Abstract
DNA-damaging agents include first-line drugs such as platinum (cisplatin, carboplatin), topoisomerase inhibitors (etoposide, doxorubicin), and replication inhibitors (cytarabine, gemcitabine). Despite their wide and long usage, there is no clinically available biomarker to predict responses to these drugs. Schlafen 11 (SLFN11), a putative DNA/RNA helicase, recently emerged as a dominant determinant of sensitivity to these drugs by enforcing the replication block in response to DNA damage. Since the clinical importance of SLFN11 is implicated, a comprehensive analysis of SLFN11 expression across human organs will provide a practical resource to develop the utility of SLFN11 in the clinic. In this study, we established a scoring system of SLFN11 expression by immunohistochemistry (IHC) and assessed SLFN11 expression in ~ 700 malignant as well as the adjacent non-tumor tissues across 16 major human adult organs. We found that the SLFN11 expression is tissue specific and varies during tumorigenesis. Although The Cancer Genome Atlas (TCGA) is a prevailing tool to assess gene expression in various malignant and normal tissues, our IHC data exhibited obvious discrepancy from the TCGA data in several organs. Importantly, SLFN11-negative tumors, potentially non-responders to DNA-damaging agents, were largely overrated in TCGA because TCGA samples are a mixture of infiltrating immune cells, including T cells, B cells, and macrophages, which have strong SLFN11 expression. Thus, our study reveals the significance of immunohistochemical procedures for evaluating expression of SLFN11 in patient samples and provides a robust resource of SLFN11 expression across adult human organs.
Keywords: DNA damage, DNA repair, Drug resistance, Immunohistochemistry, SLFN11
Introduction
Possibly the oldest unmet need in cancer therapy is that there are still no clinically available predictive biomarkers for widely used DNA-damaging anti-cancer agents including platinum derivatives since the 1960s, topoisomerase inhibitors since the 1990s, and replication inhibitors since the 1960s. However, the recent development of DNA-sequencing and omics analyses of cancer cell line data and human tissue data has led to the discovery of unappreciated but important genes for cancer therapy. The discovery of Schlafen 11 (SLFN11) as a causal and most dominant genomic determinant of response to DNA-damaging agents is a representative success of the recent application of bioinformatics to the omic data of cancer cell lines [3, 23]. The correlation and causality of SLFN11 have been consolidated by cumulative reports from independent institutions with various models [1, 4, 5, 7, 9, 11, 13–15, 17, 20, 21]. SLFN11, a nuclear protein belonging to the Schlafen family of mammalian proteins, has a putative helicase motif and RPA binding site at the C-terminus [13, 15]. We recently reported that SLFN11 augments the toxicity of DNA-damaging agents by inducing lethal replication blocks, which have been linked to the putative helicase activity of SLFN11 [16].
According to human cancer cell line databases and a human tissue database, SLFN11 exhibits a broad range of expression across various cancer types, which suggests the usefulness of SLFN11 expression as a common predictive biomarker of DNA-damaging agents [18]. Indeed, high SLFN11 expression has been implicated as a predictor of response to platinum-based chemotherapy in ovarian cancer, small cell and non-small cell lung cancers, colorectal cancer, and Ewing sarcoma [4, 5, 22, 23]. Extending the clinical usefulness of SLFN11 requires the establishment of a handy and robust method or detecting SLFN11 reliably from patient samples. Furthermore, a comprehensive analysis of SLFN11 expression across human organs will be required as a practical resource to fully develop the clinical utility of SLFN11.
Although a couple of public databases are currently available from which information on SLFN11 expression in human organs can be obtained, it should be kept in mind that RNA-seq data from non-microdissected tissue samples are contaminated with inflammatory cells [6, 12] rich in SLFN11 expression and that optimal antibodies should be used for IHC analysis. In this study, we report an optimal IHC method and establish a resource of SLFN11 expression with ~ 700 malignant and adjacent normal tissues across 16 human organs. These comprehensive data on SLFN11 expression can provide beneficial information to treat patients with DNA-damaging agents for cancer therapy.
Materials and methods
Cell lines and generation of SLFN11-deleted cells.
All cell lines were maintained as described previously [19]. SLFN11-deleted cells in the MKN45 cell line were generated by CRISPR/Cas9 methods, with details as described previously [15].
Tissue samples
Formalin-fixed paraffin-embedded blocks were obtained from the archives of the National Hospital Organization Kure Medical Center and Chugoku Cancer Center. All samples were obtained with patient consent, and this study was approved by the Ethics Committee of Kure Medical Center and Chugoku Cancer Center (Kure, Japan, no. 2019–36) and conformed to the ethical guidelines of the Declaration of Helsinki. The non-tumor area was evaluated adjacent to the tumor area. The details of the cases are described in Table 1.
Table 1.
