Abstract
Nonpoint sources of pollution that contribute fecal bacteria to surface waters have proven difficult to identify. Knowledge of pollution sources could aid in restoration of the water quality, reduce the amounts of nutrients leaving watersheds, and reduce the danger of infectious disease resulting from exposure to contaminated waters. Patterns of antibiotic resistance in fecal streptococci were analyzed by discriminant and cluster analysis and used to identify sources of fecal pollution in a rural Virginia watershed. A database consisting of patterns from 7,058 fecal streptococcus isolates was first established from known human, livestock, and wildlife sources in Montgomery County, Va. Correct fecal streptococcus source identification averaged 87% for the entire database and ranged from 84% for deer isolates to 93% for human isolates. To field test the method and the database, a watershed improvement project (Page Brook) in Clarke County, Va., was initiated in 1996. Comparison of 892 known-source isolates from that watershed against the database resulted in an average correct classification rate of 88%. Combining all animal isolates increased correct classification rates to ≥95% for separations between animal and human sources. Stream samples from three collection sites were highly contaminated, and fecal streptococci from these sites were classified as being predominantly from cattle (>78% of isolates), with small proportions from waterfowl, deer, and unidentified sources (≈7% each). Based on these results, cattle access to the stream was restricted by installation of fencing and in-pasture watering stations. Fecal coliforms were reduced at the three sites by an average of 94%, from prefencing average populations of 15,900 per 100 ml to postfencing average populations of 960 per 100 ml. After fencing, <45% of fecal streptococcus isolates were classified as being from cattle. These results demonstrate that antibiotic resistance profiles in fecal streptococci can be used to reliably determine sources of fecal pollution, and water quality improvements can occur when efforts to address the identified sources are made.
Many surface waters and groundwaters in the mid-Atlantic region of the United States are contaminated by fecal pollution (12). This contamination results in increased health risks to persons exposed to the water, degradation of recreational and drinking water quality, and nutrient loss from watersheds to surface waters, such as the Chesapeake Bay. Nonpoint sources of pollution that contribute fecal bacteria to surface waters have proven very difficult to accurately identify. Knowledge of pollution sources could aid in the restoration of the water quality, reduce the amounts of nutrients leaving watersheds, and reduce the danger of infectious disease resulting from exposure to contaminated waters. According to Environmental Protection Agency’s National Watershed Database 305b report for Virginia (2a), fecal coliform bacteria are the most widespread problem in rivers and streams, and agriculture and pasture land contribute much of the fecal coliform bacteria in Virginia’s waters. The Environmental Protection Agency’s report is typical of those from other states in the region as well.
While fecal coliforms are the most widely used bacterial indicator of water quality, there are good reasons to use fecal streptococci to determine sources of pollution. There are some potential sources (e.g., composted animal and poultry litter and advanced-treatment class B biosolids) where it is difficult to detect and isolate fecal coliforms while there is no difficulty in isolating fecal streptococci (7). Fecal coliforms would not be suitable for identifying contamination from these types of materials. While antibiotic resistance patterns have been used in the past, with variable success, to determine sources of fecal coliforms, such patterns do appear to have more potential with fecal streptococci (16). Lastly, fecal streptococci tend to persist longer in the environment than fecal coliforms, and while this may limit their usefulness as indicators of recent water contamination, a fecal organism with a longer survival time can be an advantage when collecting isolates for source determination (10, 13, 14).
Several attempts to develop methods to determine sources of fecal pollution have been made, and to date most have not proven useful. These include the ratio of fecal coliforms to fecal streptococci (4, 14), source-specific bacteriophages (15), differences in the species composition of fecal streptococci among various types of animals (2), and patterns of antibiotic resistance in fecal coliforms (9, 11). Simmons (12) successfully used fatty acid profiles and DNA fingerprinting in Escherichia coli to determine nonpoint fecal coliform sources in tidal inlets in the Chesapeake Bay. Other molecular procedures, such as random amplified polymorphic DNA analysis (1), have recently been developed for fingerprinting microbial genomes. The potential to identify individual strains of different bacteria by genetic profiles indicates that molecular approaches may also be suitable for source differentiation of fecal bacteria (3, 8).
While antibiotic resistance patterns may have some use in identifying sources of fecal coliforms (11), the potential use of such patterns for fecal streptococci currently appears to be more feasible (7, 16). Intrinsic antibiotic resistance and resistance patterns have been widely used in bacterial identification, but such patterns have yet to be proven as suitable for determining sources of fecal organisms (5, 6, 10). Wiggins (16) first demonstrated the potential for this approach by successfully using antibiotic resistance patterns in fecal streptococci and discriminant analysis (DA) to differentiate between human and animal sources and between certain types of animal sources.
The objectives of this project were (i) to validate the method described by Wiggins (16) with a larger database of known-source isolates from a wider geographical region and (ii) to use this method in a watershed project to identify fecal pollution sources.
MATERIALS AND METHODS
Sources of isolates.
Isolates from 147 samples from six known sources in Montgomery County, Va., were collected throughout 1995 and 1996 to build a source database from beef cattle, dairy cattle, deer, chickens, humans, and waterfowl (geese and ducks). For each animal source, samples were collected from fresh feces. The cattle and chicken samples were obtained from the Virginia Polytechnic Institute and State University farms, deer samples were collected from a nearby national forest recreational area containing a large deer population, and waterfowl samples were from resident flocks that frequent a pond on the campus. Human samples were collected from experimental domestic wastewater treatment systems from individual homes. After collection, all samples were placed on ice in coolers and processed within 6 h.
Isolation of fecal streptococci.
