Abstract
Nanolipoprotein particles (NLPs), also called “nanodiscs”, are discoidal particles with a patch of lipid bilayer corralled by apolipoproteins. NLPs have long been of interest due to both their utility as membrane-model systems into which membrane proteins can be inserted and solubilized and their physiological role in lipid and cholesterol transport via HDL and LDL maturation, which are important for human health. Serial femtosecond crystallography (SFX) at X-ray free electron lasers (XFELs) is a powerful approach for structural biology of membrane proteins, which are traditionally difficult to crystallize as large single crystals capable of producing high-quality diffraction suitable for structure determination. To facilitate understanding of the specific role of two apolipoprotein/lipid complexes, ApoA1 and ApoE4, in lipid binding and HDL/LDL particle maturation dynamics and develop new SFX methods involving NLP membrane protein encapsulation, we have prepared and crystallized homogeneous populations of ApoA1 and ApoE4 NLPs. Crystallization of empty NLPs yields semi-ordered objects that appear crystalline and give highly anisotropic and diffuse X-ray diffraction, similar in characteristics to fiber diffraction. Several unit cell parameters were approximately determined for both NLPs from these measurements. Thus, low-background, sample conservative methods of delivery are critical. Here we implemented a fixed target sample delivery scheme utilizing the Roadrunner fast-scanning system and ultra-thin polymer/graphene support films, providing a low-volume, low-background approach to membrane protein SFX. This study represents initial steps in obtaining structural information for ApoA1 and ApoE4 NLPs and developing this system as a supporting scaffold for future structural studies of membrane proteins crystalized in a native lipid environment.
Keywords: lipoprotein, nanodisc, serial femtosecond crystallography, XFELs, fixed target delivery
1. Introduction
The Nanolipoprotein particles (NLPs), or “nanodiscs”, are nanoscale complexes with apolipoproteins forming a corralled, discoidal bilayer.1 There are many biotechnological applications of NLPs, including the use of NLPs for structural characterization of transmembrane proteins, which have been historically intractable to X-ray crystallography and other structural techniques due to their low solubility and tendency to aggregate. Using membrane mimetics that solubilize and facilitate functional studies of membrane proteins via X-ray scattering, CryoEM, or X-ray crystallography has been extensively demonstrated by us and others.2–15 Discoidal lipoproteins, which are closely related to NLPs, represent a critical physiological transition state from lipid-free apolipoprotein to spherical HDL and LDL particles during HDL/LDL maturation, and thus play a role in the risk of cardiovascular disease, protection against atherosclerosis, and amyloid related diseases due to their key role in reverse cholesterol transport (RCT).16, 17
