Abstract
A novel drug to treat SARS-CoV-2 infections and hydroxyl chloroquine analogue, (E)-2,6-bis(4-chlorophenyl)-3-methyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine (BCMTP) compound has been synthesized in one pot reaction. The novel compound BCMTP has been characterized by FT-IR, 1H-NMR, 13C-NMR and single-crystal X-ray diffraction patterns. Crystal packing is stabilized by C8-H8A•••Cl10i, C41-H41•••Cl1ii and N1-H1A•••Cl6iii intermolecular hydrogen bonds. From the geometrical parameters, it is observed that the piperidine ring adopts chair conformation. Hirshfeld surface analysis was carried out to quantify the interactions and an interaction energy analysis was done to study the interactions between pairs of molecules. The geometrical structure was optimized by density functional theory (DFT) method at B3LYP/6-31G (d, p) as the basic set. The smaller binding energy value provides the higher reactivity of BCMTP compound than hydroxyl chloroquine and was corrected by high electrophilic and low nucleophilic reactions. The stability and charge delocalization of the molecule were also considered by natural bond orbital (NBO) analysis. The HOMO-LUMO energies describe the charge transfer which takes place within the molecule. Molecular electrostatic potential has also been analysed. Molecular docking studies are implemented to analyse the binding energy of the BCMTP compound against standard drugs such as the crystal structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES and SARS-CoV-2 main protease with an unliganded active site (2019-nCoV, corona virus disease 2019, COVID-19) and found to be considered having better antiviral agents. Molecular dynamics simulation was performed for COVID-19 main protease (Mpro: 6WCF/6Y84) to understand the elements governing the inhibitory effect and the stability of interaction under dynamic conditions.
Keywords: Piperidin-4-one, X-ray, DFT, Hirshfeld surface analysis, Molecular docking and molecular dynamic simulation
Graphical abstract
1. Introduction
Heterocyclic compounds play a vital role in biological processes and are widespread as natural products. Synthetically produced heterocycles designed by organic chemists are used, for instance, as Agrochemicals and pharmaceuticals and play an important role in human life. Among the family of heterocyclic compounds, nitrogen containing heterocyclic compounds, especially piperidine-4-ones presumably gaining considerable importance owing to varied biological properties such as antibacterial [1], antifungal [2], antiviral [3], anti-tumor [4], analgesic [5], anti-inflammatory, local anesthetic [6], Central Nervous System (CNS) and depressant activities [7]. The relative chemical shift order of equatorial and axial protons in the normal chair conformation of cyclohexane and its derivatives (deq>dax) are considered as caused by the magnetic anisotropic effect of the C-C single bonds. The influence of substituent's on the chemical shifts of protons attached to the adjacent carbons has been studied in detail [8], [9], [10].
Hydrazones are organic compounds containing R2C=NNHR group, synthesized by heating the substituted hydrazines with aldehyde and ketones in solvents such as ethanol, methanol etc. Hydrazone compound has diverse heteroatoms in the structure and are used in coordination chemistry as multidentated ligands for the synthesis of metal complexes through pharmaceutical and catalytic properties [11], [12]. In modern days, arylhydrazones are studied and found considered as the best p-type semiconductors among the semiconductors due to rapid charge transport, high photosensitivity, simple synthesis and low cost in its structure. Thus, hole-transport properties are present in arylhydrazones and are used as organic semiconductors in electrophotographic, memory storage, photovoltaic devices and organic light emitting diodes [13], [14], [15], [16], [17], [18]. Hydrazone compound are the current interest in many chemical processes such as sensors, pharmaceutical activities and nonlinear optical materials due to its high value in hyperpolarizability [19], [20]. Therefore, the presence of N-N and C=N bonds in the skeleton of hydrazones have so much attraction from various researchers as they possess many applications in pharmaceutical, biological and various applications in industrial chemistry such as anti-inflammatory, antioxidant, antitumor, anti-tuberculosis, analgesic, antiviral, anticancer, antimicrobial and insecticidal activities [21], [22], [23], [24], [25], [26], [27], [28], [29].
In our current study, we developed a novel (E)-2,6-bis(4-chlorophenyl)-3-methyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine (BCMTP) compound. The BCMTP compound is structurally characterized by FT-IR, 1H-NMR, 13C-NMR, single crystal X-ray diffraction, hirshfeld Conceptualization, Supervision, Investigation, Methodology, Resources, Formal analysis, Data curation, Writing - original draftsurface analysis, molecular docking studies and density functional theory using B3LYP/6-31G(d,p) level to investigate the intra-molecular structure via natural bond orbital (NBO), molecular electrostatic potential (MEP), frontier molecular orbital (FMO) analysis. The single crystal x-ray crystallography revealed that the BCMTP compound was crystallized in the triclinic system with space group P-1. In addition, a detailed BCMTP graphical image of the intermolecular interactions is displayed by Hirshfeld surface analysis. Molecular docking and molecular dynamic simulation studies determined the intermolecular interactions between synthesised BCMTP compound and receptor molecules.
2. Experimental section
2.1. FT-IR and FT-Raman measurement
The FT-IR spectrum of the synthesized BCMTP was measured in the 4000-400 cm−1 region using on AVATAR-330 FT-IR spectrophotometer (Thermo Nicolet). FT-IR spectrum was recorded in Department of Chemistry, Annamalai University, Annamalainagar. The FT- Raman spectrum of the title compound was recorded on BRUKER: RFS27 spectrometer operating at laser 100mW in the spectral range of 4000-50 cm−1. FT-Raman spectral measurements were carried out from Sophisticated Analytical Instrument Facility (SAIF), Indian Institute of Technology (IIT), Chennai.
2.1.1. Synthesis of 2,6-bis(4-chlorophenyl)-3,3-dimethylpiperidin-4-one (BCDP)
The parent 2,6-bis(4-chlorophenyl)-3,3-dimethylpiperidin-4-one were prepared by condensing 3-methylbutan-2-one, 4-chlorobenzaldehyde and ammonium acetate in warm ethanol in the ratio of 1:2:1, respectively, which afforded the formation of 2,6-bis(4-chlorophenyl)-3,3-dimethylpiperidin-4-one. The crude products formed were filtered and washed with an ethanol-ether (1:5) mixture to yield 2,6-bis(4-chlorophenyl)-3,3-dimethylpiperidin-4-one (BCDP) [30] and recrystallized from chloroform to obtain the pure compound.
2.1.2. Synthesis of (E)-2,6-bis(4-chlorophenyl)-3,3-dimethyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine (BCMTP)
To the boiling solution of the 2,6-bis(4-chlorophenyl)-3,3-dimethylpiperidin-4-one (25.0mmol), react with 2,4,6-trichlorophenyl hydrazine (25.0mmol) in ethanol and a few drops of acetic acid were added and refluxed for 2-4 h. After completion of the reaction, the reaction mixture was cooled to room temperature. The precipitate was filtered and washed with petroleum ether. The final product was recrystallized from ethanol by slow evaporation and harvested the pure crystal of (E)-2,6-bis(4-chlorophenyl)-3,3-dimethyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine.
2.1.3. Spectral value
2.1.3.1. (E)-2,6-bis(4-chlorophenyl)-3-methyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine
Yield: 74%, m.p. 160–165, White Solid: IR (KBr, cm−1); νmax3330-3315 (N−H st), 2850-3078 (C−H st), 1589 (C N st). 1H NMR (400 MHz, CDCl3); δ (ppm) 1.03 (s, 3H, H-3e), 1.14 (s, 3H, H-3a), 1.81 (s, 1H, NH), 2.31 (t,1H, H-5a), 2.98 (dd, 1H, H-5e), 3.77 (s, 1H, H-2a), 3.91 (dd, 1H, H-6a), 7.00 (s, 1H, Hydrazine) 7.29-7.52 (m, 10H, aromatic protons). 13C NMR (100 MHz, CDCl3); δ (ppm) 43.0 (C-3), 158.4 (C-4), 31.5 (C-5), 60.5 (C-6), 70.0 (C-2), 21.1, 22.5 (C-3 alkyl carbons), 121.5–142.7 (aromatic and ipso carbons). Calculated m/z: 541.02 (100.0%), 543.02 (64.2%), 539.03 (62.6%), 542.03 (27.3%), 545.02 (20.6%), 544.02 (18.0%), 540.03 (17.1%), 546.02 (5.6%), 543.03 (3.6%), 547.01 (3.3%), 541.03 (2.4%), 545.03 (2.3%), 542.02 (1.1%), Calculated elemental Analysis: C, 55.43; H, 4.09; Cl, 32.72; N, 7.76.
2.2. X-ray crystallographic analysis
By slow evaporation technique the crystal was grown in ethyl acetate solvent. Diffraction data were collected on a Bruker D8 Quest diffractometer using graphite monochromated MoKα radiation (λ = 0.71073Å) at 300.2 (2) K with crystal size of 0.160 × 0.150 × 0.100 mm. Data were corrected for absorption effects using the multi-scan method (SADABS). The frames were integrated with the Bruker SAINT Software package using a narrow-frame algorithm. The structure was solved by Apex 3 software. The structure was solved and refined using the Bruker SHELXTL software package and successive Fourier difference synthesis and refined by full-matrix least-square procedure on F2 with anisotropic thermal parameters. All non-hydrogen atoms were refined (SHELXL-2018/3) and placed at chemically acceptable positions. Crystallographic data have been deposited with the Cambridge Crystallographic Data Centre as supplementary publication number CCDC 2127016. Copies of the data can be obtained free of charge via http://www.ccdc.cam.ac.uk or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: þ44 1223 336 033: or e-mail: deposit@ccdc.cam.ac.uk.