Samples were suspended and diluted in saline buffer (8.5 g of NaCl, 0.3 g of KH2PO4, and 0.6 g of Na2HPO4 per liter [pH 7.3]) and filtered through a 0.45-μm-pore-size filter (type GN-6; Gelman Sciences). The filters were transferred to a 50-mm petri dish containing m-Enterococcus Agar (BBL) and incubated for 24 to 48 h at 37°C. After incubation, individual red-pigmented colonies were picked with sterile toothpicks, transferred to 96-microwell plates containing 0.2 ml of Enterococcosel broth (BBL), and incubated for another 24 to 48 h at 37°C. Those wells that exhibited growth and formed a black color after incubation in Enterococcosel broth were counted as positive (16).
Biochemical patterns (antibiotic resistance and other tests).
Thirteen antibiotics (Sigma) were evaluated: the five reported by Wiggins (16), plus amoxicillin, ampicillin, chloramphenicol, erythromycin, neomycin sulfate, rifampin, tetracycline, and vancomycin hydrochloride. The antibiotics were added from filter-sterilized stock solutions in water (ampicillin, halofuginone, neomycin, oxytetracycline, and streptomycin), water-ethanol at a 1:1 ratio (chloramphenicol, chlorotetracycline, erythromycin, salinomycin, tetracycline, and vancomycin), or water-methanol at a 1:1 ratio (amoxicillin and rifampin) to autoclaved and cooled Trypticase soy agar (BBL) at initial concentrations of 5, 10, 20, 40, 60, 80, and 100 μg/ml (6). The isolates were transferred with a 48-prong replica plater (Sigma) from the Enterococcosel-containing microwells to a set of Trypticase soy agar plates containing the various concentrations of each antibiotic to be tested and to a control plate containing no antibiotic. The plates were incubated at 37°C for 24 h, and growth of each isolate on each concentration of every antibiotic was determined. An isolate was considered resistant to a given concentration of antibiotic if growth comparable to that of the controls occurred on that plate. Any isolates that did not grow on the control plates (containing no antibiotic) or that were esculin negative were not used in the analysis. In addition to antibiotic resistance, isolates were tested for growth in brain heart infusion (BHI) broth containing 6.5% NaCl, for starch hydrolysis on BHI agar containing soluble starch, and for growth in BHI broth at 45°C (4).
Statistical analysis. (i) DA.
Data on the ability of each of the known-source isolates to grow in the presence of each concentration of each antibiotic and for other tests (starch hydrolysis, growth in 6.5% NaCl, and growth at 45°C) were analyzed with SAS (version 6.12; SAS Institute Inc.) by using the procedure DISCRIM (prior probabilities, equal; covariance matrix, pooled). Each analysis produced a classification rule where the average rate of correct classification (ARCC) for each analysis was determined by averaging the percentages of correctly classified isolates for each source as described by Wiggins (16). The DA procedure first builds a database for each known source (humans and beef cattle, etc.) and then compares each set of isolates from an unknown source against the database of known sources and classifies each isolate into one of the possible sources.
(ii) CA.
Data for each of the known-source isolates were analyzed with SAS-JMP (version 3.2.2; SAS Institute Inc.) by using Ward’s hierarchical procedure, where the distance between any two clusters is the analysis of variance sum of squares added over all variables. Cluster analysis (CA) involves clustering, a technique of grouping together variables (isolates) that have similar values. CA builds a database where all known-source isolates are grouped into a cluster by source. Ward’s method joins clusters to maximize the likelihood of a fit. The CA procedure produces a dendrogram that groups identical isolates within a set and then builds a cluster database from these sets. Unknown-source isolates are placed in the most likely cluster based on source identification and are readily visible within the dendrogram.
Watershed study.
The Page Brook watershed is located in Clarke County, Va., a rural county with an agriculture-based economy located approximately 80 km west of Washington, D.C. (Fig. 1). The watershed is characterized by karst topography with wooded tracts and farms and includes one predominant stream, Page Brook, that is approximately 5.9 km in length from origin to confluence with another stream and entry into an adjacent down-gradient watershed. Page Brook drains a small watershed of roughly 1,980 ha; the stream has been periodically monitored by state officials and was reported to be contaminated with fecal bacteria and nitrates (7). Possible sources of contamination included domestic livestock (mostly cattle) with free access to the stream, resident populations of waterfowl (mainly Canadian geese), large wildlife populations (predominated by deer), and septic tank-subsurface absorption systems from 127 residences. Water samples were collected monthly from November 1996 through February 1999 and consisted of both surface water (stream) and groundwater (residential-well) samples (usually 10 to 12 samples each). Samples were transported to the laboratory on ice packs in coolers and assayed within 24 h. Fecal streptococci were isolated and characterized as described above. For monitoring purposes, all water samples were also assayed for fecal coliforms, using membrane filtration with isolation on mFC agar (BBL) and incubation at 44.5°C (water bath) for 24 h (4). Results were recorded as CFU per 100 ml. Prior to and near the end of the project, fecal samples from known sources (e.g., deer, cattle, geese, and humans [from septage trucks unloading at a wastewater treatment plant]) were also collected and assayed.
