Abstract
Aberrant activation of monomeric G-protein signaling pathways drives some of the most aggressive cancers. Suppressing these hyperactivities has been the focus of efforts to obtain targeted therapies. Polyisoprenylated methylated protein methyl esterase (PMPMEase) is overexpressed in various cancers. Its inhibition induces the death of cancer cells that harbor the constitutively active K-Ras proteins. Furthermore, the viability of cancer cells driven by factors upstream of K-Ras, such as overexpressed growth factors and their receptors or the mutationally-activated receptors, is also susceptible to PMPMEase inhibition. Polyisoprenylated cysteinyl amide inhibitors (PCAIs) were thus designed to target cancers with hyperactive signaling pathways involving the G-proteins. The PCAIs were, however, poor inhibitors of PMPMEase, with Ki values ranging from 3.7 to 20 µM. On the other hand, they inhibited cell viability, proliferation, colony formation, induced apoptosis in cells with mutant K-Ras and inhibited cell migration and invasion with EC50 values of 1 to 3 μM. HUVEC tube formation was inhibited at submicromolar concentrations through their disruption of actin filament organization. At the molecular level, the PCAIs at 2 to 5 μM depleted monomeric G-proteins such as K-Ras, RhoA, Cdc42 and Rac1. The PCAIs also deplete vinculin and fascin that are involved in actin organization and function while disrupting vinculin punctates in the process. These demonstrate a polyisoprenylation-dependent mechanism that explains the observed PCAIs’ inhibition of the proliferative, invasive and angiogenic processes that promote both tumor growth and metastasis.
Keywords: Polyisoprenylated Cysteinyl Amide Inhibitors (PCAIs), K-Ras, Polyisoprenylated methylated protein methyl esterase (PMPMEase), isoprenylation, polyisoprenylation, G-proteins, fascin, vinculin, RhoA
1. INTRODUCTION
An estimated 2% of eukaryotic proteins are polyisoprenylated [1]. Polyisoprenylation, also known as prenylation or isoprenylation, involves the biochemical modification of the proteins with trans, trans-farnesyl, or all trans-geranylgeranyl groups [2, 3]. Three enzyme-catalyzed steps are involved in the biotransformation process for the subtype of proteins with the CAAX carboxyl-terminal signal sequence whereby C is cysteine, A represents an aliphatic amino acid and X could be M, S, Q, A, C, L or E signaling for the polyisoprenylation [4, 5]. When X is M, S, Q, A or C, the proteins undergo farnesylation while L or E results in geranylgeranylation in reactions catalyzed by farnesyl transferase and geranylgeranyl transferase I, respectively [6-8]. The requirement for polyisoprenylation of Ras and related G-proteins for malignant transformation spurred scientists to develop polyisoprenylation inhibitors for anticancer therapy [9]. The use of Farnesylation Inhibitors (FIs) to block the secondary modifications resulted in the alternative geranylgeranylation of K- and N-Ras [10], culminating in the loss of the potential anticancer benefits [11]. Geranylgeranyl transferase I inhibitors have, therefore been introduced into the antitumor drug development strategy [12, 13].
Following polyisoprenylation, the AAX tripeptide of the signal sequence is cleaved by Ras Converting Enzyme 1 (RCE1), thereby exposing the carboxyl terminal cysteine. The cysteine carboxylate undergoes methylation catalyzed by polyisoprenylated protein methyltransferase (PPMTase, also known as isoprenylcysteine methyl transferase or ICMT) [8]. A demethylating enzyme (PMPMEase) has been found to hydrolyze this methyl ester [14, 15]. The charge on the terminal polyisoprenylated cysteine may be pivotal in regulating the activities of these proteins. Moreover, electrostatic forces exerted on the carboxylate likely affect the spatial orientation of the polyisoprenyl group as the proteins undergo methylation/demethylation reactions. As polyisoprenyl-binding pockets have been revealed in some proteins [16-18], these secondary modifications are essential for the protein-protein interactions and biological activities of the proteins involved. Therefore, molecules that inhibit the polyisoprenylation pathway enzymes have long been sought to modify the proteins’ functional interactions.
This then brings into focus the role of the understudied demethylating enzyme, polyisoprenylated methylated protein methyl esterase (PMPMEase) that has been a major focus of the studies in our laboratory. PMPMEase is overexpressed in cancers including pancreatic [19], prostate [20], colorectal [21] and lung [22]. Cell lines from these cancer types are all susceptible to PMPMEase inhibition with polyisoprenylated small-molecule irreversible inhibitors [20, 22, 23]. The Polyisoprenylated Cysteinyl Amide Inhibitors (PCAIs) were designed to target and inhibit this enzyme in order to mitigate the hyperactive growth signaling in various cancers [24]. This review focuses mainly on PMPMEase, the molecules that we have designed in order to target it for inhibition, and observed cancer biological and molecular changes they induce in cancer cell lines.
2. POLYISOPRENYLATED METHYLATED PROTEIN METHYL ESTERASE
2.1. Role of PMPMEase in the Polyisoprenylation Pathway
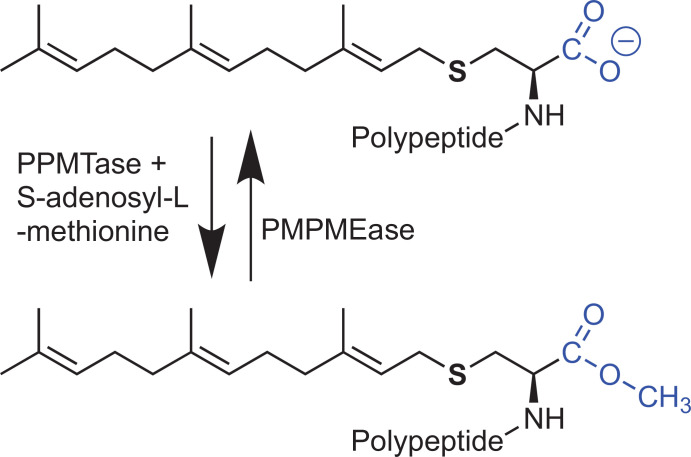
The final step of the polyisoprenylation pathway is an S-adenosyl-L-methionine (SAM)-dependent methylation reaction catalyzed by PPMTase [25-30]. The methylation converts the negatively charged carboxylate into the uncharged methyl ester (Fig. 1). It has been speculated that this net charge difference between the methylated and demethylated forms of polyisoprenylated proteins may have an impact on their functional conformations [31]. That this may be the case has been substantiated by reports of polyisoprenyl binding sites on some proteins [17, 18]. Loss of PPMTase has serious physiological consequences. For example, PPMTase knockouts in mice are lethal by mid-gestation [32], while PPMTase inactivation leads to mislocalization of K-Ras from the cell membrane [33].
Fig. (1).
Polyisoprenylation-dependent methylation and demethylation reactions. SAM-dependent methylation removes a negative charge in the vicinity of the polyisoprenyl moiety that is important for protein-protein interactions. Protein conformational changes resulting from methylation/demethylation-induced electrostatic interactions could alter the spatial orientations of the polyisoprenyl moiety relative to the polypeptide portions of the proteins, making them sterically unavailable for the protein-protein interactions.
The significance of the loss of PPMTase activity in animal physiology underscores the importance of a carefully regulated equilibrium between the methylated and demethylated forms of polyisoprenylated proteins. The methylation step is the only reversible reaction of the pathway as hydrolysis of the methyl esters regenerates the demethylated proteins. As such, this has been proposed to control polyisoprenylated protein function [29]. While this possibility has been recognized for a long time, interest in PMPMEase, the enzyme that counterbalances the effects of PPMTase has been very limited. Judging from the detrimental effects of PPMTase knockouts on mouse embryonic development [32], a pertinent question is to know whether a hyperactive PMPMEase that overwhelms PPMTase activity may be similarly impactful physiologically. Also of interest is how low PMPMEase activities may impact cell growth and animal physiology.
To begin addressing some of these questions, our earlier work sought to identify and characterize PMPMEase. The original interest in PMPMEase stemmed from the observation that excess SAM caused the carboxylmethylation of proteins similar to the heterotrimeric G-protein γ-subunits in rat brains. This resulted in Parkinson’s disease-like phenomena that were blocked by prior treatment with farnesylated compounds [34-36]. We reasoned that if excess protein methylation could cause these effects, then too little demethylation caused by low PMPMEase activities might have similar disease consequences. This led to the initial work of characterizing PMPMEase that revealed it as a serine hydrolase based on its susceptibility to irreversible serine hydrolase inhibitors [15]. This characterization as a carboxylesterase was later confirmed when it was purified from the porcine liver and the trypsin digest sequenced by tandem mass spectrometry [15] and found to be Sus scrofa carboxylesterase 1.
