Abstract
Determination of the role of methanogenic bacteria in an anaerobic ecosystem often requires quantitation of the organisms. Because of the extreme oxygen sensitivity of these organisms and the inherent limitations of cultural techniques, an accurate biomass value is very difficult to obtain. We standardized a simple method for estimating methanogen biomass in a variety of environmental matrices. In this procedure we used the thiol biomarker coenzyme M (CoM) (2-mercaptoethanesulfonic acid), which is known to be present in all methanogenic bacteria. A high-performance liquid chromatography-based method for detecting thiols in pore water (A. Vairavamurthy and M. Mopper, Anal. Chim. Acta 78:363–370, 1990) was modified in order to quantify CoM in pure cultures, sediments, and sewage water samples. The identity of the CoM derivative was verified by using liquid chromatography-mass spectroscopy. The assay was linear for CoM amounts ranging from 2 to 2,000 pmol, and the detection limit was 2 pmol of CoM/ml of sample. CoM was not adsorbed to sediments. The methanogens tested contained an average of 19.5 nmol of CoM/mg of protein and 0.39 ± 0.07 fmol of CoM/cell. Environmental samples contained an average of 0.41 ± 0.17 fmol/cell based on most-probable-number estimates. CoM was extracted by using 1% tri-(N)-butylphosphine in isopropanol. More than 90% of the CoM was recovered from pure cultures and environmental samples. We observed no interference from sediments in the CoM recovery process, and the method could be completed aerobically within 3 h. Freezing sediment samples resulted in 46 to 83% decreases in the amounts of detectable CoM, whereas freezing had no effect on the amounts of CoM determined in pure cultures. The method described here provides a quick and relatively simple way to estimate methanogenic biomass.
Methanogenesis occurs in a wide variety of anaerobic habitats, including wetlands, lake sediments, animal gastrointestinal tracts, and geothermally heated environments. The optimal temperatures for methanogenesis vary from 2°C to more than 100°C. Most methanogens can use H2 or acetate as a source of electrons for methanogenesis. In this respect, these organisms carry out the terminal step in anaerobic decomposition (41).
Thus, methanogens play an essential role in the mineralization of organic material (9, 15) due to the energetics of electron flow (10). They are directly involved in the treatment of wastewater, sewage, and solid wastes (19, 23, 24, 28–30, 32).
In ecological and environmental studies a quick, easy, reliable method for accurately estimating methanogenic biomass is often necessary. Methanogens possess several potential biomarkers, including coenzyme F420, isoprenoids, and lipid ethers in the cell membranes, as well as coenzyme M (CoM). While coenzyme F420 has been considered a molecule which could be used for biomass estimation, it suffers from the following two limitations: (i) the fact that intracellular concentrations vary up to 25-fold (14, 21, 40) and (ii) the fact that it has been found in other archaebacteria (14, 21). Quantification of unique methanogen lipids requires labor-intensive extraction and analysis procedures. CoM, one of the few naturally occurring sulfonic acids (38), was discovered (22) and identified in the mid-1970s (36). With the exception of an aerobic alkene-degrading bacterium (3), CoM is found only in methanogens; the concentration of CoM varies only by a factor of 5 in different species (5), and standard material is commercially available. CoM was recently reported to be present in Xanthobacter strains (3) participating in a highly specialized alkene oxidation pathway. While this finding indicates that CoM is not present only in methanogens, it is unlikely that the CoM present in Xanthobacter strains is quantitatively important in natural samples obtained from anaerobic environments.
The previously developed techniques for assaying CoM include a bioassay (5) and a high-performance liquid chromatography (HPLC)-based method which measures fluorescent isoindole derivatives of thiols (25). The latter method was not standardized for biomass determination. Both of these methods are cumbersome and time-consuming, require strictly anaerobic conditions, and were not designed for use with sediments.
In this study, we modified the HPLC-based procedure (25) so that we could quantify CoM within hours of sample collection without a requirement for anoxic conditions. We also standardized the technique with pure cultures so that it could be used to quantify methanogen biomass in a variety of environmental matrices.
(Some of the results have been presented previously [14a].)
MATERIALS AND METHODS
Growth of methanogens.
