Abstract
Fluorescent in situ hybridization (FISH) using rRNA-specific oligonucleotide probes has emerged as a popular technique for identifying individual microbial cells. In natural samples, however, the signal derived from fluor-labeled oligonucleotide probes often is undetectable above background fluorescence in many cells. To circumvent this difficulty, we applied fluorochrome-labeled polyribonucleotide probes to identify and enumerate marine planktonic archaea and bacteria. The approach greatly enhanced the sensitivity and applicability of FISH with seawater samples, allowing confident identification and enumeration of planktonic cells to ocean depths of 3,400 m. Quantitative whole-cell hybridization experiments using these probes accounted for 90 to 100% of the total 4′,6-diamidino-2-phenylindole (DAPI)-stained cells in most samples. As predicted in a previous study (R. Massana, A. E. Murray, C. M. Preston, and E. F. DeLong, Appl. Environ. Microbiol. 63:50–56, 1997), group I and II marine archaea predominate in different zones in the water column, with maximal cell densities of 105/ml. The high cell densities of archaea, extending from surface waters to abyssal depths, suggest that they represent a large and significant fraction of the total picoplankton biomass in coastal ocean waters. The data also show that the vast majority of planktonic prokaryotes contain significant numbers of ribosomes, rendering them easily detectable with polyribonucleotide probes. These results imply that the majority of planktonic cells visualized by DAPI do not represent lysed cells or “ghosts,” as was suggested in a previous report.
Fluorescent in situ hybridization (FISH) probes that target intracellular rRNA (e.g., phylogenetic stains; 10) have become widely used tools over the past decade (for a review, see reference 4). The approach has been successfully applied for phylogenetic identification of individual microbial cells in a number of different environmental contexts. Diverse applications of the technique continue to be developed and include the identification and quantification of specific cell types in plankton (1, 23, 27, 56), sediments (31, 48, 53), and soils (9); estimation of physiological activity via cellular rRNA content (23, 26, 39); and spatial localization of microorganisms along environmental gradients (42, 46) or in symbiotic associations (2, 7, 12–15, 38). Detection methods have included epifluorescence microscopy, laser scanning confocal microscopy (10, 54), and flow cytometry (3, 47, 55). A promising recent modification of the technique combines microautoradiography with rRNA-targeted FISH, allowing the assignment of radiotracer uptake to specific phylogenetic types (25, 36). Another advanced application includes spatial correlation of population structure measured via FISH, with the corresponding chemical environment assessed using microelectrodes (42, 45, 46).
Despite the utility of the approach, difficulties are often encountered when applying rRNA-targeted, fluorescent oligonucleotide probes and FISH to complex environmental samples. Problems encountered include variable probe binding to different rRNA target sites (16, 17), a highly autofluorescent sample or cell background, and the inherently low ribosome content of many naturally occurring cells. In aquatic samples, the proportion of cells visualized with monolabeled oligonucleotide probes can be highly variable. Some successful applications of rRNA-targeted oligonucleotide probes and FISH for visualizing picoplankton and nanoplankton have been reported (1, 19–21, 23, 27, 28, 30, 56). In many aquatic samples, however, the percentage of cells detected by oligonucleotide probes and FISH may be significantly lower than the total prokaryotic cell count (1, 19, 21, 24, 26, 27, 33, 37, 56). Probe-conferred fluorescence of many naturally occurring cells often seems to be below current detection limits using monolabeled oligonucleotides and standard epifluorescence microscopy.
A number of signal amplification methodologies have been tested and applied. Many of these are based on an enzymatic amplification step that produces a precipitating fluorescent or colored product (5, 44, 57). Although improvements in signal strength can be obtained, the penetration of enzyme complexes into prokaryote cells can be inefficient. It is usually necessary to include a very carefully controlled permeabilization step with enzymatic signal amplification approaches, balancing permeabilization with cellular integrity. Since cell wall composition varies greatly among prokaryotes, such procedures may compromise the universal applicability of this approach. In another approach, biotin-avidin systems have been applied to detect and quantify marine protists using rRNA-targeted oligonucleotides and FISH (28, 29). An alternative method to amplify a FISH signal uses multiply labeled polyribonucleotide probes (6, 32). This approach has been used to discriminate closely related Pseudomonas species and uncultivated magnetotactic bacteria (49, 52).
In this study, we modified polyribonucleotide probe protocols to quantify two abundant groups of planktonic marine archaea and bacteria. Our results extend previously reported strategies (6, 19, 32, 52) used to identify and enumerate single microbial cells with FISH. Fluor-labeled polyribonucleotide probes were developed for planktonic bacteria and two groups of planktonic archaea and tested for sensitivity and specificity. Dual-staining capabilities were developed and applied to identify and enumerate the different cell types in the same microscopic field in seawater samples. We modified FISH protocols by using multiply labeled polyribonucleotide probes for application to marine plankton samples. The technique was then successfully applied to identify and quantify archaeal and bacterial cells in seawater to depths of 3,400 m.
MATERIALS AND METHODS
Cloning of archaeal small- and large-subunit rRNA operons and genes.
Archaeal group I and II polyribonucleotide probes were synthesized via PCR amplification from plasmid or fosmid templates. The group I probe spanned the entire small-subunit rRNA and approximately 2,600 bases of the large-subunit rRNA. DNA templates for the generation of group I probes were fosmid clones with ca. 40-kb DNA inserts that contained 16S and 23S rRNA linked in an operon (43, 50). The following group I rRNA operon-containing fosmids were used for polynucleotide probe preparation: clone 101G10 (cloned from Cenarchaeum symbiosum [41]), clone 4B7 (from a 200-m Oregon coast sample [50]), and clone ANT2-74A4 (from an Antarctic marine planktonic fosmid library). The preparation of two of the fosmid DNA clone libraries has been previously described (41, 50). The third library was prepared from picoplankton collected in coastal waters off Anvers Island, Antarctica, by previously described methods (50). Fosmid templates were prepared by using Qiagen Maxi kits (Qiagen, Valencia, Calif.) and following the manufacturer’s recommendations for low-copy-number vectors.
