Abstract
Macrophages are white blood cells that play disparate roles in homeostasis and immune responses. They can reprogram their phenotypes to pro-inflammatory (M1) or anti-inflammatory (M2) states in response to their environment. About 8–15% of the macrophage transcriptome has circadian oscillations, including genes closely related to their functioning. As circadian rhythms are associated with cellular phenotypes, we hypothesized that polarization of macrophages to opposing subtypes might differently affect their circadian rhythms. We tracked circadian rhythms in RAW 264.7 macrophages using luminescent reporters. Cells were stably transfected with Bmal1:luc and Per2:luc reporters, representing positive and negative components of the molecular clock. Strength of rhythmicity, periods and amplitudes of time series were assessed using multiple approaches. M1 polarization decreased amplitudes and rhythmicities of Bmal1:luc and Per2:luc, but did not significantly affect periods, while M2 polarization increased periods but caused no substantial alterations to amplitudes or rhythmicity. As macrophage phenotypes are also altered in the presence of cancer cells, we tested circadian effects of conditioned media from mouse breast cancer cells. Media from highly aggressive 4T1 cells caused loss of rhythmicity, while media from less aggressive EMT6 cells yielded no changes. As macrophages play roles in tumors, and oncogenic features are associated with circadian rhythms, we tested whether conditioned media from macrophages could alter circadian rhythms of cancer cells. Conditioned media from RAW 264.7 cells resulted in lower rhythmicities and periods, but higher amplitudes in human osteosarcoma, U2OS-Per2:luc cells. We show that phenotypic changes in macrophages result in altered circadian characteristics and suggest that there is an association between circadian rhythms and macrophage polarization state. Additionally, our data demonstrate that macrophages treated with breast cancer-conditioned media have circadian phenotypes similar to those of the M1 subtype, and cancer cells treated with macrophage-conditioned media have circadian alterations, providing insight to another level of cross-talk between macrophages and cancer.
Keywords: macrophages, circadian rhythms, rhythmicity, cancer
Insight Box Text
Macrophages are cells of the innate immune system that play roles in homeostasis and immune responses. The circadian clock, a biological timing system that regulates functions across a 24-h cycle, has been implicated in many aspects of macrophage characteristics. Here, we combine the use of reporters for core circadian clock activities with cell biology assays and thorough computational analyses of results. Together, these elements facilitate detailed assessments of macrophage circadian rhythms and changes that occur following exposure to different stimuli. We find that macrophage oscillations are altered in dissimilar ways, and effects on different circadian components are not always comparable.
INTRODUCTION
The immune system is a network of cells, organs, and biomolecular entities that protects organisms against foreign pathogens. Multiple immune-related activities are regulated in a circadian manner, meaning that they occur with an approximately 24 h cycle. These include natural killer (NK) cell cytokine production and immunity against pathogens [1], numbers of particular white blood cell types (e.g. hematopoietic stem cells, B cells and T cells) in the blood [2] and/or lymph nodes [3], migration of hematopoietic cells, neutrophils and monocytes to tissues [2], phagocytic activity of neutrophils [4] and expression of cytolytic factors by NK cells [1, 5]. As a result, circadian time can determine immune responses to stimuli. For example, sensitivity to lipopolysaccharide (LPS, an endotoxin found in outer membranes of gram negative bacteria) [6] and inflammatory responses against Salmonella enterica serovar Typhimurium [7] have been found to be circadian, with increased responses at the start of the active phase and early rest period of mice, respectively. Mice infected with Leishmania [8] or Trichuris muris [9] pathogens in the morning showed lower parasite loads relative to those infected in the evening. Mouse models of peritoneal inflammation show more recruitment of inflammatory monocytes to the peritoneal cavity, spleen and liver in the afternoon versus the morning [10], while in mouse models of myocardial infarction, neutrophil recruitment to cardiac tissue peaks in the evening [11]. In humans, which are diurnal (as opposed to mice, which are nocturnal), immune responses can also vary in a circadian manner. Allergic symptoms are increased between midnight and the early morning [11, 12]; influenza vaccinations in the morning result in greater antibody concentrations compared to those administered in the afternoon [13], and asthma worsened lung function and resulted in increased sputnum leukocytes and eosinophils in the early morning, compared to in the late afternoon [14].
Macrophages are phagocytic white blood cells involved not only in the innate immune response, but also in organismal development, homeostasis and tissue repair [15]. Macrophages shift their functional state to respond to physiological challenges or environmental factors. Specific characteristics and roles are associated with their polarization to subtypes broadly classified as classically (M1) or alternatively (M2) activated, which are inflammatory or immune-suppressing, respectively [15, 16]. M2 macrophages may be further categorized into M2a, M2b, M2c and M2d subtypes, which arise from activation by different cytokines and have different responses. M2a macrophages are activated by IL-4 or IL-13 and lead to increased expression of IL-10, TGF-β, CCL17, CCL18 and CCL22; they possess endocytic activity and promote cell growth and tissue repair [17]. Because we are using IL-4 to polarize macrophages, exclusively resulting in the M2a subtype, we refer to these cells as being M2. Macrophages have been shown to possess their own intrinsic clocks that operate autonomously [18], with a functional core molecular circadian clock (comprised of a negative feedback loop with BMAL1 and CLOCK proteins as activators, and PER and CRY proteins as repressors) [19]. Approximately 8–15% of the macrophage transcriptome has circadian oscillations, including genes required for pathogen recognition and responses [18, 20]. Furthermore, multiple macrophage functions are circadian dependent, such as recruitment to infected tissue [12], generation of chemokines and cytokines, and phagocytosis [21, 22]. Considering that macrophages have highly divergent functions, from tissue remodeling [23] and angiogenesis [24] to generation of reactive oxygen species (ROS) [25] and phagocytosis [21, 22], it is important to address how circadian oscillations might change concomitantly with phenotype.
Previous studies have indicated that there is a link between macrophage activation and circadian rhythms and/or molecular clock elements. These evaluated the impacts of LPS (which is pro-inflammatory) and other cytokines on circadian clocks in primary, peritoneal macrophages [26] and bone marrow-derived macrophages (BMDMs) [27] derived from mPER2:luc mice, finding that immune-activating treatments affected circadian period, phase and amplitude. Curtis et al. also observed LPS-mediated effects, and assessed additional characteristics in BMDMs and peritoneal macrophages, and isolated human macrophages, at two individual time points. In this instance, Per2 and Bmal1 were evaluated; while Per2 mRNA levels and protein expression were significantly increased, those of Bmal1 decreased [28]. In separate work, downregulation of Bmal1 in mouse BMDMs increased the expression of proinflammatory cytokines, further suggesting that Bmal1 may act as an anti-inflammatory molecule in macrophages [29].
Associated with Bmal1, Curtis et al. also found that LPS significantly decreased levels of its transcriptional reporessor, REV-ERBα, but did not affect the presence of its transcriptional activator, RORα [28]. However, another study showed that stimulation with a combination of LPS and IFN-γ, which polarizes macrophages to the pro-inflammatory M1 subtype, increased REV-ERBα mRNA and protein levels in differentiated THP-1 macrophages. Concurrently, treatment with IL-4, which polarizes macrophages to the anti-inflammatory M2 subtype, resulted in REV-ERBα reduction [30]. These studies suggest that pro- and anti-inflammatory polarizations can differently affect circadian clock gene expression. However, the implications of macrophage stimulation on circadian oscillations of the positive and negative arms of the core clock are largely unknown, and have not been assessed in significant detail. Because the circadian clock is a dynamic system, it requires continuous monitoring to obtain sufficiently high resolution data for the accurate characterization of its patterns. For this reason, we have applied luciferase reporters for real-time tracking of circadian oscillations in macrophages and thorough analyses of these data to study the relationship between macrophage activation and the biological clock.
