimental models to further explore their properties and mechanisms of action, and to explore the potential of using this intercellular communication modality in the TME for molecular diagnosis and targeted therapy of tumors. In conclusion, current studies indicate that EVs is crucial in mediating the progression of liver disease and therefore can be thought as a potential therapy for HCC. With a more comprehensive understanding of EVs, more valuable references will be provided for the prevention, diagnosis and prognosis of HCC.
Author Contributions
JW: conceptualization, methodology, writing-original draft, writing-review & editing, visualization and supervision. XW: writing-original draft, formal analysis and resources. XZ: writing-original draft, formal analysis and project administration. TS: writing-original draft, software. YL: writing-original draft, data curation. WW: writing-original draft, methodology. YH: Conceptualization and Supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully thank University Sponsored Research Program of Southwest Medical University.
Glossary
| ADC | apparent diffusion coefficient |
| ADMSC | adipose-derived mesenchymal stem cells |
| AF4 | Asymmetrical flow field-flow fractionation |
| AHSG | alpha 2-HS glycoprotein |
| ALB | albumin |
| ALT | alanine aminotransferase |
| APOH | apolipoprotein H |
| ASGPR1 | asialoglycoprotein receptor |
| ASH | Alcoholic steatohepatitis |
| AST | aspartate aminotransferase |
| AUROC | Area under Receiver Operating Characteristics |
| BCA | bicinchonic acid |
| BM-MSCs | Bone marrow mesenchymal stem cells |
| CAFs | Cancer-associated fibroblasts |
| CCA | Cholangiocarcinoma |
| CCN2 | Connective tissue growth factor |
| ceRNA | competing endogenous RNA |
| CFH | Complement Factor H |
| CH | chronic hepatitis |
| CHB | chronic hepatitis B |
| CHC | chronic Hepatitis C |
| CHOP | enhancer-binding protein homologous protein |
| CK18 | Cytokeratin-18 |
| CLEC3B | C-Type Lectin Domain Family 3 Member B |
| CSCs | Cancer stem cells |
| CTGF | connective tissue growth factor |
| DCP | des-gamma-carboxy prothrombin |
| DDS | drug delivery system |
| EVs | Extracellular vesicles |
| EC | endothelial cells |
| EGF | endothelial growth factor |
| E-HCC | early-stage hepatocellular carcinoma |
| EMT | Epithelial–mesenchymal transition |
| ENO1 | Alpha-enolase |
| EpCAM | epithelial cell adhesion molecule |
| ESCRT | endosomal sorting complex required for transport |
| ELISA | enzyme linked immunosorbent assay |
| FCS | fluorescence correlation spectroscopy |
| FABP1 | fatty acid binding protein 1 |
| FGB | fibrinogen beta chain |
| FUS | focused ultrasound |
| GEVs | Glioma-derived EVs |
| GGT | glutamyl aminotransferase |
| GPC3 | glypican3 |
| HCC | hepatocellular carcinoma |
| HCV | Hepatitis C Virus |
| HDL | High-density lipoprotein particles |
| HEMs | Adult human epidermal melanocytes |
| HGF | Hepatocyte growth factor |
| HIF-1α | Hypoxia Inducible factor 1 α |
| HIF-2α | Hypoxia Inducible factor 2 α |
| HSCs | Hepatic stellate cells |
| HUVECs | Human umbilical vein endothelial cells |
| IL | nterleukin |
| ILVs | intraluminal vesicles |
| iNOS | Inducible nitric oxide synthase |
| LAMP2B | lysosomal associated membrane protein 2B |
| LC | liver cirrhosis |
| LDLT | living donor liver transplantation |
| LG3BP | galectin-3-binding protein |
| LFIA | Lateral-Flow Immunochromatographic Assay |
| MMP | Matrix metalloproteinase |
| MPs | Microparticles |
| MSC | mesenchymal stem cells |
| MVBs | multivesicular bodies |
| m/lEVs | medium/large EVs |
| MDVs | Mitochondria-Derived Vesicles |
| MAC | membrane attack complex |
| NTA | nanoparticle tracking analysis |
| NAFL | Nonalcoholic fatty liver |
| NASH | non-alcoholic steatohepatitis |
| NC | normal control |
| NVs | Nanovesicles |
| PIGR | polymeric immunoglobulin receptor |
| RBP4 | retinol binding protein 4 |
| ROS | reactive oxygen species |
| sEVs | small EVs |
| SNX9 | sorting nexin 9 |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SAH | Severe alcoholic hepatitis |
| SMAD3 | SMAD Family Member 3 |
| sPTPRG | Protein tyrosine phosphatase receptor Gamma |
| TAMs | Tumor-associated macrophages |
| TF | transferrin |
| TGF-β | Transforming growth factor |
| TME | Tumor microenvironment |
| TNFα | Tumor necrosis factor alpha |
| TSG101 | tumor susceptibility gene 101 protein |
| TRPS | tunable resistive pulse sensing |
| VEGF | Vascular endothelial growth factor |
Abbreviations
ADC, apparent diffusion coefficient; ADMSC, adipose-derived mesenchymal stem cells; AF4, Asymmetrical flow field-flow fractionation; AHSG, alpha 2-HS glycoprotein; ALB, albumin; ALT, alanine aminotransferase; APOH, apolipoprotein H; ASGPR1, asialoglycoprotein receptor; ASH, Alcoholic steatohepatitis; AST, aspartate aminotransferase; AUROC, Area under Receiver Operating Characteristics; BCA, bicinchonic acid; BM-MSCs, Bone marrow mesenchymal stem cells; CAFs, Cancer-associated fibroblasts; CCA, Cholangiocarcinoma; CCN2, Connective tissue growth factor; ceRNA, competing endogenous RNA; CFH, Complement Factor H; CH, chronic hepatitis; CHB, chronic hepatitis B; CHC, chronic Hepatitis C; CHOP, enhancer-binding protein homologous protein; CK18, Cytokeratin-18; CLEC3B, C-Type Lectin Domain Family 3 Member B; CSCs, Cancer stem cells; CTGF, connective tissue growth factor; DCP, des-gamma-carboxy prothrombin; DDS, drug delivery system; EVs, Extracellular vesicles; EC, endothelial cells; EGF, endothelial growth factor; E-HCC, early-stage hepatocellular carcinoma; EMT, Epithelial–mesenchymal transition; ENO1, Alpha-enolase; EpCAM, epithelial cell adhesion molecule; ESCRT, endosomal sorting complex required for transport; ELISA, enzyme linked immunosorbent assay; FCS, fluorescence correlation spectroscopy; FABP1, fatty acid binding protein 1; FGB, fibrinogen beta chain; FUS, focused ultrasound; GEVs, Glioma-derived EVs; GGT, glutamyl aminotransferase; GPC3, glypican3; HCC, hepatocellular carcinoma HCV, Hepatitis C Virus; HDL, High-density lipoprotein particles; HEMs, Adult human epidermal melanocytes; HGF, Hepatocyte growth factor; HIF-1α, Hypoxia Inducible factor 1 α; HIF-2α, Hypoxia Inducible factor 2 α; HSCs, Hepatic stellate cells; HUVECs, Human umbilical vein endothelial cells; IL, nterleukin; ILVs, intraluminal vesicles; iNOS, Inducible nitric oxide synthase; LAMP2B, lysosomal associated membrane protein 2B; LC, liver cirrhosis; LDLT, living donor liver transplantation; LG3BP, galectin-3-binding protein; LFIA, Lateral-Flow Immunochromatographic Assay; MMP, Matrix metalloproteinase; MPs, Microparticles; MSC, mesenchymal stem cells; MVBs, multivesicular bodies; m/lEVs, medium/large EVs; MDVs, Mitochondria-Derived Vesicles; MAC, membrane attack complex; NTA, nanoparticle tracking analysis; NAFL, Nonalcoholic fatty liver; NASH, non-alcoholic steatohepatitis; NC, normal control; NVs, Nanovesicles; PIGR, polymeric immunoglobulin receptor; RBP4, retinol binding protein 4; ROS, reactive oxygen species; sEVs, small EVs; SNX9, sorting nexin 9; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SAH, Severe alcoholic hepatitis; SMAD3, SMAD Family Member 3; sPTPRG, Protein tyrosine phosphatase receptor Gamma; TAMs, Tumor-associated macrophages; TF, transferrin; TGF-β, Transforming growth factor; TME, Tumor microenvironment; TNFα, Tumor necrosis factor alpha; TSG101, tumor susceptibility gene 101 protein; TRPS, tunable resistive pulse sensing; VEGF, Vascular endothelial growth factor.
