Abstract
Background
Diet is a key component of a species ecological niche and plays critical roles in guiding the trajectories of evolutionary change. Previous studies suggest that dietary evolution can influence the rates and patterns of species diversification, with omnivorous (animal and plant, ‘generalist’) diets slowing down diversification compared to more restricted (‘specialist’) herbivorous and carnivorous diets. This hypothesis, here termed the “dietary macroevolutionary sink” hypothesis (DMS), predicts that transitions to omnivorous diets occur at higher rates than into any specialist diet, and omnivores are expected to have the lowest diversification rates, causing an evolutionary sink into a single type of diet. However, evidence for the DMS hypothesis remains conflicting. Here, we present the first test of the DMS hypothesis in a lineage of ectothermic tetrapods—the prolific Liolaemidae lizard radiation from South America.
Results
Ancestral reconstructions suggest that the stem ancestor was probably insectivorous. The best supported trait model is a diet-dependent speciation rate, with independent extinction rates. Herbivory has the highest net diversification rate, omnivory ranks second, and insectivory has the lowest. The extinction rate is the same for all three diet types and is much lower than the speciation rates. The highest transition rate was from omnivory to insectivory, and the lowest transition rates were between insectivory and herbivory.
Conclusions
Our findings challenge the core prediction of the DMS hypothesis that generalist diets represent an ‘evolutionary sink’. Interestingly, liolaemid lizards have rapidly and successfully proliferated across some of the world’s coldest climates (at high elevations and latitudes), where species have evolved mixed arthropod-plant (omnivore) or predominantly herbivore diets. This longstanding observation is consistent with the higher net diversification rates found in both herbivory and omnivory. Collectively, just like the evolution of viviparity has been regarded as a ‘key adaptation’ during the liolaemid radiation across cold climates, our findings suggest that transitions from insectivory to herbivory (bridged by omnivory) are likely to have played a role as an additional key adaptation underlying the exceptional diversification of these reptiles across extreme climates.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12862-022-02028-3.
Keywords: Comparative phylogeny, Dietary evolution, Net diversification rate, Specialist, Generalist, Macroevolutionary sink, Liolaemidae
Background
The evolutionary radiation of animal lineages can often be influenced by the adaptive diversification of their diets [1–5]. In fact, lineage proliferations triggered by ecological opportunity—vacant niche space—are largely determined by the advent of a novel resource, or a wide array of them, that promotes niche expansions that lead to intraspecific adaptive diversification [6–8] and ecological speciation [1, 9–11]. As these processes of diversification unfold, the emergence of new species can be facilitated by partition of niche space via transitions from generalist to specialist diets. Along this axis of dietary adaptive transitions, a range of biological components of species are also affected, including their spatial distribution, their position within trophic networks, the nature of life history trade-offs that balance energetic budgets, and their chances of persistence under rapidly changing environments [5, 9, 12–15]. Collectively, therefore, the interactions among these ecological phenomena are expected to influence processes of speciation and extinction, and thus, of diversification rates within lineages [5, 16].
A growing interest in establishing the role of dietary evolution in lineage diversification has led to the emergence of the intriguing hypothesis that omnivory (the consumption of both animals and plants) can drag species to an evolutionary sink [17, 18]. This ‘dietary macroevolutionary sink’ (DMS) hypothesis suggests that dietary specialists have an ecological advantage over omnivorous species given that the former are adapted to efficiently exploit a narrow set of resources, whereas the latter perform less efficiently across a wide range of different resources (a ‘jack of all trades is a master of none’ mechanism [17]). Consistent with this prediction, analyses performed in endotherms reveal that species with a more specialized diet, such as herbivory or insectivory, undergo significantly higher diversification rates than omnivores, in which extinction rates are higher and speciation rates lower [17, 18]. However, evolutionary transitions from a specialist to a generalist diet occur at much higher rates than in other directions [17, 18]. Therefore, this hypothesis posits that omnivory is a ‘macroevolutionary sink’ [17, 18]. Yet, evidence for the DMS hypothesis remains contested. For example, a study conducted on fish lineages revealed that omnivory is associated with the highest net diversification and transition rates [19]. In addition, the hypothesis’ core predictions oppose the classical early theory that diversification tends to transition from generalist ancestors to specialist descendants, leading niche specialists towards an evolutionary "dead end" as a result of major constrains involved in the return to an omnivorous diet [20–22].
Despite the critical role that dietary evolution plays in our understanding of the proliferation of biodiversity, the lack of studies across a wider range of lineages prevents a robust assessment of the generality of its predicted evolutionary outcomes. In fact, although reptiles (avian and non-avian) represent the most species-diverse lineage among modern tetrapods [23, 24], and their diets have been shown to be linked to processes of evolutionary radiation across contrasting environments [25–27], the DMS hypothesis remains untested among ectothermic tetrapods. Most studies on the diet of ectothermic tetrapods are limited to describing lists of items consumed by species [28–32]. Only a few studies have addressed the evolution and conservatism of trophic strategies at phylogenetic scales [25, 33, 34].
Reptiles offer ideal models to address hypotheses about the macroevolutionary links between trophic transitions and lineage diversification. These vertebrates span the whole range of the dietary spectrum, with a dominant tendency for animal consumption relative to a much lower frequency of herbivory [35]. Only a few families of lizards have strictly herbivorous species, most of which are large bodied and restricted to tropical regions [27, 36]. A remarkable exception to this ‘rule of reptilian herbivory’ is the South American lizard family Liolaemidae [25]. These lizards have rapidly diversified across a range of climates that mirror the climatic range occupied by all living lizards combined [37, 38]. As a result, liolaemids have evolved a wide range of dietary adaptations from strictly herbivores and carnivore specialists, to broadly generalists that even include cannibalism [39, 40]. Espinoza et al. [25] analyzed the recurrence and faster rate of herbivory in liolaemid lizards, revealing that these species break the ecophysiological rules of reptilian herbivory because they are small bodied and live in cool climates. Another feature of liolaemids is the variability of its three genera, with Liolaemus standing as one of nature’s most prolific adaptive radiations [37, 41, 42], to the genus Ctenoblepharys represented by a single species [43]. A recent study by Olave et al. [44], showed vast differences in diversification rates within the family’s genera, but the underlying factors of these differences remain elusive. One possibility is that some traits allow the use of resources more efficiently, triggering species diversification [45]. For instance, dietary niches might be a key factor driving species diversification [17–19], helping to explain the extreme variation in both diversification rates and dietary niches in the liolaemid family, especially if the effect of other possible traits such as habitat or parity mode is ruled out [46, 47].
