Abstract
A hallmark feature of lentiviruses, which separates them from other members of the retrovirus family, is their ability to infect non-dividing cells by traversing the nuclear pore complex. The viral determinant that mediates HIV-1 nuclear import is the viral capsid (CA) protein, which forms the conical core protecting the HIV-1 genome in a mature virion. Recently, a series of novel approaches developed to monitor post-fusion events in infection have challenged previous textbook models of the viral life cycle, which envisage reverse transcription and disassembly of the capsid core as events that complete in the cytoplasm. In this review, we summarize these recent findings and describe their implications on our understanding of the spatiotemporal staging of HIV-1 infection with a focus on the nuclear import and its implications in other aspects of the viral lifecycle.
During infection, fusion of the viral and plasma membranes delivers the conical capsid core, which contains the viral genome and replicative proteins, into the cytoplasm. To complete infection, the HIV-1 ribonucleoprotein complex (RNP) must 1) initiate and complete the reverse transcription of its RNA genome into double stranded DNA 2) traffic through the cytoplasm and traverse the nuclear pore complex (NPC) 3) integrate the reverse transcribed provirus into genomic DNA of the target cell, all while 4) avoiding detection by host factors which are designed to sense foreign DNA and other viral determinants [1, 2]. To appreciate the recent changes in thinking surrounding these steps of infection, it is first necessary to consider that all of these key aspects of infection are mediated or minimally dependent on the CA, which assembles during viral maturation to form the HIV-1 core. CA is comprised of two domains - an amino terminal domain (CANTD) and a carboxy terminal domain (CACTD), connected by a flexible linker. The CANTD between adjacent CA monomers interacts to form hexameric or pentameric rings of CA which themselves assemble to form the conical capsid core of HIV-1 [3–10]. For clarity in this discussion, we will use CA to describe the capsid protein and the term “core” to describe an intact or apparently intact capsid assembly. It is the viral core that houses the viral RNA genome bound by the nucleocapsid (NC) protein along with the viral enzymes reverse transcriptase (RT), and integrase (IN). Prior to integration the core must disassemble, a process termed uncoating, to allow the preintegration complex (PIC) the ability to access genomic DNA in the nucleus. It is known that reverse transcription is dependent on the stability of the viral core, and mutations in CA which destabilize the core prevent the generation of reverse transcription products [11]. This is consistent with the idea that the discontinuous nature of reverse transcription requires a constrained environment to allow RT and the RNA genome to generate a complete, double stranded DNA copy of the RNA genome.
CA has also emerged as the critical mediator of HIV-1 nuclear import in cells [1]. Seminal experiments from Yamashita and Emmerman demonstrated that replacing the CA of murine leukemia virus (MLV), a retrovirus that cannot infect non-dividing cells and therefore requires cell division to complete infection, with the CA of HIV-1 generates a virus capable of infecting non-dividing cells [12, 13], establishing CA as the viral determinant that allows HIV-1 to enter the nucleus of non-dividing cells. Since these studies, it has been demonstrated that assembled CA engages nucleoporins (Nups) and other host proteins required for nuclear import [1]. CA also mediates an interaction with CPSF6, which influences the intra-nuclear trafficking and ultimately the integration site during infection [14–17]. Critically, interactions with Nups and CPSF6 require assembled CA, as these proteins interact with a binding pocket formed by an interaction between individual monomers in hexameric CA assemblies [18–20] .
As the appreciation for the multifaceted role CA plays during infection has increased, so to have the number of approaches designed to monitor the fate of CA and other components of the RNP during infection. Recently, these tools have begun to converge on ideas that have forced a reconsideration of dogmas inherent to the fields of nuclear import and HIV-1 infection. Below, we attempt to provide insight into the evidence and thinking that established these dogmas and describe the findings that have placed them on unexpectedly shaky ground.
Dogma 1: HIV-1 completes reverse transcription in the cytoplasm
Virtually every review or textbook schematic of the HIV-1 lifecycle published prior to 2020 includes a depiction of a complete PIC entering the nucleus. A PIC is defined as a replicative intermediate of HIV-1 infection in which reverse transcription has generated a double stranded DNA copy of the HIV-1 genome that is capable of integrating into target DNA. Indeed, at the time of this writing, the NIAID website describing the HIV-1 lifecycle still has such a depiction [21]. This dogma was codified based on two complementary concepts. First, early studies of PICs and PIC activity found that PICs could be isolated in large amounts from the cytoplasm of T cell lines [22–25]. Second, until very recently, virtually all studies and models of the uncoating process suggested that the core disassembled, as least partially, in the cytoplasm or at the nuclear pore complex prior to nuclear import [1]. Given the notion that viral core integrity is required to ensure the association of RT with its RNA template, the ability to isolate PICs from the cytoplasm jibed attractively with studies of uncoating that suggested substantial loss of CA occurring prior to nuclear import, but presumably after the completion of reverse transcription.
