ABSTRACT
The global amphibian declines are compounded by infections with members of the Ranavirus genus such as Frog Virus 3 (FV3). Premetamorphic anuran amphibians are believed to be significantly more susceptible to FV3 while this pathogen targets the kidneys of both pre- and postmetamorphic animals. Paradoxically, FV3-challenged Xenopus laevis tadpoles exhibit lower kidney viral loads than adult frogs. Presently, we demonstrate that X. laevis tadpoles are intrinsically more resistant to FV3 kidney infections than cohort-matched metamorphic and postmetamorphic froglets and that this resistance appears to be epigenetically conferred by endogenous retroviruses (ERVs). Using a X. laevis kidney-derived cell line, we show that enhancing ERV gene expression activates cellular double-stranded RNA-sensing pathways, resulting in elevated mRNA levels of antiviral interferon (IFN) cytokines and thus greater anti-FV3 protection. Finally, our results indicate that large esterase-positive myeloid-lineage cells, rather than renal cells, are responsible for the elevated ERV/IFN axis seen in the tadpole kidneys. This conclusion is supported by our observation that CRISPR-Cas9 ablation of colony-stimulating factor-3 results in abolished homing of these myeloid cells to tadpole kidneys, concurrent with significantly abolished tadpole kidney expression of both ERVs and IFNs. We believe that the manuscript marks an important step forward in understanding the mechanisms controlling amphibian antiviral defenses and thus susceptibility and resistance to pathogens like FV3.
IMPORTANCE Global amphibian biodiversity is being challenged by pathogens like the Frog Virus 3 (FV3) ranavirus, underlining the need to gain a greater understanding of amphibian antiviral defenses. While it was previously believed that anuran (frog/toad) amphibian tadpoles are more susceptible to FV3, we demonstrated that tadpoles are in fact more resistant to this virus than metamorphic and postmetamorphic froglets. We showed that this resistance is conferred by large myeloid cells within the tadpole kidneys (central FV3 target), which possess an elevated expression of endogenous retroviruses (ERVs). In turn, these ERVs activate cellular double-stranded RNA-sensing pathways, resulting in a greater expression of antiviral interferon cytokines, thereby offering the observed anti-FV3 protection.
KEYWORDS: amphibian immunity, antiviral responses, endogenous retroviruses, interferon response
INTRODUCTION
Infections by members of the genus Ranavirus (family Iridoviridae), such as Frog Virus 3 (FV3), pose a serious threat to global amphibian biodiversity (1, 2). Tadpoles of anuran (frog/toads) amphibians are thought to be significantly more susceptible to ranaviruses than the adult frogs of the respective species (3–5). This notion is contradicted by our repeated findings that tadpoles of the anuran Xenopus laevis frog bear significantly lower FV3 loads across several tissues, including their kidneys, which are a central FV3 target (6–8). Indeed, the weeks that it takes for experimentally infected tadpoles to succumb to FV3 infections negate the notion that tadpoles are devoid of effective antiviral defenses (9). Moreover, our observations that Xenopus laevis tadpoles mount what appear to be effective but distinct antiviral interferon (IFN) responses to FV3 (6, 8, 10) from adult frogs only begins to hint at the potential differences by which amphibians of distinct developmental stages deal with ranavirus infections.
In general, type I and III interferon (IFN) cytokines represent the cornerstone of vertebrate antiviral immunity. Xenopus amphibians (X. laevis and Xenopus tropicalis) encode diverse repertoires of these mediators; unique to those in bony fish, reptiles, birds, or mammals (11). Across vertebrates, both type I and III IFN expression is controlled by IFN regulatory factors (IRFs) downstream of pattern recognition receptors (12), such as those that detect double-stranded RNAs (dsRNAs) produced during viral replication (13). In turn, endogenous retroviruses (ERVs), which comprise a significant proportion of higher vertebrate genomes, also appear to play intimate roles in dictating IFN expression in mammals (14, 15) at least in part through the production of dsRNA intermediates (16). In mammals, ERVs are highly expressed during early development but are much more stringently controlled or are entirely silenced in adult tissues by means of epigenetic modifications such as DNA methylation (17). This epigenetic silencing of ERVs may be counteracted by DNA methylation inhibitors (DNMTis), such as 5-azacytidine (Aza), resulting in robust IFN responses both in higher (human cancer cell lines) and lower vertebrates (zebrafish) (16, 18, 19). Together, this information suggests that ERVs play notable roles in regulating antiviral immunity across vertebrata. While a handful of putative X. laevis ERVs are present in GenBank, Xen1 is the only ERV characterized to date within X. laevis and X. tropicalis genomes (20).
Here, we resolved the discrepancy between FV3-mediated tadpole mortalities despite relatively low tissue viral loads by demonstrating that tadpole kidney ERV-mediated IFN responses protect these animals from FV3 until the onset of metamorphosis when the ERV-elicited IFN expression wanes.
RESULTS
Tadpole kidney resistance to FV3 is marked by intrinsic antiviral protection.
Past studies compared tadpole FV3 susceptibilities with those of much older and larger adult frogs (6, 21, 22). To minimize size and cohort effects on our results, we infected tadpoles and postmetamorphic froglets from corresponding cohorts (from the same breeding but presenting distinct stages of development) with the same dose of FV3 (2.5 × 105 PFU) and examined their kidney virus loads shortly after infection. As expected, tadpoles possessed significantly lower FV3 viral particles in their kidneys than cohort-matched froglets (Fig. 1A). Concurrently, tadpole kidneys had significantly lower FV3 DNA copies than either metamorphosing or postmetamorphic froglets (Fig. 1B), suggesting that anti-FV3 protection is lost during metamorphosis.
FIG 1.
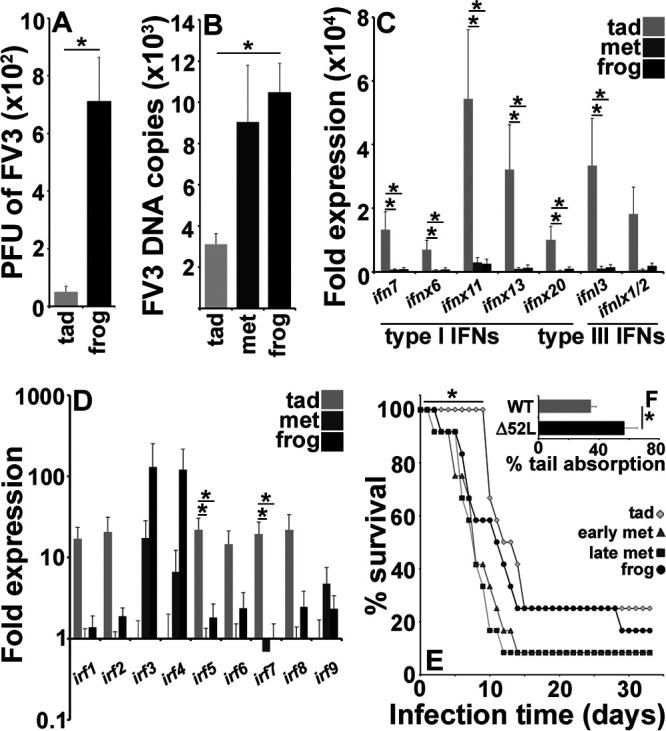
Tadpole kidney resistance to FV3 is marked by intrinsic antiviral protection. (A) Plaque assay analysis of FV3-infected tadpole and froglet kidneys (n = 6) at 6 h postinfection (hpi). (B) FV3 DNA viral loads in X. laevis tadpoles, metamorphs (NF 64), and froglets (n = 5/developmental stage). (C and D) IFN (C) and IRF (D) gene expression in healthy tadpole (NF 54) and froglet kidneys (n = 6/developmental stage). (E) Survival of tadpole (NF 54), early (NF 58) and late (NF 64) metamorphs, and froglets (n = 12/developmental stage) following FV3 infections (2.5 × 105 PFU of FV3/animal). (F) Percent tail absorption in metamorphosing tadpoles infected with WT- or Δ52L-FV3 for 4 days (2.5 × 105 PFU of FV3/animal; n = 9 per group). Results in A to D and F are means + SEM. Gene expression (C and D) was assessed relative to the gapdh endogenous control. Asterisks (*) above lines indicate statistical differences between treatment groups indicated by those lines.
Since type I and III IFNs play key roles in antiviral defenses of all vertebrates (11), including X. laevis (8, 10), we examined whether the type I and III IFN expression in tadpole, metamorphic and postmetamorphic froglet kidneys accounted for the observed differences in FV3 susceptibility. Indeed, tadpole kidneys had significantly greater transcript levels of most examined type I and III IFNs compared with metamorphs or froglets (Fig. 1C; see Fig. S1A in the supplemental material). The IFN genes expressed at greater levels in tadpole kidneys included an intron-containing type I IFNs, ifn7; intronless type I IFNs, ifnx6, ifnx11, ifnx13, and ifnx20; and an intron-containing type III IFN, ifnl3; but excluded the intronless type III IFN, ifnlx1/2 (Fig. 1C, Fig. S1A).