Summary of Schlafen 11 (SLFN11) expression in non-tumors and tumors
| Organ and diagnosis | Number | 1+ | 2+ | 3+ | SLFN11 (+) n (%) |
|---|---|---|---|---|---|
|
| |||||
| Brain | |||||
| Normal | 15 | 7 | 8 | 0 | 15 (100) |
| Meningioma | 20 | 0 | 3 | 17 | 20 (100) |
| Schwannoma | 6 | 0 | 0 | 6 | 6 (100) |
| Glioblastoma | 30 | 2 | 8 | 10 | 20 (67) |
| Lung | |||||
| Alveolar epithelium | 31 | 8 | 12 | 11 | 31 (100) |
| Glandular epithelium | 31 | 8 | 7 | 9 | 24 (77) |
| Squamous cell carcinoma | 31 | 5 | 3 | 4 | 12 (42) |
| Adenocarcinoma | 30 | 4 | 3 | 11 | 18 (40) |
| Breast | |||||
| Normal | 32 | 1 | 0 | 0 | 1 (3) |
| Invasive ductal carcinoma | 35 | 4 | 6 | 7 | 17 (49) |
| Invasive lobular carcinoma | 16 | 3 | 1 | 0 | 4 (25) |
| Esophagus | |||||
| Normal | 34 | 21 | 10 | 0 | 31 (91) |
| Squamous cell carcinoma | 34 | 10 | 10 | 8 | 28 (82) |
| Stomach | |||||
| Normal | 40 | 8 | 6 | 4 | 18 (45) |
| Adenocarcinoma | 40 | 4 | 11 | 11 | 26 (65) |
| Colon | |||||
| Normal | 39 | 0 | 0 | 0 | 0 (0) |
| Adenocarcinoma | 39 | 0 | 0 | 0 | 0 (0) |
| Liver | |||||
| Normal | 31 | 15 | 9 | 0 | 24 (77) |
| Hepatocellular carcinoma | 31 | 9 | 9 | 4 | 22 (71) |
| Bile duct | |||||
| Normal | 20 | 6 | 2 | 0 | 8 (40) |
| Adenocarcinoma | 20 | 8 | 1 | 0 | 9 (45) |
| Pancreas | |||||
| Normal | 34 | 0 | 0 | 0 | 0 (0) |
| Adenocarcinoma | 34 | 3 | 4 | 1 | 8 (24) |
| Kidney | |||||
| Normal | 44 | 44 | 0 | 0 | 44 (100) |
| Clear cell renal cell carcinoma | 41 | 4 | 13 | 20 | 37 (98) |
| Papillary renal cell carcinoma | 9 | 2 | 0 | 0 | 2 (22) |
| Chromophobe renal cell carcinoma | 6 | 0 | 0 | 0 | 0 (0) |
| Urinary bladder | |||||
| Normal | 30 | 0 | 0 | 0 | 0 (0) |
| Invasive urothelial carcinoma | 34 | 13 | 7 | 3 | 23 (68) |
| Prostate | |||||
| Normal | 32 | 0 | 0 | 0 | 0 (0) |
| Adenocarcinoma | 32 | 1 | 0 | 0 | 1 (3) |
| Uterine cervix | |||||
| Squamous epithelium | 16 | 5 | 7 | 2 | 14 (89) |
| Glandular epithelium | 13 | 5 | 2 | 4 | 11 (69) |
| Squamous cell carcinoma | 20 | 1 | 4 | 10 | 15 (75) |
| Adenocarcinoma | 13 | 1 | 3 | 5 | 9 (75) |
| Uterine corpus | |||||
| Normal | 30 | 13 | 1 | 0 | 14 (85) |
| Endometrioid carcinoma | 26 | 4 | 6 | 8 | 18 (69) |
| Serous carcinoma | 4 | 0 | 2 | 1 | 3 (75) |
| Ovary | |||||
| Fallopian tube | 15 | 9 | 5 | 1 | 15 (100) |
| Serous carcinoma | 18 | 1 | 5 | 6 | 12 (67) |
| Clear cell carcinoma | 7 | 1 | 2 | 3 | 6 (86) |
| Endometrioid carcinoma | 3 | 0 | 0 | 2 | 2 (67) |
| Mucinous carcinoma | 9 | 1 | 2 | 0 | 3 (33) |
| Bone and soft tissue | 31 | 3 | 7 | 13 | 23 (74) |
| Osteosarcoma | 6 | 0 | 0 | 4 | 4 |
| Chondrosarcoma | 1 | 0 | 0 | 0 | 0 |
| Leiomyosarcoma | 5 | 1 | 2 | 1 | 4 |
| Liposarcoma | 9 | 0 | 3 | 4 | 7 |
| Myxofibrosarcoma | 4 | 0 | 1 | 3 | 4 |
| Spindle cell sarcoma | 2 | 1 | 0 | 0 | 1 |
| Pleomorphic sarcoma | 1 | 0 | 0 | 1 | 1 |
| Synovial sarcomas | 2 | 0 | 1 | 0 | 1 |
| Clear cell sarcoma | 1 | 1 | 0 | 0 | 1 |
Antibodies
Details of the information concerning antibodies (clone, lot no., company, dilution) are summarized in Supplementary Table 1.
Immunohistochemistry
Our step-by-step IHC protocol for SLFN11 is presented in Supplementary Table 2. Negative controls were created by omitting the addition of the primary antibody.
Scoring of SLFN11 expression for IHC samples
SLFN11 expression in the nucleus of the main component of each non-neoplastic tissue (epithelium, fatty cell, brain) or tumor cells was evaluated either as positive or negative. Three surgical pathologists (N.S., D.T., and K.K.) independently determined the positive ratio of SLFN11 IHC without knowledge of the clinical and pathological parameters or patient outcome. The pathologists specified 10 high power fields with the highest SLFN11 positive ration in each IHC section, and the average values of ratio of positivity were calculated. According to the average values of ratio of positivity, SLFN11 IHC scores were considered 1+ (1–10%), 2+ (11–50%), or 3+ (51–100%). Inter-observer differences were resolved by consensus review at a double-headed microscope after independent reviews.