In this study, we analyze a large-scale dataset spanning 185 species of the Liolaemidae family distributed across a wide range of ecological environments and climates, to address the DMS hypothesis of macroevolutionary diversification mediated by dietary transitions, and to assess the role that diet evolution has played in the diversification of this prolific lizard radiation. Therefore, if the DMS hypothesis holds in reptiles, we expect to observe a higher rate of net diversification in specialized (herbivory and insectivory) than generalist (omnivory) diets, and we predict that evolutionary transitions from a specialized to an omnivorous diet will occur at much higher rates than in other directions.
Results
Ancestral diet state reconstruction
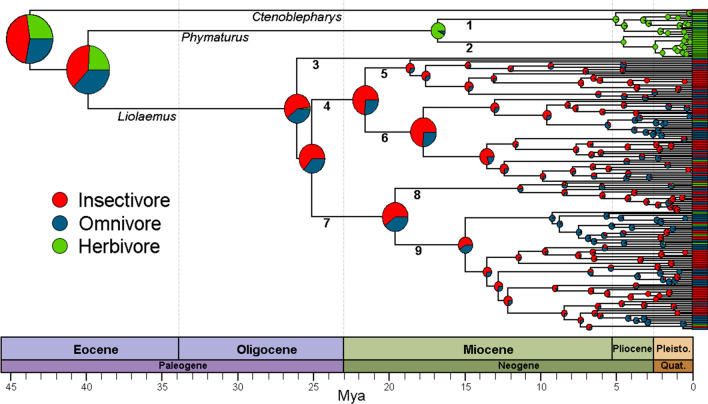
Based on trait state reconstruction (Fig. 1) with the Stochastic Character Mapping (SCM) method, the stem ancestor of the Liolaemidae family was more likely an insectivore (highest probability, p = 0.45, compared to an herbivore with p = 0.27, or an omnivore with p = 0.28). The common ancestor of Phymaturus + Liolaemus, show a high probability to have been an omnivore or insectivore (p = 0.37 and p = 0.39, respectively). Herbivory originated ~ 17 to 39 million years ago, and has remained predominantly invariable within Phymaturus. Among Liolaemus species, insectivory predominated in the earliest ancestors, herbivory has converged on nine occasions within the clade, and in all of them the insectivorous ancestor went through an omnivorous transition. Clades with both herbivory and omnivory diets increased their diversification from the Pliocene (a little more than 5 Mya) onwards.
Fig. 1.
Ancestral reconstruction of dietary diversification throughout the Liolaemidae evolutionary history (pie charts at nodes represent posterior probabilities of each diet class), averaged across 100 trees. (1) Phymaturus palluma group; (2) Phymaturus patagonicus group; (3) Liolaemus walkeri group; (4) Liolaemus subgenus; (5) Liolaemus nigromaculatus section; (6) Liolaemus chiliensis section; (7) Eulaemus subgenus; (8) Liolaemus lineomaculatus series; (9) Liolaemus montanus series.
Effect of diet type on diversification dynamics
The diversification analysis using Several Examined and Concealed States-dependent Speciation and Extinction (SecSSE) shows that the best supported model is a diet-dependent speciation rate, with independent extinction rates, no hidden states and varying transitions rates among diet types (Table 1). Under this model, herbivores have almost three times higher speciation rates (λ = 0.453) compared to insectivores (λ = 0.167), whereas omnivores have an intermediate rate (λ = 0.285). Extinction is invariable across the phylogeny, with a value of μ = 0.014. In all cases, models not including hidden states were better supported, suggesting that the effect of diet on diversification is not spurious (Table 1). Similarly, models allowing variation of transition rates among character states had better support than models assuming equal rates between character states (Table 1).
Table 1.
Model comparison for independent and dependent diet diversification, including models with hidden traits or with equal rates of transition
| Model type | Hidden trait (A/B) | Equal transition rates (Q) | Speciation rates (λi) | Extinction rates (μi) | Transition rates (Qi) | Sum of params | LogLik | AICc |
|---|---|---|---|---|---|---|---|---|
| Diet independent speciation and extinction rates | No | Yes | 1 | 1 | 1 | 3 | − 675.3 | 1356.7 |
| No | No | 1 | 1 | 6 | 8 | − 659.5 | 1335.9 | |
| Yes | Yes | 1 | 1 | 1 | 3 | − 701.2 | 1408.6 | |
| Yes | No | 1 | 1 | 18 | 20 | − 701.2 | 1447.6 | |
| Diet-dependent speciation and independent extinction rate | No | Yes | 3 | 1 | 1 | 5 | − 675.1 | 1360.5 |
| No | No | 3 | 1 | 6 | 10 | − 655.5 | 1332.2 | |
| Yes | Yes | 6 | 1 | 1 | 8 | − 857.8 | 1732.4 | |
| Yes | No | 6 | 1 | 18 | 25 | − 788.0 | 1634.2 | |
| Independent speciation and diet-dependent extinction rates | No | Yes | 1 | 3 | 1 | 5 | − 670.5 | 1351.2 |
| No | No | 1 | 3 | 6 | 10 | − 656.8 | 1335.0 | |
| Yes | Yes | 1 | 6 | 1 | 8 | − 794.2 | 1598.7 | |
| Yes | No | 1 | 6 | 18 | 25 | − 795.2 | 1648.5 | |
| Diet-dependent speciation and extinction rates | No | Yes | 3 | 3 | 1 | 7 | − 663.9 | 1342.5 |
| No | No | 3 | 3 | 6 | 12 | − 659.5 | 1344.8 | |
| Yes | Yes | 6 | 6 | 1 | 13 | − 839.1 | 1706.4 | |
| Yes | No | 6 | 6 | 18 | 30 | − 805.6 | 1683.4 |
Values in bold indicate the best model
Number of inferred rates for speciation (λ), extinction (μ) and transition (Q) are specified in each case, and LogLik and AICc values are shown
Diet-dependent speciation using MuSSE
To better account for uncertainty in parameter estimates, we repeated the best model suggested by SecSSE using a Multistate Speciation and Extinction Model (MuSSE) analysis. This model confirms previous findings that herbivores had the highest speciation rates, followed by omnivores, and insectivores had the lowest rate (Fig. 2a). As the extinction rate is the same for all three diet types (Fig. 2b), net diversification rates follow the same patterns as speciation rates, from highest in herbivores to lowest in insectivores. Differences in both speciation and diversification rates are statistically significant in both cases. Speciation Likelihood Ratio Test (LRT) (X2 = 175.21, p < 0.0001), Net diversification LRT (X2 = 452.63, p < 0.0001).
Fig. 2.