2.2. The Nature of PMPMEase
To begin to understand the role of PMPMEase in cellular functions, specific inhibitors were needed for this purpose. Using a trans, trans-farnesylated substrate to screen chromatographic fractions for PMPMEase activity, a range of substrates was developed to probe the active site of the enzyme to aid in the design of high-affinity inhibitors. These studies revealed that PMPMEase has a 320-fold higher affinity for trans, trans-farnesylated and all trans-geranylgeranylated substrates compared to substrates with shorter alkyl groups [37]. This was consistent with numerous x-ray crystallographic studies on human Carboxylesterase 1 (hCE1), revealing the hydrophobic nature and flexibility of the active site [38-41], possibly for binding the polyisoprenyl group and accommodating the varied polypeptide portions of endogenous substrates, respectively. These crystallographic data have contributed immensely in the docking analyses of substrates and inhibitors-binding interactions studies [23, 24, 42].
3. DESIGNING SPECIFIC INHIBITORS OF PMPMEASE
3.1. Sulfonyl Fluorides as Mechanism-based Inactivators
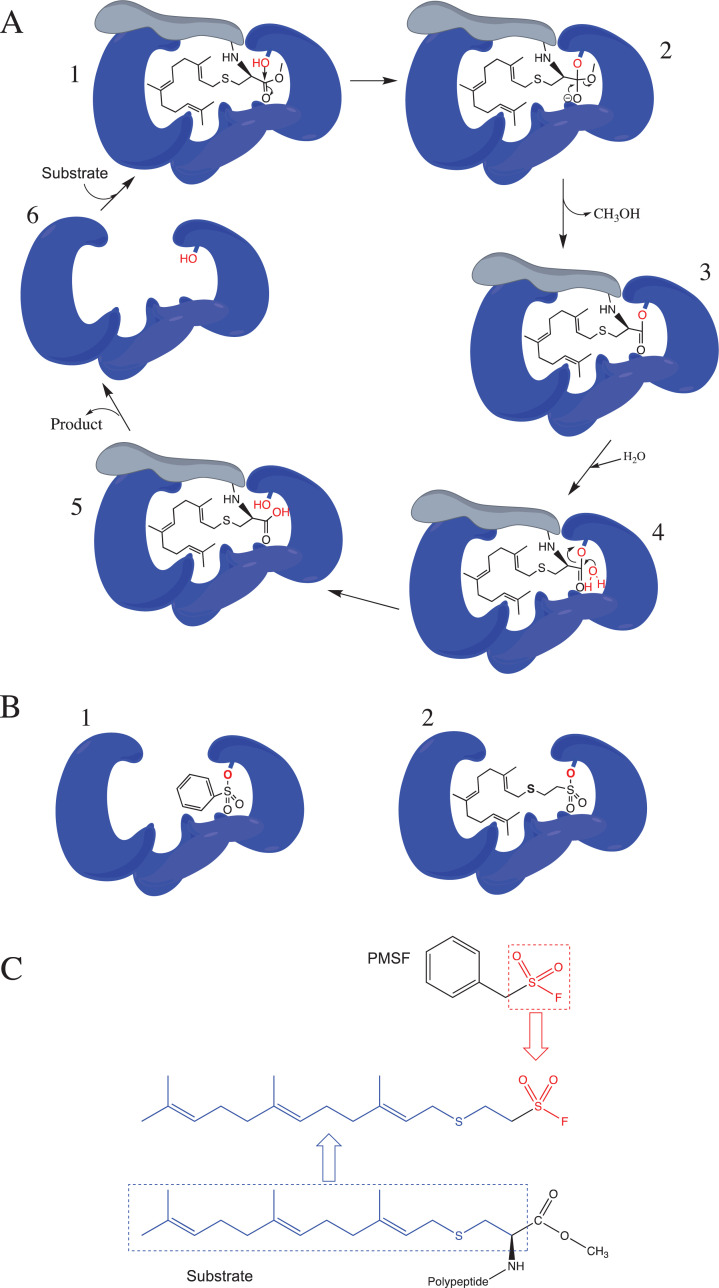
Significant insights into the design of specific high-affinity inhibitors of PMPMEase came from the substrate kinetics studies that demonstrated the requirement for the polyisoprenyl moiety for both selectivity and high-affinity interactions [37, 42]. The polyisoprenyl moiety has thus been an essential component of all the compounds that have been designed to target PMPMEase for inhibition. Using the polyisoprenyl group as the principal component of inhibitor design that defines selectivity for PMPMEase, other bioisosteric functional groups were then used to complete the inhibitor design. Given that PMPMEase is inhibited irreversibly by phenylmethylsulfonyl fluoride (PMSF) and organophosphorus compounds, the initial group of inhibitors incorporated the sulfonyl fluoride moiety as a reactive “warhead”. Since the inhibition mechanism relies on the inhibitors acting as pseudo-substrates, the “warheads” were precisely spaced from the polyisoprenyl moiety as the susceptible carboxyl esters of the substrates. The high-affinity interactions of the polyisoprene with the hydrophobic pockets of the binding site implies that the “warheads” were in close proximity to the catalytic residues for mechanism-based inactivation to occur (Fig. 2). This approach led to the sulfonyl fluoride irreversible inhibitors of PMPMEase [23]. The most effective of the sulfonyl fluorides, L-28, incorporated the sulfonyl fluoride moiety as a bioisosteric replacement of the carbonyl moiety of the substrates (Fig. 2) [23, 43].
Fig. (2).
PMPMEase enzymatic mechanism and the design of the irreversible polyisoprenylated sulfonyl fluoride inhibitor of PMPMEase. (A) Upon substrate binding (step 1), PMPMEase employs the catalytic serine residue to attack the carbonyl carbon of the susceptible ester bond (step 2), resulting in a transient acylated enzyme (step 3). This transition state is rapidly reversed by a water molecule (step 4), resulting in deacylation (step 5) and recovery of the enzyme (step 6). (B) Water is not a sufficiently strong nucleophile to displace the sulfonated enzyme. PMSF is thus an irreversible inhibitor of PMPMEase (1), a notion that was used in the design of the polyisoprenylated sulfonyl inhibitor, L-28 (2) according to the schematic (C). The polyisoprenyl moiety confers affinity towards the enzyme by the substrates and inhibitors. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.2. Polyisoprenylated Cysteinyl Amide Inhibitors (PCAIs)
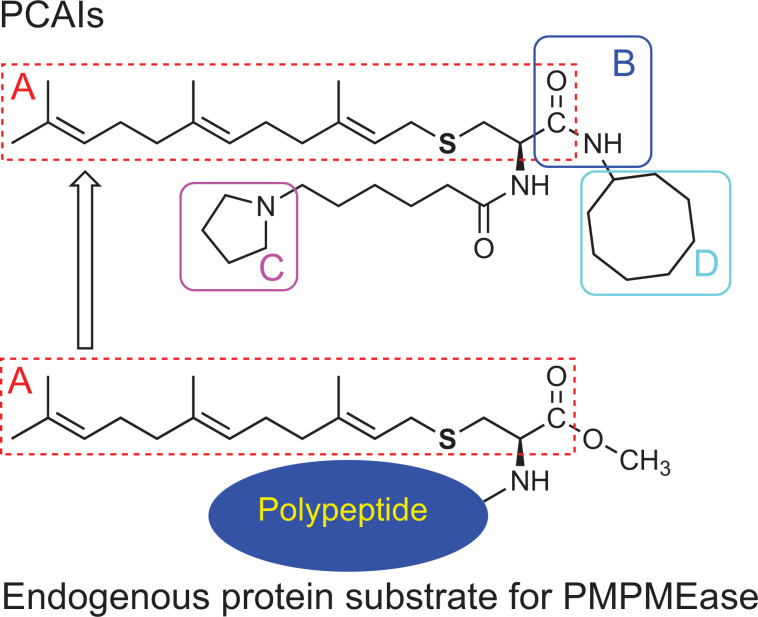
Lack of aqueous solubility and moisture sensitivity are two important drawbacks that we encountered in the use of sulfonyl fluorides to study the role of PMPMEase in cells and animals. The lack of polar/ionizable groups limited the solubility in aqueous media while the fluoride ion is easily displaced by water resulting in a sulfonic acid product that lacks the ability to react with the active site catalytic serine hydroxyl group. The shortcomings led us to design the PCAIs as potential reversible PMPMEase inhibitors. An amide bond was used as a more stable bioisostere of the ester group in the substrates [24]. Given the essential nature of the polyisoprenyl moiety for high-affinity interactions and selectivity towards the enzyme, it also constituted an essential element of the PCAIs despite its hydrophobic nature. This requirement for the polyisoprenyl moiety for efficient metabolism by PMPMEase is shared with PPMTase [28]. In order to mitigate the hydrophobicity imparted by the polyisoprenyl and the cycloalkyl groups (denoted respectively by “A” and “D” in Fig. (3)), a tertiary amine moiety was incorporated as an ionizable appendage to improve the aqueous solubility of the PCAIs. It was envisaged that these appendage groups are unlikely to sterically hinder the interactions of the PCAIs either with PMPMEase or other protein targets since it replaces the bulkier polypeptide portions of the endogenous protein substrates. When tested for their inhibition of PMPMEase and cancer cell viabilities, the PCAIs were found to be better at inhibiting cell viability than the enzyme, thereby suggesting the potential involvement of PCAIs targets other than PMPMEase in the cancer cells as discussed in section 3.5.