Our recent studies of HIV-1 nuclear import yielded data that was difficult, if not impossible, to reconcile with the idea that HIV-1 completes reverse transcription in the cytoplasm [26]. In these studies, we relied on an inducible blockade of nuclear import to understand the spatiotemporal staging of nuclear import in relation to other steps of the viral lifecycle. We observed that most of the infectious viral inoculum entered the nucleus within 2–3 hours of infection in T cells and macrophages [26]. This was surprising, with respect to the dogma that reverse transcription completes in the cytoplasm, given that reverse transcription takes much longer than 2–3 hours to complete in T cells, and in macrophages can take multiple days to complete [27–32]. Indeed, in our study, infection remained sensitive to reverse transcription inhibitors for hours after the inoculum had become insensitive to nuclear pore blockade [26], leading us to conclude that reverse transcription completed in the nucleus. This observation has now been corroborated using other experimental approaches. Selyutina et al have used biochemical fractionation approaches to show the accumulation of incomplete products of reverse transcription accumulating in the nuclear compartment [33]. Francis et al similarly used fluorescent imaging approaches to monitor the nuclear import of the viral RNP and observed that reverse transcription continues to occur following nuclear import of the RNP [34]. Collectively, these studies suggest that models envisioning the completion of reverse transcription completing in the cytoplasm require revision. These data are also relevant to the consideration of the state of the viral core during and after nuclear import, as now discussed.
Models of HIV-1 uncoating and their implications on nuclear import of HIV-1
Models of HIV-1 uncoating have been changing for the last decade, as increasingly sophisticated approaches have been developed to monitor the state of the viral core during infection. Perhaps owing to the fact that HIV-1 cores are relatively unstable, compared to MLV cores, the concept of HIV-1 uncoating began with the thinking that uncoating happened relatively soon after fusion [24, 35, 36]. This thinking has gradually shifted towards views that the capsid core, and minimally some amount of assembled CA, persist longer during infection [26, 33], and some data suggest the viral RNP is housed in a relatively intact core until moments before integration [37–39]. Unlike the case of reverse transcription in the cytoplasm, however, a consensus view of HIV-1 uncoating has not yet been achieved, and data from various studies from different groups supporting models in which CA is lost from the core prior to arriving at the NPC [40], at the NPC during the nuclear import process [41, 42] or once inside the nucleus [37–39, 43].
Nuclear Import Model 1: Core disassembly or remodeling prior to nuclear import
The nature of HIV-1 infection and the uncoating process has made uncoating a relatively difficult question to address experimentally. Measurements of uncoating have tended to rely on both fluorescent imaging and biochemical approaches. Both approaches are limited by the fact that only a small fraction of a viral inoculum that enters a cell may go on to productively infect that cell. Although both approaches have been used to make observations that have generally stood the test of time, neither is ideally suited to assess the relative amount of CA associated with an individual RNP. Generally, studies that stain RNPs for capsid have detected changes in the amount of CA stain present in populations of RNPs, and our lab and others have used this approach to characterize changes in the amount of detectable CA stain following the depletion of knockout of various host factors [44–46]. However, such changes in CA stain are open to interpretation, as changes in the ability of cores to associate with specific host factors, such as CPSF6 or CypA, might reasonably alter the amount of CA accessible epitopes available for antibody staining, as recently demonstrated [15, 16]. Similarly, while biochemical assays are able to monitor differences in core stability following experimental perturbation, the nature of these experiments do not allow one to determine the number of CA molecules present on a viral RNP following infection. However, despite these limitations, such approaches have been useful in characterizing changes in virion populations during infection, and have supported the notion that CA is lost from the viral RNP in the cytoplasm of infection. Perhaps the strongest data to support this model has been provided by Mamede et al. which used a labeling approach where GFP is trapped in the viral core and loss of GFP is interpreted as loss of core integrity [40]. Using this approach, the authors could follow the fate of individual viral particles over time, and by titrating the number of viral particles per cell in their experiments, observe instances where individual virion signal led to infection of the target cell. These studies observed two distinct reductions of GFP content during infection. One reduction of signal was observed soon after fusion, likely representing the fusion event and loss of GFP not trapped within the core, followed by a second reduction in signal in the cytoplasm during infection, which is interpreted to be a change in core integrity during infection. Notably, virions exhibiting this behavior were found to go on to productively infect the cell [40]. In the same study, the authors directly labeled CA through insertion of a tetracytsteine tag that can be labeled with a FlAsH reagent [47] and observed a partial loss of CA signal in the cytoplasm [40].
Other data supporting uncoating occurring prior to nuclear import suggests that CA loss occurs following arrival at the NPC. Electron microscopy has observed apparently intact cores at the NPC [39]. Francis et al used live cell imaging to indirectly monitor CA disassembly by labeling viral cores with DsRed-labeled CypA, which binds with high affinity to assembled and unassembled CA [41, 42]. The authors observed that viral particles docked at the nuclear envelope lost their CypA- DsRed signal, and this loss was interpreted as loss of CA. However, it remains unclear whether the loss of CypADsRed represents loss of viral CA, as the displacement of CypA by TNPO1 may occur, as suggested in the study by Fernandez et al [48]. Collectively, these studies support a mechanism for RNP translocation through the NPC that involves partial loss of viral CA either in the cytoplasm or at the NPC (Fig 1A).