We examined whether and which other antiviral and innate immune genes were expressed at greater levels in the tadpole, as opposed to metamorphosing or postmetamorphosing frogs. In addition to the IFN genes, tadpoles possessed markedly higher but variable kidney expression levels of several other antiviral genes (Fig. S1B). They included antiviral genes coding for IFN-induced protein with tetratricopeptide repeats-1b (ifit1b), ifit5, IFN-induced transmembrane protein-1 (ifitm1), IFN-induced GTPase binding protein (mx1), adenosine deaminase acting on RNA-1 (adar1), and apolipoprotein B mRNA editing catalytic polypeptide-like-2 (apobec2; significantly so for ifit1b and adar1). The antiviral gene coding for the radical S-adenosyl methionine domain-containing protein (rsad) was expressed at similar levels across the examined X. laevis developmental stages. Tadpole kidneys also exhibited greater mRNA levels of the suppressor of cytokine signaling-3 (socs3) and the proinflammatory cytokine tumor necrosis factor (tnf) (Fig. S1C). The anti-inflammatory cytokine interleukin-10 (il10) was also expressed at substantially but not significantly greater levels in tadpole kidneys, whereas the mRNA levels for the dsRNA sensor retinoic acid inducible gene-I (rigi) were greater (not significantly so) in metamorphs and froglets (Fig. S1C) Conversely, metamorphic and postmetamorphic animals had significantly greater gene expression of the nuclear factor kappa B (nfkb) transcription factor, which is associated with proinflammatory responses in mammals (Fig. S1C).
IFN expression is regulated by IRFs (12), and we observed that tadpole kidneys had significantly greater mRNA levels of irf1 than metamorphic but not postmetamorphic frogs. Tadpoles also possessed significantly more robust kidney expression of irfs 5 and 7 than both metamorphic and postmetamorphic animals (Fig. 1D). Interestingly, the gene expression of irfs 3 and 4 increased with X. laevis development, albeit not significantly so (Fig. 1D).
In a comparison of animal survival following FV3 challenge (2.5 × 105 PFU), virus-infected tadpoles took longer to succumb to FV3 infections than early-metamorphic, late-metamorphic, or postmetamorphic froglets (Fig. 1E), in line with the respective kidney FV3 loads and antiviral gene expression of these respective developmental stages (Fig. 1A to D). Moreover, the decrease in tadpole survival coincided with the onset of metamorphosis.
Corticosteroid hormones (CHs) are critical to transitioning amphibian tadpoles into metamorphosis (23), while FV3 encodes a β-hydroxysteroid dehydrogenase homolog (52L), which is thought to be important to viral immune evasion (24). Hydroxysteroid dehydrogenase enzymes are typically involved in steroid biosynthesis and metabolism (25). Because our findings indicated antiviral protection of tadpole kidneys decreased with onset of metamorphosis, we reasoned that the FV3 52L confers evasion of host immunity by promoting metamorphosis. To test this hypothesis, we infected tadpoles with wild-type (WT) and mutant (Δ52L) FV3 and examined tail absorption as a measure of metamorphosis. As anticipated, the tadpoles infected with 52L-defective FV3 metamorphosed significantly slower than WT FV3-infected animals (Fig. 1F), suggesting that FV3 has coevolved to circumvent the greater tadpole kidney antiviral resistance by promoting their metamorphosis and thus diminished kidney antiviral status.
The antiviral state within tadpole kidneys correlates with elevated ERV expression.
Because elevated IFN expression has been linked to ERVs (14, 15), we next examined whether the heightened expression of IFN genes within X. laevis tadpoles correlated with ERV expression. While all examined ERVs were elevated in tadpole kidneys compared with those in metamorphic and postmetamorphic froglets, the Xen1 ERV was expressed at significantly greater levels in the kidneys of tadpoles compared to metamorphs or froglets (Fig. 2A). Our in silico analyses indicated that all examined ERV transcripts generate dsRNA structures (Fig. 2A).
FIG 2.
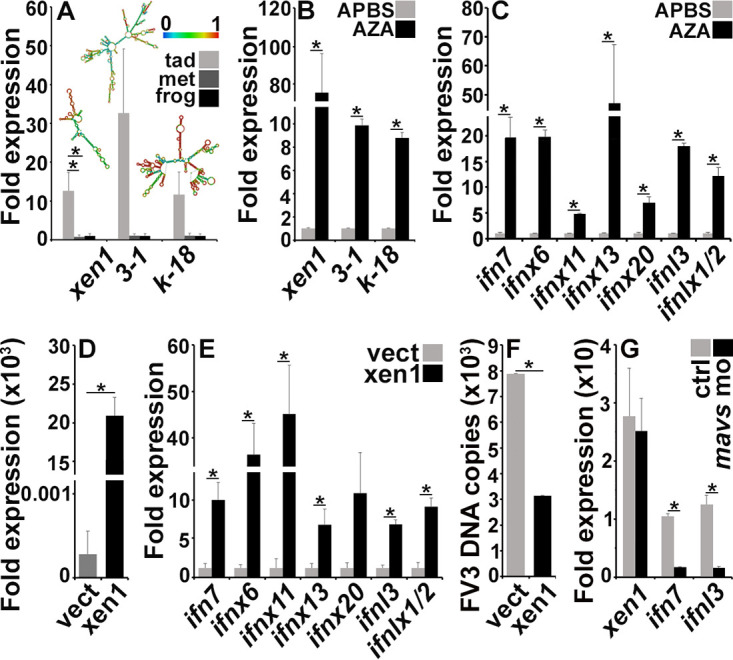
The antiviral state within tadpole kidneys is conferred by elevated ERV expression. (A) Quantitative analysis of ERV gene expression in kidneys of healthy tadpoles (NF 54), metamorphic animals (NF 64), and froglets (n = 6/stage). (B and C) ERV (B) and IFN (C) gene expression in A6 cells treated with the Aza DNA methylation inhibitors (dissolved in APBS) or solvent control (APBS n = 6/treatment group). (D and E) Xen1 (D) and IFN (E) gene expression in A6 cells transfected with a Xen1 expression construct or an empty expression vector (vect; n = 6). (F) FV3 DNA loads in empty plasmid- and Xen1 construct-transfected A6 cells 24 h after FV3 infection (MOI of 0.5; n = 6). (G) Xen1, IFN7, and IFNL3 gene expression in A6 cells overexpressing Xen1, transfected with anti-MAVS morpholino (n = 6). Gene expression (A to E and G) was assessed relative to the gapdh endogenous control. Results are means + SEM. Asterisks (*) above lines indicate statistical differences between treatment groups indicated by those lines.
Metamorphosis may be experimentally induced in amphibians through exposure to exogenous thyroid hormones such as T3 (23). X. laevis tadpole exposure to T3 resulted in a significant decrease in their kidney expression of Xen1 (Fig. S1C).
To confirm the link between the X. laevis kidney expression of ERVs and antiviral IFN genes, we first treated an X. laevis kidney-derived (A6) cell line with the DNA methylation inhibitor 5-azacytidine (Aza), which has been shown to activate epigenetically silenced ERVs (16), and found it resulted in significantly elevated A6 cell ERV expression (Fig. 2B) concomitant with an increased expression of most examined IFN genes (Fig. 2C). To verify the causal relationship between kidney cell ERV and IFN gene expression, we next overexpressed Xen1 in A6 cells (Fig. 2D), which also resulted in significantly increased A6 cell expression of most examined IFN genes (Fig. 2E) and greater anti-FV3 resistance (Fig. 2F). Overexpression of Xen1 in A6 cells also resulted in a significantly elevated expression of tnf and irfs 1, 2, 5, 6, 7, and 8 (see Fig. S2A in the supplemental material). We did not detect irf 3, 4, or 9 gene expression in A6 cells under any of the examined conditions.