2.3. Hirshfeld surface analysis
The stability of crystal structure mainly depends on the intermolecular interactions. The Hirshfeld surface analysis of the title compound was carried out with the help of Crystal Explorer 3.1 program to figure out and investigate the surface over the normalized contact distance (dnorm), which depends on contact distances to the closest atoms outside (de) and inside (di) the surface index [31], [32]. Hirshfeld surface analysis provides colour pictorial representation of inter-contact in the crystal structure with 2D fingerprint plots of the BCMTP compound.
2.4. Computational method
The complete geometrical optimized structure of the BCMTP compound based on the crystal data were operated on a personal computer using Gaussian 09 software package and calculated for the molecular structure, optimized geometry, frontier molecular orbital, molecular electrostatic potential (MEP), natural bond orbital (NBO), HOMO-LUMO energy, hyperpolarizability and Mullikan atomic charges of the targeted compound by using Lee-Parr correlation functional (B3LYP) method with 6-31G (d, p) as the base level in Gauss view 5.0 software program [33], [34], [35].
2.5. Molecular docking studies
The most current widespread program for predicting the protein-ligand interactions is the molecular docking studies [36]. The MDS tells about the interaction between the drug and DNA-molecules by placing a small molecule into the binding site. The accurate molecular docking studies were carried out by using Argus lab 4.0 and discovery studio 4.5 were the molecular graphics program for calculating and displaying feasible docking of the two modes of protein between enzyme and DNA molecules. The crystal structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES and SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19 [37] were found from Protein Data Bank (http://www.rcsb.org).
2.6. Molecular dynamics (MD) simulation
To perform the MD simulation, the best conformer was selected from docking analysis based on the intermolecular interactions and docking score values. The topology files for all the complexes were prepared by AMBERTOOLS20 with AMBER19ffSB force field using LEAP module [38]. The system setup was built with a TIP3P water model with 10Å distance on each side of the water box; and the system was neutralized by adding Cl−/Na+ ions [39]. Further the entire complex systems were minimized with steepest descent and conjugate gradient with 500 and 1500 steps respectively. The annealing process was done with 0 to 310K temperatures and 500 picoseconds (ps) NVT ensembles. Equilibration was performed by NPT ensembles over 500 ps duration. The MD production was performed up to 50ns for each complex using NPT ensembles with 310K temperature and 1 bar pressure by Langevin thermostat and Berendsen barostat method [40]. The MD trajectories were collected for each 2 femto seconds time using VMD software from the production output files [41].
3. Result and discussions
(E)-2,6-bis(4-chlorophenyl)-3-methyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine derivatives according to the synthetic sequences of reactions illustrated in Scheme 1 . The structures of the synthesized BCMTP compound is established on the basis of FT-IR, FT-Raman, 1H-NMR and 13C-NMR spectral techniques and compound BCMTP was successfully crystallized and its structure was determined by single-crystal X-ray diffraction analysis.
Scheme 1.
Synthesis of (E)-2,6-bis(4-chlorophenyl)-3,3-dimethyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine (BCMTP).
3.1. FT-IR and FT-Raman spectral analysis
The FT-IR and Raman spectrum of the compound BCMTP showed that the presence of C=N stretching frequency observed at 1589.91 and 1690.11 cm−1 confirm the formation of hydrazones [42]. However, the absence of carbonyl stretching frequency around 1720cm−1 has also confirmed the formation of the target compound. A collection of bands observed in the region 3330-3315 (IR) and 3300-3310cm−1 (Raman) is due to the presence of –NH group in piperidine and hydrazine moiety. The aromatic and aliphatic C-H stretching frequencies observed in the region of 3078-2850 cm−1 (IR) and 3000-2950 cm−1 (Raman). The C=C weak stretching frequencies is observed in the region of 1663-1700 cm−1 in IR but in Raman the strong frequency observed at 1500-1600 cm−1. The asymmetric and symmetric CH2 stretching vibrations are normally appear in the region 3100–2900 cm−1 [43]. The methyl and methylene asymmetric stretching medium band is observed at 1487 cm−1 in IR and 1480 cm−1 in Raman. In C-Cl strong stretching band observed in IR and Raman in the region of 466-503 and 520-530 cm−1. We have confirmed the formation of BCMTP compound from the observed frequencies in IR and Raman spectrum. The FT-IR and Raman spectrum of compound BCMTP is shown in Fig. S1. (Supplementary Material).
3.2. 1H NMR and 13C NMR spectral analysis
The 1H NMR signals are assigned based on their position, multiplicity and integral values. In general, the aromatic protons resonated in the downfield region around at 7ppm due to the magnetic anisotropic effect. In 1H NMR spectrum of compound, the signals appeared in the range of 7.29-7.52ppm corresponding to ten protons integral values are assigned to the aromatic protons of the phenyl groups at C-2, C-6 and also to the phenyl protons of the 2,4,6-trichlorophenyl hydrazine moiety. The doublet of doublet appeared in the higher frequency region at 3.91 ppm with a vicinal coupling constant value of J 5a, 6a =12 & J 5e, 6a = 3.2 Hz is assigned to H-6a proton of piperidine ring system. The sharp singlet observed at 3.77ppm with one proton integral value is assigned for H-2a proton. Therefore, deshielded signal at 3.91ppm and the shielded signal at 3.77ppm are assigned to benzylic protons of H-6a and H-2a. However, shielding of benzylic proton at C-2 is attributed to the substituent effect (+I effect) of alkyl groups present at C-3 position. A doublet of doublet appeared at 2.98 ppm (3J6a, 5e=14 & 2J5a, 5e = 3.2) corresponding to one proton integral value is assigned for H-5e proton. Consequently the H-5a proton appeared as triplet with one proton integral at 2.31 ppm (3J6a, 5a=14 & 2J5a, 5e=12.4). For the synthesized compound, the chemical shift value of H-5e is greater than of H-5a proton. Also, C-5 is a lower chemical shift than C-3. These observations suggested that the configuration about C(4) = N bond is E. In such a configuration, the C(5)-H5e bond is polarized. Hence, H-5e gets a partial positive charge and C-5 acquires a partial negative charge. The positive charge on H-5e deshields and the partial negative charge on C-5 shields it and H-5a. However, shielding of syn α-carbon is more pronounced than that of anti α-carbon. This is due to the interaction of the H-5e proton and the proton of nitrogen bearing 2,4,6-trichlorophenyl hydrazine moiety. From this statement, the higher frequency signal at 2.98ppm is assigned to H-5e proton.
Generally, the signals due to the aromatic carbons can be very readily distinguished from that of other carbons. Similarly, the aromatic ipso carbons showed their resonances at a further downfield region compared to other aromatic carbons. In 13C-NMR spectrum, the carbon signals appeared at 121.5-142.7ppm is assigned to aromatic carbons and aromatic ipso carbons. The low intense signals observed at 158.4 ppm are assigned to the C=N carbon atom of the piperidine ring system. Further two low intense signals at 142.7 and 139.3 ppm are due to ipso carbons of the phenyl rings at C-2′ and C-6′ carbon atom. There are four high-frequency signals observed in the aliphatic region. The signals observed at 70.0 and 60.5 ppm are assigned to C-2 and C-6 carbons. Similarly, the signals that appeared at 43.0 and 31.5 ppm are assigned to C-3 and C-5 carbons of the piperidine ring system. The alkyl substituted carbons at C-3 axial and equatorial positions of the methyl carbons are appeared at 21.1 and 22.5 ppm respectively. The 1H-NMR and 13C-NMR spectrum of compound BCMTP is shown in Fig. S2 and S3 (Supplementary Material).
3.3. Crystal and molecular structure
The ORTEP and packing diagram of compound BCMTP are given in Figs. 1 and 2 respectively. The crystal and structure refinement data are provided in Table 1 . Hydrogen bonding parameters belonging to the molecule are shown in Table 2 . Selected bond distances, bond angles and torsion angles are given in Table 3 . Compound BCMTP crystallizes in the triclinic system with the space group P-1. The unit cell dimensions observed are a = 11.766 (8) Å, b = 13.701 (9) Å, c = 16.101 (11) Å. α = 92.646(12)°, β =101.004° (12), γ=90.831(7)° and a unit cell contains four molecules. Crystal packing is stabilized by C8-H8A•••Cl10i, C41-H41•••Cl1ii, N1-H1A•••Cl6iii intermolecular hydrogen bonds. From the geometrical parameters, it is observed that the piperidine ring adopts chair conformation. The observed torsion angles of BCMTP revealed that the substituents in the piperidine ring are equatorially oriented. The configuration about >C=N- bond is anti- with respect to the alkyl substituent at C3 carbon. The C(9)-N(2) bond distance is 1.273(3) Å and it specifies double bonded nature. The distance associated with C-N single bond in the heterocyclic ring are C(11)-N(1) = 1.455(3) Å and C(7)-N(1) = 1.456(3) Å respectively. Two phenyl groups present in the piperidine are equatorially oriented as evidenced by their dihedral angles [C(6)-C(7)-C(8)-C(9) = -174.4(2); C(12)-C(11)-N(1)-C(7) =168.1(2) and C(6)-C(7)-N(1)-C(11) = -173.1(2); C(9)-C(10)-C(11)-C(12) = 176.8(2)].
Fig. 1.
ORTEP of compound BCMTP.
Fig. 2.
Packing diagram of BCMTP.
Table 1.