References
- 1. Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing Profiles of Cancer Burden Worldwide and in China: A Secondary Analysis of the Global Cancer Statistics 2020. Chin Med J (Engl) (2021) 134:783–91. doi: 10.1097/CM9.0000000000001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer Statistics 2020. CA Cancer J Clin (2020) 70:7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 3. Mu XM, Wang W, Jiang YY, Feng J. Patterns of Comorbidity in Hepatocellular Carcinoma: A Network Perspective. Int J Environ Res Public Health (2020) 17(9):3108. doi: 10.3390/ijerph17093108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular Carcinoma. Nat Rev Dis Primers (2021) 7:6. doi: 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 5. Chen R, Xu X, Tao Y, Qian Z, Yu Y. Exosomes in Hepatocellular Carcinoma: A New Horizon. Cell Commun Signal (2019) 17:1. doi: 10.1186/s12964-018-0315-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang J, Wei F, Schafer C, Wong DT. Detection of Tumor Cell-Specific mRNA and Protein in Exosome-Like Microvesicles From Blood and Saliva. PLoS One (2014) 9:e110641. doi: 10.1371/journal.pone.0110641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, et al. Malignant Effusions and Immunogenic Tumour-Derived Exosomes. Lancet (2002) 360:295–305. doi: 10.1016/S0140-6736(02)09552-1 [DOI] [PubMed] [Google Scholar]
- 8. Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, et al. Proteomic Analysis of Exosomes Isolated From Human Malignant Pleural Effusions. Am J Respir Cell Mol Biol (2004) 31:114–21. doi: 10.1165/rcmb.2003-0238OC [DOI] [PubMed] [Google Scholar]
- 9. Revenfeld AL, Baek R, Nielsen MH, Stensballe A, Varming K, Jorgensen M. Diagnostic and Prognostic Potential of Extracellular Vesicles in Peripheral Blood. Clin Ther (2014) 36:830–46. doi: 10.1016/j.clinthera.2014.05.008 [DOI] [PubMed] [Google Scholar]
- 10. Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S, et al. Extracellular Vesicles From Blood Plasma: Determination of Their Morphology, Size, Phenotype and Concentration. J Thromb Haemost (2014) 12:614–27. doi: 10.1111/jth.12554 [DOI] [PubMed] [Google Scholar]
- 11. Salih M, Zietse R, Hoorn EJ. Urinary Extracellular Vesicles and the Kidney: Biomarkers and Beyond. Am J Physiol Renal Physiol (2014) 306:F1251–1259. doi: 10.1152/ajprenal.00128.2014 [DOI] [PubMed] [Google Scholar]
- 12. Zonneveld MI, Brisson AR, van Herwijnen MJ, Tan S, van de Lest CH, Redegeld FA, et al. Recovery of Extracellular Vesicles From Human Breast Milk Is Influenced by Sample Collection and Vesicle Isolation Procedures. J Extracell Vesicles (2014) 3. doi: 10.3402/jev.v3.24215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pieters BC, Arntz OJ, Bennink MB, Broeren MG, van Caam AP, Koenders MI, et al. Commercial Cow Milk Contains Physically Stable Extracellular Vesicles Expressing Immunoregulatory TGF-Beta. PLoS One (2015) 10:e0121123. doi: 10.1371/journal.pone.0121123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, et al. Beaming and Droplet Digital PCR Analysis of Mutant Idh1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids (2013) 2:e109. doi: 10.1038/mtna.2013.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginestra A, Miceli D, Dolo V, Romano FM, Vittorelli ML. Membrane Vesicles in Ovarian Cancer Fluids: A New Potential Marker. Anticancer Res (1999) 19:3439–45. [PubMed] [Google Scholar]
- 16. Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive Properties of Ovarian Cancer Ascites-Derived Membrane Vesicles. Cancer Res (2004) 64:7045–9. doi: 10.1158/0008-5472.CAN-04-1800 [DOI] [PubMed] [Google Scholar]
- 17. Webber J, Yeung V, Clayton A. Extracellular Vesicles as Modulators of the Cancer Microenvironment. Semin Cell Dev Biol (2015) 40:27–34. doi: 10.1016/j.semcdb.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 18. Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular Nanovesicle-Mediated microRNA Transfer: A Mechanism of Environmental Modulation of Hepatocellular Cancer Cell Growth. Hepatology (2011) 54:1237–48. doi: 10.1002/hep.24504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tu T, Budzinska MA, Maczurek AE, Cheng R, Di Bartolomeo A, Warner FJ, et al. Novel Aspects of the Liver Microenvironment in Hepatocellular Carcinoma Pathogenesis and Development. Int J Mol Sci (2014) 15:9422–58. doi: 10.3390/ijms15069422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev (2018) 118:1917–50. doi: 10.1021/acs.chemrev.7b00534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borrelli DA, Yankson K, Shukla N, Vilanilam G, Ticer T, Wolfram J. Extracellular Vesicle Therapeutics for Liver Disease. J Control Release (2018) 273:86–98. doi: 10.1016/j.jconrel.2018.01.022 [DOI] [PubMed] [Google Scholar]
- 22. Wu P, Zhang B, Ocansey DKW, Xu W, Qian H. Extracellular Vesicles: A Bright Star of Nanomedicine. Biomaterials (2021) 269:120467. doi: 10.1016/j.biomaterials.2020.120467 [DOI] [PubMed] [Google Scholar]
- 23. Raposo G, Stoorvogel W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J Cell Biol (2013) 200:373–83. doi: 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thery C, Zitvogel L, Amigorena S. Exosomes: Composition. Biogenesis Function Nat Rev Immunol (2002) 2:569–79. doi: 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- 25. Colombo M, Raposo G, Thery C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol (2014) 30:255–89. doi: 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 26. Manickam DS. Delivery of Mitochondria Via Extracellular Vesicles - A New Horizon in Drug Delivery. J Control Release (2022) 343:400–7. doi: 10.1016/j.jconrel.2022.01.045 [DOI] [PubMed] [Google Scholar]
- 27. Thery C, Ostrowski M, Segura E. Membrane Vesicles as Conveyors of Immune Responses. Nat Rev Immunol (2009) 9:581–93. doi: 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- 28. Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J Extracell Vesicles (2018) 7:1535750. doi: 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun B, Zhai S, Zhang L, Sun G. The Role of Extracellular Vesicles in Podocyte Autophagy in Kidney Disease. J Cell Commun Signal (2021) 15:299–316. doi: 10.1007/s12079-020-00594-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cabeza L, Perazzoli G, Pena M, Cepero A, Luque C, Melguizo C, et al. Cancer Therapy Based on Extracellular Vesicles as Drug Delivery Vehicles. J Control Release (2020) 327:296–315. doi: 10.1016/j.jconrel.2020.08.018 [DOI] [PubMed] [Google Scholar]
- 31. Zhang X, Zhang H, Gu J, Zhang J, Shi H, Qian H, et al. Engineered Extracellular Vesicles for Cancer Therapy. Adv Mater (2021) 33:e2005709. doi: 10.1002/adma.202005709 [DOI] [PubMed] [Google Scholar]
- 32. Poon IKH, Parkes MAF, Jiang L, Atkin-Smith GK, Tixeira R, Gregory CD, et al. Moving Beyond Size and Phosphatidylserine Exposure: Evidence for a Diversity of Apoptotic Cell-Derived Extracellular Vesicles In Vitro. J Extracell Vesicles (2019) 8:1608786. doi: 10.1080/20013078.2019.1608786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jingu K, Miyoshi M, Arichi E, Ohmagari J, Wada S, Uehara S, et al. [Interstitial Radiotherapy of Carcinoma of the Tongue-Comparison Between Jacobson-Yamamoto Grading and Prognosis]. Gan No Rinsho (1989) 35:1587–90. [PubMed] [Google Scholar]
- 34. Dai J, Shupp AB, Bussard KM, Keller ET. Extracellular Vesicles and Bone-Associated Cancer. Curr Osteoporos Rep (2021) 19:223–9. doi: 10.1007/s11914-021-00668-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ahmadzada T, Kao S, Reid G, Clarke S, Grau GE, Hosseini-Beheshti E. Extracellular Vesicles as Biomarkers in Malignant Pleural Mesothelioma: A Review. Crit Rev Oncol Hematol (2020) 150:102949. doi: 10.1016/j.critrevonc.2020.102949 [DOI] [PubMed] [Google Scholar]
- 36. Nazarenko I. Extracellular Vesicles: Recent Developments in Technology and Perspectives for Cancer Liquid Biopsy. Recent Results Cancer Res (2020) 215:319–44. doi: 10.1007/978-3-030-26439-0_17 [DOI] [PubMed] [Google Scholar]
- 37. Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, et al. Exosomes Reflect the Hypoxic Status of Glioma Cells and Mediate Hypoxia-Dependent Activation of Vascular Cells During Tumor Development. Proc Natl Acad Sci USA (2013) 110:7312–7. doi: 10.1073/pnas.1220998110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang T, Gilkes DM, Takano N, Xiang L, Luo W, Bishop CJ, et al. Hypoxia-Inducible Factors and RAB22A Mediate Formation of Microvesicles That Stimulate Breast Cancer Invasion and Metastasis. Proc Natl Acad Sci USA (2014) 111:E3234–42. doi: 10.1073/pnas.1410041111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hara M, Yanagihara T, Hirayama Y, Ogasawara S, Kurosawa H, Sekine S, et al. Podocyte Membrane Vesicles in Urine Originate From Tip Vesiculation of Podocyte Microvilli. Hum Pathol (2010) 41:1265–75. doi: 10.1016/j.humpath.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 40. Yang B, Feng X, Liu H, Tong R, Wu J, Li C, et al. High-Metastatic Cancer Cells Derived Exosomal miR92a-3p Promotes Epithelial-Mesenchymal Transition and Metastasis of Low-Metastatic Cancer Cells by Regulating PTEN/Akt Pathway in Hepatocellular Carcinoma. Oncogene (2020) 39:6529–43. doi: 10.1038/s41388-020-01450-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Pace AL, Tumino N, Besi F, Alicata C, Conti LA, Munari E, et al. Characterization of Human Nk Cell-Derived Exosomes: Role of DNAM1 Receptor In Exosome-Mediated Cytotoxicity Against Tumor. Cancers (Basel) (2020) 12(3):661. doi: 10.3390/cancers12030661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ventimiglia LN, Alonso MA. Biogenesis and Function of T Cell-Derived Exosomes. Front Cell Dev Biol (2016) 4:84. doi: 10.3389/fcell.2016.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clayton A, Turkes A, Dewitt S, Steadman R, Mason MD, Hallett MB. Adhesion and Signaling by B Cell-Derived Exosomes: The Role of Integrins. FASEB J (2004) 18:977–9. doi: 10.1096/fj.03-1094fje [DOI] [PubMed] [Google Scholar]
- 44. Schorey JS, Bhatnagar S. Exosome Function: From Tumor Immunology to Pathogen Biology. Traffic (2008) 9:871–81. doi: 10.1111/j.1600-0854.2008.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He C, Zheng S, Luo Y, Wang B. Exosome Theranostics: Biology and Translational Medicine. Theranostics (2018) 8:237–55. doi: 10.7150/thno.21945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang N, Li S, Li G, Zhang S, Tang X, Ni S, et al. The Role of Extracellular Vesicles in Mediating Progression, Metastasis and Potential Treatment of Hepatocellular Carcinoma. Oncotarget (2017) 8:3683–95. doi: 10.18632/oncotarget.12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lapitz A, Arbelaiz A, Olaizola P, Aranburu A, Bujanda L, Perugorria MJ, et al. Extracellular Vesicles in Hepatobiliary Malignancies. Front Immunol (2018) 9:2270. doi: 10.3389/fimmu.2018.02270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luzio JP, Gray SR, Bright NA. Endosome-Lysosome Fusion. Biochem Soc Trans (2010) 38:1413–6. doi: 10.1042/BST0381413 [DOI] [PubMed] [Google Scholar]