ERVs have been shown to elicit IFN responses through the production of dsRNA and activation of dsRNA sensors (16). To examine whether the Xen1 (ORF1) overexpression in A6 cells resulted in increased innate immune gene expression due to the production of dsRNA, we synthesized Xen1 ORF1 RNA in vitro and transfected this RNA into A6 cells. To confirm that any observed effects resulting from this transfection were due to the double-stranded nature of the Xen1 RNA, we also transfected A6 cells with Xen1 ORF1 RNA that had been treated with RNase III, which specifically cleaves dsRNA (26). As anticipated, A6 cells transfected with the Xen1 ORF1 RNA exhibited elevated expression of ifn7, ifnl3, and tnf (significantly so for ifn7 and ifnl3), whereas A6 cells transfected with RNaseIII-treated Xen1 RNA did not show discernible gene expression differences from the mock-transfected controls (Fig. S2B). The mitochondrial antiviral signaling protein (MAVS) serves as a converging point in cell signaling from dsRNA sensors (27). Thus, we used antisense morpholinos to knock down the expression of MAVS in Xen1-overexpressing A6 cells to determine if such pathways may be mediating the increases in innate immune gene expression seen in these Xen1-overexpressing cells. While morpholino-mediated inhibition of MAVS in Xen1-overexpressing A6 cells did not affect Xen1 expression, it did decrease IFN expression by these cells (Fig. 2G). Together, this finding further supports our hypothesis that Xen1-mediated IFN expression is conferred at least in part through the recognition of dsRNA formed by the Xen1 transcripts.
Esterase-positive myeloid cells correlate with kidney ERV and IFN expression.
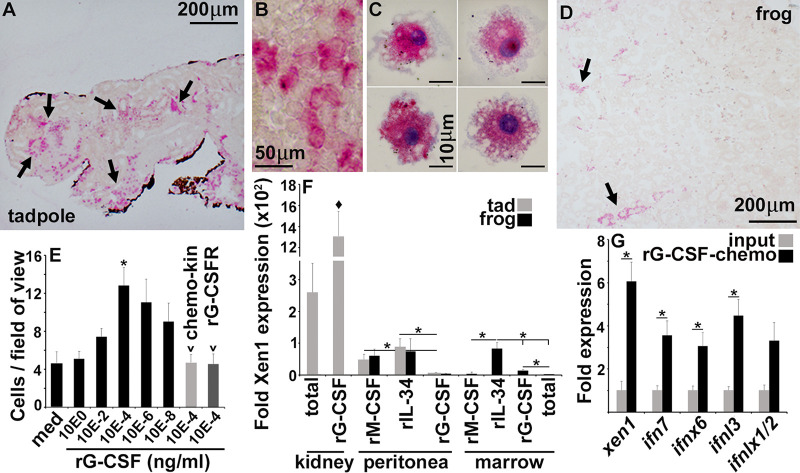
We previously showed that administering a recombinant form of the colony-stimulating factor 3 (rCSF3; granulocyte colony-stimulating factor [G-CSF]): principal neutrophil growth factor ([28]) to X. laevis tadpoles enhanced their anti-FV3 protection (29). While mammalian bone marrow is responsible for the production and storage of myeloid cells, such as granulocytes (30), amphibian tadpoles do not begin to form bone marrow until late into metamorphosis (31). Presumably, tadpoles must instead rely on other tissues, such as their kidneys, for myelopoiesis and myeloid cell storage (32, 33). Accordingly, we reasoned that kidney-resident myeloid cells may play a role in the observed ERV/IFN axis. Our histological analyses of specific esterase (granulocyte/monocyte marker) activity in tadpole and froglet kidneys confirmed that tadpoles possessed substantially greater numbers of kidney-resident esterase-positive myeloid cells than postmetamorphic froglets (Fig. 3A to D). These esterase-positive cells were relatively large (20 to 25 μm), were primarily mononuclear, and possessed extensive vacuolation and membrane ruffling (Fig. 3B and C). Using dispersed tadpole kidney cells in chemotaxis assays against increasing concentrations of rCSF3, we confirmed tadpole kidneys indeed possessed myeloid-lineage rCSF3-responsive cells (Fig. 3E). Moreover, these kidney myeloid cells responded to rCSF3 through rCSF3 gradient-dependent chemotaxis rather than random gradient-independent migration, known as chemokinesis (labeled as “chemo-kin” in Fig. 3E) and in a CSF3 receptor-dependent manner that could be ablated with the addition of an excess of soluble rCSF3 receptor (rCSF3R) (Fig. 3E).
FIG 3.
Esterase-positive myeloid cells are responsible for the high kidney ERV and IFN expression. Tadpole (NF 54) (A and B) and froglet (D) kidneys were stained and examined for the relative presence of esterase-positive (granulocyte/monocyte marker) cells. (C) Esterase-positive myeloid cells from tadpole kidneys. The results in A to D are representative of histological analyses of kidneys from 6 different tadpoles or froglets (n = 6/stage). (E) Tadpole kidney cell suspensions were subjected to chemotaxis assays against rCSF3 (100 to 10−8 ng/mL), using 5 × 105 cells per well of chemotaxis chambers (n = 4 to 8). Chemokinesis (chemo-kin) was assessed by examining migration with 104 ng/mL of rCSF3 in lower and upper chemotaxis chambers (n = 6). The role of kidney cell CSF3R in the rCSF3-mediated chemotaxis was examined by assessing migration in the absence or presence of 5 μg/mL of a soluble (extracellular portion of) rCSF3R (n = 6). (F) Comparison of Xen1 gene expression in tadpole rCSF3-recruited kidney cells (n = 6) relative to other X. laevis myeloid cells (n = 5). (G) Xen1 and IFN gene expression in total and rCSF3-chemoattracted tadpole kidney cells (n = 6). The results in E to G are means + SEM. Gene expression (F and G) was assessed relative to the gapdh endogenous control. Arrows in (A) and (D) indicate esterase-positive cells. Asterisks (*) above lines indicate statistical differences between treatment groups indicated by those lines. The diamond symbol (♦) indicates statistical difference from all other groups.
We showed previously that frog bone marrow-derived macrophages differentiated with interleukin-34 (IL-34) but not with colony-stimulating factor 1 (CSF1; macrophage colony-stimulating factor [M-CSF]) are important producers of antiviral IFNs (34). Presently, we compared the gene expression of Xen1 in total tadpole kidney cells, the rCSF3-chemo-attracted kidney myeloid cells, and several X. laevis myeloid populations (Fig. 3F). The tadpole kidney cells that migrated toward rCSF3 possessed strikingly more robust Xen1 gene expression than tadpole or frog peritoneum-derived (rIL-34 or rCSF1) macrophages or (rCSF3) granulocytes or adult frog bone marrow-derived macrophage (rIL-34- or rCSF1-differentiated) or granulocyte (rCSF3-differentiated) cultures (Fig. 3F). Freshly isolated bone marrow cells possessed the lowest Xen1 mRNA levels. Notably, compared with total kidney cells, these kidney-resident myeloid cells expressed significantly greater levels of Xen1 and IFN genes (Fig. 3G), suggesting these cells are responsible for a large proportion of the tadpole kidney ERV/IFN expression.
Tadpole myeloid cells are responsible for their kidney ERV and IFN gene expression.
Since we previously observed that rCSF3 enhanced tadpole anti-FV3 protection (28), we reasoned that one of the mechanisms by which this growth factor would be able to skew tadpole antiviral efficacies is by altering their kidney myeloid cell pools. As anticipated, tadpoles administered with rCSF3 possessed significantly greater numbers of esterase-positive cells in their kidneys than animals injected with a recombinant control (r-ctrl) (Fig. 4A and B). Moreover, rCSF3-treated tadpole kidneys exhibited significantly greater Xen1 and IFN gene expression than control tadpole kidneys (Fig. 4C). This finding confirmed that these rCSF3-responsive esterase-positive myeloid cells are important contributors to the tadpole ERV/IFN axis.
FIG 4.