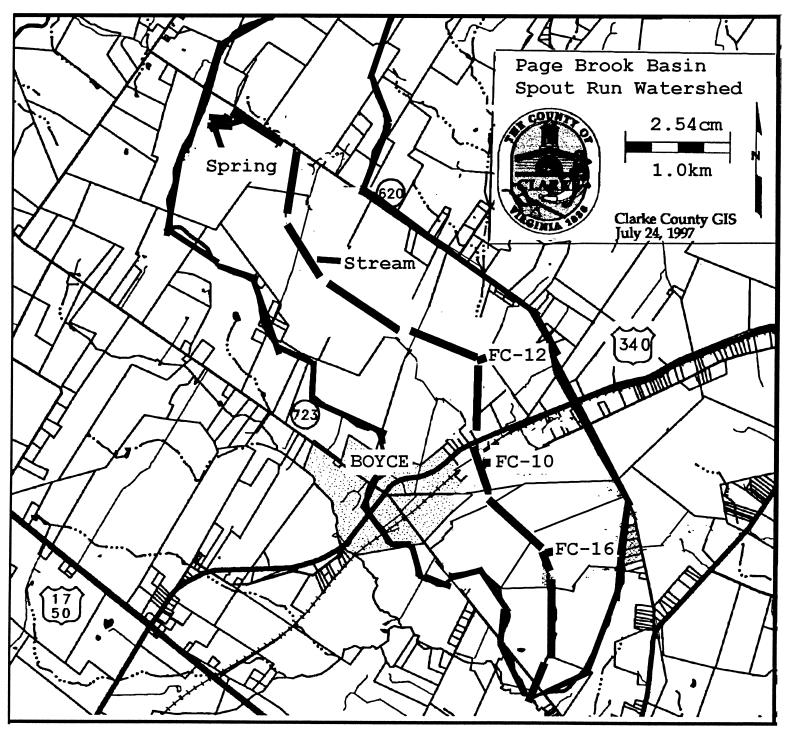
FIG. 1.
Page Brook basin (30 km2, inside the heavy lines) showing the location of Page Brook stream (heavy segmented line) and the 3.2-km impaired stream segment extending from FC-12 to FC-16.
RESULTS
Database development.
All isolates were gram-positive cocci, and almost all (98% or more) grew at 45°C and in the presence of 6.5% NaCl. Cultures with these characteristics that were isolated on m-Enterococcus agar and that hydrolyzed esculin in Enterococcosel broth were classified as fecal streptococci (4). The best separation of isolates by source was obtained with the following six antibiotics at the indicated concentrations (Table 1): chlortetracycline hydrochloride (40 and 60 μg/ml), erythromycin (7 and 15 μg/ml), neomycin sulfate (10 and 40 μg/ml), oxytetracycline hydrochloride (10, 30, 60, and 100 μg/ml), streptomycin sulfate (10, 30, 45, 60, and 100 μg/ml), and tetracycline (15, 30, and 100 μg/ml). Starch hydrolysis, growth in 6.5% NaCl, and incubation at 45°C did not enhance the level of separation over that achieved with antibiotic resistance. The other seven antibiotics did not increase the level of isolate separation and were not tested further.
TABLE 1.
Patterns of antibiotic resistance of fecal streptococci from known sources
| Drug and concn (μg/ml) | % Resistant isolates from each sourcea
|
|||||
|---|---|---|---|---|---|---|
| Beef cows (n = 1,398) | Chickens (n = 824) | Dairy cows (n = 728) | Deer (n = 1,245) | Humans (n = 1,579) | Waterfowl (n = 1,284) | |
| Chlortetracycline | ||||||
| 40 | 1 | 90 | 17 | 0 | 78 | 1 |
| 60 | 0 | 68 | 8 | 0 | 39 | 0 |
| Erythromycin | ||||||
| 7 | 1 | 89 | 4 | 0 | 90 | 1 |
| 15 | 0 | 77 | 0 | 0 | 76 | 0 |
| Neomycin | ||||||
| 10 | 2 | 100 | 100 | 0 | 54 | 0 |
| 40 | 0 | 98 | 88 | 0 | 4 | 0 |
| Oxytetracycline | ||||||
| 10 | 90 | 100 | 100 | 38 | 98 | 97 |
| 30 | 80 | 100 | 88 | 0 | 84 | 45 |
| 60 | 12 | 100 | 81 | 0 | 56 | 5 |
| 100 | 0 | 95 | 76 | 0 | 14 | 0 |
| Streptomycin | ||||||
| 10 | 96 | 100 | 100 | 59 | 100 | 97 |
| 30 | 73 | 96 | 96 | 2 | 96 | 79 |
| 45 | 25 | 89 | 50 | 2 | 91 | 31 |
| 60 | 3 | 73 | 20 | 0 | 82 | 12 |
| 100 | 1 | 65 | 1 | 0 | 25 | 0 |
| Tetracycline | ||||||
| 15 | 81 | 100 | 80 | 0 | 87 | 2 |
| 30 | 13 | 100 | 79 | 0 | 81 | 1 |
| 100 | 0 | 98 | 67 | 0 | 12 | 0 |
n, total number of isolates from each source.
The chicken, dairy cow, and human isolates exhibited the widest range of antibiotic resistance, while the beef cow, deer, and waterfowl isolates exhibited the narrowest (Table 1). The chicken and human isolates expressed similar patterns of resistance to all six antibiotics, but the chicken isolates were resistant to higher concentrations. The dairy cow isolates demonstrated resistance to all antibiotics except erythromycin, and resistance patterns were similar to those of chicken and human isolates. The beef cow isolates exhibited some resistance to three antibiotics (oxytetracycline, streptomycin, and tetracycline), but at low levels. The waterfowl isolates were similar to the beef cow isolates but lacked resistance to tetracycline, while the deer isolates exhibited low levels of resistance to just oxytetracycline and streptomycin.