Archaeal group II polyribonucleotide probes were prepared from clones derived from PCR-amplified 16S and 23S rRNA genes recovered from this group. The 23S and 16S rRNA gene clones were prepared separately. (Available data suggest that 16S and 23S rRNA genes of group II archaea are not linked in an operon, similar to Thermoplasma acidophilum [data not shown].) The group II 16S rRNA gene clones used for probe generation (SB95-74, SB95-75, and SB95-76) were isolated in a previous study (34). Group II 23S rRNA genes were recovered from surface water plankton samples containing a large proportion of group II archaea. Amplifications were performed on a Perkin-Elmer 9700 thermal cycler by using Taq Plus Precision (Stratagene, La Jolla, Calif.), following the manufacturer’s recommendations, and using primers LSU190F and LSU2445aR (Table 1). After an initial denaturation step of 3 min at 92°C, the thermal cycling parameters were denaturation at 92°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min for a total of 35 cycles. The resulting PCR products were purified by phenol-chloroform extraction (1:1) and spin dialysis in Centricon 100 units (Amicon, Beverly, Mass.). Purified amplicons were subsequently treated with cloned Pfu polymerase (Stratagene) at 72°C for 15 min in 1× cloned Pfu PCR buffer (Stratagene) by following the manufacturer’s recommendations. Amplicons were then cloned into the pCRBlunt vector (Invitrogen, Carlsbad, Calif.) by following the manufacturer’s recommendations. 23S rRNA clones were bidirectionally sequenced by using infrared dye-labeled primers and a Licor automated DNA sequencer. Sequences were aligned within a database of 23S rRNA sequences by using ARB (51). Subsequent bootstrap distance analyses were performed by using PAUP, version 4.0.0d55, Unix version, in conjunction with the GCG package (Genetics Computer Group, Inc., Madison, Wis.). The logdet distance correction was used for evolutionary distance estimation with a transition/transversion ratio of 2.0 empirical base frequencies, random taxon addition, and 1,000 bootstraps of the data set to assign confidence to branches.
TABLE 1.
Oligonucleotide PCR primers used in this study
| Primer | Sequencea | Targetb | E. coli position | Reference |
|---|---|---|---|---|
| Ar20-F | TTC CGG TTG ATC CYG CCR G | Archaeal SSU | 6–25 | 34 |
| GILSUT7-R | GCC AGT GAA TTG TAA TAC GAC TCA CTA TAG GGC GCT TCC ATC AGG CAG AG | Group I LSU | 2610–2623 | This study |
| LSU190-F | GAA YTG AAR CAT CTY AGT A | Prokaryote LSU | 190–207 | This study |
| LSU2445a-R | CCC YGG GGT ARC TTT TCT ST | Archaeal LSU | 2432–2447 | This study |
| Eub27f | AGA GTT TGA TCC TGG CTC AG | Bacterial SSU | 8–27 | 18 |
| LSUT71933e-R | GCC AGT GAA TTG TAA TAC GAC TCA CTA TAG GGA CCC GAC AAG GAA TTT CGC | Bacterial LSU | 1933–1951 | This study |
| Ar20T7-F | GCC AGT GAA TTG TAA TAC GAC TCA CTA TAG GGT TCC GGT TGA TCC YGC CRG | Archaeal SSU | 6–25 | This study |
| Eub27T7-F | GCC AGT GAA TTG TAA TAC GAC TCA CTA TAG GGA GAG TTT GAT CCT GGC TCA G | Bacterial SSU | 8–27 | This study |
| G1LSU-R | CGC TTC CAT CAG GCA GAG | Group I LSU | 2610–2623 | This study |
| LSU1933e-R | ACC CGA CAA GGA ATT TCG C | Bacterial LSU | 1933–1951 | 4 |
The T7 RNA polymerase promoter sequence is in boldface type.
SSU, small subunit; LSU, large subunit.
Preparation of DNA templates for transcription reactions.
DNA templates used for transcription reactions were prepared by PCR amplification with the Taq Plus Precision enzyme mixture (Stratagene) by following the manufacturer’s recommendations. The templates and primers used to generate T7 promoter-containing amplicons for use in subsequent transcription reactions are shown in Tables 1 and 2. After an initial denaturation step of 3 min at 94°C, the thermal cycling parameters were denaturation at 92°C for 30 s and annealing at 56°C for 30 s. An extension time of 3 min at 72°C was used to generate eubacterial, group I, and negative control probe templates. An extension time of 1 min at 72°C was used to generate group II probe templates (23S or 16S rRNAs; Table 2). A total of 30 PCR cycles were used to generate all of the T7 promoter-containing amplicons. The resulting amplicons, each containing a T7 promoter region, were purified by phenol-chloroform extraction (1:1) and subsequent spin dialysis in Centricon 100 units (Amicon).
TABLE 2.
Primers and templates used to generate amplicons for transcription reactions and optimized hybridization conditions for FISH
| Primer pair | DNA templates | Probe targeta | FISH conditionsb |
|---|---|---|---|
| Ar20-F + G1-LSUT7-R | Fosmids 101G10,c 4B7,d 74A4e | Group 1 planktonic archaeal SSU + LSU | 70 (65, 45) |
| Ar20-F + M13-F | Plasmidsf SB95-74, SB95-75, SB95-76 | Group 2 planktonic archaeal SSU | 70 (65, 45) |
| LSU190-F + M13-F | Plasmid G2lsu-1A10g | Group 2 planktonic archaeal LSU | 70 (65, 45) |
| Eub27-F + LSU1933e-R | Picoplankton DNA extracts | Planktonic bacteria | 50 (55, 45) |
| Ar20T7-F + G1-LSU | Fosmids 101G10, 4B7, 74A4 | Negative control | 50 (55, 45) |
| Eub27T7-F + 1933e-R | Picoplankton DNA extracts | Negative control | 50 (55, 45) |
Generation of fluorescently labeled polyribonucleotide probes.
Polyribonucleotide probes were generated from PCR amplicons that contained a T7 RNA polymerase promoter at the 5′ end. The oligonucleotide primer sequences, primer pairs, and DNA templates used in PCR amplifications are shown in Tables 1 and 2. Polyribonucleotide probes were synthesized by using a protocol modified from that of Chee et al. (8). Transcription reactions were performed with Ampliscribe T7 RNA polymerase (Epicentre, Madison, Wis.) by using a modification of the manufacturer’s suggested protocol. Diethyl pyrocarbonate (DEPC)-treated water and precautions for eliminating potential RNase contamination were used throughout the procedure. To generate fluorescein-containing transcripts, transcription reaction mixtures (100-μl total volume) contained 0.5 mM ATP, 0.5 mM GTP, 0.25 mM UTP, 0.25 mM CTP, 0.14 mM fluorescein-12-CTP (NEN, Boston, Mass.), 0.14 mM fluorescein-12-UTP (Boehringer), 10 mM dithiothreitol, 1× Ampliscribe Reaction Buffer, 10 μl of Ampliscribe T7 RNA polymerase enzyme solution, and approximately 2.5 μg of T7 promoter-containing amplicon (DNA template; see above). Following transcription reactions, the fluorescent RNA products were purified by spin dialysis in DEPG-treated H2O and adjusted to a volume of 100 to 200 μl. For hydrolysis of transcripts, MgCl2 was added to a final concentration of 30 mM and the reaction mixtures were heated at 90°C for 5 min. After a 5-min hydrolysis time, the samples were placed on ice and the average size of fragmented transcripts was estimated by agarose gel electrophoresis in comparison to known standards. When necessary, the hydrolysis was resumed for an additional 2 to 5 min at 90°C to attain average fragment sizes of ≤100 nucleotides. Reaction mixtures were subsequently reassayed by agarose gel electrophoresis. Reactions were terminated by addition of 50 mM Na2EDTA. The hydrolysis reactions were monitored by agarose gel electrophoresis in 0.5× Tris-borate-EDTA in 2.5% (wt/vol) Nusieve 3:1 (FMC Bioproducts, Rockland, Maine) gels. An RNA marker (Sigma, St. Louis, Mo.) was included to estimate the sizes of the hydrolysis products. Following electrophoresis, gels were examined for fluorescent products by using a flat-bed fluorescence scanner (FluorImager; Molecular Dynamics, Sunnyvale, Calif.). Gels were subsequently stained with Sybr Green 2 (FMC Bioproducts) and visualized by fluorescence scanning, and the average RNA hydrolysate size was estimated by comparison to RNA size standards.