Figure 1: Models of HIV-1 Nuclear import:
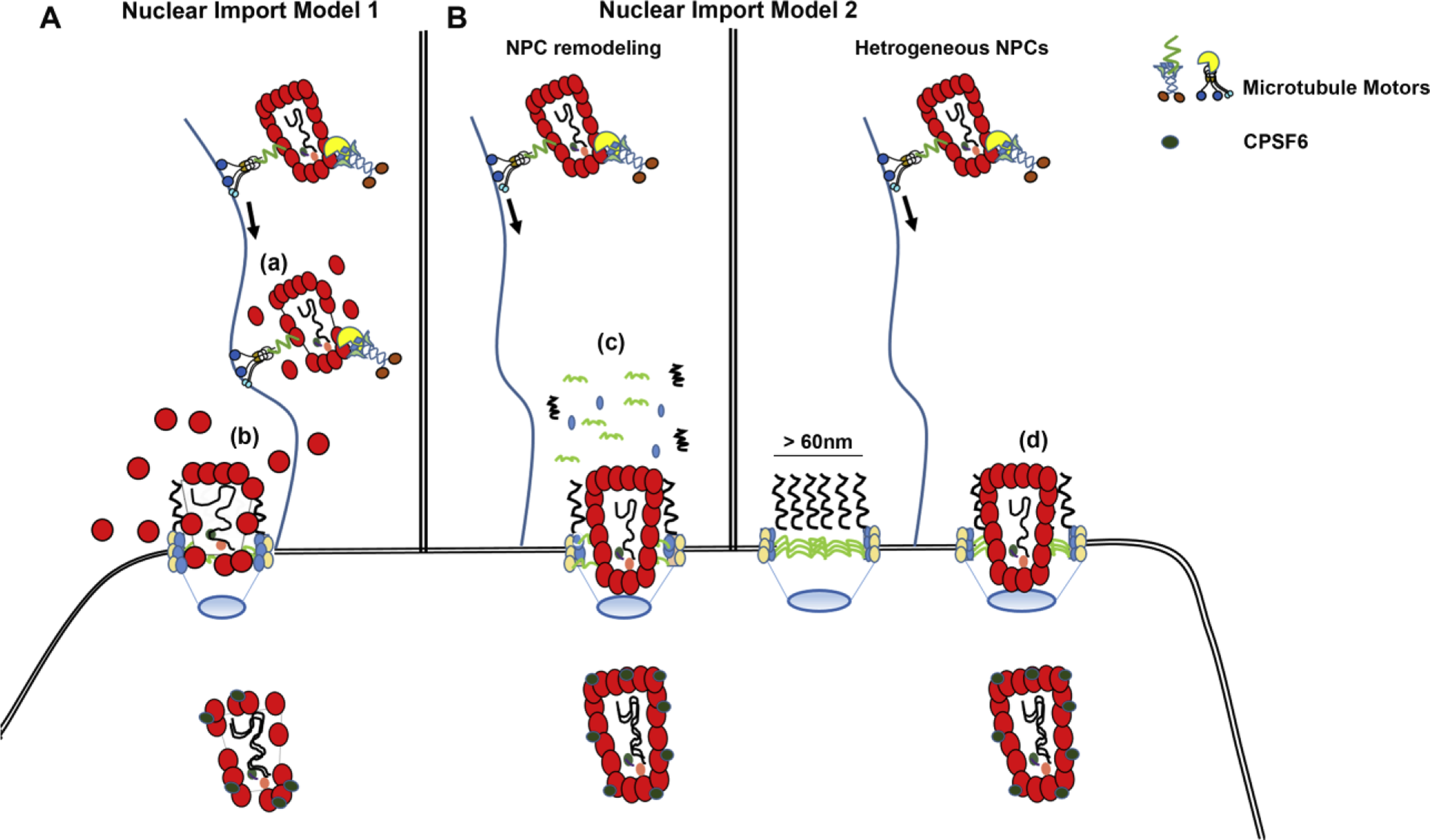
Following envelope-mediated fusion, the viral capsid core is released into the cytoplasm. (A) Model 1: Viral core undergoes disassembly enroute to the NPC (a) or at the NPC (b) allowing nuclear import. (B) Model 2: Nuclear import of intact HIV-1 cores through the NPC through (c) NPC remodeling whereby Nups are displaced from the NPC increasing the diameter of the central pore channel allowing nuclear import of intact cores. (d) Presence of heterogeneous NPCs in infected cells of varying sizes that can accommodate different sized cargos. Nuclear import mediated through docking at an NPC with central channel diameter that exceeds the diameter of an intact core.
