ABSTRACT
Adeno-associated viruses (AAVs) are being developed as clinical gene therapy vectors. One issue undermining their broad use in the clinical setting is the high prevalence of circulating antibodies in the general population capable of neutralizing AAV vectors. Hence, there is a need for AAV vectors that can evade the preexisting immune response. One possible source of human naive vectors are AAVs that do not disseminate in the primate population, and one such example is serpentine AAV (SAAV). This study characterizes the structural and biophysical properties of the SAAV capsid and its receptor interactions and antigenicity. Single particle cryo-electron microscopy (cryo-EM) and thermal stability studies were conducted to characterize the SAAV capsid structure at pH 7.4, 6.0, 5.5, and 4.0, conditions experienced during cellular trafficking. Cell binding assays using Chinese hamster ovary (CHO) cell lines identified terminal sialic acid as the primary attachment receptor for SAAV similar to AAV1, 4, 5, and 6. The binding site of sialic acid to the SAAV capsid was mapped near the 2-fold axis toward the 2/5-fold wall, in a different location than AAV1, 4, 5, and 6. Towards determining the SAAV capsid antigenicity native immunodot blots showed that SAAV evades AAV serotype-specific mouse monoclonal antibodies. However, despite its reptilian origin, it was recognized by ~25% of 50 human sera tested, likely due to the presence of cross-reactive antibodies. These findings will inform future gene delivery applications using SAAV-based vectors and further aid the structural characterization and annotation of the repertoire of available AAV capsids.
IMPORTANCE AAVs are widely studied therapeutic gene delivery vectors. However, preexisting antibodies and their detrimental effect on therapeutic efficacy are a primary challenge encountered during clinical trials. In order to circumvent preexisting neutralizing antibodies targeting mammalian AAV capsids, serpentine AAV (SAAV) was evaluated as a potential alternative to existing mammalian therapeutic vectors. The SAAV capsid was found to be thermostable at a wide range of environmental pH conditions, and its structure showed conservation of the core capsid topology but displays high structural variability on the surface. At the same time, it binds to a common receptor, sialic acid, that is also utilized by other AAVs already being utilized in gene therapy trials. Contrary to the initial hypothesis, SAAV capsids were recognized by one in four human sera tested, pointing to conserved amino acids around the 5-fold region as epitopes for cross-reacting antibodies.
KEYWORDS: AAV, adeno-associated virus, antibody, capsid, cryo-EM, gene therapy, human sera, parvovirus, receptor, sialic acid
INTRODUCTION
Adeno-associated viruses (AAVs) are ssDNA, non-enveloped, small (~26 nm diameter), T = 1, viruses of the family Parvoviridae, genus Dependoparvovirus (1). Dependoparvoviruses have been isolated from a wide range of mammals including human (2), nonhuman primate (3), cow (4), goat (5), pig (6), sea lion (7), fox (8), rodents (9), and bat (10). In addition, members of this genus have also been found in birds (11) and reptiles (12, 13). The structures of many AAV serotype, variant, and isolate capsids have been determined by X-ray crystallography and/or cryo electron microscopy (cryo-EM) (14–30). All AAV capsid structures display common features such as a channel at the 5-fold axis, protrusions around the 3-fold axis, and depressions at the 2-fold axis and around the 5-fold channel (31). The capsids are composed of three overlapping structural viral proteins (VPs), the minor proteins VP1 (~80 kDa) and VP2 (~65 kDa), and the major protein VP3 (~60 kDa). A total of 60 VPs are incorporated in a capsid at an approximate ratio of 1:1:10, respectively (32). The amino acid sequence of VP3 is shared with both minor capsid proteins, while VP2 has an N-terminal region that it shares with VP1 (VP1/2 common region). The VP1/2 common region contains nuclear localization motifs that have been shown to play a role in targeting of the incoming capsid toward the nucleus following infection of a cell with an AAV (33). In addition, VP1 contains a unique N-terminal sequence, termed the VP1 unique region (VP1u), that contains a phospholipase A2 domain that is required for escaping the endosomes during trafficking through the endo-lysosomal pathway (34). The N termini of both VP1 and VP2 are believed to be located in the interior of the capsid and following a structural rearrangement become externalized upon acidification of the endosome during cellular trafficking (35). This externalization was suggested to occur through the channel at the 5-fold axes of the capsids (36). Due to the intrinsic disorder as well as the lower copy number of the N termini of VP1 and VP2 to date, their structures are unknown. Similarly, the first ~15 N-terminal residues of VP3 are not ordered in the determined AAV capsid structures. The core of the ordered VP3 common region of the AAVs contains a jelly roll motif with an eight stranded antiparallel β-sheet, plus an additional βA-strand, antiparallel to βB, which form the interior surface of the capsid (32). Also common to AAV capsid structures is an α-helix located near the 2-fold axis. The core β-sheets and α-helical region are structurally conserved among the Dependoparvoviruses. Between the β-strands large loops are inserted that form the exterior capsid surface topology. In contrast to the core, these surface loops display high structural variability that have been defined as variable regions (VRs) (17). A total of nine VRs (VR-I to -IX) have been described. The VRs of the AAVs are responsible for the phenotypic differences of the AAVs. They have been associated with glycan and protein receptor attachment (31, 32, 37), transduction (38–40), and antigenicity (41–44).
The AAVs are widely studied for their use as therapeutic gene delivery vectors to treat a large variety of monogenetic diseases (45). Currently all the vectors used in clinical trials and approved gene therapy biologics are based on primate AAVs. Despite the nonpathogenic character of the viruses, a significant percentage of the human population (40-70%) are seropositive for neutralizing antibodies that target primate AAVs (46) due to prior exposure. These antibodies can potentially target the viral capsids and lead to vector inactivation resulting in a loss of treatment efficacy. In order to circumvent this preexisting immunity, capsids of viruses may be utilized that are not prevalent in the human population. Squamate dependoparvovirus 1, also known as serpentine adeno-associated virus (SAAV), was isolated from Python regius and Boa constrictor snakes (12), and the capsid displays low sequence identity to the primate AAVs (54–59%).
In this study, the capsid structure of SAAV was determined by cryo-electron microscopy (cryo-EM) and 3D image reconstruction at pH 7.4 to 3.25 Å resolution. Comparisons of the SAAV capsid structure to AAV1, AAV2, and AAV5 show significant differences in the surface loops while the core of the capsid remains structurally conserved. Nonetheless, SAAV was found to bind to a common glycan receptor, sialic acid, that is also utilized by other AAV serotypes such as AAV1, 4, 5, and 6. The sialic acid binding site was mapped to the 2/5-fold wall near the 2-fold axis. Following cellular attachment, the viral capsid encounters lower pH conditions during endosomal trafficking. Here, we show that the SAAV capsid structure remains largely unchanged at pH 6.0, 5.5, and 4.0, compared to pH 7.4, with some structural rearrangements of the N termini and 5-fold channel. In an effort to characterize the antigenicity of SAAV, the capsid was probed with a panel of AAV-specific monoclonal antibodies (MAbs) directed against the AAV serotypes and various human serum samples. While SAAV was not recognized by any of the MAbs, ~25% of human sera reacted with SAAV capsids, likely due to the presence of cross-reactive antibodies. This could hamper the utilization of SAAV based vectors for future gene delivery applications but may also help to identify the cross-reactive epitopes of different AAVs to further optimize vectors with antibody escape properties.
RESULTS AND DISCUSSION
SAAV structure determination.
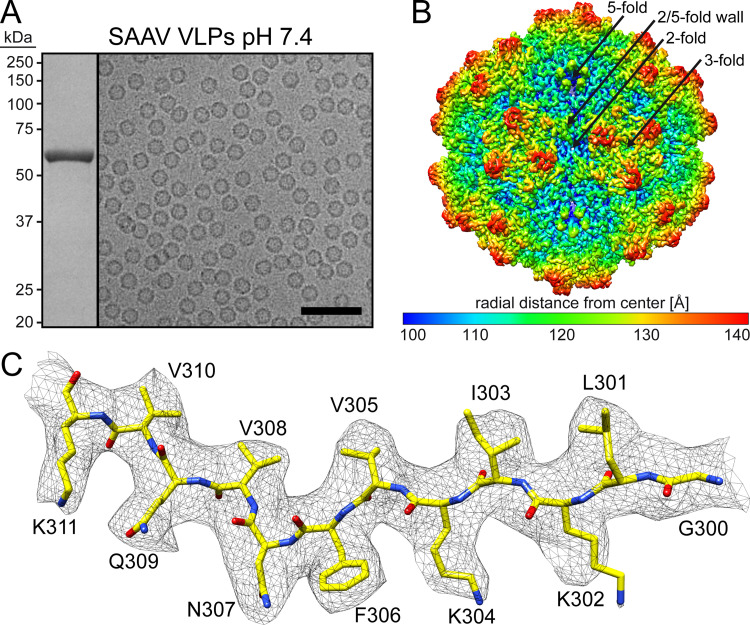
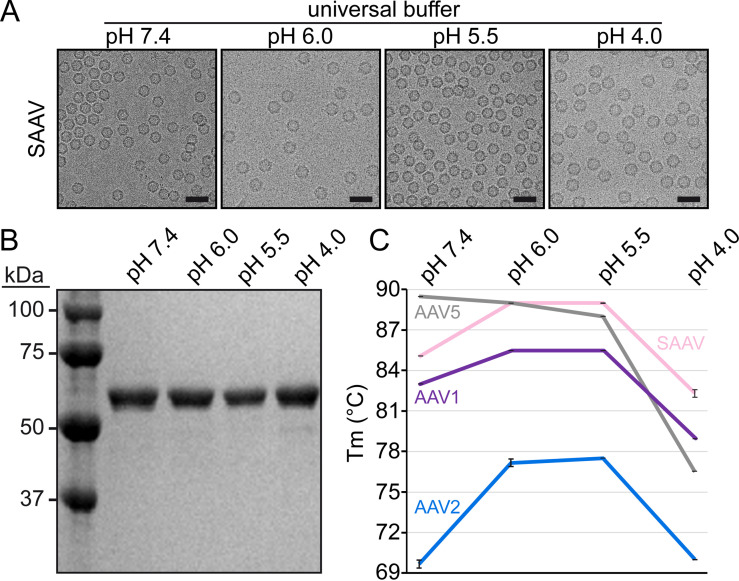
Following production of SAAV virus-like particles (VLPs) in Sf9 cells by recombinant baculoviruses and purification by sucrose cushion and gradient ultracentrifugation with subsequent anion-exchange chromatography the purified fraction contained a major band consistent with SAAV VP3, migrating at ~60 kDa, by SDS-PAGE (Fig. 1A). Negative stain EM (not shown) and cryo-EM micrographs showed intact and pure particles of approximately 25 nm in diameter (Fig. 1A). Thus, the SAAV VLPs were deemed suitable for structure determination by cryo-EM, and data collection was supported by the NIH “Southeastern Center for Microscopy of MacroMolecular Machines (SECM4)” project. For 3D-image reconstruction of the SAAV capsid, a total of 73,613 individual particles were utilized resulting in a resolution of 3.25 Å, based on an FSC threshold of 0.143 (Table 1). The reconstructed SAAV capsid map displayed the characteristic surface features also seen in other AAVs such as channels at the icosahedral 5-fold axes, three protrusions surrounding the 3-fold axes, and depressions at the 2-fold and depressions surrounding the 5-fold axes, which are separated by a raised region termed 2/5-fold wall (Fig. 1B). However, the 3-fold protrusions were less “spiky” compared to AAV2 and more similar to those of AAV5 (14, 16). The SAAV capsid map displayed well-ordered side chain densities (Fig. 1C) for most of the amino acid residues, and a model for SAAV VP3 was built starting from alanine 206 at the N-terminus to leucine 726 at the C-terminus. This N-terminal alanine is equivalent to alanine 218 in AAV1 and AAV2 and alanine 208 in AAV5. The start of structural ordering at the N-terminus is comparable to the other AAVs, with AAV1 and AAV2 showing one additional and AAV5 displaying one less ordered residue relative to SAAV (14, 16, 23). The final pH 7.4 SAAV multimerized capsid model (60mer) was refined and evaluated relative to the cryo-reconstructed map using Phenix, resulting in a correlation coefficient of 0.863 (Table 1).