- 49. Kowal J, Tkach M, Thery C. Biogenesis and Secretion of Exosomes. Curr Opin Cell Biol (2014) 29:116–25. doi: 10.1016/j.ceb.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 50. van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: A Common Pathway for a Specialized Function. J Biochem (2006) 140:13–21. doi: 10.1093/jb/mvj128 [DOI] [PubMed] [Google Scholar]
- 51. Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, et al. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct From Apoptotic Vesicles. J Immunol (2001) 166:7309–18. doi: 10.4049/jimmunol.166.12.7309 [DOI] [PubMed] [Google Scholar]
- 52. Hemler ME. Tetraspanin Proteins Mediate Cellular Penetration, Invasion, and Fusion Events and Define a Novel Type of Membrane Microdomain. Annu Rev Cell Dev Biol (2003) 19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609 [DOI] [PubMed] [Google Scholar]
- 53. Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, et al. Molecular Characterization of Dendritic Cell-Derived Exosomes. Selective Accumulation of the Heat Shock Protein Hsc73. J Cell Biol (1999) 147:599–610. doi: 10.1083/jcb.147.3.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hemler ME. Targeting of Tetraspanin Proteins–Potential Benefits and Strategies. Nat Rev Drug Discov (2008) 7:747–58. doi: 10.1038/nrd2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hurley JH. The ESCRT Complexes. Crit Rev Biochem Mol Biol (2010) 45:463–87. doi: 10.3109/10409238.2010.502516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide Triggers Budding of Exosome Vesicles Into Multivesicular Endosomes. Science (2008) 319:1244–7. doi: 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- 57. Savina A, Fader CM, Damiani MT, Colombo MI. Rab11 Promotes Docking and Fusion of Multivesicular Bodies in a Calcium-Dependent Manner. Traffic (2005) 6:131–43. doi: 10.1111/j.1600-0854.2004.00257.x [DOI] [PubMed] [Google Scholar]
- 58. Hugel B, Martinez MC, Kunzelmann C, Freyssinet JM. Membrane Microparticles: Two Sides of the Coin. Physiol (Bethesda) (2005) 20:22–7. doi: 10.1152/physiol.00029.2004 [DOI] [PubMed] [Google Scholar]
- 59. Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (Armms) at Plasma Membrane by Recruitment of TSG101 Protein. Proc Natl Acad Sci USA (2012) 109:4146–51. doi: 10.1073/pnas.1200448109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-Like Vesicles, and Apoptotic Bodies. J Neurooncol (2013) 113:1–11. doi: 10.1007/s11060-013-1084-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, et al. Membrane Vesicles, Current State-of-the-Art: Emerging Role of Extracellular Vesicles. Cell Mol Life Sci (2011) 68:2667–88. doi: 10.1007/s00018-011-0689-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiang L, Paone S, Caruso S, Atkin-Smith GK, Phan TK, Hulett MD, et al. Determining the Contents and Cell Origins of Apoptotic Bodies by Flow Cytometry. Sci Rep (2017) 7:14444. doi: 10.1038/s41598-017-14305-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Minciacchi VR, You S, Spinelli C, Morley S, Zandian M, Aspuria PJ, et al. Large Oncosomes Contain Distinct Protein Cargo and Represent a Separate Functional Class of Tumor-Derived Extracellular Vesicles. Oncotarget (2015) 6:11327–41. doi: 10.18632/oncotarget.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Medeiros T, Myette RL, Almeida JR, Silva AA, Burger D. Extracellular Vesicles: Cell-Derived Biomarkers of Glomerular and Tubular Injury. Cell Physiol Biochem (2020) 54:88–109. doi: 10.33594/000000207 [DOI] [PubMed] [Google Scholar]
- 65. Ullah M, Kodam SP, Mu Q, Akbar A. Microbubbles Versus Extracellular Vesicles as Therapeutic Cargo for Targeting Drug Delivery. ACS Nano (2021) 15:3612–20. doi: 10.1021/acsnano.0c10689 [DOI] [PubMed] [Google Scholar]
- 66. Abudoureyimu M, Zhou H, Zhi Y, Wang T, Feng B, Wang R, et al. Recent Progress in the Emerging Role of Exosome in Hepatocellular Carcinoma. Cell Prolif (2019) 52:e12541. doi: 10.1111/cpr.12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Phinney DG, Di Giuseppe M, Njah J, Sala E, Shiva S, St Croix CM, et al. Mesenchymal Stem Cells Use Extracellular Vesicles to Outsource Mitophagy and Shuttle Micrornas. Nat Commun (2015) 6:8472. doi: 10.1038/ncomms9472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, Meckes DG, Jr. Proteomic Profiling of NCI-60 Extracellular Vesicles Uncovers Common Protein Cargo and Cancer Type-Specific Biomarkers. Oncotarget (2016) 7:86999–7015. doi: 10.18632/oncotarget.13569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dong LF, Kovarova J, Bajzikova M, Bezawork-Geleta A, Svec D, Endaya B, et al. Horizontal Transfer of Whole Mitochondria Restores Tumorigenic Potential in Mitochondrial DNA-Deficient Cancer Cells. Elife (2017) 6:e22187. doi: 10.7554/eLife.22187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jang SC, Crescitelli R, Cvjetkovic A, Belgrano V, Olofsson Bagge R, Sundfeldt K, et al. Mitochondrial Protein Enriched Extracellular Vesicles Discovered in Human Melanoma Tissues can be Detected in Patient Plasma. J Extracell Vesicles (2019) 8:1635420. doi: 10.1080/20013078.2019.1635420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. D'Souza A, Burch A, Dave KM, Sreeram A, Reynolds MJ, Dobbins DX, et al. Microvesicles Transfer Mitochondria and Increase Mitochondrial Function in Brain Endothelial Cells. J Control Release (2021) 338:505–26. doi: 10.1016/j.jconrel.2021.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Todkar K, Chikhi L, Desjardins V, El-Mortada F, Pepin G, Germain M. Selective Packaging of Mitochondrial Proteins Into Extracellular Vesicles Prevents the Release of Mitochondrial Damps. Nat Commun (2021) 12:1971. doi: 10.1038/s41467-021-21984-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Todkar K, Chikhi L, Germain M. Mitochondrial Interaction With the Endosomal Compartment in Endocytosis and Mitochondrial Transfer. Mitochondrion (2019) 49:284–8. doi: 10.1016/j.mito.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 74. Islam MN, Das SR, Emin MT, Wei M, Sun L, Westphalen K, et al. Mitochondrial Transfer From Bone-Marrow-Derived Stromal Cells to Pulmonary Alveoli Protects Against Acute Lung Injury. Nat Med (2012) 18:759–65. doi: 10.1038/nm.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hough KP, Trevor JL, Strenkowski JG, Wang Y, Chacko BK, Tousif S, et al. Exosomal Transfer of Mitochondria From Airway Myeloid-Derived Regulatory Cells to T Cells. Redox Biol (2018) 18:54–64. doi: 10.1016/j.redox.2018.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin YH, Chu YD, Lim SN, Chen CW, Yeh CT, Lin WR. Impact of an MT-RNR1 Gene Polymorphism on Hepatocellular Carcinoma Progression and Clinical Characteristics. Int J Mol Sci (2021) 22(3):1119. doi: 10.3390/ijms22031119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, et al. Extracellular Vesicle-Based Therapeutics: Natural Versus Engineered Targeting and Trafficking. Exp Mol Med (2019) 51:1–12. doi: 10.1038/s12276-019-0223-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat Cell Biol (2019) 21:9–17. doi: 10.1038/s41556-018-0250-9 [DOI] [PubMed] [Google Scholar]
- 79. Xu Y, Feng K, Zhao H, Di L, Wang L, Wang R. Tumor-Derived Extracellular Vesicles as Messengers of Natural Products in Cancer Treatment. Theranostics (2022) 12:1683–714. doi: 10.7150/thno.67775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Abdelaal AM, Kasinski AL. Ligand-Mediated Delivery of RNAi-based Therapeutics for the Treatment of Oncological Diseases. NAR Cancer (2021) 3:zcab030. doi: 10.1093/narcan/zcab030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mayor S, Pagano RE. Pathways of Clathrin-Independent Endocytosis. Nat Rev Mol Cell Biol (2007) 8:603–12. doi: 10.1038/nrm2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Doherty GJ, McMahon HT. Mechanisms of Endocytosis. Annu Rev Biochem (2009) 78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- 83. Costa Verdera H, Gitz-Francois JJ, Schiffelers RM, Vader P. Cellular Uptake of Extracellular Vesicles Is Mediated by Clathrin-Independent Endocytosis and Macropinocytosis. J Control Release (2017) 266:100–8. doi: 10.1016/j.jconrel.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 84. Conner SD, Schmid SL. Regulated Portals of Entry Into the Cell. Nature (2003) 422:37–44. doi: 10.1038/nature01451 [DOI] [PubMed] [Google Scholar]
- 85. Mulcahy LA, Pink RC, Carter DR. Routes and Mechanisms of Extracellular Vesicle Uptake. J Extracell Vesicles (2014) 3. doi: 10.3402/jev.v3.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, et al. Cancer Cell-Derived Microvesicles Induce Transformation by Transferring Tissue Transglutaminase and Fibronectin to Recipient Cells. Proc Natl Acad Sci USA (2011) 108:4852–7. doi: 10.1073/pnas.1017667108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat Commun (2011) 2:180. doi: 10.1038/ncomms1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Martinez MC, Tesse A, Zobairi F, Andriantsitohaina R. Shed Membrane Microparticles From Circulating and Vascular Cells in Regulating Vascular Function. Am J Physiol Heart Circ Physiol (2005) 288:H1004–1009. doi: 10.1152/ajpheart.00842.2004 [DOI] [PubMed] [Google Scholar]
- 89. Morel O, Toti F, Hugel B, Freyssinet JM. Cellular Microparticles: A Disseminated Storage Pool of Bioactive Vascular Effectors. Curr Opin Hematol (2004) 11:156–64. doi: 10.1097/01.moh.0000131441.10020.87 [DOI] [PubMed] [Google Scholar]
- 90. Hirsova P, Ibrahim SH, Krishnan A, Verma VK, Bronk SF, Werneburg NW, et al. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology (2016) 150:956–67. doi: 10.1053/j.gastro.2015.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Burnley-Hall N, Willis G, Davis J, Rees DA, James PE. Nitrite-Derived Nitric Oxide Reduces Hypoxia-Inducible Factor 1alpha-Mediated Extracellular Vesicle Production by Endothelial Cells. Nitric Oxide (2017) 63:1–12. doi: 10.1016/j.niox.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 92. Bister N, Pistono C, Huremagic B, Jolkkonen J, Giugno R, Malm T. Hypoxia and Extracellular Vesicles: A Review on Methods, Vesicular Cargo and Functions. J Extracell Vesicles (2020) 10:e12002. doi: 10.1002/jev2.12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, et al. Role of HIF-1alpha in Hypoxia-Mediated Apoptosis, Cell Proliferation and Tumour Angiogenesis. Nature (1998) 394:485–90. doi: 10.1038/28867 [DOI] [PubMed] [Google Scholar]