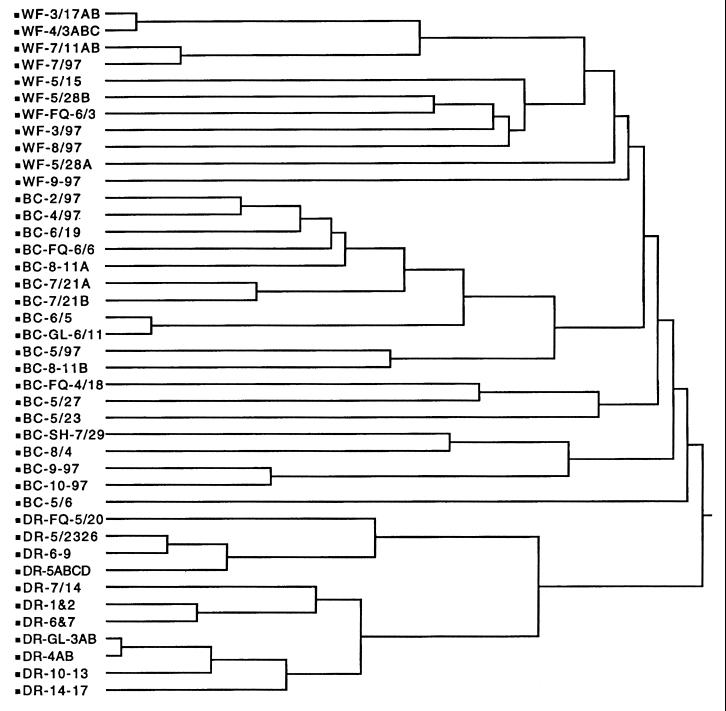
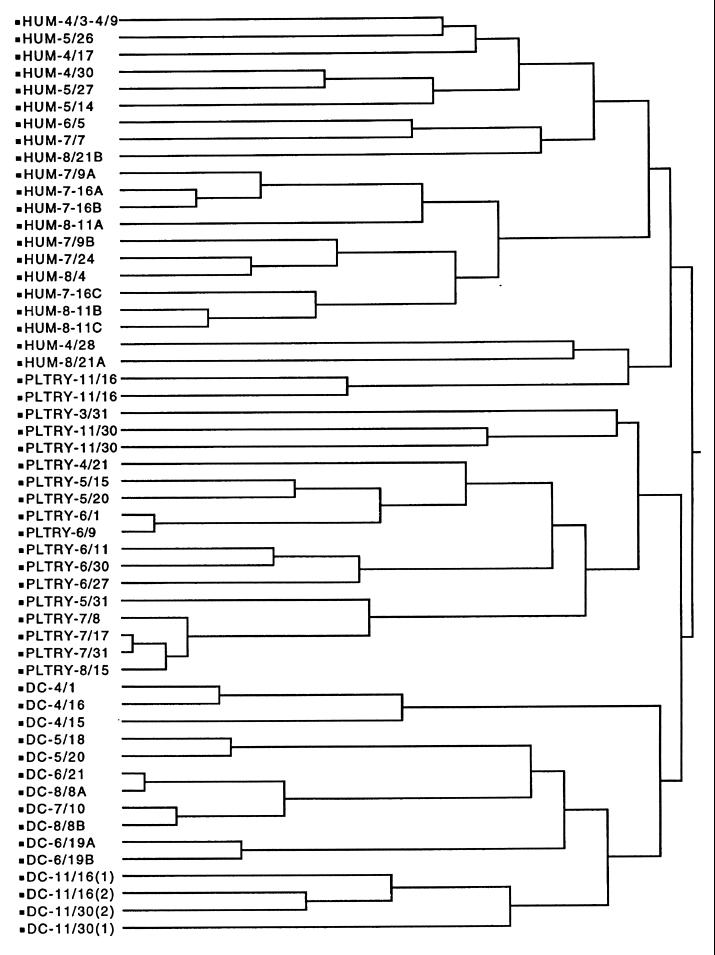
By using DA on 7,058 known isolates, the average correct classification rates varied from 85% for the chicken isolates to 93% for the human isolates (Table 2). The most common misclassifications were between human and chicken isolates and between beef cow and waterfowl isolates. CA divided the isolates into two large subclusters based on high levels of antibiotic resistance (chicken, dairy cow, and human isolates [Fig. 2]) and low levels of antibiotic resistance (beef cow, deer, and waterfowl isolates [Fig. 3]). There was excellent separation between the dairy cow and chicken isolate clusters and between the dairy cow and human isolate clusters (Fig. 2). While the chicken and human isolate clusters were separate, there was some overlap, as indicated by the small subcluster that contained isolates from both sources. This small subcluster contained the same isolates that were misclassified in Table 2. There was also excellent separation between the deer and beef cow isolate clusters and between the dairy cow and waterfowl isolate clusters (Fig. 3). While there was separation between the beef cow and wildlife isolate clusters, the wildlife isolate cluster was actually a subcluster within the larger beef cow isolate cluster. This was a reflection of the close similarity of antibiotic resistance patterns between the beef cow and wildlife isolates (Table 1) and the misclassification between the two sources (Table 2). The visual display of six distinct clusters based on source (Fig. 2 and 3) and the high rates of correct classification (Table 2) demonstrated that the database was acceptable for further validation with both known- and unknown-source isolates from a different geographical region.
TABLE 2.
Rates of correct classification (DA) for known-source database isolatesa
| Source and no. of samples | No. of isolates | No. correctly identified | Rate of correct classification (%)b | Largest source and % of misclassification |
|---|---|---|---|---|
| Beef cows, 29 | 1,398 | 1,202 | 86 | Waterfowl, 9 |
| Chickens, 17 | 824 | 700 | 85 | Humans, 7 |
| Dairy cows, 15 | 728 | 633 | 87 | Chickens, 4 |
| Deer, 26 | 1,245 | 1,045 | 84 | Beef cows, 6 |
| Humans, 33 | 1,579 | 1,468 | 93 | Chickens, 4 |
| Waterfowl, 27 | 1,284 | 1,155 | 90 | Beef cows, 7 |
| Total | 7,058 | 6,203 |
For example, the first row of data shows that 14% of the beef cow isolates were not correctly classified and that 9% were misclassified as waterfowl isolates.
The ARCC was 87%.
FIG. 2.