On account of their distinct and opposing functions, and the results of prior studies, we hypothesized that polarization of macrophages to pro- and anti-inflammatory subtypes would affect their circadian rhythms differently. To this end, we generated stable reporter cell lines assessing positive (Bmal1) and negative (Per2) components of the core circadian clock, using RAW 264.7 macrophages. We tracked their oscillations via real-time luminometry following polarization to M1 and M2 subtypes using standard cytokine treatments (M1 via LPS or a combination of LPS/IFN-γ, M2 via IL-4). Polarization states of macrophages were verified by evaluating levels of M1- and M2-associated markers via RT-PCR. M1 polarization resulted in decreased amplitudes and rhythmicities of both Bmal1:luc and Per2:luc reporters, but did not significantly affect periods. On the other hand, M2 polarization resulted in increased periods, but not significant alterations to amplitudes or rhythmicity. It is also important to note that responses of Bmal1:luc and Per2:luc were not always similar. While the mechanisms for this outcome are unknown, it has been previously observed in macrophages and other cellular models [28, 31–34].
Because macrophages play major roles in cancers, which can also affect their phenotypes, we assessed the effects of cancer cell-conditioned media on macrophage oscillations. Exposure to media from 4T1 cells, generally considered highly aggressive, resulted in significant loss of rhythmicity, while media from EMT6 (less aggressive) cells yielded no detectable changes. Finally, considering the impacts of macrophages on cancers, including in terms of aggression, we sought to determine whether macrophages could affect circadian rhythms of cancer cells. Following treatment with RAW 264.7-conditioned media, human osteosarcoma cells (U2OS-Per2:luc) showed reduced circadian rhythmicity and period, and enhanced circadian amplitudes. Taken together, our results suggest that macrophage polarization, and therefore function, is linked to circadian oscillations, and changes therein, and put forth that it is important to assess these oscillations with sufficient attention to detail to understand the changes occurring.
MATERIALS AND METHODS
Cell culture
RAW 264.7 murine macrophage, and 4T1 and EMT6 murine mammary carcinoma cells were obtained from the ATCC; 293 T human embryonic kidney cells were obtained from the Jerry Laboratory (Dept. of Veterinary and Animal Sciences, UMass Amherst); U2OS human osteosarcoma cells were obtained from the Wadsworth laboratory (Dept. of Biology, UMass Amherst). RAW 264.7, EMT6 and 4T1 cells were maintained in high glucose DMEM (Gibco), supplemented with 10% fetal bovine serum (FBS; Corning), 1% penicillin–streptomycin (Gibco) and 1% l-glutamine (Gibco). 293 T cells were maintained in DMEM/F12 media (Gibco), supplemented with 10% FBS, 1% penicillin–streptomycin and 0.015 mg/ml gentamicin (Gibco). U2OS cells were maintained in DMEM, supplemented with 10% FBS, 1% penicillin–streptomycin, 1% l-glutamine, 1% non-essential amino acids (NEAA; HyClone) and 1% sodium pyruvate (Gibco). Experiment-specific media preparations are indicated below. All cells were incubated at 37°C under 5% CO2 atmosphere, unless otherwise noted.
Lentiviral transfections
The generation of Bmal1:luc and Per2:luc plasmids have been described previously [35], as has their transfection into U2OS cells [31]. Subsequent stable transfections of these reporters into RAW 264.7 cells were performed similarly to as in other studies [31, 36]. Briefly, 3 ×106 293 T cells were seeded in 60-mm culture dishes and transiently transfected with 3 μg psPAX2 packaging plasmid, 2 μg pMD2.G envelope plasmid and 5 μg Bmal1:luc or Per2:luc plasmid constructs using Lipofectamine3000 (Thermo Fisher Scientific). After 48 h incubation, lentiviral particles were harvested and filtered through a 0.45-μm membrane (Thermo Fisher Scientific). About 9 ml lentivirus-containing supernatant was combined with 9 ml RAW 264.7 growth media containing 10 μg/ml polybrene (Sigma). Cells were seeded in T25 culture flasks at 2 × 105 cells/ml and incubated under standard conditions until 70–80% confluence was reached. At this stage, culture media were removed and 6 ml lentivirus-containing media were added. After 2 days of infection, media were replaced with selection media (DMEM with all growth supplements plus 4 μg/ml puromycin (Gibco)). The selection media were changed once every 3 days for 4 weeks.
Cell synchronization
For RAW 264.7 cell synchronization, cells were seeded in 35-mm culture dishes at 2 × 105 cells/ml and incubated for approximately 24 h. Media were then aspirated, cells washed with phosphate-buffered saline (PBS; Gibco) and synchronized by subjecting to starvation conditions by adding starvation media (L-15 media (Gibco) with 1% penicillin–streptomycin and 1% l-glutamine) and incubating for 18 h. Starvation media were removed and replaced with L-15 media containing only 100 nM dexamethasone (Sigma-Aldrich) for 2 h. Both starvation and synchronization were carried out at 37°C under 5% CO2 atmosphere. For U2OS cell synchronization, cells were seeded in 35 mm culture dishes at 2 × 105 cells/ml and incubated for approximately 24 h. Media were then aspirated and replaced with U2OS culture media containing 100 nM dexamethasone, followed by incubation for 2 h at 37°C under 5% CO2 atmosphere.
Cell treatments with cytokines and conditioned media
Lipopolysaccharide (LPS; Sigma-Aldrich), IFN-γ (BD Biosciences) and IL-4 (BioLegend) were prepared in PBS at concentrations of 1000, 100 or 200 and 100 μg/ml, respectively, and stored at −20°C as single-use aliquots. For cell treatments, cytokines were serially diluted in PBS to achieve the desired concentration, maintaining a final PBS concentration of 0.2% in cultures. Cytokines in PBS were added to bioluminescence recording media (for luminometry) or the equivalent lacking luciferin (for RT-PCR assays). For LPS-only treatments, concentrations are as indicated; for LPS/IFN-γ, 5 ng/ml of LPS and 12 ng/ml of IFN-γ were used; for IL-4 50 ng/ml was used. The RAW 264.7 recording media were prepared by dissolving powdered DMEM (Sigma-Aldrich) in Millipore-purified water (18.2 Ω resistance) to give a final concentration of 11.25 mg/ml, followed by sterile filtration using a 0.2-μm filter (Thermo Fisher Scientific), supplemented with 5% FBS, 1% HEPES (HyClone), 1% penicillin–streptomycin, 1% l-glutamine, 1% sodium pyruvate (Gibco) and 150 μg/ml d-luciferin (Pierce; not added for RT-PCR). The cytokine-containing solutions were mixed well and added to cells as designated following synchronization and removal of synchronization media. Cells were then incubated at 36.5°C under ambient atmosphere for the duration of the respective experiment.
Conditioned media derived from EMT6 and 4T1 cells were generated as follows. Cells were plated and grown to confluence in T175 flasks using culture conditions described above. Then, growth media were removed, cells were washed with PBS and media were replaced with conditioning media (powdered DMEM in Millipore-purified water (18.2 Ω resistance) to 13.5 g/ml, sterile filtered using a 0.2-μm filter, supplemented with 1% FBS, 1% HEPES, 1% penicillin–streptomycin and 1% sodium pyruvate). Cells were incubated at 36.5°C under ambient atmosphere for 72 h, after which media were removed, filtered via a 0.45-μm filter and stored at −20°C in single-use aliquots. Following synchronization, for experiments involving treatment with 4T1- or EMT6-conditioned media (and controls), media were removed and replaced from all samples, which received 50% RAW 264.7 bioluminescence recording media (with or without luciferin, depending on experiment), and 50% of one of the following: for non-treated (NT) control, conditioning media with 5% FBS; for FBS control, conditioning media; for 4T1 or EMT6, conditioned media from the designated breast cancer cell line. Independent of treatment, each sample had a final concentration of 1% l-glutamine and 150 μg/ml d-luciferin, which were supplemented as needed.