For labeling of transcripts with the cyanine dye CY-3, transcription reactions (120-μl total volume) contained 4.2 mM ATP, 6.3 mM GTP, 6.3 mM UTP, 6.3 mM CTP, 2.1 mM N6-(6-aminohexyl)ATP (Gibco Bethesda Research Laboratories, Gaithersburg, Md.), 10 mM dithiothreitol, 1× Ampliscribe Reaction Buffer, 12 μl of Ampliscribe T7 RNA polymerase enzyme solution, and 2 to 5 μg of T7 promoter-containing amplicon (see above). Following the transcription reaction, the reaction mixture was purified by gel filtration on Micro BioSpin columns (Bio-Rad, Hercules, Calif.) equilibrated with DEPC-treated H2O and the purified transcripts were mixed 1:1 (vol/vol) with 250 mM sodium carbonate buffer, pH 9.2. The sample was next mixed with dried reactive CY-3 monofunctional dye (Fluorolink; Amersham, Chicago, Ill.) by following the manufacturer’s recommendations, and reaction mixtures were incubated for 1 h at room temperature. Labeled transcripts were separated from unbound dye by gel filtration on Micro BioSpin columns (Bio-Rad) equilibrated with DEPC-treated H2O. Labeled transcripts were hydrolyzed as described above for the fluorescein-labeled transcripts. Unlabeled RNA, used as a competitor probe in some experiments, was synthesized by using the Ampliscribe kit (Epicentre) and following the manufacturer’s recommendations.
Sample collection, processing, and storage.
Following initial tests with cells of C. symbiosum, experiments were performed on formalin-fixed seawater samples collected on polycarbonate filters. For whole-cell hybridization with seawater samples, we modified the protocol of Glöckner et al. (19) by adjusting the hybridization conditions for polynucleotide probes. Seawater samples were collected with a rosette sampler at a series of offshore stations near Moss Landing, Calif. The sites sampled, total bottom depth, and miles offshore were, respectively, as follows: C1, 250 m, 2.6 miles; M1, 1,097 m, 10.8 miles; M2, 1,645 m, 26.5 miles; 67-90, 4,424 m, 177 miles. Seawater samples were fixed in 3.7% (wt/vol) formalin overnight at 4°C. A total of 3 to 10 ml of seawater (depending on the depth of origin) was filtered onto a 25-mm-diameter, 0.2-μm-pore-size polycarbonate GTTP filter (Millipore, Bedford, Mass.) under a vacuum of <5 mm Hg. The vacuum was released, and 1 ml of 2% (wt/vol) NaCl–50% (vol/vol) ethanol was placed on the filter. After a 1-min incubation, the ethanol solution was filtered through completely and the filters were dried. Filters were stored in petri slides (Millipore) at −20°C. This storage method was tested for periods of as long as 1 year with no apparent hybridization signal loss.
FISH.
Fluorescent hybridizations were performed in the inverted lids of 12-well polystyrene culture dishes. Filters were quartered with a razor blade, and the sections were placed face up on the inverted lid of a culture dish. Approximately 20 μl of preheated hybridization solution was placed on each filter, and 100 ng of hydrolyzed, fluor-labeled polynucleotide probe was subsequently added. The hybridization solution contained 50 to 70% (vol/vol) formamide (depending on the experiment; see below), 10% (wt/vol) dextran sulfate, 0.01% (wt/vol) poly(A), and 5× SET (1× SET is 150 mM NaCl, 1 mM Na2EDTA, 20 mM Tris-HCl, pH 7.8). A round 18-mm-diameter coverslip was placed over the filter, and the culture wells were placed over the lid to seal the individual hybridization mixtures. The whole assembly was then placed in a sealed container containing a small beaker with 5 ml of 5× SET to maintain humidity. Hybridization mixtures were incubated overnight in a hybridization oven (Robbins Scientific, Sunnyvale, Calif.) at the temperatures specified below. After overnight incubation, the filters were removed and placed in 5 ml of a wash solution containing 0.2× SET and 50% (vol/vol) formamide. The filters were washed for 2 h at the appropriate temperature (depending on the experiment; see below). The filters were subsequently incubated in 1× PBS (145 mM NaCl, 8.7 mM Na2HPO4, 1.5 mM NaH2PO4, pH 7.4) containing DAPI (4′,6-diamidino-2-phenylindole) at 1 μg/ml for 2 min at room temperature and quickly rinsed in 1× PBS. Filters were placed on slides and covered with approximately 10 μl of Citifluor solution (Citifluor, London, United Kingdom), and a coverslip.
A variety of conditions were tested to optimize the signal strength and specificity of each probe. The main variables tested were formamide concentration in the hybridization buffer, hybridization temperature, and wash temperature. Subsequent to these tests, the standard conditions for hybridization formamide concentration, hybridization temperature, and wash temperature indicated in Table 2 were employed. Following hybridizations, the standard wash buffer used for all of the probes consisted of 0.2× SET and 50% (vol/vol) formamide.
Filters were viewed immediately but could be used for several days after mounting when stored in the dark at 4°C. Filters were viewed with an Axiophot 2 microscope equipped with an HBO 100-W mercury lamp and a 100× Plan-APO objective (Zeiss, Thornwood, N.Y.). The following filter sets were used for fluorescence microscopy: fluorescein, a D480/30 exciter filter, a 505DCLP beam splitter, and a D535/40 barrier filter (Chroma Technology Corp., Brattleboro, Vt.); CY-3, an HQ545/30 exciter filter, a Q565LP beam splitter, and an HQ610/75 barrier filter (Chroma Technology Corp.); DAPI, a D360/40 exciter filter, a 400DCLP beam splitter, and a GG420 barrier filter (Chroma Technology Corp.). Images were captured with a Spot SP100 cooled digital color charge-coupled device camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.).