FIG 1.
SAAV VLPs cryo-EM structure. (A) SDS-PAGE of the purified SAAV VLPs at pH 7.4 with a band at ~60 kDa equivalent to the size of VP3 and example cryo-electron micrograph. Scale bar: 100 nm. (B) The capsid surface density maps contoured at a sigma (σ) threshold level of 2. The map is radially colored (blue to red) according to distance to the particle center, as indicated by the scale bar below. The icosahedral 2-, 3-, and 5-fold axes are indicated on the capsid map. (C) The modeled SAAV residues of the βD strand are shown inside their density map. The amino acid residues are shown as stick representation and colored according to atom type: C, yellow; O, red; N, blue. Panel B and C were generated using UCSF-Chimera (79).
TABLE 1.
Summary of data collection, image processing, and refinement statistics
| Parameter | pH 7.4 | pH 6.0 | pH 5.5 | pH 4.0 | 3’SLN | 6’SLN |
|---|---|---|---|---|---|---|
| Micrographs | 390 | 1,435 | 2,002 | 1,217 | 1,693 | 1,806 |
| Defocus range (μm) | 0.9–3.3 | 1.0–3.0 | 0.–2.8 | 1.0–3.0 | 1.5–2.6 | 1.2–2.4 |
| Electron dose (e-/Å2) | 62 | 60 | 34 | 60 | 34 | 34 |
| Frames per micrograph | 30 | 50 | 30 | 50 | 30 | 30 |
| Pixel size (Å/pixel) | 1.24 | 1.09 | 1.09 | 1.09 | 1.09 | 1.09 |
| Particles used for final map | 73,613 | 55,586 | 402,005 | 149,155 | 96,418 | 269,981 |
| Resolution (Å) | 3.25 | 2.73 | 2.14 | 2.66 | 3.50 | 3.50 |
| Model refinement statistics | ||||||
| Map CC | 0.866 | 0.863 | 0.870 | 0.850 | 0.844 | 0.842 |
| RMSD bond (Å) | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| RMSD angle (°) | 0.79 | 0.82 | 0.82 | 0.83 | 0.71 | 0.72 |
| All-atom clash score | 5.02 | 6.67 | 4.02 | 6.29 | 5.68 | 5.98 |
| Ramachandran (%) | ||||||
| Favored | 98.1 | 98.2 | 98.4 | 98.4 | 97.8 | 98.0 |
| Allowed | 1.9 | 1.8 | 1.6 | 1.6 | 2.2 | 2.0 |
| Unfavored | 0 | 0 | 0 | 0 | 0 | 0 |
| Rotamer outliers (%) | 0 | 0 | 0 | 0 | 0 | 0 |
| C-β deviation (%) | 0 | 0 | 0 | 0 | 0 | 0 |
Structural comparison of SAAV to other AAVs.
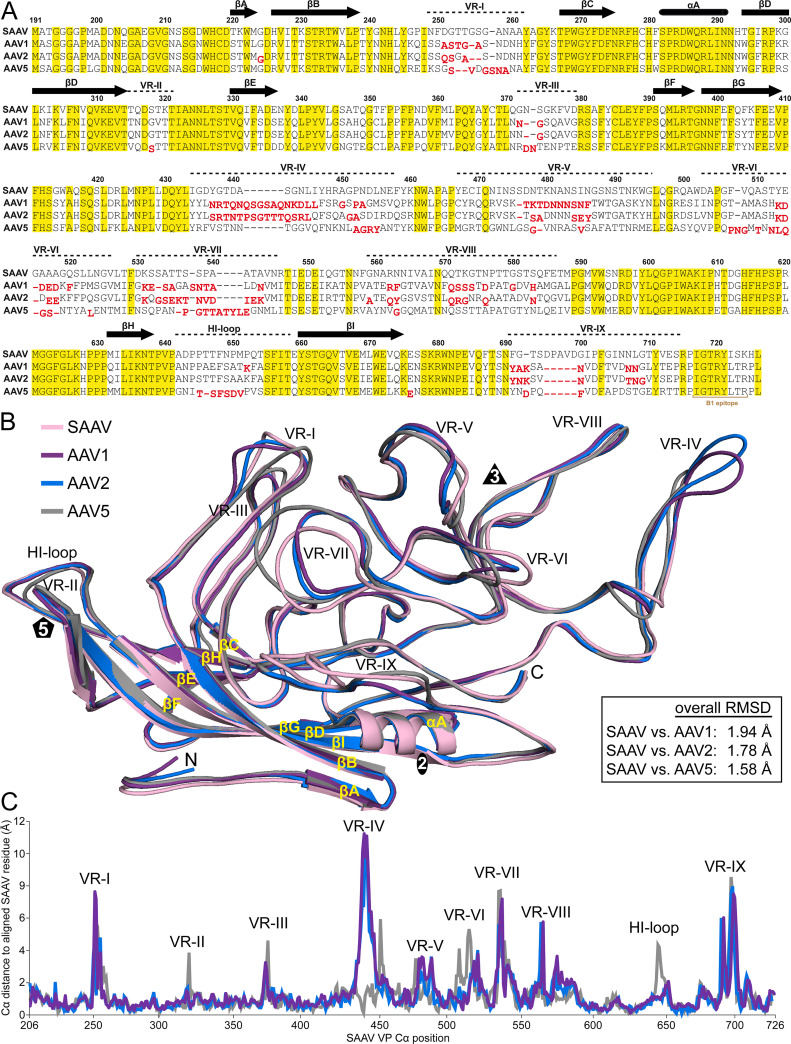
The amino acid sequence identity of SAAV for the ordered VP structure compared to other AAVs is low, e.g., 57% versus AAV1, 55% versus AAV2, and 53% versus AAV5. However, similar to other known AAV capsid structures (14–30), the SAAV VP3 topology conserves the core βA-strand, the eight-stranded antiparallel β-barrel (βB-βI), and α-helix (Fig. 2A and B), which are superposable to the AAV1 (PDB-ID 3NG9), AAV2 (1LP3), and AAV5 (3NTT) VP monomers with Cα RMSDs ranging from 0.60 to 0.66 Å. The majority of sequence conservation is located in or near the β-barrel and α-helix (Fig. 2A), whereas only 8–15% of the surface amino acids are conserved. Structural variability of the SAAV VP is mostly observed in the exterior surface loops within the previously defined VRs (Fig. 2A and B) (17). The VRs with the highest structural variability are VR-I, VR-IV, VR-VII, and VR-IX (Fig. 2B and C). The SAAV VR-I, located at the 2/5-fold wall, possesses a single amino acid insertion compared to AAV1 and two insertions compared to AAV2 and AAV5 (Fig. 2A) with a Cα distance of the aligned VPs of up to ~8 Å for AAV1, ~5 Å for AAV2, and ~6 Å for AAV5, respectively (Fig. 2C). The large structural variability of VR-IV is mainly caused by a 6 amino acid insertion in AAV1 and AAV2 when compared to SAAV and AAV5, resulting in Cα displacements of up to ~11 Å (Fig. 2A and C). The VR-IV is the loop responsible for the “spiky” appearance of the AAV2 3-fold protrusions. Thus, sharing a similar loop length to AAV5 explains the similar appearance of the SAAV capsid. While SAAV’s VR-IV loop is similar to AAV5, differences are observed at the base of the loop where SAAV is more like AAV1 and AAV2 (Fig. 2B). The other two loops contributing to the 3-fold protrusions, VR-V and VR-VIII, show less variability. In the case of SAAV’s VR-VIII, the highest Cα distance difference of up to 6 Å is observed at the base of the loop instead of the apex compared to AAV1, 2, and 5. VR-VII, located on the side of the 3-fold protrusions, adopts a different conformation in SAAV with up to 8 Å in Cα distance variation compared to the other AAVs despite the absence of insertions or deletions relative to AAV1 and AAV2 (Fig. 2B and C). SAAV’s VR-IX is unique, due to a 5 amino acid insertion compared to AAV1, 2, and 5, generating an additional loop on the SAAV surface (Fig. 2A and B). None of the other AAV serotypes or rhesus isolates possess any insertions in VR-IX indicating a possible reptile-specific adaptation of the capsid, especially since bearded dragon parvovirus, another reptile dependoparvovirus, also displays a similar insertion in VR-IX (13). VR-IX is located next to VR-I at the 2/5-fold wall and flanks the 2-fold axis. In contrast to VR-I and VR-IX, SAAV’s VR-III, which is also located at the 2/5-fold wall, only shows minimal structural variability to the other AAVs (Fig. 2B). The least structural variability to the primate AAVs is seen around the 5-fold axis, which includes VR-II and the HI-loop with the exception of AAV5, which possesses a single amino acid deletion in the HI-loop (Fig. 2A and B). The absence of significant structural differences in these loops are likely due to the essential common functions of the AAVs, such as DNA packaging and VP1u externalization (36, 47). The overall Cα RMSD of the SAAV VP structure to the other AAVs ranges from 1.6 to 1.9 Å, which is comparable to a BtAAV-10HB, another recently determined nonprimate AAV (30).
FIG 2.