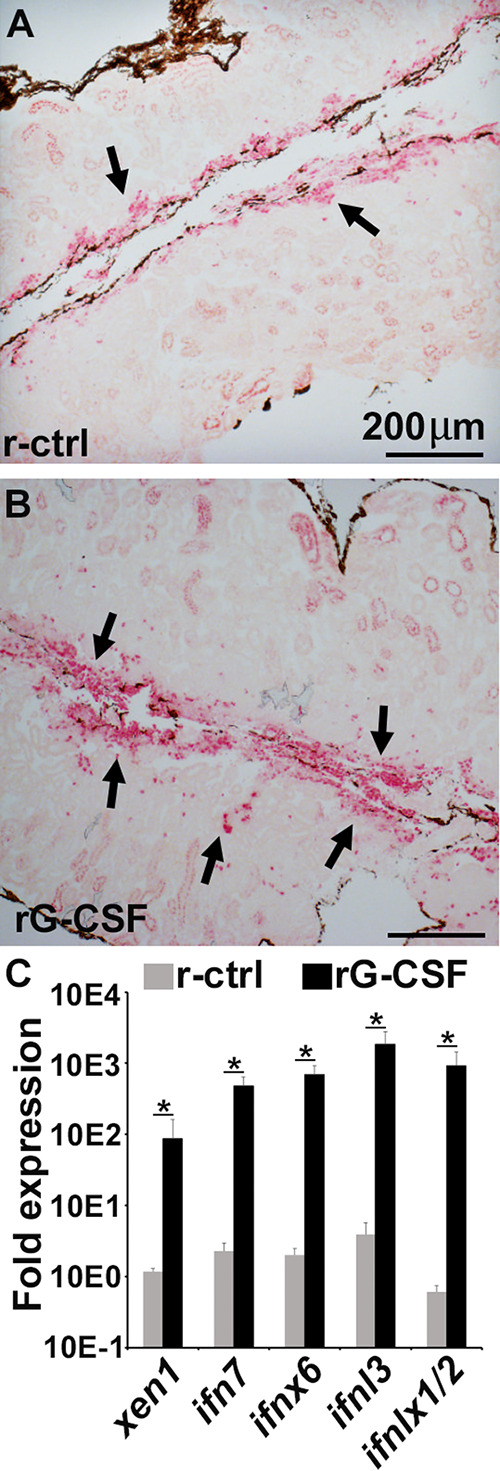
Tadpoles injected with rCSF3 possess more esterase-positive myeloid cells in their kidneys, concurrent with greater Xen1 and IFN gene expression. Tadpoles (NF 54) were injected i.p. with rCSF3 (2 μg/animal) or the recombinant control (r-ctrl), and 24 h later, their kidneys were examined for the proportions of esterase-positive cells (A and B) and Xen1 and IFN gene expression (C). Images in A and B are representative of results derived from 6 individual animals per treatment group (n = 6). The results in C are means + SEM, n = 6. Gene expression (C) was assessed relative to the gapdh endogenous control. Xen1 and IFN gene expression was normalized against the baseline (r-ctrl) expression for the respective genes. Arrows in (A) and (B) indicate esterase-positive cells. Asterisks (*) above lines indicate statistical differences between treatment groups indicated by those lines.
Retention of ERV/IFN-expressing myeloid cells within tadpole kidneys depends on CSF3.
Since tadpole kidney esterase-positive cells are responsive to rCSF3, their development, homing, and/or retention in this tissue likely also depends on CSF3. To test this idea, we used CRISPR-Cas9 to generate transgenic X. laevis with disrupted gcsf.s (Δcsf3.s) or csf3.l (Δcsf3.l) alleles (Fig. 5A). Compared with their cohort controls, Δcsf3.s and Δcsf3.l tadpoles possessed reduced numbers of esterase-positive cells in their kidneys (Fig. 5B to D), and these decreases corresponded with significantly diminished kidney gene expression of ERVs (and csf3) (Fig. 5E) concurrent with significantly reduced IFN transcript levels in these tissues (Fig. 5F).
FIG 5.
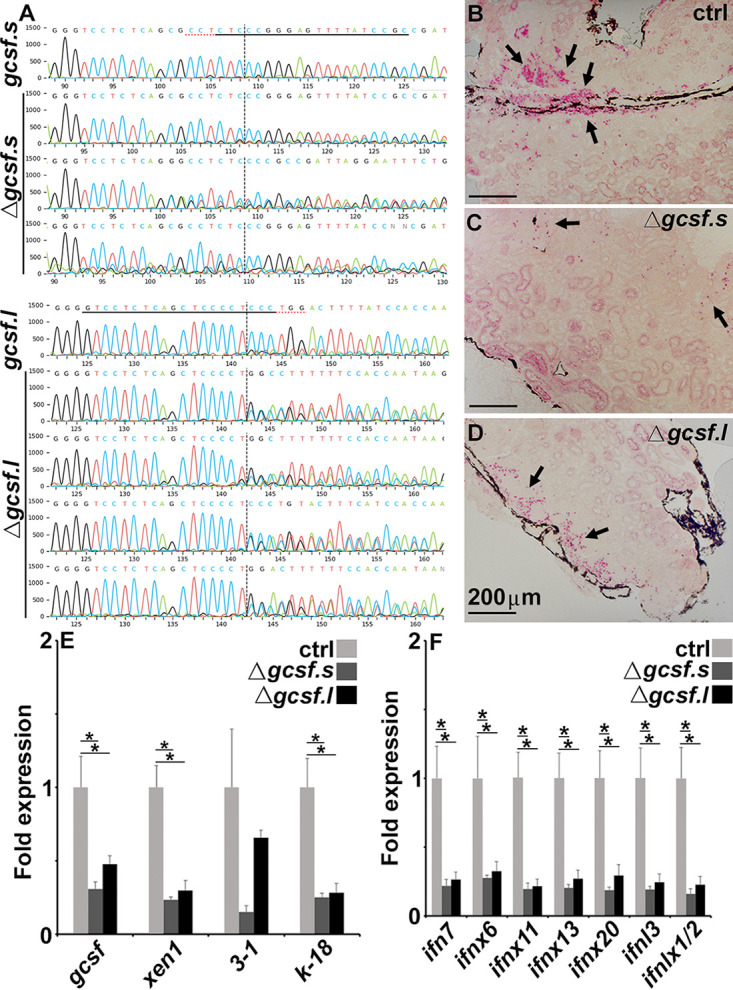
F0 transgenic tadpoles bearing CRISPR-Cas9-induced mutations in csf3.s or csf3.l possess fewer esterase positive myeloid cells in their kidneys, concurrent with lower Xen1 and IFN gene expression therein. (A) Sequenced csf3 loci from cohort controls and CRISPR-Cas9-altered F0 tadpoles. The underlined sequences indicate the guide RNA sequences, the red underlines denote the protospacer adjacent motif (PAM) sites, and the vertical dotted lines represent the cut sites. Esterase-positive cells in the kidneys of cohort controls (B), Δcsf3.s (C), and Δcsf3.s (D) F0 tadpoles. Images in C and D are representative of 3 and 4 tadpoles, respectively. Analysis of CSF3, Xen1, EVR3 to 1, and ERV k-18 (E) and IFN gene expression in cohort controls (N = 6), Δcsf3.s (N = 3), and Δcsf3.s (N = 4) F0 tadpoles (F). The results in E and F are means + SEM. Gene expression (E and F) was assessed relative to the gapdh endogenous control. Arrows in (B-D) indicate esterase-positive cells. Asterisks (*) above lines indicate statistical differences between treatment groups indicated by those lines.
Together, our findings suggest that large, esterase-positive myeloid cells are homed/retained in tadpole kidneys via CSF3, which is presumably produced by the kidney cells and/or other resident immune cells (see Fig. S3 in the supplemental material). These myeloid cells possess broad ERV expression, which we speculate results in the activation of intracellular antiviral (pathogen pattern recognition [PRR]) receptors that signal through MAVS to upregulate IFN gene expression. The subsequent production of antiviral IFNs by these myeloid cells results in a heightened antiviral state within tadpole kidneys, thus offering anti-FV3 protection.
DISCUSSION
We do not yet know the extent to which endogenous retroviruses have shaped animal immune defenses, but a growing body of literature indicates ERVs are intimately linked with the regulatory networks of IFN responses (14) across vertebrate species. Our present findings suggest the intrinsically heightened antiviral capacities within X. laevis tadpole kidneys are conferred by resident esterase-positive myeloid cells, which express elevated ERV and IFN genes. Moreover, our work indicates that akin to what has been see in mammals (35), X. laevis ERV-mediated IFN expression results from the recognition of double-stranded ERV RNAs. Presumably, this ERV-mediated IFN augmentation has been co-opted into vertebrate immune systems. In support of this notion, it was interesting to see that adult X. laevis bone marrow-derived IL-34 macrophages, which are prominently involved in X. laevis antiviral IFN responses (34), also possessed significantly greater Xen1 expression than other bone marrow-derived cell types. Moreover, autocrine antiviral priming of cells through the constitutive production of antiviral IFNs is a well-accepted phenomenon (36–38) and it stands to reason that the production and immune recognition of ERV dsRNA may be a prominent mechanism by which this priming is maintained.
Antiviral IFN gene expression is regulated largely by IFN regulatory factors (IRFs) and NF-κB (39). The promoter regions of distinct IFN genes vary in the binding sites for these transcription factors (40, 41). In mammals, IRFs 1, 3, 5, and 7 are linked with positive regulation of IFN gene activation (39). Of these, the transcripts for IRFs 1, 5, and 7 were significantly more abundant in tadpole kidneys, whereas the expression of kidney IRF3 and NF-κB increased with X. laevis development. While X. laevis encodes considerably different repertoires of IFN genes to those seen in mammals (11), it is also notable that tadpoles and adult frogs upregulate distinct IFN genes following FV3 infections (8, 10). These disparate IRF/NF-κB expression levels in tadpole and adult kidneys may account at least in part for the differences in tadpole and adult IFN gene expression at baseline and following viral infections.
Our results suggest that the heightened tadpole kidney IFN expression and intrinsic antiviral protection are linked to esterase-positive myeloid cells residing within this tissue and their ERV expression. It is notable that peritoneum-derived tadpole and froglet macrophages and granulocytes did not substantially differ in their ERV expression. Moreover, while the bone marrow-derived IL-34-macrophages exhibited broader ERV expression than other bone marrow-derived cell types, the tadpole kidney rCSF3-chemotaxed cells possessed ERV expression that was magnitudes greater than even these cells.