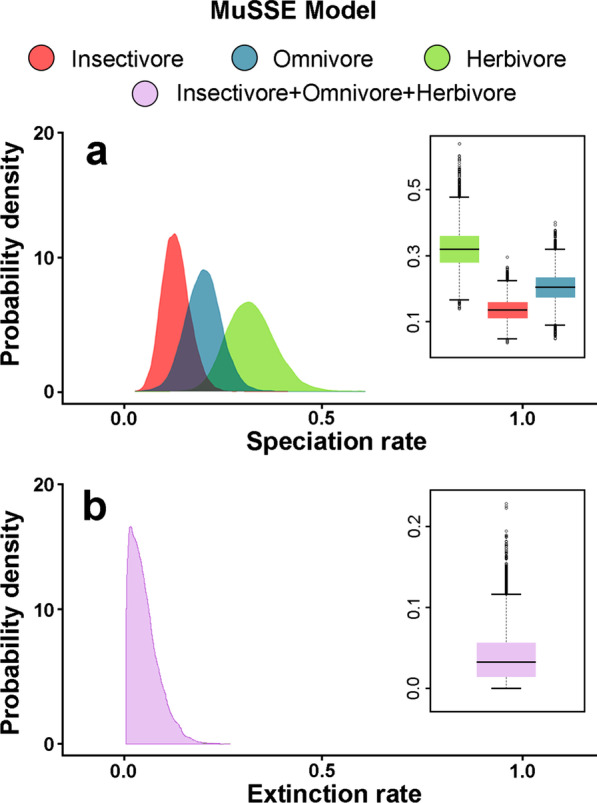
Distribution patterns of evolutionary rates across diets of the liolaemid family. Probability densities from the MuSSE model for a speciation rates associated with different dietary types, herbivory (green), omnivory (blue), and insectivory (red), and for b extinction rates across all the trees. Box plots within panels show the variation of rates in quartiles within and across diets. Different diet types show significant differences as compared with EMMs adjusted by means of the Tukey as a post-hoc test
Transition rates among diet types
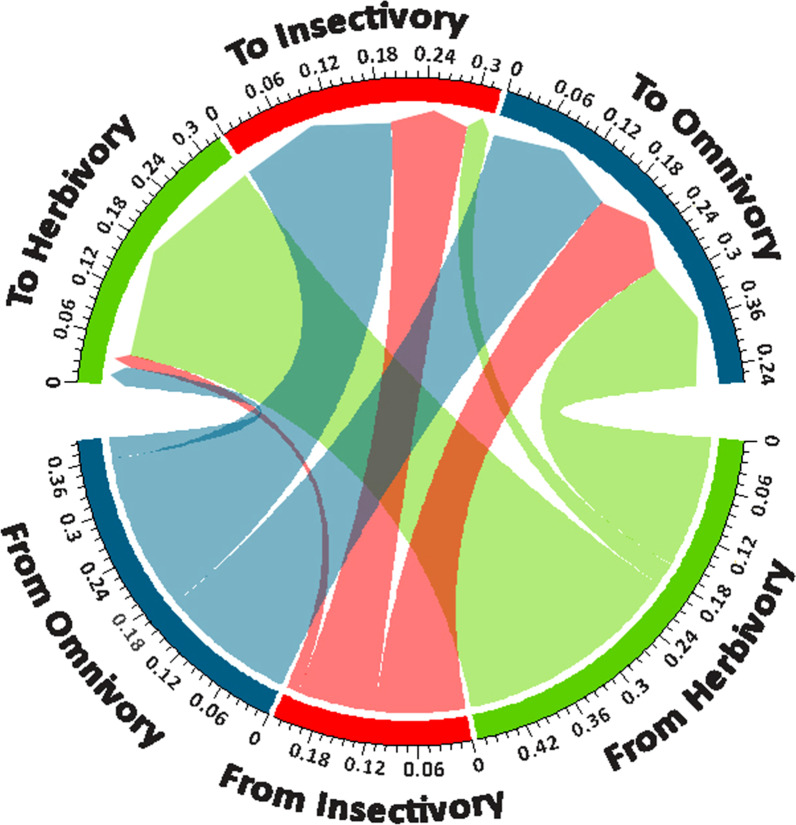
In the MuSSE model, herbivory has the highest trait conservatism and transition rate estimates to other diets (0.52—i.e., the mean value of the posterior distribution of the corresponding rate) (lower half, Fig. 3). From this diet, almost 31% of transitions (0.16) go into omnivory, and only 6% (0.03) to insectivory. The remaining 61% (0.32) are conserved, that is, lineages maintained as herbivores. In omnivory, trait conservatism and transition rates estimated to other diets is 0.43, where 46% (0.20) retain this form of diet, 46% (0.20) transition to insectivory, representing the highest rate of transition between diets; and only 7% (0.03) transition to herbivory. Insectivory has the lowest trait conservatism and transition rate (0.27) to other diets, where 51% (0.11) transition to omnivory, 39% (0.14) remain as insectivory, and only 7% (0.02) change to herbivory; this transition is the lowest among diet types. All rates in trait conservatism and dietary transitions show statistically significant differences based on the EMMs adjusted by Tukey method and the LRT from the GLM analysis (X2 = 305.5, p < 0.0001).
Fig. 3.
Trait conservatism and transition rates estimated across dietary states, where values are total cumulative rates for each diet type. The lower half of the circle shows the proportion that each diet contributes to trait conservatism (within the same diet) and transition to other diet types, and the upper half of the circle shows the contribution that each diet receives from transition events, whether is from its own dietary state (a trait conservatism process) or from other diets during all the speciation events on the phylogenetic tree
In general, more specialized diets (i.e., insectivory and herbivory) have switched more often into omnivory or remain in their own state across time than they have switched between each other (upper half, Fig. 3). Herbivory is more supported by trait conservation than by transitions with other diets. Omnivory, however, contributes more to insectivory by transition than the conservation of the insectivorous trait itself, this process has allowed the insectivorous diet to exist throughout the liolaemid evolutionary history.
Discussion
Our study provides the first test of the DMS hypothesis—that evolutionary transitions in diet influence diversification of animal lineages—performed in ectothermic tetrapods. Using the prolific Liolaemidae lizard radiation, our evidence challenges the hypothesis’s core prediction that omnivory acts as a macroevolutionary sink. In contrast with this prediction, we observed that omnivory ranks second in net diversification rate and contributes to insectivory through the highest evolutionary transitions. Although the net diversification rate in insectivores is the lowest among diet types, insectivory is present in more species than herbivory or omnivory (84, 33, 68 species, respectively). These findings challenge the preconception that traits with high net diversification rates have high species diversity [44, 48].