Dendrogram showing cluster formation from the database of known-source isolates from waterfowl (WF), beef cattle (BC), and deer (DR). The remaining descriptor for each line is a code for sampling date and location. Individual isolates with the same antibiotic resistance patterns were pooled to simplify the number of entries in the dendrogram.
FIG. 3.
Dendrogram showing cluster formation from the database of known-source isolates from humans (HUM), chickens (PLTRY), and dairy cattle (DC). The remaining descriptor for each line is a code for sampling date and location. Individual isolates with the same antibiotic resistance patterns were pooled to simplify the number of entries in the dendrogram.
Database validation: Page Brook watershed.
With DA, the ARCC for the 892 isolates from known sources within the Page Brook watershed ranged from 85% for beef cow isolates to 93% for human isolates (Table 3). There were no chicken or dairy cow sources within the watershed. There were no isolates that were not identified with a source, regardless of whether the source was correct. The most common misclassifications were between beef cow and waterfowl isolates and between deer and beef cow isolates. Near the end of the project, correct classification rates for 642 known-source isolates were lower by 5 to 7% but were still in a very acceptable range (ARCC was 82% [Table 3]). Since the most important goal was to differentiate between human and animal sources, all animal sources were pooled (Table 4). This pooling of all animal sources (with DA) improved the rates of correct classification for both the known-source database (human isolates, 96%; isolates from all animals, 98%) and the known-source isolates from the Page Brook watershed (human isolates, 95%; isolates from all animals, 96%).
TABLE 3.
ARCC for known-source isolates from Page Brook watersheda
| Sampling period (mo/yr) and source | No. of isolates | No. correctly identified | Rate of correct classification (%) | Largest source and % of misclassification |
|---|---|---|---|---|
| 10/96–11/96b | ||||
| Beef cows | 226 | 192 | 85 | Waterfowl, 8 |
| Deer | 214 | 184 | 86 | Beef cows, 7 |
| Humans | 257 | 239 | 93 | Waterfowl, 4 |
| Waterfowl | 195 | 174 | 89 | Beef cows, 5 |
| Total | 892 | 789 | ||
| 8/98–9/98c | ||||
| Beef cows | 193 | 151 | 78 | Waterfowl, 9 |
| Deer | 131 | 106 | 81 | Beef cows, 10 |
| Humans | 152 | 131 | 86 | Waterfowl, 8 |
| Waterfowl | 166 | 136 | 82 | Beef cows, 9 |
| Total | 642 | 524 |
For example, the first row of data shows that 15% of the beef cow isolates were incorrectly classified and that 8% were misclassified as waterfowl isolates. At least four different samples were collected for each known source.
Prior to watershed project. The ARCC was 88%.
Near completion of watershed project. The ARCC was 82%.
TABLE 4.
ARCC with all animal sources pooled
Over the 28 months of sampling, well samples were almost uniformly negative for fecal coliforms and fecal streptococci (data not shown). There were a few well samples that were occasionally positive for fecal coliforms, but always at low numbers of organisms (<10 CFU/100 ml). For fecal coliforms in the stream samples over the first 12 months of sampling, six sampling locations were usually negative, three were usually positive (but at relatively low levels [<100 CFU/100 ml]), and three (PB10, PB12, and PB16) were high, especially during the period from August to October 1997 (Table 5). The sampling location for PB16 was approximately 2.4 km downstream from PB10, and PB12 was roughly 0.8 km upstream of PB10. These three sites defined a 3.2-km impaired stream segment based on high fecal coliform numbers. The next sampling location upstream (PB29) of these three was positive for fecal coliforms for 15 of the 28 monthly samples but yielded <10 CFU/100 ml for 14 of the 15 positive samples.
TABLE 5.
Fecal coliform and fecal streptococcus populations from the three most contaminated sites of Page Brook watershed
| Sampling period, mo/yr (no. of samples) | Avg no. of colonies/100 ml
|
|||||
|---|---|---|---|---|---|---|
| PB10
|
PB12
|
PB16
|
||||
| Fecal coliforms | Fecal streptococci | Fecal coliforms | Fecal streptococci | Fecal coliforms | Fecal streptococci | |
| 11/96–4/97 (6) | 160 | 17 | 80 | 20 | 40 | 11 |
| 5/97–7/97 (3) | 190 | 88 | 6,015 | 428 | 185 | 5 |
| 8/97–10/97 (3) | 3,103 | 260 | 42,400 | 565 | 2,347 | 435 |
| 11/97–4/98 (6) | 211 | 56 | 497 | 44 | 53 | 21 |
| 5/98–7/98 (3) | 306 | 356 | 1,043 | 391 | 488 | 464 |
| 8/98–10/98 (3) | 347 | 381 | 1,596 | 582 | 934 | 583 |
| 11/98–2/99 (4) | 15 | 10 | 2,002 | 436 | 807 | 148 |
Sites PB10, PB12, and PB16 all involved farms where Page Brook passed through pastures that contained cattle herds with unrestricted access to the stream (Fig. 1). Both fecal coliform and fecal streptococcus populations over the first 12 months of sampling were low during cool weather (November 1996 to April 1997) and much higher during warm weather (May to July 1997 and August to October 1997), when cattle were commonly found in the stream. During cool weather in the second year of sampling (November 1997 to April 1998), cattle access to the stream was restricted by installation of fences with either in-pasture watering devices or stream access points for watering. Reducing stream access resulted in much lower fecal coliform numbers over the warm periods of the second year (May to July 1998 and August to October 1998 [Table 5]).