Western blot analyses
For Western blot analyses, cells were lysed as described previously [19]. Immunocomplexes were visualized with an ECL Plus Western Blot Detection System (Amersham Biosciences, Piscataway, NJ, USA). β-Actin (Sigma-Aldrich) was detected as a loading control.
Analysis of TCGA data
Batch effects-normalized RNA-Seq mRNA expression data from TCGA Pan-Cancer Atlas were downloaded from the UCSC Xena Functional Genomics Explorer (https://xenabrowser.net/). Expression levels for tumor and normal tissues are log2(x + 1)-transformed batch effects-normalized values. The normalization methodology is described at https://www.synapse.org/#!Synapse:syn4976363.
Results
Validation of anti-SLFN11 antibodies for IHC
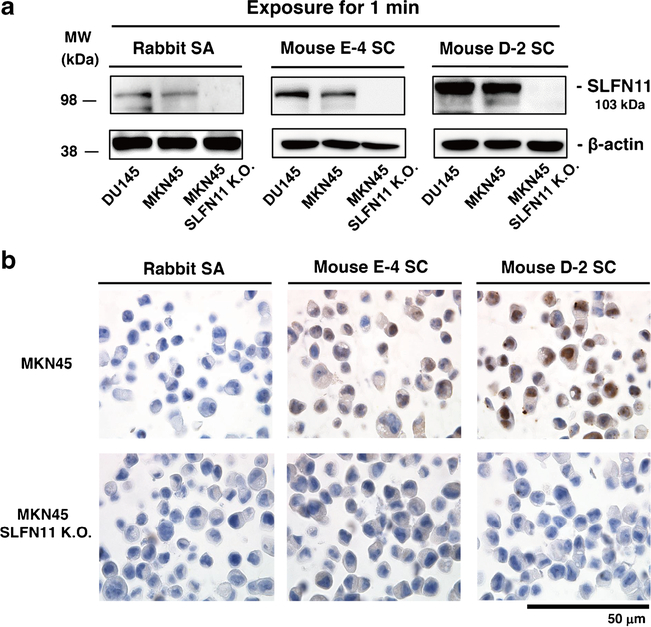
Two recent IHC studies have been conducted on SLFN11 expression using different antibodies [2, 11]. However, the staining patterns of SLFN11 seemed different depending on the antibodies used. Because SLFN11 exclusively localizes in the nucleus in cultured cells, staining of cytoplasmic regions is likely to be non-specific, which, for example, is observed with melanoma samples in The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000172716-SLFN11/pathology) with rabbit anti-SLFN11 antibody (Sigma-Aldrich, HPA023030). We compared sensitivity and specificity for SLFN11 of three commercially available anti-SLFN11 antibodies by Western blotting and IHC. We used the human prostate cancer DU145 cell line having high SLFN11 expression [23] and the human gastric cancer MKN45 cell line having SLFN11 expression comparable to that of DU145 (Fig. 1a). To evaluate the non-specificity of the antibodies, we generated and used SLFN11-deleted cells in MKN45 (MKN45 SLFN11-K.O.) (Fig. 1a). All three antibodies successfully detected SLFN11 by Western blotting without an obvious nonspecific band at ~ 100 kDa. The intensity was strongest with mouse D-2 SC (Fig. 1a). For the IHC, rabbit SA did not draw out any nuclear staining whereas mouse D-2 SC provided the strongest nuclear staining in MKN45. Mouse E-4 SC provided nuclear staining in MKN45 but weaker than that with mouse D-2 SC (Fig. 1b). In MKN45 SLFN11-K.O., none of the antibodies provided positivity (Fig. 1b). From these results, we decided to use the mouse D-2 SC antibody (hereafter named D-2 antibody) for further studies.
Fig. 1.