Crystal data and structure refinement details of BCMTP.
| Parameters | BCMTP |
|---|---|
| CCDC No. | 2127016 |
| Chemical formula | C25H22Cl5N3 |
| Mr | 541.70 |
| Crystal system, space group | Triclinic, P-1 |
| Temperature (K) | 296 |
| a, b, c (Å) | 11.766 (8), 13.701 (9), 16.101 (11) |
| α, β, γ (°) | 92.646 (12), 101.004 (12), 90.831 (7) |
| V (Å3) | 2544 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| µ (mm−1) | 0.59 |
| Crystal size (mm) | 0.30 × 0.30 × 0.25 |
| Data collection Diffractometer | Bruker kappa apex2 CCD Diffractometer |
| Absorption correction | Multi-scan SADABS (Bruker, 2012) |
| Tmin, Tmax | 0.833, 0.871 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 26615, 11589, 9309 |
| Rint | 0.082 |
| (sin θ/λ)max (Å−1) | 0.650 |
|
Refinement R[F2 > 2σ(F2)], wR(F2), S |
0.062, 0.166, 1.07 |
| No. of reflections | 11589 |
| No. of parameters | 615 |
| No. of restraints | 4 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.82, −0.67 |
Table 2.
Hydrogen bonds for BCMTP [A and deg.].
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| C8—H8A···Cl10i | 0.97 | 2.82 | 3.515 (3) | 129 |
| C41—H41···Cl1ii | 0.93 | 2.88 | 3.741 (4) | 154 |
| N1—H1A···Cl6iii | 0.90 (1) | 2.80 (2) | 3.652 (3) | 160 (3) |
Symmetry codes: (i) x, y−1, z; (ii) −x+1, −y+1, −z+1; (iii) −x+2, −y+1, −z+1.
Table 3.
Selected bond length (A)˚, bond angle (°) and torsional angles (°) of the BCMTP by XRD.
| Bond Angle | Exp | Dihedral angle | Exp |
|---|---|---|---|
| C7—N1 | 1.456 (3) | C6—C1—C2—C3 | 0.0 (4) |
| C9—N2 | 1.273 (3) | C1—C2—C3—C4 | 0.3 (4) |
| C11—N1 | 1.455 (3) | C1—C2—C3—Cl1 | −179.6 (2) |
| C28—Cl6 | 1.728(3) | C2—C3—C4—C5 | −0.2 (4) |
| C15—Cl2 | 1.743 (3) | Cl1—C3—C4—C5 | 179.7 (2) |
| C20—N3 | 1.391 (3) | C3—C4—C5—C6 | −0.1 (4) |
| N1—H1A | 0.896 (10) | C4—C5—C6—C1 | 0.4 (4) |
| N2—N3 | 1.414 (3) | C4—C5—C6—C7 | 178.9 (2) |
| N3—H3A | 0.896 (10) | C2—C1—C6—C5 | −0.4 (4) |
| C21—Cl3 | 1.729 (3) | C2—C1—C6—C7 | −178.8 (2) |
| C23—Cl4 | 1.727 (3) | C5—C6—C7—N1 | −55.3 (3) |
| C25—Cl5 | 1.721 (3) | C1—C6—C7—N1 | 123.1 (2) |
| C3—Cl1 | 1.733 (3) | C5—C6—C7—C8 | 64.0 (3) |
| Bond Angle | C1—C6—C7—C8 | −117.6 (3) | |
| C20—C21—Cl3 | 119.1 (2) | N1—C7—C8—C9 | −53.9 (3) |
| C20—C25—Cl5 | 120.6 (2) | C6—C7—C8—C9 | −174.4 (2) |
| C24—C25—Cl5 | 117.7 (2) | C7—C8—C9—N2 | −130.2 (3) |
| N1—C7—C6 | 109.7 (2) | C7—C8—C9—C10 | 50.4 (3) |
| N1—C7—C8 | 107.5 (2) | N2—C9—C10—C19 | −108.6 (3) |
| N1—C7—H7 | 109.2 | C8—C9—C10—C19 | 70.9 (3) |
| N2—C9—C8 | 126.3 (2) | N2—C9—C10—C18 | 11.6 (3) |
| N2—C9—C10 | 116.6 (2) | C8—C9—C10—C18 | −169.0 (2) |
| N1—C11—C12 | 109.4 (2) | N2—C9—C10—C11 | 131.4 (2) |
| N1—C11—C10 | 109.6 (2) | C8—C9—C10—C11 | −49.1 (3) |
| N1—C11—H11 | 108.1 | C19—C10—C11—N1 | −64.1 (3) |
| C25—C20—N3 | 122.3 (3) | C18—C10—C11—N1 | 174.5 (2) |
| N3—C20—C21 | 121.3 (3) | C9—C10—C11—N1 | 54.2 (3) |
| C11—N1—C7 | 112.6 (2) | C19—C10—C11—C12 | 58.5 (3) |
| C11—N1—H1A | 110 (2) | C18—C10—C11—C12 | −62.9 (3) |
| C7—N1—H1A | 105 (2) | C9—C10—C11—C12 | 176.8 (2) |
| C9—N2—N3 | 116.0 (2) | N1—C11—C12—C17 | 33.3 (3) |
| C20—N3—N2 | 113.0 (2) | C10—C11—C12—C17 | −89.5 (3) |
| C20—N3—H3A | 115 (3) | N1—C11—C12—C13 | −145.6 (2) |
| N2—N3—H3A | 120 (3) | C10—C11—C12—C13 | 91.6 (3) |
| C4—C3—Cl1 | 119.1 (2) | C17—C12—C13—C14 | 1.7 (4) |
| C2—C3—Cl1 | 119.3 (2) | C11—C12—C13—C14 | −179.4 (3) |
| C14—C15—Cl2 | 119.1 (3) | C12—C13—C14—C15 | −1.8 (5) |
| C16—C15—Cl2 | 119.1 (3) | C13—C14—C15—C16 | 0.8 (5) |
| C24—C23—Cl4 | 119.2 (3) | C13—C14—C15—Cl2 | −178.7 (2) |
| C22—C23—Cl4 | 119.1 (2) | C14—C15—C16—C17 | 0.3 (5) |
| C22—C21—Cl3 | 117.9 (2) | Cl2—C15—C16—C17 | 179.8 (3) |
| Dihedral angle | C13—C12—C17—C16 | −0.5 (4) | |
| C22—C23—C24—C25 | 0.0 (5) | C11—C12—C17—C16 | −179.4 (3) |
| Cl4—C23—C24—C25 | 178.8 (2) | C15—C16—C17—C12 | −0.5 (5) |
| N3—C20—C25—C24 | −178.2 (3) | C25—C20—C21—C22 | 0.3 (4) |
| C21—C20—C25—C24 | −1.0 (4) | N3—C20—C21—C22 | 177.6 (3) |
| N3—C20—C25—Cl5 | −0.8 (4) | C25—C20—C21—Cl3 | −178.7 (2) |
| C21—C20—C25—Cl5 | 176.5 (2) | N3—C20—C21—Cl3 | −1.4 (4) |
| C23—C24—C25—C20 | 0.9 (4) | C20—C21—C22—C23 | 0.5 (5) |
| C23—C24—C25—Cl5 | −176.7 (2) | Cl3—C21—C22—C23 | 179.5 (2) |
| C12—C11—N1—C7 | 168.1 (2) | C21—C22—C23—C24 | −0.7 (5) |
| C10—C11—N1—C7 | −66.8 (3) | C21—C22—C23—Cl4 | −179.5 (2) |
| C6—C7—N1—C11 | −173.1 (2) | C10—C9—N2—N3 | 176.9 (2) |
| C8—C7—N1—C11 | 65.0 (3) | C25—C20—N3—N2 | −56.6 (4) |
| C8—C9—N2—N3 | −2.4 (4) | C21—C20—N3—N2 | 126.3 (3) |
| C9—N2—N3—C20 | 179.1 (3) | ||
3.4. Hirshfeld surface analysis
The HSA was operated to examine the intermolecular short and long contacts which can be figured out as visual spots on the surface of the molecule. In the Hirshfeld surface, the quantities de and di represents the external and internal distance from a point on the HSA to the closest atoms. Table 4 shows Hirshfeld surface and 2D molecular fingerprint provides detail information about molecular interaction in the compound BCMTP are shown in Figs. 3 and 4 . On the surface, the different colours designate the differences in electrostatic potential. In the two-dimensional fingerprint plots, the highest contribution in the BCMTP compound is 38.7% due to H...Cl contacts and followed by H...H, C...Cl, Cl...Cl, N...H, C...C, C...N and N...Cl interactions contributing 29.9%, 7.6%, 2.6%, 1.6%, 1.3%, 1.2% and 0.1% respectively. [44], [45], [46].
Table 4.
Quantification of intercontacts in BCMTP compound.
| Entry | Type | Intercontacts (%) |
|---|---|---|
| 1 | C...H | 25.2%, |
| 2 | N...H | 9%, |
| 3 | S...H | 6.9% |
| 4 | C...N | 5.2% |
| 5 | O...H | 4.6% |
| 6 | N...N | 3.2% |
| 7 | C...C | 1.4% |
Fig. 3.
The Hirshfeld surface of the BCMTP compound with a variety of properties mapped onto the surface. Hirshfield surface mapped on its natural range; Distance internal to the surface, di; Distance External to the surface, de; Shape index S, Curvedness.
Fig. 4.
The two-dimensional fingerprint plots of the BCMTP compound, showing (A) all interactions, and (B) H…Cl, (C) H...H, (D) C...Cl, (E) Cl...Cl, (F) N...H,(G) C...C, (H) C...N and (I) N...Cl interactions[de and di represent the distances from a point on the HS to the nearest atoms outside (external) and inside (internal) the surface respectively].