Total prokaryotic cell densities were estimated by DAPI staining and direct epifluorescence microscopic counting (22). To quantify probe-positive cells, five fields of approximately 50 to 150 cells per field were counted for each probe in each sample. A negative control probe (the complement of either the archaeal probe or the bacterial probe) was included for every sample and used to calculate the final probe-positive cell concentration. The fraction of probe-positive cells was calculated as the ratio of the number of probe-positive cells (archaeal group I, archaeal group II, or bacteria) to the total number of DAPI-stained cells after background subtraction of the control probe counts for each probe treatment. The cell densities of group I archaea, group II archaea, and bacteria were calculated by multiplying the fraction of probe-positive cells by the total prokaryotic cell concentration estimated from direct epifluorescence microscopic counts.
Quantitative rRNA blotting experiments.
Collection of picoplankton samples, nucleic acid extractions, and quantitative slot blotting experiments were performed as previously described (34, 35). The relative abundance of eucaryal, bacterial, and archaeal rRNAs in nucleic acid extracts was estimated by using oligonucleotide probe hybridization as previously described (34, 35). Nucleic acids were immobilized on nylon membranes (Hybond-N; Amersham) and then hybridized with 32P-labeled, rRNA-targeted oligonucleotide probes. The binding of each group-specific probe was quantified relative to that of a universal probe and normalized to the relative response of each probe to known standards.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been submitted to the GenBank database and assigned accession no. AF198456 and AF198457.
RESULTS
Cloning and characterization of rRNA genes and operons.
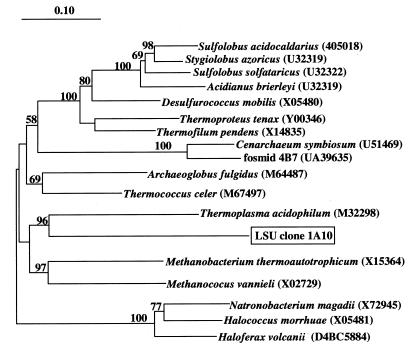
The archaeal 23S rRNA genes amplified from surface seawater were not related to group I archaeal 23S rRNA genes, consistent with previous observations of low group I archaeal abundance in surface waters (34). The archaeal 23S rRNA gene fragment on clone 1A10 was most closely affiliated with the Euryarchaota and specifically related to T. acidophilum (Fig. 1). This phylogenetic placement is nearly identical to that of group II archaeal 16S rRNA genes (11, 34), including the specific affiliation with T. acidophilum. To verify that the 23S rRNA gene contained on clone 1A10 was derived from a group II archaeon, we performed dual-hybridization experiments with a fluorescein-labeled group II 16S rRNA probe and a CY-3-labeled group II 23S rRNA probe (derived from clone 1A10; Table 2). The results showed that the same population of cells that bound the group II 16S probe also bound the 1A10-derived, 23S rRNA-targeted probe (Fig. 2). In combination, the ecological, phylogenetic, and FISH data provided strong evidence that the 23S rRNA gene contained on clone 1A10 was indeed derived from group II planktonic archaea. Consequently, we used 23S rRNA clone 1A10 to generate group II rRNA probes in subsequent experiments.
FIG. 1.
Phylogenetic position of the planktonic euryarchaeal 23S rRNA gene contained on clone G2lsu-1A10. The number at each bifurcation represents the percentage of 1,000 bootstrap resamplings that yielded the branching pattern appearing to the right of the value. The scale bar represents the estimated number of fixed mutations per nucleotide position.
FIG. 2.
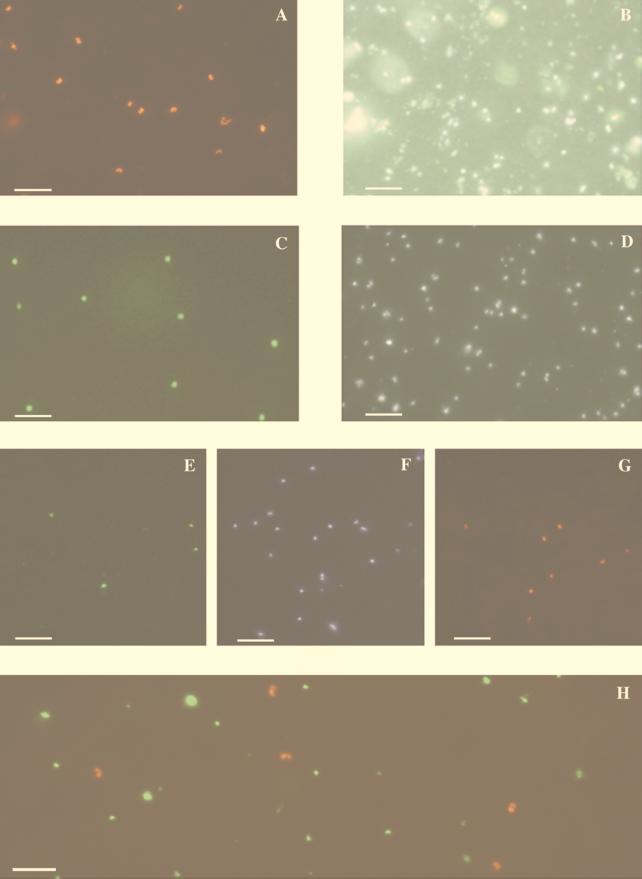
Surface seawater sample from Monterey Bay hybridized with a fluorescein-labeled 23S group II archaeal probe and a CY-3-labeled 16S group II archaeal probe (Table 2). Images of the same field were captured by using the fluorescein filter set (A), the CY-3 filter set (B), and the DAPI filter set (C). Bars, 5 μm.
Synthesis of rRNA-targeted, fluorescent polyribonucleotide probes.
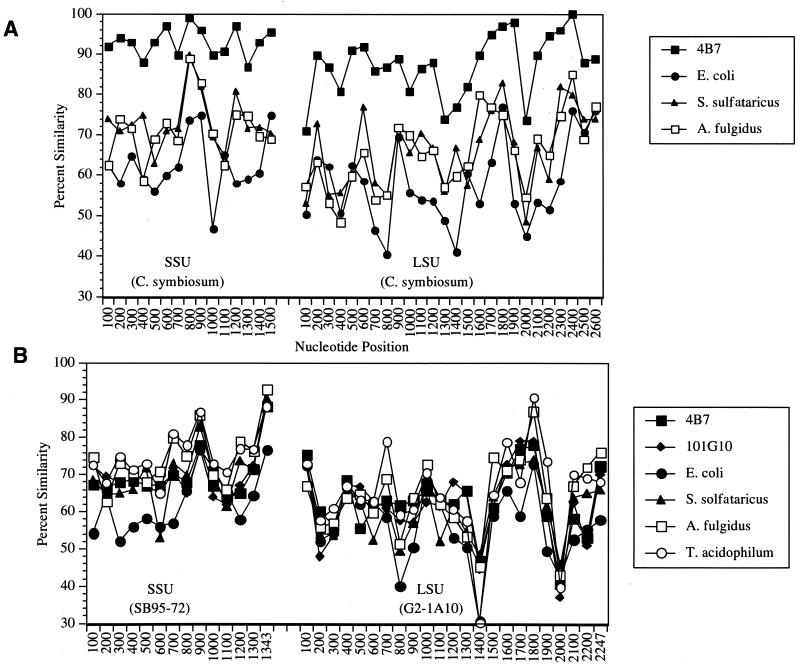
The large evolutionary distance separating group I and group II archaea from other archaeal and bacterial groups facilitated the application of polyribonucleotide probes for their identification. Plots of the unrestricted sequence similarity of templates used to generate the archaeal probes versus homologues from other archaeal and bacterial groups are shown in Fig. 3. The 23S and 16S rRNA genes of the group I archaeon C. symbiosum are highly similar to those of group 1 marine planktonic archaea, ≥90% over most 100-nucleotide segments (Fig. 3A). The similarity of C. symbiosum to other archaea and bacteria is much lower, on average, ≤75% over most regions of the 23S and 16S rRNAs. The large evolutionary distance separating group II archaea from other known archaea and bacteria is evident in Fig. 3B. The group II 16S rRNAs are ≤75% similar to those of other archaea and bacteria over most of the molecule. Group II 23S rRNAs are ≤70% similar, over most of the molecule, to other known and cultivated and uncultivated microorganisms.