Methanococcus thermolithotrophicus DSM 2095 and Methanococcus voltae DSM 1537 were grown as previously described (34, 39). Methanobacterium thermoautotrophicum Marburg (= DSM 2133), Methanospirillum hungatei GP1 (= DSM 1101), and Methanosarcina barkeri Fusaro (= DSM 804) were cultured as described by Daniels et al. (13). Methanosarcina barkeri was grown on N2-CO2-methanol, H2-CO2, and N2-CO2-acetate. Methanolobus tindarius DSM 2278 was grown as previously described (20) by using 0.4% methanol added from a sterile stock solution. Methanosaeta concilli DSM 3671 was grown as described by Patel (30). The most-probable-number (MPN) assays used were three-tube assays to a 10−8 dilution. The media used for MPN assays performed with pure methanogenic cultures were the media described above for the organisms. For MPN assays performed with sediment samples (1), the medium used was medium 1 prepared as previously described (4). Direct cell counts were obtained by using a hemocytometer. All experiments were performed twice in duplicate. The data are presented as averages with standard deviations where appropriate.
Methanogen cultures (100 ml) were grown to the late log phase, harvested by centrifugation at 2,700 × g for 20 min at 5 to 10°C, and resuspended in 10 ml of sterile medium. To determine levels of CoM over time, organisms were grown in 2-liter batch cultures, and 100-ml portions of the cultures were harvested at intervals. The protein contents of pure cultures were determined by using the bicinchoninic acid protein assay (Pierce Chemical Co.).
HPLC equipment and solvent system.
A Beckman model 157 fluorescence detector (equipped with a 338-nm excitation filter and a 450-nm emission filter) was used to quantitate CoM. The HPLC analysis was performed by using an Econosphere C18 column. The mobile phase was 50 mM sodium acetate buffer (pH 5.7)–acetonitrile (70:30) flowing at a rate of 1 ml/min with a 20-μl injection loop. The isoindole derivative of CoM (calculated mass, 301 Da) was identified by using a liquid chromatograph-mass spectrophotometer (LC-MS) (Hewlett-Packard Series 1100) equipped with an Econosphere C18 column. The mobile phase was 10 mM ammonium acetate buffer (pH 5.7)–acetonitrile (70:30) flowing at a rate of 1 ml/min. Both a standard containing 100 μM CoM and an extract of hydrocarbon-contaminated sediment were tested and analyzed in the electrospray-negative mode for the mass spectrophotometer and scanned from 100 to 500 mass units. The retention times for CoM with both solvent systems were determined by using synthetic CoM.
Analysis of CoM and samples.
All reagents were purchased from Sigma, and standards and stock solutions were prepared fresh daily. A 1 mM CoM stock solution was prepared by dissolving CoM in 50 mM acetate–1 mM EDTA buffer (pH 4.5). All dilutions were prepared with 1% tri-(N)-butylphosphine in 2-propanol (1% TBP). The reagents used for derivatization of CoM were 20 μl of o-phthalaldehyde (20 mg/ml of methanol) per ml of sample and 20 μl of ethanolamine (20 μl/ml of boric acid buffer [pH 9.0]) per ml of sample. Standards were allowed to derivatize aerobically for 5 min at room temperature.
A variety of thiols were tested to determine whether common thiols interfered with quantification of CoM. Each thiol was prepared as a 1 mM stock solution and diluted with 1% TBP prior to derivatization.
The linearity of the assay was determined by diluting the 1 mM CoM stock solution with 1% TBP. The concentrations of the resulting standards ranged from 0.1 to 100 μM, which corresponded to 2 to 2,000 pmol of CoM injected.
To extract CoM from cells, 0.5 ml of cells was mixed with 1 ml of 1% TBP, and the preparation was incubated in Eppendorf tubes for 1 h at room temperature. Samples were then centrifuged at 12,000 × g for 2 min. The supernatant (1 ml) was derivatized as described above and was analyzed by HPLC.
The effect of freezing on the extractability of CoM was determined by using cultures of Methanosarcina barkeri (acetate grown), Methanococcus thermolithotrophicus, Methanococcus voltae, and Methanobacterium thermoautotrophicum Marburg. Cultures were harvested, and aliquots were immediately assayed to determine their CoM contents as described above or were stored frozen at −60°C for 3 days in Eppendorf tubes. After 3 days, samples were thawed at room temperature and assayed as described above.
A variety of environmental matrices were sampled and assayed to determine their CoM contents. These matrices included sediments from a landfill leachate-contaminated aquifer (2, 7, 8) a hydrocarbon-contaminated aquifer (6, 11, 12, 35), and a campus pond, as well as sewage sludge. To analyze environmental samples, 5 g was mixed with 2 ml of 1% TBP and incubated for 1 h at room temperature to extract the CoM.