Interaction between nascent HDL/LDL discs and lecithin cholesterol acyltransferase (LCAT) leads to conversion of cholesterol to cholesteryl ester and apolipoprotein mediated formation of the spherical particles.18–23 Current structural models of the disc suggest an anti-parallel apolipoprotein chain orientation surrounding the lipid bilayer patch that can adjust to accommodate a number of discrete particle diameters based on the protein to lipid ratio. Computational modeling has been performed in the presence of phospholipids24 and several NMR solution structures exist.25, 26,27 The apolipoprotein-mediated lipid binding properties of HDL/LDL and the importance of specific sub-populations of nascent HDL implies control by the apolipoprotein over the maturation process.28, 29 Therefore, a mechanistic understanding of HDL/LDL maturation and its role in disease states requires an understanding of how proteins involved in HDL maturation, such as LCAT, interact with the disc and of how apolipoprotein-lipid and apolipoprotein-cholesterol/ cholesteryl ester interaction controls final particle size. We have optimized protocols to generate homogenous preparations of NLPs1, 24, 30 for structural studies confirmed by small angle x-ray scattering (SAXS) and small angle neutron scattering (SANS).10, 31, 32
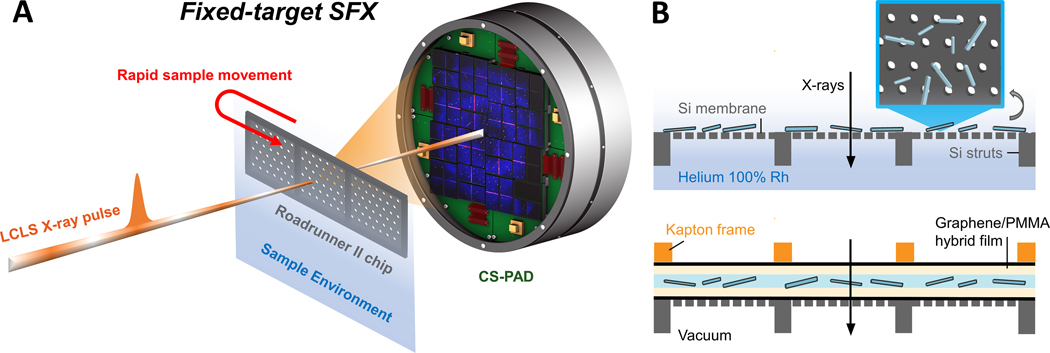
Several X-ray crystallographic studies of lipid-bound apolipoproteins at low resolution have been published that contain diffuse features or weak diffraction and report a high degree of susceptibility to radiation damage even at cryogenic temperature.33, 34 Serial femtosecond crystallography (SFX) combined with low-background fixed-target sample delivery (Figure 1) is well suited to mitigate fast crystal decay due to radiation damage. The ultrashort, high brightness pulses from XFELs, such as the Linac Coherent Light Source (LCLS), can capture damage-free,35 single-pulse diffraction images from biological micro- and nano-objects at room temperature, including protein microcrystals36, 37 and weakly diffracting objects like virus particles,38, 39 2-dimensional protein crystals,40–42 or protein fibrils.43 Using the SFX approach, high-resolution macromolecular structures can be derived from thousands of individual diffraction patterns obtained sequentially from continuously replenished microcrystals.44–46 A fixed target sample delivery approach can drastically reduce the amount of sample required for experiments limited by protein or crystallization yield by minimizing crystal loss to, for example, jet flow, while enabling the measurement of samples with heterogeneous mixtures of crystal size. The “Roadrunner” fast scanning fixed target system developed by the Centre for Free-Electron Laser Science (CFEL) utilizes fast stages synchronized with the X-ray pulse arrival time such that X-rays shots align with the pores of a micropatterned silicon chip47 at the 120 Hz repetition rate of LCLS.48 To enable fixed target SFX in-vacuum, we have recently demonstrated the use of large-area few-layer graphene (FLG) in conjunction with polymer thin-films, as enclosing layers to maintain sample hydration for room temperature studies, while imparting mechanical robustness and only minimally adding to the X-ray scatter background.49
Figure 1.
Scheme illustrating A) SFX data collection and the fixed target sample delivery approach using B) crystal deposition onto a second-generation Roadrunner II micropatterned Si chips. Cross sections (not to scale) of chips used at MFX at near 100% relative humidity (above) and the Graphene/PMMA enclosed chips used at CXI in vacuum (below) are shown.