4. DFT study
The BCMTP compound was performed in Gaussian 03 software program by using B3LYP at 6-31G (d,p) as the basic theory of the program. From the functional group in this compound, electron donor and electron acceptor are noticeable for the wavelength and oscillator. In the targeted compound, the bond length observed in C10 – C6, C2 – C11, C4 – Cl9, C3 – N12, H13 – N12, N12 – N20, N20 – C17, C14 – N29, H30 – 29N, C15 – N29, Cl52 – C48 and C51l – C38 are 1.76 (Å), 1.76 (Å), 1.75 (Å), 1.47 (Å), 0.99 (Å), 1.39 (Å), 1.29 (Å), 1.48 (Å), 1.00 (Å), 1.47 (Å), 1.75(Å) and 1.75 (Å) respectively. The bond angle in C10l – C6 – C1,C3 – N12 – H13, C4 – C3 – N12, H13 – N12 – N20, N12 – N20 – C17, C16 – C17 – N20, C18 – C14 – N29, C46 – C48 – C52l, C31 – C14 – N29, C44 – C48 – Cl52 and C36 – C38 – Cl51 are examined at 120.08°, 110.06°, 120.69°, 109.50°, 122.95°, 123.64°, 122.86°, 120.05°, 118.27°, 120.07° and 120.09° respectively whereas the dihedral angle at 8H – 5C – 6C – 10Cl, 4C – 5C – 6C – 10Cl, 2C – 1C – 6C – 10Cl, 12N – 3C – 2C – 11Cl, 33C – 36C – 38C – 51Cl, 3C – 12N – 20N – 17C, 12N – 20N – 17C – 18C, 15C – 16C – 17C – 20N, 5C – 4C – 3C – 12N, 4C – 3C – 12N – 20N, 14C – 18C – 17C – 20N, 30H – 29N – 14C – 31C, 54H – 15C – 29N – 30H and 54H – 15C – 29N – 14C are spotted at 0.72°, -0.72°, 179.14°, 1.20°, 179.85°, -158.91°, 171.01°, -164.78°, 178.49°, 147.47°, 137.76°, -172.39° and 68.27° respectively [47], [48], [49]. From the calculated geometrical parameters it is observed that the piperidine ring adopts chair conformation. The optimized geometry structure of BCMTP compound is revealed in Fig. 5 . The DFT selected bond parameters are given in Table 5 .
Fig. 5.
Optimization structure of BCMTP.
Table 5.
Selected bond length (A)˚, bond angle (°) and torsional angles (°) of the BCMTP compound by DFT.
| Bond Length | Cal | Bond Angle | Cal |
|---|---|---|---|
| 10C-6C | 1.76 | 8H-5C-6C-10Cl | 0.72 |
| 2C-11C | 1.76 | 7H-1C-6C -10Cl | -0.72 |
| 4C-9Cl | 1.75 | 4C-5C-6C-10Cl | -179.3 |
| 3C-12N | 1.47 | 2C-1C-6C-10Cl | 179.14 |
| 13H-12N | 0.99 | 7H-1C-2C-11Cl | -0.05 |
| 12N-20N | 1.39 | 6C-1C-2C-11Cl | -179.92 |
| 20N-17C | 1.29 | 4C-3C-2C -11Cl | -178.98 |
| 14C 29N | 1.48 | 12N-3C-2C-11Cl | 1.20 |
| 30H-29N | 1.00 | 1C-2C-3C-12N | -178.64 |
| 15C-29N | 1.47 | 2C-3C-12N-13H | -152.77 |
| 52Cl-48C | 1.75 | 4C-3C -12N-13H | 27.41 |
| 51Cl-38C | 1.75 | 9Cl-4C-3C-12N | -1.48 |
| Bond Angle | 5C-4C-3C-12N | 178.49 | |
| 10Cl-6C-1C | 120.08 | 4C-3C-12N-13H | 27.41 |
| 10C- 6C-5C | 119.92 | 4C-3C-12N-20N | 147.47 |
| 1C-2C-11Cl | 193.75 | 42C-44C-48C-52Cl | -179.50 |
| 3C-2C-11Cl | 120.08 | 49H-44C-48C-52Cl | 0.50 |
| 5C-4C-9Cl | 119.95 | 50H-46H-48C-52Cl | -0.18 |
| 3C-4C-9Cl | 119.96 | 40H-36C-38C-51Cl | -0.12 |
| 3C-12N-13H | 110.06 | 33C-36C-38C-51Cl | 179.85 |
| 4C-3C-12N | 120.69 | 32C-34C-38C-51Cl | -179.88 |
| 2C-3C-12N | 119.71 | 39H-34C-38C-51Cl | 0.25 |
| 13H-12N- 20N | 109.50 | 3C-12N-20N-17C | -158.91 |
| 12N-20N-17C | 122.95 | 13H-12N-20N-17C | -38.50 |
| 16C-17C-20N | 123.64 | 12N-20N-17C-18C | 171.01 |
| 30H-29N-15C | 109.08 | 12N-20N-17C-16C | -8.75 |
| 54H-15C-29N | 109.71 | 15C-16C-17C-20N | -164.78 |
| 53H-14C-29N | 89.56 | 14C-18C-17C-20N | 137.76 |
| 31C-14C-29N | 118.27 | 17C-18C-14C-29N | 25.21 |
| 18C-14C-29N | 122.86 | 18C-14C-29N-30H | -98.54 |
| 46C-48-52Cl | 120.05 | 18C-14C-29N-15C | 22.01 |
| 44C-48C-52Cl | 120.07 | 17C-16C-15C-29N | 32.87 |
| 34C-38C-51Cl | 120.02 | 30H-29N-14C-31C | 75.62 |
| 36C-38C-51Cl | 120.09 | 32C-31C-14C-29N | -79.44 |
| 43C-41C-15C-29N | 29.65 | ||
| 54H-15C-29N-30H | -172.39 | ||
| 54H-15C-29N-14C | 68.27 | ||
Natural bond orbital predicts the inter-molecular and intra-molecular charge transfer from donor to acceptor through a single-double bond conjugated organic molecules. The dipole moment (µ), polarizability (α) and hyperpolarizability (β0) are performed in the title compound by using DFT/6-31(d, p) in Gaussian 09 software program. The stabilization energy (E2) values determined the hyper conjugative interactions and charge transfers by the orbital overlap between π-π* (C44-C48) with stabilization energy 237.72kj/mol. The greater value of E(2) gives more interactions from electron donors to electron acceptors in the system. In NBO analysis, it is observed that the higher value of dipole moment, polarizability and hyperpolarizability contributes more stability to the NLO activity [50], [51]. The whole details of intensive interactions are shown in Table 6 .
Table 6.
Significant delocalization energies of second order perturbation theory analysis of Fock matrix in NBO for compound BCMTP.
| Type | Donor | Acceptor | E2KJ/Mol |
|---|---|---|---|
| π* - π* | C44 -C48 | C43 - C46 | 237.72 |
| π* - π* | C36-C38 | C32-C34 | 225.56 |
| π* -π* | C44-C48 | C41-C42 | 190.49 |
| n - π* | N12-N20 | C3-C4 | 23.02 |
| π - π* | C41-C42 | C44-C48 | 22.41 |
| n - π* | N12-N20 | C17-N20 | 20.42 |
| π - π* | C43-C46 | C44-C48 | 20.08 |
| π - π* | C3-C4 | C1-C2 | 16.68 |
| σ -σ* | C17-C18 | N12-N20 | 7.17 |
| σ -σ* | C33-C36 | C38-Cl51 | 4.88 |
| σ -σ* | C15-C16 | C17-N20 | 3.96 |
| σ -σ* | C25-H28 | C16-C17 | 2.97 |
| π - σ* | C17-N20 | C18-H55 | 2.64 |
| σ -σ* | C41-C43 | C46-H50 | 1.99 |
| π*- σ* | C41-C42 | C15-N29 | 1.04 |
| π - σ* | C31-C33 | C14-N29 | 0.51 |
The frontier molecular orbital's can calculate the qualitative excitation properties and electron transport in the given scheme. The energy of frontier molecular orbital's is the Highest Occupied Molecular Orbital and the Lowest Unoccupied Molecular Orbital. The HOMO energy clarifies the ability to donate an electron while LUMO energy also has the capacity to accept an electron. The Highest Occupied Molecular Orbital (HOMO) and the Lowest Unoccupied Molecular Orbital (LUMO) play a vital factor in the chemical reactivity, bioactivity, electrical and optical properties in the target compound. The HOMO and LUMO energy of the title compound are calculated at the B3LYP method in 6-31g (p, d) as the basis set of the theory. The energy values of the ligand and its compound are EHOMO, EHOMO-1, EHOMO-2, levels are -0.24, -0.25, -0.26eV and ELUMO LUMO, LUMO+ 1, LUMO+2 levels are -0.00 eV, -0.00 eV, -0.00eV respectively. The main energy gap between the HOMO and LUMO of the ligand are 0.24eV, 0.25eV and 0.26eV respectively. The molecule with small energy gap is more polarizability and gives low kinetic stability which is known as soft molecule. The soft molecules explained the resistance towards the electron cloud and polarization of chemical systems [52], [53], [54]. The atomic orbital mechanisms of the frontier molecular orbital are shown in Fig. 6 .
Fig. 6.
Frontier Molecular Orbitals HOMO-i and LUMO + i where i = 1,2 with their orbital energies (eV) and orbital energy gaps (eV).