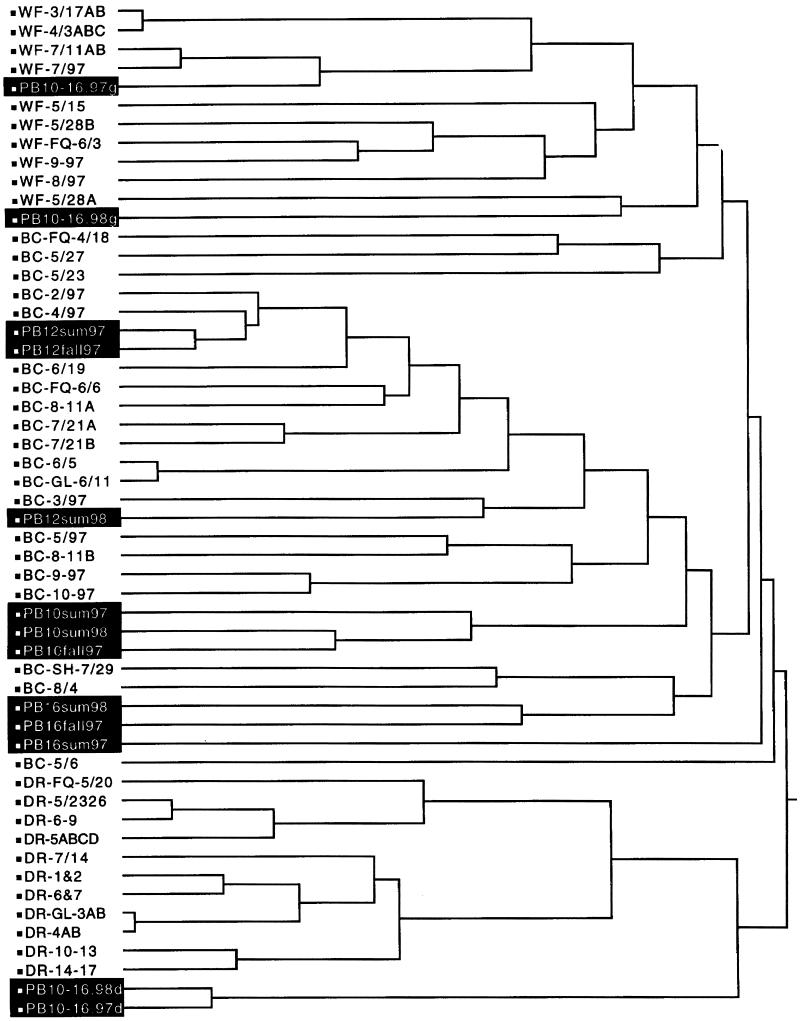
The decision to reduce cattle access to the stream was made based on source identification of the 4,615 unknown-source isolates (Table 6 and Fig. 4). No isolates were classified as coming from humans over the entire course of the study. With DA, for the combined warm seasons during the first year (May to October 1997), 78 to 86% of the fecal streptococci were identified as being from beef cattle, with the remainder divided between deer and waterfowl. With CA (May to October 1997 [Fig. 4]), all sets of unknown-source isolates were grouped in beef cow, deer, and waterfowl isolate clusters. Some isolates were placed outside of the clusters by CA, and these were tabulated and reported as unidentified isolates (Table 6). These results formed the rationale for reducing cattle access to the stream by fencing during the cool season in the second year (November 1997 to April 1998). Restricting stream access resulted in 50% (or greater) reductions in the percentage of isolates identified as being from beef cattle during the second-year warm season (May to October 1998) compared to the first-year warm season (May to October 1997 [Table 6]). With the reduction in beef cow isolates, deer and waterfowl isolates were more numerous and there appeared to be a modest increase in the percentage of unidentified isolates.
TABLE 6.
Source identification of unknown-source isolates from the three most contaminated sites of Page Brook watershed
| Sampling period (mo/yr) | Sampling site | No. of isolatesa | Source identification (%)
|
||||
|---|---|---|---|---|---|---|---|
| Beef cow | Deer | Human | Waterfowl | None (unidentified) | |||
| 5/97–10/97 | PB10 | 481 | 81 | 11 | 0 | 4 | 4 |
| PB12 | 555 | 86 | 6 | 0 | 5 | 3 | |
| PB16 | 551 | 78 | 5 | 0 | 8 | 9 | |
| 11/97–4/98 | PB10 | 304 | 58 | 16 | 0 | 18 | 8 |
| PB12 | 312 | 64 | 11 | 0 | 19 | 6 | |
| PB16 | 345 | 51 | 13 | 0 | 24 | 12 | |
| 5/98–10/98 | PB10 | 492 | 38 | 23 | 0 | 24 | 15 |
| PB12 | 469 | 44 | 19 | 0 | 26 | 11 | |
| PB16 | 480 | 37 | 25 | 0 | 21 | 17 | |
| 11/98–12/98 | PB10 | NDb | |||||
| PB12 | 321 | 48 | 13 | 0 | 21 | 18 | |
| PB16 | 305 | 33 | 21 | 0 | 22 | 24 | |
Isolates collected from monthly samples.
ND, not determined; too few isolates were recovered.
FIG. 4.
Dendrogram showing cluster formation from the Page Brook isolates (in black) within the database of known-source isolates from waterfowl (WF), beef cattle (BC), and deer (DR) (Fig. 2). The remaining descriptor for each line is a code for sampling date and location. Individual isolates with the same antibiotic resistance patterns were pooled to simplify the number of entries in the dendrogram.
Limiting cattle access to the stream also reduced the number of fecal coliforms in the second-year warm season (August to October 1998) by 88.8% (PB10), 96.2% (PB12), and 60.3% (PB16) compared to the number in the first-year warm season (August to October 1997); the highest fecal coliform counts were recorded during these warm seasons (Table 7). When the counts of fecal coliforms for the periods from May to July 1997 and May to July 1998 were compared, the reduction was found to be significant only for PB12 (82.7% reduction). Comparing the numbers for the periods from November 1997 to February 1997 and from November 1998 to February 1999 revealed a significant reduction only for PB10 (93.9% reduction).
