ABSTRACT
Interferons (IFNs) are cytokines that induce a global change in the cell to establish antiviral immunity. We previously demonstrated that human adenovirus (HAdV) exploits IFN-induced viral repression to persist in infected cells. Although this in vitro persistence model has been described, the mechanism behind how persistent HAdV infection is established is not well understood. In this study, we demonstrate that IFN signaling is essential for viral repression and promoting persistent infection. Cyclin-dependent kinase 4 (CDK4), an antagonist of retinoblastoma (Rb) family proteins, was shown to disrupt the viral repression induced by IFNs. Consistent with this result, knockout of the Rb family proteins pRb, p107, and/or p130 drastically reduced the effect of IFNs on viral replication. The pRb protein specifically contributed the greatest effect to IFN inhibition of viral replication. Interestingly, IFNs did not impact pRb through direct changes in protein or phosphorylation levels. Cells treated with IFNs continued to cycle normally, consistent with observations that persistently infected cells remain for long periods of time in the host and in our in vitro persistent infection model. Finally, we observed that histone deacetylase (HDAC) inhibitors activated productive viral replication in persistently infected cells in the presence of IFN. Thus, HDACs, specifically class I HDACs, which are commonly associated with Rb family proteins, play a major role in the maintenance of persistent HAdV infection in vitro. This study uncovers the critical role of pRb and class I HDACs in the IFN-induced formation of a repressor complex that promotes persistent HAdV infections.
IMPORTANCE Adenoviruses are ubiquitous viruses infecting more than 90% of the human population. HAdVs cause persistent infections that may lead to serious complications in immunocompromised patients. Therefore, exploring how HAdVs establish persistent infections is critical for understanding viral reactivation in immunosuppressed individuals. The mechanism underlying HAdV persistence has not been fully explored. Here, we provide insight into the contributions of the host cell to IFN-mediated persistent HAdV infection. We found that HAdV-C5 productive infection is inhibited by an Rb-E2F-HDAC repressor complex. Treatment with HDAC inhibitors converted a persistent infection to a lytic infection. Our results suggest that this process involves the noncanonical regulation of Rb-E2F signaling. This study provides insight into a highly prevalent human pathogen, bringing a new level of complexity and understanding to the replicative cycle.
KEYWORDS: adenovirus, interferon, persistent infection, retinoblastoma protein
INTRODUCTION
Interferons (IFNs) are a family of cytokines that have pleiotropic effects, including antiviral, antiproliferative, and antitumor activities, as well as immunomodulatory effects on the innate and adaptive immune responses (1). All three types of IFNs (types I, II, and III) classically signal through IFN receptors, Janus kinases (JAKs), and signal transducer and activator of transcription (STAT) proteins. The binding of IFNs to their cognate receptors triggers STAT protein activation via JAK-mediated phosphorylation. Phosphorylated STAT proteins may homo- and heterodimerize in the cytoplasm and are transported into the nucleus. Once STAT-protein dimers enter the nucleus, the transcription of hundreds of interferon-stimulated genes (ISGs) is induced, which ultimately results in global cell changes and an antiviral state (2, 3). Adenoviruses (AdVs) block the IFN response by inhibiting cytoplasmic signaling pathways, the activation of ISGs, as well as the functions of specific ISG products (4).
Human AdVs (HAdVs) are highly prevalent pathogens that infect >90% of the population in the first few years of life (5). They are largely associated with mild illness in healthy individuals but can lead to severe and life-threatening disease in immunocompromised populations (6). HAdVs establish persistent infections, and several clinical studies have demonstrated that persistence can last for several months or longer (7–9). Persistent HAdV infections are most evident in the allogeneic transplantation setting, where viral reactivation seemed to be the predominant cause of illness in patients rather than de novo infection (6). Although these persistent infections are well documented, it remains unclear how they are established and maintained.
The immediate early E1A proteins are the first viral gene products expressed following infection and are critical for the induction of viral early gene expression and all subsequent aspects of viral replication (10). Investigation of the molecular basis for HAdV persistence using a novel cell culture model (11) revealed an interplay between IFN signaling and an IFN-responsive repressor element in the E1A gene enhancer region. IFN-induced repression of E1A expression was associated with an evolutionarily conserved E2F binding site and repressor complexes containing the retinoblastoma (Rb) family of tumor suppressors (11). Mutation of the conserved E2F binding site abolished the repressive effect that IFNs had on HAdV E1A expression and viral replication (11). By targeting the expression of the E1A immediate early gene, IFNs can abrogate the expression of HAdV gene products that have anti-IFN activities.
The Rb family proteins pRb, p107, and p130, termed the pocket proteins, are well known for their role in regulating the cell cycle, specifically the transition through the restriction point in G1 phase (12). Rb family proteins bind to E2F transcription factors and repress gene transcription. The pocket proteins physically associate with histone deacetylases (HDACs), specifically class I HDACs, to exert repressive activity by regulating chromatin acetylation and preventing access of the transcriptional machinery to promoters (13, 14). To overcome this inhibition, cyclin-dependent kinases (CDKs) hyperphosphorylate Rb family members, which releases them from E2Fs, allows transcriptional activation, and promotes cell cycle progression. CDK activity, in turn, is regulated in several ways to prevent inappropriate cell cycle progression. This includes the regulation of cyclin-dependent kinase inhibitors (CKIs), CDK phosphorylation, and cyclin levels (15, 16).
Despite a clear role for pocket proteins in the IFN-mediated repression of E1A expression, the molecular mechanism(s) and implications for the establishment of HAdV persistence remained unclear. In this study, we demonstrate that canonical IFN signaling is essential for HAdV persistence in normal human cells. We found that CDK4 is an antagonist of viral persistence, but it is not directly affected by IFN treatment. Through a series of single-, double-, and triple-gene knockouts, we found that the pocket proteins, most prominently pRb, play a major role in the IFN-mediated repression of HAdV early gene expression and DNA replication. Here, we show that IFN treatment does not alter the expression of E2F-Rb or cyclin/CDK/CKI proteins, nor does the phosphorylation status of pRb and CDK2/CDK4 change. Rather, IFNs appear to regulate HAdV chromatin since the inhibition of class I HDACs reverts a persistent infection to a lytic infection despite the presence of IFNs. We show that IFNs do not inhibit the association of cellular histone H3 with the HAdV genome and that IFNs may induce the condensation of viral chromatin via viral protein VII. Collectively, these results support a model in which IFN-induced HAdV persistence is regulated by Rb-E2F-HDAC repressor complexes that modulate the viral chromatin structure.
RESULTS
JAK/STAT signaling is essential for IFN inhibition of HAdV-C5 replication.
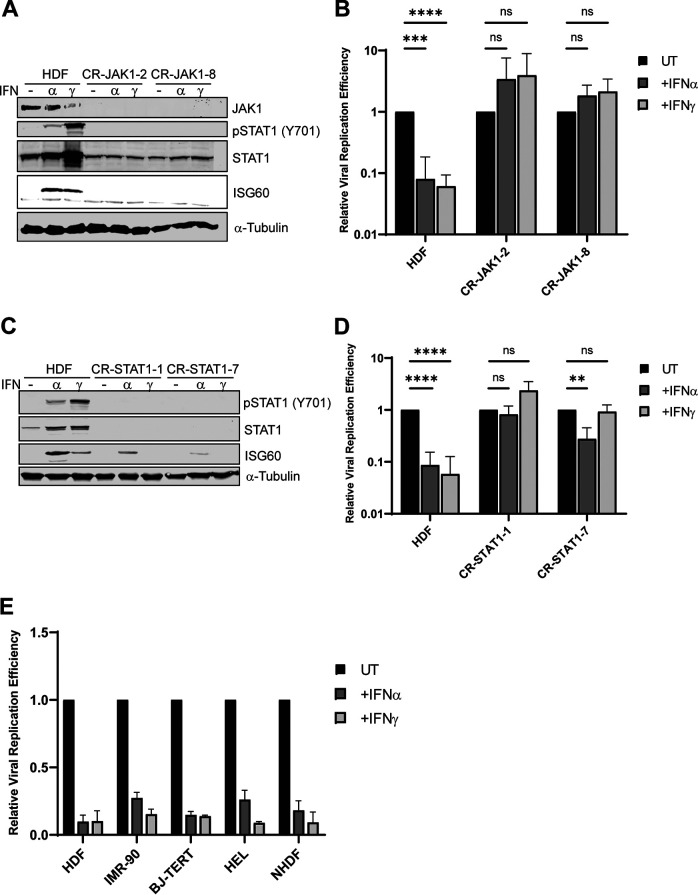
We previously demonstrated that IFNs inhibit the replication of wild-type HAdV-C5 in normal human diploid fibroblasts (NHDFs) (HDF cells), promoting persistent viral infection (11). To evaluate if canonical IFN signaling is essential for this inhibition, JAK1 was knocked out using a CRISPR-Cas9 strategy in HDF cells. Western blot analysis confirmed the ablation of JAK1 expression and the loss of STAT1 phosphorylation and ISG60 (IFIT3) induction following treatment with type I and type II IFNs, IFN-α and IFN-γ, respectively (Fig. 1A). HAdV-C5 replication was inhibited >10-fold in parental HDF cells treated with IFN-α or IFN-γ (Fig. 1B). In contrast, there was a complete restoration of viral replication in JAK1 knockout cells in the presence of IFNs (Fig. 1B). As both IFN-α and IFN-γ transduce signals using STAT1, we asked if IFN-induced viral repression was dependent on STAT1. STAT1 was knocked out using a CRISPR-Cas9 strategy in HDF cells (Fig. 1C). Although STAT1 expression was eliminated, the induction of ISG60 was still observed with IFN-α treatment although to a lower level than in parental cells, indicating the use of a noncanonical IFN-α signaling pathway; ISG60 expression was not induced in IFN-γ-treated STAT1 knockout cells (Fig. 1C). There was a complete restoration of viral replication with IFN-γ treatment and a partial restoration of viral replication with IFN-α treatment in STAT1 knockout cells, with clonal variation (Fig. 1D). The partial effect with IFN-α may be due to signal transduction and ISG activation by a STAT2 dimer (3). Finally, we analyzed four other sources of normal fibroblast cells, immortalized and nonimmortalized, for IFN repression of HAdV-C5 replication. A significant decrease in viral replication was observed in all IFN-treated normal fibroblasts examined (Fig. 1E). These results stress the importance of intact IFN signaling for the inhibition of HAdV replication by IFNs and the conservation of this activity in normal human cells.