- 94. Shen Z, Sun J, Shao J, Xu J. Ultraviolet B Irradiation Enhances the Secretion of Exosomes by Human Primary Melanocytes and Changes Their Exosomal miRNA Profile. PLoS One (2020) 15:e0237023. doi: 10.1371/journal.pone.0237023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Aubertin K, Silva AK, Luciani N, Espinosa A, Djemat A, Charue D, et al. Massive Release of Extracellular Vesicles From Cancer Cells After Photodynamic Treatment or Chemotherapy. Sci Rep (2016) 6:35376. doi: 10.1038/srep35376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sims PJ, Faioni EM, Wiedmer T, Shattil SJ. Complement Proteins C5b-9 Cause Release of Membrane Vesicles From the Platelet Surface That Are Enriched in the Membrane Receptor for Coagulation Factor Va and Express Prothrombinase Activity. J Biol Chem (1988) 263:18205–12. doi: 10.1016/S0021-9258(19)81346-7 [DOI] [PubMed] [Google Scholar]
- 97. Pilzer D, Gasser O, Moskovich O, Schifferli JA, Fishelson Z. Emission of Membrane Vesicles: Roles in Complement Resistance, Immunity and Cancer. Springer Semin Immunopathol (2005) 27:375–87. doi: 10.1007/s00281-005-0004-1 [DOI] [PubMed] [Google Scholar]
- 98. Tang K, Zhang Y, Zhang H, Xu P, Liu J, Ma J, et al. Delivery of Chemotherapeutic Drugs in Tumour Cell-Derived Microparticles. Nat Commun (2012) 3:1282. doi: 10.1038/ncomms2282 [DOI] [PubMed] [Google Scholar]
- 99. Gao T, Guo W, Chen M, Huang J, Yuan Z, Zhang Y, et al. Extracellular Vesicles and Autophagy in Osteoarthritis. BioMed Res Int (2016) 2016:2428915. doi: 10.1155/2016/2428915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Eltom S, Dale N, Raemdonck KR, Stevenson CS, Snelgrove RJ, Sacitharan PK, et al. Respiratory Infections Cause the Release of Extracellular Vesicles: Implications in Exacerbation of Asthma/COPD. PLoS One (2014) 9:e101087. doi: 10.1371/journal.pone.0101087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Sheybani ND, Batts AJ, Mathew AS, Thim EA, Price RJ. Focused Ultrasound Hyperthermia Augments Release of Glioma-derived Extracellular Vesicles With Differential Immunomodulatory Capacity. Theranostics (2020) 10:7436–47. doi: 10.7150/thno.46534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, et al. Techniques Used for the Isolation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey. J Extracell Vesicles (2016) 5:32945. doi: 10.3402/jev.v5.32945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-Mediated Transfer of mRNAs and microRNAs Is a Novel Mechanism of Genetic Exchange Between Cells. Nat Cell Biol (2007) 9:654–9. doi: 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 104. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat Cell Biol (2008) 10:1470–6. doi: 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lane RE, Korbie D, Trau M, Hill MM. Purification Protocols for Extracellular Vesicles. Methods Mol Biol (2017) 1660:111–30. doi: 10.1007/978-1-4939-7253-1_10 [DOI] [PubMed] [Google Scholar]
- 106. Thietart S, Rautou PE. Extracellular Vesicles as Biomarkers in Liver Diseases: A Clinician's Point of View. J Hepatol (2020) 73:1507–25. doi: 10.1016/j.jhep.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 107. Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Res Int (2018) 2018:8545347. doi: 10.1155/2018/8545347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and Characterization of Exosomes From Cell Culture Supernatants and Biological Fluids. Curr Protoc Cell Biol (2006) Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- 109. Vidal M, Mangeat P, Hoekstra D. Aggregation Reroutes Molecules From a Recycling to a Vesicle-Mediated Secretion Pathway During Reticulocyte Maturation. J Cell Sci (1997) 110(Pt 16):1867–77. doi: 10.1242/jcs.110.16.1867 [DOI] [PubMed] [Google Scholar]
- 110. Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R. Co-Isolation of Extracellular Vesicles and High-Density Lipoproteins Using Density Gradient Ultracentrifugation. J Extracell Vesicles (2014) 3. doi: 10.3402/jev.v3.23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc Natl Acad Sci USA (2016) 113:E968–977. doi: 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of Protein, microRNA, and mRNA Yields Using Different Methods of Urinary Exosome Isolation for the Discovery of Kidney Disease Biomarkers. Kidney Int (2012) 82:1024–32. doi: 10.1038/ki.2012.256 [DOI] [PubMed] [Google Scholar]
- 113. Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, et al. Comparison of Serum Exosome Isolation Methods for microRNA Profiling. Clin Biochem (2014) 47:135–8. doi: 10.1016/j.clinbiochem.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 114. Gholizadeh S, Shehata Draz M, Zarghooni M, Sanati-Nezhad A, Ghavami S, Shafiee H, et al. Microfluidic Approaches for Isolation, Detection, and Characterization of Extracellular Vesicles: Current Status and Future Directions. Biosens Bioelectron (2017) 91:588–605. doi: 10.1016/j.bios.2016.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, Nieuwland R. Single-Step Isolation of Extracellular Vesicles by Size-Exclusion Chromatography. J Extracell Vesicles (2014) 3. doi: 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ, Heusermann W, et al. Ultrafiltration With Size-Exclusion Liquid Chromatography for High Yield Isolation of Extracellular Vesicles Preserving Intact Biophysical and Functional Properties. Nanomedicine (2015) 11:879–83. doi: 10.1016/j.nano.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 117. Lozano-Ramos I, Bancu I, Oliveira-Tercero A, Armengol MP, Menezes-Neto A, Del Portillo HA, et al. Size-Exclusion Chromatography-Based Enrichment of Extracellular Vesicles From Urine Samples. J Extracell Vesicles (2015) 4:27369. doi: 10.3402/jev.v4.27369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gamez-Valero A, Monguio-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borras FE. Size-Exclusion Chromatography-based Isolation Minimally Alters Extracellular Vesicles' Characteristics Compared to Precipitating Agents. Sci Rep (2016) 6:33641. doi: 10.1038/srep33641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kreimer S, Ivanov AR. Rapid Isolation of Extracellular Vesicles From Blood Plasma With Size-Exclusion Chromatography Followed by Mass Spectrometry-Based Proteomic Profiling. Methods Mol Biol (2017) 1660:295–302. doi: 10.1007/978-1-4939-7253-1_24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sitar S, Kejzar A, Pahovnik D, Kogej K, Tusek-Znidaric M, Lenassi M, et al. Size Characterization and Quantification of Exosomes by Asymmetrical-Flow Field-Flow Fractionation. Anal Chem (2015) 87:9225–33. doi: 10.1021/acs.analchem.5b01636 [DOI] [PubMed] [Google Scholar]
- 121. Kang D, Oh S, Ahn SM, Lee BH, Moon MH. Proteomic Analysis of Exosomes From Human Neural Stem Cells by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. J Proteome Res (2008) 7:3475–80. doi: 10.1021/pr800225z [DOI] [PubMed] [Google Scholar]
- 122. Yoo CE, Kim G, Kim M, Park D, Kang HJ, Lee M, et al. A Direct Extraction Method for microRNAs From Exosomes Captured by Immunoaffinity Beads. Anal Biochem (2012) 431:96–8. doi: 10.1016/j.ab.2012.09.008 [DOI] [PubMed] [Google Scholar]
- 123. Reategui E, van der Vos KE, Lai CP, Zeinali M, Atai NA, Aldikacti B, et al. Engineered Nanointerfaces for Microfluidic Isolation and Molecular Profiling of Tumor-Specific Extracellular Vesicles. Nat Commun (2018) 9:175. doi: 10.1038/s41467-017-02261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhao Z, Yang Y, Zeng Y, He M. A Microfluidic ExoSearch Chip for Multiplexed Exosome Detection Towards Blood-Based Ovarian Cancer Diagnosis. Lab Chip (2016) 16:489–96. doi: 10.1039/c5lc01117e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics (2017) 7:789–804. doi: 10.7150/thno.18133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rho J, Chung J, Im H, Liong M, Shao H, Castro CM, et al. Magnetic Nanosensor for Detection and Profiling of Erythrocyte-Derived Microvesicles. ACS Nano (2013) 7:11227–33. doi: 10.1021/nn405016y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lee K, Shao H, Weissleder R, Lee H. Acoustic Purification of Extracellular Microvesicles. ACS Nano (2015) 9:2321–7. doi: 10.1021/nn506538f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Serrano-Pertierra E, Oliveira-Rodriguez M, Matos M, Gutierrez G, Moyano A, Salvador M, et al. Extracellular Vesicles: Current Analytical Techniques for Detection and Quantification. Biomolecules (2020) 10(6):824. doi: 10.3390/biom10060824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Comfort N, Cai K, Bloomquist TR, Strait MD, Ferrante AW, Jr., Baccarelli AA. Nanoparticle Tracking Analysis for the Quantification and Size Determination of Extracellular Vesicles. J Vis Exp (2021) (169):10.3791/62447. doi: 10.3791/62447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Carnell-Morris P, Tannetta D, Siupa A, Hole P, Dragovic R. Analysis of Extracellular Vesicles Using Fluorescence Nanoparticle Tracking Analysis. Methods Mol Biol (2017) 1660:153–73. doi: 10.1007/978-1-4939-7253-1_13 [DOI] [PubMed] [Google Scholar]
- 131. Osteikoetxea X, Balogh A, Szabo-Taylor K, Nemeth A, Szabo TG, Paloczi K, et al. Improved Characterization of EV Preparations Based on Protein to Lipid Ratio and Lipid Properties. PLoS One (2015) 10:e0121184. doi: 10.1371/journal.pone.0121184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mihaly J, Deak R, Szigyarto IC, Bota A, Beke-Somfai T, Varga Z. Characterization of Extracellular Vesicles by IR Spectroscopy: Fast and Simple Classification Based on Amide and CH Stretching Vibrations. Biochim Biophys Acta Biomembr (2017) 1859:459–66. doi: 10.1016/j.bbamem.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 133. Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, et al. Obstacles and Opportunities in the Functional Analysis of Extracellular Vesicle RNA - an ISEV Position Paper. J Extracell Vesicles (2017) 6:1286095. doi: 10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Devhare PB, Ray RB. Extracellular Vesicles: Novel Mediator for Cell to Cell Communications in Liver Pathogenesis. Mol Aspects Med (2018) 60:115–22. doi: 10.1016/j.mam.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Xiong W, Sun LP, Chen XM, Li HY, Huang SA, Jie SH. Comparison of microRNA Expression Profiles in HCC-derived Microvesicles and the Parental Cells and Evaluation of Their Roles in HCC. J Huazhong Univ Sci Technol Med Sci (2013) 33:346–52. doi: 10.1007/s11596-013-1122-y [DOI] [PubMed] [Google Scholar]