Structural alignment of SAAV, AAV1, AAV2, and AAV5 VPs. (A) Structure-based sequence alignment, except for the VP3 N termini prior to the first structurally ordered amino acid. The VP3 residue numbers indicated above the amino acid sequence are based on SAAV. Secondary structure elements such as β-strands and α-helices are shown as black arrows and black cylinders, respectively. The positions of the VRs are indicated above the sequence. Amino acids highlighted in yellow indicate sequence identity among all four viruses. Amino acids whose Cα atoms are further than 2 Å apart when superposed onto SAAV are shown offset and in red below the aligned residues. (B) Structural superposition of SAAV (pink), AAV1 (purple), AAV2 (blue), and AAV5 (gray) shown as ribbon diagrams. The position of β-strand A-I, α-helix A, the N- and C-terminus, and the icosahedral 2-, 3-, and 5-fold axes are labeled. This figure was generated using PyMol (86). (C) Cα-Cα distance plot for SAAV compared to AAV1 (purple), AAV2 (blue), and AAV5 (gray). The positions of the VRs are indicted.
SAAV binds to terminal sialic acids.
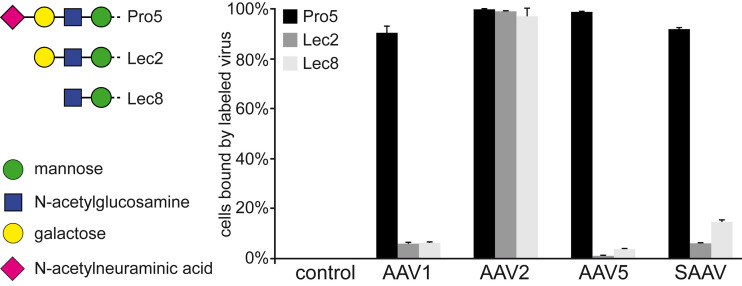
One of the functions of the capsid is to attach to a receptor of the host cell. Many AAVs utilize glycan receptors such as terminal sialic acid (AAV1, AAV4, AAV5, and AAV6), galactose (AAV9), and heparan sulfate proteoglycans (AAV2, AAV3, AAV6, and AAV13), which have been described as cell attachment factors (48–53). In order to determine whether SAAV utilizes these common glycans for cell attachment, a heparin binding assay and a cell binding assay using differential glycan presenting CHO cell lines were conducted. For the heparin binding assay, AAV2 was used as a positive control that bound to heparin-conjugated beads. In contrast, SAAV did not bind to these beads (data not shown), thus likely excluding heparan sulfate proteoglycan as a receptor for SAAV. For the cell binding assay, the CHO-Pro5 cell line that displays terminal sialic acid, the mutant Lec2 cell line that displays terminal galactose, and the mutant Lec8 cell line that displays terminal N-acetylglucosamine were utilized. The Lec2 and Lec8 cell lines are derived from the parental CHO-Pro5 cell line resulting from mutations in specific genes required for glycan biosynthesis (54). Binding of fluorescently-labeled capsids to the cells was determined by a FACS-based assay. The AAV2 capsid was used as a positive control for the three cell lines since heparan sulfate proteoglycan is present on all CHO cell lines. AAV1 and AAV5 showed robust binding to the Pro5 cells but not to the sialic acid deficient Lec2 and Lec8 cell lines (Fig. 3). These AAVs are known sialic acid binders and thus used as benchmark for this receptor type. Indeed, SAAV showed a very similar binding pattern as AAV1 and AAV5, strongly indicating that terminal sialic acid is a receptor for SAAV. Also, terminal galactose could be excluded as a SAAV receptor as it would have shown robust preferential binding to the Lec2 cell line, as seen previously for AAV9 (52).
FIG 3.
SAAV binds to terminal sialic acid. Fluorescent-labeled VLPs were incubated with CHO Pro5, Lec2, and Lec8 cells as described in the methods and the percentage of cells with fluorescent signals determined by a FACS-based assay. AAV2 was tested as a positive control for all cell lines as it binds to heparan sulfate proteoglycan. AAV1 and AAV5 were included as sialic acid binding controls.
The SAAV sialic acids binding site is located at the 2/5-fold wall.
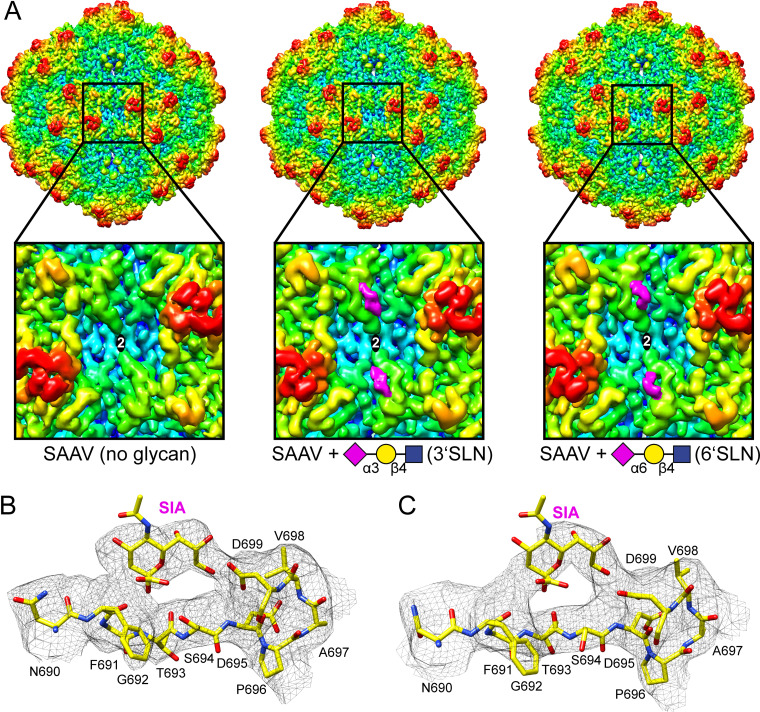
Both the sialic binding pockets in the AAV1 and AAV5 capsid have been identified (55, 56). The AAV1 capsid binds sialic acid at the side of the 3-fold protrusion toward the 2/5-fold wall, making contact with N447, V473 (both VR-IV), and N500 (VR-V) (55), whereas AAV5 binds sialic acids near the center of the 3-fold protrusion with residues M569, Y585, and L587 (all in VR-VIII) being the important contact residues (56). For AAV4 the sialic acid binding pocket has not been fully confirmed yet, but AAV4 capsid variants with amino acid substitutions in VR-V (K492E, K503E) and VR-VIII (G581D, Q583E, N585S) prevent cell binding and hemagglutination (57), also pointing to the 3-fold region as the sialic acid binding pocket. The SAAV capsid does not conserve any of these contact residues in structurally equivalent positions, suggesting an alternative binding site for sialic acid. In order to determine the sialic acid binding site 3’SLN (NeuNAcα2-3Galβ1-4GlcNAc) or 6’SLN (NeuNAcα2-6Galβ1-4GlcNAc) was added to SAAV VLPs at 10-fold excess per potential binding site and cryo-EM data collected on these complexes. Both glycans with alternative sialic acid linkages were tested as some AAVs such as AAV4 bind to α2-3-linked sialic acids but not α2-6-linked sialic acids (53). In contrast, other AAVs such as AAV1, AAV5, and AAV6 bind to both types of sialic acids (48). The SAAV-glycan complexes were reconstructed to 2.30 Å (3’SLN) and 2.14 Å (6’SLN) resolution from 96,418 and 269,981 capsids, respectively. The resulting density maps showed very well-ordered amino acid side chain densities, and the model generated from the 3.25 Å resolution map fitted well without significant structural deviations. At this high resolution some additional disordered densities on the capsid surface were observed for both complex maps. For better comparison to the lower resolution SAAV map, an earlier iteration of the reconstruction process (at 3.5 Å resolution) for both complex maps was used (Fig. 4A). In these maps, the extra density at the 2/5-fold wall is clearly visible when the SAAV capsids were complexed with 3’SLN or 6’SLN. This density is similar in profile in both maps and is located near VR-IX, and potential contact residues include F691, G692, D695, and D699 (Fig. 4B and C). Both glycans are trisaccharides, but only the terminal sialic acid is expected to bind to the capsid whereas the attached galactose and N-Acetyl glucosamine will likely be flexible and not observed in the icosahedral reconstructed cryo-EM structure. Thus, the densities near VR-IX were interpreted as sialic acids (Fig. 4B and C) which indicate that SAAV binds both α2-3- and α2-6-linked sialic acids, similar to AAV1, AAV5, and AAV6 (48). As the sialic acid binding pockets of the other AAVs are located around the 3-fold protrusions, SAAVs’ sialic acid binding pocket around the 2-fold is unique. The capsids of minute virus of mice (MVM), another parvovirus of a different genus, also bind to sialic acid near the 2-fold symmetry axis (58). However, its binding mode is different from SAAV as it binds to sialic acid utilizing residues in VR6 (equivalent to VR-VI of the AAVs) (59) that are located away from the 2/5-fold wall.
FIG 4.
Determination of the SAAV sialic acid binding site. (A) Capsid surface density maps of SAAV without glycan, SAAV with 3’SLN, and SAAV with 6’SLN contoured at a sigma (σ) threshold level of 1. The maps are radially colored (blue to red) according to distance to the particle center, as indicated by the scale bar in Fig. 1 A closeup of the 2-fold region for each map is shown below. The position of the sialic acid binding site is indicated with a pink oval. The missing density for the map without any glycans is indicated by a white oval. (B and C) Amino acid residues 690–699 including a sialic acid (SIA) molecule inside the density map at a sigma (σ) threshold level of 1.0 depicted as a black mesh. The model is shown in stick representation and the atoms are colored: C, yellow; O, red; N, blue. This figure was generated with Chimera (79).
SAAV capsids gain stability at low pH.
Following the attachment to the target cell and receptor-mediated endocytosis, the viral capsid undergoes continuous acidification as it traffics through the endosomal/lysosomal pathway. These changes in pH are believed to trigger rearrangements in the capsid resulting in the externalization of the VP1u and VP1/2 common region. In order to characterize the SAAV capsid at the conditions of the early endosome (pH 6), the late endosome (pH 5.5), and lysosome (pH 4), the VLPs were dialyzed to the pH of these environments. Cryo-EM micrographs showed intact capsids for all the tested conditions (Fig. 5A), and no indication of degradation of the VPs was visible by SDS-PAGE (Fig. 5B). To further analyze the stability of the capsids, the melting temperatures (Tm) of the SAAV capsid were determined at the different pHs by differential scanning fluorimetry (DSF) and compared to AAV1, 2, and 5. The Tm of SAAV (in universal buffer) at pH 7.4 was determined to be ~85°C (Fig. 5C). At the lower pHs of 6.0 and 5.5, the SAAV capsid exhibited increased stability with a Tm of ~89°C. However, pH 4.0 resulted in an abrupt reduction of capsid stability with a Tm of ~82°C. The same trend of stability at the different pH conditions was also observed for AAV1 and AAV2 (Fig. 5C). However, AAV5 did not follow this trend. The highest Tm for AAV5 was seen at physiological pH of 7.4 at ~89.5°C, then steadily decreasing as the pH is reduced to ~89.0°C at pH 6.0, ~88.0°C at pH 5.5, and ~76.5°C at pH 4.0, respectively (Fig. 5C). The Tms reported here for AAV1, AAV2, and AAV5 follow the same trend recently described for these viruses in citrate-phosphate buffer (60). Variations of the Tm at the identical pH are caused by the buffer conditions (61).