FIG 1.
JAK/STAT signaling is essential for IFN inhibition of HAdV-C5 replication. (A) HDF cells were depleted of JAK1 using CRISPR-Cas9. Cells (clones 4 and 14) were treated with IFN-α or IFN-γ for 24 h or left untreated. JAK1 and STAT1 protein levels were analyzed by Western blotting, and IFN signaling was analyzed using antibodies to detect phosphorylated STAT1(Y701) and ISG60 expression. α-Tubulin levels were used as a loading control. (B) HDF and HDF-JAK1 knockout cells (clones 2 and 8) were treated with IFN-α or IFN-γ for 24 h or left untreated (UT) and then infected with HAdV-C5 at 25 p/cell. Cells were harvested at 5 and 48 hpi, and viral replication efficiency was quantified by qPCR. Viral DNA copy numbers were first normalized to GAPDH, and the fold increase in the viral copy number was then calculated by normalizing the amount of viral DNA present at 48 h to the amount present at 5 h. Finally, the samples that were left untreated were set to a value of 1, and the values for the treated samples were determined relative to the untreated value. (C) HDF cells were depleted of STAT1 using CRISPR-Cas9. Cells (clones 1 and 7) were treated with IFN-α or IFN-γ for 24 h or left untreated. STAT1 protein levels were analyzed by Western blotting, and IFN signaling was analyzed using antibodies to detect phosphorylated STAT1(Y701) and ISG60 expression. (D) HDF and HDF-STAT1 knockout cells were treated with IFN-α or IFN-γ for 24 h or left untreated and then infected with HAdV-C5 at 25 p/cell. Cells were harvested at 5 and 48 hpi, and viral replication efficiency was quantified by qPCR as described above. (E) HDF, IMR-90, BJ-TERT, HEL, and NHDF cells were treated with IFN-α or IFN-γ for 24 h or left untreated and then infected with HAdV-C5 at 25 p/cell. Cells were harvested at 5 and 48 hpi, and viral replication efficiency was quantified by qPCR as described above. All values are plotted as means ± SD (n = 3) (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant).
CDK4 antagonizes the establishment of persistent HAdV-C5 infection.
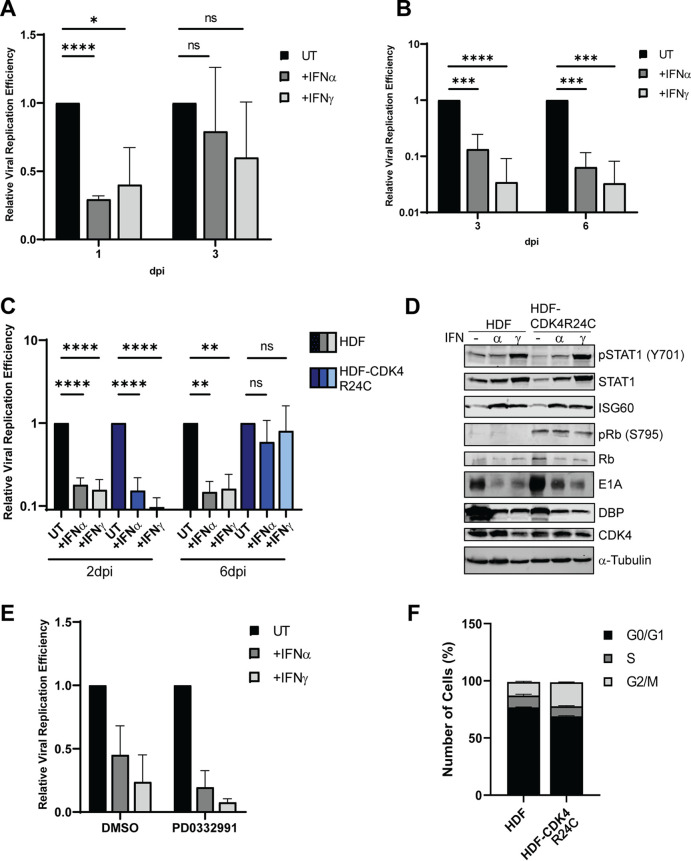
Following our previous observation that HAdV-C5 replication was inhibited by IFNs in normal human bronchial epithelial cells (NHBECs) (11), we sought to test the establishment of persistent infection in epithelial cell lines. In telomerase reverse transcriptase (TERT)-immortalized normal human bronchial epithelial cells (HBEC3-KT), IFNs repressed HAdV-C5 replication minimally at 1 day postinfection (dpi) and not in a statistically significant manner at 3 dpi (Fig. 2A). The same trend was observed in TERT-immortalized small airway epithelial cells (HSAEC1-KT): IFN repression of HAdV-C5 replication was not sustained at 3 dpi compared to 1 dpi (data not shown). To examine if the inability to establish persistence in these cells was related to the epithelial cell type, we tested primary normal epithelial cells and found that these cells could support persistent infection, as seen through IFN-mediated repression of viral replication at 3 and 6 dpi (Fig. 2B). We noted that the immortalized epithelial cells used in these experiments contained active CDK4 (17). This observation is consistent with our previous study that showed a critical role for the Rb-E2F axis in IFN-mediated HAdV persistence (11), as Rb family proteins are CDK4 substrates (12). To determine if CDK4 influenced the ability of IFNs to inhibit HAdV replication in HBEC3-KT cells, we utilized a CDK4/6 specific inhibitor, palbociclib (PD0332991). Inhibition of CDK4 in the presence of IFNs resulted in greater inhibition of viral replication than in cells treated with IFNs alone (Fig. 2E). This result supports the idea that inhibition of CDK4 leads to decreased phosphorylation of Rb and, subsequently, more intact Rb-E2F repressor complexes.
FIG 2.
CDK4 antagonizes the establishment of persistent HAdV-C5 infection. (A) HBEC3-KT cells were treated with IFN-α or IFN-γ for 24 h or left untreated and then infected at 100 p/cell with HAdV-C5. Cells were harvested at 4 hpi and at 1 and 3 dpi, and viral replication efficiency was quantified by qPCR as described in Fig. 1 legend. (B) Primary HBECs were treated with IFN-α or IFN-γ for 24 h or left untreated and then infected at 5 p/cell with HAdV-C5. Cells were harvested at 4 hpi and at 3 and 6 dpi, and viral replication efficiency was quantified by qPCR as described in Fig. 1 legend. (C and D) HDF cells were transduced with a lentivirus to express CDK4(R24C). HDF and HDF-CDK4 cells were treated with IFN-α or IFN-γ for 24 h or left untreated and then infected at 25 p/cell with HAdV-C5. (C) Cells were harvested at 5 hpi and at 2 and 6 dpi, and viral replication efficiency was quantified by qPCR as described above. (D) Whole-cell extracts from cells at 6 dpi were analyzed by Western blotting for STAT1 and phospho-STAT1(Y710), ISG60, Rb and phospho-Rb-S795, AdV E1A and DBP, and CDK4. (E) HBEC3-KT cells were treated with IFN-α or IFN-γ for 24 h or left untreated. Twelve hours prior to infection, cells were treated with DMSO or palbociclib (PD0332991) (2 μM) and then infected at 100 p/cell with HAdV-C5. Cells were harvested at 4 and 24 hpi, and viral replication efficiency was quantified by qPCR, as described above. Values are plotted as means ± SD (n = 2). (F) HDF and HDF-CDK4 cells were analyzed via flow cytometry for DNA content, as described above. All values are plotted as means ± SD unless otherwise stated (n = 3) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
To investigate further whether CDK4 antagonizes HAdV persistence in immortalized epithelial cells, we expressed a mutated form of CDK4, CDK4R24C (18), that is refractory to inhibition by p16(INK4a) in HDF cells. There were no changes in HAdV-C5 replication between HDF and HDF-CDK4R24C cells at 2 dpi following IFN treatment, but a defect in the establishment of persistent infection was observed by 6 dpi in CDK4-expressing cells (Fig. 2C and D). CDK4R24C-expressing cells still responded to IFN normally: phospho-STAT1 and ISG60 expression were still induced at 6 dpi (Fig. 2D). HDF-CDK4R24C cells showed minor differences in cell cycle profiles compared to the parental cells (Fig. 2F). This is expected as it has been shown that mammalian cells continue to cycle without CDK4 as other CDKs can compensate (19, 20). HDF-CDK4R24C cells showed increased phospho-Rb levels compared to the parental HDF cells (Fig. 2D), and since phosphorylation disrupts Rb-E2F complexes, these data suggest that CDK4 antagonizes HAdV persistence by phosphorylation and disruption of Rb repressor complexes.