Other studies of HIV-1 nuclear import have pointed to a core remodeling event as preceding nuclear import of the RNP. The Di Nunzio group combined TEM and immunogold labeling for CA to examine RNPs inside and outside of the nucleus in human lymphocytes [49]. The authors observed CA present in both complexes. However, the authors observed a different pattern of immunogold staining and core morphologies in nuclear RNPs. Viral DNA decorated with multiple CA proteins resembling a pearl necklace-like shape was observed during and following nuclear entry of HIV-1. A recent study from the Bishop lab also supports the concept of core remodeling. This study used cores which were hyperstabilized by cysteine crosslinking to determine that core hyperstabilization trapped the RNP at the NPC, unable to enter the nucleus [50]. Such remodeling events are reported to mediate nuclear import of host protein complexes as in the case of messenger ribonucleoprotein particles (mRNP) which has an overall dimension of ∼50nm. Using electron microscope tomography, it has been shown that these mRNP particles traverse the NPC by rearranging into rod like structures thereby decreasing the diameter to approximately half of the original dimension [51, 52].
Taken together, these studies suggest that uncoating or remodeling of the viral core occurs before nuclear import. However, a primary challenge in integrating these studies to something approaching consensus is in understanding the degree of connection between core remodeling and uncoating, as well as any differences between the two. Generally, studies and approaches examining uncoating, which attempt to measure a loss of CA from the RNP, are generally not capable of measuring core remodeling, and vice versa, although it is possible studies examining core remodeling or uncoating are measuring similar phenomena. In point of this fact, biophysical and biochemical studies have observed that the process of reverse transcription can induce both CA remodeling and uncoating. Rankovic and colleagues have used atomic force microscopy to demonstrate that the initiation of RT induces pressure inside the core that progressively remodels the core architecture [53, 54]. Similarly, a recent study from Christiansen and colleagues used purified HIV-1 virions and an impressive cell free system to observe viral cDNA loops to projecting out from partially uncoated cores, which led them to suggest the rupture of the viral capsid can occur due to the increased pressure induced by the nascent dsDNA genome generated inside the core [55]. These observations generally align with cell biology experiments which have observed that reverse transcription promotes uncoating, as measured by CA staining [56] or sensitivity to a CA targeting restriction factor TRIM-Cyp [57]. The emergence of approaches that allow biophysical interrogation of cores by AFM [53, 54] and recapitulation of HIV-1 replication in cell free systems [55], as well as approaches that can facilitate correlative light electron microscopy (CLEM) to interrogate viral RNPs in cells during infection [49], will hopefully allow an integrated view of how core uncoating and remodeling are connected and how cellular and host factors drive these phenomena.
It is important to note that even models which suggest CA disassembly occurs in the cytoplasm or prior to nuclear import must also accommodate some amount of assembled CA traversing the NPC with the RNP. Numerous structural and functional studies demonstrate that both Nup153 and CPSF6 bind to the binding pocket created by adjacent CA monomers in assembled CA hexamers [18–20]. As Nup153 forms the nucleoplasmic basket of the NPC and CPSF6 has been shown to influence intra-nuclear trafficking and integration site selection during infection [14–16], it stands to reason that not all assembled CA is lost from the RNP during nuclear import, even if some fraction of the original core is uncoated or remodeled during this step of infection.
Nuclear Import Model 2: Nuclear import of intact cores
Recently, a provocative and potentially dogma disrupting alternative to the above models, has emerged in studies by two different groups, both of which suggest that intact cores enter the nucleus during infection [37–39]. In addition to forcing the reconsideration of models suggesting these events occur prior to nuclear import, such a model is dogma defying because of the dimensions of the HIV-1 core, which is 60 nm wide. This is around 50% larger than the size limitations for nuclear import established in previous, oft-cited studies [58–61]. However, strong data from two sets of studies suggests that this may indeed be the case. Burdick et al first reported the possibility that intact HIV-1 cores enter the nucleus during infection [37]. To monitor core integrity, the authors used GFP labeled CA to monitor the fate of viral cores, thereby overcoming the limitations of antibody staining mentioned above. Using live cell imaging, they observed loss of GFP signal only inside the nucleus and close to integration sites, thus arriving at the conclusion that viral core remains largely intact following nuclear import and uncoating occurs before viral integration [37]. The same group also used a labeling approach similar to Mamede et al [40] using free GFP as a marker for core integrity, but reported results discordant with the Mamede study, observing the loss of GFP signal only inside the nucleus and shortly before integration [43]. Other studies by the Kräusslich group used correlative light and electron microscopy (CLEM) to observe intact or nearly intact CA inside the nucleus [38, 39]. Zila et al., using the A77V, a CPSF6 deficient CA mutant, visualized intact cores inside the NPC, with the narrow end pointing towards the central channel [39]. A more recent study from the same group using wildtype (WT) virus reports similar findings as before but additionally observed conical and elongated structures resembling open cores inside the nucleus [38]. Thus, these reports suggest that disassembly occurs following nuclear import, with the Muller et al study [38] suggesting disassembly of CA lattice through localized physical disruption rather than cooperative disassembly.