FIG. 3.
Similarity plots comparing rRNA sequences of templates used to generate probes to homologous regions in other bacteria or archaea. Each data point represents the unrestricted sequence similarity value along a 100-nucleotide stretch. Nonoverlapping similarity values were calculated in 100-nucleotide sequence segments along the length of the 16S and 23S genes. (A) Group I archaea (C. symbiosum) rRNA compared to homologous regions of other bacteria and archaea. (B) Group II 16S (SB95-72) and 23S (G2lsu-1A10) rRNAs compared to homologous regions of other bacteria and archaea. The respective accession numbers of the small-subunit (SSU) and large-subunit (LSU) rRNA sequences used are as follows: 4B7, U39635 and AF198456; 101G10, AF083071 and AF083071; Escherichia coli, U00006 and U00006; Sulfolobus solfataricus, X03235 and U32322; Archaeoglobus fulgidus, X05567 and M64487; T. acidophilum, M38637 and M32298.
Transcription reactions incorporating fluorescein-12-CTP and fluorescein-12-UTP yielded the expected full-length products, in yields ranging from 5 to 10 μg of RNA per 100-μl transcription reaction mixture. Since the fluor-labeled RNA transcripts were greater than 1 kb in size, a hydrolysis step was employed to fragment them to an average size of ≤100 nucleotides. The hydrolysis step facilitated probe penetration into fixed cells and may also improve the uniformity and specificity of hybridization (8). Fragmentation of large transcripts to an average size of around 100 nucleotides has been reported to be essential for strong hybridization signals using FISH (6). In addition, 23S rRNA-targeted probes of >200 nucleotides used for FISH may incompletely penetrate cells, as indicated by a halo of fluorescence surrounding the periphery of cells hybridized with these large RNA probes (52).
Time series for transcript hydrolysis were performed to optimize conditions for generation of probe fragments with an average size of ≤100 nucleotides. It was important to monitor the reactions after a 5-min hydrolysis time to carefully control the average fragment size. When necessary, hydrolysis was resumed for an additional 2 to 5 min at 90°C. For 4.2-kb transcripts, 10 min of hydrolysis at 90°C was sufficient for the generation of fragments of ≤100 nucleotides. To fragment shorter full-length RNA transcripts, it was necessary to reduce the total hydrolysis time to between 5 and 7.5 min. By using this approach, it was possible to reproducibly generate fluor-labeled polyribonucleotide probes of approximately 100 nucleotides for use in FISH experiments. Small variations in the average size of the hydrolyzed transcripts did not noticeably affect their hybridization properties or the FISH signal.
Optimization of hybridization conditions for polyribonucleotide probes.
Initial experiments to determine optimal hybridization conditions for specificity and sensitivity were performed with fluorescein-labeled, group I-targeted polyribonucleotide probes. Formalin-fixed, macerated tissues from the marine sponge Axinella mexicana that contained large numbers of the archaeon C. symbiosum (40, 41) were used to initially assess hybridization conditions. The variables examined included formamide concentration in the hybridization buffer, hybridization temperature, and wash temperature. Comparison of the polynucleotide probe results to results obtained with 16S rRNA-targeted oligonucleotide probes verified the specificity of these probes for their intended targets (40). The group I probes clearly discriminated archaeal symbionts from contaminating bacteria in the sponge tissue and, at high stringencies, did not cross-react with formalin-fixed preparations of cultivated bacteria or archaea (data not shown; 40). The fluorescence intensity obtained with polyribonucleotide probes was much greater than that observed with singly labeled oligonucleotide probes. We estimate at least a 10- to 50-fold increase in sensitivity with the multiply labeled probes compared to that obtained with single-fluorochrome-labeled oligonucleotides (data not shown).
The group I probe bound a morphologically homogeneous population of cells in seawater samples. We could not detect the archaeal cells in the same seawater samples by using singly fluorescein-labeled or CY-3-labeled oligonucleotides and standard epifluorescence microscopy (data not shown). The group I archaeal shape and staining pattern (Fig. 4) were nearly identical to those of cells previously observed in Antarctic picoplankton (35) and C. symbiosum (41) visualized by using multiple oligonucleotide probes. The group I probe-binding cells displayed strong fluorescence at both poles, as well as an unstained central region, giving them a peanut-like shape (Fig. 4A, B, and H). The central portion, devoid of bound probe, corresponded to the DAPI-staining nucleoid region within these cells (41). Dual hybridization experiments demonstrated that group I archaeal, group II archaeal, and bacterial polynucleotide probes did not cross-react with the same cell population (Fig. 4H). CY-3-labeled and fluorescein-labeled group I polynucleotide probes applied to the same sample bound to a morphologically identical cell population and yielded identical group I cell densities (data not shown). We routinely used fluorescein-labeled negative control probes that comprised the reverse complement of the group I archaea or bacterial probes (Table 2). Background counts with these negative control probes were consistently low, typically representing <1% of the total epifluorescence direct counts.
FIG. 4.
Epifluorescence micrographs of picoplankton visualized with polynucleotide probes and FISH on polycarbonate filters. (A and B) Seawater sample, collected from a 200-m depth at a nearshore station in Monterey Bay, hybridized with the CY-3-labeled group I probe and viewed by using the CY-3 (A) or the DAPI (B) filter set. (C and D) Surface seawater sample, from a nearshore station in Monterey Bay, hybridized with the fluorescein-labeled 23S rRNA group II probe and viewed with the fluorescein (C) or the DAPI (D) filter set. (E to G) A 100-m sample, from an offshore station in Monterey Bay, dually hybridized with the fluorescein-labeled 23S group II probe and the CY-3-labeled group I probe. The sample was viewed with the fluorescein (E), DAPI (F), and CY-3 (G) filter sets. (H) Seawater sampled at an 80-m depth at 177 miles offshore of Moss Landing, Calif. The sample was dually hybridized with the fluorescein-labeled bacterial probe and the CY-3-labeled group I probe. Images were captured independently by using the fluorescein or CY-3 filter set, and the separate images were overlaid in Adobe PhotoShop. Scale bars, 5 μm.