Conditioned media from RAW 264.7 cells were prepared similarly to above, using RAW 264.7 bioluminescence recording media (without luciferin) containing 1% FBS as conditioning media. After synchronization, for experiments involving treatment of U2OS cells with RAW 264.7 conditioned media (or controls), U2OS media were removed from all samples and replaced as follows. For NT controls, samples received 100% U2OS bioluminescence recording media (described below). Other treatments used 50% U2OS bioluminescence recording media, and 50% of one of the following: for FBS control, RAW 264.7 conditioning media; for conditioned samples, RAW 264.7 conditioned media. U2OS bioluminescence recording media were prepared by dissolving powdered DMEM (Sigma-Aldrich) in Millipore-purified water (18.2 Ω resistance) to give a final concentration of 11.25 g/ml, followed by sterile filtration using a 0.2-μm filter, supplemented with 5% FBS, 1% HEPES (HyClone), 0.25% penicillin–streptomycin, 0.35 g/l sodium bicarbonate (Fisher Scientific) and 150 μg/ml d-luciferin. Independent of treatment, each sample had a final concentration of 0.35 g/l sodium bicarbonate and 150 μg/ml d-luciferin, which were supplemented as needed.
Bioluminescence recording and analysis of time series
Following synchronization and addition of cytokines or conditioned media, dishes were sealed with 40-mm sterile cover glass using silicon vacuum grease and subjected to a LumiCycle 32 System (Actimetrics) for monitoring at 36.5°C for 5–7 days. Bioluminescence signals were measured every 10 min.
Bioluminescence recordings were pre-processed to exclude the initial 24-h transient and spikes [32]. Recordings were considered arrhythmic outliers and excluded from further analysis if their range (maximum minus minimum over time) was less than one-third that of the median range for that reporter and treatment. To assess the strength of rhythmicity for each rhythmic recording, a quadratic trend was removed, and then three measures were computed: the relative power of the band in the power spectral density corresponding to periods of 16–32 h (RelPow), the rhythmicity index computed as the height of the third peak of the correlogram (RI) [37], and the maximum value of the chi square periodorgram (MaxQp) [38]. To assess circadian period and amplitude for each rhythmic recording, the average of a 24-h moving window was removed, resulting in detrended data with 12 h of data eliminated from the beginning and end of the recordings, and then the average of a 3-h moving window was used to smooth the data. The resulting time series (t = 36 to 132 h) were fit to a damped cosine curve [32] for one estimate of the period and amplitude. Furthermore, phase markers (peak, trough, mean-crossings) were identified to compute additional measures: the period as estimated by the difference in marker times from cycle to cycle, and the amplitude as measured by the difference in peak and trough heights for cycle 1 or cycle 2 [32].
RESULTS
M1 and M2 polarization conditions differently alter the circadian rhythms of RAW 264.7 macrophages
To evaluate how circadian rhythms change in opposing macrophage subtypes, it is critical to generate data with sufficient resolution to facilitiate detailed analyses of oscillations. To track circadian rhythms in a time-resolved manner, with frequent sampling over the course of multiple circadian cycles, we opted to use luciferase reporters for promoter activity. Here, we stably transfected RAW 264.7 macrophage cells with a reporter for a positive (Bmal1) and a negative (Per2) component of the core circadian clock, yielding RAW 264.7-Bmal1:luc and RAW 264.7-Per2:luc cells, respectively (Fig. S1). LPS is an endotoxin found in outer membranes of gram negative bacteria, and results in M1 responses in RAW 264.7 macrophages [39, 40]. IL-4 is secreted by T helper 2 (TH2) cells [16, 41] and cancers [42], yielding M2 responses. To investigate circadian alterations that occur in M1 and M2 subtypes, RAW 264.7 reporter cells were polarized under the following conditions: for the M1 subtype, 5, 20 or 50 ng/ml of LPS, and a combination of 5 ng/ml of LPS and 12 ng/ml of IFN-γ (LPS/IFN-γ) were used; for the M2 subtype, 50 ng/ml of IL-4 was used [43]. To confirm cell polarization following treatments, we carried out RT-PCR to assess the presence of M1- and M2-specific markers (Fig. S2). As in other studies, polarization to the M1 subtype using the aforementioned conditions resulted in enhanced levels of the M1 markers Tnf-α (Fig. S2A) [39, 44] and iNos (Fig. S2B) [40, 44], which increased with LPS concentration and LPS/IFN-γ. Under these conditions the M2-specific marker CD206 was down-regulated. In contrast, when RAW 264.7 macrophages were polarized to the M2 subtype via IL-4, the expression of M1 markers Tnf-α and iNos decreased, as expected [43] while M2-associated CD206 increased (Fig. S2C).
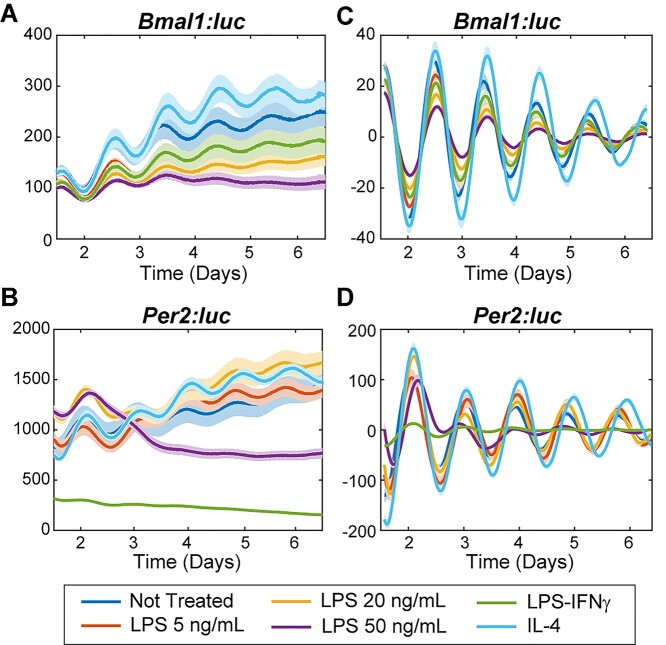
Circadian oscillations based on Bmal1:luc and Per2:luc signals were tracked via real-time luminometry. Raw and detrended circadian data of M1 polarized cells show that average bioluminescence levels of Bmal1:luc reporters are lower (raw data shown in Fig. 1A, Fig. S3A; detrended data shown in Fig. 1C, Fig. S4A) while those of Per2:luc reporters vary (raw data shown in Fig. 1B, Fig. S3B; detrended data shown in Fig. 1D, Fig. S4B), compared to NT samples in the respective RAW 264.7 reporter cell lines. In contrast, raw and detrended circadian data of M2 polarized cells show increased average bioluminescence of both Bmal1:luc (raw data shown in Fig. 1A, Fig. S3A; detrended data shown in Fig. 1C, Fig. S4A) and Per2:luc reporters (raw data shown in Fig. 1B, Fig. S3B; detrended data shown in Fig. 1D, Fig. S4B) compared to respective NT samples in the same RAW 264.7 reporter cell lines.
Figure 1.
Bioluminescence rhythms of Bmal1 and Per2 promoter activities following cytokine treatments in RAW 264.7-Bmal1:luc and –Per2:luc cells. Shown are raw (A,B) and detrended, smoothed (C,D) time series averaged across replicates (N = 8) for each treatment. The standard error is shown with a light envelope around the mean; in some instances, this is too small to be visualized. The raw and detrended data with individual replicates can be found in Fig. S3 and S4, respectively.