Dogma 2: NPCs do not allow the transport of cargoes larger than 40 nm
Although “core remodeling” may also explain the discord between the dimensions of the viral core and size limitations of nuclear import [58–61], date from other studies also suggest that nuclear import of an intact core might be possible in the absence of a remodeling event. One possibility is that the core temporarily changes dimensions while traversing the nuclear pore. However, given the EM tomograms of nuclear cores with intact, conical morphology observed in the Kräusslich papers [38, 39] , the core would need to revert back to its original shape following such an event. It is also possible that NPCs can conditionally accommodate cargo exceeding the established size limitation, either by remodeling of the NPC, or by a population of pores capable of accommodating cargo of the dimensions of the core (Fig 1B). With respect to the possibility of NPC remodeling, we have observed relocalization of Nup358, a cytoplasmic FG Nup to the cytoplasm during infection [62]. In our more recent study, we also observed cytoplasmic relocalization of Nup62, the central FG Nup following WT infection [26]. Interestingly the localization of Nup153, a nucleoplasmic Nup did not change following infection [62] suggesting this infection induced relocalization remodels the NPC structure rather than a complete breakdown of the complex. This pattern of Nup relocalization we observed was strikingly similar to another study that looked at the nuclear entry of adenovirus [63]. In this study, the authors observed relocalization of Nups to the cytoplasm following adenovirus infection and the permeability of the NPC was increased following infection. The proposed model involves adenovirus docking to the cytoplasmic side of the NPC, which then triggers relocalization of NPC components and requires the plus end microtubule motor kinesin-1 during this process. In this study, this relocalization of NPC constituents induced by adenovirus infection increased the permeability barrier allowing the movement of large cargoes, consistent with the notion that NPC remodeling allows for the trafficking of larger cargo. This model suggested for adenovirus nuclear entry matched our findings, where apart from the Nup relocalization we observed following HIV-1 infection, this phenotype was also dependent on KIF5B, the kinesin-1 motor. To our knowledge, this type of Nup relocalization has not been reported outside of the study of viruses. However, it is tempting to speculate that these viruses, and perhaps others, are exploiting an uncharacterized cellular response to NPCs that have become clogged with cargos that exceed the size limitation of nucleocytoplasmic transport.
Dogma 3: NPCs are homogenous
Another possibility is that the cells might contain NPCs that can accommodate an intact core (Fig 1B). A number of studies support the notion that HIV-1 can utilize distinct nuclear import pathways for entry, which may actually be different types of NPCs. The first line of evidence came from the KewalRamani lab which showed that virus with capsid bearing the N74D mutation, which disrupts binding to CPSF6, exhibited a differential requirement of Nups during infection compared to WT CA [64]. A similar observation was also shown for the CypA deficient mutant P90A [65]. Kane et al explored the functional interaction between HIV-1 CA, Nups, MX2, and CypA and observed variability in NPC composition among different cell types, and suggested the existence of several Nup-dependent pathways for HIV-1 nuclear entry [66]. In support, we observe the CA mutants N74D and P90A were comparatively insensitive to a Nup62 mediated nuclear pore blockade in cells that potently block infection by wild type CA [26]. This suggests the presence of NPCs that do not harbor Nup62, and therefore the existence of a heterogeneous population of NPC in cells. NPCs have been long considered homogeneous in their composition and size. Now with improved techniques that can resolve NPCs with greater resolution and in their native environment, the concept of homogeneous pores is slowly fading and studies supporting heterogeneity in the composition and size of NPCs are reported in the literature. A study that utilized atomic force microscopy to evaluate the structure of isolated Xenopus laevis oocyte nuclear envelopes describes variability among nuclear pore sizes [67]. A more recent study that used cryo-focused –ion-beam milling technique in DLD-1, a colorectal adenocarcinoma cell line to obtain structural model of human NPCs in their native environment observed NPCs with central channel dimensions of 57nm [68] which is substantially wider than the previously reported dimensions [69–72]. The authors suggest that the cellular environment plays a considerable influence on NPC dimensions and architecture. Although this study provides strong evidence refuting the concept of homogeneous NPCs, this observation does not completely align with the previous observations from independent groups that showed fluorescently labeled dextran or gold particles exceeding 40kDa are unable to bypass the central NPC channel [58, 73–75]. However, the increased central channel dimension fits with the study from Zila et al where the authors observed the width of the central channel that contain docked viral cores has a diameter of ∼64nm [39] that suggest the presence of NPC with central pore channel that is wide enough to fit an intact core. However, with cells reported to harbor more than 1000 NPCs [76, 77], it remains to be elucidated how HIV-1 can find the preferred type of NPC for import into the nucleus. One possibility, based on a study that looked at the nuclear entry of adenovirus [78] is a motor protein assisted cytoplasmic exploration. During HIV-1 infection, both the microtubule motors through their respective adaptor proteins, which is BICD2 for dynein, and FEZ1 for kinesin-1 is shown to mediate HIV-1 trafficking to the nuclear import sites [79–81]. Thus, a cytoplasmic exploration of incoming capsids through a coordinated effort with the motor proteins would possibly allow docking at the proper NPC for import.