Cells that bound the group II probe had a different morphology than group I cells, generally a coccoid shape that uniformly stained with the polynucleotide probe throughout the entire cell (Fig. 4C and D). Dual hybridization with a fluorescein-labeled group II 23S probe and a CY-3-labeled group I probe showed two discrete cell populations with no cross-hybridization between them (Fig. 4E, F, and G). The group II cells had a characteristic DAPI staining compared to other cell types, with the DAPI-staining region appearing more diffuse and weakly stained than in group I archaea or bacteria. This phenomenon may be an artifact of the fixation or hybridization treatment but might also reflect specific characteristics of group II archaeal cell walls or cytoplasm. There was generally no cross-reaction between the group I, group II, and eubacterial probes—each recognized a discrete and unique cell population (Fig. 4E to H). In rare cases, a weak cross-reaction between eubacteria and the group II probe was observed. This cross-reactivity could be eliminated by the inclusion of unlabeled bacterial polyribonucleotide probe in the fluorescein-labeled group II probe hybridization mixtures (data not shown).
Estimation of group I and II archaeal cell densities in ocean waters.
The FISH assays were highly reproducible. The mean and standard error for replicate experiments, performed at a variety of depths at station M2, are shown in Fig. 5. We also checked reproducibility by routinely performing dual hybridizations with a CY-3-labeled group I probe, included in the fluorescein-labeled group II hybridizations. Dual hybridizations using the CY-3-labeled group I archaeal probe consistently yielded group I cell densities that were identical to those obtained with hybridizations using the fluorescein-labeled group I probe alone. To independently assess the reliability of the FISH method, cell densities of group I and II archaea determined by FISH were compared with the relative rRNA abundance derived from radiolabeled oligonucleotide probe hybridization experiments (Fig. 6). In general, the FISH results and the estimates of relative rRNA abundance were in good agreement. As a further indicator of reliability, the sum of archaeal group I, group II, and bacterial cells determined by FISH generally accounted for the majority of the DAPI-stained cells in seawater samples (Table 3). In most samples, 90% or more of the total DAPI-staining cells were accounted for by the combined total derived from the three probes (Table 3).
FIG. 5.
Group I archaea and bacterial cell concentrations at various depths, determined by polyribonucleotide probe hybridization and FISH and performed in triplicate. Error bars represent standard errors, and where not visible, they are smaller than the symbols. Methods are described in the text.
FIG. 6.
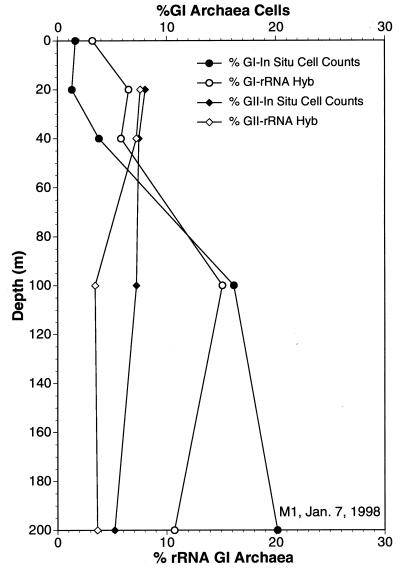
Cell densities of group I and II archaea determined by polyribonucleotide probe hybridization (Hyb) and FISH, compared to the percentage of rRNA from each group in the same sample estimated by quantitative oligonucleotide probe hybridization. Methods are described in the text.
TABLE 3.
Percentages of total epifluorescent cell counts enumerated by FISH probes
| Sample | Depth (m) | Cell density (104/ml)
|
% of total cells via FISH | |||
|---|---|---|---|---|---|---|
| Total counts (DAPI) | FISH group I | FISH group II | FISH bacteria | |||
| 34498 M2 | 0 | 147 | 10.1 | 18.7 | 113 | 96.6 |
| 34498 M2 | 10 | 120 | 6.9 | 13.2 | 98.8 | 98.9 |
| 34498 M2 | 20 | 143 | 7.0 | 13.8 | 107 | 89.2 |
| 34498 M2 | 30 | 149 | 7.8 | 16.7 | 132 | 105 |
| 34498 M2 | 40 | 140 | 8.6 | 14.6 | 112 | 96.6 |
| 34498 M2 | 60 | 95.5 | 11.7 | 7.5 | 73.2 | 96.8 |
| 34498 M2 | 80 | 53.9 | 11.6 | 7.9 | 36.4 | 104 |
| 34498 M2 | 100 | 45.6 | 10.6 | 4.9 | 31.6 | 103 |
| 34498 M2 | 150 | 42.0 | 7.9 | 3.9 | 31.3 | 102 |
| 34498 M2 | 200 | 34.0 | 6.7 | 2.2 | 24.0 | 96.7 |
| 34498 M2 | 500 | 18.6 | 4.3 | 1.7 | 14.2 | 108 |
| 34498 M2 | 1,000 | 13.8 | 4.4 | 1.8 | 3.9 | 73.3 |
| 18198 M2 | 0 | 131 | 1.8 | 10.8 | 111 | 95.1 |
| 18198 M2 | 20 | 132 | 0.8 | 14.7 | 128 | 108 |
| 18198 M2 | 40 | 98.1 | 8.2 | 10.7 | 74.7 | 95.4 |
| 18198 M2 | 60 | 78.1 | 17.4 | 9.1 | 50.5 | 98.6 |
| 18198 M2 | 80 | 61.4 | 15.1 | 4.5 | 30.0 | 80.8 |
| 18198 M2 | 100 | 57.3 | 11.6 | 2.9 | 39.0 | 93.3 |
| 18198 M2 | 150 | 47.6 | 12.4 | 2.1 | 22.0 | 76.7 |
| 18198 M2 | 200 | 37.2 | 9.9 | 0.9 | 22.1 | 88.5 |
| 18198 M2 | 500 | 18.6 | 5.6 | 0.9 | 8.8 | 82.1 |
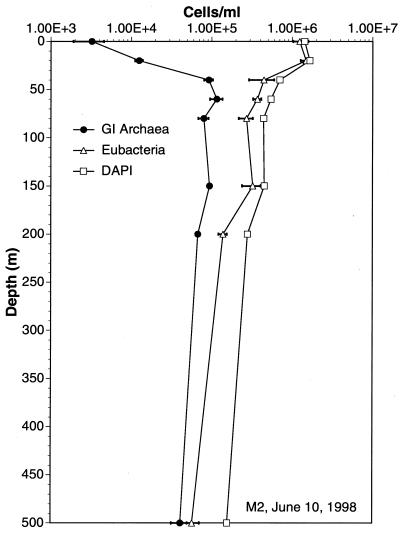
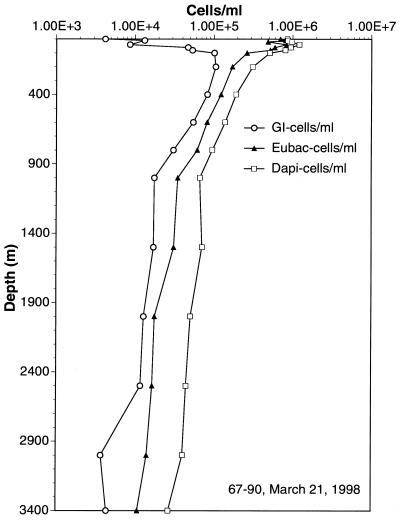
Group I archaeal cell numbers were generally lower in surface waters and increased with depth as total prokaryotic cell numbers decreased. At greater depths, group I archaeal cell densities were quite high, approximately 105/ml (Fig. 5 and 6 and Table 3). In most samples, group I archaea in Monterey Bay reached maximal cell densities at depths of ≥60 m (Fig. 5 and 6), a distribution strikingly similar to that observed in previous studies of rRNA abundances in the Santa Barbara Channel (34). Also in agreement with previous studies of rRNA relative abundance, group II archaeal cell numbers exceeded those of group I archaea at depths of less than 40 m (Fig. 6; Table 3). The polynucleotide probes could be used successfully even at great depths (Fig. 7; Table 3). Group I archaeal abundances were continuously high below 80 m, and these archaea made up a substantial fraction of the total cell population at 3,400 m. Group I and II archaeal cells were generally smaller and dimmer at great depths than those found at the depth with the cell density maximum (data not shown).