We showed recently that tadpoles respond to intestinal FV3 infections by recruiting esterase-positive, IFN-expressing, and IL-34-dependent mononuclear phagocytes into this tissue (7). Considering that in addition to polymorphonuclear granulocytes, the CSF3 receptor (CSF3R) is also expressed on mammalian monocytes (42) and X. laevis IL-34-macrophages (43), we anticipate that the presently described rCSF3-responsive mononuclear phagocytes residing within the tadpole kidneys share features with the frog antiviral IL-34-macrophages. While our past work suggests that tadpoles fail to engage IL-34-macrophages during their kidney anti-FV3 responses (44), perhaps tadpoles already possess sufficient IL-34-macrophage-like cells within their kidneys before even encountering this pathogen. In turn, these tadpole myeloid population(s) likely adopt tissue-specific features that are either not shared with other X. laevis myeloid lineages and/or are incurred upon entering these respective tissues and/or residing therein. Indeed, X. laevis possesses other myeloid lineage cells that share characteristics with but are overall unique to conventional mammalian phagocytes (45).
In addition to granulopoiesis, the mammalian CSF3 also serves to mobilize stem cells from bone marrow into blood circulation (42). Additionally, administration of CSF3 to mammals may also increase the content and diversity of bone marrow progenitor cells (46). While, to our knowledge, hematopoietic progenitors are not known to possess substantial esterase activity or antiviral capacities, pluripotent cells do express substantial ERV levels (47). Thus, it is possible some of the ERV transcripts detected in the X. laevis tadpole kidney reflect the hematopoietic cells therein. However, it is notable that the adult frog bone marrow cells, which possess granulocyte and macrophage precursors (48, 49), exhibited the lowest ERV expression from any examined X. laevis cell type. This finding suggests that at least in adult frogs, pluripotency does not equate with heightened ERV expression.
The demands of amphibian physiology on processes such as hematopoiesis and immune defenses are relatively less well understood than those of mammals. As such, it is difficult to speculate whether the ERV/IFN-expressing kidney-resident cells described here coevolved with pathogens like FV3 to serve as immune sentinels or whether they are stored there akin to what is seen with the mammalian bone marrow. In support of the former notion, neither peritoneum-derived tadpole or adult or adult bone marrow-derived myeloid cells possessed comparable features to these tadpole esterase-positive kidney-resident cells. Possibly, this/these immune population(s) is/are unique to tadpole kidneys where these cells facilitate immune surveillance and defenses. The fact that FV3 has evolved a mechanism to expedite metamorphosis, which coincides with the loss of this kidney antiviral state, also supports the notion that these tadpole-resident myeloid cells function as mediators of antiviral immunity.
Generally, juvenile (premetamorphic) amphibians are thought to be more susceptible to ranaviruses than postmetamorphic animals (50). Notably, ranavirus-associated die-off events have often been reported in animals approaching or during metamorphosis (51–53). The onset and progression of amphibian metamorphosis are controlled by thyroid hormones and corticosteroids, culminating in an energetically costly process that coincides with naturally induced immune suppression (23, 54). Anuran amphibians undergoing metamorphosis were proposed to be especially susceptible to ranaviruses, coinciding with ranavirus-mediated die-offs in the summer months when anuran species are typically metamorphosing (51–53). Developmental and epidemiological data both support the notion that at least in some anuran species, as tadpoles progress toward metamorphosis their ranavirus susceptibility increases (55). Our findings underline these converging ideas at the molecular levels. While some of our findings are correlative at this stage, we definitively and consistently observe that tadpoles bear lower FV3 loads than adult frogs (8, 10, 44, 56). As kidneys of all stages of X. laevis represent central FV3 targets, it is notable that tadpoles bear lower kidney FV3 loads, concurrent with a greater expression of antiviral IFNs. Our in vitro data indicate that an overexpression of the Xen1 (ORF1) ERV is sufficient to offer anti-FV3 protection, presumably resulting from Xen1 dsRNA-mediated activation of antiviral receptors and culminating in the observed increases in IFN gene expression. Moreover, we see that in vivo, ERV and IFN expression are linked to esterase-positive tadpole kidney-resident myeloid cells, which are present in drastically lower numbers in kidneys of postmetamorphic animals. Enrichment of these myeloid cells results in greater tadpole kidney ERV/IFN expression while their depletion results in decreases in ERV/IFN expression. It is thus pertinent that the tadpole kidney ERV/IFN expression decreases with metamorphosis (and following T3 treatment), concurrent with their increased susceptibility to FV3. Thus, it is perhaps not surprising that FV3 appears to have a potential strategy for speeding up tadpole metamorphosis (via 52L gene), thereby presumably forcing these animals to diminish their kidney antiviral protection. And while the β-hydroxysteroid dehydrogenase is thought to modulate metamorphosis by altering corticosteroid metabolism, FV3 possesses nearly 100 genes, of which the functions of the majority are unknown (57). As such, FV3 may have evolved additional means for expediting tadpole metamorphosis, possibly involving thyroid hormones. It will be most invaluable to learn with future studies the physiological reasons behind why these antiviral myeloid cells populate tadpole kidneys, why their numbers are decreased during and postmetamorphosis, and how this ties into the coevolution of amphibians and ranaviruses, beyond X. laevis and FV3.
While to our knowledge Xen1 is the only ERV described within the X. laevis genome, a number of ERVs have been described in other amphibian species, including the related X. tropicalis (58). It will be interesting to learn with further studies which of these have been conserved across amphibian evolution and how these respective ERVs contribute to amphibian immunity and physiology.
A greater understanding of the physiological and evolutionary mechanisms that shaped the immune defenses of organisms, such as amphibians, will grant new avenues to better the ecological health of such animals and concomitantly offer new windows into the evolution of our own antimicrobial defenses.
MATERIALS AND METHODS
Animal husbandry, infections, and tissue collections.
All animals were purchased from Xenopus 1 (Dexter, MI) and reared in-house under strict laboratory conditions in accordance with IACUC regulations (approval number 15-024).
For T3 treatments, tadpoles (Stage NF 54) were reared in water containing T3 (10 nM, final concentration) or solvent control (NaOH) for 5 days. The water was changed daily for the 5-day duration of the experiments. At day 5, the animals were scarified and analyzed for ERV expression and viral load analysis.
Tadpoles (NF stage 54; n = 6) and froglets (n = 6) were injected intraperitoneally (i.p.) with 2-μg total of rCSF3 or equal volumes of the vector control in amphibian phosphate-buffered saline (APBS; 100 mM sodium chloride, 8 mM sodium phosphate, and 1.5 mM potassium phosphate [pH 7.7]) using finely pulled glass needles. At 24 h following the injections, the animals were scarified, and their kidneys were isolated for histology and expression analyses.
For all infection studies, tadpoles (NF stage 54), early (NF stage 58) and late metamorphs (NF stage 64), and froglets were inoculated intraperitoneally with 250,000 PFU of FV3 (n = 6 per group for kidney FV3 analyses and n = 12 per group for survival studies).
For tadpole kidney cell isolations, whole kidneys were incubated in Liberase (0.1 mg/mL; Roche Diagnostics) diluted with APBS for 30 min at 27°C and then washed 1× with APBS. The suspensions were passed subsequently through 70-μm cell strainers (VWR, Radnor, PA) to clear debris. The viability of the isolated kidney cell was confirmed by Trypan blue exclusion.
For tail regression studies, tadpoles (n = 9 per group) were inoculated i.p. with 250,000 PFU of WT- or Δ52L-FV3 and tails measured at the start of the experiment and 4 days after infection.
Identification of X. laevis ERVS.
To date, Xen1 (National Center for Biotechnology Information [NCBI] accession no. AJ506107.1) is the only characterized ERV in the X. laevis genome. Gene synteny analysis was performed to identify other, uncharacterized ERVs, including endogenous retrovirus group K member 18 Pol protein-like (ERVK-18; XR_005962535.1) and endogenous retrovirus group 3 member 1 Env polyprotein-like (ERV3-1; XR_005962535.1). The RNA Fold Web Server was used to predict possible X. laevis ERV dsRNA.
FV3 stocks and infections.
WT FV3 was propagated in baby hamster kidney (BHK-21) cells by inoculating fresh BHK-21 cultures with FV3 (multiplicity of infection [MOI], 0.1 PFU of FV3 per cell), and cultures were maintained at 30°C and 5% CO2 until they were completely lysed (approximately 5 days). The culture lysates were subjected to ultracentrifugation to obtain supernatant with FV3 using 30% sucrose. The supernatant was resuspended in APBS, and a plaque assay analysis on BHK-21 cells was used for determining the viral titers. Δ52L-FV3 was a kind gift from Jacques Robert. The production and characterization of this virus has been described (24). Briefly, the Δ52L-FV3 showed similar replication kinetics to WT FV3 in a mammalian and a fish-derived cell line but resulted in delayed and reduced infected tadpole mortalities, compared with WT FV3 (24).