Concluding remarks
Over the last year, significant progress has been made to understand many of the post entry stages of the HIV-1 life cycle. Better techniques that allows these post entry events to be monitored with greater accuracy, and these have fundamentally altered the ways in which we think about HIV-1 infection and nuclear import. Understanding this stage of the viral life cycle remains a priority as it represents an excellent target for therapeutic intervention. Several antiviral compounds that can bind HIV-1CA and abrogate infection have been identified and a few of these compounds that include PF74, GS-CA1 and GS-6207 bind CA to the same binding pocket as Nup153/CPSF6 (reviewed in [82]). Also, antiviral sensors myxovirus resistance protein 2 (MxB) [83–85] and SUN proteins [86, 87] that are present at the nuclear envelope have been shown to recognize viral CA as the pathogen-associated molecular pattern (PAMP). Therefore, more direct approaches such as the inducible NPC blockade assay and super-resolution imaging to monitor HIV-1 nuclear import soon will improve our understanding of the spatiotemporal staging of HIV-1 nuclear entry.
Acknowledgement
This work was funded by National Institute of Allergy and Infectious Diseases (NIAID) grants R21 AI139009 and R01 AI11626946 to EMC.
Footnotes
Declaration of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1. Campbell EM and Hope TJ, HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol, 2015. 13(8): p. 471–83. **This review describes the post entry role of HIV- capsid and how capsid interaction with various host factors modulates early HIV-1 infection. Data that argues for and against the various models of HIV-1 uncoating along with the techniques used to measure HIV-1 uncoating are described in this review article.
- 2.Yin X, et al. , Sensor Sensibility-HIV-1 and the Innate Immune Response. Cells, 2020. 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs JA, et al. , Structural organization of authentic, mature HIV-1 virions and cores. EMBO J, 2003. 22(7): p. 1707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deshmukh L, et al. , Structure and dynamics of full-length HIV-1 capsid protein in solution. J Am Chem Soc, 2013. 135(43): p. 16133–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganser BK, et al. , Assembly and analysis of conical models for the HIV-1 core. Science, 1999. 283(5398): p. 80–3. [DOI] [PubMed] [Google Scholar]
- 6.Ganser-Pornillos BK, Cheng A, and Yeager M, Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell, 2007. 131(1): p. 70–9. [DOI] [PubMed] [Google Scholar]
- 7.Gres AT, et al. , STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science, 2015. 349(6243): p. 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattei S, et al. , The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science, 2016. 354(6318): p. 1434–1437. [DOI] [PubMed] [Google Scholar]
- 9.Pornillos O, et al. , X-ray structures of the hexameric building block of the HIV capsid. Cell, 2009. 137(7): p. 1282–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pornillos O, Ganser-Pornillos BK, and Yeager M, Atomic-level modelling of the HIV capsid. Nature, 2011. 469(7330): p. 424–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forshey BM, et al. , Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol, 2002. 76(11): p. 5667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamashita M and Emerman M, Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol, 2004. 78(11): p. 5670–8. **The findings from this work placed HIV-1 capsid as the critical determinant mediating HIV- infection in nondividing cells.
- 13.Yamashita M, et al. , Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog, 2007. 3(10): p. 1502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Achuthan V, et al. , Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe, 2018. 24(3): p. 392–404 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin CR, et al. , Direct Visualization of HIV-1 Replication Intermediates Shows that Capsid and CPSF6 Modulate HIV-1 Intra-nuclear Invasion and Integration. Cell Rep, 2015. 13(8): p. 1717–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis AC, et al. , HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Nat Commun, 2020. 11(1): p. 3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, et al. , CPSF6-Dependent Targeting of Speckle-Associated Domains Distinguishes Primate from Nonprimate Lentiviral Integration. mBio, 2020. 11(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya A, et al. , Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A, 2014. 111(52): p. 18625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matreyek KA, et al. , Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog, 2013. 9(10): p. e1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AJ, et al. , CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog, 2012. 8(8): p. e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NIAID. HIV-1 Replication Cycle. 2018; Available from: https://www.niaid.nih.gov/diseasesconditions/hiv-replication-cycle.
- 22.Farnet CM and Haseltine WA, Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci U S A, 1990. 87(11): p. 4164–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farnet CM and Haseltine WA, Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol, 1991. 65(12): p. 6942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller MD, Farnet CM, and Bushman FD, Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol, 1997. 71(7): p. 5382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MD, Wang B, and Bushman FD, Human immunodeficiency virus type 1 preintegration complexes containing discontinuous plus strands are competent to integrate in vitro. J Virol, 1995. 69(6): p. 3938–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dharan A, et al. , Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat Microbiol, 2020. 5(9): p. 1088–1095. **For more than a decade, HIV-1 RT was considered to be completed prior to nuclear import. This study from our lab using an inducible nuclear pore blockade assay demonstrates that HIV-1 RT remains active for hours following viral nuclear import. Also using the capsid binding molecule PF74, our study demonstrates the presence of an assembled CA inside the nucleus.