Our results suggest that insectivory is likely to have been promoted by omnivory, and favored by a low extinction rate. Similar patterns have been observed in birds [17] where transitions from omnivory to insectivory occur at higher rates than its own speciation and net diversification rates, resulting in the most species-rich diet. Therefore, omnivory plays an important role in the transition across specialists by acting as an evolutionary stepping-stone, rather than as a macroevolutionary sink, as predicted by the DMS hypothesis [17, 18]. Similarly, a study on fish suggested that trophic versatility can sustain high diversification rates, relax interspecific competition, and facilitate the local co-occurrence of ecologically similar species [19]. We observed this versatility to be the case in Liolaemus lizards—the second most species-rich genus of lizards in the Neotropics, and which spans all types of diets. In fact, the rapid adaptive radiation undergone by Liolaemus across a remarkable range of environments throughout South America has been suggested to be associated with their exceptional adaptive versatility in multiple components of their phenotypes [8, 37, 38, 42, 44, 49–52].
Our observation that herbivory is associated with the highest rates of speciation, aligns with the patterns of speciation rate observed by Olave et al. [44] in Phymaturus. However, in this study, high extinction rates in Phymaturus rank this group second in net diversification rate [44]. This may be related to the fact that this genus is dominated by strong niche conservatism with species sharing multiple key ecological and life history traits, including their adaptation to cold climates, their occupation of rocky outcrops, their herbivorous diets, viviparous reproductions, and extreme low fecundity [26, 46, 49, 53, 54]. Conversely, herbivory in Liolaemus unlike Phymaturus evolved convergently (nine times), and in all cases the insectivore ancestor had to undergo an omnivorous transition; thus, it is likely that the common ancestor of both Phymaturus and Liolaemus was probably an omnivore.
Herbivory in lizards
Herbivory in lizards is uncommon relative to other diet types, found in only 5% of species [55], A key explanation for such rarity is that plant-consumption provides poorly nutritious, and difficult to digest food resource [56]. For example, herbivores must be highly efficient to maintain a constant high body temperature, and form symbiotic associations with bacteria, fungi, and protozoa [25, 57–59]. Remarkably, Liolaemidae contains the highest prevalence of herbivory among living reptiles [25, 35] (33 species in this study), a dietary specialization that our results show to have the highest speciation rates. These findings may be linked to the direct effects that The Andes are believed to have exerted on the evolutionary radiation of liolaemid lizards, triggered by the vast-scale emergence of ecological opportunity that the active uplifts of the mountains provided over the past ~ 20–30 million years [26, 37, 38, 42]. Given that another feature that comes with Andean organisms is their predominantly small ranges [60], as it is the case with liolaemid lizards [26, 37, 42, 61], their evolutionary history tightly linked to the rapidly changing topography (and climate) of these mountains is likely to have led to active episodes of extinctions at the same time.
The patterns in transition rates we found between diets are contrary to the hypothesis that specialization is a “dead end” [19, 62–64]. Moreover, transition rates from specialists to omnivores are similar to the net diversification rates of specialists. In fact, the transition rate from insectivory to omnivory is greater than the net diversification rate of insectivory, which suggests that specialized diets may have a high probability of transitioning to another diet type also. Herbivory contributes more species to omnivory than insectivory, a result that was also found in mammals [18] and birds [17], and the transition rate from omnivory to insectivory is higher than to herbivory, which is similar in birds [17]. Perhaps because it is physiologically easier for an herbivore to process protein again, than for an insectivore to change the digestive tract to receive and process plant material, which is difficult to digest because of the low levels of essential nutrients [65, 66]. Plants often defend themselves with toxins, and associations with microbes or symbiotic nematodes are necessary to improve digestion [57, 58]. Diet specialization depends on the degree of interactions between intrinsic traits of individuals and ecological contexts [67], and some clades with highly specialized diets will hardly transition to another type of diet. However, with our results we show that there is a certain degree of specialization that still has the possibility of transitioning to completely opposite diets through an intermediate step.
Paleoecological history
During the mid-Eocene, southern South America recorded a more seasonal climate and open areas dominated by grasslands appeared, probably associated with a moisture gradient [68–70]. This environmental change has been proposed to be linked to a shift in species dietary composition in mammals, where insectivores switched to an herbivorous diet [69, 71]. Similarly, a detailed analysis of the content in a Eocene bird fossil revealed the presence of plant remains, concluding that this species had a facultative herbivorous diet [72]. The genus Phymaturus, which consists of almost exclusively herbivores, diverged in the late Eocene ~ 40 Mya, and it is estimated to have originated in Patagonia, and central Andes [37], an area with a habitat type that is consistent with the presence of open areas dominated by grasslands in the mid Eocene [70, 73]. For this reason, like mammals and birds, it is possible that the appearance of open areas dominated by grasslands in the Eocene triggered the transition to an herbivorous diet in Liolaemidae.
Conclusions
Our findings challenge the core prediction of the DMS hypothesis that generalist diets represent an "evolutionary sink". Unlike other groups, omnivory in liolaemids plays an important role in diversification and transition from other diets (i.e., insectivory and herbivory). Potentially as a consequence of this versatility, this family has rapidly and successfully proliferated in a variety of climates throughout its altitudinal and latitudinal range in southern South America. Therefore, just like the evolution of viviparity in liolaemids has been regarded as a "key adaptation" underlying the prolific radiation of these reptiles across cold climates, our findings suggest that transitions through omnivory are likely to have played an important role as an additional key adaptation that has facilitated their rapid evolution across such extreme environments.
Methods
Taxon sampling and phylogenetic tree
To perform phylogenetic analyses, we used the calibrated tree in Esquerré et al. [37], based on six nuclear (B1D, EXPH5, KIF24, MXRA5, PLRL, PNN) and four mitochondrial loci (cytb, 12S, ND2, ND4) as molecular markers. Their tree is based on a GTR + G for the best gene partitioning scheme and substitution model. Fossils representing the earliest record of the Eulaemus clade in the Early Miocene were used to place a mean prior on the tree height of this subgenus. The tree covers almost 70% (1 Ctenoblepharys, 188 Liolaemus, 35 Phymaturus) of the current species of Liolaemidae [23]. For more details on the time-calibrated phylogenetic tree see Esquerré et al. [37].
Diet data compilation
We compiled dietary data for 185 liolaemid species (33 herbivores, 84 insectivores, and 68 omnivores). This accounts for 55% of representativeness of the family (Additional file 1: Table S1). We based our diet data compilation on Meiri [55], which contains an extensive revision of lizard life history traits, and many of these publications include the type of diet without any other details. One exception is Espinoza et al. [25], who made one of the first classifications of diet types based on proportion of consumed items. We found this to be very helpful, however their classification of the omnivore (11–50% volumetric proportion of plant matter in the diet) and herbivore groups (70–100%) left an unclassified gap between 50 and 70% of plant matter, making the boundary between these two diet groups uncertain [37]. Subsequent studies adopted this classification adjusting omnivory to 11–50% and herbivory to > 50% [55, 74], but we believe that in this arrangement, omnivory is underestimated and herbivory is overestimated. Therefore, we considered an insectivorous species when up to 10% plant matter was found in the stomach content, omnivory is better represented between 11 and 75% of plant matter, and that a fundamental plant diet (herbivorous) is > 75% [75]. Using these criteria, a more detailed literature revision, and personal data (i.e., records of stomach contents) gathered by one of us (DPD), we found 8 species that needed to be changed from herbivore to omnivore (see details in Additional file 1: Table S1).