For all infection studies, tadpoles, metamorphs, and postmetamorphic froglets were infected by intraperitoneal (i.p.) injections with 250,000 PFU of FV3 in 10 μL of APBS. Control animals were mock infected by i.p. injections with APBS (not containing FV3). Animals were euthanized by tricaine mesylate overdose (tadpoles, 1%; adult frogs, 5%); kidney tissues were excised, immediately flash frozen in TRIzol reagent (Invitrogen) over dry ice and, stored at −80°C until RNA isolation. FV3-infected kidneys of tadpoles and adults were also subjected to plaque-forming assays as described previously (24).
To compare animal survival (n = 12/stage), tadpoles (NF stage 54), early (NF stage 58) and late metamorphs (NF stage 64), and postmetamorphic froglets were infected i.p. with 250,000 PFU of FV3 using finely pulled glass needles, and the survival was monitored thereafter daily for 1 month.
For in vitro infection studies, transfected A6 cells were infected at a multiplicity of infection (MOI) of 0.5 PFU of FV3/A6 cells for 24 h, at 27°C with 5% CO2. Subsequently, the cells were collected in TRIzol for DNA isolation and viral load analysis.
Production of recombinant X. laevis myeloid growth factors.
The X. laevis recombinant (r)CSF1 rIL-34 rCSF3 and a soluble (extracellular portion of) rCSF3R were produced by cloning the sequences representing the signal peptide-cleaved transcripts of the respective cytokines into the pMIB/V5 His A insect expression vector (Invitrogen). These constructs were transfected into Sf9 insect cells (cellfectin II; Invitrogen). Recombinant protein production was confirmed by Western blot, and the positive transfectants were selected using 10 μg/mL blasticidin. The expression cultures were scaled up as 500-mL liquid cultures, grown for 5 days, and pelted, and the supernatants were collected. They were dialyzed overnight at 4°C against 150 mM sodium phosphate, concentrated against polyethylene glycol flakes (8 kDa) at 4°C, dialyzed overnight at 4°C against 150 mM sodium phosphate, and passed through Ni-nitrilotriacetic acid (NTA) agarose columns (Qiagen). Columns were washed with 2 times with 10 volumes of high stringency wash buffer (0.5% Tween 20, 50 mM sodium phosphate, 500 mM sodium chloride, and 100 mM imidazole) and 5 times with 10 volumes of low stringency wash buffer (as above, but with 40 mM imidazole). Recombinant cytokines were eluted using 250 mM imidazole and were confirmed by Western blot against the V5 epitopes on the proteins, and the protein concentrations were determined by Bradford protein assays (Bio-Rad). Halt protease inhibitor cocktail (containing 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride [AEBSF], aprotinin, bestatin, E-64, leupeptin, and pepstatin A; Thermo Scientific) was added to the purified proteins, which were then stored at −20°C in aliquots until use.
The recombinant control (r-ctrl) was generated by transfecting an empty pMIB/V5 His A insect expression vector into Sf9II cells and isolating and processing the resulting cell supernatants akin to and in parallel with the recombinant cytokine production.
Generation of X. laevis bone marrow- and peritoneum-derived myeloid cell subsets.
All cell cultures were established using Iscove’s modified Dulbecco's medium supplemented with fetal bovine serum (10%); X. laevis serum (0.25%); primatone (2.5%); insulin (Sigma); nonessential amino acids (Sigma); and a cocktail of penicillin G (100 U/mL), streptomycin (100 mg/mL), and gentamycin (10 mg/mL), buffered with sodium bicarbonate (pH 7.7) and diluted to amphibian osmolarity. All cultures were maintained at 27°C with 5% CO2 and 95% relative humidity.
Bone marrow cells were isolated from adult, mixed-sex X. laevis (approximately 1 year old). To this end, animals were euthanized in 5% tricaine mesylate followed by cervical dislocation. Their femurs were aseptically removed, and cells flushed out of the bones using ice-cold APBS under sterile conditions. White and red blood cells were separated using differential centrifugation against 51% Percoll. Bone marrow cells were enumerated using trypan blue live/dead exclusion, and cells were seeded at a density of 105 cells/well in the presence of 250 ng/mL of rCSF1, rIL-34, or rCSF3. After 3 days of culture, cells were treated again with the respective cytokines. The cells were collected after 5 days of culture and processed for gene expression analyses.
Tadpole and froglet CSF1-/IL-34 macrophages and CSF3 granulocytes were generated by injecting tapoles and froglets i.p. with 1 or 5 μg (respectively) of the respective recombinant cytokines. Based on previous studies, after 1 (rCSF3) or 3 (rCSF1/rIL-34) days of injection, the peritoneal leukocytes enriched for the respective myeloid subsets were isolated by peritoneal lavage with ice-cold APBS. The cells were processed for gene expression analyses.
For inhibition of DNA methylation experiments, A6 cells were seeded at 10,000 cells/well in 96-well plates. The cells were treated with 500 nM 5-azacytidine (Aza; Sigma, St. Louis, MO) or APBS (solvent control) for 72 h, and DNA and RNA were isolated at 7 days following removal of the drug.
Construction of the plasmid overexpressing Xen1-ORF1 and transfection in A6 cells.
Xen-1 is a provirus with a complete proviral genome of 10,207 bp in length, organized as LTR-ORF1, ORF2, gag, pol, and env-LTR. The two open reading frames (ORFs) have been suggested to encode hypothetical proteins with no similarity found with proteins of known functions. Xen1 cDNA corresponding to the open reading frame (ORF1) was cloned into a pcDNA3.1 (+) vector using primers designed by NEBuilder for Gibson assembly reactions. The resulting clones were verified using sequencing. The positive clones were then transfected into A6 cells using Lipofectamine LTX reagent with Plus reagent (Invitrogen, Carlsbad, CA) following manufacturer’s instructions.
Morpholino targeting of X. laevis mitochondrial antiviral signaling protein.
Translation-blocking Morpholino (mo), targeting the conserved region of all isoforms of the X. laevis MAVS (5′-AACTTGTCTTCAGCAAAGCCCATTC-3′), was designed by and purchased from GeneTools, LLC, and introduced into A6 cells using the Endo-Porter polyethylene glycol (PEG) reagent according to the manufacturer’s directions (GeneTools, LLC).
Chemotaxis assays.
The chemotaxis and chemokinesis assays were performed as described previously (7, 59) using 100 to 10−8 ng/mL of rCSF3 and 5 × 104 kidney cells per well.
All chemotaxis assays were performed using blind well Boyden chambers (NeuroProbe, USA). The lower wells were filled with 100 to 10−8 ng/mL of rCSF3 in complete medium. The wells were overlaid with chemotaxis filters (5-μm pore size; Neuro Probe), and the top wells were attached and filled with 5 × 104 tadpole kidney cells/well in medium. Chemokinesis (gradient-independent migration) experiments employed 10−4 ng/mL of rCSF3 (optimal chemotaxis dose) in both upper and lower chemotaxis chambers, thereby abolishing any rCSF3 gradients across lower and upper chemotaxis chambers. To confirm the role of the CSF3R in kidney cell migration toward rCSF3, kidney cells were coincubated with 5 μg/mL of a soluble (extracellular portion of) rCSF3R (to compete for cellular CSF3-CSF3R binding), and 10−4 ng/mL of rCSF3 was added to lower chambers. For all chemotaxis experiments, the chemotaxis chambers were incubated at 27°C and 5% CO2 for 3 h. After this step, the contents of the top wells were aspirated, and the top sides of the filters were gently wiped with cotton swabs. The filters were then stained with Giemsa (Gibco, Thermo Fisher), mounted bottom side up onto microscope slides, and examined by microscopy. For each filter, cells present in 10 random fields of view were enumerated with a 40× objective. The cells that had migrated to rCSF3 in the lower chambers were collected in TRIzol for gene expression analyses.
RNA and DNA isolation.
For all experiments, tissues and cells were homogenized by passage through progressively higher gauge needles in TRIzol reagent (Invitrogen), flash frozen on dry ice, and stored at −80°C until RNA and DNA isolation. RNA isolation was performed using TRIzol (Invitrogen) according to the manufacturer’s directions. DNA was isolated from the TRIzol following RNA isolation. In brief, following phase separation and extraction of RNA, the remaining TRIzol layer was mixed with back extraction buffer (4 M guanidine thiocyanate, 50 mM sodium citrate, and 1 M Tris [pH 8.0]), centrifuged to isolate the DNA containing aqueous phase. The DNA was precipitated overnight with isopropanol, pelted by centrifugation, washed with 70% ethanol, and resuspended in Tris-EDTA (TE; 10 mM Tris [pH 8.0] and 1 mM EDTA) buffer. DNA was then purified by phenol-chloroform extraction and resuspended in molecular-grade water.