- 136. Lemoinne S, Thabut D, Housset C, Moreau R, Valla D, Boulanger CM, et al. The Emerging Roles of Microvesicles in Liver Diseases. Nat Rev Gastroenterol Hepatol (2014) 11:350–61. doi: 10.1038/nrgastro.2014.7 [DOI] [PubMed] [Google Scholar]
- 137. Kornek M, Popov Y, Libermann TA, Afdhal NH, Schuppan D. Human T Cell Microparticles Circulate in Blood of Hepatitis Patients and Induce Fibrolytic Activation of Hepatic Stellate Cells. Hepatology (2011) 53:230–42. doi: 10.1002/hep.23999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Jaffar Ali D, He C, Xu H, Kumaravel S, Sun B, Zhou Y, et al. Microvesicles Mediate Sorafenib Resistance in Liver Cancer Cells Through Attenuating p53 and Enhancing FOXM1 Expression. Life Sci (2021) 271:119149. doi: 10.1016/j.lfs.2021.119149 [DOI] [PubMed] [Google Scholar]
- 139. Li H, Sun L, Chen X, Xiong W, Hu D, Jie S. Microvesicle microRNA Profiles and Functional Roles Between Chronic Hepatitis B and Hepatocellular Carcinoma. Clin Transl Oncol (2014) 16:315–21. doi: 10.1007/s12094-013-1078-1 [DOI] [PubMed] [Google Scholar]
- 140. Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, et al. miR-34a Inhibits Migration and Invasion by Down-Regulation of c-Met Expression in Human Hepatocellular Carcinoma Cells. Cancer Lett (2009) 275:44–53. doi: 10.1016/j.canlet.2008.09.035 [DOI] [PubMed] [Google Scholar]
- 141. Li D, Jia H, Zhang H, Lv M, Liu J, Zhang Y, et al. TLR4 Signaling Induces the Release of Microparticles by Tumor Cells That Regulate Inflammatory Cytokine IL-6 of Macrophages Via microRNA Let-7b. Oncoimmunology (2012) 1:687–93. doi: 10.4161/onci.19854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang W, Zhao P, Xu XL, Cai L, Song ZS, Cao DY, et al. Annexin A2 Promotes the Migration and Invasion of Human Hepatocellular Carcinoma Cells In Vitro by Regulating the Shedding of CD147-harboring Microvesicles From Tumor Cells. PLoS One (2013) 8:e67268. doi: 10.1371/journal.pone.0067268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-Derived Exosomal miR-1247-3p Induces Cancer-Associated Fibroblast Activation to Foster Lung Metastasis of Liver Cancer. Nat Commun (2018) 9:191. doi: 10.1038/s41467-017-02583-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular Carcinoma-Derived Exosomal miRNA-21 Contributes to Tumor Progression by Converting Hepatocyte Stellate Cells to Cancer-Associated Fibroblasts. J Exp Clin Cancer Res (2018) 37:324. doi: 10.1186/s13046-018-0965-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology (2019) 70:241–58. doi: 10.1002/hep.30607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA Tuc339. Int J Mol Sci (2018) 19(10):2958. doi: 10.3390/ijms19102958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 Promotes Proliferation and Invasion in Hepatocellular Carcinoma by Directly Inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun (2018) 502:515–21. doi: 10.1016/j.bbrc.2018.05.208 [DOI] [PubMed] [Google Scholar]
- 148. Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. Lncrna FAL1 Promotes Cell Proliferation and Migration by Acting as a CeRNA of miR-1236 in Hepatocellular Carcinoma Cells. Life Sci (2018) 197:122–9. doi: 10.1016/j.lfs.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 149. Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal Circfblim1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3564 [DOI] [PubMed] [Google Scholar]
- 150. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three Isoforms of Exosomal circPTGR1 Promote Hepatocellular Carcinoma Metastasis Via the miR449a-MET Pathway. EBioMedicine (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Mao X, Zhou L, Tey SK, Ma APY, Yeung CLS, Ng TH, et al. Tumour Extracellular Vesicle-Derived Complement Factor H Promotes Tumorigenesis and Metastasis by Inhibiting Complement-Dependent Cytotoxicity of Tumour Cells. J Extracell Vesicles (2020) 10:e12031. doi: 10.1002/jev2.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular Carcinoma Cell-Secreted Exosomal Microrna-210 Promotes Angiogenesis In Vitro and In Vivo . Mol Ther Nucleic Acids (2018) 11:243–52. doi: 10.1016/j.omtn.2018.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, et al. Hepatoma Cell-Secreted Exosomal microRNA-103 Increases Vascular Permeability and Promotes Metastasis by Targeting Junction Proteins. Hepatology (2018) 68:1459–75. doi: 10.1002/hep.29920 [DOI] [PubMed] [Google Scholar]
- 154. Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, et al. CD90+ Liver Cancer Cells Modulate Endothelial Cell Phenotype Through the Release of Exosomes Containing H19 Lncrna. Mol Cancer (2015) 14:155. doi: 10.1186/s12943-015-0426-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zhou Y, Zhu Y, Fan X, Zhang C, Wang Y, Zhang L, et al. NID1, a New Regulator of EMT Required for Metastasis and Chemoresistance of Ovarian Cancer Cells. Oncotarget (2017) 8:33110–21. doi: 10.18632/oncotarget.16145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Mao X, Tey SK, Yeung CLS, Kwong EML, Fung YME, Chung CYS, et al. Nidogen 1-Enriched Extracellular Vesicles Facilitate Extrahepatic Metastasis of Liver Cancer by Activating Pulmonary Fibroblasts to Secrete Tumor Necrosis Factor Receptor 1. Adv Sci (Weinh) (2020) 7:2002157. doi: 10.1002/advs.202002157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Yukawa H, Suzuki K, Aoki K, Arimoto T, Yasui T, Kaji N, et al. Imaging of Angiogenesis of Human Umbilical Vein Endothelial Cells by Uptake of Exosomes Secreted From Hepatocellular Carcinoma Cells. Sci Rep (2018) 8:6765. doi: 10.1038/s41598-018-24563-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Dai W, Wang Y, Yang T, Wang J, Wu W, Gu J. Downregulation of Exosomal CLEC3B in Hepatocellular Carcinoma Promotes Metastasis and Angiogenesis Via AMPK and VEGF Signals. Cell Commun Signal (2019) 17:113. doi: 10.1186/s12964-019-0423-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin HM, et al. Vps4A Functions as a Tumor Suppressor by Regulating the Secretion and Uptake of Exosomal microRNAs in Human Hepatoma Cells. Hepatology (2015) 61:1284–94. doi: 10.1002/hep.27660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, et al. Exosome-Transmitted Long Non-Coding RNA Senp3-EIF4A1 Suppresses the Progression of Hepatocellular Carcinoma. Aging (Albany NY) (2020) 12:11550–67. doi: 10.18632/aging.103302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-Transmitted Circular RNA Hsa_Circ_0051443 Suppresses Hepatocellular Carcinoma Progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022 [DOI] [PubMed] [Google Scholar]
- 162. Cao L, Xie B, Yang X, Liang H, Jiang X, Zhang D, et al. Mir-324-5p Suppresses Hepatocellular Carcinoma Cell Invasion by Counteracting Ecm Degradation Through Post-Transcriptionally Downregulating ETS1 and SP1. PLoS One (2015) 10:e0133074. doi: 10.1371/journal.pone.0133074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Huang X, Sun L, Wen S, Deng D, Wan F, He X, et al. RNA Sequencing of Plasma Exosomes Revealed Novel Functional Long Noncoding RNAs in Hepatocellular Carcinoma. Cancer Sci (2020) 111:3338–49. doi: 10.1111/cas.14516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, et al. Loss of Exosomal miR-320a From Cancer-Associated Fibroblasts Contributes to HCC Proliferation and Metastasis. Cancer Lett (2017) 397:33–42. doi: 10.1016/j.canlet.2017.03.004 [DOI] [PubMed] [Google Scholar]
- 165. Basu S, Bhattacharyya SN. Insulin-Like Growth Factor-1 Prevents miR-122 Production in Neighbouring Cells to Curtail Its Intercellular Transfer to Ensure Proliferation of Human Hepatoma Cells. Nucleic Acids Res (2014) 42:7170–85. doi: 10.1093/nar/gku346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zhang J, Shan WF, Jin TT, Wu GQ, Xiong XX, Jin HY, et al. Propofol Exerts Anti-Hepatocellular Carcinoma by Microvesicle-Mediated Transfer of miR-142-3p From Macrophage to Cancer Cells. J Transl Med (2014) 12:279. doi: 10.1186/s12967-014-0279-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, et al. The Significance of Exosomes in the Development and Treatment of Hepatocellular Carcinoma. Mol Cancer (2020) 19:1. doi: 10.1186/s12943-019-1085-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, et al. Serum Exosomal miR-638 Is a Prognostic Marker of HCC Via Downregulation of VE-cadherin and ZO-1 of Endothelial Cells. Cancer Sci (2021) 112:1275–88. doi: 10.1111/cas.14807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer Cell-Derived Exosomal circUHRF1 Induces Natural Killer Cell Exhaustion and may Cause Resistance to anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19:110. doi: 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Pardee AD, Butterfield LH. Immunotherapy of Hepatocellular Carcinoma: Unique Challenges and Clinical Opportunities. Oncoimmunology (2012) 1:48–55. doi: 10.4161/onci.1.1.18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Lipinski S, Tiemann K. Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor-Immune Interactions. Int J Mol Sci (2021) 22(7):3374. doi: 10.3390/ijms22073374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New Insights Into Cancer Immunoediting and Its Three Component Phases–Elimination, Equilibrium and Escape. Curr Opin Immunol (2014) 27:16–25. doi: 10.1016/j.coi.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zheng W, Yang Y, Sequeira RC, Bishop CE, Atala A, Gu Z, et al. Effects of Extracellular Vesicles Derived From Mesenchymal Stem/Stromal Cells on Liver Diseases. Curr Stem Cell Res Ther (2019) 14:442–52. doi: 10.2174/1574888X14666190308123714 [DOI] [PubMed] [Google Scholar]
- 174. Seo KW, Sohn SY, Bhang DH, Nam MJ, Lee HW, Youn HY. Therapeutic Effects of Hepatocyte Growth Factor-Overexpressing Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells on Liver Fibrosis in Rats. Cell Biol Int (2014) 38:106–16. doi: 10.1002/cbin.10186 [DOI] [PubMed] [Google Scholar]
- 175. Lai L, Chen J, Wei X, Huang M, Hu X, Yang R, et al. Transplantation of MSCs Overexpressing HGF Into a Rat Model of Liver Fibrosis. Mol Imaging Biol (2016) 18:43–51. doi: 10.1007/s11307-015-0869-x [DOI] [PubMed] [Google Scholar]