Rb family proteins play an important role in IFN-mediated repression of HAdV replication.
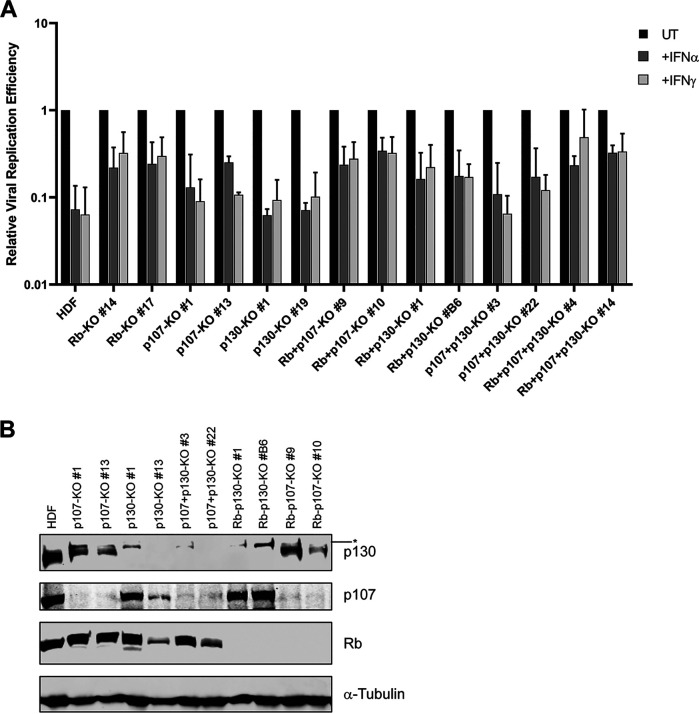
To investigate further the role of Rb family proteins in HAdV persistence, we knocked out all three Rb family proteins (pRb, p107, and p130 triple knockout [TKO]) using a CRISPR-Cas9 strategy. HAdV-C5 replication was only ~2- to 3-fold decreased with IFN treatment in TKO cells, whereas viral replication was reduced >15-fold by IFNs in parental HDF cells (Fig. 3A). Rb family protein knockout was verified by Western blotting (Fig. 3B). The viral life cycle was also restored in HDF-TKO cells as seen by the augmented expression of the viral immediate early protein E1A and the early DNA binding protein (DBP) (Fig. 3B). These results were not due to defective IFN signaling as STAT1 phosphorylation was still induced in HDF-TKO cells (Fig. 3B). We generated HDF knockout cells with every possible combination of Rb family protein knockout (Fig. 4B). All knockout cell lines lacking pRb supported viral replication in the presence of IFNs to a greater extent than in cell lines expressing pRb. Viral replication was reduced only ~3- to 5-fold in Rb, Rb/p130, and Rb/p107 knockout cells treated with IFN, whereas the knockout of p107, p130, or p107/p130 resulted in an ~15-fold decrease in viral replication in response to IFN, an effect similar to that seen in the parental HDF cells (Fig. 4A). Analysis of viral DNA replication (Fig. 3C) and viral protein expression (E1A and DBP) (Fig. 3D) identified pRb as the major contributor to the repression of AdV replication by IFN. Surprisingly, we did not detect any changes in the pRb phosphorylation status at sites associated with CDK4 following IFN treatment using a number of phosphospecific antibodies to sites of pRb phosphorylation (Fig. 3E). We also found that there were no changes in the total levels of any of the three Rb family members following IFN treatment of HDF cells (Fig. 3F).
FIG 3.
Rb family proteins play an important role in IFN-mediated repression of HAdV-C5 replication. (A and B) HDF Rb-p107-p130 triple-knockout cells (clones 4 and 14) were pretreated with IFN-α or IFN-γ for 24 h or left untreated and then infected at 25 p/cell with HAdV-C5. Viral replication was quantified by qPCR as described in Fig. 1 legend. (A), and p130, p107, Rb, E1A, DBP, STAT1, and phospho-STAT1(Y701) protein expression was analyzed by Western blotting (B). An asterisk is used to identify a nonspecific, cross-reactive protein that becomes visible when p130 is knocked out. (C and D) HDF Rb knockout cells (clones 14 and 17) were treated with IFN-α or IFN-γ for 24 h or left untreated, infected with HAdV-C5, and analyzed as described above for panels A and B. (E and F) HDF cells were treated with IFN-α or IFN-γ for 48 h or left untreated. Rb and phospho-Rb-S608, -S795, -S780, and -S807/S811; STAT1 and phospho-STAT1; and p130 and p107 protein levels were analyzed by Western blotting.
FIG 4.
Rb is the major contributor to IFN-mediated repression of HAdV-C5 replication. (A) HDF cells and HDF cells knocked out for the indicated Rb family members were treated with IFN-α or IFN-γ for 24 h or left untreated and then infected at 25 p/cell with HAdV-C5. Cells were collected at 5 and 48 hpi, and viral replication efficiency was quantified by qPCR, as described in Fig. 1 legend. Values are plotted as means ± SD (n = 3). (B) Western blot analysis of Rb family protein ablation in HDF knockout cells. Cell extracts were prepared from the different knockout cell lines listed and analyzed for the expression of Rb, p107, and p130 proteins. An asterisk is used to identify a nonspecific, cross-reactive protein that becomes visible when p130 is knocked out.
Cell cycle regulators are unaltered by IFN treatment in HDF cells.
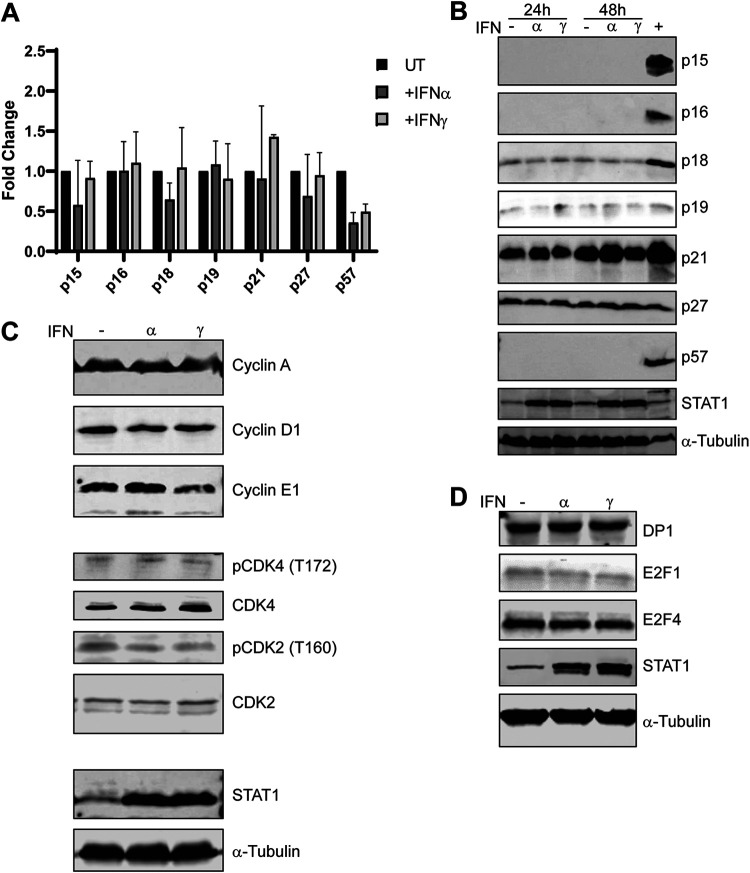
We examined the expression of cell cycle regulators following IFN treatment. Cyclin kinase inhibitors (CKIs) are upregulated in response to IFNs in various established cell lines (21–25). We analyzed CKI mRNA and protein levels in HDF cells with and without IFN treatment. To prevent cellular changes due to contact inhibition, low-confluence cells were used in this series of experiments. We observed that both CKI mRNA levels 24 h after IFN treatment (Fig. 5A) and protein levels 24 and 48 h after treatment (Fig. 5B) were unaffected by IFNs. These results imply that there is no direct inhibition of a CDK via an IFN-induced CKI. We analyzed cyclin protein levels and activating phosphorylation sites on CDKs in HDF cells following IFN treatment. The protein levels of the CDK4 binding partner cyclin D remained unaltered following IFN treatment; the same result was also found with cyclins A and E1 (Fig. 5C). Similarly, activating phosphorylation of CDK4 at Tyr172 and CDK2 at T160 by CDK-activating kinase (CAK) was unaltered by IFN treatment (Fig. 5C). Finally, we found that the protein levels of E2F1, E2F4, and E2F dimerization partner DP1 remained unaltered by IFN treatment (Fig. 5D). We conclude that IFN treatment does not directly induce Rb-E2F repressor complex formation via the upregulation of CKIs, downregulation of cyclins, or inactivation of CDK2 or CDK4.