Quantitative gene expression analyses.
Toward quantitative gene expression analyses, total extracted RNAs (500 ng/sample) were reverse transcribed using iScript cDNA synthesis kits (Bio-Rad, Hercules, CA) per manufacturer’s instructions. All expression analyses were performed using the delta^delta threshold cycle (CT) method, compared with the GAPDH as an endogenous control.
For FV3 copy number analyses, an FV3 standard curve was generated by serially diluting an FV3 vDNA Pol (ORF 60R) fragment cloned into the pGEM-T plasmid. This FV3 DNA Pol standard curve was used in absolute quantitative PCRs (qPCRs), assessing “absolute” FV3 DNA copies per 500 ng of input DNA from respective samples of interest. All qPCR assays were performed using the iTaq universal SYBR green Supermix (Bio-Rad Laboratories), and all experiments were analyzed via the CFX96 real-time system (Bio-Rad Laboratories, Hercules, CA) and the Bio-Rad CFX manager software (SDS). All primers were validated prior to use, and all primer sequences are listed in Table S1 in the supplemental material.
Synthesis of Xen1 ORF1 RNA, RNase III treatment, and A6 cell transfections.
The Xen1 ORF1 was cloned into the pGEM T Easy vector (Promega), and the RNA was synthesized using T7 RNA polymerase (Sigma) according to the manufacturer’s instructions. The RNA was purified using RNA purification columns (Zymo Research). Some of this RNA was subjected to RNase III digestion (Invitrogen) according to the manufacturer’s instructions and repurified. A6 cells were seeded into individual wells of 96-well plates at 2.5 × 104 cells per well and the following day mock-transfected (transfection reagent alone) or transfected with the RNase III digested and the undigested Xen1 ORF1 RNA (100 ng/well; 4 wells per treatment group, n = 4). After 24 h, the cells were collected in TRIzol (Invitrogen) and RNA isolation, cDNA synthesis, and qPCR analysis were performed as described above.
Histological analyses of X. laevis kidneys.
Upon isolating X. laevis kidneys, they were immediately fixed in 10% neutral buffered formalin (VWR) for a minimum of 24 h before these tissues were submitted for processing, embedding in paraffin, and sectioning (5 μm) by the GWU Pathology Core. Sections were processed for and stained using a chloroacetate esterase (Leder; Sigma) kit according to the manufacturer’s instructions and optimized to frog tissues.
CRISPR-Cas9 editing of csf3.s and csf3.l genes.
CRISPR direct (http://crispr.dbcls.jp) and Indelphi (https://www.crisprindelphi.design/) were used to design individual single guide RNAs (sgRNAs) to the coding region of X. laevis gcsf.l and gcsf.s, ensuring unique hits. To create a DNA template for subsequent sgRNA production, a modified universal reverse primer was used in conjunction with a gene-specific forward primer containing a T7 polymerase promoter (see Table S3 in the supplemental material). The GG nucleotides were added to the 5′ ends of forward primers for improved mutagenic activity (8). sgRNAs were synthesized by in vitro transcription of the sgRNA PCR template using the T7 MEGAscript kits (Ambion). Prior to injections, guide cocktails were prepared, including specific sgRNAs (750 pg/nL), Cas9 protein (1 ng/μL), and Texas red RNA tracer and control cocktail without sgRNA. The Δcsf3.s and Δcsf3.l F0 founders were produced by injecting respective guide cocktails into the animal view at the 1-cell embryo stage of X. laevis J strains to generate the mutants. The control F0 founders were generated using the same process. A day after injections, embryos fluorescing the tracer were sorted out. Of them, 6 embryos were genotyped for ensuring the mutating efficiency (see Table S4 in the supplemental material). The remaining guide/control-injected embryos were raised to generate F0 mutant and control tadpoles (NF stage 54). The kidneys from these animals were excised and processed for RNA/DNA isolation and histology.
Statistical analyses.
All the statistical analyses, including independent t tests, one-way analysis of variance (ANOVA), post hoc Tukey’s-test, and log-rank test for survival curve analysis were performed using IBM SPSS statistics 27. A probability level of <0.05 was considered significant.
ACKNOWLEDGMENTS
We thank Jacques Robert and the Xenopus laevis immunology resource for providing the mutant Δ52L-FV3 and conversations with Emily Le Sage, which precipitated the Δ52L-FV3 work. We thank Katherine Chiappinelli for guidance in Aza-treatment experiments. We thank Erik Rodriguez for generously providing us with the pcDNA3.1 (+) expression vector.
This work was supported by grants from NSF-IOS 1749427 to L.G. and NIH R24OD030008 and P40OD010997 to M.E.H.
We thank the anonymous reviewers, whose astute comments and suggestions helped improve the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Leon Grayfer, Email: leon_grayfer@gwu.edu.
Rozanne M. Sandri-Goldin, University of California, Irvine
REFERENCES
- 1.Chinchar VG, Hyatt A, Miyazaki T, Williams T. 2009. Family Iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol 328:123–170. 10.1007/978-3-540-68618-7_4. [DOI] [PubMed] [Google Scholar]
- 2.Williams T, Barbosa-Solomieu V, Chinchar VG. 2005. A decade of advances in iridovirus research. Adv Virus Res 65:173–248. 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- 3.Hoverman JT, Gray MJ, Miller DL. 2010. Anuran susceptibilities to ranaviruses: role of species identity, exposure route, and a novel virus isolate. Dis Aquat Organ 89:97–107. 10.3354/dao02200. [DOI] [PubMed] [Google Scholar]
- 4.Landsberg JH, Kiryu Y, Tabuchi M, Waltzek TB, Enge KM, Reintjes-Tolen S, Preston A, Pessier AP. 2013. Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Dis Aquat Organ 105:89–99. 10.3354/dao02625. [DOI] [PubMed] [Google Scholar]
- 5.Reeve BC, Crespi EJ, Whipps CM, Brunner JL. 2013. Natural stressors and ranavirus susceptibility in larval wood frogs (Rana sylvatica). Ecohealth 10:190–200. 10.1007/s10393-013-0834-6. [DOI] [PubMed] [Google Scholar]
- 6.Grayfer L, De Jesus Andino F, Robert J. 2015. Prominent amphibian (Xenopus laevis) tadpole type III interferon response to the frog virus 3 ranavirus. J Virol 89:5072–5082. 10.1128/JVI.00051-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauser K, Singer J, Hossainey MRH, Moore T, Wendel ES, Yaparla A, Kalia N, Grayfer L. 2021. Amphibian (Xenopus laevis) tadpoles and adult frogs differ in their antiviral responses to intestinal Frog Virus 3 infections. Front Immunol 12:737403. 10.3389/fimmu.2021.737403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendel ES, Yaparla A, Koubourli DV, Grayfer L. 2017. Amphibian (Xenopus laevis) tadpoles and adult frogs mount distinct interferon responses to the Frog Virus 3 ranavirus. Virology 503:12–20. 10.1016/j.virol.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Gantress J, Maniero GD, Cohen N, Robert J. 2003. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology 311:254–262. 10.1016/S0042-6822(03)00151-X. [DOI] [PubMed] [Google Scholar]
- 10.Wendel ES, Yaparla A, Melnyk MLS, Koubourli DV, Grayfer L. 2018. Amphibian (Xenopus laevis) tadpoles and adult frogs differ in their use of expanded repertoires of type I and type III interferon cytokines. Viruses 10:372. 10.3390/v10070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Y, Jennings J, Gong Y, Sang Y. 2019. Xenopus interferon complex: inscribing the amphibiotic adaption and species-specific pathogenic pressure in vertebrate evolution? Cells 9:67. 10.3390/cells9010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6:644–658. 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 13.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat Immunol 7:131–137. 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 14.Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351:1083–1087. 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandi N, Tramontano E. 2018. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol 9:2039. 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Budhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Chan TA, Baylin SB, Strick R. 2017. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 169:361. 10.1016/j.cell.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 17.Eiden MV. 2008. Endogenous retroviruses—aiding and abetting genomic plasticity. Cell Mol Life Sci 65:3325–3328. 10.1007/s00018-008-8493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chernyavskaya Y, Mudbhary R, Zhang C, Tokarz D, Jacob V, Gopinath S, Sun X, Wang S, Magnani E, Madakashira BP, Yoder JA, Hoshida Y, Sadler KC. 2017. Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development 144:2925–2939. 10.1242/dev.147629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, O'Brien C, De Carvalho DD. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162:961–973. 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kambol R, Kabat P, Tristem M. 2003. Complete nucleotide sequence of an endogenous retrovirus from the amphibian, Xenopus laevis. Virology 311:1–6. 10.1016/s0042-6822(03)00263-0. [DOI] [PubMed] [Google Scholar]