- 176. Fiore EJ, Bayo JM, Garcia MG, Malvicini M, Lloyd R, Piccioni F, et al. Mesenchymal Stromal Cells Engineered to Produce IGF-I by Recombinant Adenovirus Ameliorate Liver Fibrosis in Mice. Stem Cells Dev (2015) 24:791–801. doi: 10.1089/scd.2014.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Ko SF, Yip HK, Zhen YY, Lee CC, Lee CC, Huang CC, et al. Adipose-Derived Mesenchymal Stem Cell Exosomes Suppress Hepatocellular Carcinoma Growth in a Rat Model: Apparent Diffusion Coefficient, Natural Killer T-Cell Responses, and Histopathological Features. Stem Cells Int (2015) 2015:853506. doi: 10.1155/2015/853506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Jiang W, Tan Y, Cai M, Zhao T, Mao F, Zhang X, et al. Human Umbilical Cord MSC-Derived Exosomes Suppress the Development of CCl4-Induced Liver Injury Through Antioxidant Effect. Stem Cells Int (2018) 2018:6079642. doi: 10.1155/2018/6079642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Bruno S, Collino F, Deregibus MC, Grange C, Tetta C, Camussi G. Microvesicles Derived From Human Bone Marrow Mesenchymal Stem Cells Inhibit Tumor Growth. Stem Cells Dev (2013) 22:758–71. doi: 10.1089/scd.2012.0304 [DOI] [PubMed] [Google Scholar]
- 180. Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, et al. Exosomes Derived From miR-122-modified Adipose Tissue-Derived MSCs Increase Chemosensitivity of Hepatocellular Carcinoma. J Hematol Oncol (2015) 8:122. doi: 10.1186/s13045-015-0220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Gilbert CA, Ross AH. Cancer Stem Cells: Cell Culture, Markers, and Targets for New Therapies. J Cell Biochem (2009) 108:1031–8. doi: 10.1002/jcb.22350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Zhao Y, Bao Q, Renner A, Camaj P, Eichhorn M, Ischenko I, et al. Cancer Stem Cells and Angiogenesis. Int J Dev Biol (2011) 55:477–82. doi: 10.1387/ijdb.103225yz [DOI] [PubMed] [Google Scholar]
- 183. Shiozawa Y, Nie B, Pienta KJ, Morgan TM, Taichman RS. Cancer Stem Cells and Their Role in Metastasis. Pharmacol Ther (2013) 138:285–93. doi: 10.1016/j.pharmthera.2013.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Seo N, Akiyoshi K, Shiku H. Exosome-Mediated Regulation of Tumor Immunology. Cancer Sci (2018) 109:2998–3004. doi: 10.1111/cas.13735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Su C, Zhang J, Yarden Y, Fu L. The Key Roles of Cancer Stem Cell-Derived Extracellular Vesicles. Signal Transduct Target Ther (2021) 6:109. doi: 10.1038/s41392-021-00499-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Koren E, Fuchs Y. The Bad Seed: Cancer Stem Cells in Tumor Development and Resistance. Drug Resist Update (2016) 28:1–12. doi: 10.1016/j.drup.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 187. Wang Z, Zoller M. Exosomes, Metastases, and the Miracle of Cancer Stem Cell Markers. Cancer Metastasis Rev (2019) 38:259–95. doi: 10.1007/s10555-019-09793-6 [DOI] [PubMed] [Google Scholar]
- 188. Al-Sowayan BS, Al-Shareeda AT, Alrfaei BM. Cancer Stem Cell-Exosomes, Unexposed Player in Tumorigenicity. Front Pharmacol (2020) 11:384. doi: 10.3389/fphar.2020.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Scioli MG, Terriaca S, Fiorelli E, Storti G, Fabbri G, Cervelli V, et al. Extracellular Vesicles and Cancer Stem Cells in Tumor Progression: New Therapeutic Perspectives. Int J Mol Sci (2021) 22(19):10572. doi: 10.3390/ijms221910572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Domenis R, Cesselli D, Toffoletto B, Bourkoula E, Caponnetto F, Manini I, et al. Systemic T Cells Immunosuppression of Glioma Stem Cell-Derived Exosomes Is Mediated by Monocytic Myeloid-Derived Suppressor Cells. PLoS One (2017) 12:e0169932. doi: 10.1371/journal.pone.0169932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhang D, Li D, Shen L, Hu D, Tang B, Guo W, et al. Exosomes Derived From Piwil2induced Cancer Stem Cells Transform Fibroblasts Into Cancerassociated Fibroblasts. Oncol Rep (2020) 43:1125–32. doi: 10.3892/or.2020.7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Alzahrani FA, El-Magd MA, Abdelfattah-Hassan A, Saleh AA, Saadeldin IM, El-Shetry ES, et al. Potential Effect of Exosomes Derived From Cancer Stem Cells and MSCs on Progression of DEN-Induced HCC in Rats. Stem Cells Int (2018) 2018:8058979. doi: 10.1155/2018/8058979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 Polarization. Eur J Pharmacol (2020) 877:173090. doi: 10.1016/j.ejphar.2020.173090 [DOI] [PubMed] [Google Scholar]
- 194. Biswas SK, Mantovani A. Orchestration of Metabolism by Macrophages. Cell Metab (2012) 15:432–7. doi: 10.1016/j.cmet.2011.11.013 [DOI] [PubMed] [Google Scholar]
- 195. Cheng L, Wang Y, Huang L. Exosomes From M1-Polarized Macrophages Potentiate the Cancer Vaccine by Creating a Pro-inflammatory Microenvironment in the Lymph Node. Mol Ther (2017) 25:1665–75. doi: 10.1016/j.ymthe.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Wang Y, Wang B, Xiao S, Li Y, Chen Q. miR-125a/b Inhibits Tumor-Associated Macrophages Mediated in Cancer Stem Cells of Hepatocellular Carcinoma by Targeting CD90. J Cell Biochem (2019) 120:3046–55. doi: 10.1002/jcb.27436 [DOI] [PubMed] [Google Scholar]
- 197. Biswas SK, Mantovani A. Macrophage Plasticity and Interaction With Lymphocyte Subsets: Cancer as a Paradigm. Nat Immunol (2010) 11:889–96. doi: 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- 198. Pu J, Xu Z, Nian J, Fang Q, Yang M, Huang Y, et al. M2 Macrophage-Derived Extracellular Vesicles Facilitate CD8+T Cell Exhaustion in Hepatocellular Carcinoma Via the miR-21-5p/YOD1/YAP/beta-catenin Pathway. Cell Death Discov (2021) 7:182. doi: 10.1038/s41420-021-00556-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol (2004) 25:677–86. doi: 10.1016/j.it.2004.09.015 [DOI] [PubMed] [Google Scholar]
- 200. Liu Y, Tan J, Ou S, Chen J, Chen L. Adipose-Derived Exosomes Deliver miR-23a/b to Regulate Tumor Growth in Hepatocellular Cancer by Targeting the VHL/HIF Axis. J Physiol Biochem (2019) 75:391–401. doi: 10.1007/s13105-019-00692-6 [DOI] [PubMed] [Google Scholar]
- 201. Koeck ES, Iordanskaia T, Sevilla S, Ferrante SC, Hubal MJ, Freishtat RJ, et al. Adipocyte Exosomes Induce Transforming Growth Factor Beta Pathway Dysregulation in Hepatocytes: A Novel Paradigm for Obesity-Related Liver Disease. J Surg Res (2014) 192:268–75. doi: 10.1016/j.jss.2014.06.050 [DOI] [PubMed] [Google Scholar]
- 202. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA Secreted From Adipocytes Promotes the Growth of Hepatocellular Carcinoma by Targeting Deubiquitination-Related USP7. Oncogene (2019) 38:2844–59. doi: 10.1038/s41388-018-0619-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 203. Obata Y, Kita S, Koyama Y, Fukuda S, Takeda H, Takahashi M, et al. Adiponectin/T-Cadherin System Enhances Exosome Biogenesis and Decreases Cellular Ceramides by Exosomal Release. JCI Insight (2018) 3(8):e99680. doi: 10.1172/jci.insight.99680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Kita S, Maeda N, Shimomura I. Interorgan Communication by Exosomes, Adipose Tissue, and Adiponectin in Metabolic Syndrome. J Clin Invest (2019) 129:4041–9. doi: 10.1172/JCI129193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, et al. Cancer-Associated Fibroblasts Promote Hepatocellular Carcinoma Progression Through Downregulation of Exosomal Mir-150-3p. Eur J Surg Oncol (2021) 47:384–93. doi: 10.1016/j.ejso.2020.08.002 [DOI] [PubMed] [Google Scholar]
- 206. Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 Suppresses Tumorigenicity and Regulates G1/S Transition of Human Hepatocellular Carcinoma Cells. Hepatology (2009) 50:113–21. doi: 10.1002/hep.22919 [DOI] [PubMed] [Google Scholar]
- 207. Wang R, Zhao N, Li S, Fang JH, Chen MX, Yang J, et al. MicroRNA-195 Suppresses Angiogenesis and Metastasis of Hepatocellular Carcinoma by Inhibiting the Expression of VEGF, VAV2, and CDC42. Hepatology (2013) 58:642–53. doi: 10.1002/hep.26373 [DOI] [PubMed] [Google Scholar]
- 208. Meng W, Hao Y, He C, Li L, Zhu G. Exosome-Orchestrated Hypoxic Tumor Microenvironment. Mol Cancer (2019) 18:57. doi: 10.1186/s12943-019-0982-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Patton MC, Zubair H, Khan MA, Singh S, Singh AP. Hypoxia Alters the Release and Size Distribution of Extracellular Vesicles in Pancreatic Cancer Cells to Support Their Adaptive Survival. J Cell Biochem (2020) 121:828–39. doi: 10.1002/jcb.29328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Thomas SN, Liao Z, Clark D, Chen Y, Samadani R, Mao L, et al. Exosomal Proteome Profiling: A Potential Multi-Marker Cellular Phenotyping Tool to Characterize Hypoxia-Induced Radiation Resistance in Breast Cancer. Proteomes (2013) 1:87–108. doi: 10.3390/proteomes1020087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Yu Y, Min Z, Zhou Z, Linhong M, Tao R, Yan L, et al. Hypoxia-Induced Exosomes Promote Hepatocellular Carcinoma Proliferation and Metastasis Via miR-1273f Transfer. Exp Cell Res (2019) 385:111649. doi: 10.1016/j.yexcr.2019.111649 [DOI] [PubMed] [Google Scholar]
- 212. Ramteke A, Ting H, Agarwal C, Mateen S, Somasagara R, Hussain A, et al. Exosomes Secreted Under Hypoxia Enhance Invasiveness and Stemness of Prostate Cancer Cells by Targeting Adherens Junction Molecules. Mol Carcinog (2015) 54:554–65. doi: 10.1002/mc.22124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Matsuura Y, Wada H, Eguchi H, Gotoh K, Kobayashi S, Kinoshita M, et al. Exosomal Mir-155 Derived From Hepatocellular Carcinoma Cells Under Hypoxia Promotes Angiogenesis in Endothelial Cells. Dig Dis Sci (2019) 64:792–802. doi: 10.1007/s10620-018-5380-1 [DOI] [PubMed] [Google Scholar]
- 214. Chen X, Zhou J, Li X, Wang X, Lin Y, Wang X. Exosomes Derived From Hypoxic Epithelial Ovarian Cancer Cells Deliver microRNAs to Macrophages and Elicit a Tumor-Promoted Phenotype. Cancer Lett (2018) 435:80–91. doi: 10.1016/j.canlet.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 215. Wang LP, Lin J, Ma XQ, Xu DY, Shi CF, Wang W, et al. Exosomal DLX6-AS1 From Hepatocellular Carcinoma Cells Induces M2 Macrophage Polarization to Promote Migration and Invasion in Hepatocellular Carcinoma Through microRNA-15a-5p/CXCL17 Axis. J Exp Clin Cancer Res (2021) 40:177. doi: 10.1186/s13046-021-01973-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 216. Rong L, Li R, Li S, Luo R. Immunosuppression of Breast Cancer Cells Mediated by Transforming Growth Factor-Beta in Exosomes From Cancer Cells. Oncol Lett (2016) 11:500–4. doi: 10.3892/ol.2015.3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le Cam E, et al. Hypoxic Tumor-Derived Microvesicles Negatively Regulate NK Cell Function by a Mechanism Involving TGF-beta and miR23a Transfer. Oncoimmunology (2016) 5:e1062968. doi: 10.1080/2162402X.2015.1062968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Pant S, Hilton H, Burczynski ME. The Multifaceted Exosome: Biogenesis, Role in Normal and Aberrant Cellular Function, and Frontiers for Pharmacological and Biomarker Opportunities. Biochem Pharmacol (2012) 83:1484–94. doi: 10.1016/j.bcp.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Faber W, Sharafi S, Stockmann M, Denecke T, Sinn B, Puhl G, et al. Long-Term Results of Liver Resection for Hepatocellular Carcinoma in Noncirrhotic Liver. Surgery (2013) 153:510–7. doi: 10.1016/j.surg.2012.09.015 [DOI] [PubMed] [Google Scholar]