Phylogenetic comparative methods
Ancestral diet state reconstruction
To reconstruct ancestral diet states and evaluate their historical shifts across the evolution of Liolaemidae, we implemented Stochastic Character Mapping (SCM) [76] on the Maximum Clade Credibility (MCC) Phylogenetic Tree using the make.simmap function from the phytools package [77] within the statistical environment R [78]. SCM is a Bayesian approach that generates a posterior probability distribution, based on Maximum Likelihood (ML), of the ancestral states of diet and their transition times across the branches of the MCC tree by way of Markov Chain Monte Carlo (MCMC) [79]. Before running the SCM, we selected the best evolutionary model for character distribution across the tree by comparing three different models: (1) an equal-rates model “ER”, where a single parameter governs all transition rates, (2) a symmetric model “SYM”, where forward and reverse transitions share the same parameter, and (3) an all-rates-are different model “ARD”, where each rate is a unique parameter. Models were ran using the FitMk function from phytools [77]. Finally, we selected “SYM” as the best model according to the Akaike Information Criterion (AIC). All models were built using 100 simulated trees.
Effect of diet type on diversification dynamics
To test the influence of diet on species diversification dynamics, we compared different models of diversification using Several Examined and Concealed States-dependent Speciation and Extinction, using the SecSSE R package [47]. This method allows to fit different character-dependent speciation and extinction, where rates may vary across different character states. In addition, one can also include “hidden” traits that could influence diversification independently of the trait of interest. The values of each speciation (λ) and extinction (μ) rates are estimated simultaneously with transition rates among character states (Q) by maximum likelihood approach. In our case, we tested the hypothesis that different diets (herbivore, insectivore, omnivore) have different rates of speciation and extinction. To do so, we fitted four types of models: (i) λ and μ are independent of the diet type, (ii) λ depends on diet while μ is independent, (iii) λ is independent while μ depends on the diet type and (iv) both λ and μ can vary with diet type. These four combinations where then repeated with and without constraining Q to be equal between character states or allowing rates to vary. In addition, all models were repeated while including a hidden trait with two states (A/B), to assess whether associations between diet and diversification might be spurious. Therefore, we compared a total of 16 different models combining diet-dependent and independent λ and μ, equal or different Q and including or not hidden states. When concealed (i.e., hidden) states are included, dual transitions are set to zero, so we do not allow transitions from diet and hidden states happening simultaneously. SecSSE models also allow to account for differences in species sampling among character states. In our case, our study includes 185 species with known phylogenetic relationships and diet out of 338 species in the clade [23], with a sampling proportion (F) for each diet as Fherbivore = 0.46, Finsectivore = 0.58 and Fomnivore = 0.57. The models were set to run for 50,000 iterations and all models reached convergence. The fit of each model was compared using the AICc criteria.
Diet-dependent speciation using MuSSE
In order to better account for parameter uncertainty in the speciation and extinction rates, we repeated the best model from the SecSSE analysis using a Bayesian approach, implemented in the Multi-State Speciation and Extinction (MuSSE) models from the diversitree R package [78, 80]. We ran a model where speciation rates may vary among character states (i.e., diet types), maintaining a single extinction rate among states. The character is modeled as evolving under a constant rate Markov model of evolution (mcmc function). Models were ran using 10,000 iterations through a Markov Chain Monte Carlo (MCMC) process, using an exponential prior and a tuning parameter for the sampler (w) that was pre-calibrated with 100 Bayesian parameter estimation MCMCs. To test for differences in speciation, extinction, diversification, and transition rates among diet strategies, we used independent GLMs with a gaussian distribution and identity link function. Parameter estimates of the models were evaluated for statistical significance based on LRT. Models considered the estimated rate as the response variable and diet with three levels (insectivore, herbivore and omnivore) as the independent variable. Finally, we conducted EMMs adjusted by means of the Tukey method [81] as post-hoc comparison using the emmeans package [82].
Supplementary Information
Additional file 1: Table S1. Species, diet, and bibliography consulted.
Acknowledgements
We would like to thank Lizette Siles and two anonymous reviewers for comments that greatly improved the manuscript.
Abbreviations
- SecSSE
Several examined and concealed states-dependent speciation and extinction
- MCC
Maximum clade credibility
- DMS
Dietary macroevolutionary sink
- Mya
Million years ago
- LRT
Likelihood ratio test
- MuSSE
Multistate Speciation and Extinction Model
- MCMC
Markov Chain Monte Carlo
- SCM
Stochastic character mapping
- ML
Maximum likelihood
- ER
Equal-rates model
- SYM
Symmetric model
- ARD
All-rates-are different model
- AIC
Akaike information criterion
- GLM
General linear models
- EMMs
Estimated marginal means
Author contributions
MO and RSR designed the study. DPD contributed his own data. MO, RSR, and FS ran the analyses. MO wrote the first draft and all the authors contributed to the final version. Each author read and approved the final manuscript.
Funding
This study has no funding resources.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its Additional information files].