- 21.Grayfer L, De Jesus Andino F, Robert J. 2014. The amphibian (Xenopus laevis) type I interferon response to frog virus 3: new insight into ranavirus pathogenicity. J Virol 88:5766–5777. 10.1128/JVI.00223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robert J, Grayfer L, Edholm ES, Ward B, De Jesus Andino F. 2014. Inflammation-induced reactivation of the ranavirus Frog Virus 3 in asymptomatic Xenopus laevis. PLoS One 9:e112904. 10.1371/journal.pone.0112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rollins-Smith LA. 1998. Metamorphosis and the amphibian immune system. Immunol Rev 166:221–230. 10.1111/j.1600-065x.1998.tb01265.x. [DOI] [PubMed] [Google Scholar]
- 24.Andino FDJ, Grayfer L, Chen G, Chinchar VG, Edholm E-S, Robert J. 2015. Characterization of Frog Virus 3 knockout mutants lacking putative virulence genes. Virology 485:162–170. 10.1016/j.virol.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swarbrick M, Zhou H, Seibel M. 2021. Mechanisms in endocrinology: local and systemic effects of glucocorticoids on metabolism: new lessons from animal models. Eur J Endocrinol 185:R113–R1129. 10.1530/EJE-21-0553. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson AW. 2014. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip Rev RNA 5:31–48. 10.1002/wrna.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hur S. 2019. Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37:349–375. 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin KR, Wong HL, Witko-Sarsat V, Wicks IP. 2021. G-CSF - A double edge sword in neutrophil mediated immunity. Semin Immunol 54:101516. 10.1016/j.smim.2021.101516. [DOI] [PubMed] [Google Scholar]
- 29.Koubourli VD, Wendel ES, Yaparla A, Ghaul JR, Grayfer L. 2017. Immune roles of amphibian (Xenopus laevis) tadpole granulocytes during Frog Virus 3 ranavirus infections. Dev Comp Immunol 72:112–118. 10.1016/j.dci.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Metzemaekers M, Gouwy M, Proost P. 2020. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell Mol Immunol 17:433–450. 10.1038/s41423-020-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estefa J, Tafforeau P, Clement AM, Klembara J, Niedźwiedzki G, Berruyer C, Sanchez S. 2021. New light shed on the early evolution of limb-bone growth plate and bone marrow. Elife 10:e51581. 10.7554/eLife.51581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank G. 1988. Granulopoiesis in tadpoles of Rana esculenta. Survey of the organs involved. J Anat 160:59–66. [PMC free article] [PubMed] [Google Scholar]
- 33.Frank G. 1989. Granulopoiesis in tadpoles of Rana esculenta. Ultrastructural observations on the developing granulocytes and on the development of eosinophil granules. J Anat 163:97–105. [PMC free article] [PubMed] [Google Scholar]
- 34.Yaparla A, Popovic M, Grayfer L. 2018. Differentiation-dependent antiviral capacities of amphibian (Xenopus laevis) macrophages. J Biol Chem 293:1736–1744. 10.1074/jbc.M117.794065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassiotis G, Stoye JP. 2016. Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol 16:207–219. 10.1038/nri.2016.27. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton JA. 2008. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol 8:533–544. 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 37.Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J 17:6660–6669. 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel SN, Fertsch D. 1984. Endogenous interferon production by endotoxin-responsive macrophages provides an autostimulatory differentiation signal. Infect Immun 45:417–423. 10.1128/iai.45.2.417-423.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Kim TK, Maniatis T. 1997. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell 1:119–129. 10.1016/S1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 41.Ryals J, Dierks P, Ragg H, Weissmann C. 1985. A 46-nucleotide promoter segment from an IFN-alpha gene renders an unrelated promoter inducible by virus. Cell 41:497–507. 10.1016/s0092-8674(85)80023-4. [DOI] [PubMed] [Google Scholar]
- 42.Demetri GD, Griffin JD. 1991. Granulocyte colony-stimulating factor and its receptor. Blood 78:2791–2808. 10.1182/blood.V78.11.2791.bloodjournal78112791. [DOI] [PubMed] [Google Scholar]
- 43.Yaparla A, Koubourli D, Popovic M, Grayfer L. 2020. Exploring the relationships between amphibian (Xenopus laevis) myeloid cell subsets. Dev Comp Immunol 113:103798. 10.1016/j.dci.2020.103798. [DOI] [PubMed] [Google Scholar]
- 44.Grayfer L, Robert J. 2014. Divergent antiviral roles of amphibian (Xenopus laevis) macrophages elicited by colony-stimulating factor-1 and interleukin-34. J Leukoc Biol 96:1143–1153. 10.1189/jlb.4A0614-295R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neely HR, Guo J, Flowers EM, Criscitiello MF, Flajnik MF. 2018. “Double-duty” conventional dendritic cells in the amphibian Xenopus as the prototype for antigen presentation to B cells. Eur J Immunol 48:430–440. 10.1002/eji.201747260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deotare U, Al-Dawsari G, Couban S, Lipton JH. 2015. G-CSF-primed bone marrow as a source of stem cells for allografting: revisiting the concept. Bone Marrow Transplant 50:1150–1156. 10.1038/bmt.2015.80. [DOI] [PubMed] [Google Scholar]
- 47.Santoni FA, Guerra J, Luban J. 2012. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 9:111. 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaparla A, Reeves P, Grayfer L. 2019. Myelopoiesis of the amphibian Xenopus laevis is segregated to the bone marrow, away from their hematopoietic peripheral liver. Front Immunol 10:3015. 10.3389/fimmu.2019.03015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaparla A, Wendel ES, Grayfer L. 2016. The unique myelopoiesis strategy of the amphibian Xenopus laevis. Dev Comp Immunol 63:136–143. 10.1016/j.dci.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 50.Cullen BR, Owens L. 2002. Experimental challenge and clinical cases of Bohle iridovirus (BIV) in native Australian anurans. Dis Aquat Organ 49:83–92. 10.3354/dao049083. [DOI] [PubMed] [Google Scholar]
- 51.Green DE, Converse KA. 2005. Diseases of frogs and toads. In Majumdar SK, Huffman JE, Brenner FJ, Panah AI (ed), Wildlife diseases: landscape epidemiology, spatial distribution and utilization of remote sensing technology. Pennsylvania Academy of Science, Easton, Pennsylvania. [Google Scholar]
- 52.Greer AL, Berrill M, Wilson PJ. 2005. Five amphibian mortality events associated with ranavirus infection in south central Ontario, Canada. Dis Aquat Organ 67:9–14. 10.3354/dao067009. [DOI] [PubMed] [Google Scholar]
- 53.Speare R, Smith JR. 1992. An iridovirus-like agent isolated from the ornate burrowing frog Limnodynastes ornatus in northern Australia. Dis Aquat Org 14:51–57. 10.3354/dao014051. [DOI] [Google Scholar]
- 54.Sachs LM, Buchholz DR. 2019. Insufficiency of thyroid hormone in frog metamorphosis and the role of glucocorticoids. Front Endocrinol (Lausanne) 10:287. 10.3389/fendo.2019.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warne RW, Crespi EJ, Brunner JL. 2011. Escape from the pond: stress and developmental responses to ranavirus infection in wood frog tadpoles. Funct Ecol 25:139–146. 10.1111/j.1365-2435.2010.01793.x. [DOI] [Google Scholar]
- 56.Grayfer L, Robert J. 2015. Distinct functional roles of amphibian (Xenopus laevis) colony-stimulating factor-1- and interleukin-34-derived macrophages. J Leukoc Biol 98:641–649. 10.1189/jlb.4AB0315-117RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majji S, Thodima V, Sample R, Whitley D, Deng Y, Mao J, Chinchar VG. 2009. Transcriptome analysis of Frog virus 3, the type species of the genus Ranavirus, family Iridoviridae. Virology 391:293–303. 10.1016/j.virol.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grau JH, Poustka AJ, Meixner M, Plotner J. 2014. LTR retroelements are intrinsic components of transcriptional networks in frogs. BMC Genomics 15:626. 10.1186/1471-2164-15-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hauser K, Popovic M, Yaparla A, Koubourli DV, Reeves P, Batheja A, Webb R, Forzán MJ, Grayfer L. 2020. Discovery of granulocyte-lineage cells in the skin of the amphibian Xenopus laevis. FACETS 5:571–597. 10.1139/facets-2020-0010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S4, Figures S1-S3. Download jvi.00634-22-s0001.pdf, PDF file, 0.7 MB (728.5KB, pdf)