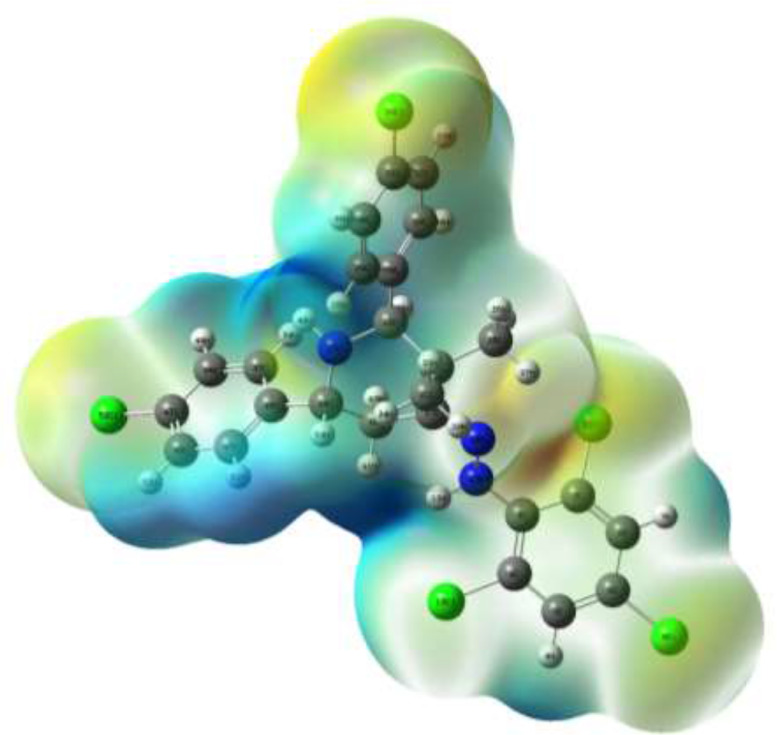
The electrostatic potential is one of a great tool for interpreting and predicting the reactive charges towards electrophilic and nucleophilic reaction and one of the indicators of biological investigation and interaction towards hydrogen bonding. The diagram of MEP shows the most probable reactive side of the molecule for interaction with electrophilic and nucleophilic charges. The BCMTP compound of molecular electrostatic potential was calculated at the DFT/B3LYP method with 6-31G (d, p) as the basic level. The negative region represents the white colour due to the presence of electrophilic reactivity while the positive region represents the pink colour due to the nucleophilic reaction as shown in Fig. 7 . The maximum electrostatic potential arises due to the positive region in the presence of the nitrogen atom and hydrogen atoms which are nucleophilic. The result of MEP provides detailed indication for the biological activity of the BCMTP compound [55], [56].
Fig. 7.
Molecular electrostatic potential of BCMTP.
The atomic charge value of the synthesized BCMTP compound was calculated with the help of Lee-Parr correlation functional (B3LYP) method at 6-31 G (d, p) as the basic level of theory. The atomic distribution of positive and negative charges in the particle and the bond length of the molecule changed between the atoms. The carbon atom which is attached to C21, N19, N12, Cl39 and Cl53 has a negative charge. Besides, all the hydrogen atoms which are attached to nitrogen atoms have a positive charge [ 57 ]. The calculated Mullikan atomic charge of the BCMTP compound is shown in Table 7 .
Table 7.
Mulliken atomic charges of BCMTP.
| Atom | Charge | Atom | Charge |
|---|---|---|---|
| 1 C | -0.09 | 29 C | -0.08 |
| 2 C | -0.06 | 30 C | -0.09 |
| 3 C | 0.26 | 31 C | -0.09 |
| 4 C | -0.11 | 32 C | -0.09 |
| 5 C | -0.08 | 33 C | -0.13 |
| 6 C | -0.08 | 34 C | 0.08 |
| 7 H | 0.14 | 35 H | 0.11 |
| 8 H | 0.13 | 36 H | 0.11 |
| 9 Cl | 0.01 | 37 H | 0.11 |
| 10 Cl | 0.02 | 38 H | 0.10 |
| 11 Cl | 0.02 | 39 Cl | -0.02 |
| 12 N | -0.44 | 40 H | 0.25 |
| 13 H | 0.27 | 41 H | 0.11 |
| 14 C | 0.01 | 42 H | 0.13 |
| 15 C | 0.02 | 43 C | -0.09 |
| 16 C | -0.01 | 44 C | -0.09 |
| 17 C | 0.30 | 45 C | -0.09 |
| 18 C | -0.21 | 46 C | 0.06 |
| 19 N | -0.54 | 47 C | -0.13 |
| 20 N | -0.37 | 48 C | -0.09 |
| 21 C | -0.31 | 49 H | 0.11 |
| 22 H | 0.12 | 50 H | 0.10 |
| 23 H | 0.12 | 51 H | 0.12 |
| 24 H | 0.11 | 52 H | 0.11 |
| 25 C | -0.31 | 53 Cl | -0.02 |
| 26 H | 0.11 | 54 H | 0.13 |
| 27 H | 0.13 | 55 H | 0.09 |
| 28 H | 0.10 |
5. Molecular docking studies
The molecular docking analysis of (E)-2,6-bis(4-chlorophenyl)-3-methyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine (BCMTP) ligand with COVID-19/6WCF and COVID-19/6Y84 receptors were operated [58], [59]. For structure-based drug design, molecular docking is extremely essential. The particular treatment for COVID-19 is not available up to date, so by researchers many anti-retroviral drugs against COVID-19 were reported and existing such as Atazanavir, Darunavir ritonavir, lopinavir, oseltamivir, remdesivir, chloroquine and hydroxychloroquine [58]. The molecular docking mechanism between ligand (BCMTP) and the 6WCF receptor was investigated and evaluated. 6WCF is the Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES41. In the binding mode, the compound was attractively bound to 6WCF via conventional hydrogen bond, Pi-Pi stacked, Alkyl and Pi-alkyl hydrophobic interactions. The residues ILE25, ALA154, PRO225 and VAL49 were binding with chlorine atom in the 1,3,5-trichlorobenzene ring with distance 3.45Å, 2.2Å, 4.05Å and 4.59Å by conventional hydrogen bond. The residue LYS158 is binding in chlorobenzene ring with distance 2.02Å through alkyl bond. The residue PHE256 is binding with a piperidine ring with two bond distances 4.10Å and 4.44Å by Pi-Pi stacked and Pi-alkyl interactions. While, the standard drug hydroxychloroquine is enclosed with Pi-donor hydrogen bond, mixed alkyl and Pi-alkyl hydrophobic interactions such as PHE156 (1.78Å), VAL49(2.88Å), ILE23 (3.21Å), PRO125 (3.63 Å), LEU126 (4.72Å), LEU160 (3.43Å), LEU164 (4.40Å), PRO136 (4.28Å) and VAL155 (2.99Å, 3.64Å, 4.28Å) amino acids with different distances. The binding energy of compound BCMTP is -11.108kcal/mol while the standard drug (hydroxychloroquine) is -8.96kcal/mol. Docked 2D and 3D images of compound BCMTP and standard drug (hydroxychloroquine) with 6WCF receptor were shown in Fig. 8 .
Fig. 8.
3D and 2D View of interactions of BCMTP and standard drug with Covid-19/6WCF receptor.
Secondly, the interactions between BCMTP ligand and the 6Y84 receptor are as follows. 6Y84 is the COVID-19 main protease with a un-liganded active site. SARS-CoV-2 main protease has a vital role in the processing of polyprotein that is translated from viral RNA, and the protease is considered as key for viral survival and growth. The compound was well bound to 6Y84 via carbon hydrogen bond, conventional hydrogen bond, halogen, Pi-donor hydrogen bond, Pi-sigma, Pi-Pi stacked, alkyl and pi-alkyl hydrophobic interactions. The conventional hydrogen bond and halogen bond interactions of the amino acids GLN107 (3.14Å) and GLN100 (2.55Å) are binding with 1,3,5-trichlorobenzene ring. PHE294 (4.93Å), (4.11Å), (5.33Å), PRO293 (4.14Å), (5.23Å) and THR292 (2.53Å) is well bound with chlorobenzene and piperidine moiety in different distances by Pi-Pi stacked, Pi-alkyl and alkyl interactions. The residue ILE249 (3.81Å, 3.23Å) is attached to chlorobenzene and shows piperidine bond interactions. But the standard hydroxychloroquine is having less binding energy due to fewer interactions of carbon hydrogen bond between ligand and receptor. The binding energy of compound BCMTP is -11.25kcal/mol while the standard drug (hydroxychloroquine) is -8.68kcal/mol. The docking of 2D and 3D images of target compound and standard drug (hydroxychloroquine) with 6Y84 receptor were shown in Fig. 9 . From the above molecular docking results, it can be concluded that the synthesised BCMTP molecules can be considered as potential agents against COVID-19/6Y84-6WCF receptors.
Fig. 9.
3D and 2D View of interactions of BCMTP and standard drug with Covid-19/6Y84 receptor.
6. Molecular dynamics simulation study
The binding stability of t both piperidin and hydroxychloroquine drug molecules with the active site of the ADP-ribose phosphatase and main protease of SARS-CoV-2 were studied by performing 50ns MD simulations. The RMSD (Root Means Square Deviation) plot affirms the stability of both molecules with each protein during the MD simulations. Also, RMSF (Root Means Square Fluctuation) plots show conformational modifications of the amino acid residues of both proteins with the drug molecules during the entire MD simulations.