To maximize solvent recovery, the sediment was weighed and placed into a 10-ml syringe with its plunger removed and a piece of Whatman filter paper placed in the bottom of the syringe barrel. The syringe was set into a hole cut in the lid of a capped 50-ml centrifuge tube (Nalgene). After centrifugation (27,000 × g), the syringe and sediment were discarded, and 1 ml of the supernatant in the centrifuge tube was assayed as described above.
Adsorption of CoM to sediments was tested by adding 100 μl of a 0.1 to 50 μM CoM solution to 2 g of sediment that previously had been determined to contain no detectable CoM. The CoM-spiked sediment was incubated aerobically at room temperature for 3 h, and this was followed by extraction and derivatization as described above.
To determine the completeness of CoM extraction, environmental samples were subjected to an additional extraction procedure. Sediment samples from the leachate-contaminated aquifer and pond sediment were assayed to determine their CoM contents, and then they were reextracted and assayed again.
The effect of freezing on CoM extractability in environmental samples was determined. Landfill aquifer sediment, hydrocarbon-contaminated sediment, pond sediment, and sewage sludge (5 g each) were all analyzed, and an additional 5 g of each was set aside and frozen at −60°C for 3 days in a stoppered 10-ml syringe. After 3 days, samples were thawed at room temperature, and the amount of CoM was determined.
RESULTS
Assay development and validation.
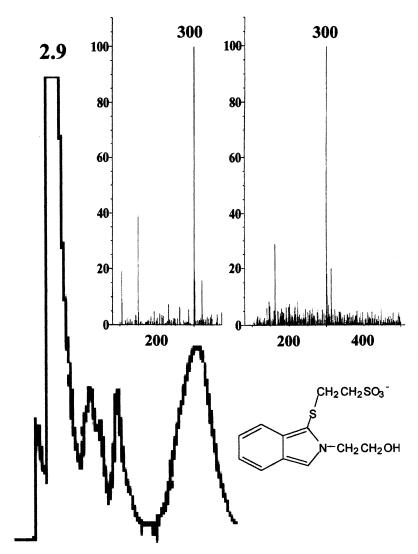
Using the HPLC method resulted in an isolated peak at approximately 3.6 min when the sample was dissolved in water and at 2.9 min when the sample was dissolved in isopropanol. The peak was confirmed to be the isoindole derivative of CoM by mass spectroscopy (Fig. 1). The detection limit was 2 pmol, as determined by an analysis of standards. With both pure methanogen cultures and environmental samples several other peaks were produced. However, it was not difficult to resolve the CoM peak. All of the other thiols tested eluted at different times than CoM (Table 1). The assay proved to be linear with synthetic CoM (2 to 2,000 pmol of CoM), pure methanogen cultures (0.5 to 5.0 ml), and environmental samples (0.5 to 5.0 g). Synthetic CoM added to environmental samples was almost completely recovered, and the amount of CoM mirrored the synthetic CoM regression line, indicating that there was no interference from sediments and that extracellular CoM did not adsorb to sediments.
FIG. 1.
HPLC chromatogram of derivatized CoM (retention time, 2.9 min) from the hydrocarbon-contaminated aquifer. The buffer system used for both HPLC and LC-MS was ammonium acetate-acetonitrile (70:30). (Inset) LC-MS profile of the isoindole derivative of standard CoM (left panel) and the corresponding profile of the environmental sample (right panel). Both spectra show that the ionic molecular weight was 300, which is consistent with the structure of the fluorescent CoM derivative shown.
TABLE 1.
Retention times of various thiols when the HPLC procedure is used
| Compound | Retention time (min) |
|---|---|
| Sulfide | 2.75 |
| CoM | 2.90 |
| Thiophenol | 3.40 |
| 1-Octanethiol | 3.46 |
| Thioglycolate | 3.90 |
| Thiosulfate | 3.97 |
| Glutathione | 4.10 |
| Heptylmercaptan | 4.92 |
| Mercaptosuccinic acid | 6.53 |
| Thiosalycilic acid | 7.05 |
| 2-Bromoethanesulfonic acid | 8.49 |
| 1-Pentanethiol | 8.96 |
| 2-Mercaptoethanol | 10.63 |
| Cyclopentylmercaptan | 10.78 |
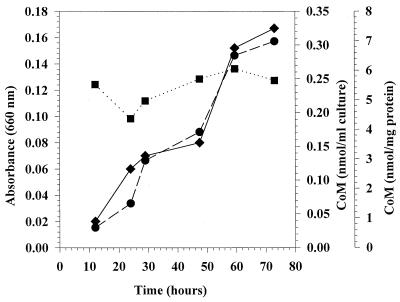
Methanobacterium thermoautotrophicum Marburg and Methanospirillum hungatei were both analyzed during growth. With Methanobacterium thermoautotrophicum Marburg, the amount of total CoM (CoM per milliliter of culture) increased with time from 0.03 to 0.31 nmol of CoM as the absorbance at 600 nm increased from 0.02 to 0.17. However, when the values were normalized for protein content, there was not a significant change in the amount of CoM per milligram of protein over time (Fig. 2). Similar results were obtained with Methanospirillum hungatei (data not shown). This indicated that the amounts of CoM per milligram of protein were stable, and consequently, the growth phase of the cells may not substantially influence CoM levels in natural samples.