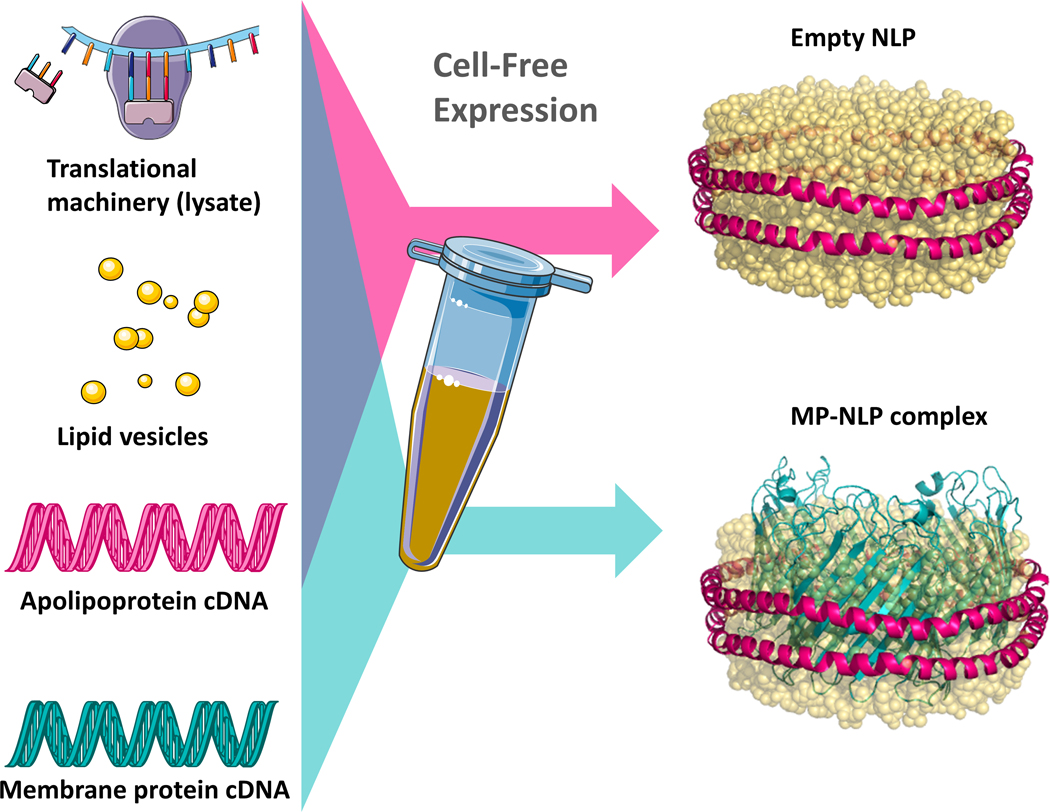
To understand the structure and function of lipoprotein particles and NLPs, we have optimized crystallization protocols for multiple types of lipid-bound apolipoproteins (ApoA1 and ApoE4) to be subject to SFX at LCLS. Our lipid-bound apolipoproteins are produced via cell-free expression (Figure 2). Cell free expression is an optimal route to assembly of NLPs and scaffold supported membrane proteins of interest due to its open architecture and elimination of the need for detergent solubilization or extensive purification.50–53 This provides a path forward for understanding the formation of mature spherical forms of HDL/LDL particles and constitutes a critical step in validating membrane protein-NLP complexes as a platform for structural studies.
Figure 2.
Cell-free expression and self-assembly of empty NLPs (pink arrow) and NLPs with embedded transmembrane proteins (teal arrow).
2. Materials and Methods
2.1. Cell-free expression, preparation, and characterization of ApoA1 and ApoE4 nanolipoprotein particles
Preparative 1 ml reactions were carried out using the Rabbit Biotech RTS 500 ProteoMaster E. coli HY Kit for protein expression and purification as previously described.32 In brief, lyophilized reaction components (E. coli lysate, reaction mixture, amino acid mixture, and methionine) were dissolved in reconstitution buffer and combined as specified by the manufacturer. A total of 1 μg of plasmid DNA, either the truncated forms of apolipoprotein A-I (Δ1–49), or apolipoprotein E-4 (22k), was added to the lysate mixture along with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) vesicles. Small unilamellar vesicles (SUV) below 100 nm in size of DMPC were prepared by probe-sonicating a 20 mg/ml aqueous solution of DMPC until optical clarity was achieved. The SUVs were added to the cell-free reaction at a concentration of 2 mg/ml. The reactions were incubated at 37 °C overnight for approximately 18 hrs. Immobilized nickel affinity chromatography was used to isolate the His-tagged apolipoproteins according to the manufacturer’s protocol (Roche Molecular Diagnostics). Elution fractions were assayed with SDS-PAGE and peak elution fractions containing the protein of interest were pooled and buffer-exchanged overnight into 20 mM ammonium bicarbonate pH 7.0 using a 10 kDa molecular weight sieve. The pooled protein was then concentrated with 10 kDa molecular weight cutoff spin concentrators to 5 mg/ml in preparation for crystallization experiments and stored in 20 mM ammonium bicarbonate pH 7.0. Dynamic Light Scattering (DLS) measurements were performed on diluted samples of the ApoA1 and ApoE4 discs with a Zetasizer Nano Particle Size Analyzer (Malvern Panalytical).