FIG 5.
Analysis of the SAAV capsid stability at different pH conditions. (A) Cryo-EM micrographs of the SAAV capsids dialyzed to indicated pH buffer. (B) SDS-PAGE analysis of the dialyzed samples. (C) SAAV, AAV1, AAV2, and AAV5 capsid stability analysis by differential scanning fluorimetry. The melting temperatures were determined at pH 7.4, 6.0, 5.5, and 4.0. Results are displayed as mean ± standard deviation (n = 3).
Low pH conditions result in rearrangements of the N-terminus and the 5-fold channel.
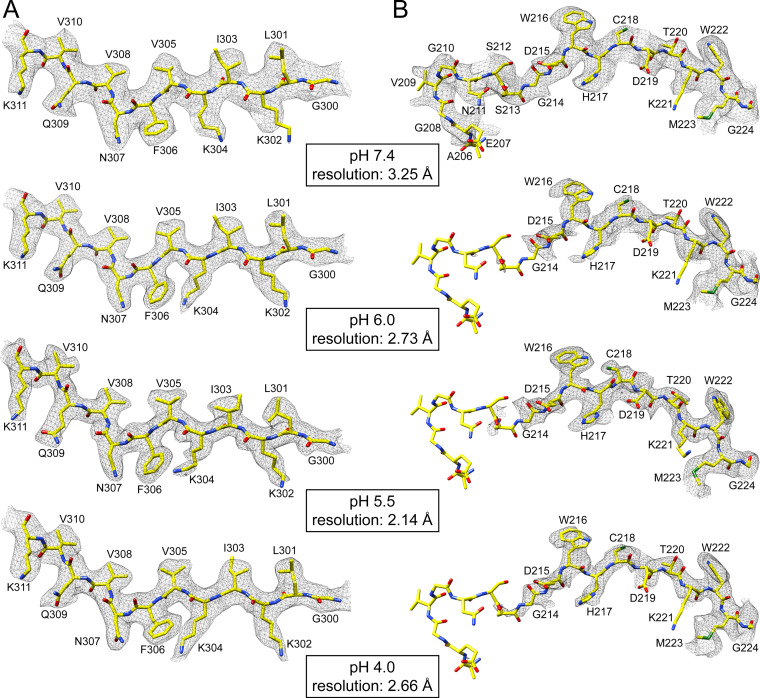
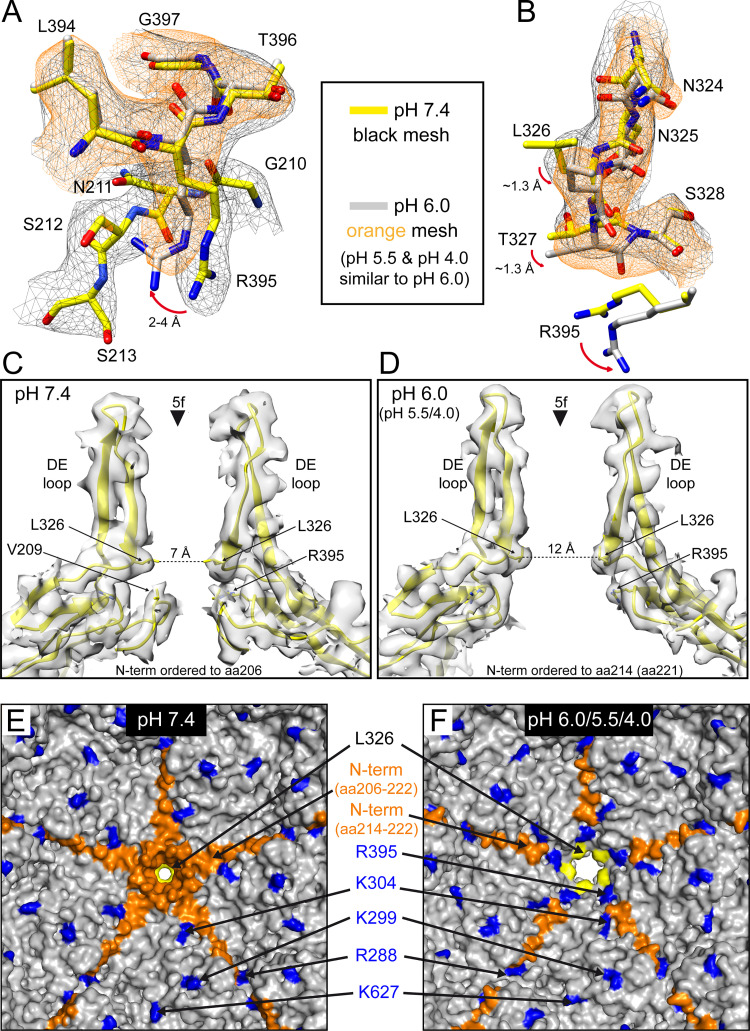
The observation that the SAAV capsid remains intact at low pH conditions allowed the determination of their capsid structures. For this purpose, the SAAV VLPs were dialyzed to these pH conditions and vitrified on EM grids, and data were collected. A total of 55,586 particles at pH 6.0, 402,005 particles at pH 5.5, and 149,155 particles at pH 4.0 were 3D-reconstructed to resolutions of 2.73, 2.14, and 2.66 Å, respectively (Table 1). The reconstructed SAAV maps did not show any significant differences on the surface of the capsid (not shown) compared to the SAAV capsid at pH 7.4 (Fig. 1B). The refined atomic SAAV model at pH 7.4 was docked into the three pH maps and where necessary, structures were refined to fit the density, resulting in correlation coefficients similar to the pH 7.4 structure of 0.863 (pH 6.0), 0.870 (pH 5.5), and 0.850 (pH 4.0), respectively (Table 1). All the pH maps showed well-ordered amino acid side chain densities (Fig. 6A) allowing a reliable comparison to the pH 7.4 structure. As indicated by the similar capsid surface projections, the overall VP structures are very similar to Cα RMSDs of 0.46 Å (pH 7.4 versus 6.0), 0.39 Å (pH 7.4 versus 5.5), and 0.45 Å (pH 7.4 versus 4.0). However, a major difference at all pH conditions was observed for the N-terminus of the VP and at the base of the 5-fold channel. While structural ordering of the VP monomer at pH 7.4 starts at amino acid 206, density for the first ordered residue was not observed until amino acid 214 at pH 6.0, pH 5.5, and pH 4.0 (Fig. 6B). A similar disordering of the N-terminus was previously reported for two AAV2 capsid variants (AAV2-L336C and -R432A) whose capsid structures were determined by cryo-EM (24, 25). In the case of the AAV2-L336C variant, structural ordering started at glycine 226 (25) which is equivalent to the ordering seen for glycine 214 in the low pH SAAV maps (Fig. 6B). In the AAV2-R432A variant the first ordered residue was serine 232 (24), which aligns to threonine 220 of SAAV. These AAV2 variants were also shown to display both genome packaging defects and altered VP1u externalization properties (47, 62). Furthermore, in both these AAV2 variant structures the side chain of arginine 404 moves into the available space previously occupied by the N-terminus. The same rearrangements can be seen in the SAAV capsid at R395, which is the structural equivalent position to AAV2 R404. At pH 7.4 R395 of SAAV adopts the same side chain orientation compared to R404 in AAV2 (Fig. 7A). At the lower pH conditions the orientation of the arginine side chain shifts radially outwards by 4 Å, identical to R404 in AAV2-L336C and AAV2-R432A, and moves into the space where the N-terminus was ordered at pH 7.4. In close proximity to R395 a second observed shift occurs in the main-chain at the base of the DE loop at low pH conditions, which results in the leucine 326 side chain rotating in a sideways manor (Fig. 7B). These shifts of the main-chain were also observed in the AAV2-L336C and -R432A capsid structures (24, 25). Leucine 326 in SAAV, which is equivalent to L336 in AAV2 and L172 in MVM, represents the narrowest point of the 5-fold channel and has been suggested as a “gate-keeper” to prevent premature genome ejection (63). The rotation of the leucine side chain results in a widening of the 5-fold channel from ~7 Å (at pH 7.4) to ~12 Å (at pH 6.0/5.5/4.0) (Fig. 7C and D), which was also observed in the AAV2-L336C structure (25). This leucine residue in the 5-fold channel is conserved across the parvoviruses (32) and is involved in a conserved hydrophobic interaction to valine 209 in SAAV or V221 in AAV2. A likely mechanism could be that low pH conditions disrupt this hydrophobic interaction, a state that was also simulated by the leucine-to-cysteine variant, which leads to the N-terminus to become more flexible, thereby allowing R395 and the base of the DE-loop to move, resulting in the widening of the 5-fold pore (Fig. 7C to F) for the externalization of the VP1u and VP1/2 common region (36). Since the conserved leucine-valine pair in the 5-fold channel has been suggested to be involved in genome retention (63), it is also possible that a genome release is triggered by the loss of interaction between the residues at low pH conditions. It remains to be seen whether SAAV capsids indeed eject their genome under these conditions, as the VLPs used in this study were free from packaged genomes. If confirmed, this would limit purification methods for potential SAAV based vectors to neutral conditions only.
FIG 6.
The SAAV capsid structure at different pH conditions. (A) The modeled SAAV residues of the βD strand are shown inside their density map for pH 7.4, 6.0, 5.5, and 4.0. Of note is the alternative side chain orientation of K304 in pH 7.4 versus the low pH conditions. (B) The modeled SAAV residues near the N-terminus (206–224) are shown inside their density map for pH 7.4, 6.0, 5.5, and 4.0. While structural ordering begins with residue 206 in pH 7.4, residues 206–213 are disordered in pH 6.0, 5.5, and 4.0. The amino acid residues are shown as stick representation and colored according to atom type: C, yellow; O, red; N, blue. Panels B and C were generated using UCSF-Chimera (79).
FIG 7.
The low pH conditions result in structural rearrangements at the base of the 5-fold channel. (A) The modeled SAAV residues 210–213 and 394–397 are shown inside their density map for pH 7.4 (black mesh) and pH 6.0 (orange mesh). The side chain orientation of R395 shifts at low pH conditions and moves into the space of the disordered N-terminus. (B) The modeled SAAV residues 324–328 are shown inside their density map as in A. The movement of the arginine side chain (here shown without its density mesh) affects the base of the DE loop above with a slight shift of the main-chain and the movement of the L326 side-chain. (C and D) Shown are cross sections of two VP monomers inside their density maps at the 5-fold symmetry axis for pH 7.4 (C) and pH 6.0 (D). The positions of critical residues are indicated, as well as the distance of the leucine residues in the SAAV model at their narrowest point within the 5-fold channel. Panels A–D were generated using UCSF-Chimera (79). (E and F) Shown are interior surface representations based on the refined SAAV models at pH 7.4 (E) or pH 6.0 (F) conditions viewed down the 5-fold axis from the interior of the capsid. Residues with structural variability or alternative side-chain conformations compared to pH 7.4 are highlighted. Panels E and F were generated using PyMol (86).