FIG. 2.
Absorbance (●), CoM content per milliliter of culture (⧫), and CoM content normalized for culture protein content (■) in a Methanobacterium thermoautotrophicum Marburg culture growing on hydrogen.
CoM contents of cultures and sediments.
Pure cultures of methanogens were compared to determine the variation in CoM levels among different organisms, and the data suggest that CoM levels may vary up to sevenfold (Table 2). Based on the environmental samples tested, all of the samples contained measurable amounts of CoM (Table 3), and sewage sludge had the highest CoM content (on a per-gram basis), 0.09 nmol of CoM/g.
TABLE 2.
CoM levels in various methanogens and CoM levels per cell
| Organism | Amt of CoM
|
Direct count procedure
|
MPN estimation procedure
|
|||
|---|---|---|---|---|---|---|
| nmol/mg of protein | nmol/ml | No. of cells/ml | fmol of CoM/cell | No. of cells/ml | fmol of CoM/cell | |
| Methanolobus tindarius | 11.49 | 0.56 | ||||
| Methanospirillum hungatei | 19.41 | 0.52 | 7.40 × 105 | 0.70 | 1.1 × 106 | 0.47 |
| Methanosarcina barkeri (grown on N2-CO2-methanol) | 41.11 | 1.97 | 4.60 × 106 | 0.43 | ||
| Methanosarcina barkeri (grown on N2-CO2-acetate) | 6.13 | 0.70 | 1.80 × 106 | 0.39 | ||
| Methanosarcina barkeri (grown on H2-CO2) | 18.21 | 1.78 | ||||
| Methanosaeta concilli | 6.59 | 0.38 | ||||
| Methanococcus voltae | 23.75 | 1.45 | 6.20 × 106 | 0.23 | ||
| Methanococcus thermolithotrophicus | 41.09 | 2.59 | 5.15 × 106 | 0.50 | 7.5 × 106 | 0.34 |
| Methanobacterium thermoautotrophicum Marburg | 7.48 | 0.40 | 2.85 × 106 | 0.14 | 1.1 × 107 | 0.36 |
TABLE 3.
CoM levels in various sediments and CoM levels per cell for sediments
| Environmental sample | Depth (ft) | Amt of CoM
|
MPN estimation procedure
|
||
|---|---|---|---|---|---|
| nmol/g of sediment | nmol/ml | No. of cells/ml | fmol of CoM/cell | ||
| Pond sediment | 0.02 | 0.06 | 1.5 × 105 | 0.40 | |
| Landfill sediment | 0.01 | 0.02 | 1.1 × 105 | 0.18 | |
| Sewage sludge | 0.09 | 0.22 | 4.6 × 105 | 0.48 | |
| Hydrocarbon-contaminated sediment | 2.5 | 0.00 | NDa | ND | |
| 3.0 | 0.02 | 0.05 | 7.5 × 104 | 0.67 | |
| 4.0 | 0.08 | 0.20 | 3.3 × 105 | 0.47 | |
| 5.5 | 0.07 | 0.18 | 2.2 × 105 | 0.65 | |
| 7.5 | 0.08 | 0.20 | 3.6 × 105 | 0.43 | |
| 8.5 | 0.07 | 0.19 | 2.7 × 105 | 0.55 | |
| 10.0 | 0.07 | 0.17 | 1.6 × 105 | 0.74 | |
ND, not determined.
Extraction efficiency of CoM assay.
Several cultures and anoxic sediments were examined to determine whether a second extraction increased the recovery of CoM. For pure cultures, 87.8 to 96.8% of the total CoM detectable was recovered during the first extraction, while the values for environmental samples ranged from 91.8 to 100% during the first extraction. The results suggest that only a single extraction is necessary to recover the majority of the CoM present.
Cell lysis with a French press.