FIG. 7.
Densities of group I archaea and bacterial cells at various depths, determined by polyribonucleotide probe hybridization and FISH. The sampling site was 177 miles offshore of Moss Landing, Calif. Methods are described in the text. Eubac, eubacterial.
DISCUSSION
In this study, we modified and extended the use of multiply labeled polyribonucleotide probes to identify and enumerate uncultivated planktonic archaeal groups. After optimization of probe synthesis and hybridization, the technique proved much more sensitive than FISH using single oligonucleotides, consistent with previous findings. The inclusion of a hydrolysis step after probe syntheses resulted in efficient probe penetration into bacterial and archaeal cells. Dual-staining approaches using probes labeled with different fluorochromes allowed simultaneous identification of different target groups and provided better assessment of the binding specificity of each probe. Negative control probes consistently yielded few or no fluorescently labeled cells, further indicating the high signal-to-noise ratio obtained with the approach. As previously reported for oligonucleotides and FISH (19), hybridization on polycarbonate filters was efficient and this approach facilitated rapid processing and quantitative analysis of multiple aquatic samples. In seawater samples, target archaeal groups that could not be reliably visualized or quantified with oligonucleotide probes were easily and routinely visualized and quantified with the polyribonucleotide probes.
The approach described here differs from those used in previous studies in several respects. Unlike in previous studies (49, 52), we used large rRNA sequence tracts to generate specific probes. This was possible because the groups to be distinguished (group I archaea, group II archaea, and bacteria) are so evolutionarily distant from one another. Polyribonucleotide probes with greater specificity could easily be generated by targeting more variable regions, particularly in 23S rRNA (49, 52). Unlike some previous studies, ours also used a hydrolytic step to generate shorter probe fragments from long transcripts. When using this protocol, it is important to carefully control the hydrolysis of the probe after initial synthesis. If the probes are hydrolyzed for too long, they could be reduced to the size of short oligonucleotides, altering their binding specificity or rendering them incapable of binding under the stringency conditions used.
We are currently applying these polynucleotide probes in ecological studies of marine archaeal and bacterial spatial and temporal variability. These data provide a much higher degree of reliability and resolution than is possible by using quantitative rRNA blots, fluorescent oligonucleotides and FISH, or PCR experiments. Preliminary results suggest that high archaeal cell densities are a common feature of the world’s oceans. If this is true, then archaea appear to represent a very large and significant fraction of the picoplankton biomass in the world’s oceans.
Polynucleotide probes targeting planktonic bacteria consistently yielded a high signal intensity for a wide variety of morphotypes that accounted for the majority of cells enumerated by epifluorescence direct counts in seawater. Significantly, the bacterial and archaeal probe-positive cells accounted for a majority percentage (>90%) of the total cells enumerated by DAPI staining. Even in deep seawater samples, most of the DAPI-stained cells appear to contain significant amounts of rRNA readily detectable by the polyribonucleotide probe and FISH protocol reported here. These results suggest that the majority of the marine picoplankton visualized by DAPI staining does not represent lysed cells or ghosts devoid of nucleic acids, as was suggested in a previous report (58). Our results strongly suggest that the majority of marine picoplankton cells visualized by epifluorescence direct counts are intact and ribosome replete and so represent viable and, quite possibly, physiologically active cells.
ACKNOWLEDGMENTS
This work was supported by a grant to MBARI from the David and Lucille Packard Foundation and NSF grant OCE95-29804 to E.F.D.