Fig. (3).
Enzyme-substrate kinetics-based design of the PCAIs. The portion of the substrate, including the polyisoprenyl moiety (A) was maintained in the design while the ester functional group was replaced with the bioisosteric amide bond (B). The potential hydrophobicity of the PCAIs was mitigated by the inclusion of the ionizable side chain amines (C) that renders them aqueous-soluble making it possible for the non-pharmacophore A-B-D to be left intact. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.3. Polyisoprenylated Proteins in Receptor Signaling and Cancer
In order to understand the potential role of the PCAIs against cancers, it is imperative to examine (1) the relevance of polyisoprenylated proteins in signaling pathways, (2) cancer drivers that appear upstream of and including the polyisoprenylated proteins and (3) protein-protein interactions that depend on the polyisoprenyl moieties of the proteins. Polyisoprenylated proteins are very important signaling molecules in various signaling pathways. Receptor tyrosine kinases require these proteins for transmitting various signals from extracellular growth factors into the cell. For example, the epidermal growth factor receptors require Ras as a downstream signaling mediator for cell proliferation, survival and cell cycle progression. Vascular endothelial growth factor receptors require proteins such as RhoA, Cdc42 and Rac1 for regulating actin polymerization into filopodia, lamellipodia and stress fibers necessary for maintaining cell shape and movement [44]. Cell movement is central to metastasis, the most devastating aspect of cancer since it promotes cell migration and invasion as well as cell reorganization during angiogenesis [45]. The latter is not only required for providing adequate nutrient supplies to fast-growing tumors but provides conduits for tumor cells to escape to form secondary tumors at distant sites.
3.4. Polyisoprenylated Protein-associated Cancer Drivers
Aberrant levels of expression and/or mutations resulting in hyperactive proteins that promote cancers have been reported for extracellular growth factors, their receptors and the downstream signaling intermediates that include polyisoprenylated proteins. Summarized in Table 1 are the incidence rates of some of the aberrant polyisoprenylated proteins in various cancers. Overexpression results in hyperactivities and increased signaling for cancer-promoting effects. Even more devastating are mutations that abolish the GTPase activities of some of the monomeric G-proteins. K-Ras mutations, in particular, have been reported to occur in over 50 and 90% of colorectal and pancreatic cancers, respectively, as well as to lesser extents in other cancers (Table 1). For example, while the percentage of lung cancer patients with hyperactive G-proteins may be relatively small, the fact that lung cancer afflicts a lot more people each year compared to other cancers, implies that hyperactive G-proteins is still a major problem in lung cancer.
Table 1.
Some prominent G-protein cancer drivers and their incidence rates in human cancers. K-Ras, RhoA, Rac1 and Cdc42 are depleted in cancer cells treated with PCAIs resulting in loss of cancer progression phenomena such as cell viability, cytoskeletal organization, cell shape, motility and invasion, consistent with loss of their biological functions [24, 46-51].
| Protein | Cancer type | Aberrations (type of alteration) | Incidence rates (%) | References |
| K-Ras | Pancreatic cancer | Mutation | 90 | [52] |
| Non-small cell lung cancer | Mutation | 30-35 | [52] | |
| Breast | Mutation | 5 | [53] | |
| Colorectal cancer | Mutation | 30-54 | [52, 54] | |
| Eleven most common cancers | Mutation | 14.3 | [55] | |
| N-Ras | Human cutaneous melanoma | Mutation | 94 | [56] |
| Acute myelogenous leukemia (AML) | Mutation | 59 | [57] | |
| H-Ras | Bladder urothelial | Mutations | 57 | [58] |
| RhoA | Colon | Overexpression | 95 | [59] |
| Lung | Overexpression | 95 | [59] | |
| RAC1 | Breast | Overexpression | 70 | [59, 60] |
| Mutation | 50 | [61, 62] | ||
| Lung | Overexpression | 50 | [63] | |
| Melanoma | Mutation | 5 | [56] | |
| Rac2 | Melanoma | Mutation | 10 | [56] |
| Cdc42 | Breast | Overexpression | 95 | [59] |
| Colorectal | Overexpression | 60 | [64] | |
| Melanoma | Mutation | 5 | [56] |
The extremely low picomolar dissociation constants for the G-protein-GTP complexes imply that finding molecules that can effectively compete with and displace the GTP from the binding sites has been a huge challenge [65]. Polyisoprenylation pathway modifications are important not only for functional localization but also for the protein-protein interactions that govern their translocation, additional functional regulation and stability to proteases [66, 67]. The methyltransferase activity that generates the carboxyl methyl esters on the terminal polyisoprenylated cysteine is countered by PMPMEase that hydrolyzes the methyl ester substrates (Fig. 1). This has been demonstrated using small molecule polyisoprenylated substrates [14, 15, 37, 42] and RhoA [68].
3.5. Polyisoprenylation-dependent Protein-protein Interactions With Chaperone Proteins
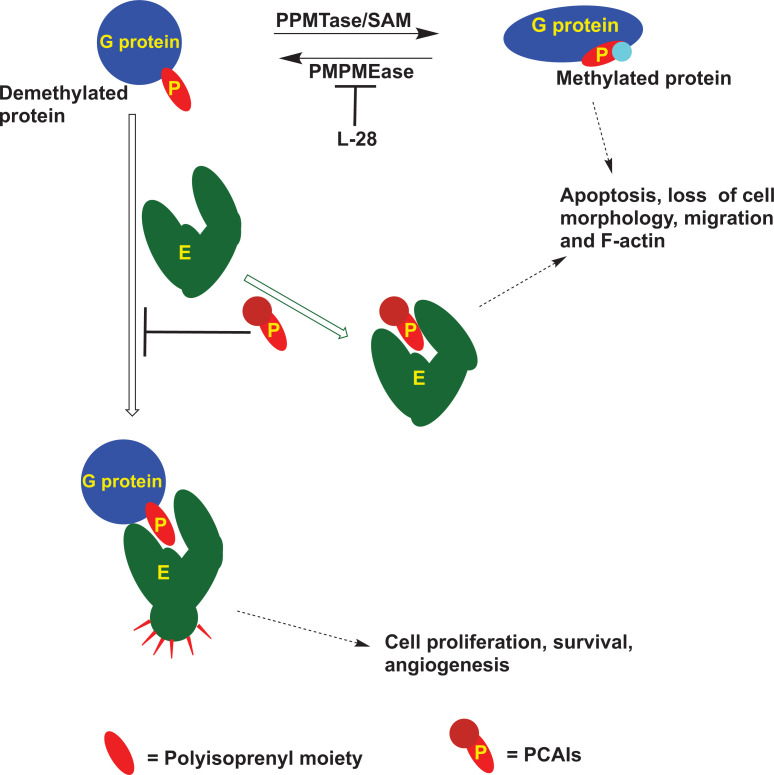
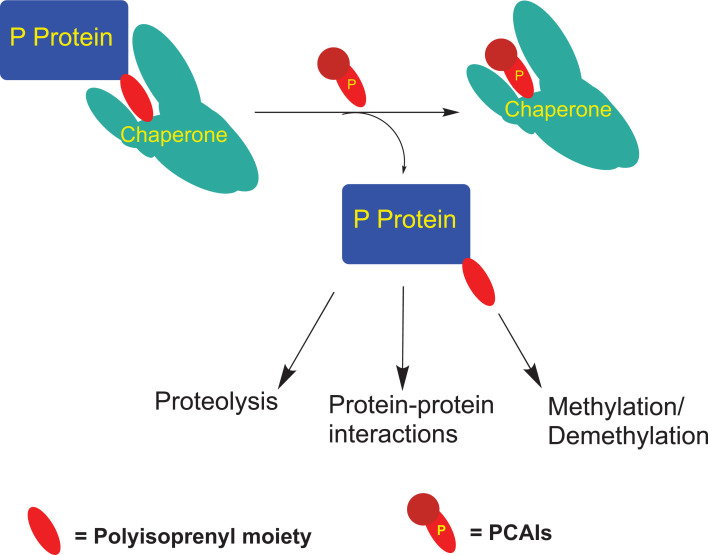
Adding to the complexity by which the monomeric G-proteins are regulated, are studies showing their complexation with other proteins in a polyisoprenylation-dependent manner. These include proteins such as Rho-GDIs, polyisoprenylated Rho acceptor protein 1 (PRA1), calmodulin (CaM) and Galectin 3. Rho-GDIs, PRA1, 14-3-3 proteins and CaM are reported to solubilize the polyisoprenylated proteins from cell membranes into the cytoplasm, thereby diminishing the interactions with cell surface receptors required for their activation. Various studies have demonstrated that interactions of the different G-proteins with these chaperone proteins as well as the shuttling back and forth to the plasma membrane, are dynamic [69, 70]. It is unclear at this moment whether the PCAIs have any effects on the interactions of G-proteins with these chaperone proteins. If so, proteins with the polyisoprenyl-binding pockets would constitute targets for the PCAIs (Fig. 4). If that were the case, it would be interesting to know what the consequences of PCAIs treatment on (1) RTKs signaling, (2) effector interactions with the polyisoprenylated proteins, (3) proteolytic stability of the affected G-proteins and (4) the cellular responses, and which of these cellular responses would contravene cancer progression (Fig. 5).