2.2. High-throughput crystallization screening
ApoA1 and ApoE4 NLP solutions at 5 mg/ml protein concentration in 20 mM ammonium bicarbonate were submitted to the high throughput crystallization screen at the High-Throughput Screening Laboratory at the Hauptman-Woodward Medical Research Institute in Buffalo, New York. This facility makes use of automated liquid handling and imaging systems coordinated through remote-accessible database to quickly set up and record the outcomes of 1,536 unique microbatch-under-oil crystallization screening experiments per submission. 400 nl crystallization drops were dispensed under paraffin oil and monitored over six weeks via optical imaging. All wells were imaged with Ultraviolet Two-Photon Excited Fluorescence (UV-TPEF) to identify biological crystalline objects via tryptophan fluorescence and second-order nonlinear optical imaging of chiral crystals (SONICC) at 28 days.
2.3. Preparation of NLP crystals for SFX experiments
Selected crystal hits identified in the high throughput screen (Table S1) were independently reproduced in microbatch-under-oil experiments (Table 1). NLP solutions concentrated to 5 mg/ml in protein were mixed in 1 μl quantities with several volume ratios of each cocktail (typically 2:1, 1:1, and 1:2) in Terasaki-style microbatch trays and covered with a thin layer of paraffin oil. Wells were incubated at 21°C and monitored for crystal growth with optical microscopy over a four-week period. Several crystallization conditions showing apparent crystal growth were selected and optimized to maximize crystal number, density, and uniformity for SFX experiments. Crystals for SFX experiments were grown in a large number of replicate wells in microbatch trays under paraffin oil using crystallization cocktails summarized in Table 1, harvested from individual wells, and pooled to yield at least a 50 μl volume of crystal slurry for deposition onto the micropatterned chips. For conditions with low crystal density, the slurry was concentrated two-fold with gentle spin centrifugation.
Table 1.
Compositional characteristics, crystal sizes, and approximate crystal densities for crystallization conditions reproduced at LLNL and chosen to optimize for SFX experiments to maximize reproducibility, crystal uniformity, and crystal density for ApoA1 and ApoE4. Boxed conditions were measured in SFX experiments at LCLS.
| Cocktail number | Buffer concentration and pH | PEG precipitant concentration | Salt additive concentration | Length range, μm (Average) | Estimated density, #crystals/μl | |
|---|---|---|---|---|---|---|
| ApoE4 NLPs | M1328 | 0.1M Tris pH 8.5 | 22.6% w/v PEG 4000 | 0.2M Lithium Sulfate monohydrate | 50–140 (85) | 73 |
|
| ||||||
| M318 | 0.1M Sodium Citrate pH 5.6 | 16% w/v PEG 4000 | 0.1M Ammonium Phosphate-dibasic | 50–150 (70) | 17.5 | |
| M548 | 0.2M Tris pH 8.5 | 35% w/v PEG 4000 | 0.2M Lithium Sulfate monohydrate | 25–50 (42) | 8 | |
| M750 | 0.1M Tris pH 8.5 | 40% w/v PEG 1000 | 0.2M Lithium Sulfate monohydrate | |||
| M752 | 0.1M Tris pH 8.5 | 37.5% w/v PEG 1000 | 0.3M Lithium Sulfate monohydrate | |||
|
| ||||||
| S1022 | 0.1M Sodium Citrate pH 5.5 | 20% w/v PEG 1000 | 0.1M Lithium sulfate monohydrate | 40–130 (52) | 74 | |
|
| ||||||
| S1438 | --- | 22.6% w/v PEG 4000 | 0.2M Sodium Citrate tribasic dihydrate | |||
| ApoA1 NLPs | S714 | 0.1M Tris pH 8.0 | 20% w/v PEG 1000 | 0.1M Magnesium chloride-hexahydrate | ||
| S789 | 0.1M CAPS, pH 10 | 20% w/v PEG 1000 | 0.1M Lithium chloride | |||
| S816 | 0.1M CAPS pH 10 | 20% w/v PEG 1000 | 0.1M Sodium bromide | 80–200 (120) | 84 | |
| S816–2 | 0.1M CAPS pH 10 | 25% w/v PEG 1000 | 0.1M Sodium bromide | 20–140 (58) | 229 | |
| S816–3 | 0.1M CAPS pH 10 | 30% w/v PEG 1000 | 0.1M Sodium bromide | 30–100 (62) | 198 | |
|
| ||||||
| S816–4 | 0.1M CAPS pH 10 | 35% w/v PEG 1000 | 0.1M Sodium bromide | 20–160 (75) | 92 | |
|
| ||||||
| S831 | 0.1M CAPS pH 10 | 20% w/v PEG 1000 | 0.1M Lithium sulfate-monohydrate | |||
2.4. Fixed target preparation and sample deposition