In addition to these changes at the N-terminus and at the 5-fold channel, multiple basic residues were observed with different side chain orientations at the low pH conditions compared to pH 7.4. These residues include R288, K299, K304, and K627, which are conserved among the AAV serotype and correspond to R298, K309, K314, and K640 in AAV2. Mutation of these residues to alanine in AAV2 resulted in assembly defects except for K640A, which displayed a wild-type-like phenotype (64). Interestingly, all of these amino acids are located on the interior surface of the AAV capsid (Fig. 7E and F). K304 (Fig. 6A) is in close proximity to the ordered N-terminus at the low pH conditions and may change its orientation as a result. Another possibility could be that these basic residues are involved in the interaction with the packaged, negatively charged genome. The movements of the lysine or arginine thus may be an adaptation to prepare the capsid at low pH conditions to release the genome. None of these residues are near the known nucleotide binding pocket of the AAVs (18–20, 22, 65). This pocket is also conserved in SAAV but in the absence of a packaged genome of the VLPs did not show any ordered nucleotides. In a previous study, genome-containing AAV8 particles were analyzed at low pH conditions, which resulted in a loss of the ordered nucleotide compared to AAV8 particles at neutral pH conditions (65). However, none of the observed structural rearrangements of the N-terminus, including the widening of the 5-fold pore, were seen with either genome-containing AAV8 capsids or AAV9-VLPs at the same pH conditions (65, 66), indicating that the effects may be independent of the package genome and possibly specific to SAAV.
SAAV is not recognized by AAV-specific MAbs but by some human sera.
To test the antigenicity of the SAAV, a series of MAbs, detecting conformational epitopes on intact capsids of various AAV serotypes were probed against the SAAV capsid in a native dot blot assay. While the MAbs recognized the previously described AAV serotypes, ADK1a bound to AAV1 and 6 (67), A20 to AAV2 and AAV3 (68), ADK4 to AAV4 (67), ADK5b to AAV5 (67), ADK6 to AAV6 (69), ADK8 to AAV3 and AAV8 (69), and ADK9 to AAV9 (69), none of these antibodies recognized SAAV capsids (data not shown). The majority of these antibodies bind to or near the 3-fold protrusions or the 2/5-fold wall (70) that are formed by VR-I, -III, -IV, -V, -VI, VII, -VIII, and -IX, which displayed the largest structural and amino acid sequence variability of the SAAV capsid compared to AAV1, AAV2, and AAV5 (Fig. 2). Thus, the observed inability of the MAbs to bind to the SAAV capsid was not surprising.
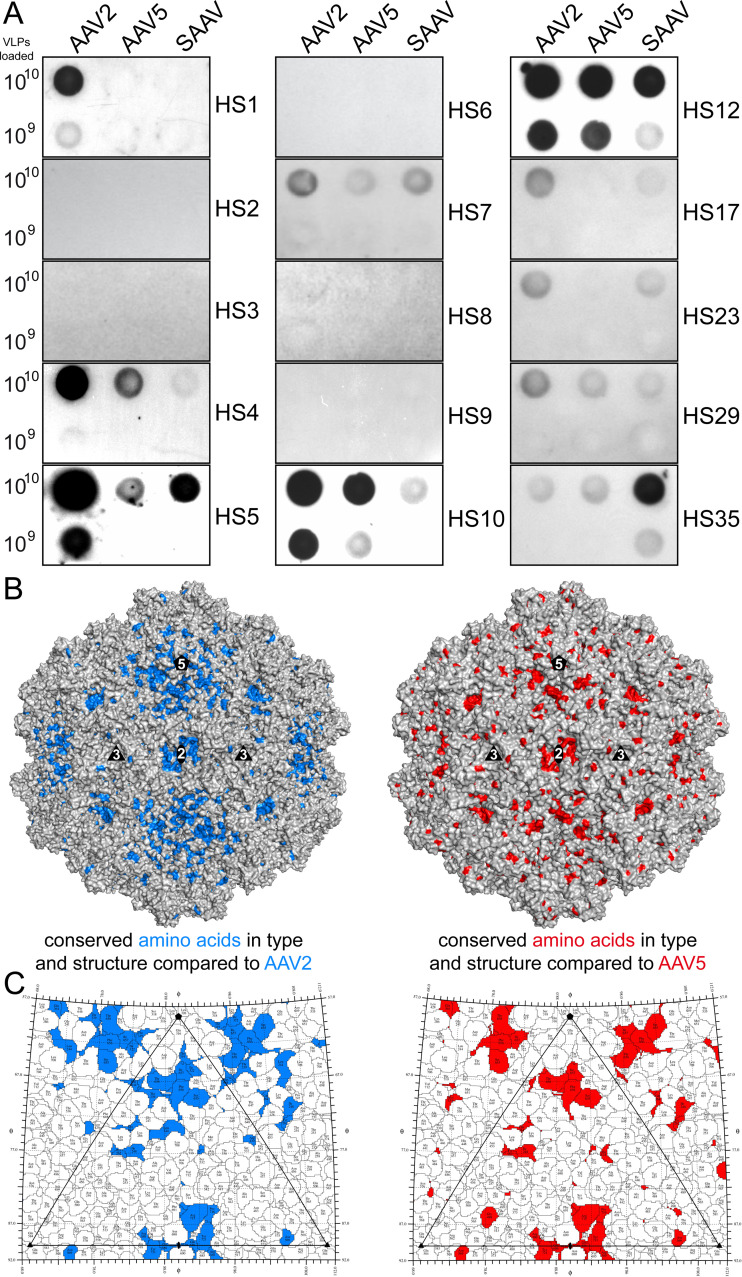
To further characterize the antigenicity, the SAAV capsids were probed with a panel of 40 different human serum (HS) samples alongside with AAV2 and AAV5. The vast majority (75%) of the analyzed human sera did not recognize any of the three AAV capsids. AAV2 capsids were detected by 25% (n = 10) of the human serum samples (Fig. 8A). This percentage is much lower compared to previous studies that reported seroprevalences of anti-AAV2 antibodies of up to ~70% (46, 71). While variations of seroprevalence of geographically different donor populations are possible, another factor is the lower detection limit of native dot blots compared to ELISAs used in other studies, and samples with low antibody titers against AAV capsids remain undetected. Compared to AAV2, SAAV is detected with 23% (n = 9) and AAV5 with 18% (n = 7) of the human serum samples (Fig. 8A). However, all sera reacting with SAAV or AAV5 also react with AAV2. This indicates the presence of potential cross-reactive antibodies, especially since the signal intensities toward SAAV are generally lower compared to AAV2 with the exception of HS35 (Fig. 8A). While the majority of the SAAV capsid surface is structurally different with many amino acid substitutions, the region around the 5-fold symmetry axis and in the 2-fold depression show some conservation compared to AAV2 and AAV5 (Fig. 8B and C). To date, no antibody capable of binding inside the 2-fold depression has been found for any of the AAVs or any other parvovirus (70), likely due to the size of an Fab. Thus, as the immune response is polyclonal targeting different regions of the capsids, it is possible that some antibodies generated against AAV2 capsids are also capable of binding to the SAAV and AAV5 capsids around the 5-fold region. The fact that the 2- and 5-fold region possess these conserved residues in the otherwise very different capsid surface of AAV2, AAV5, and SAAV may indicate common functions such as genome packaging, VP1u externalization, and possibly transcriptional activation (47, 72). Thus, changing these residues in any of these virus capsids may be detrimental to AAV infectivity for vector development.
FIG 8.
Dot immunoblot analysis of SAAV against human sera. (A) Representative native dot immunoblots of AAV2, AAV5, and SAAV against human sera with 1010 or 109 loaded VLPs. Native blots for human sera 1–10, and additional sera positive for SAAV, are shown. (B) Surface representation of the SAAV capsid, generated using PyMol (86), with conserved residues highlighted in blue (vs. AAV2) or red (vs. AAV5) that are structurally in equivalent positions (<1 Å distance) to AAV2 or AAV5, respectively. (C) Roadmap projections generated using RIVEM (87) of the icosahedral asymmetric unit highlighting the conserved surface amino acids as in B.
Conclusions.
With 55% amino acid sequence identity of the ordered VP structure compared to AAV2, SAAV has one of the most divergent capsids known to date in the genus Dependoparvovirus. Nonetheless, the core capsid topology is conserved, and structural variability is restricted to the capsid surface. Despite these differences, it binds to a common glycan receptor, sialic acid, that is also utilized by other AAV serotypes (AAV1, 4, 5, and 6), but at an alternative position of the capsid. With the intention to utilize SAAV as a potential alternative to existing mammalian therapeutic vectors, the thermostability and antigenicity of the SAAV capsid were evaluated. The SAAV capsid was found to be highly thermostable, comparable to AAV5, at a wide range of environmental pH conditions but with rearrangement at the VP N termini following the exposure to low pH conditions. This is consistent with the suggested dynamics of this region needed for the externalization of the enzymatic PLA2 domain located within VP1u during endosomal trafficking (36). Due to the significant sequence and structural differences on the capsid surface, it was expected that SAAV would be antigenically silent when encountering antibodies targeting primate AAVs, thus circumventing potential neutralization. Indeed, monoclonal antibodies developed against the different AAV serotypes were unable to bind to SAAV. However, despite its reptilian origin, SAAV is recognized by 23% of human sera, likely due to the presence of cross-reactive antibodies that might bind the 5-fold region of the capsid. While this could hamper the utilization of SAAV based vectors for future gene delivery applications, it could help to identify the cross-reactive epitopes of different AAVs to further optimize vectors with antibody escape properties.
MATERIALS AND METHODS
Cell culture.
Spodoptera frugiperda insect (Sf9) cells (ATCC CRL-1711) were grown in suspension culture using Sf-900 II Serum-Free Media (Thermo Fisher), supplemented with 1% Antibiotic-Antimycotic (100X, Thermo Fisher). Cells were grown at 28°C in a shaking incubator. Adherent cultures of the Chinese Hamster Ovary (CHO) cell line variants Pro5 (ATCC CRL-1781), Lec2 (ATCC CRL-1736), and Lec8 (ATCC CRL-1737) were grown in Minimum Essential Medium Alpha + Glutamax (GIBCO) supplemented with 10% Fetal Bovine Serum, and 1% Antibiotic-Antimycotic. Cells were incubated in a 37°C stationary incubator in 5% CO2.
Expression and purification of SAAV virus-like particles.