Fig. (4).
Logical representation of the effects of treating cells with PMPMEase irreversible inhibitor, L-28 and PCAIs, and the possible interactions resulting in such effects. Some of the protein-protein interactions of the polyisoprenylated proteins are dependent on the polyisoprenyl moiety (P) [16]. The PCAIs possibly interfere with those interactions between the G-proteins and their effector (E) dependent on the polyisoprenyl cysteine, thereby disrupting the functional interactions and the subsequent events [46, 47, 50, 51]. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Fig (5).
Possible interactions of PCAIs with polyisoprenylated protein chaperones. PCAIs may competitively disrupt the well-defined polyisoprenylation-dependent interactions of polyisoprenylated proteins (P protein) with chaperone proteins [16], resulting in their dislodgement such that their localizations, functions, biotransformation and breakdown patterns become altered. This is exemplified by the phosphorylation of the hypervariable region of K-Ras4B that is inhibited by its binding to CaM, thereby controlling its polyisoprenylation-dependent interactions [87] and depletion of K-Ras [50], RhoA, Rac1 and Cdc42 [48]. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.6. Effects of PCAIs on Signaling Pathways
RTKs overexpression and/or gain in function mutations contribute to the progression of various neoplasms (Table 2). To mitigate the excessive signaling, monoclonal antibody (MABs) biologics are directed at the extracellular domains of the RTKs and/or their ligands to suppress signaling. Kinase inhibitors such as erlotinib target the intracellular mutated tyrosine kinase domains that drive some cancers. These targeted therapies are, however, ineffective in cancers that harbor tumor-promoting changes in downstream mediators such as Ras [88, 89]. The PCAIs targeting of polyisoprenylated protein interactomes is potentially beneficial at treating tumors that harbor G-protein hyperactivities as well as those of RTKs and other upstream drivers (Fig. 6).
Table 2.
Some prominent growth factors and receptor tyrosine kinases rely on monomeric G-proteins such as Ras and Rho to drive cancer progression. Using the PCAIs to uncouple the functional protein-protein interactions involving G-proteins downstream of the receptors might curtail the excessive signaling from the upstream receptors as K-Ras, Rho, Rac1 and Cdc42 that drive cancer progression are depleted in PCAIs-treated cancer cells [48, 50, 51, 71].
| Protein | Cancer type | Aberrations | Incidence rate (%) | References |
| Epidermal growth factor receptor, EGFR (ErbB-1) | Pancreatic cancer | Overexpression | 90 | [72] |
| Glial tumors | Overexpression | 50 | [73] | |
| Non-small cell lung cancer | Amplification and mutation | 10 | [73, 74] | |
| Non-small cell lung cancer | Overexpression | 40-80 | [75] | |
| Breast | Mutation | 60 | [53] | |
| Head and Neck | Amplification | 7 | [76] | |
| Colon cancer | Amplification | 6 | [77] | |
| Ovarian cancer | Overexpression | 64 | [78] | |
| HER2 (ErbB-2) | Breast cancers | Overexpression | 20-25 | [79] |
| Colon cancer | Overexpression | 15.5 | [80] | |
| ErbB-3 | Lung | Overexpression | 50-70 | [81] |
| Breast | Overexpression | 50-70 | [81] | |
| Colon | Overexpression | 50-70 | [81] | |
| ErbB-4 | Colon | Overexpression | 22 | [81] |
| IGF1R | Breast | Amplification | 10 | [81] |
| Melanoma | Amplification | 3 | [81] | |
| Prostate cancer risk | Amplification | 4.3 | [82] | |
| IGF2R | Breast | Mutations | 40 | [83] |
| Hepatocellular | Mutations | 80 | [84] | |
| Colorectal | Mutations | 5 | [85] | |
| bFGF | Prostate | Overexpressed | 83 | [86] |
| VEGF | Colon | Overexpression | 55.5 | [80] |
| Ovarian cancer | Overexpression | 25 | [78] |
Fig. (6).
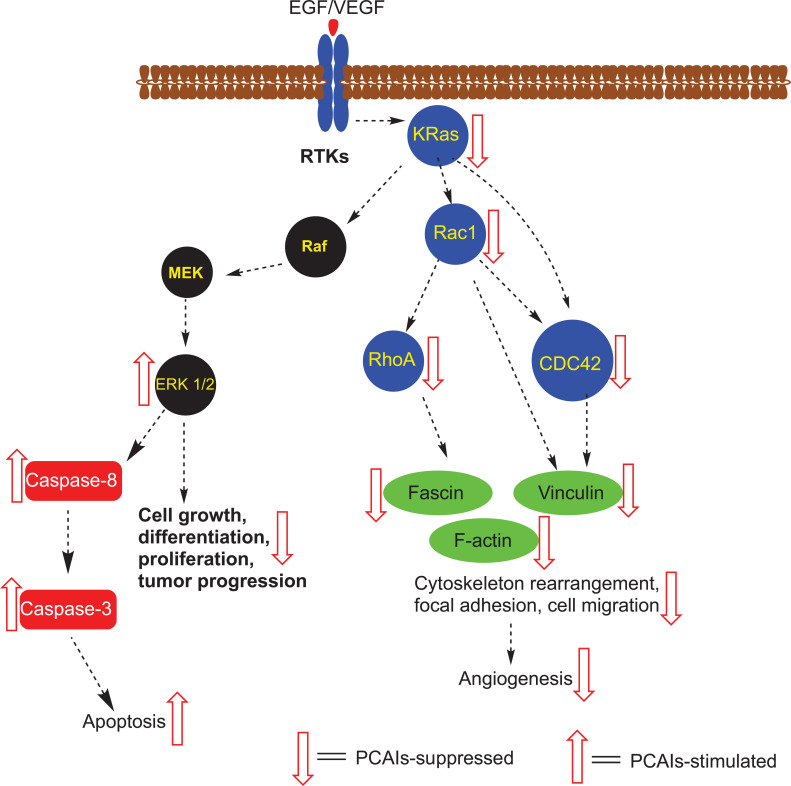
Cellular processes inhibited by the PCAIs suggest interactions with receptor tyrosine kinase (RTKs) signal transduction pathway intermediates. Polyisoprenylated proteins (blue circles) are the most likely direct targets of the PCAIs as they are depleted from cancer cells treated with the analogs. Fascin and vinculin that contribute to the formation of focal adhesion complexes are also depleted. In combination with the inhibition of F-actin, processes that depend on the filamentous actin cytoskeleton and focal adhesion, such as cell migration and angiogenesis, are largely abolished at low to submicromolar concentrations of the PCAIs. Cancers with such drivers as the growth factors EGF and VEGF and the RTKs, which occur upstream of the polyisoprenylated proteins, are also prone to inhibition by virtue of their dependence on the G-proteins as signaling mediators. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
The Ras-Raf-MEK-ERK signaling cascade appears to be susceptible to PCAIs treatment as K-Ras has been demonstrated to be depleted while ERK 1/2 phosphorylation was stimulated 48 h after treatment with PCAIs [50]. It is uncertain at this moment whether the depletion of these monomeric G-proteins is due to suppression of their synthesis or that PCAIs-induced dissociation from their chaperones leaves them exposed and vulnerable to proteolytic degradation. The depletion of K-Ras following treatment with the PCAIs is consistent with its role as an oncogenic protein and the apoptotic effects of the PCAIs on cancer cells harboring wild type as well as the mutant constitutively active K-Ras [50]. Although the stimulation of ERK 1/2 phosphorylation is counter to expectations given the PCAIs-induced depletion of K-Ras, numerous studies have linked increased ERK activity to the extrinsic apoptotic pathway involving caspase-8 activation (reviewed in [90]).