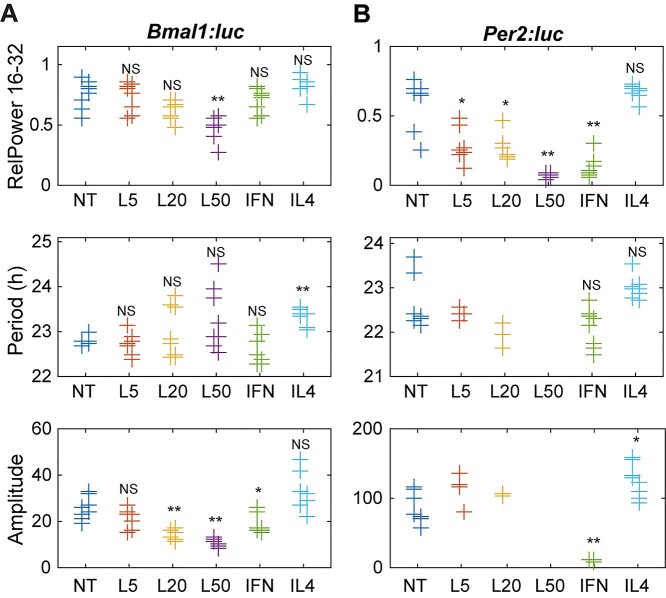
Circadian rhythmicity, period and amplitude were differently altered in the M1 and M2 polarization states, with dissimilar Bmal1:luc and Per2:luc responses. The strength of rhythmicity for each time series was assessed by three methods: the relative power in the circadian (16–32 h) band of the power spectral density (RelPow), the rhythmicity index computed as the height of the third peak of the correlogram (RI), and the maximum value of the chi-squared periodorgram (MaxQp). M1 polarization with 5 and 20 ng/ml of LPS significantly decreased only Per2:luc rhythmicity, while 50 ng/ml of LPS reduced rhythmicity of both Bmal1:luc and Per2:luc reporters (Fig. 2 top, Fig. S5). Like the lower concentration LPS treatments, the combination of LPS/IFN-γ significantly decreased rhythmicity only in the Per2:luc reporter (Fig. 2B top, Fig. S5B), compared to respective NT samples. In contrast, M2 polarization with IL-4 significantly increased the rhythmicity only in the Per2:luc reporter when measured using MaxQp, but not using other methods (RI, RelPow).
Figure 2.
Circadian parameters for (A) Bmal1 and (B) Per2 oscillations following cytokine treatments. Shown are measures of strength of rhythmicity (top), period (middle), and amplitude (bottom). The measure of rhythmicity is the relative power in the 16- to 32-h band of the power spectral density. The period and amplitude are estimated by fitting each time series to a damped cosine curve. Data points are color-coded by dose; within each, data is separated by experiment (those slightly to the left are from the first, and those to the right are from the second). The distribution of measures for each treatment is compared to that of the non-treated samples using a randomization test for difference in means (NS indicates “not significant,” *p < 0.05, **p < 0.01, and no indicator above a treatment indicates that there were too few data points for the test to have sufficient power). Time series that did not fit well to a damped cosine (GOF < 0.9) were excluded from period and amplitude evaluations. NT = non-treated; L5 = 5 ng/mL LPS; L20 = 20 ng/mL LPS; L50 = 50 ng/mL LPS; IFN = 5 ng/mL LPS and 12 ng/mL IFN-γ.
Circadian periods were also affected by polarization. M1 polarization with LPS altered the period of RAW 264.7 cells differently depending on treatment. Decreased periods were observed following LPS treatments with 5 ng/ml (Bmal1:luc, mean-crossing up), 20 ng/ml (Per2:luc, trough-to-trough, mean CWT period), and 50 ng/ml (Per2:luc, mean CWT period) (Fig. S6). However, increased periods were observed following 50 ng/ml LPS (Per2:luc, mean-crossing up, peak-to-peak) (Fig. 2A, Fig. S6). Furthermore, period values exhibited wider spreads with increasing LPS concentration in both reporters, which may be attributed to lower rhythmicity. In contrast, M2 polarization via IL-4 treatment increased periods in both reporters when measured by mean-crossing up and down, and trough-to-trough (Fig. S6), but this period enhancement was only found in the Bmal1:luc reporter when determined by DC Fit, peak-to-peak and mean CWT methods (Fig. 2A middle, Fig. S6A). No significant period reductions were found in any of the methods used. For samples with reduced rhythmicity (time series that did not fit well to a damped cosine (GOF < 0.9)), no period was calculated.
Lastly, amplitudes were altered depending on polarization treatment. M1 polarization with 20 and 50 ng/ml of LPS, and the combination of LPS/IFN-γ significantly reduced the amplitudes of Bmal1:luc reporters in RAW 264.7 cells when analyzed using DC fit amplitude and peak1-trough1 (Fig. 2A bottom, Fig. S7A), while the peak2-trough2 method showed significant period reductions for the 20 and 50 ng/ml LPS treatments, but not LPS/IFN-γ (Fig. S7A). Amplitude reductions were observed for LPS/IFN-γ (DC fit amplitude, peak-to-trough) and 50 ng/ml LPS (peak2-trough2) conditions in Per2:luc reporter (Fig. 2B bottom, Fig. S7B). No significant amplitude increases were observed under any M1 polarization conditions for either Bmal1 or Per2. In contrast, amplitude enhancement was observed in IL-4-treated Per2:luc samples via the DC fit method (Fig. 2B bottom, Fig. S7B), and no significant amplitude reductions were observed under M2 polarization conditions. Where samples showed reduced rhythmicity (e.g. LPS-treated Raw 264.7-Per2:luc), no amplitudes were calculated.
Exposure to breast cancer-conditioned media alters circadian rhythms of RAW 264.7 cells
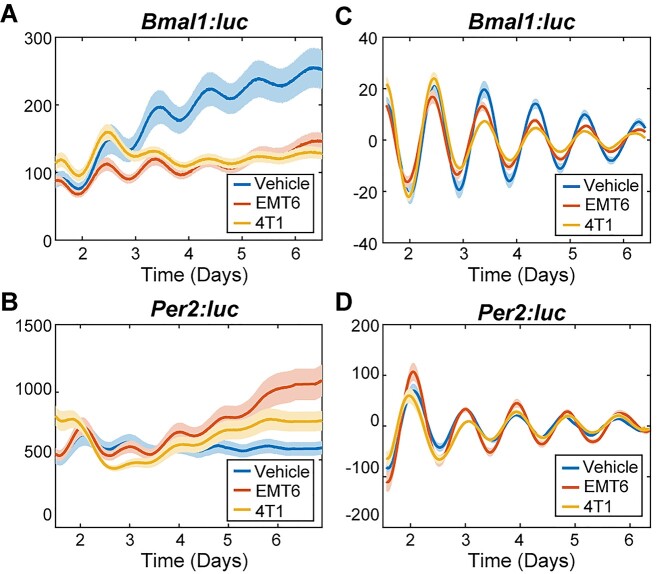
Previous studies have shown that macrophages play critical roles in the dissemination of cancer cells [45], and that some cancers can polarize macrophages toward the M2 subtype [46]. These studies imply that there is a cross-talk between macrophages and the tumor microenvironment. Hence, we hypothesized that cancer cells would also affect the circadian rhythms of macrophages. To test this, first we assessed the effects of conditioned media derived from 4T1 or EMT6 mouse breast cancer cells on M1 and M2 marker levels in RAW 264.7 cells via RT-PCR. We observed that both 4T1- and EMT6-conditioned media increased the expression of the M1 markers Tnf-α and iNos, and decreased the expression of the M2 marker CD206 (Fig. S8). Then, we tracked circadian rhythms of RAW 264.7 Bmal1:luc and Per2:luc reporters exposed to these conditioned media samples. Raw and detrended circadian data of both EMT6- and 4T1-conditioned media-treated samples showed that average bioluminescence of Bmal1:luc reporters were lower (raw data shown in Fig. 3A, Fig. S9A; detrended data shown in Fig. 3C, Fig. S10A) while Per2:luc reporters varied in their levels (raw data shown in Fig. 3B, Fig. S9B; detrended data shown in Fig. 3D, Fig. S10B), compared to respective NT samples in the same RAW 264.7 reporter cell lines.