Validation of anti-SLFN11 antibodies. a Western blots in DU145 (prostate), MKN45 (gastric), and MKN45 SLFN11-deleted (SLFN11 K.O.) cell lines with the indicated antibodies. b Immunohistochemical analysis for SLFN11 with the indicated anti-SLFN11 antibodies in MKN45 and MKN45 SLFN11 K.O. cell lines. Scale bars are 50 μm in the enlarged images. MW, molecular weight; Rabbit SA, rabbit anti-SLFN11 antibody (#H117570, Sigma-Aldrich); Mouse E-4 SC, mouse anti-SLFN11 antibody (E-4, #sc-374,339, Santa Cruz); Mouse D-2 SC, mouse anti-SLFN11 antibody (D-2, #sc-515,071, Santa Cruz)
Diversity of SLFN11 expression among organs and dynamic change of SLFN11 expression during tumorigenesis
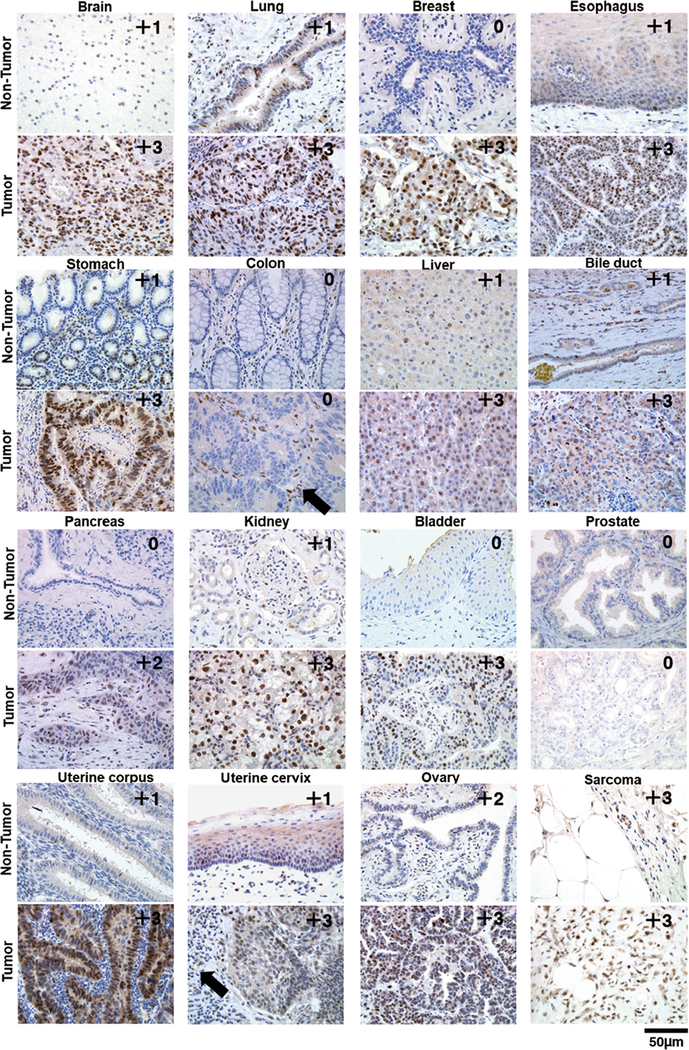
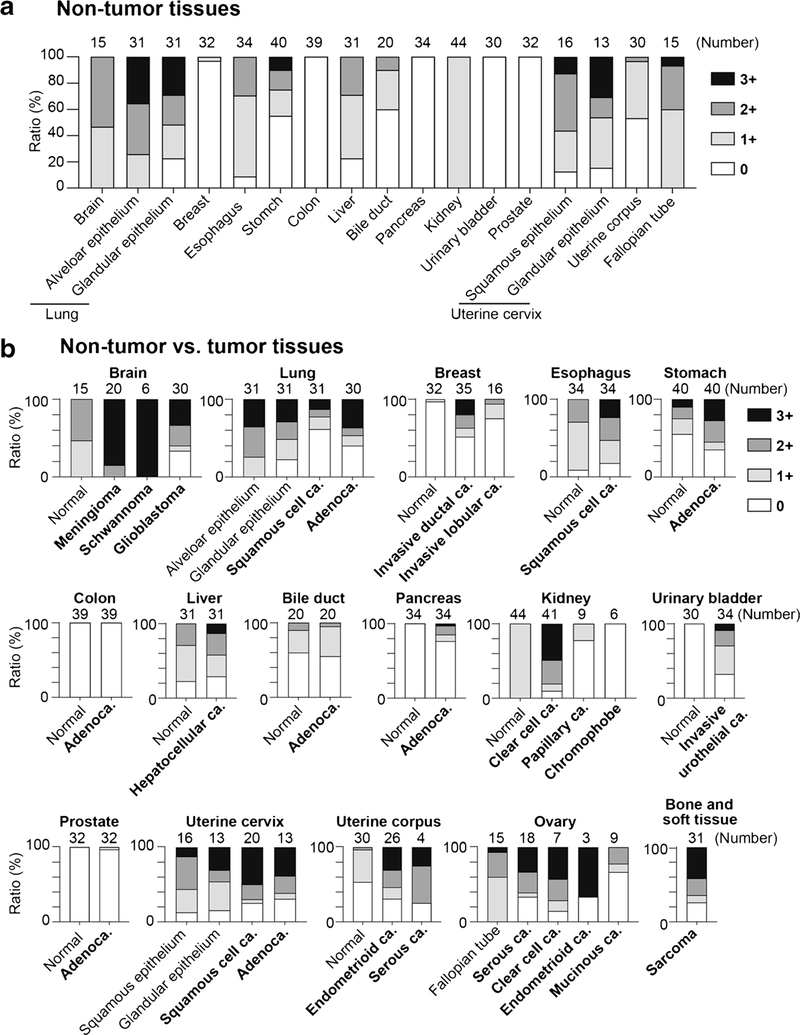
To establish the resource of SLFN11 expression profiles across human tissues, we performed IHC for ~ 700 malignant and adjacent non-tumor tissues across 16 major human adult organs and scored them as 0/1+/2+/3+ according to the ratio (%) of SLFN11-positive cells in the main components. Representative expression patterns of SLFN11 in non-tumor and tumor tissues of major organs are shown in Fig. 2 with their respective scores. The representative images of tumor tissues with different scores are shown in Supplementary Fig. S1. SLFN11 was predominantly detected in the nucleus in all positive samples, confirming our established IHC protocol with the mouse D-2 antibody. The scores for all samples are provided in Table 1 and plotted in Fig. 3. Notably, the positivity of SLFN11 in non-tumor tissues exhibited broad diversity with scores ranging from ~ 0 to ~ 100% across the organs (Fig. 3a).
Fig. 2.
Representative images of immunohistochemistry (IHC) for non-tumor and tumor regions in the indicated organs. The pairs of non-tumor and tumor samples of each organ are not always from identical patients. IHC scores are annotated at the top right of each panel. The black arrows indicate inflammatory cells surrounding the main components
Fig. 3.