6.1. Intermolecular interactions of compound BCMTP with 6WCF/6Y84 receptors
The binding energy value of synthesized BCMTP molecule is -11.10kcal/mol with 6WCF receptors and forms the expected conventional hydrogen bonding interactions with the active site amino acid VAL41 (1.29Å), this complex was chosen for further analysis. In molecular dynamic simulation the BCMTP molecule forms various types of interactions (pi-donor hydrogen bond, halogen bond, pi-cation, pi-pi stacked, alkyl and pi-alkyl interactions) with 6WCF receptors. The ASN37 (4.74Å) form pi-donor hydrogen bonding interactions with 2,4,6-trichlorophenyl hydrazine moiety in MD. The residue ALA35 (5.37Å) interacts with the chlorine atom of the phenyl hydrazine by halogen bond. Pi-cation and pi-pi stacked interactions formed with C-6 and C-2 phenyl group through the amino acids LYS41 (2.74Å) and TYR39 (5.56Å). Alkyl and pi-alkyl interactions also formed with 2,4,6-trichlorophenyl hydrazine moiety and C-2 aromatic phenyl group by the residues of PHE129 (6.41Å), ALA47 (4.15Å), ILE128 (4.84Å) and LYS99 (6.19Å) with different distances. But the standard hydroxychloroquine drug molecule forms pi-pi stacked interaction with active site amino acid PHE129 (5.09Å) with quinoline moiety. The amino acids ALA36 (4.08 Å), ALA47 (4.76 Å) and HIS42 (4.27Å) formed Alkyl and pi-alkyl interactions with chlorine atom. ILE128 (6.00Å) and ALA47 (4.65Å) residues formed alkyl and pi-alkyl bonding interactions with a quinoline ring containing methyl and ethyl substituent. The 2D representation of their intermolecular interactions and surface view of standard drug and BCMTP with 6WCF receptor are given in Fig. 10 .
Fig. 10.
The 2D representation of their intermolecular interactions and surface view of standard drug (A, B) and BCMTP (C, D) with 6WCF receptor.
The ligand pose energy value of BCMTP molecule is -11.25kcal/mol with 6Y84 receptors and is formed by carbon hydrogen bonding interactions with the active site amino acid PRO9 (2.20Å) with chlorine atom of 2,4,6-trichlorophenyl hydrazine moiety. In MD simulation the BCMTP molecule forms various types of interactions (Van der Waals interactions, Halogen bond, pi-sigma, pi-pi stacked, alkyl and pi-alkyl interactions) with 6Y84 receptors. The MET6 forms pi-sigma bonding interactions with 2,4,6-trichlorophenyl hydrazine moiety with distance 3.56Å. The pi-pi stacked bond also formed with C-6 phenyl group through the amino acids PHE8 (6.32Å). Alkyl and pi-alkyl interactions also formed with methyl moiety present in C-3 carbon atom and chlorine atom of C-6 aromatic phenyl group through the amino acids ARG4 (3.20Å) and PHE8 (6.76Å) with different distances. In hydroxychloroquine the carbon hydrogen bond formed with oxygen atom of ethanolic OH and ethyl moiety by PRO108 (3.20Å) and ASP245 (4.45Å). GLY109 (4.94Å) is formed amide pi-stacked bonding interaction with a quinoline ring. The amino acid ILE249 (5.60), (4.84), (4.95), HIS246 (4.21), VAL202 (6.21), PHE294 (6.22) also formed alkyl and pi-alkyl interactions with hydroxycholroquine moiety with different distances. As compared to synthesized BCMTP molecule, the standard hydroxychloroquine drug has moderate interactions with low bond distances. In molecular dynamic simulation studies the amino acid residues, mutation number and bond distances are changed from docking results due to the non-super imposable of the structure. The molecular dynamic simulation results revealed that the synthesized BCMTP molecule has greater intermolecular interactions and more stability with 6WCF/6Y84 receptors than standard drug. The 2D representation of their intermolecular interactions and surface view of standard drug and BCMTP with 6Y84 receptor are given in Fig. 11 .
Fig. 11.
The 2D representation of their intermolecular interactions and surface view of standard drug (A, B) and BCMTP (C,D) with 6Y84 receptor.
6.2. Root mean square deviation (RMSD)
To understand the stability of all complexes, RMSD plots were generated using MD trajectories as shown in Fig. 12 . The complexes which have hydroxychloroquine and piperidin with ADP-ribosephosphatase shows deviation between the range 1.5 to 2.0 Å and it confirms the high stability of the both complexes during the MD simulation. But the complexes which have hydroxychloroquine and piperidin with main protease shows the deviation in the average range 1 to 3 Å as uneven, although both deviations are under the acceptable range and these plots still confirms the stability during the MD simulation. In these two complexes, ligand RMSD of both hydroxychloroquine and piperidin shows high range modifications unlike with ADP-ribosephosphatase. However, the overall deviation of the complexes shows the stability of the complexes during the MD simulations is stable.
Fig. 12.
RMSD plots for the complexes (a) Hydroxychloroquine-ADP-ribosephosphatase, (b) Piperidin-ADP-ribosephosphatase, (c) Hydroxychloroquine-main protease, and (d) Piperidin-main protease. In these plots, Protein (Red) and Ligand (Black).
6.3. Root means square fluctuation (RMSF)
Root means square fluctuation (RMSF) has been plotted to understand fluctuations of the individual amino acids of each complex during the MD simulations. In all the cases, N and C terminal indicates larger fluctuations due to their start and end sequence placing in the protein structure as nearly 4 to 7Å range averagely. Also some high RMSF values are observed during the simulations for the residues located in loop regions of all the complexes. Remaining the side chain residues shows the less fluctuation values which are under the acceptable range (less than 4Å). Among these results confirms the structural confirmations of the complexes during the MD simulations. RMSF plots for the complexes are given in Fig. 13 .
Fig. 13.
RMSF plots for the complexes (a) Hydroxychloroquine-ADP-ribosephosphatase, (b) Piperidin-ADP-ribosephosphatase, (c) Hydroxychloroquine-main protease, and (d) Piperidin-main protease.
7. Conclusion
In conclusion, we have designed, synthesized and characterized (E)-2,6-bis(4-chlorophenyl)-3-methyl-4-(2-(2,4,6-trichlorophenyl)hydrazono)piperidine derivatives (BCMTP). This synthesized compound BCMTP exhibits in chair conformation with equatorial orientation of two phenyl groups at C-2, C-6 and methyl group at C-3 carbon. The single crystal XRD study of BCMTP also evidenced E configuration about the C=N bond and the chair conformation of the piperidine ring. Hirshfeld surface analysis was performed to observe better intermolecular interactions in the crystal packing of BCMTP. The stabilization energy (E2) values determined the hyper conjugative interactions and charge transfers by the orbital overlap between π-π* (C44-C48) with stabilization energy 237.72kj/mol. The possible intermolecular and intra-molecular transitions inside the BCMTP molecule were studied by NBO analysis. The energy gap between HOMO and LUMO is given by 0.24eV labelled that the BCMTP compound is suitable for chemical stability and the transfer of electrons from the ground state to the excited state of the molecules. The MEP diagram of the BCMTP molecule visualized its electrophilic and nucleophilic regions. The binding energy values of compound BCMTP is -11.10 and -11.25kcal/mol while the standard drug hydroxychloroquine is -8.96 and -8.68kcal/mol with 6WCF/6Y84 receptor. Molecular dynamics simulation and molecular docking studies of compound BCMTP into the binding site of ADP ribose phosphatase of NSP3 and main protease with an unliganded active site from SARS-CoV-2 exhibited the probable interactions of BCMTP with the binding site of the SARS-CoV-2 main protease. Molecular docking and molecular dynamics simulation results have shown that the BCMTP compound can be considered as a potential antiviral agent against COVID-19/ 6Y84-6WCF receptors.
CRediT authorship contribution statement
L. Athishu Anthony: Conceptualization, Investigation, Methodology, Resources, Formal analysis, Data curation, Writing – original draft. D. Rajaraman: Conceptualization, Supervision, Investigation, Methodology, Resources, Formal analysis, Data curation, Writing – original draft. G. Sundararajan: Software. M. Suresh: Software. P. Nethaji: Software. R. Jaganathan: Software. Kumaradhas Poomani: Software.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgement
The authors are grateful to the Annamalai University, Annamalainagar- 605 803, Tamil Nadu, India for providing the instrument facilities.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2022.133483.