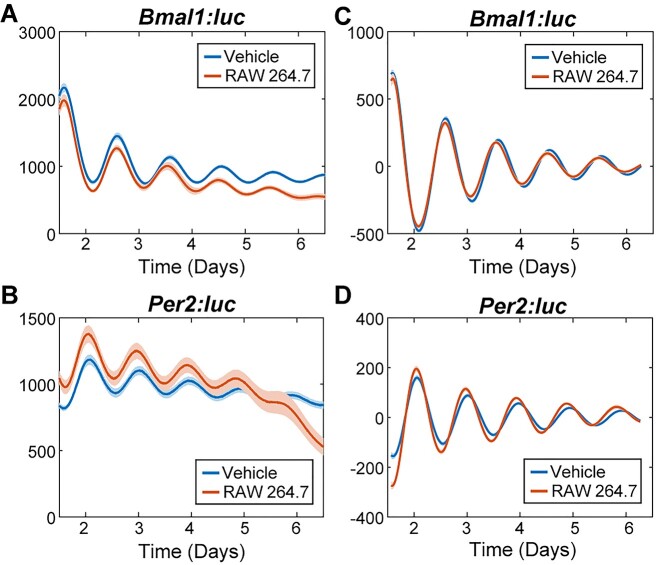
Figure 3.
Bioluminescence rhythms of Bmal1 and Per2 promoter activities following macrophage exposure to cancer-conditioned media. Shown are raw (A,B) and detrended, smoothed (C,D) time series averaged across replicates (N = 12) for each treatment. The standard error is shown with a light envelope around the mean; in some instances, this is too small to be visualized. The raw data and detrended data with individual replicates can be found in Fig. S9 and S10, respectively.
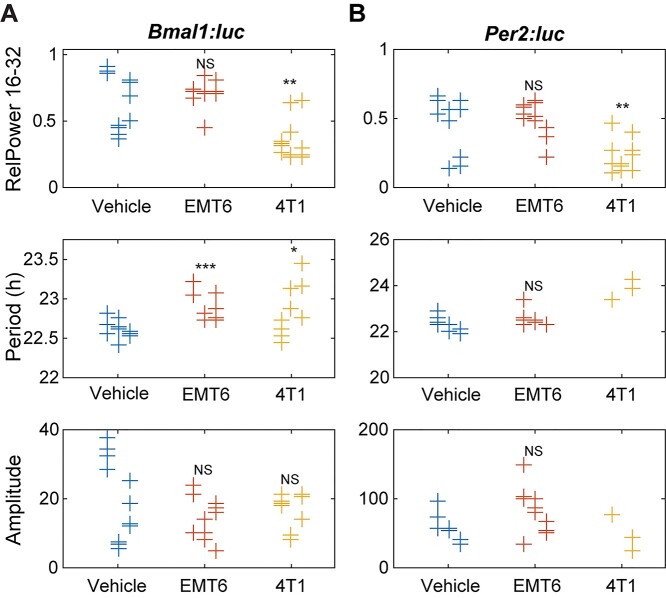
Conditioned media derived from the more aggressive 4T1 breast cancer cell line were found to elicit greater circadian effects than that from EMT6 cells, including with regard to reductions in circadian rhythmicity and period distributions (Fig. 4 and Figs S11, S12). Rhythmicity was determined via multiple methods, all of which, except for the rhythmicity index, showed significant decreases for both reporters in 4T1-conditioned media-treated cells (Fig. 4, Fig. S11). Concurrently, treatment with EMT6 breast cancer-conditioned media resulted in no significant alterations in rhythmicity by any method used except for the rhythmicity index (Fig. 4, Fig. S11). For additional circadian evaluations (e.g. period and amplitude assessments), samples that had periods outside of the 16–32-h circadian period range, or had a damped cosine fit with GOF < 0.9 were considered arrhythmic and excluded.
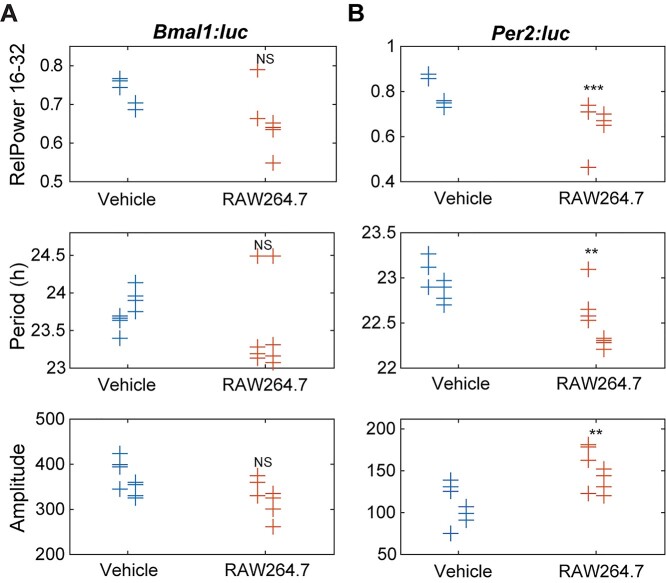
Figure 4.
Circadian parameters for (A) Bmal1 and (B) Per2 oscillations following exposure to cancer cell-conditioned media. Shown are measures of strength of rhythmicity (top), period (middle), and amplitude (bottom). The measure of rhythmicity is the relative power in the 16- to 32-h band of the power spectral density. The period and amplitude are estimated by fitting each time series to a damped cosine curve. Data points are color-coded by dose; within each, data is separated by experiment (those to the left are from the first, in the middle are from the second, and to the right are from the third). The distribution of measures for each treatment is compared to that of the vehicle using a randomization test for difference in means (NS indicates “not significant,” *p < 0.05, **p < 0.01, and no indicator above a treatment indicates that there were too few data points for the test to have sufficient power). Time series that did not fit well to a damped cosine (GOF < 0.9) were excluded from period and amplitude evaluations. (Vehicle = FBS control, EMT6 = EMT6 cell-conditioned media, 4T1 = 4T1 cell-conditioned media).
Periods were also affected by conditioned media treatments, with Bmal1 and Per2 being dissimilarly affected in some instances (especially in the effects of EMT6 media treatments). Overall, 4T1-conditioned media-treated macrophages showed enhanced periods of Bmal1:luc and Per2:luc. Bmal1:luc had longer periods as determined by the DC Fit, mean-crossing up and peak-to-peak methods; however, it showed a period reduction using the mean-crossing down method (Fig. S12A). Per2:luc periods were determined to increase via mean-crossing down, peak-to-peak and mean CWT period methods; no decreases were found (Fig. S12B). EMT6-conditioned media-treated samples showed significant period enhancements in the Bmal1:luc reporter using multiple methods (DC fit, mean-crossing up, peak-to-peak and trough-to-trough; Fig. 4A, Fig. S12A), while the Per2:luc reporter showed only significant period increases using peak-to-peak and mean CWT period methods, which were also to a lesser degree (Fig. 4B, Fig. S12B). No period reductions for either reporter were observed following EMT-conditioned media treatment, using any tests. It is also noteworthy that the overall distributions of period values for the 4T1-conditioned media-treated samples were wider than those of controls and EMT6-conditioned media-treated samples.
4T1 cell-conditioned media-treated macrophage reporter cells were also affected in terms of amplitude changes, although these were not as substantial as the effects on other circadian characteristics. Both Bmal1:luc and Per2:luc oscillations showed significantly reduced amplitudes following 4T1-conditioned media treatment using the peak2-trough2 method (Fig. S13). However, the other approaches used did not result in statistically significant changes. No amplitude evaluations showed effects on either Bmal1:luc or Per2:luc signals for cells treated with EMT6-conditioned media. (Fig. 4, Fig. S13).