FIG 5.
Cell cycle regulators are unaltered by IFN treatment in HDF cells. (A) RNA was isolated from HDF cells that had been treated with IFN-α or IFN-γ for 24 h or left untreated. RNA was converted to cDNA, and levels of CKI mRNAs (p15, p16, p18, p19, p21, p27, and p57) were quantified by qPCR as described in Fig. 1 legend. The results were normalized to GAPDH mRNA levels, and fold changes in CKI levels were determined relative to the untreated sample and plotted as means ± SD (n = 3). (B) Cell extracts from HDF cells were treated with IFN-α or IFN-γ for 24 h or 48 h or left untreated and analyzed by Western blotting for levels of CKI proteins p15, p16, p18, p19, p21, p27, and p57. (C and D) Cell extracts from HDF cells were treated with IFN-α or IFN-γ for 48 h or left untreated and analyzed by Western blotting for levels of cyclins A, D1, and E1; CDK4; phospho-CDK4(T172); CDK2; phospho-CDK2(T160); DP1; E2F1; and E2F4.
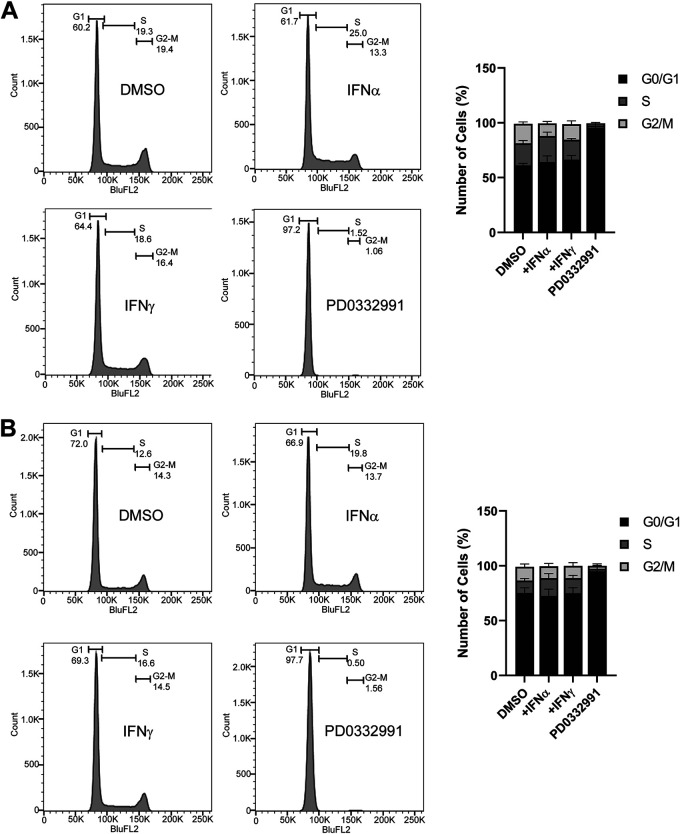
HDF cells cycle normally despite IFN treatment.
Since Rb proteins play a major role in controlling the cell cycle, we investigated if IFN treatment alters the cell cycle in HDF cells in a way that was not CKI or cyclin related (12). We hypothesized that IFNs may cause a delayed or prolonged G1 phase since Rb-E2F repressor complexes regulate the G1-to-S-phase transition (16). We previously demonstrated that persistent HAdV-C5 infection for >100 days could be established in HDF cells treated with IFN-γ (11). Thus, we believed that cells would not be arrested in G1 phase by IFNs but possibly could exhibit a prolonged G1 phase. To assess this, the cell cycle profiles of HDF cells were examined in control cells and 24 h after treatment with dimethyl sulfoxide (DMSO), IFN-α, IFN-γ, or the CDK4 inhibitor PD0332991 as a G1 arrest control. We found that there were no significant changes in the cycling of HDF cells following IFN treatment (Fig. 6A). The same was true 48 h after IFN treatment (Fig. 6B). Cells treated with PD0332991 showed significant G1 arrest, as expected. The results with IFN treatment may reflect the compensatory roles that CDK4 and CDK6 play with each other as CDK4-null mice and CDK knockout cell lines have been described to have no changes in cell cycling (19, 20). We conclude that IFNs do not cause a direct change to the cell cycle in HDF cells.
FIG 6.
HDF cells cycle normally despite IFN treatment. HDFs were treated with IFN-α, IFN-γ, palbociclib (PD) (2 μM), or control DMSO for 24 h (A) or 48 h (B). Cells were washed, fixed, stained with propidium iodide (PI), and analyzed by flow cytometry for DNA content. Cells were gated for single cells, for pulse shape, and then for PI signal histograms, and cell cycle profiles were determined, as shown. Values are plotted as means ± SD (n = 3). The bar graphs represent summaries of the results.
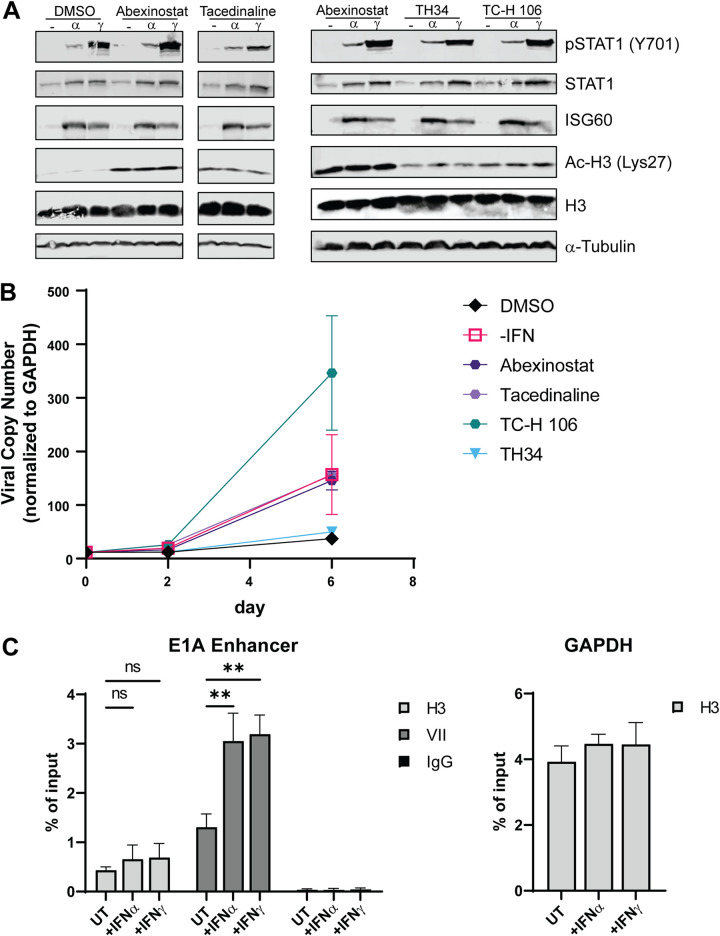
Class I HDACs are essential for HAdV-C5 persistent infection in HDF cells.