Ratio of immunohistochemistry (IHC) scores of SLFN11 in non-tumor and tumor tissues across various organs. a Bar graphs plotting the ratio (%) of IHC scores of Table 1 for non-tumor tissues of the indicated organs. b Bar graphs plotting the ratio (%) of IHC scores of Table 1 for the indicated organs in non-tumor and tumor tissues. The number of samples is annotated at the top of each bar. The key to the IHC score colors is shown to the right of the graphs
The expression of SLFN11 is mostly regulated epigenetically [5, 17, 21], and SLFN11-inactivation by genetic mutation has rarely been found until now [13]. Hence, the expression of SLFN11 can be dynamic and affected by various environmental factors during tumor development. The comparison between non-tumor vs tumor tissues shown in Fig. 3b reveals that colon and prostate tissues are consistently SLFN11-negative in non-tumor and tumor tissues. The ratio of SLFN11 scores was most consistent in the bile duct. In other organs, there was a tendency for the population with 3+ positivity to be higher in tumor tissues compared to non-tumor tissues, suggesting that SLFN11 expression can be highly activated during tumorigenesis, consistent with its high expression in Ewing’s sarcoma [1, 12]. By contrast, SLFN11 expression was inactivated in tumors compared to non-tumors for glioblastoma, lung tissues, papillary renal cell carcinoma, and chromophobe renal cell carcinoma. These results show that SLFN11 expression is highly dynamic and very different between non-tumor and tumor tissues in some organs. These comprehensive SLFN11 expression analyses provide clinically important information about which organs to focus on when using SLFN11 as a predictive biomarker for DNA-damaging agents.
Discrepancy in SLFN11 expression between our IHC and the TCGA database
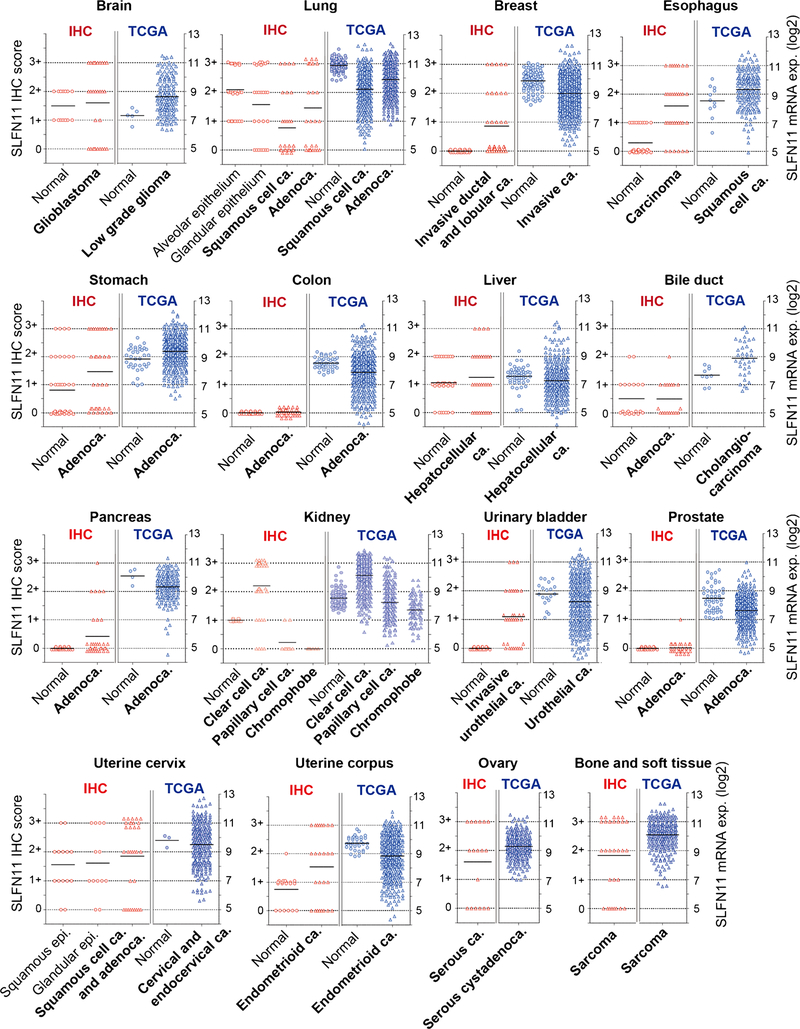
TCGA consortium provides a Pan-Cancer Atlas dataset including gene expression for ~ 11,000 tumors from 33 of the most prevalent forms of cancer (https://portal.gdc.cancer.gov/). Although there is no question about the usefulness of the TCGA database, the staining pattern of SLFN11 by IHC raised some concerns about the tumor specificity of SLFN11 evaluation by TCGA. In addition to the main component of each tissue, inflammatory or stromal cells surrounding the main compartments sometimes exhibit robust nuclear staining of SLFN11, with examples indicated by the black arrows in Fig. 2 and Supplementary Fig. S1. Because RNA-seq samples of TCGA include a mixture of tumor cells and surrounding cells, SLFN11 expression levels could be influenced by contamination from the sub-component cells. Hence, we plotted the expression levels of SLFN11 in TCGA and the scores of SLFN11 IHC in parallel to visualize the difference in their ranges (Fig. 4). Although there is no direct way to compare RNA-seq data and IHC scores, we can reasonably compare attributes of their distributions. In the brain and liver, for instance, the TCGA and IHC distributions similarly span a wide range of values for tumor tissues, whereas matched normal tissues largely have mean values comparable to the tumor ones. However, in other tissues such as breast and pancreas tissues, there is a huge discrepancy between TCGA and IHC distributions for both normal and tumor tissues. The narrow, near-baseline distribution with IHC relative to the broader and more substantial expression in TCGA is especially apparent in the colon and prostate tissues. These results are clinically important because SLFN11-negative tumors by IHC are potentially non-responsive to DNA-damaging agents, yet they could be overrated by tissue RNA-seq.