RAW 264.7-conditioned media affect the circadian rhythms of U2OS cells
Macrophages have different responses to and interactions with cancers. In some cases, macrophages facilitate the metastasis and invasion of breast cancer cells [47, 48] while in others, they reduce cell proliferation [49, 50]. Here, we investigated whether and how conditioned media derived from macrophages affect osteosarcoma cells, which have diminished growth following macrophage exposure, and their circadian outputs. We used the human osteosarcoma cell line U2OS, which is a well-established model for circadian studies [33, 51, 52] to evaluate the effects of macrophage-conditioned media on circadian oscillations of cancer cells. While the cell lines are derived from different species, interactions between the two have been previously confirmed [53]. We used U2OS cells, stably transfected with Bmal1:luc and Per2:luc luciferase reporters [31], and treated them with conditioned media harvested from naïve RAW 264.7 cells. The circadian oscillations of U2OS-Bmal1:luc and –Per2:luc were subsequently tracked via luminometer for 5–7 days.
Raw and detrended circadian data of RAW 264.7-conditioned media-treated U2OS cells showed that the average bioluminescence levels of Bmal1:luc reporters were lower (raw data shown in Fig. 5A, Fig. S14A; detrended data shown in Fig. 5C, Fig. S15A) while Per2:luc reporter bioluminescence levels were higher (raw data shown in Fig. 5B, Fig. S14B; detrended data shown in Fig. 5D, Fig. S15B), compared to respective NT samples, in the same U2OS reporter cell line.
Figure 5.
Bioluminescence rhythms of Bmal1 and Per2 promoter activities in U2OS cells following exposure to conditioned media from macrophages. Shown are raw (A,B) and de-trended, smoothed (C,D) time series averaged across replicates (N = 8) for each treatment. The standard error is shown with a light envelope around the mean; in some instances, this is too small to be visualized. The raw and detrended data with individual replicates can be found in Fig. S14 and S15, respectively.
The Per2:luc reporter showed significantly decreased rhythmicity in the RelPow 16–32 h and rhythmicity index methods while the Bmal1:luc reporter did not show any significant rhythmicity alterations (Fig. 6, Fig. S16). No samples showed increased rhythmicity. Period was also altered due to the conditioned media treatment. The Per2:luc reporter showed significantly decreased periods in the DC fit, mean-crossing up and peak-to-peak methods while the Bmal1:luc reporter did not show any significant period alterations (Fig. 6, Fig. S17). No samples showed increased period. Overall, the conditioned media-treated samples showed wider distributions of period values compared to NT samples. Furthermore, the Per2:luc reporter showed significantly increased amplitudes across all methods used (DC fit mean damped amplitude, peak1- trough1 and peak2-trough2), while Bmal1:luc was not significantly affected in terms of amplitude (Fig. 6, Fig. S18). In follow-up experiments, we also evaluated U2OS cell treatments with conditioned media from M1- and M2-polarized macrophages; however, there were no changes in the results obtained and thus are not presented.
Figure 6.
Circadian parameters of (A) Bmal1 and (B) Per2 in U2OS cells following exposure to macrophage conditioned media. Shown are measures of strength of rhythmicity (top), period (middle), and amplitude (bottom). The measure of rhythmicity is the relative power in the 16- to 32-h band of the power spectral density. The period and amplitude are estimated by fitting each time series to a damped cosine curve. Data points are color-coded by dose; within each, data is separated by experiment (those slightly to the left are from the first and to the right are from the second). The distribution of measures for each treatment is compared to that of the vehicle using a randomization test for difference in means (NS indicates “not significant,” ***p < 0.001, **p < 0.01, and no indicator above a treatment indicates that there were too few data points for the test to have sufficient power). Time series that did not fit well to a damped cosine (GOF < 0.9) were excluded from period and amplitude evaluations. (Vehicle = FBS control, RAW264.7 = RAW 264.7 cell-conditioned media).
DISCUSSION
Macrophages have diverse functions that range from pro-inflammatory (e.g. generation of reactive oxygen and nitrogen species) to immune-suppressing (e.g. tissue remodeling and angiogenesis) activities. To produce the appropriate responses to physiological or environmental challenges, macrophages are plastic—they can be polarized to broadly M1 (pro-inflammatory) or M2 (anti-inflammatory) subtypes [15, 16]. Previous studies have shown that 8–15% of the macrophage transcriptome, including genes involved in pathogen recognition and responses, has circadian oscillations [18, 20]. Multiple functions and characteristics of macrophages have also been shown to occur in circadian manner, such as recruitment to infected tissue [12], generation of chemokines and cytokines, and phagocytosis [21, 22]. Based on the distinct, opposing functions of the two general macrophage subtypes, and macrophages’ inherent associations with circadian rhythms, we hypothesized that the oscillations of circadian genes are differently altered depending on stimuli.
Here, we studied the relationship between macrophage polarization and core clock oscillations by tracking the signals of Bmal1:luc and Per2:luc reporters in RAW 264.7 cells exposed to various stimuli, using real-time luminometry. We performed two series of experiments in this regard: first, we used cytokines to polarize cells to standard immune-stimulating (M1) and—suppressing (M2) states; then, we exposed macrophages to conditioned media from two different murine breast cancer cell types, 4T1 (more aggressive) and EMT6 (less aggressive). We found that opposing polarizations of macrophages differentially altered their circadian rhythms, and that the circadian alterations observed following conditioned media treatments are similar to those of M1 polarized macrophages.
It has been previously shown that mRNA/protein levels of core circadian clock genes in macrophages are altered under pro-inflammatory conditions [26, 28]. LPS treatment significantly reduced Bmal1 mRNA levels in peritoneal macrophages [26], mouse BMDM and peritoneal macrophages, and human macrophages and peripheral blood mononuclear cells (PBMCs) [28]. In our own study, we similarly saw that LPS-treated Bmal1:luc reporters have lower bioluminescence levels compared to NT samples in both raw (Fig. 1A, Fig. S3A) as well as detrended (Fig. 1B, Fig. S4A) data, suggesting that M1 polarization with LPS weakens Bmal1 circadian rhythms. It has also been shown that treatment with a high concentration of LPS significantly increased Per2 mRNA levels in mouse BMDMs [28]. However, a luminometry study carried out with mPer2:luc mouse-derived peritoneal macrophages showed that 5 ng/ml LPS reduced, but 20 and 100 ng/ml LPS increased non-detrended signal intensities for a portion of the experiment (24–54 h) compared to control [26], while another study showed that 50 ng/ml LPS reduced non-detrended rhythms in mouse BMDMs over four days [27]. Likewise, our data showed (over 24–72 h) that 5 ng/ml reduced raw bioluminescence rhythms of Per2:luc in RAW 264.7 cells, but (over 24 h to 1 week) 20 ng/ml and (over 24–72 h) 50 ng/ml LPS resulted in increases (Fig. 1B and Fig. S3B). However, after ~3 days, we saw increased raw bioluminescence levels for 5 ng/ml LPS and reduced bioluminescence for 50 ng/ml LPS-treated samples compared to NT.
For LPS/IFN-γ-treated samples, reduced bioluminescence was observed, compared to NT, for the entire time-series (Fig. 1B and Fig. S3B), as seen previously with mPer2:luc BMDM [27]. Our data showed that circadian amplitudes were reduced when cells were exposed to LPS or LPS/IFN-γ (Fig. 2 and Fig. S7), which were also similar to previous studies showing that LPS or LPS/IFN-γ treatments can reduce the amplitudes of peritoneal macrophages [26] and BMDMs [27] in mPer2:luc mice. While LPS or LPS/ IFN-γ treatments showed no significant period effects in mPer2:luc BMDM [27], and in our study (Fig. 2 and Fig. S6), LPS treatment has been shown to induce a subtle lengthening of period in mPer2:luc peritoneal macrophages [26]. We also observed that M1 polarization with 5 and 20 ng/ml of LPS, and a combination of LPS/IFN-γ significantly decreased Per2:luc rhythmicity, while 50 ng/ml of LPS reduced rhythmicity of both Bmal1:luc and Per2:luc reporters (Fig. 2, Fig. S5). However, other studies have not assessed cellular rhythmicity.