TABLE 7.
Fecal coliform populations from the three most contaminated sites of Page Brook watershed
| Sampling site | No. of fecal coliforms/100 ml in samples taken during the indicated period (mo/yr)a
|
% Reduction between avgs | |||||
|---|---|---|---|---|---|---|---|
| Before cattle access was limited
|
After cattle access was limited
|
||||||
| Avg | Minimum | Maximum | Avg | Minimum | Maximum | ||
| 5/97–7/97 | 5/98–7/98 | ||||||
| PB10 | 190 A | 10 | 280 | 306 A | 17 | 530 | None |
| PB12 | 6,015 A | 910 | 11,120 | 1,043 B | 160 | 350 | 82.7 |
| PB16 | 185 A | 0 | 370 | 488 A | 34 | 780 | None |
| 8/97–10/97 | 8/98–10/98 | ||||||
| PB10 | 3,103 A | 410 | 4,800 | 347 B | 190 | 610 | 88.8 |
| PB12 | 42,400 A | 7,900 | 72,000 | 1,596 B | 260 | 4,100 | 96.2 |
| PB16 | 2,347 A | 610 | 4,400 | 934 B | 290 | 1,700 | 60.3 |
| 11/97–2/98 | 11/98–2/99 | ||||||
| PB10 | 245 A | 170 | 370 | 15 B | 0 | 20 | 93.9 |
| PB12 | 1,475 B | 10 | 3,400 | 2,002 B | 10 | 3,900 | None |
| PB16 | 175 A | 0 | 280 | 807 A | 10 | 2,280 | None |
Average (arithmetic mean) numbers in each row followed by different letters indicate a significant difference obtained by using Duncan’s multiple-range test (P < 0.01).
DISCUSSION
As described by Wiggins (16), antibiotic resistance patterns of isolates of fecal streptococci, analyzed with DA, was a suitable method to differentiate and identify sources of fecal pollution in water. However, the five antibiotics and concentrations evaluated by Wiggins did not provide adequate separation of isolates from known sources, so it was necessary to test a wider range of antibiotics and concentrations in order to find those that did provide levels of separation that were as high as possible. Both DA and CA were suitable statistical procedures for analyzing antibiotic resistance patterns. While either procedure could be satisfactorily used alone, the advantage of using both is mainly in the additional confidence generated when the two methods provide the same answers.
The advantage of using DA is that percentages of isolates from different sources are provided and rates of correct classification can easily be determined (Tables 2 and 3). The advantage of using CA is that it provides a dendrogram that shows how well the separations by source are occurring, and the degree of relatedness between isolates from different sources is readily apparent. DA was essential with the samples from streams where isolates from multiple sources occurred, as it was critical to know the proportions contributed by each source. However, DA cannot create a category for unidentified isolates, so those clustered as unidentified by CA were either incorrectly classified by DA or had a novel resistance pattern that CA could not resolve (Table 6). CA was especially useful in testing different antibiotics and concentrations while developing the database of known-source isolates. Dendrograms were generated with hundreds of different combinations of antibiotics and concentrations until the specific combination that clustered each set of isolates in the database within the correct source was found (Fig. 2 and 3).
The conventional tests that were evaluated made it more difficult to adequately separate isolate clusters by source and were discontinued. Growth in the presence of 6.5% NaCl is used to separate the enterococcus group (all positive) from Streptococcus bovis and Streptococcus equinus (both negative), while starch hydrolysis is associated with just S. bovis. Many isolates from all known sources were positive for both starch hydrolysis and growth in 6.5% NaCl, and this caused difficulty with CA and DA in adequately separating isolates by source.
The high rate of correct classification for the known-source isolates from Page Brook was an important result, as it meant that the larger known-source database could be successfully used with isolates from a different geographical region (Table 3). This close fit may have been a fortuitous result, since it is reasonable to expect that antibiotic resistance could vary considerably among isolates from widely different areas. Other areas could also include potential sources of pollution that were not included in the database (e.g., dogs, horses, sheep, swine, and beavers). The best approach in using antibiotic resistance profiles should always be to first test the isolate database with some known-source isolates whenever a new region is considered for determining sources. If these provide suitable levels of correct classification, then the database will not need to be altered. If suitable classification levels are not obtained, there are two options to explore. One is to alter the database by not including some of the antibiotics or concentrations in DA or CA to try to find some combination that provides acceptable levels of correct classification (this could also involve adding new antibiotics). The second option is to build a new database composed entirely of known-source isolates from the new geographical area.
For a database to be able to correctly classify bacteria in a polluted stream, accuracy of classification (precision) is important but not sufficient alone. The database must also contain enough isolates to be representative of the organism being classified. It is not really a question of a specific number of isolates needed to provide better source identification (higher ARCCs) but rather a question of “representativeness” of the database. One could have a 100% ARCC with one isolate from each known source (independent of how many antibiotics were used), but that one isolate would probably not be very representative of all the possible isolates of that type. At this point, perhaps the best approach to determine if a database is representative is to regularly add samples (groups of known-source isolates) to an existing database. If the ARCC (and/or the individual correct classifications) do not change appreciably (up or down) as new samples are added, then the library should be representative. In our experience, the database of known sources will require a few hundred isolates per source before that point is reached (Table 3).