Methanosarcina barkeri (N2-CO2-methanol), Methanospirillum hungatei, Methanobacterium thermoautotrophicum Marburg, Methanococcus thermolithotrophicus, and Methanococcus voltae were examined to determine if cell lysis increased the total recovery of CoM. Cells were lysed by using a French pressure cell at 38,000 lb/in2. CoM was extracted from cells and cell extracts in the usual manner. For all four methanogens tested the results were similar (data not shown).
Effect of freezing on CoM.
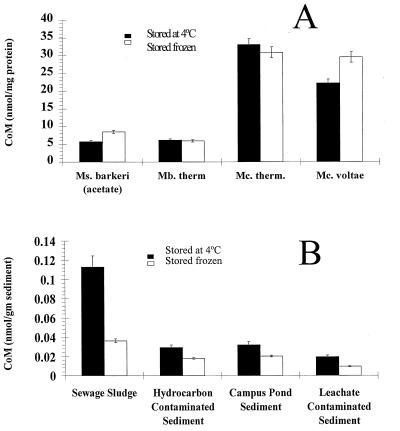
Pure cultures and anoxic sediments were examined to determine if freezing had an effect on the recovery of CoM. With all of the pure cultures tested there was not a significant loss of CoM after freezing (Fig. 3A). In contrast to the pure cultures, with all of the sediments there was a significant decrease in the amount of detectable CoM after samples were frozen and thawed (Fig. 3B).
FIG. 3.
(A) Cultures of methanogens tested before and after freezing at −60°C. There was no decrease in the amount of CoM detected in cells due to freezing. (B) Environmental samples tested before and after freezing at −60°C. There were 46 to 83% decreases in the amount of detectable CoM depending on the matrix. Bars indicate standard errors. Abbreviations: Ms., Methanosarcina; Mb. therm, Methanobacterium thermoautotrophicum; Mc. therm., Methanococcus thermolithotrophicus; Mc., Methanococcus.
Amount of CoM per cell.
The amount of CoM per cell was determined by both an MPN assay and a direct cell counting procedure. The amounts of CoM per cell were comparable, and the overall values for the organisms were similar (Table 2). The amounts determined by the direct cell count procedure ranged from 0.14 fmol of CoM/cell in Methanobacterium thermoautotrophicum to 0.70 fmol of CoM/cell in Methanospirillum hungatei, and the average was 0.40 ± 0.20 fmol of CoM/cell. The corresponding MPN-based values ranged from 0.34 fmol of CoM/cell in Methanococcus thermolithotrophicus to 0.47 fmol of CoM/cell in Methanospirillum hungatei, and the average was 0.39 ± 0.07 fmol of CoM/cell.
The amounts of CoM per cell were calculated for sediment from a campus pond, a landfill leachate-contaminated aquifer, sewage sludge, and six different depths in a sediment core obtained from the hydrocarbon-contaminated site (Table 3). In the sediment core obtained from the hydrocarbon-contaminated site, the values ranged from 0.43 to 0.74 fmol of CoM/cell. The values for the other three samples ranged from 0.18 to 0.48 fmol of CoM/cell. An overall average of 0.41 ± 0.17 fmol of CoM/cell was calculated by using the average for the hydrocarbon-contaminated site (0.59 fmol of CoM/cell) and the values for the other three samples.
DISCUSSION
For many applications in environmental microbiology, accurate estimates of microbial population size are beneficial. Methanogens are important in many anoxic environments and carry out the terminal electron-accepting process in a variety of ecosystems. Because of this, an accurate estimate of methanogen biomass is often desirable.
Previously, several marine thiols, including CoM, were quantified by using an HPLC-based technique involving precolumn derivatization (25). In a more recent study, this technique was used to quantitate thiols in pore water but not in sediments (37). This is not the first report of the use of HPLC for thiol detection. Two forms of CoM, mesna (sodium 2-mercaptoethanesulfonate) and dimesna (2-mercaptoethanesulfonate disulfate), were quantitated by using HPLC-based methods with postcolumn derivatization and colorimetry (18, 33). The difference between the postcolumn methods used previously and the detection procedure used in this study is that two steps were involved in the former methods; to separate the thiol of interest from others compounds, derivatization and quantitation were performed separately, which required additional labor. More recently, CoM was detected by using a fluorescent precolumn derivatization procedure, followed by reverse-phase HPLC (16). This procedure is similar to the procedure which we used but differs in that synthesis of the derivatizing reagents is difficult and time-consuming, whereas the reagents required for our procedure are readily available.