SAAV virus-like particles (VLPs) were produced using the baculovirus expression system in Sf9 cells. The SAAV capsid gene encoding VP3 was cloned into the pFastBac1 vector in order to produce a recombinant baculovirus via standard Bac-to-Bac protocols (Invitrogen) (73). For SAAV production the Sf9 cells were infected with the recombinant Baculovirus, and the infected cells harvested 3 days postinfection by pelleting at 2,000 rpm for 10 min at 4°C in a JA-10 rotor (Beckman). The medium supernatant was mixed with 10% wt/vol PEG8000 (poly-ethylene glycol 8000), stirred at 4°C overnight, and spun at 9,000 rpm in a JA-10 rotor for 90 min to generate the PEG pellet. The cell and PEG pellets were resuspended in 1× PBS supplemented with 2.5 mM KCl and 1 mM MgCl2 and subjected to three freeze/thaw cycles using liquid nitrogen and a 37°C water bath. After the last thaw step, benzonase (25 units per mL) was added to the sample and incubated for 1 h in a 37°C water bath. The lysate was clarified by centrifugation at 10,000 rpm for 15 min at 4°C in a JA-20 rotor (Beckman). Purification of the SAAV VLPs was carried out using a sucrose cushion and sucrose density gradient. For the sucrose cushion, 5 mL of 20% (wt/vol) sucrose in TNET (50 mM Tris HCl, 100 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, pH 8) was pipetted under the supernatant in an ultracentrifugation tube and spun at 45,000 rpm in a Ti70 rotor (Beckman) for 3 h at 4°C. The pelleted sample was manually resuspended in TNTM buffer (25 mM Tris HCl, 100 mM NaCl, 2 mM MgCl2, 0.2% Triton X-100, pH 8) and incubated overnight at 4°C for further resuspension. The sample was centrifuged at 10,000 rpm for 10 min at 4°C in a JA-20 rotor to remove insoluble material. The clarified supernatant was subjected to a 10–40% (wt/vol) step sucrose gradient with centrifugation at 35,000 rpm for 3 h at 4°C in an SW40 Ti rotor (Beckman) with slow acceleration and deceleration. Sample fractions were recovered using needle puncture, diluted in 20 mM Tris-HCl (pH 8.5), 15 mM NaCl, and further purified by anion-exchange chromatography using prepacked HiTrap Q HP column (GE Healthcare). After washing with 20 mM Tris-HCl (pH 8.5), 15 mM NaCl, 1 mL fractions were eluted with a linear gradient of 20 mM Tris-HCl (pH 8.5), 500 mM NaCl. Fractions containing SAAV VP3 were identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which were then dialyzed into 20 mM Tris-HCl, 300 mM NaCl, 2 mM MgCl2, pH 7.4 or universal buffer (20 mM HEPES, 20 mM morpholineethanesulfonic acid, 20 mM sodium acetate, 150 mM NaCl, 5 mM CaCl2) adjusted to pH 7.4, 6.0, 5.5, or 4.0, respectively. The concentration was determined based on UV absorbance of 280 nm with an extinction coefficient of 1.7 M−1 cm−1. The purified virus-like particles were concentrated to ~1 mg/mL using Apollo concentrators (Orbital Biosciences).
VLP Sample purity and integrity.
The purity and integrity of the sample were confirmed by SDS-PAGE and negative-stain electron microscopy (EM), respectively. For the SDS-PAGE analysis, the sample was incubated with 1× Laemmle Sample Buffer (Bio-Rad) with 10% vol/vol β-mercaptoethanol and boiled for 5 min at 100°C. The denatured proteins were applied to a 10% SDS-polyacrylamide gel and run at 80 V. After the run, the gel was washed three times with distilled water (diH2O) and stained with GelCode Blue Protein Safe stain (Invitrogen) for 1 h. The gel was destained with diH2O prior to imaging using a GelDoc EX system (Bio-Rad). For negative stain EM, 5 μL of the sample was incubated with glow-discharged CF400-CU carbon coated 400 mesh copper grids (Electron Microscopy Sciences) for 2 min and washed in three 15 μL droplets of water. Excess water was blotted with filter paper (Whatman), and the grid was stained with filtered 2% uranyl acetate. Excess stain was blotted, and grids were imaged using a Tecnai G2 Spirit Transmission Electron Microscope (FEI) operating at 120 kV.
Vitrification and cryo-electron microscopy data collection.
Three μL of the SAAV VLPs in their respective pH buffers or in complex with 3’SLN or 6’SLN (received from the Center for Functional Glycomics Reagent Bank) were applied to glow discharged C-flat holey carbon-coated grids (Protochips Inc) and vitrified using the Vitrobot Mark IV (FEI) automatic plunge-freezing system. The sample was incubated on the grids at 4°C and 95% humidity for 3.0 s prior to blotting using filter paper and plunging into an ethane slush, cooled with liquid nitrogen, for vitrification. The grids were maintained at liquid nitrogen temperatures until data collection. The particle distribution and ice quality of the grids were screened in-house using an FEI Tecnai G2 F20-TWIN microscope (FEI Co.) operated under low-dose conditions (200 kV, ~20e−/Å2). The data collection for high resolution cryo-EM was performed at the Florida State University (FSU) Biological Science Imaging Resource Core or the University of California, Los Angeles (UCLA) using the Titan Krios electron microscope. The microscopes were operated at 300 kV and data were collected on a DE-20 (pH 7.4) or K3 (pH 6.0, pH 5.5, pH 4.0, 3’SLN, and 6’SLN) (Gatan) direct electron detector camera. During data collection at FSU, a total dose of 60 to 62 e−/Å2 was utilized for 30 to 50 movie frames per micrograph. The Titan Krios electron microscope at UCLA was equipped with a Gatan Energy Filter (GIF), and a slit width of 20 eV was set for energy filter. Movies were recorded with the K3 camera operating in counting mode with dose rate of the electron beam set to 27 electrons per physical pixel per second on camera. An accumulated dose of 34e- per Å2 on the sample was fractionated into a movie stack of 30 image frames. The movie frames collected on the DE-20 detector were aligned using the DE_process_frames software package (Direct Electron) without dose weighting as previously described (74, 75). MotionCor2 was used for aligning the movie frames collected on the K3 detector with dose weighting (76). The pH 7.4, pH 6.0, and pH 4.0 data sets were collected as part of the NIH “Southeastern Center for Microscopy of MacroMolecular Machines (SECM4)” project. The pH 5.5 data set was collected as part of the National Institutes of Health (NIH) “West/Midwest Consortium (WMC) for High-Resolution Cryo Electron Microscopy” project.
Data processing and 3D image reconstruction.
For the three-dimensional image reconstruction of the SAAV data sets, the cisTEM software package was utilized (77). Briefly, the aligned micrographs were imported into the program and their contrast transfer function (CTF) parameters estimated. The CTF information was used to eliminate micrographs of poor quality. This was followed by automatic particle picking using a particle radius of 125 Å. This set of particles was subjected to 2D classification that eliminated non-VLP particles (ice and debris) from the automatic picking process. Following 2D classification, SAAV particles were reconstructed using default settings. This included the ab initio 3D model generation, auto refinement, and density map sharpening with a pre-cutoff B-factor value of -90 Å2, and variable post-cutoff B-factor values 0, 20, and 50 Å2. The sharpened density maps were inspected in the Coot and Chimera applications (78, 79). The −90 Å2/0 Å2 sharpened maps were used for assignment of the amino acid main- and side-chains. The resolutions of the cryo-reconstructed maps were estimated based on a Fourier Shell Correlation (FSC) of 0.143.
Model building.
The Swiss Model online server was used to generate an in silico model of the SAAV VP monomer based on the amino acid sequence (80). A 60mer of this model was generated using ViperDB (81) and docked into the cryo-EM density map with Chimera using the “Fit in Map” option (79). Voxel size was adjusted to maximize the correlation coefficient. The EMAN2 subroutine e2proc3d.py was implemented to resize maps based on best fit parameters as determined by correlation coefficients from Chimera (79, 82) and converted to the CCP4 format using MAPMAN (83). The main- and side-chains of the SAAV model were manually refined in Coot using the real-space refinement tool (78). The SAAV model was further automatically refined using PHENIX, which also provided the final refinement statistics (Table 1) (84).
Structural comparison.
For structural comparison, the SAAV VP model was superposed in Coot onto the previously determined VP structures of AAV1 (PDB ID 3NG9), AAV2 (PDB ID 1LP3), and AAV5 (PDB ID 3NTT) (14, 16, 23) to obtain the distances of the aligned Cα positions. Distances between nonoverlapping Cα positions, due to residue deletion/insertions, were measured using the distance tool in Coot. Regions of two or more adjacent amino acids with ≥2.0 Å difference in superposed VP Cα positions were considered to be structurally diverse and assigned to previously described VRs.
Fluorescent labeling of AAV VLPs.
SAAV, AAV1, AAV2, and AAV5 VLPs were fluorescently labeled using the DyLight 488 Antibody Labeling Kit (Thermo Fisher). Following a modified version of the manufacturer’s specifications, 40 μL of borate buffer (0.67 M, pH 8.5) was added to the VLPs at a concentration >0.5 mg/mL, and the entire volume transferred to the DyLight Reagent vial as per manufacturer’s suggestion. The samples were mixed and incubated, protected from light, for 1 h at RT. Following this, an additional 20 μL of borate buffer was added to the reaction and incubated for 30 min. Unbound fluorescent molecules were removed from the samples via three rounds of dialysis into 1× PBS. The success of the labeling procedure was confirmed by SDS-PAGE showing fluorescent VP bands when viewed under UV light.
Cell binding assay.
CHO variant cell lines Pro-5, Lec-2, and Lec-8 were passaged to 50% confluence into 15 cm plates 24 h prior to conducting the experiment. On the day of the cell binding assay, cells at 90–100% confluence were detached from the plate by addition of 2 mL 0.5 M EDTA to the medium of a 15-cm plate. Cells were transferred into a 50-mL conical tube and centrifuged in a benchtop centrifuge for 5 min at 500 rpm. The supernatant was removed, and the pellet was resuspended in 6 mL of prechilled unsupplemented MEM. The cells were counted under a microscope using a hemocytometer, diluted to 5 × 105 cells/mL, aliquoted to 500-μL fractions, and prechilled for 30 min at 4°C. Each tube of cells was then incubated with the fluorescently labeled VLPs at an MOI of 106 under constant rotation and protected from light for 3 to 4 h at 4°C. Following the incubation, the cells were pelleted at 2,000 rpm for 10 min in a benchtop centrifuge and the supernatant discarded. Unbound VLPs were removed by washing the cells with 300 μL prechilled 1× PBS followed by centrifugation. Pellets were resuspended in 300 μL 1× PBS and analyzed utilizing a FACS Calibur (BD Biosciences). All experiments were conducted in triplicate. The FSC Express5 software suite (BD Biosciences) was used to analyze the raw data.
SAAV capsid low pH characterization.