We thank Oded Beja, Marcelino Suzuki, Victoria Orphan, Grieg Steward, and Christa Schleper for advice, suggestions, and encouragement. We also thank the officers and crew of the Point Sur and Point Lobos for able assistance.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Gloeckner F O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Springer N, Ludwig W, Gortz H D, Schleifer K H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann R I, Zarda B, Stahl D A, Schleifer K H. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1992;58:3007–3011. doi: 10.1128/aem.58.9.3007-3011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauman J G, Bentvelzen P. Flow cytometric detection of ribosomal RNA in suspended cells by fluorescent in situ hybridization. Cytometry. 1988;9:517–524. doi: 10.1002/cyto.990090602. [DOI] [PubMed] [Google Scholar]
- 7.Cary S C, Giovannoni S J. Transovarial inheritance of endosymbiotic bacteria in clams inhabiting deep-sea hydrothermal vents and cold seeps. Proc Natl Acad Sci USA. 1993;90:5695–5699. doi: 10.1073/pnas.90.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 9.Christensen H, Hansen M, Sorensen J. Counting and size classification of active soil bacteria by fluorescence In situ hybridization with an rRNA oligonucleotide probe. Appl Environ Microbiol. 1999;65:1753–1761. doi: 10.1128/aem.65.4.1753-1761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 11.DeLong E F, Wu K Y, Prezelin B B, Jovine R V. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 12.Distel D L, DeLong E F, Waterbury J B. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl Environ Microbiol. 1991;57:2376–2382. doi: 10.1128/aem.57.8.2376-2382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubilier N, Giere O, Distel D L, Cavanaugh C M. Characterization of chemoautotrophic bacterial symbionts in a gutless marine worm Oligochaeta, Annelida) by phylogenetic 16S rRNA sequence analysis and in situ hybridization. Appl Environ Microbiol. 1995;61:2346–2350. doi: 10.1128/aem.61.6.2346-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Embley T M, Finlay B J, Thomas R H, Dyal P L. The use of rRNA sequences and fluorescent probes to investigate the phylogenetic positions of the anaerobic ciliate Metopus palaeformis and its archaeobacterial endosymbiont. J Gen Microbiol. 1992;138:1479–1487. doi: 10.1099/00221287-138-7-1479. [DOI] [PubMed] [Google Scholar]
- 15.Fenchel T, Ramsing N B. Identification of sulphate-reducing ectosymbiotic bacteria from anaerobic ciliates using 16S rRNA binding oligonucleotide probes. Arch Microbiol. 1992;158:394–397. doi: 10.1007/BF00276298. [DOI] [PubMed] [Google Scholar]
- 16.Frischer M E, Floriani P J, Nierzwicki-Bauer S A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can J Microbiol. 1996;42:1061–1071. doi: 10.1139/m96-136. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 20.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karner M, Fuhrman J A. Determination of active marine bacterioplankton—a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemp P F, Lee S, LaRoche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee N, Nielsen P H, Andreasen K H, Juretschko S, Nielsen J L, Schleifer K H, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S, Kemp P F. Single-cell RNA content of natural marine planktonic bacteria measured by hybridization with multiple 16S rRNA-targeted fluorescent probes. Limnol Oceanogr. 1994;39:869–879. [Google Scholar]
- 27.Lee S, Malone C, Kemp P F. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar Ecol Prog Ser. 1993;101:193–201. [Google Scholar]
- 28.Lim E L, Amaral L A, Caron D A, DeLong E F. Application of rRNA-based probes for observing marine nanoplanktonic protists. Appl Environ Microbiol. 1993;59:1647–1655. doi: 10.1128/aem.59.5.1647-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim E L, Caron D A, Delong E F. Development and field application of a quantitative method for examining natural assemblages of protists with oligonucleotide probes. Appl Environ Microbiol. 1996;62:1416–1423. doi: 10.1128/aem.62.4.1416-1423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim E L, Dennett M R, Mark R, Caron D A. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol Oceanogr. 1999;44:37–51. [Google Scholar]
- 31.Llobet-Brossa E, Rossell-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig W, Dorn S, Springer N, Kirchhof G, Schleifer K H. PCR-based preparation of 23S rRNA-targeted group-specific polynucleotide probes. Appl Environ Microbiol. 1994;60:3234–3244. doi: 10.1128/aem.60.9.3236-3244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manz W, Szewzyk U, Ericsson P, Amann R, Schleifer K H, Stenstrom T A. In situ identification of bacteria in drinking water and adjoining biofilms by hybridization with 16S and 23S rRNA-directed fluorescent oligonucleotide probes. Appl Environ Microbiol. 1993;59:2293–2298. doi: 10.1128/aem.59.7.2293-2298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray A E, Preston C M, Massana R, Taylor L T, Blakis A, Wu K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2895. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouverney C C, Fuhrman J A. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl Environ Microbiol. 1999;65:1746–1752. doi: 10.1128/aem.65.4.1746-1752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pernthaler J, Glöckner F O, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polz M F, Distel D L, Zarda B, Amann R, Felbeck H, Ott J A, Cavanaugh C M. Phylogenetic analysis of a highly specific association between ectosymbiotic, sulfur-oxidizing bacteria and a marine nematode. Appl Environ Microbiol. 1994;60:4461–4467. doi: 10.1128/aem.60.12.4461-4467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston C M. Ph.D. thesis. Santa Barbara: University of California; 1998. [Google Scholar]
- 41.Preston C M, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsing N B, Kühl M, Jørgensen B B. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schleper C, DeLong E F, Preston C M, Feldman R A, Wu K Y, Swanson R V. Genomic analysis reveals chromosomal variation in natural populations of the uncultured psychrophilic archaeon Cenarchaeum symbiosum. J Bacteriol. 1998;180:5003–5009. doi: 10.1128/jb.180.19.5003-5009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schonhuber W, Fuchs B, Juretschko S, Amann R. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schramm A, Larsen L H, Revsbech N P, Amann R I. Structure and function of a nitrifying biofilm as determined by microelectrodes and fluorescent oligonucleotide probes. Water Sci Technol. 1997;36:263–270. [Google Scholar]
- 46.Schramm A, Larsen L H, Revsbech N P, Ramsing N B, Amann R, Schleifer K H. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon N, LeBot N, Marie D, Partensky F, Vaulot D. Fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes to identify small phytoplankton by flow cytometry. Appl Environ Microbiol. 1995;61:2506–2513. doi: 10.1128/aem.61.7.2506-2513.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spring S, Lins U, Amann R, Schleifer K H, Ferreira L C, Esquivel D M, Farina M. Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch Microbiol. 1998;169:136–147. doi: 10.1007/s002030050553. [DOI] [PubMed] [Google Scholar]
- 50.Stein J L, Marsh T L, Wu K Y, Shizuya T, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckman N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. [Online.] Munich, Germany: Department of Microbiology, Technische Universität München; 1998. http://www.mikro.biologie.tu-muenchen.de . [10 February 1999, last date accessed.] [Google Scholar]
- 52.Trebesius K, Amann R I, Ludwig W, Mulegger K, Schleifer K H. Identification of whole fixed bacterial cells with nonradiactive 23S rRNA-targeted polynucleotide probes. Appl Environ Microbiol. 1994;60:3228–3235. doi: 10.1128/aem.60.9.3228-3235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner M, Amann R, Lemmer H, Schleifer K H. Probing activated sludge with oligonucleotides specific for proteobacteria—inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner M, Assmus B, Hartmann A, Hutzler P, Amann R. In situ analysis of microbial consortia in activated sludge using fluorescently labeled, rRNA-targeted oligonucleotide probes and confocal scanning laser microscopy. J Microsc. 1994;176:181–187. doi: 10.1111/j.1365-2818.1994.tb03513.x. [DOI] [PubMed] [Google Scholar]
- 55.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 56.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarda B, Amann R, Wallner G, Schleifer K H. Identification of single bacterial cells using digoxigenin-labeled, rRNA-targeted oligonucleotides. J Gen Microbiol. 1991;137:2823–2830. doi: 10.1099/00221287-137-12-2823. [DOI] [PubMed] [Google Scholar]
- 58.Zweifel U L, Hagström Å. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Appl Environ Microbiol. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]