Fig. 4.
Parallel comparison of immunohistochemistry (IHC) scores and TCGA data of SLFN11 expression across 16 organs. IHC scores (0/1+/2+/3+) from Table 1 (left) and SLFN11 expression levels from TCGA Pan-Cancer Atlas data (right) are plotted in parallel for the indicated organs and tissues. mRNA expression levels are log2(x + 1)-transformed batch effects-normalized values. Each point represents a patient sample, and circular points represent normal tissue whereas triangular points represent tumor tissues. Averages are shown with black bars
Expression of SLFN11 in inflammatory cells
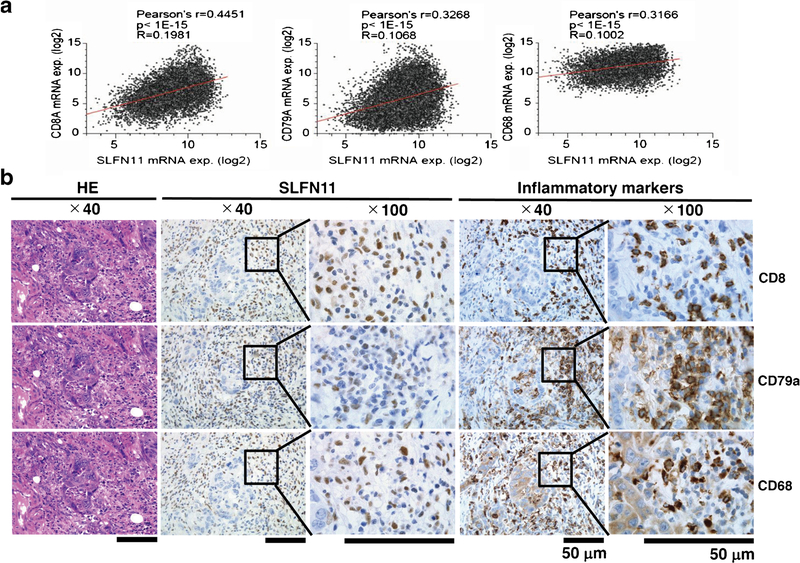
To assess the possibility of tumor contamination due to SLFN11 expression from inflammatory cells in tissue RNA-seq samples, we analyzed the correlation between the expression of SLFN11 and CD8A, CD79A, and CD68, which are representative cell surface markers of T cells, B cells, and macrophages, respectively. We found significant correlations between the expression of SLFN11 and all three markers in the Pan-Cancer Atlas dataset (Fig. 5a), indicating that the SLFN11 expression in tissue RNA-seq is influenced by inflammatory cells regardless of the origin of the tissues. To further validate the precise distribution of expression of SLFN11, we examined the expression of SLFN11 and CD8A, CD79A, and CD68 cell surface markers by IHC using two sequential tissue sections. We found that some of the SLFN11-positive inflammatory cells showed robust expression of the cell surface markers of either CD8, CD79a, or CD68 (Fig. 5b and Supplementary Fig. S2). Overall, our study emphasizes the significance of evaluation by IHC with mouse D-2 antibody, rather than by tissue RNA-seq, to precisely determine the expression of SLFN11.
Fig. 5.
Expression of SLFN11 in inflammatory cells. a Plots of SLFN11 expression (x axis) and the indicated inflammatory cell markers (CD8A, left; CD79A, centre; CD68, right) expression (y axis) in TCGA Pan-Cancer Atlas dataset. Statistical analysis results are shown above each panel. b Sequential tissue sections of colorectal cancer were analyzed with HE staining (left) and immunohistochemistry (IHC) for SLFN11 (centre) and IHC for inflammatory markers (right top, CD8; right middle, CD79A; right bottom, CD68) (Original magnification: × 40 and × 100). Scale bars are 50 μm in the enlarged images
Discussion
Although there is a pressing need to evaluate the expression of SLFN11 precisely in patient samples, prior to our study, there have been no reports examining tissue specificity or diversity of the expression of SLFN11 in a wide variety of tissue sections. In the present study, we set up the methods of IHC for SLFN11 and established a resource of expression patterns and ratios of SLFN11 using ~ 700 non-tumor and tumor tissues of major human organs. We also showed by in silico analysis and IHC that inflammatory cells have robust expression of SLFN11, which raises the importance of using IHC rather than tissue RNA-seq to evaluate SLFN11 expression in patient samples. One of the crucial findings of this study is that the expression pattern of SLFN11 revealed by IHC spans a broad range and exhibits tissue specificity. Some organs including colon and prostate showed almost no expression of SLFN11, in either non-tumor or tumor tissues, whereas other organs such as brain and lung showed varying levels of expression of SLFN11 in both tissue types. These findings highlight that tumors in such tissues with a wide range of SLFN11 expression are likely to be the most suitable for examining SLFN11 expression because such evaluation could be tested to select appropriate drug regimens for patients.