SFX experiments took place over two 6-hour Protein Crystal Screening beamtimes at the MFX and CXI endstations at LCLS using micropatterned silicon-based fixed targets for sample delivery (Figure 1). Micro-patterned single crystalline silicon chips were prepared at CFEL or commercially manufactured by Finnlitho (Joensuu, Finland) following the design principles of second generation Roadrunner chips described previously.54 Each chip (32.7 mm x 12 mm) used at MFX was comprised of a 200 μm thick frame with three 10 mm x 10 mm square areas of Si “windows” thinned to 10 μm and supported by 100 μm wide struts in between the windows (Figure S1). These windows were patterned further with a hexagonal dense pattern of 15 μm pores spaced 100 μm apart, yielding >40,000 pores per chip that could hold crystal samples. Chips used at CXI had a similar design but had a slightly different window configuration and pore spacing (18 × 5 array of 1.5 mm x 1.5 mm windows with 50 μm pore spacing).
Experiments were conducted in humidified atmosphere, Roadrunner III system at MFX, or in vacuum using the vacuum-compatible Roadrunner IV system in the 0.1 μm in vacuo sample environment of CXI, necessitating two approaches to chip preparation and sample deposition. For humidified environment experiments at MFX, Finnlitho chips were loaded with 50 μl of freshly crystallized NLP microcrystal suspension by pipetting and spreading the crystal slurry onto the flat side of the chip and wicking away excess mother liquor from the opposite side to aid in drawing microcrystals into the chip pores. The loaded chips were then immediately transferred to the Roadrunner sample chamber flushed with humidified helium (near 100 % humidity).
Preparation of the graphene-polymer film “sandwich” used to preserve sample hydration in vacuum at CXI was performed as described in Shelby and Gilbile et. al., 2020.49 A silicon chip was covered on the flat side by a film comprised of few-layer graphene (FLG) and 40 nm thick polymethyl methacrylate (PMMA) and approximately 20 μl of a two-fold concentrated S816–4 ApoA1 microcrystal slurry was carefully pipetted onto the film. A Kapton frame with a secondary PMMA-FLG film was then carefully aligned with the chip and placed on top of the microcrystal solution to aid in solution spread over the chip area by capillary action. The edges of the sandwiched sample were sealed by application of a thin layer of vacuum grease to prevent dehydration. Chips prepared with ApoA1 NLP microcrystals were tested for hydration retention in a vacuum chamber at room temperature prior to the SFX experiment. Figure S2 shows intact crystals after 30 minutes in the vacuum chamber.
2.5. SFX experiments
SFX experiments were conducted at an X-ray energy of 7.5 keV and 9.5 keV with a beam size at the sample of 120 nm x 170 nm full width half maximum (FWHM) and 3 μm x 3 μm FWHM for experiments at CXI55 and MFX (humidified atmosphere), respectively. During each sample scan, chips were translated through the beam at the full 120 Hz repetition rate of LCLS such that the X-ray pulse arrival was synchronized spatially with the patterned micropores and diffraction images were recorded shot-by-shot by a Cornell-SLAC Pixel Array Detector (CSPAD).56 The design and operation of the Roadrunner fast scanning system is described in detail elsewhere.47–49, 57 58 NLP samples were measured with between 3 % and 10 % beam transmission (at CXI) and between 20 and 30% beam transmission (at MFX) rather than the full X-ray flux of 4.5 mJ/pulse due to concerns regarding damage to the chip resulting from the lower-intensity “wings” of the X-ray beam around the central focus spot (~1 % of the total intensity). All chips were assessed for damage immediately following measurement with optical microscopy. The nominal pulse duration was 40 fs for both experiments. During MFX experiments, an aluminum attenuator plate covered the low-resolution area of the detector to a resolution of ~6 Å.