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grant BR, Grant PR. What Darwin’s finches can teach us about the evolutionary origin and regulation of biodiversity. Bioscience. 2003;53(10):965–975. doi: 10.1641/0006-3568(2003)053[0965:WDFCTU]2.0.CO;2. [DOI] [Google Scholar]
- 2.Losos JB. Lizards in an evolutionary tree. Ecology and adaptive radiation of anoles. Berkeley: University of California Press; 2009. [Google Scholar]
- 3.Pigot AL, Sheard C, Miller ET, Bregman TP, Freeman BG, Roll U, Seddon N, Trisos CH, Weeks BC, Tobias JA. Macroevolutionary convergence connects morphological form to ecological function in birds. Nat Ecol Evol. 2020;4(2):230–239. doi: 10.1038/s41559-019-1070-4. [DOI] [PubMed] [Google Scholar]
- 4.Price T. Speciation in birds. Colorado: Roberts & Company Publishers; 2008. [Google Scholar]
- 5.Schluter D. The ecology of adaptive radiation. New York: Oxford University Press; 2000. [Google Scholar]
- 6.Bolnick DI, Doebeli M. Sexual dimorphism and adaptive speciation: two sides of the same ecological coin. Evolution. 2003;57(11):2433–2449. doi: 10.1111/j.0014-3820.2003.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 7.Bolnick DI, Svanbäck R, Fordyce JA, Yang LH, Davis JM, Hulsey CD, Forister ML. The ecology of individuals: incidence and implications of individual specialization. Am Nat. 2003;161(1):1–28. doi: 10.1086/343878. [DOI] [PubMed] [Google Scholar]
- 8.Pincheira-Donoso D, Tregenza T, Butlin RK, Hodgson DJ. Sexes and species as rival units of niche saturation during community assembly. Glob Ecol Biogeogr. 2018;27:593–603. doi: 10.1111/geb.12722. [DOI] [Google Scholar]
- 9.Nosil P. Ecological speciation. New York: Oxford University Press; 2012. [Google Scholar]
- 10.Coyne JA, Orr HA. Speciation. Massachusetts: Sinauer Associates Sunderland; 2004. [Google Scholar]
- 11.Dumont ER, Dávalos LM, Goldberg A, Santana SE, Rex K, Voigt CC. Morphological innovation, diversification and invasion of a new adaptive zone. Proc R Soc B. 2012;279:1797–1805. doi: 10.1098/rspb.2011.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catoni C, Peters A, Schaefer HM. Life history trade-offs are influenced by the diversity, availability and interactions of dietary antioxidants. Anim Behav. 2008;76:1107–1119. doi: 10.1016/j.anbehav.2008.05.027. [DOI] [Google Scholar]
- 13.Vizueta J, Macías-Hernández N, Arnedo MA, Rozas J, Sánchez-Gracia A. Chance and predictability in evolution: the genomic basis of convergent dietary specializations in an adaptive radiation. Mol Ecol. 2019;28:4028–4045. doi: 10.1111/mec.15199. [DOI] [PubMed] [Google Scholar]
- 14.Angilletta MJ., Jr . Thermal adaptation a theoretical and empirical synthesis. New York: Oxford University Press; 2009. [Google Scholar]
- 15.Bolnick DI, Svanbäck R, Araújo MS, Persson L. Comparative support for the niche variation hypothesis that more generalized populations also are more heterogeneous. Proc Natl Acad Sci. 2007;104(24):10075–10079. doi: 10.1073/pnas.0703743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoder JB, Clancey E, Des Roches S, Eastman JM, Gentry L, Godsoe W, Hagey TJ, Jochimsen D, Oswald BP, Robertson J, et al. Ecological opportunity and the origin of adaptive radiations. J Evol Biol. 2010;23:1581–1596. doi: 10.1111/j.1420-9101.2010.02029.x. [DOI] [PubMed] [Google Scholar]
- 17.Burin G, Kissling WD, Guimaraes PR, Sekercioglu CH, Quental TB. Omnivory in birds is a macroevolutionary sink. Nat Commun. 2016;7:11250. doi: 10.1038/ncomms11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price SA, Hopkins SSB, Smith KK, Roth VL. Tempo of trophic evolution and its impact on mammalian diversification. Proc Natl Acad Sci. 2012;109(18):7008–7012. doi: 10.1073/pnas.1117133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gajdzik L, Aguilar-Medrano R, Frédérich B. Diversification and functional evolution of reef fish feeding guilds. Ecol Lett. 2019;22(4):572–582. doi: 10.1111/ele.13219. [DOI] [PubMed] [Google Scholar]
- 20.Kelley ST, Farrell BD. Is specialization a dead end? The phylogeny of host use in Dendroctonus bark beetles (Scolytidae) Evolution. 1998;52(6):1731–1743. doi: 10.1111/j.1558-5646.1998.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 21.Simpson GG. The major features of evolution. New York: Columbia University Press; 1953. [Google Scholar]
- 22.Cope ED. The primary factors of organic evolution. Chicago: The Open Court Publishing Company; 1896. [Google Scholar]
- 23.Uetz P, Freed P, Hošek J: The reptile database. In., Nov/06/2021 edn; 2022.
- 24.Roll U, Feldman A, Novosolov M, Allison A, Bauer AM, Bernard R, Böhm M, Castro-Herrera F, Chirio L, Collen B, et al. The global distribution of tetrapods reveals a need for targeted reptile conservation. Nat Ecol Evol. 2017 doi: 10.1038/s41559-017-0332-2:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Espinoza RE, Wiens JJ, Tracy CR. Recurrent evolution of herbivory in small, cold-climate lizards: breaking the ecophysiological rules of reptilian herbivory. Proc Natl Acad Sci. 2004;101(48):16819–16824. doi: 10.1073/pnas.0401226101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reaney AM, Saldarriaga-Córdoba M, Pincheira-Donoso D. Macroevolutionary diversification with limited niche disparity in a species-rich lineage of cold-climate lizards. BMC Evol Biol. 2018;18:1–12. doi: 10.1186/s12862-018-1133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iverson JB. Adaptations to herbivory in iguanine lizards. In: Burghardt GM, Rand AS, editors. Iguanas of the world their behavior, ecology, and conservation. New Jersey: Noyes Publications; 1982. pp. 60–76. [Google Scholar]
- 28.Olivera-Jara DA, Aguilar CA. Dieta de la lagartija neotropical Liolaemus polystictus (Squamata: Liolaemidae) de los andes centrales, Huancavelica, Perú. Rev Peru Biol. 2020;27(3):339–348. doi: 10.15381/rpb.v27i3.18680. [DOI] [Google Scholar]
- 29.Castillo GN, Acosta JC, Blanco GM. Trophic analysis and parasitological aspects of Liolaemus parvus (Iguania: Liolaemidae) in the Central Andes of Argentina. Turk J Zool. 2019;43:277–286. doi: 10.3906/zoo-1812-33. [DOI] [Google Scholar]