The present study suggests the plasticity of SLFN11 expression during tumourigenesis. In breast, pancreas, and urinary bladder tissues, expression of SLFN11 was low in the normal tissues whereas its expression was found in 20–70% of the tumor tissues. In contrast, SLFN11 expression was largely suppressed in lung squamous cell carcinoma compared to normal alveolar epithelium. Additionally, the population with 3+ positivity was overall higher in tumor tissues compared to non-tumor tissues. As SLFN11 expression is mostly regulated epigenetically [5, 17, 18, 21], SLFN11 expression will be convertible in both directions. Indeed, erasing of promoter DNA or histone methylation by 5-azacitidine, by inhibitors of EZH2, or with HDAC inhibitors can reactivate SLFN11 expression, leading to re-sensitization to DNA-damaging agents in cultured cells and mouse xenograft models [5, 17, 21]. This information implies that tumors have or easily develop heterogeneity of SLFN11 expression, although the mechanisms of regulation are mostly unknown except for the finding of FLI1 and ETS transcription factors as direct transcriptional activators of SLFN11 [20]. Because the regulation of SLFN11 expression is druggable by epigenetic modulators [5, 17, 21], clinical trials may be warranted to reactivate SLFN11 and sensitize tumors to DNA-damaging agents.
Previous studies have suggested the functional role of SLFN11 in immune pathways and the expression of SLFN11 in inflammatory or stroma cells, which can be supported by the fact that the expression of SLFN11 was induced by cytokines including IFN-β and IFN-γ [8, 10, 14]. One report examined the correlation between the expression levels of representative markers of T cells and SLFN11 in breast cancer [8]. However, the detailed regulation of SLFN11 expression in inflammatory or stroma cells has not been clarified. Indeed, not all of the inflammatory or stromal cells are positive for SLFN11, examples of which are seen in Fig. 2 in non-tumor and tumor tissues of the stomach and uterine corpus. Contrastingly, colon tissues with highly SLFN11-positive inflammatory or stroma cells in non-tumor and tumor tissues are shown in Fig. 2. These cells highly expressing SLFN11 in their subcomponents can confound the interpretation of expression levels by bulk tissue RNA-seq. The significance of SLFN11 expression in the stroma of some tumors is notable, and further studies are warranted to establish its potential significance, namely, in the context of immune checkpoint modulators.
In summary, we developed a new resource establishing the staining property of SLFN11 in a wide variety of sections of normal and tumor tissues from organs of adult humans. We anticipate that more retrospective or prospective studies from independent facilities and research groups will be conducted to verify the usefulness of SLFN11 expression as a predictive biomarker for DNA-damaging agents in cancer patients. For that purpose, the present resource provides numerous items of practical importance, such as the rigid IHC protocol, and it raises noteworthy issues relating to sources of SLFN11 expression. Furthermore, our study provides important insights into the issues of drug resistance and disease recurrence and suggests a strategy to overcome drug resistance through reactivation of SLFN11 expression.
Supplementary Material
Acknowledgments
We thank Mr. Shinichi Norimura for his technical assistance. This work was carried out with kind cooperation from the Research Center for Molecular Medicine of the Faculty of Medicine of Hiroshima University. We also thank the Analysis Center of Life Science of Hiroshima University for the use of their facilities.
Funding information
This work was supported by Grants-in-Aid for Scientific Research (JP15H04713 and JP16K08691 to W.Y., JP16H06999 to N.S.) and (19H03505 to J.M.), Challenging Exploratory Research (26670175, JP16K15247 to W.Y.) from the Japan Society for the Promotion of Science, AMED (Japan Agency for Medical Research and Development) Project for Cancer Research and Therapeutic Evolution (to J.M.), and a research grant from The Uehara Memorial Foundation (to J.M.). Y.P. and V.R. are supported by the Center for Cancer Research, the Intramural program of the US National Cancer Institute, NIH (Z01 BC 006150).
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflicts of interest.
Ethics approval This study was approved by the Ethics Committee of Kure Medical Center and Chugoku Cancer Center (Kure, Japan, no. 2019–36) and conformed to the ethical guidelines of the Declaration of Helsinki.
Consent to participate All samples were obtained with patient consent.
Code availability Statistical differences were evaluated using the Pearson’s test. Statistical analyses were conducted primarily using GraphPad Prism software (GraphPad Software Inc.).
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00428-020-02840-6) contains supplementary material, which is available to authorized users.