X-ray images were analyzed in near real-time for an estimation of hit rate using OnDA59. Cheetah60 was used to find crystal hits. ADXV61 was used to visualize hits and to measure inter-reflection spacing by integrating signal intensity over ± 3pixels along lattice lines.
3. Results and Discussion
3.1. Cell-free expression
Cell-free expression and assembly of ApoA1 and ApoE4 was selected as a preparative strategy for crystallization. This is based on both its established capabilities to produce high yields of homogeneously sized NLPs and to facilitate future structural studies of membrane proteins through co-translational techniques that enable facile insertion of membrane proteins into the NLP bilayer.62 Previous studies have extensively characterized the particle size and homogeneity resulting from our cell free assembly. Samples were characterized with: size exclusion chromatography (SEC), DLS, negative stain TEM, and SAXS/SANS.27, 32, 53, 62–66 The yields of protein ranged from 3–4 mg of apolipoprotein, which were typical for a 1 ml cell-free reaction. Based on SDS-PAGE analysis the purified protein was greater than 98% homogeneous. To assess NLP formation and size distribution, we ran DLS, which confirmed that the pooled Ni-NTA affinity purified NLPs were homogenously distributed in size, centered at 11.5 nm and 32.0 nm for ApoA1 and ApoE4 NLPs respectively (Figure S3). NLP preparations were deemed sufficient for crystallization screens based on both the high yield and purity.
3.2. Crystallization Optimization
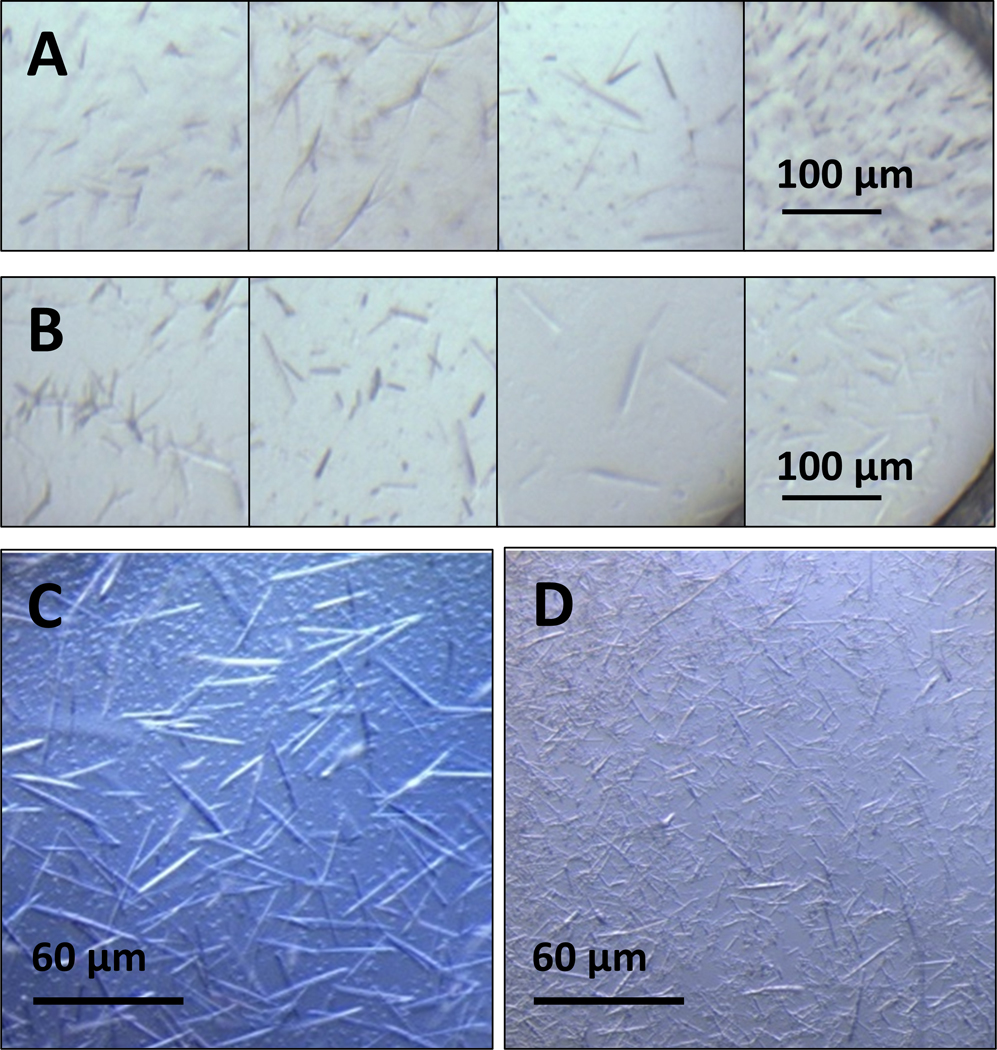
Initial high throughput crystallization screens subjected both NLP types to a wide range of crystallization conditions to obtain crystal hits for further optimization. Screens designed for both soluble proteins67 and membrane proteins68 were applied for a total of 2976 unique cocktails. The screens included incomplete factorial experiments consisting of PEGs and other polymer precipitants of varying molecular weight, salts, and a variety of pHs and buffer conditions, as well as commercially available screens from Hampton Research (including PEGRx HT, PEG Ion HT, Crystal Screen HT, Index, Salt Rx HT, Silver Bullet with PEG3350 pH 6.8 precipitants, Grid Screen Ammonium Sulfate, Slice pH, Ionic Liquids, and Polymer Screen) and Molecular Dimensions (MemGold screen) used as received or lightly modified.67, 68 Hits were identified from manual scoring of optical imaging of individual wells at day one and subsequently at weekly intervals. Potential hits