Weakened circadian amplitudes observed in cells polarized to greater extents toward M1 states (e.g. via 50 ng/ml LPS) may be caused by the generation of reactive oxygen species (ROS) in these macrophages, which was also found previously [26]. M1 polarization increases expression of ROS producing genes (e.g. Tnf-α and iNos). Amplitudes and rhythmicity in M1 cells can also be affected by increased levels of the Bmal1 repressor, REV-ERB, which are present following treatment with LPS and IFN-γ [30]. Under M2 polarization (described further below), such ROS generating genes are down-regulated (Fig. S2), and enhanced amplitudes were observed (Figs 1 and 2). However, another possible reason for decreased Bmal1 oscillations is the production of proinflammatory miRNA-155 in response to LPS treatment. Curtis et al. showed that miR-155 levels are inversely correlated with those of Bmal1, which possesses two miR-155 binding sites in its promoter, and are unaffected following LPS treatment in the presence of an miR-155 antagomir [28].
While the circadian effects of M2 polarized macrophages have not been as thoroughly studied, M2 polarization/IL-4 (20 ng/ml) treatment has been shown to decrease mRNA and protein levels of REV-ERBα (a Bmal1 transcriptional repressor) in differentiated THP-1 macrophages [30]. Such reduction of REV-ERBα should increase levels of Bmal1; we also found increased levels of raw bioluminescence for the Bmal1:luc reporter (Fig. 1A) upon M2 polarization in our study. The increased Bmal1 should also drive Per2 expression, which we also observed via amplitude enhancement in Per2:luc (Fig. 2B, Fig. S7), and was also found by Chen et al. [26]. However, while that study did not find any effects on period, we observed increased periods in both reporters following M2 polarization (Fig. 2, Fig. S6). Furthermore, we also found that M2 polarization significantly increased rhythmicity of the Per2:luc reporter (Fig. 2, Fig. S5).
There is significant evidence to indicate cross-talk between macrophages and tumor microenvironments, but the contributions of macrophages also depends on cancer type [45, 46, 54, 55]. Similarly, macrophage responses to conditioned media can vary. For example, conditioned media derived from high tumor grade MDA-MB-231 (human breast cancer) cells has been shown to upregulate both M1 and M2 markers of human CD14+ macrophages [46], while murine BMDMs exposed to 4T1 breast cancer conditioned media, and splenocytes cocultured with 4T1 cells expressed higher levels of M1 marker genes including Tnf-α [54, 55]. 4T1 cell-conditioned media have been shown to contain significant amounts of IFN-γ [55], which can itself polarize macrophages to the M1 subtype [56, 57]. However, the effects of tumor microenvironments on macrophage circadian rhythms are unknown. To evaluate the interactions between the two, we treated RAW 264.7-Bmal1:luc and -Per2:luc macrophages with EMT6 (less aggressive) and 4T1 (highly aggressive) murine mammary carcinoma [58] derived-conditioned media and analyzed their circadian effects. Our data showed that the more aggressive 4T1 breast cancer-conditioned media reduced macrophage rhythmicity akin to M1 polarization conditions, which was not surprising given the similar marker profile as determined by RT-PCR (Fig. S8). On the other hand, the less aggressive EMT6-derived conditioned media did not affect rhythmicity, but instead yielded significantly altered periods. Altogether our data suggests that mouse breast cancer-conditioned media polarizes macrophages M1 subtype as a defense mechanism to protect the host against the cancer.
As mentioned above, macrophages can result in positive or negative effects following interaction with cancers. Previous studies show that macrophages promote metastasis and invasion of breast cancer cells [47, 48], while others show that macrophages inhibit osteosarcoma cell growth via inhibition of cell proliferation and increased phagocytosis [49, 50, 53]. Considering the previously investigated effects of macrophage-conditioned media on cancer cells, and the interactions between macrophages and osteosarcomas, we wished to evaluate whether the former might also influence circadian rhythms of the latter. We treated human osteosarcoma reporter cell lines, U2OS-Bmal1:luc and -Per2:luc with conditioned media harvested from naïve RAW 264.7 cells. We observed reduction in rhythmicity and amplitude enhancement of the U2OS oscillations. As circadian rhythms are found to be increasingly disrupted with disease severity and oncogenic characteristics [36] and even subtle renormalization of disrupted rhythms can reduce oncogenic features [33], our data highlight another connection between circadian oscillations and cancer, which should be studied further.
Taken together, we show that the circadian rhythms of macrophages are influenced by their polarization states. While M1 polarization was associated with significant loss of rhythmicity, M2 polarization resulted in amplitude and period enhancements. We also found that macrophage circadian effects translate to cultures with cancer cell-conditioned media, where M1-like marker characteristics were accompanied by M1-like circadian oscilations. Finally, we show that conditioned media of macrophages enhance the circadian rhythms of another cancer cell type (U2OS), which macrophages act against in oncogenic environments. This raises a hypothesis that macrophages can alter the circadian rhythms of tumors and cancer cell types to affect their oncogenic features. In the future, this relationship should be examined with other cancer models, including to determine whether macrophages negatively affect the circadian oscillations of cancer cells/types with which they interact to facilitate disease. In terms of macrophages themselves, it is of interest to study whether the lack of rhythmicity in inflammatory macrophages is due to loss of synchrony by performing single-cell experiments, and to assess whether this circadian divergence confers benefits in terms of immune response and host defense.
Supplementary Material
Acknowledgements
We would like to thank Tanya Leise (Mathematics & Statistics, Amherst College) for helpful discussions regarding analyses.
Contributor Information
Sujeewa S Lellupitiyage Don, Department of Chemistry, University of Massachusetts Amherst, Amherst, MA, USA.
Javier A Mas-Rosario, Molecular and Cellular Biology Graduate Program, University of Massachusetts Amherst, Amherst, MA, USA.
Hui-Hsien Lin, Department of Chemistry, University of Massachusetts Amherst, Amherst, MA, USA.
Evelyn M Nguyen, Mount Holyoke College, South Hadley, MA, USA.
Stephanie R Taylor, Department of Computer Science, Colby College, Waterville, ME, USA.
Michelle E Farkas, Department of Chemistry, University of Massachusetts Amherst, Amherst, MA, USA; Molecular and Cellular Biology Graduate Program, University of Massachusetts Amherst, Amherst, MA, USA.
Funding
This work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM143016. J.A.M.-R. was supported by a fellowship from the Chemistry-Biology Interface Training Program (National Research Service Award [T32 GM008515]) from the National Institutes of Health; H.-H.L. was supported by a University of Massachusetts Amherst Chemistry-Biology Interface (CBI) training fellowship as part of the same program.
Conflict of interest statement
The authors declare that there is no conflict of interest.