In order to simulate the conditions the SAAV capsid encounters during the endo-lysosomal trafficking, the SAAV, AAV1, AAV2, and AAV5 VLPs were dialyzed into universal buffer at pH 7.4, 6.0, 5.5, and 4.0. Their capsid stability was determined by differential scanning fluorimetry as described previously (61). Briefly, 22.5 μL of the VLPs from each pH condition were mixed with 2.5 μL SYPRO Orange dye (1% vol/vol; Life Technologies). Samples were loaded into a 96-well PCR plate in a Bio-Rad MyiQ2 Real-Time PCR instrument and were subjected to a temperature gradient (30°C–99°C) with an increase of 0.5°C per 30 s. The capsid melting temperature was calculated from a plot of fluorescence emission versus temperature, and statistical significance was calculated using standard deviation and standard error, with n = 3. The dialyzed SAAV capsids were also used for structural characterization by cryo-EM as described above.
Native immuno dot blot.
The cross-reactivity of SAAV capsid with monoclonal antibodies against conformational epitopes of AAV serotypes 1 to 9 was tested using native dot blots. Fifty μL of virus (50 ng/μL) was applied to a nitrocellulose membrane using a dot blot manifold, which allowed excess fluid to be drawn through the membrane by vacuum filtration. Blots were blocked overnight at 4°C in 10% wt/vol in 1× PBS + 0.05% Tween 20. The purified mouse IgG primary antibodies were diluted 1:500 for ADK1a, ADK4, and ADK8, 1:700 for ADK9, 1:1000 for A20 and ADK5b, and 1:3000 for ADK6 and B1 in 5% wt/vol milk in 0.05% Tween 20-PBS and incubated with the nitrocellulose membrane for 90 min. The membranes were subjected to 3 × 10 min washes in 0.05% Tween 20-PBS prior to secondary antibody incubation. Anti-mouse horseradish peroxidase-conjugated secondary antibody (GE Healthcare) was diluted 1:10,000 in sterile-filtered 1× PBS, and incubated with the membranes for 1 h at RT. The membranes were washed as described above prior to development in a 1:1 solution of peroxide solution and luminol (ThermoFisher) with the signals detected on X-ray film. To further determine the antigenicity of the SAAV capsids, 40 individual human sera from healthy donors (Valley Biomedical, Winchester, VA, USA) were tested against AAV2, AAV5, and SAAV VLPs, as described previously (85).
Data availability.
The SAAV cryo-EM reconstructed density maps and models built for the capsids at different pH conditions and in complex with sialic acids were deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/emdb/) with accession numbers EMD-26390 (Protein Data Bank [PDB] ID 7U94) (pH 7.4), EMD-26391 (PDB ID 7U95) (pH 6.0), EMD-26392 (PDB ID 7U96) (pH 5.5), EMD-26393 (PDB ID 7U97) (pH 4.0), EMD-26394 (3’SLN), and EMD-26395 (6’SLN).
ACKNOWLEDGMENTS
We thank the late Mavis Agbandje-McKenna for initiating this research project. The study was funded by the NIH grant R01 NIH GM082946 (to M.A.-M. and R.M.). The authors also thank the UF-ICBR Electron microscopy core for access to electron microscopes utilized for cryo-electron micrograph screening. In addition, the authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057, 1S10OD018111, and 1U24GM116792), NSF (DBI-1338135), and CNSI at UCLA. Data collection at Florida State University through the Southeastern Consortium for Microscopy of MacroMolecular Machines (SECM4) was made possible by NIH grants S10OD018142-01, S10RR025080-01, and U24GM116788.
Contributor Information
Robert McKenna, Email: rmckenna@ufl.edu.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Cotmore SF, Agbandje-McKenna M, Canuti M, Chiorini JA, Eis-Hubinger AM, Hughes J, Mietzsch M, Modha S, Ogliastro M, Penzes JJ, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, ICTV Report Consortium . 2019. ICTV virus taxonomy profile: Parvoviridae. J Gen Virol 100:367–368. 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantel-Schaal U, Zur Hausen H. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52–63. 10.1016/0042-6822(84)90271-x. [DOI] [PubMed] [Google Scholar]
- 3.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 78:6381–6388. 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt M, Katano H, Bossis I, Chiorini JA. 2004. Cloning and characterization of a bovine adeno-associated virus. J Virol 78:6509–6516. 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbetman AE, Lochrie M, Zhou S, Wellman J, Scallan C, Doroudchi MM, Randlev B, Patarroyo-White S, Liu T, Smith P, Lehmkuhl H, Hobbs LA, Pierce GF, Colosi P. 2005. Novel caprine adeno-associated virus (AAV) capsid (AAV-Go.1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J Virol 79:15238–15245. 10.1128/JVI.79.24.15238-15245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bello A, Chand A, Aviles J, Soule G, Auricchio A, Kobinger GP. 2014. Novel adeno-associated viruses derived from pig tissues transduce most major organs in mice. Sci Rep 4:6644. 10.1038/srep06644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Shan T, Wang C, Cote C, Kolman J, Onions D, Gulland FM, Delwart E. 2011. The fecal viral flora of California sea lions. J Virol 85:9909–9917. 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodewes R, van der Giessen J, Haagmans BL, Osterhaus AD, Smits SL. 2013. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J Virol 87:7758–7764. 10.1128/JVI.00568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lochrie MA, Tatsuno GP, Arbetman AE, Jones K, Pater C, Smith PH, McDonnell JW, Zhou SZ, Kachi S, Kachi M, Campochiaro PA, Pierce GF, Colosi P. 2006. Adeno-associated virus (AAV) capsid genes isolated from rat and mouse liver genomic DNA define two new AAV species distantly related to AAV-5. Virology 353:68–82. 10.1016/j.virol.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Li J, Liu Y, Shi Z, Liu H, Wei Y, Yang L. 2019. Bat adeno-associated viruses as gene therapy vectors with the potential to evade human neutralizing antibodies. Gene Ther 26:264–276. 10.1038/s41434-019-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossis I, Chiorini JA. 2003. Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J Virol 77:6799–6810. 10.1128/jvi.77.12.6799-6810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farkas SL, Zádori Z, Benkő M, Essbauer S, Harrach B, Tijssen P. 2004. A parvovirus isolated from royal python (Python regius) is a member of the genus Dependovirus. J Gen Virol 85:555–561. 10.1099/vir.0.19616-0. [DOI] [PubMed] [Google Scholar]
- 13.Penzes JJ, Pham HT, Benko M, Tijssen P. 2015. Novel parvoviruses in reptiles and genome sequence of a lizard parvovirus shed light on Dependoparvovirus genus evolution. J Gen Virol 96:2769–2779. 10.1099/vir.0.000215. [DOI] [PubMed] [Google Scholar]
- 14.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci USA 99:10405–10410. 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lerch TF, Xie Q, Chapman MS. 2010. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology 403:26–36. 10.1016/j.virol.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govindasamy L, Dimattia MA, Gurda BL, Halder S, McKenna R, Chiorini JA, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. 2013. Structural insights into adeno-associated virus serotype 5. J Virol 87:11187–11199. 10.1128/JVI.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, Agbandje-McKenna M. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol 80:11556–11570. 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, Aslanidi G, Byrne B, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. 2007. Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol 81:12260–12271. 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halder S, Van Vliet K, Smith JK, Duong TT, McKenna R, Wilson JM, Agbandje-McKenna M. 2015. Structure of neurotropic adeno-associated virus AAVrh.8. J Struct Biol 192:21–36. 10.1016/j.jsb.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikals K, Nam HJ, Van Vliet K, Vandenberghe LH, Mays LE, McKenna R, Wilson JM, Agbandje-McKenna M. 2014. The structure of AAVrh32.33, a novel gene delivery vector. J Struct Biol 186:308–317. 10.1016/j.jsb.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burg M, Rosebrough C, Drouin LM, Bennett A, Mietzsch M, Chipman P, McKenna R, Sousa D, Potter M, Byrne B, Jude Samulski R, Agbandje-McKenna M. 2018. Atomic structure of a rationally engineered gene delivery vector, AAV2.5. J Struct Biol 203:236–241. 10.1016/j.jsb.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mietzsch M, Barnes C, Hull JA, Chipman P, Xie J, Bhattacharya N, Sousa D, McKenna R, Gao G, Agbandje-McKenna M. 2020. Comparative analysis of the capsid structures of AAVrh.10, AAVrh.39, and AAV8. J Virol 94:e01769-19. 10.1128/JVI.01769-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. 2010. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol 84:12945–12957. 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drouin LM, Lins B, Janssen M, Bennett A, Chipman P, McKenna R, Chen W, Muzyczka N, Cardone G, Baker TS, Agbandje-McKenna M. 2016. Cryo-electron microscopy reconstruction and stability studies of the wild type and the R432A variant of adeno-associated virus type 2 reveal that capsid structural stability is a major factor in genome packaging. J Virol 90:8542–8551. 10.1128/JVI.00575-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan YZ, Aiyer S, Mietzsch M, Hull JA, McKenna R, Grieger J, Samulski RJ, Baker TS, Agbandje-McKenna M, Lyumkis D. 2018. Sub-2 Å Ewald curvature corrected structure of an AAV2 capsid variant. Nat Commun 9:3628. 10.1038/s41467-018-06076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett A, Keravala A, Makal V, Kurian J, Belbellaa B, Aeran R, Tseng YS, Sousa D, Spear J, Gasmi M, Agbandje-McKenna M. 2019. Structure comparison of the chimeric AAV2.7m8 vector with parental AAV2. J Struct Biol 209:107433. 10.1016/j.jsb.2019.107433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiMattia MA, Nam HJ, Van Vliet K, Mitchell M, Bennett A, Gurda BL, McKenna R, Olson NH, Sinkovits RS, Potter M, Byrne BJ, Aslanidi G, Zolotukhin S, Muzyczka N, Baker TS, Agbandje-McKenna M. 2012. Structural insight into the unique properties of adeno-associated virus serotype 9. J Virol 86:6947–6958. 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaelber JT, Yost SA, Webber KA, Firlar E, Liu Y, Danos O, Mercer AC. 2020. Structure of the AAVhu.37 capsid by cryoelectron microscopy. Acta Crystallogr F Struct Biol Commun 76:58–64. 10.1107/S2053230X20000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther CM, Brun MJ, Bennett AD, Ho ML, Chen W, Zhu B, Lam M, Yamagami M, Kwon S, Bhattacharya N, Sousa D, Evans AC, Voss J, Sevick-Muraca EM, Agbandje-McKenna M, Suh J. 2019. Protease-activatable adeno-associated virus vector for gene delivery to damaged heart tissue. Mol Ther 27:611–622. 10.1016/j.ymthe.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mietzsch M, Li Y, Kurian J, Smith JK, Chipman P, McKenna R, Yang L, Agbandje-McKenna M. 2020. Structural characterization of a bat Adeno-associated virus capsid. J Struct Biol 211:107547. 10.1016/j.jsb.2020.107547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol Biol 807:47–92. 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 32.Mietzsch M, Penzes JJ, Agbandje-McKenna M. 2019. Twenty-five years of structural parvovirology. Viruses 11:362. 10.3390/v11040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grieger JC, Snowdy S, Samulski RJ. 2006. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J Virol 80:5199–5210. 10.1128/JVI.02723-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol 83:973–978. 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 35.Kronenberg S, Bottcher B, von der Lieth CW, Bleker S, Kleinschmidt JA. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J Virol 79:5296–5303. 