Previous observations using patient samples showed that HDAC inhibitors (HDACi) reactivated persistent HAdV infection to a lytic infection (26). We tested the potential role of HDACs in the IFN-mediated repression of HAdV replication. HDACs not only physically associate with Rb family proteins (27–29) but also are integral to IFN signaling (30). HDACs fall into four broad classes (I to IV) based on amino acid sequence conservation (31). We analyzed the effects of HDACi that target different combinations of HDACs. These included abexinostat (HDACs 1, 2, 3, 6, 8, and 10), TH34 (HDACs 6, 8, and 10), tacedinaline (HDACs 1, 2, and 3), and TC-H106 (HDACs 1, 2, and 3). Due to the essential role of HDACs in IFN signaling in certain cells, we performed experiments to determine if IFN signaling was intact in HDF cells treated with HDAC inhibitors. Cells were treated with an HDACi for 1 h prior to the addition of IFNs and for 24 h after IFN addition. We examined IFN signaling and histone H3 acetylation by Western blotting (Fig. 7A). These results demonstrated that HDACi did not inhibit STAT1 phosphorylation or the induction of ISG60 expression, but they did induce the acetylation of H3-Lys27. HDF cells that were persistently infected for 30 days with HAdV-C5 in the presence of IFN-γ were untreated or treated with different HDACi. We found that HDACi, specifically HDAC class I inhibitors, could switch persistently infected cells back to lytic viral replication at levels similar to those observed when IFN was removed from the medium (Fig. 7B). This supports the idea that HDACs, specifically class I HDACs, are required for Rb-E2F repressor complex function and persistent HAdV-C5 infection induced by IFN-γ. To determine if there were changes to the viral chromatin structure in the presence of IFNs, we performed chromatin immunoprecipitations (ChIPs) with infected HDF cells in the presence and absence of IFNs (Fig. 7C). We analyzed the association of histone H3 with the HAdV-C5 genome plus and minus IFN treatment. There were similar levels of H3 association with the E1A enhancer region in IFN-treated and untreated HDF cells at 18 h postinfection (hpi), a time point that is just prior to the onset of viral DNA replication in HDF cells (Fig. 7C). H3 association with the cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter region was not affected by IFN treatment. Surprisingly, the level of protein VII at the E1A enhancer region in IFN-treated cells increased 2-fold compared to that in untreated cells (Fig. 7C). These results show that IFNs do not inhibit the ability of histone H3 to associate with the HAdV genome and suggest IFNs may promote the condensation of viral chromatin with protein VII.
FIG 7.
Class I HDACs are essential for HAdV-C5 persistent infection in HDF cells. HDF cells were treated with the indicated HDAC inhibitors. (A) Treatment began 1 h prior to the addition of IFNs. Cells were harvested 24 h after IFN treatment and analyzed by Western blotting for STAT1 and phospho-STAT1(Y701), ISG60, histone H3, and acetyl-H3-Lys27 [Ac-H3(Lys27)]. (B) Persistent HADV-C5 infection of HDF cells was established by continuous treatment of infected cells with IFN-γ, as described previously (11). At 27 dpi, cells were seeded into new plates in triplicate in the presence of IFN-γ, and day 0 samples were taken from the split. After letting the cells settle for 3 h, IFN-γ was removed from one set, and IFN-γ was maintained with all other cells with the addition of the DMSO control, 200 nM PCI24781, 10 μM TH34, 4 μM TC-H106, 2 μM abexinostat, or 1 μM tucidinostat. The medium was replaced with the same treatments 3 days after the split. HAdV-C5 DNA replication was quantified by qPCR at days 0, 2, and 6, as described above. (C) Enrichment of histone H3 and protein VII at the E1A enhancer region. HDF cells treated with IFNs for 24 h or left untreated were infected with dl309 at 200 p/cell. At 18 hpi, ChIP was performed using antibodies against H3 or protein VII. Precipitated DNA was quantified by qPCR using oligonucleotides that detect the E1A enhancer or the cellular GAPDH gene. All values are plotted as means ± SD (n = 3) (**, P < 0.01).
DISCUSSION
Adenoviruses establish both lytic and persistent infections (5, 6). Understanding the mechanism and key players in controlling the establishment of persistent infections will improve our ability to manage them. We previously demonstrated that type I and II IFNs induce persistent HAdV-C5 infection in an in vitro model using normal human fibroblasts (11). Persistent HAdV-C5 infections could be maintained in the presence of IFN-γ for more than 3 months with viral replication and virus production reduced ~100-fold in the cultures (11). Here, we confirm that canonical IFN signaling is essential for the repression of HAdV-C5 replication in HDF cells by IFN-α and IFN-γ and that IFNs repress AdV replication in a number of other primary and immortalized human fibroblasts (Fig. 1). In contrast, we were unable to establish a persistent HAdV-C5 infection in immortalized normal human bronchial and small airway epithelial cells (Fig. 2A and data not shown), although this could be accomplished using primary human bronchial epithelial cells (Fig. 2B), suggesting that the epithelial nature of the cells did not influence the results. Rather, the notable difference between these epithelial cells is the expression of active CDK4 in the immortalized lines (17). Indeed, the constitutive expression of active CDK4 in HDF cells inhibited the ability to establish a persistent HAdV-C5 infection in the presence of IFN-α or IFN-γ (Fig. 2C), even though CDK4 did not affect IFN signaling in these cells (Fig. 2D). The ability of CDK4 to block the establishment of a persistent HAdV infection in vitro is consistent with the role of Rb family proteins in this process.
Rb family proteins are well-documented transcriptional repressors. Rb family proteins assemble large multiprotein complexes at E2F-regulated promoters. HDACs are recruited to these complexes, which repress gene expression through epigenetic changes (27–29). Through interaction with E2F transcription factors bound at their cognate binding sites, they inhibit the transcription of target genes (12). The ~20-bp target in the E1A enhancer region of IFN-mediated repression of HAdV-C5 E1A expression contains an E2F binding site that is conserved among different HAdV species (11). Our data confirm the vital role that Rb family proteins play in the inhibition of HAdV-C5 replication by IFNs (Fig. 3A and B) and especially the important role of the pRb protein in this process since even the single knockout of the Rb gene greatly diminished IFN repression of HAdV-C5 replication (Fig. 3C and D). Even in the absence of IFN signaling, the conserved E2F site in the E1A enhancer downregulates E1A expression, but this negative effect is greatly augmented by IFN stimulation (11). Despite the results described above with CDK4 expression, we did not observe any detectable changes in pRb phosphorylation at CDK4-associated sites with IFN treatment (Fig. 3E) or any notable changes in the levels of the Rb family member E2F1, E2F4, or DP1; the levels of CDK2 and CDK4 and their activated phosphorylated forms; or the levels of any cyclin kinase inhibitors or cyclin A, D1, or E1 (Fig. 3F and Fig. 5). This was further reflected in cell cycle analyses where IFN treatment did not arrest or delay the cell cycle in HDF cells (Fig. 6). Thus, we conclude that Rb family proteins, and especially pRb, are vital for IFN inhibition of HAdV-C5 replication and the establishment of persistent infection but not in a classical cell cycle-dependent manner. Although Rb may be modified at a site that we did not explore, we speculate that IFN regulation of the Rb-E2F axis involves noncanonical Rb signaling (32). The noncanonical Rb pathway promotes histone modification and regulates transcription in a manner that is independent of the effects of Rb on the cell cycle. Specifically, Rb may interact with E2F family members in different ways such that Rb phosphorylation may inhibit binding to some E2Fs but not to others (33, 34). Furthermore, the cell cycle-inhibitory activity of Rb is not dependent on its ability to recruit HDACs. While HDACs play an important role in Rb-mediated transcriptional repression, they are dispensable for the regulation of certain genes that regulate the cell cycle (35). That HDACs are dispensable for Rb-mediated cell cycle arrest is consistent with our observation that our cells cycle normally in the presence of IFNs (Fig. 6).