We modified the previously described method (25) by adding 1% TBP. The 1% TBP acts as a reducing agent which prevents oxidation of CoM-SH to a homodisulfide of CoM-S-S-CoM or a heterodisulfide of CoM-S-S-HTP (7-mercaptoheptanoylthreonine phosphate) (26, 27) as the cells are lysed in isopropanol. This means that the assay can be conducted without stringent anoxic conditions. The CoM in several pure cultures of methanogens and environmental samples was quantified. The protocol can be accomplished within 3 h of sample collection. With this technique, an accurate estimate of the methanogenic population at a given site may be obtained. This conclusion is based on several findings, including the detection limit (2 pmol of CoM) and the linear response of the assay for both the synthetic cofactor and CoM obtained from pure cultures. The assay was found to be applicable to environmental samples. An HPLC-based method for CoA quantitation (17) produced similar results, suggesting that a similar technique may be developed for other organisms.
Although our technique has many advantages over direct quantitation of methanogens with the culturing technique, it clearly has some limitations. The major limitation is the fact that the technique cannot distinguish between viable and nonviable cells. Oxic samples involved in oxidation of alkenes could also contain significant numbers of nonmethanogenic organisms which contain CoM used in the degradation reactions. We do not believe that these issues should restrict the utility of this procedure for the quantitation of methanogens.
Our experiments revealed that the CoM concentration increased as the absorbance of pure cultures increased over time but that the amount of CoM per milligram of protein was constant. Thus, the CoM content per cell varied little with the growth phase. The same trend was found for coenzyme F420 in methanogens (31). We observed that the amount of CoM per milligram of protein varied by up to a factor of 7 among organisms, which was comparable to the factor of 5 associated with the previously described CoM bioassay (5). The amounts of CoM per milligram of protein were consistent in the current study compared to the bioassay (5). Methanosarcina barkeri (grown on methanol) contained 41.1 nmol of CoM/mg of protein, compared to the 44.4 nmol of CoM/mg of protein found with the bioassay, while Methanobacterium thermoautotrophicum contained 7.5 nmol of CoM/mg of protein, compared to the 6.7 nmol of CoM/mg of protein found with the bioassay. The Methanospirillum hungatei data were not as consistent; this organism contained 19.4 nmol of CoM/mg of protein, compared to the 3.9 nmol of CoM/mg of protein reported previously (5).
While there was no decrease in the ability to detect CoM in pure cultures due to freezing, there were 46 to 83% decreases in the environmental samples. While the reasons for this are not clear, the data suggest that it is not advisable to store environmental samples in the freezer prior to the assay.
The CoM biomass determined by the HPLC-based assay and the CoM biomass determined by the direct cell count or MPN assay agreed well. The average values obtained were as follows: 0.40 ± 0.20 fmol of CoM/cell with the direct cell count procedure and 0.39 ± 0.07 fmol of CoM/cell with the MPN method for pure cultures and 0.41 ± 0.17 fmol of CoM/cell for environmental samples. The similarity of the values obtained by using MPN data for environmental samples indicates that a reference value for methanogens is possible. This would allow quick and accurate estimation of methanogen populations directly from the HPLC-based CoM values.
Based on the data obtained in this study, there is at most a threefold difference in the amount of CoM per cell in environmental samples. Also, if a reference value of 0.41 fmol of CoM/cell were used, the laborious MPN assay would no longer be required; this would allow reasonably accurate estimation of a methanogen population in the time required to complete the assay.
The inherent advantage of the procedure described here over previously described procedures is that the CoM content per cell has been determined for a variety of environmental matrices. This procedure provides a quick and simple aerobic method for detecting the cofactor as a biomarker and thus quantifying methanogen biomass.
ACKNOWLEDGMENTS
This work was supported by grants from DOE and EPA Epscor.
We thank Melanie Mormile for her initial investigations during this study, Kevin Kropp for his invaluable assistance in confirming the identity of the CoM derivative with the LC-MS, and Richard Sparling for his gift of Methanococcus thermolithotrophicus and Methanococcus voltae.