- 27.Srivastava KK, et al. , Human immunodeficiency virus type 1 NL4–3 replication in four T-cell lines: rate and efficiency of entry, a major determinant of permissiveness. J Virol, 1991. 65(7): p. 3900–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Larsson R, et al. , Replication of patient isolates of human immunodeficiency virus type 1 in T cells: a spectrum of rates and efficiencies of entry. Proc Natl Acad Sci U S A, 1992. 89(6): p. 2223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray JM, Kelleher AD, and Cooper DA, Timing of the components of the HIV life cycle in productively infected CD4+ T cells in a population of HIV-infected individuals. J Virol, 2011. 85(20): p. 10798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes M, Zhang F, and Bieniasz PD, Single-Cell and Single-Cycle Analysis of HIV-1 Replication. PLoS Pathog, 2015. 11(6): p. e1004961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomaras GD, et al. , CD8+ T cell-mediated suppressive activity inhibits HIV-1 after virus entry with kinetics indicating effects on virus gene expression. Proc Natl Acad Sci U S A, 2000. 97(7): p. 3503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bejarano DA, et al. , Detailed Characterization of Early HIV-1 Replication Dynamics in Primary Human Macrophages. Viruses, 2018. 10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selyutina A, et al. , Nuclear Import of the HIV-1 Core Precedes Reverse Transcription and Uncoating. Cell Rep, 2020. 32(13): p. 108201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francis AC, et al. , HIV-1 Uncoating and Nuclear Import Precede the Completion of Reverse Transcription in Cell Lines and in Primary Macrophages. Viruses, 2020. 12(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bukrinsky MI, et al. , Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci U S A, 1993. 90(13): p. 6125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fassati A and Goff SP, Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol, 2001. 75(8): p. 3626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burdick RC, et al. , HIV-1 uncoats in the nucleus near sites of integration. Proc Natl Acad Sci U S A, 2020. 117(10): p. 5486–5493. **In this study, the authors monitored HIV-1 uncoating by direct labeling of HIV-1 CA with GFP. By monitoring GFP in living infected cells, loss of signal was observed only inside the nucleus and close to the region of integration. Based on this observation the authors report that HIV-1 capsid remains largely intact during and after nuclear import and uncoating occurs close to the viral integration sites inside the nucleus.
- 38.Muller TG, et al. , HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. Elife, 2021. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zila V, et al. , Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell, 2021. 184(4): p. 1032–1046 e18. **This work combining correlative light and elctron microscopy with subtomogram averaging to monitor the structural status of HIV-1 complexes in infected T cells. The authors detected intact cone shaped capsids at the NPC and inside the nucleus of infected cells. This study also observed the width of the central NPC channel is wide enough for an intact cone shaped capsid to accomplish nuclear import.
- 40. Mamede JI, et al. , Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc Natl Acad Sci U S A, 2017. 114(34): p. E7169-E7178. *By using an intravirion fluid phase marker to moitor CA integrity, this work monitored HIV-1 uncoating through live cell imaging in infected cell lines and primary cells. By monitoring the loss of the marker in their experiments, the authors report viral uncoating starts in the cytoplasm of infected cells.
- 41.Francis AC, et al. , Time-Resolved Imaging of Single HIV-1 Uncoating In Vitro and in Living Cells. PLoS Pathog, 2016. 12(6): p. e1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Francis AC and Melikyan GB, Single HIV-1 Imaging Reveals Progression of Infection through CA-Dependent Steps of Docking at the Nuclear Pore, Uncoating, and Nuclear Transport. Cell Host Microbe, 2018. 23(4): p. 536–548 e6. *HIV-1 uncoating at the nuclear pore complex was reported in this study where the authors incorporated CypA-DsRed into the viral particles to label the viral CA. By monitoring the loss of CypA-DsRed in live cell imaging experiments, the authors report early cytoplasmic uncoating promotes protesomal degradation of viral complexes and loss of CypA-DsRed occurs once the viral CA docks at the nuclear envelope.