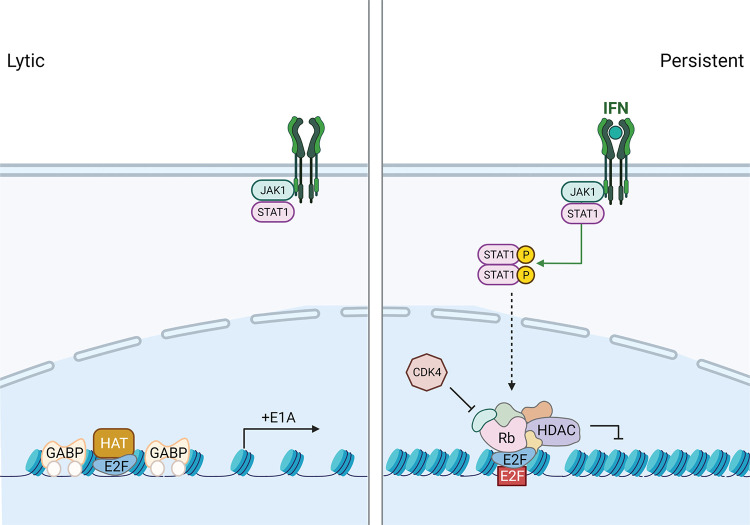
It has been demonstrated that the requirement of HDACs for Rb-mediated repression is promoter specific (35). From our results using HDAC inhibitors (Fig. 7), we suggest that E1A expression is regulated by IFNs in an HDAC-specific manner. It is well established that chromatin formation regulates the latent-to-lytic infection switch in herpesvirus-infected cells, including herpes simplex virus 1 (HSV-1), human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), and Kaposi’s sarcoma-associated herpesvirus (KSHV) (36–39). This has been particularly well characterized with HSV-1 where naked viral DNA within the virion may be rapidly chromatinized following infection into repressive heterochromatin if this process is not antagonized by viral proteins such as ICP0 (40). The removal of repressive heterochromatin from the HSV-1 genome correlates with the activation of viral gene expression. Conversely, quiescent and latent HSV-1 genomes are associated with the assembly of repressive heterochromatin on the viral genome (38). A recent study has shown that persistent species C adenoviruses were found in tonsil samples and that following treatment of tonsillar lymphocytes with HDAC inhibitors, virus replication was reactivated (26). These compounds promoted H3 acetylation and association with viral early gene promoters, uncovering a role for histone acetylation in AdV reactivation. We believe that the same idea holds true in our in vitro HAdV-C5 persistent infection model in HDF cells where class I HDAC inhibitors were found to augment viral DNA replication in IFN-γ-treated cells (Fig. 7). We propose the model for E1A regulation, with and without IFN signaling, shown in Fig. 8. In untreated cells, the E1A gene is actively expressed via the activating transcription factors GA binding protein (GABP) and E2F binding to the E1A enhancer region (41–43). The data presented here support the epigenetic regulation of the adenoviral genome in response to IFN treatment and the formation of a repressive Rb-E2F-HDAC complex at the E1A enhancer. This repression significantly reduces E1A expression and, consequently, all aspects of the AdV replication cycle to promote persistent HAdV infection. It remains unclear what drives Rb-E2F-HDAC complex formation in the context of IFN signaling.
FIG 8.
Schematic diagram of lytic and persistent HAdV infections. In untreated HDF cells, lytic infection occurs as the activating transcription factors GABP and E2F drive the expression of E1A. In IFN-treated HDF cells, canonical IFN signaling induces the association of repressive Rb-E2F-HDAC complexes at the E1A enhancer. Due to epigenetic regulation of the adenoviral genome, E1A expression is significantly reduced, and consequently, all aspects of the HAdV replication cycle are inhibited to promote persistent infection. HAT, histone acetyltransferase.
Core protein VII is a histone-like protein that condenses the HAdV genome within the capsid and is associated with incoming viral DNA, creating a compact viral chromatin structure (44, 45). Protein VII defends the viral genome early after infection from host defenses such as the DNA damage response (46) and a danger signal response (47). There has been debate on the function of protein VII in regulating HAdV early gene transcription. Some studies have shown that it condenses the viral genome and represses viral transcription, while other studies have shown it as a necessary factor for lytic replication (48–51). Protein VII is displaced from the HAdV genome coincident with early gene expression (52). We found that IFNs increased the association of protein VII with the viral genome in comparison to untreated cells, while histone H3 association was maintained (Fig. 7C). This result may simply reflect the displacement of VII due to early gene transcription in untreated cells and the maintenance of VII with IFN treatment due to reduced viral gene expression. We do not know if protein VII and H3 bind to the same genome within a cell, if separate genomes bind to one or the other activity, or even if these represent differential interactions within different cells in the population. Protein VII was reported to be acetylated in HAdV-C5-infected cells (53). Perhaps, in HAdV persistent infection, there is chromatinization of the viral genome with protein VII alone and/or with cellular histones, and IFN signaling regulates these activities by deacetylation using an Rb-E2F-HDAC complex. The intricacies of the regulation of the HAdV chromatin structure and viral gene expression remain to be elucidated, and the analysis of this regulation in the context of IFN signaling may provide a valuable tool to dissect these processes.
MATERIALS AND METHODS
Cell culture and viruses.
Normal human diploid fibroblasts immortalized by the expression of human telomerase (HDF-TERT) (referred to as HDF cells) (54), BJ-TERT cells (55), IMR-90 cells (ATCC), HEL-299 cells (ATCC), and NHDF cells (Lonza) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) supplemented with 100 μg/mL penicillin and streptomycin. Human bronchial epithelial cells (HBEC3-KT; ATCC) and primary human bronchial epithelial cells (Lonza) were maintained in airway epithelial cell medium (ATCC) supplemented with an epithelial cell growth kit according to the manufacturer’s instructions (ATCC). Wild-type adenovirus 5 (HAdV-C5) was used in all experiments except for chromatin immunoprecipitation (ChIP), where dl309, a phenotypically wild-type HAdV-C5 (11), was used. Virus infections were performed for 1 h at 37°C at the multiplicities of infection (MOIs) described in the figure legends, followed by the removal of virus and replacement with fresh medium. For all experiments, cells were treated with IFN-α or IFN-γ (PBL Assay Sciences) at 500 U/mL or 1,000 U/mL, respectively. Cells were treated with the HDAC inhibitors TH34 (catalog number 8773), TC-H106 (catalog number 6738), and tacedinaline (catalog number 2818) from SelleckChem and abexinostat (catalog number 4098) and PD0332991 (catalog number 8316) from APExBIO at the concentrations indicated in the figure legends. For HBEC3-KT and HSAEC1-KT cells, an MOI of 100 virus particles/cell (p/cell) results in infection of 40% of the population. For HDF-TERT cells, an MOI of 25 p/cell results in infection of 10% of the population. The percentage of cells infected at different MOIs was determined by infection with an enhanced green fluorescent protein (EGFP)-expressing HAdV-C5 and flow cytometry of EGFP-positive cells at 24 h postinfection (hpi).
Viral replication assay and RT-qPCR.
Total cellular DNA was purified at 4 and 24 hpi for epithelial cells and 5 and 48 hpi for HDF cells using a Qiagen DNeasy blood and tissue kit. Both viral and cellular genome copy numbers were determined by quantitative PCR (qPCR) using primer pairs that recognize either the HAdV-C5 genome or the cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene using a DyNAmo HS SYBR green qPCR kit (Thermo). After normalizing the viral DNA copy numbers to GAPDH, the fold increase in viral copy numbers was calculated by normalizing the amount of DNA present at 24 or 48 hpi to the amount present at 4 or 5 hpi, respectively. For reverse transcription-qPCR (RT-qPCR), HDF cells were infected for 24 h, and total cellular RNA was isolated using a Qiagen RNeasy kit. Equal amounts of total RNA were used to synthesize the first-strand cDNA using SuperScript II reverse transcriptase and an oligo(dT) primer (Life Technologies). Equal amounts of cDNA were then subjected to qPCR using primer pairs that recognize individual CKI mRNAs and cellular GAPDH mRNA. CKI mRNA was normalized to the internal control GAPDH mRNA and the untreated control. The oligonucleotides used were as follows (5′ to 3′): ACCCAGAAGACTGTGGATGG and TTCTAGACGGCAGGTCAGGT for GAPDH cDNA, CCCCACACACATGCACTTACC and CCTAGTCCCAGGGCTTTGATT for GAPDH genomic DNA, GCGAAAATGGCCAAATGTTA and TAATGAGGGGGTGGAGTTTG for HAdV-C5 inverted terminal repeat (ITR) and the E1A enhancer, ACGGAGTCAACCGTTTCGGGAG and GGTCGGGTGAGAGTGGCAGG for p15 cDNA, CTCGTGATGCTACTGAGGA and GGTCGGCGCAGTTGGGCTCC for p16 cDNA, CGTCAATGCACAAAATGGATTTGG and GAATGACAGCGAAACCAGTTCGG for p18 cDNA, AGGTGGACCTGGAGACTCTCAG and TCCTCTTGGAGAAGATCAGCCG for p21 cDNA, ATAAGGAAGCGACCTGCAACCG and TTCTTGGGCGTCTGCTCCACAG for p27 cDNA, AGATCAGCGCCTGAGAAGTCGT and TCGGGGCTCTTTGGGCTCTAAA for p57 cDNA, CGGCGCTATGCTAAAATGA and GGGGGCACAGTGACAATAC for HAdV-C5 VII ChIP, and GCCATGTAGACCCCTTGAAGAG and ACTGGTTGAGCACAGGGTACTTTAT for GAPDH ChIP.
Western blot analysis.