identified with optical imaging were compared to UV-TPFF imaging at 28 days to confirm that crystalline objects were protein-containing rather than inorganic material. This scoring process identified a total of 165 and 237 potential hits for the ApoA1 and ApoE4 NLP samples respectively and are summarized in Table S1 and Figure S4. The dominant morphologies among hits for both proteins were either individual rods or needles or clusters of rods or needle-like objects (Figure 3) and the majority of hits were detected after relatively long incubation periods, typically at the three-week timepoint. Some trends were identified in the hits obtained, particularly the prevalence of mono or divalent inorganic salts, mid-range or very low molecular weight PEGs, and pH conditions at or above the isoelectric point of the apolipoprotein (5.56 and 5.65 for ApoA1 and ApoE4 respectively69) (Figure S4).
Figure 3.
Hits generated by the HWI High throughput screen for A) ApoA1 and B) ApoE4 NLPs. Wells were imaged between 2 and 4 weeks after the experiment was initiated using a custom-built optical imaging system at 21°C. Rod/rod cluster or needle/needle cluster morphologies dominate hit conditions containing varying buffer and pH conditions, salt additives, and precipitants for both ApoA1 and ApoE4 NLPs. Hits reproduced and optimized for SFX at LLNL for C) ApoA1 and D) ApoE4. Conditions pictured are C) S816–4 and D) M1328.
Of these, 10 conditions for each apolipoprotein were successfully reproduced and further optimized for SFX experiments from a subset of hits selected to explore a large chemical space and for apparent crystal quality. In general, a condition that produces a high density of uniform crystals on the scale of tens of micron, as opposed to a few crystals on the 100s of micron scale, is ideal for SFX and conditions were optimized as judged by reproducibility, crystal density, and uniformity. The most successful conditions, summarized in Table 1, chiefly consist of high concentrations of mid molecular weight PEGs (~20–40% w/v PEG 1000 or 4000) and salts with monovalent anions (Li+, Na+, NH4+). For ApoA1, conditions were primarily at pH 10 with CAPS while conditions for ApoE4 were optimized at both relatively low pH (sodium citrate, pH 5.5–5.6) and well above the ApoE4 pI (Tris pH 8.5). Crystal size distributions and number densities were measured by counting optical images of known volumes of crystal slurry for the conditions with the four highest apparent number densities for each apolipoprotein (Table 1).