- 30.Toyama KS, Junes K, Ruiz J, Mendoza A, Pérez JM. Ontogenetic changes in the diet and head morphology of an omnivorous Tropidurid lizard (Microlophus thoracicus) Zoology. 2018;129:45–53. doi: 10.1016/j.zool.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan S, Wolf AJ, Hellgren EC, Moody RW, Bogosian V., II Diet selection by a lizard ant-specialist in an urban system bereft of preferred prey. J Herpetol. 2018;52(1):79–85. doi: 10.1670/16-072. [DOI] [Google Scholar]
- 32.Gallardo GA, Barrionuevo MJ, Scrocchi GJ. Dieta de la lagartija arenícola Liolaemus laurenti (Sauria: Liolaemidae) en un bioma de desierto de Argentina. Cuadernos de Herpetología. 2018;32(1):61–66. doi: 10.31017/CdH.2018.(2017-29). [DOI] [Google Scholar]
- 33.Cruz FB, Antenucci D, Luna F, Abdala CS, Vega LE. Energetics in Liolaemini lizards: implications of a small body size and ecological conservatism. J Comp Physiol B. 2011;181:373–382. doi: 10.1007/s00360-010-0524-4. [DOI] [PubMed] [Google Scholar]
- 34.Román-Palacios C, Scholl JP, Wiens JJ. Evolution of diet across the animal tree of life. Evol Lett. 2019 doi: 10.1002/evl3.127:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitt LJ. Shifting paradigms: Herbivory and body size in lizards. Proc Natl Acad Sci. 2004;101(48):16713–16714. doi: 10.1073/pnas.0407439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clauss M, Steuer P, Müller DWH, Codron D, Hummel J. Herbivory and body size: allometries of diet quality and gastrointestinal physiology, and implications for herbivore ecology and dinosaur gigantism. PLoS ONE. 2013;8(10):e68714. doi: 10.1371/journal.pone.0068714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esquerré D, G. BI, Catullo RA, Torres-Pérez F, Keogh JS. How mountains shape biodiversity: the role of the Andes in biogeography, diversification, and reproductive biology in South America’s most species-rich lizard radiation (Squamata: Liolaemidae). Evolution 2019, 73(2):214–230. [DOI] [PubMed]
- 38.Pincheira-Donoso D, Harvey LP, Ruta M. What defines an adaptive radiation? Macroevolutionary diversification dynamics of an exceptionally species-rich continental lizard radiation. BMC Evol Biol. 2015;15:1–13. doi: 10.1186/s12862-015-0435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiménez-Robles O, De la Riva I. Cannibalism in the Andean lizard Liolaemus orientalis. Stud Neotrop Fauna Environ. 2017;52(3):244–247. doi: 10.1080/01650521.2017.1356051. [DOI] [Google Scholar]
- 40.Pincheira-Donoso D. Intraspecific predation in the Liolaemus lizard radiation: a primer. Anim Biol. 2012;62:227–287. doi: 10.1163/157075611X618219. [DOI] [Google Scholar]
- 41.Pincheira-Donoso D, Bauer AM, Meiri S, Uetz P. Global taxonomic diversity of living reptiles. PLoS ONE. 2013;8(3):1–10. doi: 10.1371/journal.pone.0059741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pincheira-Donoso D, Tregenza T, Witt MJ, Hodgson DJ. The evolution of viviparity opens opportunities for lizard radiation but drives it into a climatic cul-de-sac. Glob Ecol Biogeogr. 2013;22:857–867. doi: 10.1111/geb.12052. [DOI] [Google Scholar]
- 43.Pincheira-Donoso D, Scolaro JA, Sura P. A monographic catalogue on the systematics and phylogeny of the South American iguanian lizard family Liolaemidae (Squamata, Iguania) Zootaxa. 2008;1800:1–85. doi: 10.11646/zootaxa.1800.1.1. [DOI] [Google Scholar]
- 44.Olave M, Gonzáles Marín A, Avila LJ, Sites JW., Jr . Disparate patterns of diversification within liolaemini lizards. In: Valentín R, Carnaval AC, editors. Neotropical diversification: patterns and processes. Cham, Switzerland: Springer Nature Switzerland; 2020. pp. 765–790. [Google Scholar]
- 45.Losos JB. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am Nat. 2010;175(6):623–639. doi: 10.1086/652433. [DOI] [PubMed] [Google Scholar]
- 46.Olave M, Avila LJ, Sites JW, Morando M. How important is it to consider lineage diversification heterogeneity in macroevolutionary studies? Lessons from the lizard family Liolaemidae. J Biogeogr. 2020;47:1286–1297. doi: 10.1111/jbi.13807. [DOI] [Google Scholar]
- 47.Herrera-Alsina L, van Els P, Etienne RS. Detecting the dependence of diversification on multiple traits from phylogenetic trees and trait data. Syst Biol. 2019;68(2):317–328. doi: 10.1093/sysbio/syy057. [DOI] [PubMed] [Google Scholar]
- 48.Rabosky DL. Ecological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol Lett. 2009;12:735–743. doi: 10.1111/j.1461-0248.2009.01333.x. [DOI] [PubMed] [Google Scholar]
- 49.Abdala CS, Quinteros AS. Los últimos 30 años de estudios de la familia de lagartijas más diversa de Argentina. Actualización taxonómica y sistemática de Liolaemidae. Cuadernos Herpetol. 2014;28(2):55–82. [Google Scholar]
- 50.Pincheira-Donoso D, Hodgson DJ, Tregenza T. The evolution of body size under environmental gradients in ectotherms: why should Bergmann's rule apply to lizards? BMC Evol Biol. 2008;8:1–13. doi: 10.1186/1471-2148-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pincheira-Donoso D, Jara M, Reaney A, García-Roa R, Saldarriaga-Córdoba M, Hodgson DJ. Hypoxia and hypothermia as rival agents of selection driving the evolution of viviparity in lizards. Glob Ecol Biogeogr. 2017;26(11):1238–1246. doi: 10.1111/geb.12626. [DOI] [Google Scholar]
- 52.Schulte JA, II, Macey JR, Espinoza RE, Larson A. Phylogenetic relationships in the iguanid lizard genus Liolaemus: multiple origins of viviparous reproduction and evidence for recurring Andean vicariance and dispersal. Biol J Lin Soc. 2000;69:75–102. doi: 10.1006/bijl.1999.0346. [DOI] [Google Scholar]
- 53.Boretto JM, Cabezas-Cartes F, Ibargüengoytía NR. Slow life histories in lizards living in the highlands of the Andes Mountains. J Comp Physiol B. 2017;188(3):491–503. doi: 10.1007/s00360-017-1136-z. [DOI] [PubMed] [Google Scholar]
- 54.Scolaro JA, Ibargüengoytía NR, Pincheira-Donoso D. When starvation challenges the tradition of niche conservatism: On a new species of the saxicolous genus Phymaturus from Patagonia Argentina with pseudoarboreal foraging behaviour (Iguania, Liolaemidae) Zootaxa. 2008;1786:48–60. doi: 10.11646/zootaxa.1786.1.4. [DOI] [Google Scholar]
- 55.Meiri S. Traits of lizards of the world: variation around a successful evolutionary design. Glob Ecol Biogeogr. 2018;27(10):1168–1172. doi: 10.1111/geb.12773. [DOI] [Google Scholar]