- 220. Wang T, Zhang KH, Hu PP, Wan QS, Han FL, Zhou JM, et al. Combination of Dual Serum Fluorescence, AFP and Hepatic Function Tests Is Valuable to Identify HCC in AFP-elevated Liver Diseases. Oncotarget (2017) 8:97758–68. doi: 10.18632/oncotarget.22050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Dufour DR, Lott JA, Nolte FS, Gretch DR, Koff RS, Seeff LB. Diagnosis and Monitoring of Hepatic Injury. II. Recommendations for Use of Laboratory Tests in Screening, Diagnosis, and Monitoring. Clin Chem (2000) 46:2050–68. doi: 10.1093/clinchem/46.12.2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Fornari F, Ferracin M, Trere D, Milazzo M, Marinelli S, Galassi M, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and Mir-494, Identify Cirrhotic Patients With HCC. PLoS One (2015) 10:e0141448. doi: 10.1371/journal.pone.0141448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum Exosomal microRNAs as Novel Biomarkers for Hepatocellular Carcinoma. Exp Mol Med (2015) 47:e184. doi: 10.1038/emm.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Arbelaiz A, Azkargorta M, Krawczyk M, Santos-Laso A, Lapitz A, Perugorria MJ, et al. Serum Extracellular Vesicles Contain Protein Biomarkers for Primary Sclerosing Cholangitis and Cholangiocarcinoma. Hepatology (2017) 66:1125–43. doi: 10.1002/hep.29291 [DOI] [PubMed] [Google Scholar]
- 225. Julich-Haertel H, Urban SK, Krawczyk M, Willms A, Jankowski K, Patkowski W, et al. Cancer-Associated Circulating Large Extracellular Vesicles in Cholangiocarcinoma and Hepatocellular Carcinoma. J Hepatol (2017) 67:282–92. doi: 10.1016/j.jhep.2017.02.024 [DOI] [PubMed] [Google Scholar]
- 226. Wang W, Li H, Zhou Y, Jie S. Peripheral Blood Microvesicles Are Potential Biomarkers for Hepatocellular Carcinoma. Cancer Biomark (2013) 13:351–7. doi: 10.3233/CBM-130370 [DOI] [PubMed] [Google Scholar]
- 227. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum Exosomal microRNAs Combined With Alpha-Fetoprotein as Diagnostic Markers of Hepatocellular Carcinoma. Cancer Med (2018) 7:1670–9. doi: 10.1002/cam4.1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Shi M, Jiang Y, Yang L, Yan S, Wang YG, Lu XJ. Decreased Levels of Serum Exosomal miR-638 Predict Poor Prognosis in Hepatocellular Carcinoma. J Cell Biochem (2018) 119:4711–6. doi: 10.1002/jcb.26650 [DOI] [PubMed] [Google Scholar]
- 229. Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, et al. Serum Exosomal miR-125b Is a Novel Prognostic Marker for Hepatocellular Carcinoma. Onco Targets Ther (2017) 10:3843–51. doi: 10.2147/OTT.S140062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, et al. Exosomal miR-665 as a Novel Minimally Invasive Biomarker for Hepatocellular Carcinoma Diagnosis and Prognosis. Oncotarget (2017) 8:80666–78. doi: 10.18632/oncotarget.20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, et al. Circulating Exosomal Mir-92b: Its Role for Cancer Immunoediting and Clinical Value for Prediction of Posttransplant Hepatocellular Carcinoma Recurrence. Am J Transplant (2019) 19:3250–62. doi: 10.1111/ajt.15490 [DOI] [PubMed] [Google Scholar]
- 232. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of Serum Exosomal microRNA-21 in Human Hepatocellular Carcinoma. BioMed Res Int (2014) 2014:864894. doi: 10.1155/2014/864894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a Bona Fide microRNA Biomarker in Serum Exosomes That Predicts Hepatocellular Carcinoma Recurrence After Liver Transplantation. Br J Cancer (2015) 112:532–8. doi: 10.1038/bjc.2014.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, et al. The Exosome Encapsulated microRNAs as Circulating Diagnostic Marker for Hepatocellular Carcinoma With Low Alpha-Fetoprotein. Int J Cancer (2020) 147:2934–47. doi: 10.1002/ijc.33111 [DOI] [PubMed] [Google Scholar]
- 235. Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, et al. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics (2019) 9:1965–79. doi: 10.7150/thno.30958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNA-HEIH in Serum and Exosomes as a Potential Biomarker in the HCV-related Hepatocellular Carcinoma. Cancer Biomark (2018) 21:651–9. doi: 10.3233/CBM-170727 [DOI] [PubMed] [Google Scholar]
- 237. Xu H, Chen Y, Dong X, Wang X. Serum Exosomal Long Noncoding Rnas ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev (2018) 27:710–6. doi: 10.1158/1055-9965.EPI-17-0770 [DOI] [PubMed] [Google Scholar]
- 238. Sun L, Su Y, Liu X, Xu M, Chen X, Zhu Y, et al. Serum and Exosome Long Non Coding RNAs as Potential Biomarkers for Hepatocellular Carcinoma. J Cancer (2018) 9:2631–9. doi: 10.7150/jca.24978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, et al. Circulating Exosomal Noncoding RNAs as Prognostic Biomarkers in Human Hepatocellular Carcinoma. Int J Cancer (2019) 144:1444–52. doi: 10.1002/ijc.31931 [DOI] [PubMed] [Google Scholar]
- 240. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 Promotes Hepatocellular Carcinoma Metastasis Via Enhancing Invasiveness and Angiogenesis. J Exp Clin Cancer Res (2020) 39:20. doi: 10.1186/s13046-020-1529-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Pang Y, Zhang S, Yang H, Zhou RL. Serum LAPTM4B-35 Protein as a Novel Diagnostic Marker for Hepatocellular Carcinoma. Beijing Da Xue Xue Bao Yi Xue Ban (2021) 53:710–5. doi: 10.19723/j.issn.1671-167X.2021.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, et al. Primary Tumor-Derived Exosomes Facilitate Metastasis by Regulating Adhesion of Circulating Tumor Cells Via SMAD3 in Liver Cancer. Oncogene (2018) 37:6105–18. doi: 10.1038/s41388-018-0391-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Gorji-Bahri G, Moghimi HR, Hashemi A. RAB5A Effect on Metastasis of Hepatocellular Carcinoma Cell Line Via Altering the Pro-Invasive Content of Exosomes. Exp Mol Pathol (2021) 120:104632. doi: 10.1016/j.yexmp.2021.104632 [DOI] [PubMed] [Google Scholar]
- 244. Jiang K, Dong C, Yin Z, Li R, Mao J, Wang C, et al. Exosome-Derived ENO1 Regulates Integrin alpha6beta4 Expression and Promotes Hepatocellular Carcinoma Growth and Metastasis. Cell Death Dis (2020) 11:972. doi: 10.1038/s41419-020-03179-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Abd El Gwad A, Matboli M, El-Tawdi A, Habib EK, Shehata H, Ibrahim D, et al. Role of Exosomal Competing Endogenous RNA in Patients With Hepatocellular Carcinoma. J Cell Biochem (2018) 119:8600–10. doi: 10.1002/jcb.27109 [DOI] [PubMed] [Google Scholar]
- 246. Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, et al. Circulating Exosome-Derived Bona Fide Long Non-Coding RNAs Predicting the Occurrence and Metastasis of Hepatocellular Carcinoma. J Cell Mol Med (2020) 24:1311–8. doi: 10.1111/jcmm.14783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, et al. Purification of HCC-specific Extracellular Vesicles on Nanosubstrates for Early HCC Detection by Digital Scoring. Nat Commun (2020) 11:4489. doi: 10.1038/s41467-020-18311-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ, et al. Serum Small Extracellular Vesicle-Derived LINC00853 as a Novel Diagnostic Marker for Early Hepatocellular Carcinoma. Mol Oncol (2020) 14:2646–59. doi: 10.1002/1878-0261.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Tian XP, Wang CY, Jin XH, Li M, Wang FW, Huang WJ, et al. Erratum: Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis: Erratum. Theranostics (2021) 11:6522–3. doi: 10.7150/thno.60140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, et al. MicroRNA-500 as a Potential Diagnostic Marker for Hepatocellular Carcinoma. Biomarkers (2009) 14:529–38. doi: 10.3109/13547500903150771 [DOI] [PubMed] [Google Scholar]
- 251. Wang S, Wu J, Shen S, Geng Z, Ling Y, Peng B. Evaluation of Different Blood Circulating miRNAs for Hepatocellular Carcinoma Diagnosis. J Nanosci Nanotechnol (2020) 20:1983–8. doi: 10.1166/jnn.2020.17160 [DOI] [PubMed] [Google Scholar]
- 252. Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as Biomarkers for Hepatocellular Carcinoma in Chinese Patients With Chronic Hepatitis B Virus Infection. PLoS One (2011) 6:e28486. doi: 10.1371/journal.pone.0028486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: A Potential Marker for Hepatitis B Virus-Related Hepatocellular Carcinoma Screening. Dig Dis Sci (2012) 57:2910–6. doi: 10.1007/s10620-012-2317-y [DOI] [PubMed] [Google Scholar]
- 254. Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, et al. Circulating miR-15b and miR-130b in Serum as Potential Markers for Detecting Hepatocellular Carcinoma: A Retrospective Cohort Study. BMJ Open (2012) 2:e000825. doi: 10.1136/bmjopen-2012-000825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Peng C, Ye Y, Wang Z, Guan L, Bao S, Li B, et al. Circulating microRNAs for the Diagnosis of Hepatocellular Carcinoma. Dig Liver Dis (2019) 51:621–31. doi: 10.1016/j.dld.2018.12.011 [DOI] [PubMed] [Google Scholar]
- 256. Wittmann J, Jack HM. Serum microRNAs as Powerful Cancer Biomarkers. Biochim Biophys Acta (2010) 1806:200–7. doi: 10.1016/j.bbcan.2010.07.002 [DOI] [PubMed] [Google Scholar]
- 257. Takahashi K, Yan IK, Wood J, Haga H, Patel T. Involvement of Extracellular Vesicle Long Noncoding RNA (linc-VLDLR) in Tumor Cell Responses to Chemotherapy. Mol Cancer Res (2014) 12:1377–87. doi: 10.1158/1541-7786.MCR-13-0636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q, Zhang L. Molecular Mechanism of HEIH and HULC in the Proliferation and Invasion of Hepatoma Cells. Int J Clin Exp Med (2015) 8:12956–62. [PMC free article] [PubMed] [Google Scholar]