We have so far shown that the PCAIs induce apoptotic cell death in cancer cells harboring K-Ras mutations from pancreatic [24] and hyperactive EGFR in prostate cancer [47, 49]. They inhibit cell migration by disrupting F-actin organization, filopodia and lamellipodia, resulting in cell rounding. The effects on F-actin, leading to inhibited cell migration, also result in the inhibition of tube formation in human umbilical vein endothelial cells (HUVEC) assays for angiogenesis [46]. In in vivo analysis using chick chorioallantoic membrane and zebrafish embryos, the PCAIs inhibited angiogenesis – like phenomena at low micromolar concentrations [46]. These effects may be due in no small part to their effects on the monomeric G-proteins that regulate the cytoskeleton and cell motility. This was shown in our recent work in which treatment of cells with PCAIs resulted in significant depletion of RhoA, Cdc42 and Rac1 [48]. RhoA regulates focal adhesion, a critical event in cell migration, invasion and angiogenesis [44, 45, 91]. Its depletion somewhat explains the observed strong inhibitory effects of the PCAIs on cell migration and invasion [47, 48] as well as angiogenesis [46] that rely on focal adhesion formation to anchor cells to the extracellular matrix for traction.
Rac1 and Cdc42 are monomeric G-proteins that also play critical roles in the formation of filopodia and lamellipodia at the leading edges of migrating cells [44]. This effect was compounded by the fact that the levels of integrin α4 were suppressed by over 50% upon treatment with PCAIs. The levels of cleaved integrin α4 increased correspondingly [51]. The depletion of these proteins is therefore consistent with the antiangiogenic and anti-invasive nature of the PCAIs that underscores a potential anti-metastatic effect.
3.7. Effect of the PCAIs on Focal Adhesion Proteins
In addition to the depletion of the polyisoprenylated proteins RhoA, Rac1 and Cdc42, the PCAIs also induce the depletion of vinculin and fascin in treated cells as observed by western blotting [51]. Furthermore, vinculin depletion correlated with almost complete dissipation of vinculin punctates [51]. The intracellular immunofluorescence intensity of fascin, an actin-binding protein, decreased with the concentration of PCAIs treatment. The levels of other proteins such as α-actinin and integrin β5 were unchanged.
CONCLUSION AND FUTURE DIRECTIONS
Although the initial objective for the design and synthesis of the PCAIs was to inhibit PMPMEase and control its hyperactivities that spur cancer cell survival and proliferation, their poor inhibitory potencies against the enzyme (Ki values of 3.7 to 20 µM) compared to their superior activities against cancer cell viability was a strong indication that the latter effects are unlikely to be as a result of PMPMEase inhibition. However, the effects against cancer cell growth, proliferation, colony formation, migration and invasion, angiogenesis and disruption of F-actin organization are consistent with perturbations in polyisoprenylated protein function given the intricate signaling roles that Ras and Rho protein families play in these cellular events. The PCAIs have been demonstrated to be effective against cancer cells with hyperactive epidermal growth factor receptors (EGFR, RTKs) and constitutively active mutant K-Ras on which kinase inhibitors such as erlotinib are ineffective [88, 89]. In addition to their ability to disrupt the F-actin cytoskeleton, cell migration and invasion, as well as their inhibition of angiogenesis, implies that the PCAIs may have a multifaceted mechanism against cancer progression. They potentially could have broad clinical applications for treating cancers with hyperactive growth factors, RTKs and G-proteins such as Rho and Ras. Synthetic optimization to improve potency, pharmacological target identification, bioavailability and in vivo testing will contribute to their continuous development into a novel class of targeted agents to address the unmet therapeutic need for neoplasms involving hyperactive monomeric G-protein signaling pathways.
ACKNOWLEDGEMENTS
Declared None.
LIST OF ABBREVIATIONS
- AML
Acute Myelogenous Leukemia
- bFGF
Basic Fibroblast Growth Factor
- CaM
Calmodulin
- CAM
Chorioallantoic Membrane
- Cdc42
Cell Division Control Protein 42
- CES1
Carboxylesterase 1 or PMPMEase
- EGFR
Epidermal Growth Factor Receptor
- ERK
Extracellular Signal-regulated Kinase or Mitogen-activated Protein Kinase (MAPK)
- GAP
GTPase-activating Protein
- GDP
Guanosine Diphosphate
- GEF
Guanine Nucleotide Exchange Factor
- GF
Growth Factors
- GTP
Guanosine Triphosphate
- GTPase
Guanosine Triphosphate Hydrolase
- hCE1
Human Carboxylesterase 1
- HER2
Human Epidermal Growth Factor Receptor Type 2
- H-Ras
Harvey Rat Sarcoma
- HUVEC
Human Umbelical Vein Endothelial Cell
- IGF1R
Insulin-like Growth Factor 1 Receptor
- IGF2R
Insulin-like Growth Factor 2 Receptor
- K-Ras
Kirsten Rat Sarcoma
- MAB
Monoclonal Antibody
- MEK MAPK/ERK
Kinase or Mitogen-activated Protein Kinase Kinase (MAPKK)
- N-Ras
Neuroblastoma Rat Sarcoma
- PCAIs
Polyisoprenylated Cysteinyl Amide Inhibitors
- PMPMEase
Polyisoprenylated Methylated Protein Methyl Esterase
- PMSF
Phenylmethylsulfonyl Fluoride
- PPMTase
Polyisoprenylated Protein Methyl Transferase or Isoprenylcysteine Methyl Transferase (ICMT)
- PRA1
Polyisoprenylated Rho Acceptor Protein 1
- RAC1
Ras-related C3 Botulinum Toxin Substrate 1
- RAC2
Ras-related C3 Botulinum Toxin Substrate 2
- Raf
Rapidly Accelerated Fibrosarcoma or Mitogen-activated Protein Kinase Kinase Kinase (MAPKKK)
- RCE1
Ras Converting Enzyme 1
- RhoA
Ras Homolog Family Member A
- Rho-GDIs
Rho-dissociation Inhibitors
- RTKs
Receptor Tyrosine Kinases
- SAM
S-adenosyl-L-methionine
- VEGF
Vascular Endothelial Growth Factor
CONSENT FOR PUBLICATION
Not Applicable.