References
- 1.Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, Masrorpour F, Fan Y, Bara RO, Feng Y, Ru Y, Fujimoto J, Kundu ST, Post LE, Yu K, Shen Y, Glisson BS, Wistuba I, Heymach JV, Gibbons DL, Wang J, Byers LA (2017) Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8:28575. 10.18632/oncotarget.15338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestrero A, Bedognetti D, Ferraioli D, Franceschelli P, Labidi-Galy SI, Leo E, Murai J, Pommier Y, Tsantoulis P, Vellone VG, Zoppoli G (2017) Report on the first SLFN11 monothematic workshop: from function to role as a biomarker in cancer. J Transl Med 15:199. 10.1186/s12967-017-1296-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The cancer cell line encyclopedia enables predictive modelling of anti-cancer drug sensitivity. Nature 483:603–607. 10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng Y, Cai Y, Huang Y, Yang Z, Bai Y, Liu Y, Deng X, Wang J (2015) High SLFN11 expression predicts better survival for patients with KRAS exon 2 wild type colorectal cancer after treated with adjuvant oxaliplatin-based treatment. BMC Cancer 15:833. 10.1186/s12885-015-1840-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, Ni A, Khodos I, de Stanchina E, Nguyen T, Sage J, Campbell JE, Ribich S, Rekhtman N, Dowlati A, Massion PP, Rudin CM, Poirier JT (2017) Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 31:286–299. 10.1016/j.ccell.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao Y, Yan M, Heath BR, Lei YL, Xie Y (2019) Fast and robust deconvolution of tumor infiltrating lymphocyte from expression profiles using least trimmed squares. PLoS Comput Biol 15: e1006976. 10.1371/journal.pcbi.1006976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He T, Zhang M, Zheng R, Zheng S, Linghu E, Herman JG, Guo M (2017) Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 9:849–862. 10.2217/epi-2017-0019 [DOI] [PubMed] [Google Scholar]
- 8.Isnaldi E, Ferraioli D, Ferrando L, Brohee S, Ferrando F, Fregatti P, Bedognetti D, Ballestrero A, Zoppoli G (2019) Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer. Breast Cancer Res Treat 177:335–343. 10.1007/s10549-019-05313-w [DOI] [PubMed] [Google Scholar]
- 9.Kang MH, Wang J, Makena MR, Lee JS, Paz N, Hall CP, Song MM, Calderon RI, Cruz RE, Hindle A, Ko W, Fitzgerald JB, Drummond DC, Triche TJ, Reynolds CP (2015) Activity of MM-398, nanoliposomal irinotecan (nal-IRI), in Ewing's family tumor xenografts is associated with high exposure of tumor to drug and high SLFN11 expression. Clin Cancer Res 21:1139–1150. 10.1158/1078-0432.CCR-14-1882 [DOI] [PubMed] [Google Scholar]
- 10.Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, David M (2012) Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491:125–128. 10.1038/nature11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, de Stanchina E, Teicher BA, Riaz N, Powell SN, Poirier JT, Rudin CM (2017) PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res 23:523–535. 10.1158/1078-0432.CCR-16-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnuson AM, Kiner E, Ergun A, Park JS, Asinovski N, Ortiz-Lopez A, Kilcoyne A, Paoluzzi-Tomada E, Weissleder R, Mathis D, Benoist C (2018) Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci U S A 115:E10672–E10681. 10.1073/pnas.1810580115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzi L, Szabova L, Gordon M, Weaver Ohler Z, Sharan SK, Beshiri ML, Etemadi M, Murai J, Kelly K, Pommier Y (2019) The indenoisoquinoline TOP1 inhibitors selectively target homologous recombination deficient- and Schlafen 11-positive cancer cells and synergize with olaparib. Clin Cancer Res. 10.1158/1078-0432.CCR-19-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mezzadra R, de Bruijn M, Jae LT, Gomez-Eerland R, Duursma A, Scheeren FA, Brummelkamp TR, Schumacher TN (2019) SLFN11 can sensitize tumor cells towards IFN-gamma-mediated T cell killing. PLoS One 14:e0212053. 10.1371/journal.pone.0212053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, Pommier Y (2016) Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 7:76534–76550. 10.18632/oncotarget.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murai J, Tang SW, Leo E, Baechler SA, Redon CE, Zhang H, Al Abo M, Rajapakse VN, Nakamura E, Jenkins LMM, Aladjem MI, Pommier Y (2018) SLFN11 blocks stressed replication forks independently of ATR. Mol Cell 69:371–384.e376. 10.1016/j.molcel.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogales V, Reinhold WC, Varma S, Martinez-Cardus A, Moutinho C, Moran S, Heyn H, Sebio A, Barnadas A, Pommier Y, Esteller M (2016) Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 7:3084–3097. 10.18632/oncotarget.6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajapakse VN, Luna A, Yamade M, Loman L, Varma S, Sunshine M, Iorio F, Sousa FG, Elloumi F, Aladjem MI, Thomas A, Sander C, Kohn KW, Benes CH, Garnett M, Reinhold WC, Pommier Y (2018) CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. iScience 10:247–264. 10.1016/j.isci.2018.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto N, Naito Y, Oue N, Sentani K, Uraoka N, Oo HZ, Yanagihara K, Aoyagi K, Sasaki H, Yasui W (2014) MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci 105:236–243. 10.1111/cas.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang SW, Bilke S, Cao L, Murai J, Sousa FG, Yamade M, Rajapakse V, Varma S, Helman LJ, Khan J, Meltzer PS, Pommier Y (2015) SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in Ewing sarcoma. Clin Cancer Res 21:4184–4193. 10.1158/1078-0432.CCR-14-2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang SW, Thomas A, Murai J, Trepel JB, Bates SE, Rajapakse VN, Pommier Y (2018) Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin Cancer Res 24:1944–1953. 10.1158/1078-0432.CCR-17-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L, Song S, Liu X, Wang Y, Xu X, Hu Y, Xu J (2014) Schlafen-11 sensitizes colorectal carcinoma cells to irinotecan. Anti-Cancer Drugs 25:1175–1181. 10.1097/cad.0000000000000151 [DOI] [PubMed] [Google Scholar]
- 23.Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, Doroshow JH, Pommier Y (2012) Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A 109:15030–15035. 10.1073/pnas.1205943109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.