- 43.Li C, et al. , HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc Natl Acad Sci U S A, 2021. 118(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hulme AE, et al. , Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells. J Virol, 2015. 89(10): p. 5350–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukic Z, et al. , HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol, 2014. 88(23): p. 13613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaney MK, et al. , Distinct functions of diaphanous-related formins regulate HIV-1 uncoating and transport. Proc Natl Acad Sci U S A, 2017. 114(33): p. E6932–E6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell EM, et al. , Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J Cell Biol, 2008. 180(3): p. 549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez J, et al. , Transportin-1 binds to the HIV-1 capsid via a nuclear localization signal and triggers uncoating. Nat Microbiol, 2019. 4(11): p. 1840–1850. [DOI] [PubMed] [Google Scholar]
- 49.Blanco-Rodriguez G, et al. , Remodeling of the Core Leads HIV-1 Preintegration Complex into the Nucleus of Human Lymphocytes. J Virol, 2020. 94(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guedan A, et al. , HIV-1 requires capsid remodelling at the nuclear pore for nuclear entry and integration. PLoS Pathog, 2021. 17(9): p. e1009484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daneholt B, Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A, 2001. 98(13): p. 7012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mehlin H, Daneholt B, and Skoglund U, Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell, 1992. 69(4): p. 605–13. [DOI] [PubMed] [Google Scholar]
- 53.Rankovic S, et al. , HIV-1 uncoating occurs via a series of rapid biomechanical changes in the core related to individual stages of reverse transcription. J Virol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rankovic S, et al. , Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J Virol, 2017. 91(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen DE, et al. , Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science, 2020. 370(6513). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cosnefroy O, Murray PJ, and Bishop KN, HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology, 2016. 13(1): p. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hulme AE, Perez O, and Hope TJ, Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A, 2011. 108(24): p. 9975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pante N and Kann M, Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell, 2002. 13(2): p. 425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keminer O and Peters R, Permeability of single nuclear pores. Biophys J, 1999. 77(1): p. 217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ribbeck K and Gorlich D, Kinetic analysis of translocation through nuclear pore complexes. EMBO J, 2001. 20(6): p. 1320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohr D, et al. , Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J, 2009. 28(17): p. 2541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dharan A, et al. , KIF5B and Nup358 Cooperatively Mediate the Nuclear Import of HIV-1 during Infection. PLoS Pathog, 2016. 12(6): p. e1005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strunze S, et al. , Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe, 2011. 10(3): p. 210–23. [DOI] [PubMed] [Google Scholar]
- 64. Lee K, et al. , Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe, 2010. 7(3): p. 221–33. *Identification of CPSF6 as an HIV-1 host factor and the virus utilizing alternate pathways for nuclear import was first reported in this study. By infecting cells depeleted of specific nucleoporins with viral particles harboring either the wildtype or N74D CA, the authors observed that the CPSF6 deficient mutant N74D altered the nucleoporin requirement as observed with the wildtype virus.
- 65.Schaller T, et al. , HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog, 2011. 7(12): p. e1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kane M, et al. , Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. Elife, 2018. 7. *This eloborate study evaluated the consequence of siRNA knockdown of individual nucleoporins on the expression and function of other nucleoporins. By monitoring the ability of different NLS to permit nuclear localization of cargos in these knockdown cells, the authors suggest the presence of heterogeneous NPCs in cells.
- 67.Stanley GJ, Fassati A, and Hoogenboom BW, Atomic force microscopy reveals structural variability amongst nuclear pore complexes. Life Sci Alliance, 2018. 1(4): p. e201800142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schuller AP, et al. , The cellular environment shapes the nuclear pore complex architecture. Nature, 2021. *This study used cryo electron tomograph and samples prepared using cryo-focused-ion-beam milling to obtain structural model of NPC in their native environment. The authors observed in their model that the size of the central channel is substantially wider when compared with previous human NPC models that used purified nuclear envelopes. This study further provides evidence of heterogeneity in NPCs and how the cellular environment influence the dimesions and architecture of NPC.
- 69.Kosinski J, et al. , Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science, 2016. 352(6283): p. 363–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin DH, et al. , Architecture of the symmetric core of the nuclear pore. Science, 2016. 352(6283): p. aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bui KH, et al. , Integrated structural analysis of the human nuclear pore complex scaffold. Cell, 2013. 155(6): p. 1233–43. [DOI] [PubMed] [Google Scholar]
- 72.Eibauer M, et al. , Structure and gating of the nuclear pore complex. Nat Commun, 2015. 6: p. 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kee HL, et al. , A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol, 2012. 14(4): p. 431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma J, et al. , Self-regulated viscous channel in the nuclear pore complex. Proc Natl Acad Sci U S A, 2012. 109(19): p. 7326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Onischenko E, et al. , Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell, 2017. 171(4): p. 904–917 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dultz E and Ellenberg J, Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol, 2010. 191(1): p. 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kabachinski G and Schwartz TU, The nuclear pore complex--structure and function at a glance. J Cell Sci, 2015. 128(3): p. 423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou J, et al. , Role of kinesins in directed adenovirus transport and cytoplasmic exploration. PLoS Pathog, 2018. 14(5): p. e1007055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carnes SK, Zhou J, and Aiken C, HIV-1 Engages a Dynein-Dynactin-BICD2 Complex for Infection and Transport to the Nucleus. J Virol, 2018. 92(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dharan A, et al. , Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc Natl Acad Sci U S A, 2017. 114(50): p. E10707-E10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Malikov V, et al. , HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat Commun, 2015. 6: p. 6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saito A and Yamashita M, HIV-1 capsid variability: viral exploitation and evasion of capsidbinding molecules. Retrovirology, 2021. 18(1): p. 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goujon C, et al. , Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Z, et al. , The Interferon-Inducible MxB Protein Inhibits HIV-1 Infection. Cell Host Microbe, 2013. [DOI] [PubMed] [Google Scholar]
- 85.Kane M, et al. , MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature, 2013. 502(7472): p. 563–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schaller T, et al. , Effects of Inner Nuclear Membrane Proteins SUN1/UNC-84A and SUN2/UNC84B on the Early Steps of HIV-1 Infection. J Virol, 2017. 91(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhargava A, et al. , Inhibition of HIV infection by structural proteins of the inner nuclear membrane is associated with reduced chromatin dynamics. Cell Rep, 2021. 36(13): p. 109763. [DOI] [PubMed] [Google Scholar]