FUNDING
The study reported in this publication has been supported by the National Cancer Institute (NCI) and National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under Grant SC1CA190505 and by the National Institute on Minority Health and Health Disparities of the National Institutes of Health under Award Number U54 MD007582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Nalivaeva N.N., Turner A.J. Post-translational modifications of proteins: acetylcholinesterase as a model system. Proteomics. 2001;1(6):735–747. doi: 10.1002/1615-9861(200106)1:6<735::AID-PROT735>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Barbu V.D. [Isoprenylation of proteins: what is its role?]. C. R. Seances Soc. Biol. Fil. 1991;185(5):278–289. [PubMed] [Google Scholar]
- 3.Khosravi-Far R., Cox A.D., Kato K., Der C.J. Protein prenylation: key to ras function and cancer intervention? Cell Growth Differ. 1992;3(7):461–469. [PubMed] [Google Scholar]
- 4.Lerner S., Haklai R., Kloog Y. Isoprenylation and carboxylmethylation in small GTP-binding proteins of pheochromocytoma (PC-12) cells. Cell. Mol. Neurobiol. 1992;12(4):333–351. doi: 10.1007/BF00734934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitten G.T., Nigg E.A. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J. Cell Biol. 1991;113(1):13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowell C.A., Kowalczyk J.J., Lewis M.D., Garcia A.M. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J. Biol. Chem. 1997;272(22):14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F.L., Casey P.J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 8.Hurwitz H.I., Casey P.J. Prenylation of CaaX-type proteins: Basic principles through clinical applications. Curr. Top. Membr. 2002;52:531–550. doi: 10.1016/S1063-5823(02)52021-4. [DOI] [Google Scholar]
- 9.Jiang H., Zhang X., Chen X., Aramsangtienchai P., Tong Z., Lin H. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 2018;118(3):919–988. doi: 10.1021/acs.chemrev.6b00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whyte D.B., Kirschmeier P., Hockenberry T.N., Nunez-Oliva I., James L., Catino J.J., Bishop W.R., Pai J.K. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997;272(22):14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 11.Berndt N., Hamilton A.D., Sebti S.M. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer. 2011;11(11):775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazi A., Carie A., Blaskovich M.A., Bucher C., Thai V., Moulder S., Peng H., Carrico D., Pusateri E., Pledger W.J., Berndt N., Hamilton A., Sebti S.M. Blockade of protein geranylgeranylation inhibits Cdk2-dependent p27Kip1 phosphorylation on Thr187 and accumulates p27Kip1 in the nucleus: implications for breast cancer therapy. Mol. Cell. Biol. 2009;29(8):2254–2263. doi: 10.1128/MCB.01029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karasic T.B., Chiorean E.G., Sebti S.M., O’Dwyer P.J. A Phase I study of GGTI-2418 (geranylgeranyl transferase I inhibitor) in patients with advanced solid tumors. Target. Oncol. 2019;14(5):613–618. doi: 10.1007/s11523-019-00661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oboh O.T., Lamango N.S. Liver prenylated methylated protein methyl esterase is the same enzyme as Sus scrofa carboxylesterase. J. Biochem. Mol. Toxicol. 2008;22(1):51–62. doi: 10.1002/jbt.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamango N.S. Liver prenylated methylated protein methyl esterase is an organophosphate-sensitive enzyme. J. Biochem. Mol. Toxicol. 2005;19(5):347–357. doi: 10.1002/jbt.20100. [DOI] [PubMed] [Google Scholar]
- 16.Wang M., Casey P.J. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 2016;17(2):110–122. doi: 10.1038/nrm.2015.11. [DOI] [PubMed] [Google Scholar]
- 17.Gosser Y.Q., Nomanbhoy T.K., Aghazadeh B., Manor D., Combs C., Cerione R.A., Rosen M.K. C-terminal binding domain of Rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature. 1997;387(6635):814–819. doi: 10.1038/42961. [DOI] [PubMed] [Google Scholar]
- 18.Schmohl M., Rimmele S., Pötz O., Kloog Y., Gierschik P., Joos T.O., Schneiderhan-Marra N. Protein-protein-interactions in a multiplexed, miniaturized format a functional analysis of Rho GTPase activation and inhibition. Proteomics. 2010;10(8):1716–1720. doi: 10.1002/pmic.200900597. [DOI] [PubMed] [Google Scholar]
- 19.Aguilar B.J., Nkembo A.T., Duverna R., Poku R.A., Amissah F., Ablordeppey S.Y., Lamango N.S. Polyisoprenylated methylated protein methyl esterase: a putative biomarker and therapeutic target for pancreatic cancer. Eur. J. Med. Chem. 2014;81:323–333. doi: 10.1016/j.ejmech.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poku R.A., Amissah F., Duverna R., Aguilar B.J., Kiros G.E., Lamango N.S. Polyisoprenylated methylated protein methyl esterase as a putative drug target for androgen-insensitive prostate cancer. Ecancermedicalscience. 2014;8:459. doi: 10.3332/ecancer.2014.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amissah F., Duverna R., Aguilar B.J., Poku R.A., Lamango N.S. Polyisoprenylated methylated protein methyl esterase is both sensitive to curcumin and overexpressed in colorectal cancer: implications for chemoprevention and treatment. BioMed Res. Int. 2013;2013:416534. doi: 10.1155/2013/416534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amissah F., Duverna R., Aguilar B.J., Poku R.A., Kiros G.E., Lamango N.S. Polyisoprenylated methylated protein methyl esterase overexpression and hyperactivity promotes lung cancer progression. Am. J. Cancer Res. 2014;4(2):116–134. [PMC free article] [PubMed] [Google Scholar]
- 23.Aguilar B., Amissah F., Duverna R., Lamango N.S. Polyisoprenylation potentiates the inhibition of polyisoprenylated methylated protein methyl esterase and the cell degenerative effects of sulfonyl fluorides. Curr. Cancer Drug Targets. 2011;11(6):752–762. doi: 10.2174/156800911796191015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayuk-Takem L., Amissah F., Aguilar B.J., Lamango N.S. Inhibition of polyisoprenylated methylated protein methyl esterase by synthetic musks induces cell degeneration. Environ. Toxicol. 2014;29(4):466–477. doi: 10.1002/tox.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergo M.O., Gavino B.J., Hong C., Beigneux A.P., McMahon M., Casey P.J., Young S.G. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J. Clin. Invest. 2004;113(4):539–550. doi: 10.1172/JCI200418829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majmudar J.D., Hahne K., Hrycyna C.A., Gibbs R.A. Probing the isoprenylcysteine carboxyl methyltransferase (Icmt) binding pocket: sulfonamide modified farnesyl cysteine (SMFC) analogs as Icmt inhibitors. Bioorg. Med. Chem. Lett. 2011;21(9):2616–2620. doi: 10.1016/j.bmcl.2011.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rando R.R. Chemical biology of protein isoprenylation/methylation. Biochim. Biophys. Acta. 1996;1300(1):5–16. doi: 10.1016/0005-2760(95)00233-2. [DOI] [PubMed] [Google Scholar]
- 28.Tan E.W., Pérez-Sala D., Cañada F.J., Rando R.R. Identifying the recognition unit for G protein methylation. J. Biol. Chem. 1991;266(17):10719–10722. doi: 10.1016/S0021-9258(18)99074-5. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Sala D., Tan E.W., Cañada F.J., Rando R.R. Methylation and demethylation reactions of guanine nucleotide-binding proteins of retinal rod outer segments. Proc. Natl. Acad. Sci. USA. 1991;88(8):3043–3046. doi: 10.1073/pnas.88.8.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perezsala D., Tan E.W., Rando R.R. G-Protein Methylation in Rod Outer Segments.; Investigative Ophthalmology and Visual Science, annual meeting; Sarasota, Florida . 1991. pp. 667–1427. [Google Scholar]
- 31.Philips M.R., Pillinger M.H., Staud R., Volker C., Rosenfeld M.G., Weissmann G., Stock J.B. Carboxyl methylation of Ras-related proteins during signal transduction in neutrophils. Science. 1993;259(5097):977–980. doi: 10.1126/science.8438158. [DOI] [PubMed] [Google Scholar]