Whole-cell lysates were prepared by suspending cell pellets in SDS lysis buffer (50 mM Tris-HCl [pH 6.8], 4% SDS) and boiled for 10 min. The protein concentration was determined using a Pierce bicinchoninic acid (BCA) protein assay kit. Equal amounts of proteins were resolved on an SDS-PAGE gel and then transferred to a nitrocellulose membrane. The membranes were blocked in Tris-HCl-buffered-saline (TBS) buffer containing 3% bovine serum albumin (BSA) for 1 h with rocking at room temperature. After blocking, primary antibodies against the protein of interest were added, and the mixture was incubated with rocking at 4°C overnight. Membranes were washed with TBS buffer containing 0.1% Tween 20 (TBS-T) and then incubated with IRDye 800CW-conjugated goat anti-rabbit antibody (catalog number 926-32211; Li-Cor) (1:5,000) and IRDye 680RD-conjugated goat anti-mouse antibody (catalog number 925-068071; Li-Cor) (1:5,000) for 1 h at room temperature. After three washes with TBS-T buffer, images were captured using the Odyssey CLx infrared imaging system (Li-Cor). Alternatively, horseradish peroxidase (HRP)-conjugated antibodies (Amersham) were used in conjunction with ECL Western blotting (Millipore Immobilon), and images were captured using a GE ImageQuant LAS 500 system. The antibodies used were as follows: HAdV-C5 E1A M73 (catalog number MS-588-P; NeoMarkers) (1:1,000), HAdV-C5 DBP B6-8 (56) (1:100), STAT1 (catalog number 9172; Cell Signaling Technology) (1:1,000), phospho-STAT1(Y701) (catalog number 9167; Cell Signaling Technology) (1:1,000), ISG60/IFIT3 (catalog number 112442; GeneTex) (1:1,000), α-tubulin (catalog number T5168; Millipore-Sigma) (1:10,000), JAK1 (catalog number 3344; Cell Signaling Technology) (1:1,000), Rb (catalog number 9309; Cell Signaling Technology) (1:1,000), phospho-Rb-S608 (catalog number 8147; Cell Signaling Technology) (1:1,000), phospho-Rb-S780 (catalog number 8180; Cell Signaling Technology) (1:1,000), phospho-Rb-S795 (catalog number 9301; Cell Signaling Technology) (1:1,000), phospho-Rb-S807/811 (catalog number 8516; Cell Signaling Technology) (1:1,000), CDK2 (catalog number 2546; Cell Signaling Technology) (1:1,000), phospho-CDK2 (catalog number 2561; Cell Signaling Technology) (1:1,000), CDK4 (catalog number 260; Santa Cruz Biotechnology) (1:500), phospho-CDK4 (catalog number AP0593; Abclonal) (1:1,000), p107/RBL1 (catalog number 89798; Cell Signaling Technology) (1:1,000), p130/RBL2 (catalog number 13610; Cell Signaling Technology) (1:1,000), E2F1 (catalog number 193; Santa Cruz Biotechnology) (1:1,000), E2F4 (catalog number 866; Santa Cruz Biotechnology) (1:1,000), DP1 (catalog number 53612; Santa Cruz Biotechnology) (1:1,000), p15 (catalog number 36303; Cell Signaling Technology) (1:1,000), p16 (catalog number 80772; Cell Signaling Technology) (1:1,000), p18 (catalog number 2896; Cell Signaling Technology) (1:1,000), p19 (catalog number 10272-2-AP; Protein Tech Group) (1:1,000), p21 (catalog number 2947; Cell Signaling Technology) (1:1,000), p27 (catalog number 3686; Cell Signaling Technology) (1:1,000), p57 (catalog number 2557; Cell Signaling Technology) (1:1,000), cyclin A (catalog number 06-138; Millipore-Sigma) (1:1,000), cyclin D1 (catalog number 718; Santa Cruz Biotechnology) (1:500), cyclin E1 (catalog number 103045; GeneTex) (1:1,000), H3 (catalog number 1791; Abcam) (1:1,000), and acetyl-H3-Lys27 (catalog number 8173; Cell Signaling Technology) (1:1,000).
Cell cycle analysis and flow cytometry.
For cell cycle analysis, cells were harvested via trypsinization and pelleted. After one wash with phosphate-buffered saline (PBS), cells were fixed with cold 70% ethanol added dropwise while vortexing to ensure the fixation of all cells. After at least 30 min in ethanol on ice, the cells were washed with PBS and pelleted again. Five hundred microliters of FxCycle propidium iodide (PI)/RNase staining solution (Invitrogen) was used to resuspend and dye the cells for 30 min at room temperature. Stained cells were filtered and analyzed by flow cytometry. Analysis was done on the Cytek DxP8 flow cytometer and gated using FlowJo software. The first gating was forward scatter versus side scatter for single cells, the second gating was fluor area versus fluor width to gate for stained single cells, and the final gating was histogram versus fluor. The first peak corresponds to G0/G1-phase cells, the second peak corresponds to G2-phase cells, and cells in the region between the G0/G1 and G2 peaks represent cells in S phase.
Lentivirus production and transduction.
A lentivirus vector expressing CDK4(R24C) was produced by the transfection of cells with the vector along with the pLP1, pLP2, and pLP/VSV-G plasmids (Invitrogen). The culture supernatants were harvested 3 to 4 days after transfection, filtered using Millex-HV filters (Millipore), and stored at −80°C. HDF cells were transduced with lentivirus vectors and placed under blasticidin selection 24 h after transduction. Pools of blasticidin-resistant HDF cells were used, and CDK4 expression was confirmed by Western blotting.
Generation of CRISPR-Cas9 knockout cell lines.
pLenti-CRISPR-v2 vectors containing puromycin, blasticidin, or zeocin resistance genes were obtained from Addgene and engineered to express 20-nucleotide (nt) targeting sequences for pRb (ACGATGTGAACATCGAATCA), p107 (AATTTCGTGAACGTATAGAA), and p130 (GTACGTTCTCGGAAATGTGG), respectively. Individual colonies were isolated following Lenti-CRISPR transduction and the selection of HDF-TERT cells and screened by Western blotting for gene knockout. Cell lines with two or three gene knockouts were generated sequentially with appropriate drug selection.
Chromatin immunoprecipitation.
ChIP was performed as previously described (11). For chromatin immunoprecipitation reactions, anti-histone H3 (catalog number ab1791) was purchased from Abcam, and anti-pVII was a gift from Daniel Engel, University of Virginia.
Statistical analysis.
All numerical values represent means ± standard deviations (SD). Each experiment was done in three experimental replicates, and a representative replicate is shown for each blot. Statistical significance was calculated using Student’s t test.
ACKNOWLEDGMENTS
We thank Janet Hearing, Erich Mackow, and Nancy Reich for critical review of the manuscript and members of our laboratory for informed discussions. Figure 8 was created with BioRender.com.
This work was supported by NIH grant R01CA122677 to P.H. S.S. was supported by NIH training grant T32AI007539.
Contributor Information
Patrick Hearing, Email: patrick.hearing@stonybrook.edu.
Lawrence Banks, International Centre for Genetic Engineering and Biotechnology.