- 259. Wang Y, Li Z, Xu S, Guo J. Novel Potential Tumor Biomarkers: Circular Rnas and Exosomal Circular RNAs in Gastrointestinal Malignancies. J Clin Lab Anal (2020) 34:e23359. doi: 10.1002/jcla.23359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao Y, et al. circFBLIM1 Act as a ceRNA to Promote Hepatocellular Cancer Progression by Sponging Mir-346. J Exp Clin Cancer Res (2018) 37:172. doi: 10.1186/s13046-018-0838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 Acts as the Sponge of microRNA-9 to Suppress Hepatocellular Carcinoma Progression. Hepatology (2017) 66:1151–64. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 262. Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X, et al. CircSETD3 (Hsa_Circ_0000567) Acts as a Sponge for microRNA-421 Inhibiting Hepatocellular Carcinoma Growth. J Exp Clin Cancer Res (2019) 38:98. doi: 10.1186/s13046-019-1041-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 Inhibits Growth and Metastasis in Hepatocellular Carcinoma. J Hepatol (2018) 68:1214–27. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 264. Yao T, Chen Q, Shao Z, Song Z, Fu L, Xiao B. Circular RNA 0068669 as a New Biomarker for Hepatocellular Carcinoma Metastasis. J Clin Lab Anal (2018) 32:e22572. doi: 10.1002/jcla.22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Szabo G, Momen-Heravi F. Extracellular Vesicles in Liver Disease and Potential as Biomarkers and Therapeutic Targets. Nat Rev Gastroenterol Hepatol (2017) 14:455–66. doi: 10.1038/nrgastro.2017.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes Derived From Alcohol-Treated Hepatocytes Horizontally Transfer Liver Specific miRNA-122 and Sensitize Monocytes to LPS. Sci Rep (2015) 5:9991. doi: 10.1038/srep09991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Povero D, Yamashita H, Ren W, Subramanian MG, Myers RP, Eguchi A, et al. Characterization and Proteome of Circulating Extracellular Vesicles as Potential Biomarkers for NASH. Hepatol Commun (2020) 4:1263–78. doi: 10.1002/hep4.1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Kornek M, Lynch M, Mehta SH, Lai M, Exley M, Afdhal NH, et al. Circulating Microparticles as Disease-Specific Biomarkers of Severity of Inflammation in Patients With Hepatitis C or Nonalcoholic Steatohepatitis. Gastroenterology (2012) 143:448–58. doi: 10.1053/j.gastro.2012.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Cho YE, Im EJ, Moon PG, Mezey E, Song BJ, Baek MC. Increased Liver-Specific Proteins in Circulating Extracellular Vesicles as Potential Biomarkers for Drug- and Alcohol-Induced Liver Injury. PLoS One (2017) 12:e0172463. doi: 10.1371/journal.pone.0172463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Rodriguez-Suarez E, Gonzalez E, Hughes C, Conde-Vancells J, Rudella A, Royo F, et al. Quantitative Proteomic Analysis of Hepatocyte-Secreted Extracellular Vesicles Reveals Candidate Markers for Liver Toxicity. J Proteomics (2014) 103:227–40. doi: 10.1016/j.jprot.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Lang SA, Bednarsch J, Joechle K, Amygdalos I, Czigany Z, Heij L, et al. Prognostic Biomarkers for Cholangiocarcinoma (CCA): State of the Art. Expert Rev Gastroenterol Hepatol (2021) 15:497–510. doi: 10.1080/17474124.2021.1912591 [DOI] [PubMed] [Google Scholar]
- 272. Lambrecht J, Verhulst S, Mannaerts I, Reynaert H, van Grunsven LA. Prospects in Non-Invasive Assessment of Liver Fibrosis: Liquid Biopsy as the Future Gold Standard? Biochim Biophys Acta Mol Basis Dis (2018) 1864:1024–36. doi: 10.1016/j.bbadis.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 273. Welsh JA, Scorletti E, Clough GF, Englyst NA, Byrne CD. Leukocyte Extracellular Vesicle Concentration Is Inversely Associated With Liver Fibrosis Severity in NAFLD. J Leukoc Biol (2018) 104:631–9. doi: 10.1002/JLB.5A1217-501R [DOI] [PubMed] [Google Scholar]
- 274. Cho YE, Mezey E, Hardwick JP, Salem N, Jr., Clemens DL, Song BJ. Increased Ethanol-Inducible Cytochrome P450-2E1 and Cytochrome P450 Isoforms in Exosomes of Alcohol-Exposed Rodents and Patients With Alcoholism Through Oxidative and Endoplasmic Reticulum Stress. Hepatol Commun (2017) 1:675–90. doi: 10.1002/hep4.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Balaphas A, Meyer J, Sadoul R, Morel P, Gonelle-Gispert C, Buhler LH. Extracellular Vesicles: Future Diagnostic and Therapeutic Tools for Liver Disease and Regeneration. Liver Int (2019) 39:1801–17. doi: 10.1111/liv.14189 [DOI] [PubMed] [Google Scholar]
- 276. Sukriti S, Maras JS, Bihari C, Das S, Vyas AK, Sharma S, et al. Microvesicles in Hepatic and Peripheral Vein can Predict Nonresponse to Corticosteroid Therapy in Severe Alcoholic Hepatitis. Aliment Pharmacol Ther (2018) 47:1151–61. doi: 10.1111/apt.14564 [DOI] [PubMed] [Google Scholar]
- 277. Bissonnette J, Altamirano J, Devue C, Roux O, Payance A, Lebrec D, et al. A Prospective Study of the Utility of Plasma Biomarkers to Diagnose Alcoholic Hepatitis. Hepatology (2017) 66:555–63. doi: 10.1002/hep.29080 [DOI] [PubMed] [Google Scholar]
- 278. Moratti E, Vezzalini M, Tomasello L, Giavarina D, Sorio C. Identification of Protein Tyrosine Phosphatase Receptor Gamma Extracellular Domain (sPTPRG) as a Natural Soluble Protein in Plasma. PLoS One (2015) 10:e0119110. doi: 10.1371/journal.pone.0119110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Charrier A, Chen R, Chen L, Kemper S, Hattori T, Takigawa M, et al. Exosomes Mediate Intercellular Transfer of Pro-Fibrogenic Connective Tissue Growth Factor (CCN2) Between Hepatic Stellate Cells, the Principal Fibrotic Cells in the Liver. Surgery (2014) 156:548–55. doi: 10.1016/j.surg.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. van Dongen HM, Masoumi N, Witwer KW, Pegtel DM. Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiol Mol Biol Rev (2016) 80:369–86. doi: 10.1128/MMBR.00063-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Millard M, Yakavets I, Piffoux M, Brun A, Gazeau F, Guigner JM, et al. mTHPC-loaded Extracellular Vesicles Outperform Liposomal and Free mTHPC Formulations by an Increased Stability Drug Delivery Efficiency and Cytotoxic Effect in Tridimensional Model of Tumors. Drug Delivery (2018) 25:1790–801. doi: 10.1080/10717544.2018.1513609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Schindler C, Collinson A, Matthews C, Pointon A, Jenkinson L, Minter RR, et al. Exosomal Delivery of Doxorubicin Enables Rapid Cell Entry and Enhanced In Vitro Potency. PLoS One (2019) 14:e0214545. doi: 10.1371/journal.pone.0214545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Yan F, Zhong Z, Wang Y, Feng Y, Mei Z, Li H, et al. Exosome-Based Biomimetic Nanoparticles Targeted to Inflamed Joints for Enhanced Treatment of Rheumatoid Arthritis. J Nanobiotechnol (2020) 18:115. doi: 10.1186/s12951-020-00675-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, et al. Treatment of Brain Inflammatory Diseases by Delivering Exosome Encapsulated Anti-Inflammatory Drugs From the Nasal Region to the Brain. Mol Ther (2011) 19:1769–79. doi: 10.1038/mt.2011.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, et al. Development of Exosome-Encapsulated Paclitaxel to Overcome MDR in Cancer Cells. Nanomedicine (2016) 12:655–64. doi: 10.1016/j.nano.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, et al. Exosomes as Drug Delivery Vehicles for Parkinson's Disease Therapy. J Control Release (2015) 207:18–30. doi: 10.1016/j.jconrel.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Mathiyalagan P, Sahoo S. Exosomes-Based Gene Therapy for MicroRNA Delivery. Methods Mol Biol (2017) 1521:139–52. doi: 10.1007/978-1-4939-6588-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Pascucci L, Cocce V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, et al. Paclitaxel Is Incorporated by Mesenchymal Stromal Cells and Released in Exosomes That Inhibit In Vitro Tumor Growth: A New Approach for Drug Delivery. J Control Release (2014) 192:262–70. doi: 10.1016/j.jconrel.2014.07.042 [DOI] [PubMed] [Google Scholar]
- 289. Kooijmans SAA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJA, et al. Electroporation-Induced siRNA Precipitation Obscures the Efficiency of siRNA Loading Into Extracellular Vesicles. J Control Release (2013) 172:229–38. doi: 10.1016/j.jconrel.2013.08.014 [DOI] [PubMed] [Google Scholar]
- 290. Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular Vesicles for Drug Delivery. Adv Drug Deliv Rev (2016) 106:148–56. doi: 10.1016/j.addr.2016.02.006 [DOI] [PubMed] [Google Scholar]
- 291. Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in Extracellular Vesicles. Wiley Interdiscip Rev RNA (2017) 8(4):10.1002/wrna.1413. doi: 10.1002/wrna.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Fritz JV, Heintz-Buschart A, Ghosal A, Wampach L, Etheridge A, Galas D, et al. Sources and Functions of Extracellular Small RNAs in Human Circulation. Annu Rev Nutr (2016) 36:301–36. doi: 10.1146/annurev-nutr-071715-050711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Zhang G, Huang X, Xiu H, Sun Y, Chen J, Cheng G, et al. Extracellular Vesicles: Natural Liver-Accumulating Drug Delivery Vehicles for the Treatment of Liver Diseases. J Extracell Vesicles (2020) 10:e12030. doi: 10.1002/jev2.12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Xiao Z, Liu Y, Li Q, Liu Q, Liu Y, Luo Y, et al. Evs Delivery of miR-1915-3p Improves the Chemotherapeutic Efficacy of Oxaliplatin in Colorectal Cancer. Cancer Chemother Pharmacol (2021) 88:1021–31. doi: 10.1007/s00280-021-04348-5 [DOI] [PubMed] [Google Scholar]
- 295. Guo Q, Yan J, Song T, Zhong C, Kuang J, Mo Y, et al. Microrna-130b-3p Contained in MSC-Derived Evs Promotes Lung Cancer Progression by Regulating the FOXO3/NFE2L2/TXNRD1 Axis. Mol Ther Oncolytics (2021) 20:132–46. doi: 10.1016/j.omto.2020.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Slomka A, Mocan T, Wang B, Nenu I, Urban SK, Gonzales-Carmona M, et al. Evs as Potential New Therapeutic Tool/Target in Gastrointestinal Cancer and HCC. Cancers (Basel) (2020) 12(10):3019. doi: 10.3390/cancers12103019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Jordan KR, Hall JK, Schedin T, Borakove M, Xian JJ, Dzieciatkowska M, et al. Extracellular Vesicles From Young Women's Breast Cancer Patients Drive Increased Invasion of Non-Malignant Cells Via the Focal Adhesion Kinase Pathway: A Proteomic Approach. Breast Cancer Res (2020) 22:128. doi: 10.1186/s13058-020-01363-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as Regulators, Biomarkers and Therapeutic Targets in Liver Diseases. Gut (2021) 70:784–95. doi: 10.1136/gutjnl-2020-322526 [DOI] [PubMed] [Google Scholar]
- 299. Liu H, Li B. The Functional Role of Exosome in Hepatocellular Carcinoma. J Cancer Res Clin Oncol (2018) 144:2085–95. doi: 10.1007/s00432-018-2712-7 [DOI] [PubMed] [Google Scholar]
- 300. Xue D, Zheng Y, Wen J, Han J, Tuo H, Liu Y, et al. Role of Chemokines in Hepatocellular Carcinoma (Review). Oncol Rep (2021) 45:809–23. doi: 10.3892/or.2020.7906 [DOI] [PMC free article] [PubMed] [Google Scholar]