- 32.Bergo M.O., Leung G.K., Ambroziak P., Otto J.C., Casey P.J., Gomes A.Q., Seabra M.C., Young S.G. Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J. Biol. Chem. 2001;276(8):5841–5845. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 33.Bergo M.O., Leung G.K., Ambroziak P., Otto J.C., Casey P.J., Young S.G. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J. Biol. Chem. 2000;275(23):17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- 34.Lamango N.S., Ayuk-Takem L.T., Nesby R., Zhao W.Q., Charlton C.G. Inhibition mechanism of S-adenosylmethionine-induced movement deficits by prenylcysteine analogs. Pharmacol. Biochem. Behav. 2003;76(3-4):433–442. doi: 10.1016/j.pbb.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 35.Lamango N.S., Charlton C.G. Farnesyl-L-cysteine analogs block SAM-induced Parkinson’s disease-like symptoms in rats. Pharmacol. Biochem. Behav. 2000;66(4):841–849. doi: 10.1016/S0091-3057(00)00274-4. [DOI] [PubMed] [Google Scholar]
- 36.Lamango N.S., Nesby R.A., Charlton C.G. Quantification of S-adenosylmethionine-induced tremors: a possible tremor model for Parkinson’s disease. Pharmacol. Biochem. Behav. 2000;65(3):523–529. doi: 10.1016/S0091-3057(99)00220-8. [DOI] [PubMed] [Google Scholar]
- 37.Lamango N.S., Duverna R., Zhang W., Ablordeppey S.Y. Porcine liver carboxylesterase requires polyisoprenylation for high affinity binding to cysteinyl substrates. Open Enzyme Inhib. J. 2009;2:12–27. doi: 10.2174/1874940200902010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bencharit S., Edwards C.C., Morton C.L., Howard-Williams E.L., Kuhn P., Potter P.M., Redinbo M.R. Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1. J. Mol. Biol. 2006;363(1):201–214. doi: 10.1016/j.jmb.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming C.D., Bencharit S., Edwards C.C., Hyatt J.L., Tsurkan L., Bai F., Fraga C., Morton C.L., Howard-Williams E.L., Potter P.M., Redinbo M.R. Structural insights into drug processing by human carboxylesterase 1: tamoxifen, mevastatin, and inhibition by benzil. J. Mol. Biol. 2005;352(1):165–177. doi: 10.1016/j.jmb.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 40.Redinbo M.R., Bencharit S., Potter P.M. Human carboxylesterase 1: from drug metabolism to drug discovery. Biochem. Soc. Trans. 2003;31(Pt 3):620–624. doi: 10.1042/bst0310620. [DOI] [PubMed] [Google Scholar]
- 41.Bencharit S., Morton C.L., Hyatt J.L., Kuhn P., Danks M.K., Potter P.M., Redinbo M.R. Crystal structure of human carboxylesterase 1 complexed with the Alzheimer’s drug tacrine: from binding promiscuity to selective inhibition. Chem. Biol. 2003;10(4):341–349. doi: 10.1016/S1074-5521(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 42.Duverna R., Ablordeppey S.Y., Lamango N.S. Biochemical and docking analysis of substrate interactions with polyisoprenylated methylated protein methyl esterase. Curr. Cancer Drug Targets. 2010;10(6):634–648. doi: 10.2174/156800910791859443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sierra-Fonseca J.A., Najera O., Martinez-Jurado J., Walker E.M., Varela-Ramirez A., Khan A.M., Miranda M., Lamango N.S., Roychowdhury S. Nerve growth factor induces neurite outgrowth of PC12 cells by promoting Gβγ-microtubule interaction. BMC Neurosci. 2014;15:132. doi: 10.1186/s12868-014-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nobes C.D., Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81(1):53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 45.Zankov D.P., Ogita H. Actin-tethered junctional complexes in angiogenesis and lymphangiogenesis in association with vascular endothelial growth factor. BioMed Res. Int. 2015;2015:314178. doi: 10.1155/2015/314178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nkembo A.T., Ntantie E., Salako O.O., Amissah F., Poku R.A., Latinwo L.M., Lamango N.S. The antiangiogenic effects of polyisoprenylated cysteinyl amide inhibitors in HUVEC, chick embryo and zebrafish is dependent on the polyisoprenyl moiety. Oncotarget. 2016;7(42):68194–68205. doi: 10.18632/oncotarget.11908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nkembo A.T., Salako O., Poku R.A., Amissah F., Ntantie E., Flores-Rozas H., Lamango N.S. Disruption of actin filaments and suppression of pancreatic cancer cell viability and migration following treatment with polyisoprenylated cysteinyl amides. Am. J. Cancer Res. 2016;6(11):2532–2546. [PMC free article] [PubMed] [Google Scholar]
- 48.Ntantie E., Fletcher J., Amissah F., Salako O.O., Nkembo A.T., Poku R.A., Ikpatt F.O., Lamango N.S. Polyisoprenylated cysteinyl amide inhibitors disrupt actin cytoskeleton organization, induce cell rounding and block migration of non-small cell lung cancer. Oncotarget. 2017;8(19):31726–31744. doi: 10.18632/oncotarget.15956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poku R.A., Salako O.O., Amissah F., Nkembo A.T., Ntantie E., Lamango N.S. Polyisoprenylated cysteinyl amide inhibitors induce caspase 3/7- and 8-mediated apoptosis and inhibit migration and invasion of metastatic prostate cancer cells. Am. J. Cancer Res. 2017;7(7):1515–1527. [PMC free article] [PubMed] [Google Scholar]
- 50.Nkembo A.T., Amissah F., Ntantie E., Poku R.A., Salako O.O., Ikpatt O.F., Lamango N.S. Polyisoprenylated cysteinyl amide inhibitors deplete K-Ras and induce caspase-dependent apoptosis in lung cancer cells. Curr. Cancer Drug Targets. 2019;19(10):838–851. doi: 10.2174/1568009619666190325144636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ntantie E., Allen M.J., Fletcher J., Nkembo A.T., Lamango N.S., Ikpatt O.F. Suppression of focal adhesion formation may account for the suppression of cell migration, invasion and growth of non-small cell lung cancer cells following treatment with polyisoprenylated cysteinyl amide inhibitors. Oncotarget. 2018;9(40):25781–25795. doi: 10.18632/oncotarget.25372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 2014;13(11):828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez-Muñoz A., Gallego E., de Luque V., Pérez-Rivas L.G., Vicioso L., Ribelles N., Lozano J., Alba E. Lack of evidence for KRAS oncogenic mutations in triple-negative breast cancer. BMC Cancer. 2010;10:136. doi: 10.1186/1471-2407-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neumann J., Zeindl-Eberhart E., Kirchner T., Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol. Res. Pract. 2009;205(12):858–862. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 55.Prior I.A., Hood F.E., Hartley J.L. The frequency of ras mutations in cancer. Cancer Res. 2020;80(14):2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.P., Nickerson E., Auclair D., Li L., Place C., Dicara D., Ramos A.H., Lawrence M.S., Cibulskis K., Sivachenko A., Voet D., Saksena G., Stransky N., Onofrio R.C., Winckler W., Ardlie K., Wagle N., Wargo J., Chong K., Morton D.L., Stemke-Hale K., Chen G., Noble M., Meyerson M., Ladbury J.E., Davies M.A., Gershenwald J.E., Wagner S.N., Hoon D.S., Schadendorf D., Lander E.S., Gabriel S.B., Getz G., Garraway L.A., Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr, Laird P.W., Baty J.D., Fulton L.L., Fulton R., Heath S.E., Kalicki-Veizer J., Kandoth C., Klco J.M., Koboldt D.C., Kanchi K.L., Kulkarni S., Lamprecht T.L., Larson D.E., Lin L., Lu C., McLellan M.D., McMichael J.F., Payton J., Schmidt H., Spencer D.H., Tomasson M.H., Wallis J.W., Wartman L.D., Watson M.A., Welch J., Wendl M.C., Ally A., Balasundaram M., Birol I., Butterfield Y., Chiu R., Chu A., Chuah E., Chun H.J., Corbett R., Dhalla N., Guin R., He A., Hirst C., Hirst M., Holt R.A., Jones S., Karsan A., Lee D., Li H.I., Marra M.A., Mayo M., Moore R.A., Mungall K., Parker J., Pleasance E., Plettner P., Schein J., Stoll D., Swanson L., Tam A., Thiessen N., Varhol R., Wye N., Zhao Y., Gabriel S., Getz G., Sougnez C., Zou L., Leiserson M.D., Vandin F., Wu H.T., Applebaum F., Baylin S.B., Akbani R., Broom B.M., Chen K., Motter T.C., Nguyen K., Weinstein J.N., Zhang N., Ferguson M.L., Adams C., Black A., Bowen J., Gastier-Foster J., Grossman T., Lichtenberg T., Wise L., Davidsen T., Demchok J.A., Shaw K.R., Sheth M., Sofia H.J., Yang L., Downing J.R., Eley G. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinstein J.N., Akbani R., Broom B.M., Wang W.Y., Verhaak R.G.W., McConkey D., Lerner S., Morgan M., Creighton C.J., Smith C. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fritz G., Just I., Kaina B. Rho GTPases are over-expressed in human tumors. Int. J. Cancer. 1999;81(5):682–687. doi: 10.1002/(SICI)1097-0215(19990531)81:5<682::AID-IJC2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 60.Fritz G., Brachetti C., Bahlmann F., Schmidt M., Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer. 2002;87(6):635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J.W., Campbell P.J., Stratton M.R., Futreal P.A. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schnelzer A., Prechtel D., Knaus U., Dehne K., Gerhard M., Graeff H., Harbeck N., Schmitt M., Lengyel E. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene. 2000;19(26):3013–3020. doi: 10.1038/sj.onc.1203621. [DOI] [PubMed] [Google Scholar]