REFERENCES
- 1.Adkins J P, Cornell L A, Tanner R S. Microbial composition of carbonate petroleum reservior fluids. Geomicrobiol J. 1992;10:87–97. [Google Scholar]
- 2.Adrian N R, Robinson J A, Suflita J M. Spatial variability in biodegradation rates as evidenced by methane production from an aquifer. Appl Environ Microbiol. 1994;60:3632–3639. doi: 10.1128/aem.60.10.3632-3639.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen J R, Clark D D, Krum J G, Ensign S A. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci USA. 1999;96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogenesis: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balch W E, Wolfe R S. Specificity and biological distribution of coenzyme M (2-mercaptoethanesulfonic acid) J Bacteriol. 1979;137:256–263. doi: 10.1128/jb.137.1.256-263.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker G W, Raterman K T, Fisher J B, Corgan J M, Trent G L, Brown D R, Kemp N, Sublette K L. A case study of the natural attenuation of gas condensate hydrocarbons in soil and groundwater. Appl Biochem Biotechnol. 1996;57:791–801. doi: 10.1007/BF02941759. [DOI] [PubMed] [Google Scholar]
- 7.Beeman R E, Suflita J M. Environmental factors influencing methanogenesis in a shallow anoxic aquifer: a field and laboratory study. J Ind Microbiol. 1990;5:45–58. doi: 10.1007/BF01569605. [DOI] [PubMed] [Google Scholar]
- 8.Beeman R E, Suflita J M. Microbial ecology of a shallow unconfined ground water aquifer polluted by municipal landfill leachate. Microb Ecol. 1987;14:39–54. doi: 10.1007/BF02011569. [DOI] [PubMed] [Google Scholar]
- 9.Blaut M. Metabolism of methanogens. Antonie Leeuwenhoek. 1994;66:187–208. doi: 10.1007/BF00871639. [DOI] [PubMed] [Google Scholar]
- 10.Blaut M, Muller V, Gottschalk G. Energetics of methanogenesis studied in vesicular systems. J Bioenerg Biomembr. 1992;24:529–546. doi: 10.1007/BF00762346. [DOI] [PubMed] [Google Scholar]
- 11.Borole A, Sublette K L, Raterman K T, Javanmardian M, Fisher J B. The potential for intrinsic bioremediation of BTEX hydrocarbons in soil/groundwater contaminated with gas condensate. Appl Biochem Biotechnol. 1997;63:719–730. doi: 10.1007/BF02920470. [DOI] [PubMed] [Google Scholar]
- 12.Borole A P, Sublette K L, Fisher J B, Raterman K T, McInerney M J. Mechanisms of intrinsic bioremediation of gas condensate hydrocarbons in saturated soil. Appl Biochem Biotechnol. 1996;57:817–826. [Google Scholar]
- 13.Daniels L, Belay N, Rajagopal B S. Assimilatory reduction of sulfate and sulfite by methanogenic bacteria. Appl Environ Microbiol. 1986;51:703–709. doi: 10.1128/aem.51.4.703-709.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiMarco A A, Bobik T A, Wolfe R S. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 14a.Elias D A, Krumholz L R, Tanner R S, Mormile M R, Suflita J M. Abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C.: American Society for Microbiology; 1998. Estimation of methanogen biomass via quantitation of co-enzyme M, abstr. N-90. [Google Scholar]
- 15.Garcia J L. Taxonomy and ecology of methanogens. FEMS Microbiol Rev. 1990;87:297–308. [Google Scholar]
- 16.Gatti R, Cavrini V, Roveri P, Pinzauti S. High-performance liquid chromatographic determination of aliphatic thiols with aroylacrylic acids as fluorogenic precolumn derivatization reagents. J Chromatogr. 1990;507:451–458. doi: 10.1016/s0021-9673(01)84224-2. [DOI] [PubMed] [Google Scholar]
- 17.Hermans-Lokkerbol A, van der Heijden R, Verpoorte R. Isocratic high-performance liquid chromatography of coenzyme A esters involved in the metabolism of 3S-hydroxy-3-methylglutaryl-coenzyme A detection of related enzyme activities in Catharanthus roseus plant cell cultures. J Chromatogr A. 1996;752:123–130. [Google Scholar]
- 18.James C A, Rogers H J. Estimation of mesna and dimesna in plasma and urine by high-performance liquid chromatography with electrochemical detection. J Chromatogr Biomed Appl. 1986;382:394–398. doi: 10.1016/s0378-4347(00)83550-1. [DOI] [PubMed] [Google Scholar]
- 19.Jewell W J. Anaerobic sewage treatment. Environ Sci Technol. 1987;21:14–21. [Google Scholar]
- 20.Konig H, Stetter K O. Isolation and characterization of Methanolobus tindarius, sp. nov., a coccoid methanogen growing only on methanol and methylamines. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe C. 1982;3:478–490. [Google Scholar]