References
- 1. Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol 2005;174:7618–24. [DOI] [PubMed] [Google Scholar]
- 2. Scheiermann C, Kunisaki Y, Lucas D et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity 2012;37:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Druzd D, Matveeva O, Ince L et al. Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 2017;46:120–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hriscu ML. Modulatory factors of circadian phagocytic activity. Ann N Y Acad Sci 2005;1057:403–30. [DOI] [PubMed] [Google Scholar]
- 5. Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol 2013;13:190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibbsa JE, Blaikleya J, Beesleya S et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A 2012;109:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bellet MM, Deriu E, Liu JZ et al. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci U S A 2013;110:9897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kiessling S, Dubeau-Larameé G, Ohm H et al. The circadian clock in immune cells controls the magnitude of Leishmania parasite infection. Sci Rep 2017;7:10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hopwood TW, Hall S, Begley N et al. The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci Rep 2018;8:3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nguyen KD, Fentress SJ, Qiu Y et al. Circadian gene Bmal1 regulates diurnal oscillations of Ly6Chi inflammatory monocytes. Science 2013;341:1483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schloss MJ, Horckmans M, Nitz K et al. The time-of-day of myocardial infarction onset affects healing through oscillations in cardiac neutrophil recruitment. EMBO Mol Med 2016;8:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pick R, He W, Chen CS et al. Time-of-day-dependent trafficking and function of leukocyte subsets. Trends Immunol 2019;40:524–37. [DOI] [PubMed] [Google Scholar]
- 13. Long JE, Drayson MT, Taylor AE et al. Morning vaccination enhances antibody response over afternoon vaccination: a cluster-randomised trial. Vaccine 2016;34:2679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panzer SE, Dodge AM, Kelly EAB et al. Circadian variation of sputum inflammatory cells in mild asthma. J Allergy Clin Immunol 2003;111:308–12. [DOI] [PubMed] [Google Scholar]
- 15. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wager CML, Wormley FL. Classical versus alternative macrophage activation: the Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol 2014;7:1023–35. [DOI] [PubMed] [Google Scholar]
- 17. Yao Y, Xu XH, Jin L. Macrophage polarization in physiological and pathological pregnancy. Front Immunol 2019;10:792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keller M, Mazuch J, Abraham U et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A 2009;106:21407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silver AC, Arjona A, Hughes ME et al. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun 2012;26:407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Collins EJ, Cervantes-Silva MP, Timmons GA et al. Post-transcriptional circadian regulation in macrophages organizes temporally distinct immunometabolic states. Genome Res 2021;31:171–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull 2007;30:621–6. [DOI] [PubMed] [Google Scholar]
- 22. Kitchen GB, Cunningham PS, Poolman TM et al. The clock gene Bmal1 inhibits macrophage motility, phagocytosis, and impairs defense against pneumonia. Proc Natl Acad Sci U S A 2020;117:1543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bosurgi L, Cao YG, Cabeza-Cabrerizo M et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 2017;356:1072–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo L, Akahori H, Harari E et al. CD163+ macrophages promote angiogenesis and vascular permeability accompanied by inflammation in atherosclerosis. J Clin Invest 2018;128:1106–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fortes GB, Alves LS, De Oliveira R et al. Heme induces programmed necrosis on macrophages through autocrine TNF and ROS production. Blood 2012;119:2368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Pati P, Xu Y et al. Endotoxin disrupts circadian rhythms in macrophages via reactive oxygen species. PLoS One 2016;11:e0155075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen S, Fuller KK, Dunlap JC et al. A pro- and anti-inflammatory axis modulates the macrophage circadian clock. Front Immunol 2020;11:867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Curtis AM, Fagundes CT, Yang G et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci U S A 2015;112:7231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oishi Y, Hayashi S, Isagawa T et al. Bmal1 regulates inflammatory responses in macrophages by modulating enhancer RNA transcription. Sci Rep 2017;7:7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chandra V, Mahajan S, Saini A et al. Human IL10 gene repression by rev-erbα ameliorates mycobacterium tuberculosis clearance. J Biol Chem 2013;288:10692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin H-H, Robertson KL, Bisbee HA et al. Oncogenic and circadian effects of small molecules directly and indirectly targeting the core circadian clock. Integr Cancer Ther 2020;19: 1534735420924094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin H-H, Robertson KL, Lellupitiyage Don SS et al. Chemical modulation of circadian rhythms and assessment of cellular behavior via indirubin and derivatives. Methods Enzymol 2020;639:115–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lellupitiyage Don SS, Robertson KL, Lin H-H et al. Nobiletin affects circadian rhythms and oncogenic characteristics in a cell-dependent manner. PLoS One 2020;15:e0236315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Foteinou PT, Venkataraman A, Francey LJ et al. Computational and experimental insights into the circadian effects of SIRT1. Proc Natl Acad Sci U S A 2018;115:11643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin H-H, Qraitem M, Lian Y et al. Analyses of BMAL1 and PER2 oscillations in a model of breast cancer progression reveal changes with malignancy. Integr Cancer Ther 2019;18: 1534735419836494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lellupitiyage Don SS, Lin H-H, Furtado JJ et al. Circadian oscillations persist in low malignancy breast cancer cells. Cell Cycle 2019;18:2447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levine JD, Funes P, Dowse HB et al. Signal analysis of behavioral and molecular cycles. BMC Neurosci 2002;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sokolove PG, Bushell WN. The chi square periodogram: Its utility for analysis of circadian rhythms. J Theor Biol 1978;72:131–60. [DOI] [PubMed] [Google Scholar]
- 39. Qin H, Wilson CA, Lee SJ et al. LPS induces CD40 gene expression through the activation of NF-κB and STAT-1α in macrophages and microglia. Blood 2005;106:3114–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taciak B, Białasek M, Braniewska A et al. Evaluation of phenotypic and functional stability of RAW 264.7 cell line through serial passages. PLoS One 2018;13:e0198943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gordon S. Alternative activation of macrophages. Nat Rev Immunol 2003;3:23–35. [DOI] [PubMed] [Google Scholar]
- 42. Lin Y, Xu J, Lan H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J Hematol Oncol 2019;12:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hardie J, Mas-Rosario JA, Ha S et al. Macrophage activation by a substituted pyrimido[5,4-b]indole increases anti-cancer activity. Pharmacol Res 2019;148:104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xue B, Wu Y, Yin Z et al. Regulation of lipopolysaccharide-induced inflammatory response by glutathione S-transferase P1 in RAW264.7 cells. FEBS Lett 2005;579:4081–7. [DOI] [PubMed] [Google Scholar]
- 45. Linde N, Casanova-Acebes M, Sosa MS et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 2018;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sousa S, Brion R, Lintunen M et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res 2015;17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. DeNardo DG, Barreto JB, Andreu P et al. CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009;16:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khabbazi S, Goumon Y, Parat MO. Morphine modulates interleukin-4- or breast cancer cell-induced pro-metastatic activation of macrophages. Sci Rep 2015;5:11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dumars C, Ngyuen JM, Gaultier A et al. Dysregulation of macrophage polarization is associated with the metastatic process in osteosarcoma. Oncotarget 2016;7:78343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Punzo F, Bellini G, Tortora C et al. Mifamurtide and TAM-like macrophages: effect on proliferation, migration and differentiation of osteosarcoma cells. Oncotarget 2020;11:687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Z, Yoo S, Park Y-S et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A 2012;109:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tamai TK, Nakane Y, Ota W et al. Identification of circadian clock modulators from existing drugs. EMBO Mol Med 2018;10:e8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ray M, Lee YW, Hardie J et al. CRISPRed macrophages for cell-based cancer immunotherapy. Bioconjug Chem 2018;29:445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Madera L, Greenshields A, Coombs MRP et al. 4T1 murine mammary carcinoma cells enhance macrophage-mediated innate inflammatory responses. PLoS One 2015;10:e0133385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kano A. Tumor cell secretion of soluble factor(s) for specific immunosuppression. Sci Rep 2015;5:8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Davis MJ, Tsang TM, Qiu Y et al. Macrophage M1/M2 polarization dynamically adapts to changes in cytokine microenvironments in Cryptococcus neoformans infection. MBio 2013;4:e00264–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Porcheray F, Viaud S, Rimaniol AC et al. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol 2005;142:481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ouzounova M, Lee E, Piranlioglu R et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat Commun 2017;8:14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.