- 63.Shieh D.B., Godleski J., Herndon J.E., II, Azuma T., Mercer H., Sugarbaker D.J., Kwiatkowski D.J. Cell motility as a prognostic factor in Stage I nonsmall cell lung carcinoma: the role of gelsolin expression. Cancer. 1999;85(1):47–57. doi: 10.1002/(SICI)1097-0142(19990101)85:1<47::AID-CNCR7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 64.Gómez Del Pulgar T., Valdés-Mora F., Bandrés E., Pérez-Palacios R., Espina C., Cejas P., García-Cabezas M.A., Nistal M., Casado E., González-Barón M., García-Foncillas J., Lacal J.C. Cdc42 is highly expressed in colorectal adenocarcinoma and downregulates ID4 through an epigenetic mechanism. Int. J. Oncol. 2008;33(1):185–193. doi: 10.3892/ijo.33.1.185. [DOI] [PubMed] [Google Scholar]
- 65.Rajalingam K., Schreck R., Rapp U.R., Albert S. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta. 2007;1773(8):1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 66.Haklai R., Weisz M.G., Elad G., Paz A., Marciano D., Egozi Y., Ben-Baruch G., Kloog Y. Dislodgment and accelerated degradation of Ras. Biochemistry. 1998;37(5):1306–1314. doi: 10.1021/bi972032d. [DOI] [PubMed] [Google Scholar]
- 67.Kloog Y., Cox A.D. RAS inhibitors: potential for cancer therapeutics. Mol. Med. Today. 2000;6(10):398–402. doi: 10.1016/S1357-4310(00)01789-5. [DOI] [PubMed] [Google Scholar]
- 68.Cushman I., Cushman S.M., Potter P.M., Casey P.J. Control of RhoA methylation by carboxylesterase I. J. Biol. Chem. 2013;288(26):19177–19183. doi: 10.1074/jbc.M113.467407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sperlich B., Kapoor S., Waldmann H., Winter R., Weise K. Regulation of K-Ras4B membrane binding by calmodulin. Biophys. J. 2016;111(1):113–122. doi: 10.1016/j.bpj.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weise K., Kapoor S., Werkmüller A., Möbitz S., Zimmermann G., Triola G., Waldmann H., Winter R. Dissociation of the K-Ras4B/PDEδ complex upon contact with lipid membranes: membrane delivery instead of extraction. J. Am. Chem. Soc. 2012;134(28):11503–11510. doi: 10.1021/ja305518h. [DOI] [PubMed] [Google Scholar]
- 71.Ntantie E., Fletcher J., Amissah F., Salako O.O., Nkembo A.T., Poku R.A., Ikpatt F.O., Lamango N.S. Polyisoprenylated cysteinyl amide inhibitors disrupt actin cytoskeleton organization, induce cell rounding and block migration of non-small cell lung cancer. Oncotarget. 2017;8(19):31726–31744. doi: 10.18632/oncotarget.15956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Troiani T., Martinelli E., Capasso A., Morgillo F., Orditura M., De Vita F., Ciardiello F. Targeting EGFR in pancreatic cancer treatment. Curr. Drug Targets. 2012;13(6):802–810. doi: 10.2174/138945012800564158. [DOI] [PubMed] [Google Scholar]
- 73.Wong A.J., Bigner S.H., Bigner D.D., Kinzler K.W., Hamilton S.R., Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc. Natl. Acad. Sci. USA. 1987;84(19):6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paez J.G., Jänne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., Naoki K., Sasaki H., Fujii Y., Eck M.J., Sellers W.R., Johnson B.E., Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 75.Molina J.R., Yang P., Cassivi S.D., Schild S.E., Adjei A.A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83(5):584–594. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee J.W., Soung Y.H., Kim S.Y., Nam H.K., Park W.S., Nam S.W., Kim M.S., Sun D.I., Lee Y.S., Jang J.J., Lee J.Y., Yoo N.J., Lee S.H. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2005;11(8):2879–2882. doi: 10.1158/1078-0432.CCR-04-2029. [DOI] [PubMed] [Google Scholar]
- 77.Bhargava R., Gerald W.L., Li A.R., Pan Q., Lal P., Ladanyi M., Chen B. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod. Pathol. 2005;18(8):1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 78.Ranjbar R., Nejatollahi F., Nedaei Ahmadi A.S., Hafezi H., Safaie A. Expression of vascular endothelial growth factor (VEGF) and epidermal growth factor receptor (EGFR) in patients with serous ovarian carcinoma and their clinical significance. Iran. J. Cancer Prev. 2015;8(4):e3428. doi: 10.17795/ijcp-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 80.Li Q., Wang D., Li J., Chen P. Clinicopathological and prognostic significance of HER-2/neu and VEGF expression in colon carcinomas. BMC Cancer. 2011;11:277. doi: 10.1186/1471-2407-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Normanno N., Bianco C., De Luca A., Maiello M.R., Salomon D.S. Target-based agents against ErbB receptors and their ligands: a novel approach to cancer treatment. Endocr. Relat. Cancer. 2003;10(1):1–21. doi: 10.1677/erc.0.0100001. [DOI] [PubMed] [Google Scholar]
- 82.Chan J.M., Stampfer M.J., Giovannucci E., Gann P.H., Ma J., Wilkinson P., Hennekens C.H., Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279(5350):563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 83.Hankins G.R., De Souza A.T., Bentley R.C., Patel M.R., Marks J.R., Iglehart J.D., Jirtle R.L. M6P/IGF2 receptor: a candidate breast tumor suppressor gene. Oncogene. 1996;12(9):2003–2009. [PubMed] [Google Scholar]
- 84.De Souza A.T., Hankins G.R., Washington M.K., Orton T.C., Jirtle R.L. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat. Genet. 1995;11(4):447–449. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 85.Souza R.F., Wang S., Thakar M., Smolinski K.N., Yin J., Zou T.T., Kong D., Abraham J.M., Toretsky J.A., Meltzer S.J. Expression of the wild-type insulin-like growth factor II receptor gene suppresses growth and causes death in colorectal carcinoma cells. Oncogene. 1999;18(28):4063–4068. doi: 10.1038/sj.onc.1202768. [DOI] [PubMed] [Google Scholar]
- 86.Dirix L.Y., Vermeulen P.B., Pawinski A., Prové A., Benoy I., De Pooter C., Martin M., Van Oosterom A.T. Elevated levels of the angiogenic cytokines basic fibroblast growth factor and vascular endothelial growth factor in sera of cancer patients. Br. J. Cancer. 1997;76(2):238–243. doi: 10.1038/bjc.1997.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abdelkarim H., Banerjee A., Grudzien P., Leschinsky N., Abushaer M., Gaponenko V. The hypervariable region of K-Ras4B governs molecular recognition and function. Int. J. Mol. Sci. 2019;20(22):E5718. doi: 10.3390/ijms20225718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang Y., Mackley H., Cheng H., Ajani J.A. Use of K-Ras as a predictive biomarker for selecting anti-EGF receptor/pathway treatment. Biomarkers Med. 2010;4(4):535–541. doi: 10.2217/bmm.10.74. [DOI] [PubMed] [Google Scholar]
- 89.Dempke W.C.M., Heinemann V. Ras mutational status is a biomarker for resistance to EGFR inhibitors in colorectal carcinoma. Anticancer Res. 2010;30(11):4673–4677. [PubMed] [Google Scholar]
- 90.Cagnol S., Chambard J.C. ERK and cell death: mechanisms of ERK-induced cell death--apoptosis, autophagy and senescence. FEBS J. 2010;277(1):2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 91.Singh A.V., Kishore V., Santomauro G., Yasa O., Bill J., Sitti M. Mechanical coupling of puller and pusher active microswimmers influences motility. Langmuir. 2020;36(19):5435–5443. doi: 10.1021/acs.langmuir.9b03665. [DOI] [PMC free article] [PubMed] [Google Scholar]