REFERENCES
- 1.Hoffmann HH, Schneider WM, Rice CM. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 36:124–138. 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samarajiwa SA, Forster S, Auchettl K, Hertzog PJ. 2009. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res 37:D852–D857. 10.1093/nar/gkn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazewski C, Perez RE, Fish EN, Platanias LC. 2020. Type I interferon (IFN)-regulated activation of canonical and non-canonical signaling pathways. Front Immunol 11:606456. 10.3389/fimmu.2020.606456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohn SY, Hearing P. 2019. Adenoviral strategies to overcome innate cellular responses to infection. FEBS Lett 593:3484–3495. 10.1002/1873-3468.13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khanal S, Ghimire P, Dhamoon AS. 2018. The repertoire of adenovirus in human disease: the innocuous to the deadly. Biomedicines 6:30. 10.3390/biomedicines6010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lion T. 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27:441–462. 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lion T, Baumgartinger R, Watzinger F, Matthes-Martin S, Suda M, Preuner S, Futterknecht B, Lawitschka A, Peters C, Pötschger U, Gadner H. 2003. Molecular monitoring of adenovirus in peripheral blood after allogeneic bone marrow transplantation permits early diagnosis of disseminated disease. Blood 102:1114–1120. 10.1182/blood-2002-07-2152. [DOI] [PubMed] [Google Scholar]
- 8.Lion T, Kosulin K, Landlinger C, Rauch M, Preuner S, Jugovic D, Pötschger U, Lawitschka A, Peters C, Fritsch G, Matthes-Martin S. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714. 10.1038/leu.2010.4. [DOI] [PubMed] [Google Scholar]
- 9.Kosulin K, Berkowitsch B, Matthes S, Pichler H, Lawitschka A, Pötschger U, Fritsch G, Lion T. 2018. Intestinal adenovirus shedding before allogeneic stem cell transplantation is a risk factor for invasive infection post-transplant. EBioMedicine 28:114–119. 10.1016/j.ebiom.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Stamminger T, Hearing P. 2016. E2F/Rb family proteins mediate interferon induced repression of adenovirus immediate early transcription to promote persistent viral infection. PLoS Pathog 12:e1005415. 10.1371/journal.ppat.1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berk AJ. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685. 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 12.Cobrinik D. 2005. Pocket proteins and cell cycle control. Oncogene 24:2796–2809. 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira R, Naguibneva I, Mathieu M, Ait-Si-Ali S, Robin P, Pritchard LL, Harel-Bellan A. 2001. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep 2:794–799. 10.1093/embo-reports/kve173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolov MV, Dyson NJ. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117:2173–2181. 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- 15.Baker SJ, Reddy EP. 2012. CDK4: a key player in the cell cycle, development, and cancer. Genes Cancer 3:658–669. 10.1177/1947601913478972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suski JM, Braun M, Strmiska V, Sicinski P. 2021. Targeting cell-cycle machinery in cancer. Cancer Cell 39:759–778. 10.1016/j.ccell.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. 2004. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 64:9027–9034. 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard KE, McArthur GA. 2013. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res 19:5320–5328. 10.1158/1078-0432.CCR-13-0259. [DOI] [PubMed] [Google Scholar]
- 19.Barrière C, Santamaría D, Cerqueira A, Galán J, Martín A, Ortega S, Malumbres M, Dubus P, Barbacid M. 2007. Mice thrive without Cdk4 and Cdk2. Mol Oncol 1:72–83. 10.1016/j.molonc.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malumbres M, Sotillo R, Santamaría D, Galán J, Cerezo A, Ortega S, Dubus P, Barbacid M. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118:493–504. 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Almontashiri NAM, Fan M, Cheng BLM, Chen H-H, Roberts R, Stewart AFR. 2013. Interferon-gamma activates expression of p15 and p16 regardless of 9p21.3 coronary artery disease risk genotype. J Am Coll Cardiol 61:143–147. 10.1016/j.jacc.2012.08.1020. [DOI] [PubMed] [Google Scholar]
- 22.Hobeika AC, Etienne W, Torres BA, Johnson HM, Subramaniam PS. 1999. IFN-gamma induction of p21(WAF1) is required for cell cycle inhibition and suppression of apoptosis. J Interferon Cytokine Res 19:1351–1361. 10.1089/107999099312812. [DOI] [PubMed] [Google Scholar]
- 23.Lee S-H, Kim J-W, Oh S-H, Kim Y-J, Rho S-B, Park K, Park K-L, Lee J-H. 2005. IFN-gamma/IRF-1-induced p27kip1 down-regulates telomerase activity and human telomerase reverse transcriptase expression in human cervical cancer. FEBS Lett 579:1027–1033. 10.1016/j.febslet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Mandal M, Bandyopadhyay D, Goepfert TM, Kumar R. 1998. Interferon-induces expression of cyclin-dependent kinase-inhibitors p21WAF1 and p27Kip1 that prevent activation of cyclin-dependent kinase by CDK-activating kinase (CAK). Oncogene 16:217–225. 10.1038/sj.onc.1201529. [DOI] [PubMed] [Google Scholar]
- 25.Sangfelt O, Erickson S, Castro J, Heiden T, Gustafsson A, Einhorn S, Grandér D. 1999. Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene 18:2798–2810. 10.1038/sj.onc.1202609. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Zhang M, Li J, Yang G, Huang Q, Li J, Wang H, He S, Li E. 2020. Histone deacetylase inhibitors promote latent adenovirus reactivation from tonsillectomy specimens. J Virol 94:e00100-20. 10.1128/JVI.00100-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo RX, Postigo AA, Dean DC. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463–473. 10.1016/S0092-8674(00)80940-X. [DOI] [PubMed] [Google Scholar]
- 28.Chan HM, Smith L, La Thangue NB. 2001. Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 20:6152–6163. 10.1038/sj.onc.1204793. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. 1998. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA 95:10493–10498. 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gatla HR, Muniraj N, Thevkar P, Yavvari S, Sukhavasi S, Makena MR. 2019. Regulation of chemokines and cytokines by histone deacetylases and an update on histone decetylase [sic] inhibitors in human diseases. Int J Mol Sci 20:1110. 10.3390/ijms20051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Seto E. 2016. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med 6:a026831. 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dick FA, Goodrich DW, Sage J, Dyson NJ. 2018. Non-canonical functions of the RB protein in cancer. Nat Rev Cancer 18:442–451. 10.1038/s41568-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calbó J, Parreño M, Sotillo E, Yong T, Mazo A, Garriga J, Grana X. 2002. G1 cyclin/cyclin-dependent kinase-coordinated phosphorylation of endogenous pocket proteins differentially regulates their interactions with E2F4 and E2F1 and gene expression. J Biol Chem 277:50263–50274. 10.1074/jbc.M209181200. [DOI] [PubMed] [Google Scholar]
- 34.Cecchini MJ, Dick FA. 2011. The biochemical basis of CDK phosphorylation-independent regulation of E2F1 by the retinoblastoma protein. Biochem J 434:297–308. 10.1042/BJ20101210. [DOI] [PubMed] [Google Scholar]
- 35.Siddiqui H, Solomon DA, Gunawardena RW, Wang Y, Knudsen ES. 2003. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol Cell Biol 23:7719–7731. 10.1128/MCB.23.21.7719-7731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell M, Yang W-S, Yeh WW, Kao C-H, Chang P-C. 2020. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency. Front Microbiol 11:850. 10.3389/fmicb.2020.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins-McMillen D, Kamil J, Moorman N, Goodrum F. 2020. Control of immediate early gene expression for human cytomegalovirus reactivation. Front Cell Infect Microbiol 10:476. 10.3389/fcimb.2020.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schang LM, Hu M, Cortes EF, Sun K. 2021. Chromatin-mediated epigenetic regulation of HSV-1 transcription as a potential target in antiviral therapy. Antiviral Res 192:105103. 10.1016/j.antiviral.2021.105103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata T, Sugimoto A, Inagaki T, Yanagi Y, Watanabe T, Sato Y, Kimura H. 2021. Molecular basis of Epstein-Barr virus latency establishment and lytic reactivation. Viruses 13:2344. 10.3390/v13122344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knipe DM, Raja P, Lee J. 2017. Viral gene products actively promote latent infection by epigenetic silencing mechanisms. Curr Opin Virol 23:68–74. 10.1016/j.coviro.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hearing P, Shenk T. 1983. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33:695–703. 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- 42.O’Connor RJ, Hearing P. 1991. The C-terminal 70 amino acids of the adenovirus E4-ORF6/7 protein are essential and sufficient for E2F complex formation. Nucleic Acids Res 19:6579–6586. 10.1093/nar/19.23.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruder JT, Hearing P. 1991. Cooperative binding of EF-1A to the E1A enhancer region mediates synergistic effects on E1A transcription during adenovirus infection. J Virol 65:5084–5087. 10.1128/JVI.65.9.5084-5087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirza MA, Weber J. 1982. Structure of adenovirus chromatin. Biochim Biophys Acta 696:76–86. 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- 45.Lynch KL, Gooding LR, Garnett-Benson C, Ornelles DA, Avgousti DC. 2019. Epigenetics and the dynamics of chromatin during adenovirus infections. FEBS Lett 593:3551–3570. 10.1002/1873-3468.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karen KA, Hearing P. 2011. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J Virol 85:4135–4142. 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avgousti DC, Herrmann C, Kulej K, Pancholi NJ, Sekulic N, Petrescu J, Molden RC, Blumenthal D, Paris AJ, Reyes ED, Ostapchuk P, Hearing P, Seeholzer SH, Worthen GS, Black BE, Garcia BA, Weitzman MD. 2016. A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 535:173–177. 10.1038/nature18317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson JS, Osheim YN, Xue Y, Emanuel MR, Lewis PW, Bankovich A, Beyer AL, Engel DA. 2004. Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J Virol 78:6459–6468. 10.1128/JVI.78.12.6459-6468.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostapchuk P, Suomalainen M, Zheng Y, Boucke K, Greber UF, Hearing P. 2017. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog 13:e1006455. 10.1371/journal.ppat.1006455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komatsu T, Haruki H, Nagata K. 2011. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res 39:889–901. 10.1093/nar/gkq783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakanishi Y, Maeda K, Ohtsuki M, Hosokawa K, Natori S. 1986. In vitro transcription of a chromatin-like complex of major core protein VII and DNA of adenovirus serotype 2. Biochem Biophys Res Commun 136:86–93. 10.1016/0006-291X(86)90880-6. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, Morral N, Engel DA. 2007. Transcription releases protein VII from adenovirus chromatin. Virology 369:411–422. 10.1016/j.virol.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Fedor MJ, Daniell E. 1980. Acetylation of histone-like proteins of adenovirus type 5. J Virol 35:637–643. 10.1128/JVI.35.3.637-643.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu J, Boyapati A, Rundell K. 2001. Critical role for SV40 small-t antigen in human cell transformation. Virology 290:192–198. 10.1006/viro.2001.1204. [DOI] [PubMed] [Google Scholar]
- 55.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352. 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 56.Reich NC, Sarnow P, Duprey E, Levine AJ. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480–484. 10.1016/0042-6822(83)90274-X. [DOI] [PubMed] [Google Scholar]