- 21.Lin X, White R H. Occurrence of coenzyme F420 and its γ-monoglutamyl derivative in nonmethanogenic archaebacteria. J Bacteriol. 1986;168:444–448. doi: 10.1128/jb.168.1.444-448.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride B C, Wolfe R S. A new coenzyme of methyl transfer, coenzyme M. Biochemistry. 1971;10:2317–2324. doi: 10.1021/bi00788a022. [DOI] [PubMed] [Google Scholar]
- 23.McCarty P L. One-hundred years of anaerobic treatment. In: Hughes D E, Stafford D A, Wheatley B I, Baader W, Lettinga G, Nyns E J, Verstraete W, Wentworth R L, editors. Anaerobic Digestion 1981. Proceedings of the 2nd International Symposium on Anaerobic Digestion, Travemunde, Germany, 1981. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1982. pp. 3–22. [Google Scholar]
- 24.Metcalf E. Wastewater engineering, treatment, disposal, and reuse. 3rd ed. New York, N.Y: McGraw-Hill, Inc.; 1991. [Google Scholar]
- 25.Mopper K, Delmas D. Trace determination of biological thiols by liquid chromatography and precolumn fluorometric labeling with o-phthalaldehyde. Anal Chem. 1984;56:2557–2560. doi: 10.1021/ac00277a064. [DOI] [PubMed] [Google Scholar]
- 26.Noll K M, Donelly M L, Wolfe R S. Synthesis of 7-mercaptoheptaheptanoylthreonine phosphate and its activity in the methylcoenzyme M methylreductase system. J Biol Chem. 1987;262:513–515. [PubMed] [Google Scholar]
- 27.Noll K M, Reinhart K L, Jr, Tanner R S, Wolfe R S. Structure of component B (7-mercaptoheptaheptanoylthreonine phosphate) of the methylcoenzyme M methylreductase system of Methanobacterium thermoautotrophicum. Proc Natl Acad Sci USA. 1986;83:4238–4242. doi: 10.1073/pnas.83.12.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novaes R F V. Microbiology of anaerobic digestion. Water Sci Technol. 1986;18:1–14. [Google Scholar]
- 29.Parkin G F, Owen W F. Fundamentals of anaerobic digestion of wastewater sludges. J Environ Eng. 1986;112:867–920. [Google Scholar]
- 30.Patel G B. Characterization and nutritional properties of Methanothrix concilii sp. nov., a mesophilic, acetoclastic methanogen. Can J Microbiol. 1984;30:1383–1396. [Google Scholar]
- 31.Peck M W. Changes in concentration of coenzyme F420 analogs during batch growth of Methanosarcina barkeri and Methanosarcina mazei. Appl Environ Microbiol. 1989;55:940–945. doi: 10.1128/aem.55.4.940-945.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sidau B, Shaw I C. Determination of sodium 2-mercaptoethanesulfonate by high-performance liquid chromatography using post-column reaction colorimetry or electrochemical detection. J Chromatogr Biomed Appl. 1984;311:234–238. doi: 10.1016/s0378-4347(00)84716-7. [DOI] [PubMed] [Google Scholar]
- 34.Sparling R R M, Daniels L. Source of carbon and hydrogen in methane produced from formate by Methanococcus thermolithotrophicus. J Bacteriol. 1986;168:1402–1407. doi: 10.1128/jb.168.3.1402-1407.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sublette K L, Kolhatkar R, Borole A, Raterman K T, Trent G L, Javanmardian M, Fisher J B. Intrinsic bioremediation of gas hydrocarbons. Results of over two years of ground water and soil core analysis and monitoring. Appl Biochem Biotechnol. 1997;63:823–834. doi: 10.1007/BF02920478. [DOI] [PubMed] [Google Scholar]
- 36.Taylor C D, Wolfe R S. Structure and methylation of coenzyme M (HSCH2-CH2-SO3) J Biol Chem. 1974;249:4879–4885. [PubMed] [Google Scholar]
- 37.Vairavamurthy A, Mopper K. Field method for determination of traces of thiols in natural waters. Anal Chim Acta. 1990;236:363–370. [Google Scholar]
- 38.White R H, Zhou D. Biosynthesis of coenzymes in methanogens. In: Ferry J G, editor. Methanogenesis—1993. New York, N.Y: Chapman & Hall; 1993. pp. 409–444. [Google Scholar]
- 39.Whitman W B, Ankwanda E, Wolfe R S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982;149:852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong D, Lin Z, Juck D, Terrick K A, Sparling R. Electron transfer reactions for the reduction of NADP+ in Methanosphaera stadtmanae. FEMS Microbiol Lett. 1994;120:285–290. [Google Scholar]
- 41.Zinder S H. Physiological ecology of methanogens. In: Ferry J G, editor. Methanogenesis—1993. New York, N.Y: Chapman & Hall; 1993. pp. 128–206. [Google Scholar]