Fecal coliform and fecal streptococcus populations in the stream samples reflected the activities of the cattle herds that had unrestricted access to Page Brook (Table 5). Cattle loafed in the stream on a regular basis during warm weather, and this resulted in the high counts obtained from August to October 1997 (Tables 5 and 7). Restricting access dramatically lowered fecal coliform counts during the unusually hot and dry conditions that occurred from August to October 1998. Average counts at two of the three sampling sites (PB10 and PB16 [Table 7]) were reduced to levels below recreational-water standards for Virginia (1,000 per 100 ml for any one sample). While it was not a goal of this project to reduce fecal coliform levels to below recreational-water standards, this appears to be achievable, especially as vegetation in riparian zones adjacent to the stream becomes more established over time. However, remaining below such standards may be difficult in rural areas like the Page Brook watershed, where large populations of resident Canada geese, deer, and other wildlife occur. Since the reductions in fecal bacteria between the two summers was primarily among isolates from cattle, the proportion of isolates from waterfowl, wildlife, and unknown sources appeared to increase (Table 6). These unclassified organisms were most likely from sources that were not included in the database (e.g., dogs, cats, horses, and sheep) rather than misclassified, since the rates of correct classification remained high throughout the study (Table 3). In the first warm season (May to October 1997, prior to fencing) the proportion of isolates from cattle was so high that those from other sources were difficult to find.
The results presented here affirmed the work by Wiggins (16), showing that antibiotic resistance patterns can be used with fecal streptococci to determine sources of fecal pollution in water. With the addition of CA, both statistical methods (CA and DA) provided reliable and reproducible results with a small-scale watershed validation test for fecal source identification. With current regulatory interest in the concept of total maximum daily loading (TMDLs) for streams, it may be possible through accurate source identification to develop TMDLs for fecal bacteria from specific sources (e.g., humans, livestock, or wildlife). Our results (detection of no human isolates) had a direct impact on water quality improvement in Page Brook, as local officials were able to focus restoration efforts on the actual sources (e.g., beef cattle) rather than on those that made no contribution to the water pollution. Many recreational, surface, and well waters test positive for fecal bacteria throughout the world, but efficient use of resources for water quality improvement needs to be based on accurate identification of the source(s) of the fecal pollution. If the procedures presented here can reliably and accurately identify and separate different fecal sources, as they appear to do, they can provide an important tool to those who are responsible for public health and environmental protection and are charged with reducing pollution, protecting public health, and improving water quality.
ACKNOWLEDGMENTS
We thank B. Wiggins for critical review of the manuscript. Thanks also are due to Alison Teetor, Natural Resource Planner for Clarke County, Va., for cooperation in the watershed project and sample collection.
This work was supported by Program 319 funding from the U.S. Environmental Protection Agency and the Virginia Water Resources Research Center.
REFERENCES
- 1.Berg D E, Akopyants N S, Kersulyte D. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol. 1994;5:13–24. [Google Scholar]
- 2.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 2a.Environmental Protection Agency. 1 September 1999. National Watershed Database 305b. [Online.] http://www.epa.gov/surf3/states/VA. [31 October 1999, last date accessed.]
- 3.Gillespie B E, Jayarao B M, Oliver S P. Identification of Streptococcus species by random polymorphic deoxyribonucleic acid fingerprinting. J Dairy Sci. 1997;80:471–476. doi: 10.3168/jds.S0022-0302(97)75959-9. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 5.Hagedorn C. Role of genetic variants in autecological research. In: Tate R L, editor. Microbial autecology: a method for environmental studies. New York, N.Y: John Wiley & Sons; 1986. pp. 61–74. [Google Scholar]
- 6.Hagedorn C. Spontaneous and intrinsic antibiotic resistance markers. In: Weaver R W, Angle J S, Botomley R S, editors. Methods of soil analysis. Part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America; 1994. pp. 575–592. [Google Scholar]
- 7.Hagedorn C. Proceedings of the National Onsite Wastewater Recycling Association Annual Conference. 1998. A method to determine sources of fecal pollution; pp. 112–116. [Google Scholar]
- 8.Kariuki S, Gilks C, Kimari J, Obanda A, Muyodi J, Waiyaki P, Hart C A. Genotype analysis of Escherichia coli strains isolated from children and chickens living in close contact. Appl Environ Microbiol. 1999;65:472–476. doi: 10.1128/aem.65.2.472-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaspar C W, Burgess J L, Knight I T, Colwell R R. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can J Microbiol. 1990;36:891–894. doi: 10.1139/m90-154. [DOI] [PubMed] [Google Scholar]
- 10.Kibbey H J, Hagedorn C, McCoy E L. Use of fecal streptococci as indicators of pollution in soil. Appl Environ Microbiol. 1978;35:711–717. doi: 10.1128/aem.35.4.711-717.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillai S D, Widmer V W, Maciorowski K G, Ricke S C. Antibiotic resistance profiles of Escherichia coli isolated from rural and urban environments. J Environ Sci Health Part A. 1997;32:1665–1675. [Google Scholar]
- 12.Simmons G M., Jr . Proceedings of the Interstate Seafoood Seminar. 1994. Potential sources for fecal coliforms in tidal inlets; pp. 49–67. [Google Scholar]
- 13.Sinton L W, Donnison A M, Hastie C M. Faecal streptococci as faecal pollution indicators: a review. I. Taxonomy and enumeration. N Z J Mar Freshw Res. 1993;27:101–115. [Google Scholar]
- 14.Sinton L W, Donnison A M, Hastie C M. Faecal streptococci as faecal pollution indicators: a review. II. Sanitary significance, survival, and use. N Z J Mar Freshw Res. 1993;27:117–137. [Google Scholar]
- 15.Tartera C, Lucena F, Jofre J. Human origin of Bacteroides fragilis bacteriophages present in the environment. Appl Environ Microbiol. 1989;55:2696–2701. doi: 10.1128/aem.55.10.2696-2701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiggins B A. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl Environ Microbiol. 1996;62:3997–4002. doi: 10.1128/aem.62.11.3997-4002.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]