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatakrishnan B, Yarbrough J, Domsic J, Bennett A, Bothner B, Kozyreva OG, Samulski RJ, Muzyczka N, McKenna R, Agbandje-McKenna M. 2013. Structure and dynamics of adeno-associated virus serotype 1 VP1-unique N-terminal domain and its role in capsid trafficking. J Virol 87:4974–4984. 10.1128/JVI.02524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Large EE, Silveria MA, Zane GM, Weerakoon O, Chapman MS. 2021. Adeno-associated virus (AAV) gene delivery: dissecting molecular interactions upon cell entry. Viruses 13:1336. 10.3390/v13071336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asokan A, Conway JC, Phillips JL, Li C, Hegge J, Sinnott R, Yadav S, DiPrimio N, Nam HJ, Agbandje-McKenna M, McPhee S, Wolff J, Samulski RJ. 2010. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol 28:79–82. 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulicherla N, Shen S, Yadav S, Debbink K, Govindasamy L, Agbandje-McKenna M, Asokan A. 2011. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol Ther 19:1070–1078. 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raupp C, Naumer M, Muller OJ, Gurda BL, Agbandje-McKenna M, Kleinschmidt JA. 2012. The threefold protrusions of adeno-associated virus type 8 are involved in cell surface targeting as well as postattachment processing. J Virol 86:9396–9408. 10.1128/JVI.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett AD, Wong K, Lewis J, Tseng YS, Smith JK, Chipman P, McKenna R, Samulski RJ, Kleinschmidt J, Agbandje-McKenna M. 2018. AAV6 K531 serves a dual function in selective receptor and antibody ADK6 recognition. Virology 518:369–376. 10.1016/j.virol.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurda BL, DiMattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen WJ, Muzyczka N, Olson NH, Sinkovits RS, Chiorini JA, Zolotutkhin S, Kozyreva OG, Samulski RJ, Baker TS, Parrish CR, Agbandje-McKenna M. 2013. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol 87:9111–9124. 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jose A, Mietzsch M, Smith JK, Kurian J, Chipman P, McKenna R, Chiorini J, Agbandje-McKenna M. 2019. High-resolution structural characterization of a new adeno-associated virus serotype 5 antibody epitope toward engineering antibody-resistant recombinant gene delivery vectors. J Virol 93. 10.1128/JVI.01394-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng YS, Gurda BL, Chipman P, McKenna R, Afione S, Chiorini JA, Muzyczka N, Olson NH, Baker TS, Kleinschmidt J, Agbandje-McKenna M. 2015. Adeno-associated virus serotype 1 (AAV1)- and AAV5-antibody complex structures reveal evolutionary commonalities in parvovirus antigenic reactivity. J Virol 89:1794–1808. 10.1128/JVI.02710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D, Tai PWL, Gao G. 2019. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov 18:358–378. 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. 2010. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21:704–712. 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 47.Bleker S, Sonntag F, Kleinschmidt JA. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol 79:2528–2540. 10.1128/JVI.79.4.2528-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang LY, Halder S, Agbandje-McKenna M. 2014. Parvovirus glycan interactions. Curr Opin Virol 7:108–118. 10.1016/j.coviro.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mietzsch M, Broecker F, Reinhardt A, Seeberger PH, Heilbronn R. 2014. Differential adeno-associated virus serotype-specific interaction patterns with synthetic heparins and other glycans. J Virol 88:2991–3003. 10.1128/JVI.03371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. 2006. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol 80:9093–9103. 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol 72:1438–1445. 10.1128/JVI.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K, Agbandje-McKenna M, Wilson JM. 2011. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest 121:2427–2435. 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol 75:6884–6893. 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.North SJ, Huang HH, Sundaram S, Jang-Lee J, Etienne AT, Trollope A, Chalabi S, Dell A, Stanley P, Haslam SM. 2010. Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J Biol Chem 285:5759–5775. 10.1074/jbc.M109.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang LY, Patel A, Ng R, Miller EB, Halder S, McKenna R, Asokan A, Agbandje-McKenna M. 2016. Characterization of the adeno-associated virus 1 and 6 sialic acid binding site. J Virol 90:5219–5230. 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afione S, DiMattia MA, Halder S, Di Pasquale G, Agbandje-McKenna M, Chiorini JA. 2015. Identification and mutagenesis of the adeno-associated virus 5 sialic acid binding region. J Virol 89:1660–1672. 10.1128/JVI.02503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen S, Troupes AN, Pulicherla N, Asokan A. 2013. Multiple roles for sialylated glycans in determining the cardiopulmonary tropism of adeno-associated virus 4. J Virol 87:13206–13213. 10.1128/JVI.02109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Bueno A, Rubio MP, Bryant N, McKenna R, Agbandje-McKenna M, Almendral JM. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J Virol 80:1563–1573. 10.1128/JVI.80.3.1563-1573.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mietzsch M, McKenna R, Vaisanen E, Yu JC, Ilyas M, Hull JA, Kurian J, Smith JK, Chipman P, Lasanajak Y, Smith D, Soderlund-Venermo M, Agbandje-McKenna M. 2020. Structural characterization of cuta- and tusavirus: insight into protoparvoviruses capsid morphology. Viruses 12:653. 10.3390/v12060653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lins-Austin B, Patel S, Mietzsch M, Brooke D, Bennett A, Venkatakrishnan B, Van Vliet K, Smith AN, Long JR, McKenna R, Potter M, Byrne B, Boye SL, Bothner B, Heilbronn R, Agbandje-McKenna M. 2020. Adeno-associated virus (AAV) capsid stability and liposome remodeling during endo/lysosomal pH trafficking. Viruses 12:668. 10.3390/v12060668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett A, Patel S, Mietzsch M, Jose A, Lins-Austin B, Yu JC, Bothner B, McKenna R, Agbandje-McKenna M. 2017. Thermal stability as a determinant of AAV serotype identity. Mol Ther Methods Clin Dev 6:171–182. 10.1016/j.omtm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bleker S, Pawlita M, Kleinschmidt JA. 2006. Impact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. J Virol 80:810–820. 10.1128/JVI.80.2.810-820.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian S, Organtini LJ, Grossman A, Domeier PP, Cifuente JO, Makhov AM, Conway JF, D'Abramo A, Jr, Cotmore SF, Tattersall P, Hafenstein S. 2017. Cryo-EM maps reveal five-fold channel structures and their modification by gatekeeper mutations in the parvovirus minute virus of mice (MVM) capsid. Virology 510:216–223. 10.1016/j.virol.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu P, Xiao W, Conlon T, Hughes J, Agbandje-McKenna M, Ferkol T, Flotte T, Muzyczka N. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J Virol 74:8635–8647. 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nam HJ, Gurda BL, McKenna R, Potter M, Byrne B, Salganik M, Muzyczka N, Agbandje-McKenna M. 2011. Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking. J Virol 85:11791–11799. 10.1128/JVI.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Penzes JJ, Chipman P, Bhattacharya N, Zeher A, Huang R, McKenna R, Agbandje-McKenna M. 2021. Adeno-associated virus 9 structural rearrangements induced by endosomal trafficking pH and glycan attachment. J Virol 95:e0084321. 10.1128/JVI.00843-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuck D, Kern A, Kleinschmidt JA. 2007. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J Virol Methods 140:17–24. 10.1016/j.jviromet.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 68.Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74:9281–9293. 10.1128/jvi.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sonntag F, Kother K, Schmidt K, Weghofer M, Raupp C, Nieto K, Kuck A, Gerlach B, Bottcher B, Muller OJ, Lux K, Horer M, Kleinschmidt JA. 2011. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol 85:12686–12697. 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emmanuel SN, Mietzsch M, Tseng YS, Smith JK, Agbandje-McKenna M. 2021. Parvovirus capsid-antibody complex structures reveal conservation of antigenic epitopes across the family. Viral Immunol 34:3–17. 10.1089/vim.2020.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. 2009. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199:381–390. 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aydemir F, Salganik M, Resztak J, Singh J, Bennett A, Agbandje-McKenna M, Muzyczka N. 2016. Mutants at the 2-fold interface of adeno-associated virus type 2 (AAV2) structural proteins suggest a role in viral transcription for AAV capsids. J Virol 90:7196–7204. 10.1128/JVI.00493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berger I, Poterszman A. 2015. Baculovirus expression: old dog, new tricks. Bioengineered 6:316–322. 10.1080/21655979.2015.1104433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mietzsch M, Kailasan S, Garrison J, Ilyas M, Chipman P, Kantola K, Janssen ME, Spear J, Sousa D, McKenna R, Brown K, Soderlund-Venermo M, Baker T, Agbandje-McKenna M. 2017. Structural insights into human bocaparvoviruses. J Virol 91:e00261-17. 10.1128/JVI.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spear JM, Noble AJ, Xie Q, Sousa DR, Chapman MS, Stagg SM. 2015. The influence of frame alignment with dose compensation on the quality of single particle reconstructions. J Struct Biol 192:196–203. 10.1016/j.jsb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant T, Rohou A, Grigorieff N. 2018. cisTEM, user-friendly software for single-particle image processing. Elife 7:e35383. 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 79.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 80.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–8. 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho PT, Montiel-Garcia DJ, Wong JJ, Carrillo-Tripp M, Brooks CL, 3rd, Johnson JE, Reddy VS. 2018. VIPERdb: a tool for virus research. Annu Rev Virol 5:477–488. 10.1146/annurev-virology-092917-043405. [DOI] [PubMed] [Google Scholar]
- 82.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 83.Kleywegt GJ, Jones TA. 1996. xdlMAPMAN and xdlDATAMAN—programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr 52:826–828. 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 84.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu JC, Mietzsch M, Singh A, Jimenez Ybargollin A, Kailasan S, Chipman P, Bhattacharya N, Fakhiri J, Grimm D, Kapoor A, Kučinskaitė-Kodzė I, Žvirblienė A, Söderlund-Venermo M, McKenna R, Agbandje-McKenna M. 2021. Characterization of the GBoV1 capsid and its antibody interactions. Viruses 13:330. 10.3390/v13020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Lano WL. 2002. The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA. [Google Scholar]
- 87.Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J Struct Biol 158:182–187. 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The SAAV cryo-EM reconstructed density maps and models built for the capsids at different pH conditions and in complex with sialic acids were deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/emdb/) with accession numbers EMD-26390 (Protein Data Bank [PDB] ID 7U94) (pH 7.4), EMD-26391 (PDB ID 7U95) (pH 6.0), EMD-26392 (PDB ID 7U96) (pH 5.5), EMD-26393 (PDB ID 7U97) (pH 4.0), EMD-26394 (3’SLN), and EMD-26395 (6’SLN).