- 56.Scharf I, Feldman A, Novosolov M, Pincheira-Donoso D, Das I, Böhm M, Uetz P, Torres-Carvajal O, Bauer A, Roll U, et al. Late bloomers and baby boomers: ecological drivers of longevity in squamates and the tuatara. Glob Ecol Biogeogr. 2015;24:396–405. doi: 10.1111/geb.12244. [DOI] [Google Scholar]
- 57.Carothers JH, Jaksic FM. Parasite loads and altitudinal distribution of Liolaemus lizards in the central Chilean Andes. Rev Chil Hist Nat. 2001;74:681–686. [Google Scholar]
- 58.Kohl KD, Brun A, Magallanes M, Brinkerhoff J, Laspiur A, Acosta JC, Bordenstein SR, Caviedes-Vidal E. Physiological and microbial adjustments to diet quality permit facultative herbivory in an omnivorous lizard. J Exp Biol. 2016;219:1903–1912. doi: 10.1242/jeb.138370. [DOI] [PubMed] [Google Scholar]
- 59.Pough FH. Lizard energetics and diet. Ecology. 1973;54(4):837–844. doi: 10.2307/1935678. [DOI] [Google Scholar]
- 60.Meiri S, Bauer A, Allison A, Castro-Herrera F, Chirio L, Colli G, Das I, Doan TM, Glaw F, Grismer LL, et al. Extinct, obscure or imaginary: the lizard species with the smallest ranges. Divers Distrib. 2018;24:262–273. doi: 10.1111/ddi.12678. [DOI] [Google Scholar]
- 61.Pincheira-Donoso D. Predictable variation of range-sizes across an extreme environmental gradient in a lizard adaptive radiation: evolutionary and ecological inferences. PLoS ONE. 2011;6(12):1–8. doi: 10.1371/journal.pone.0028942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Day EH, Hua X, Bromham L. Is specialization an evolutionary dead end? Testing for differences in speciation, extinction and trait transition rates across diverse phylogenies of specialists and generalists. J Evol Biol. 2016;29:1257–1267. doi: 10.1111/jeb.12867. [DOI] [PubMed] [Google Scholar]
- 63.Rojas D, Ramos Pereira MJ, Fonseca C, Dávalos LM. Eating down the food chain: generalism is not an evolutionary dead end for herbivores. Ecol Lett. 2018;21:402–410. doi: 10.1111/ele.12911. [DOI] [PubMed] [Google Scholar]
- 64.Nosil P, Mooers AØ. Testing hypotheses about ecological specialization using phylogenetic trees. Evolution. 2005;59(10):2256–2263. doi: 10.1111/j.0014-3820.2005.tb00933.x. [DOI] [PubMed] [Google Scholar]
- 65.Pincheira-Donoso D. Correlated evolution between herbivory and gastrointestinal tract in a prolific lizard adaptive radiation. Anim Biol. 2021;71:233–241. doi: 10.1163/15707563-bja10051. [DOI] [Google Scholar]
- 66.White TCR. The significance of unripe seeds and animal tissues in the protein nutrition of herbivores. Biol Rev. 2011;86:217–224. doi: 10.1111/j.1469-185X.2010.00143.x. [DOI] [PubMed] [Google Scholar]
- 67.Mori T, Nakata S, Izumiyama S. Dietary specialization depending on ecological context and sexual differences in Asiatic black bears. PLoS ONE. 2019;14(10):e0223911. doi: 10.1371/journal.pone.0223911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Romero EJ. Paleogene Phytogeography and Climatology of South America. Ann Mo Bot Gard. 1986;73(2):449–461. doi: 10.2307/2399123. [DOI] [Google Scholar]
- 69.Ortiz-Jaureguizar E, Cladera GA. Paleoenvironmental evolution of southern South America during the Cenozoic. J Arid Environ. 2006;66:498–532. doi: 10.1016/j.jaridenv.2006.01.007. [DOI] [Google Scholar]
- 70.Bellosi E, Genise JF, Zucol A, Bond M, Kramarz A, Sánchez MV, Krause M. Diverse evidence for grasslands since the Eocene in Patagonia. J S Am Earth Sci. 2021;108:103357. doi: 10.1016/j.jsames.2021.103357. [DOI] [Google Scholar]
- 71.Pascual R, Ortiz-Jaureguizar E, Prado JL. Land mammals: paradigm of Cenozoic South American geobiotic evolution. In: Arratia G, editor. Contribution of Southern South America to Vertebrate Paleontology. vol. 30. Müncher: Müncher Geowissenschaftliche Abhandlungen (A); 1996: 265–319.
- 72.Mayr G, Richter G. Exceptionally preserved plant parenchyma in the digestive tract indicates a herbivorous diet in the Middle Eocene bird Strigogyps sapea (Ameghinornithidae) Palaeontol Zeitschrift. 2011;85:303–307. doi: 10.1007/s12542-010-0094-5. [DOI] [Google Scholar]
- 73.Debandi G, Corbalán V, Scolaro JA, Roig-Juñent SA. Predicting the environmental niche of the genus Phymaturus: are palluma and patagonicus groups ecologically differentiated? Austral Ecol. 2011;37(3):392–400. doi: 10.1111/j.1442-9993.2011.02295.x. [DOI] [Google Scholar]
- 74.Semhan RV, Halloy M, Abdala CS. Diet and reproductive states in a high altitude neotropical lizard, Liolaemus crepuscularis (Iguania: Liolaemidae) S Am J Herpetol. 2013;8(2):102–108. doi: 10.2994/SAJH.D.12-00029.1. [DOI] [Google Scholar]
- 75.Montori A. Alimentación de los adultos de Euproctus asper (Dugés 1852) en la montaña media del prepirineo catalan (españa) Revista Española Herpetol. 1991;5:23–36. [Google Scholar]
- 76.Bollback JP. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinform. 2006;7:88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Revell LJ. Application phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 78.R Core Team: R: a language and environment for statistical computing. In., 4.0.3 edn. Vienna, Austria: R Foundation for Statistical Computing; 2020: "Bunny-Wunnies Freak Out".
- 79.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52(2):131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 80.FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–1092. doi: 10.1111/j.2041-210X.2012.00234.x. [DOI] [Google Scholar]
- 81.Lenth RV, Buerkner P, Herve M, Love J, Riebl H, Singmann H. Estimated Marginal Means, aka Least-Squares Means. In., 1.6.1 edn: CRAN R; 2021: obtain estimated marginal means (EMMs) for many linear, generalized linear, and mixed models. Compute contrasts or linear functions of EMMs, trends, and comparisons of slopes. Plots and other displays. Least-squares means are discussed, and the term ``estimated marginal means'' is suggested, in Searle, Speed, and Milliken (1980) Population marginal means in the linear model: an alternative to least squares means. Am Stat. 2021; 1934(1984): 1216–1221.
- 82.Searle SF, Speed FM, Milliken GA. Population marginal means in the linear model: an alternative to least squares means. Am Stat. 1980;34(4):216–221. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Species, diet, and bibliography consulted.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its Additional information files].