Appendix. Supplementary materials
References
- 1.Parthiban P., Balasubramanian S., Aridoss G., Kabilan S. Synthesis and NMR spectral studies of some 2,6-diarylpiperidin-4-one O-benzyloximes. SpectrochimicaActa Part A. 2008;70(1):1124. doi: 10.1016/j.saa.2007.07.059. [DOI] [PubMed] [Google Scholar]
- 2.Ramalingam C., Park Y.T., Kabilan S. Synthesis, stereochemistry, and antimicrobial evaluation of substituted piperidin-4-one oxime ethers. Eur. J. Med. Chem. 2006;41:683–696. doi: 10.1016/j.ejmech.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Mobio I.G., Soldatenkov A.T., Federov V.O., Ageev E.A., Sergeeva N.D., Lin S., Stashenku E.E., Prostakov N.S., Andreeva E.L. Khim Farm Zh, Synthesis and physiological activity of 2, 3, 6-triaryl-4-oxo (hydroxy, oximino, amino) piperidine. ChemInform. 1989;23:421–427. [Google Scholar]
- 4.Parthiban P., Aridoss G., Rathika P., Ramkumar V., Kabilan S. Synthesis, spectral, crystal and antimicrobial studies of biologically potent oxime ethers of nitrogen, oxygen and sulfur heterocycles. Bioorg. Med. Chem. Lett. 2009;19:2981–2985. doi: 10.1016/j.bmcl.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 5.El-Subbagh H.I., Abu-Zaid S.M., Mahran M.A., Badria F.A., Al-Obaid A.M. Synthesis and biological evaluation of certain α,β-unsaturated ketones and their corresponding fused pyridines as antiviral and cytotoxic agents. J. Med. Chem. 2000;43:2915–2921. doi: 10.1021/jm000038m. [DOI] [PubMed] [Google Scholar]
- 6.Watsona A.A., Fleet G.W.J., Asano N., Molyneux R.J., Nugh R.J. Polyhydroxylated alkaloids natural occurrence and therapeutic applications. Phytochemistry. 2001;56:265–295. doi: 10.1016/s0031-9422(00)00451-9. [DOI] [PubMed] [Google Scholar]
- 7.Aridoss G., Amirthaganesan S., Kim M.S., Kim J.T., Jeong Y.T. Synthesis, spectral and biological evaluation of some new thiazolidinones and thiazoles based on t-3-alkyl-r-2,c-6-diarylpiperidin-4-ones. Eur. J. Med. Chem. 2009;44:4199–4210. doi: 10.1016/j.ejmech.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Eliel E.L., Gianni M.H., Williams T.H., Stothers J.B. Chemical shifts in the nmr spectra of substituted cyclohexanols. Tetrahedron Lett. 1962;3(17):741–747. doi: 10.1016/s0040-4039(00)70511-9. [DOI] [Google Scholar]
- 9.Bhacca N.S., Williams D.H. Holden-Day; 1964. Applications of NMR Spectroscopy in Organic Chemistry: Illustration from the Steroid Field. [Google Scholar]
- 10.Booth H., Little J.H. Proton magnetic resonance studies of cyclic compounds-V. Tetrahedron. 1967;23(1):291–297. doi: 10.1016/s0040-4020(01)83313-5. [DOI] [Google Scholar]
- 11.Santiago P.H.O., Santiagob M.B., Martins C.H.G., Gatto C.C. Copper(II) and zinc(II) complexes with Hydrazone: Synthesis, crystal structure, Hirshfeld surface and antibacterial activity. InorganicaChimicaActa. 2020;508 doi: 10.1016/j.ica.2020.119632. doi:org/ [DOI] [Google Scholar]
- 12.Weiss D.S., Abkowitz M. Advances in organic photoconductor technology. Chem. Rev. 2010;110(1):479–526. doi: 10.1021/cr900173r. [DOI] [PubMed] [Google Scholar]
- 13.Lygaitis R., Getautis V., Grazulevicius J.V. Hole-transporting hydrazones. Chem. Soc. Rev. 2008;37(4):770–788. doi: 10.1039/b702406c. [DOI] [PubMed] [Google Scholar]
- 14.Sidir I., Y.Gii Sidir, Berber H., Demiray F. Electronic structure and optical properties of Schiff base hydrazone derivatives by solution technique for optoelectronic devices: Synthesis, experiment and quantum chemical investigation. J. Mol. Struct. 2018 doi: 10.1016/j.molstruc.2018.08.067. [DOI] [Google Scholar]
- 15.Laurinaviciute R., Ostrauskaite J., Grazulevicius J.V., Jankauskas V. Synthesis, properties, and self-polymerization of hole-transporting carbazole- and triphenylamine-based hydrazone monomers. Des. Monomers Polym. 2014;17(3):255–265. doi: 10.1080/15685551.2013.840499. http://dx.doi.org/ [DOI] [Google Scholar]
- 16.Hwang J., Moon H., Seo J., Park S.Y., Aoyama T., Wada T., Sasabe H. Synthesis and characterization of photoconducting non-linear optical polymers containing indole-benzoxazole moiety. Polymer. 2001;42(7):3023–3031. doi: 10.1016/s0032-3861(00)00662-5. [DOI] [Google Scholar]
- 17.Quirino W.G., Legnani C., Cremona M., Reyes R., Mota G.V., Weibel D.E., Rocco M.L.M. Photoluminescence, photoabsorption and photoemission studies of hydrazone thin film used as hole transporting material in OLEDs. J. Braz. Chem. Soc. 2008;19:827–876. [Google Scholar]
- 18.Sharma G.D., Suresh P., Sharma S.K., Roy M.S. Optical and electrical properties of hybrid photovoltaic devices from poly (3-phenyl hydrazonethiophene) (PPHT) and TiO2 blend films. Sol. Energy Mater. Sol. Cells. 2008;92:61–70. doi: 10.1016/j.solmat.2007.08.009. [DOI] [Google Scholar]
- 19.Meenatchi V., Siva S., Cheng L. Synthesis, crystal growth, spectroscopic characterization, Hirshfeld surface analysis and DFT investigations of novel nonlinear optically active 4-benzoylpyridine-derived hydrazine. J. Mol. Struct. 2021;1243 doi: 10.1016/j.molstruc.2021.130858. https://doi.org/ [DOI] [Google Scholar]
- 20.Khalid M., Arshad M.N., Tahir M.N., Asiri A.M., Naseer M.M., Ishaq M., Khan M.U., Shafiq Z. An efficient synthesis, structural (SC-XRD) and spectroscopic (FTIR, 1HNMR, MS spectroscopic) characterization of novel benzofuran based hydrazones: an experimental and theoretical studies. J. Mol. Struct. 2020;1216 doi: 10.1016/j.molstruc.2020.128318. [DOI] [Google Scholar]
- 21.Li Z., Xu L., Zhu L., Zhao Y., Hu T., Yin B, Liu Y., Hou Y. Design, synthesis and biological evaluation of novel pteridinone derivatives possessing a hydrazone moiety as potent PLK1 inhibitors. Bioorg. Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127329. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 22.Noma S.A.A., Erzengin M., Tunc T., Balcioglu S. Synthesis, characterization and biological assessment of a novel hydrazone as potential anticancer agent and enzyme inhibitor. J. Mol. Struct. 2019 doi: 10.1016/j.molstruc.2019.127550. https://doi.org/ [DOI] [Google Scholar]
- 23.S.Saouli I.Selatnia, B.Zouchoune A.Sid, Zendaoui S.M., Bensouici C., Bendeif El-E. Synthesis, spectroscopic characterization, crystal structure, DFT studies and biological activities of new hydrazone derivative: 1-(2,5- bis((E)-4-isopropylbenzylidene)cyclopentylidene)-2-(2,4- dinitrophenyl) hydrazine. J. Mol. Struct. 2020;1213 doi: 10.1016/j.molstruc.2020.128203. https://doi.org/ [DOI] [Google Scholar]
- 24.Sh. El-Etrawya A-A., Sherbiny F.F. Design, synthesis, biological evaluation and molecular modeling investigation of new N’-(2-Thiouracil-5-oyl) hydrazone derivatives as potential anti-breast cancer and anti-bacterial agents. J. Mol. Struct. 2021;1232 doi: 10.1016/j.molstruc.2021.129993. https://doi.org/ [DOI] [Google Scholar]
- 25.Abou-Melha K.S., Al-Hazmi G.A., Althagafi I., Alharbi A., Shaaban F., El-Metwaly N.M., El-Bindary A.A., El-Bindary M.A. Synthesis, characterization, DFT calculation, DNA binding and antimicrobial activities of metal complexes of dimedonearylhydrazone. J. Mol. Liq. 2021;334 doi: 10.1016/j.molliq.2021.116498. https://doi.org/ [DOI] [Google Scholar]
- 26.A.Noma S.A., Erzengin M., Tunc T., Balcıoglu S. Synthesis, characterization and biological assessment of a novel hydrazone as potential anticancer agent and enzyme inhibitor. J. Mol. Struct. 2019 doi: 10.1016/j.molstruc.2019.127550. https://doi.org/ [DOI] [Google Scholar]
- 27.Tariq Q-un-N., Malik S., Khan A., Naseer M.M., Khan S.U., Ashraf A., Ashraf M., Rafiq M., Mahmood K., Tahir M.N., Shafiq Z. Xanthenone-based hydrazones as potent α-glucosidase inhibitors: synthesis, solid state self-assembly and in silico studies. Bioorg. Chem. 2019;84:372–383. doi: 10.1016/j.bioorg.2018.11.053. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 28.Yang K., Yang J-Q., Luo S-H., Mei W-J., Lin J-Y., Zhan J-Q., Wang Z-Y. Synthesis of N-2(5H)-furanonyl sulfonyl hydrazone derivatives and their biological evaluation in vitro and in vivo activity against MCF-7 breast cancer cells. Bioorg. Chem. 2020 doi: 10.1016/j.bioorg.2020.104518. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 29.Tian Y., Qin M., Yang X., Zhang X., Liu Y., Guo X., Chen B. Acid-catalyzed synthesis of imidazole derivatives via N-phenylbenzimidamides and sulfoxoniumylides cyclization. Tetrahedron. 2019;75:2817–2823. [Google Scholar]
- 30.Rajaraman D., Sundararajan G., Kamaraj A., Saleem H., Krishnasamy K. Synthesis, computational and spectroscopic analysis on (E)-(4-(2-(benzo[d]thiazol-2-yl)hydrazono)-3-methyl-2,6-diphenylpiperidine-1-yl)(phenyl)methanone using DFT approach. Spectrochim. Acta. A. 2015;151:480–489. doi: 10.1016/j.saa.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 31.Ourhzif El-M., A-Thomas I., Chalard P., Khouili M., Troin Y., Akssira M., Ketatni El.M. Synthesis, characterisation, crystal structure, hirsfeld surface analysis and DFT studies of novel compounds based on the methoxynaphthalene ring. J. Mol. Struct. 2021;1244 [Google Scholar]
- 32.Jayasheela K., Al-Wahaibi L.H., Periandy S., Hassan H.M., Sebastian S., Xavier S., Daniel Joseph C., El-Emam Ali A., Attia M.I. Probing vibrational activities, electronic properties, molecular docking and Hirshfeld surfaces analysis of 4-chlorophenyl ({[(1E)3(1Himidazol1yl)1phenylpropylidene]amino}oxy)methanone: a promising anti-Candida agent. J. Mol. Struct. 2018;1159:83–95. [Google Scholar]
- 33.Foujji L.El., Bourakadi K.El., Mokhtar Essass El., Boere R.T., Qaiss A.El.K, Bouhfid R. solid-state zwitterionictautoerization of 2-((5-methyl-1H- pyrazol-3-yl)methyl)-1H-benzimidazole: synthesis, characterization, DFT calculation and docking studies. J. Mol. Struct. 2021;1235 [Google Scholar]
- 34.Verma V.A., Saundane A.R., Meti R.S., Vennapu D.R. Synthesis of novel indolo[3,2-c]isoquinoline derivatives bearing pyrimidine, piperazine rings and their biological evaluation and docking studies against COVID-19 virus main protease. J. Mol. Struct. 2021;1229 doi: 10.1016/j.molstruc.2020.129829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Athishu Anthony L., Nethaji P., Rajaraman D. Synthesis, spectral characterization, crystal examination, DFT method and molecular docking studies of 1-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-2-(thiophen-2-yl)-1Hphenanthro[9,10-d]imidazole. Chem. Data Collections. 2021;34 [Google Scholar]
- 36.Douche D., Sert Y., Brandan S.A., Kawther A.A., Bilmez B., Dege N., El Louzi A., Bougrina K., Karrouchif K., Himmi B. 5-((1H-imidazol-1-yl)methyl)quinolin-8-ol as potential antiviral SARS-CoV-2 candidate: synthesis, crystal structure, Hirshfeld surface analysis, DFT and molecular docking studies. J. Mol. Struct. 2021;1232 doi: 10.1016/j.molstruc.2021.130005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viji A., Balachandran V., Babiyana S., Narayana B., Salian V.V. FT-IR and FT-Raman investigation, quantum chemical studies, molecular docking study and antimicrobial activity studies on novel bioactive drug of 1-(2,4-Dichlorobenzyl)-3-[2-(3-(4-chlorophenyl)-5- (4-(propan-2-yl)phenyl-4,5-dihydro-1H-pyrazol-1-yl]-4-oxo-4,5- dihydro-1,3-thiazol-5(4H)-ylidence]-2,3-dihydro-1H-indol-2-one. J. Mol. Struct. 2020;1215 [Google Scholar]
- 38.Eargle J., Wright D., Luthey-Schulten Z. Multiple Alignment of protein structures and sequences for VMD. Bioinformatics. 2006;22(4):504–506. doi: 10.1093/bioinformatics/bti825. [DOI] [PubMed] [Google Scholar]
- 39.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105(43):9954–9960. doi: 10.1021/jp003020w. https://doi.org/ [DOI] [Google Scholar]
- 40.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Computat. Phys. 1977;23(3):327–341. doi: 10.1016/0021-9991(77)90098-5. https://doi.org/ [DOI] [Google Scholar]
- 41.Tian C., Kasavajhala K.A.A., Belfon K., Raguette L., Huang H.N., Migues A., Bickel J., Wang Y., Pincay J., Wu Q., Simmerling C. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 2019;16(1):528–552. doi: 10.1021/acs.jctc.9b00591. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 42.Michalska D. Wroclaw University of Technology; 2003. Raint Program. [Google Scholar]
- 43.Baas J.M.A. John Wiley; Chichester: 2010. A Guide to the Complete Interpretation of Infrared Spectra of Organic Structures. N.P.G. RoegesISBN 0-471-93998-6. Recueil Des TravauxChimiquesDes Pays-Bas, 114(3),(1994), 114–114. [DOI] [Google Scholar]
- 44.Gomathi M.A., Karnan C., Sivanesan T., Rhoda J.C., Manivannan S., Ragavendran V., Vinitha G., Prabakaran A.R. An organic benzimidazoliumbenzilate (BDBA) crystal: Structural description, spectral investigations, DFT calculations, thermal, photoluminescence, linear and nonlinear optical analysis. Chem. Phys. Lett. 2021;776 doi: 10.1016/j.molstruc.2021.131212. https://doi.org/ [DOI] [Google Scholar]
- 45.Muthu S., Ramachandran G. Spectroscopic studies (FTIR, FT-Raman and UV–Visible), normal coordinate analysis, NBO analysis, first order hyper polarizability, HOMO and LUMO analysis of (1R)-N-(Prop-2-yn-1-yl)-2,3-dihydro-1H-inden-1- amine molecule by ab initio HF and density functional methods. SpectrochimicaActa Part A. 2014;121:394–403. doi: 10.1016/j.saa.2013.10.093. [DOI] [PubMed] [Google Scholar]
- 46.Vijesh A.M., A.M.Isloor S.Telkar, Peethambar S.K., Rai S. Synthesis, characterisation and antimicrobial studies of some new pyrazole incorporated imidazole derivatives. Eur. J. Med. Chem. 2011;46:3531–3536. doi: 10.1016/j.ejmech.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 47.Wu P., Zhang X., Chen Baohua. Direct synthesis of 2,4,5-trisubstituted imidazoles and Di/Tri-substituted Pyrimidines via cycloadditions of α,β-Unsaturated Ketones/Aldehydes and N-Hydroxyl. Tetrahedron Lett. April 2019;60(16):1103–1107. [Google Scholar]
- 48.Bonin M-A, Giguere D., Roy R. N-Arylimidazole synthesis by cross-cycloaddition of isocyanides using a novel catalytic system. Tetrahedron. 2007;63:4912–4917. [Google Scholar]
- 49.Vanitha U., Elancheran R., Manikandan V., Kabilan S., Krishnasamy K. Design synthesis, characterization, molecular docking and computational studies of 3-phenyl-2-thioxoimidazolidin-4-one derivatives. J. Mol. Struct. 2021;1246 [Google Scholar]
- 50.Serdaroglu G., Sahin N. the synthesis and characterization of 1-(Allyl)-3-(2-methylbenzyl)bensimidazolium chloride: FT-IR, NMR and DFT computational investigation. J. Mol. Struct. 2018 doi: 10.1016/j.molstruc.2018.10.028. doi:https://doi.org/ [DOI] [Google Scholar]
- 51.Shankar Rao Y.B., Prasad M.V.S., Udaya Sri N., Veeraiah V. Vibratonal (FT-IR) and UV-Visible spectroscopic studies, HOMO-LUMO, NBO and MEP analysis of Benzyl(imino(1H-pyrazol-1-yl)methyl) carbamate using DFT calculations. J. Mol. Struct. 2016;1108:567–582. [Google Scholar]
- 52.Milanovic Z.B., Dimic D.S., Avdovic E.H., Milenkovic D.A., Markovic J.D., Klisuric O.R., Trifunovic S.R., Markovic Z.S. Synthesis and comprehensive spectroscopic (X-ray, NMR, FTIR, UV–Vis), quantum chemical and molecular docking investigation of 3-acetyl-4–hydroxy–2-oxo-2H-chromen-7-yl acetate. J. Mol. Struct. 2021;1225 [Google Scholar]
- 53.Kamath A., Brahman D., Chhetri S., Mc Ardle P., Sinha B. [Diaquo{bis(p-hydroxybenzoato-κ1O1)}(1-methylimidazoleκ1N1)}copper(II)]: synthesis, crystal structure, catalytic activity and DFT study. J. Mol. Struct. 2022;1247 [Google Scholar]
- 54.Shafieyoon P., Mehdipour E., Mary Y.S. Synthesis, characterization and biological investigation of glycine- based sulfonamide derivative and its complex: vibration assignment, HOMO – LUMO analysis, MEP and molecular docking. J. Mol. Struct. 2018 doi: 10.1016/j.molstruc.2018.12.06. [DOI] [Google Scholar]
- 55.Mushtaque Md., Avecilla F., Haque A., Yab Z., Rizvi M.M.A., Khan M.S. Synthesis, structural and biological activity of N-substituted 2-methyl-4-/5-nitroimidazole derivatives. J. Mol. Struct. 2019;1185:440–449. [Google Scholar]
- 56.Bouhdada M., Amane M.EL., Hamdani H.EL. Synthesis, characterization, antimicrobial activity, DFT and molecular docking studies of the N-salicylidene-glycinato and their metal mixed ligand complexes with caffeine. J. Mol. Struct. 2020 doi: 10.1016/j.molstruc.2020.129679. https://doi.org/ [DOI] [Google Scholar]
- 57.Castillo A.E., Ceballos P., Ceron M., Perez-Gutierrez E., Sosa-Rivadeneyra M., Bernal W., Thamotharan S., Siegler. M.J. Percino M.A. Spectroscopic characterization of 4,5-diphenyl-2-(2,4,5-trimethoxyphenyl)-1H-imidazole obtained from the condensation of benzyl. Experimental and DFT approach. J. Mol. Struct. 2021;1246 [Google Scholar]
- 58.Rohan Narkhede R., RameshwarCheke S., Jaya Ambhore P., Sachin Shinde D. The molecular docking study of potential drug candidates showing anti- COVID-19 activity by exploring of therapeutic targets of SARS-CoV-2. Eurasian J. Med. 2020;4(3):185–195. doi: 10.14744/ejmo.2020.31503. [DOI] [Google Scholar]
- 59.Chhetri A., Chettri S., Rai P., Mishra D., Sinha B., Brahman D. Synthesis, char- acterization and computational study on potential inhibitory action of novel azo imidazole derivatives against COVID-19 main protease. J. Mol. Struc. 2021;1225 doi: 10.1016/j.molstruc.2020.129230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.