3.3. SFX Experiments
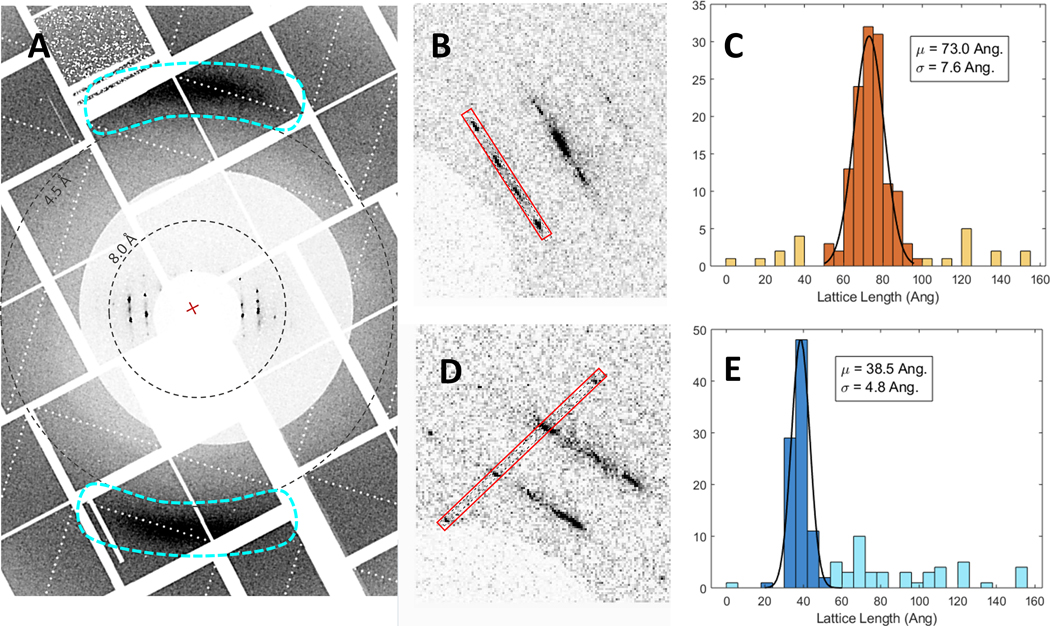
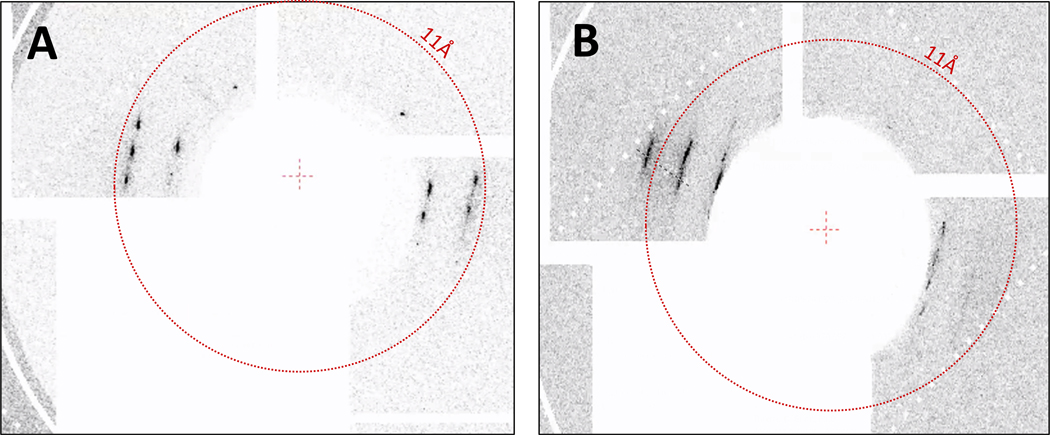
Crystals from three conditions were measured in the humidified helium environment at MFX, S816–4 (ApoA1), S1022 (ApoE4) and S1328 (ApoE4) (Table 1, boxed conditions). CSPAD images from ~42,900 shots were recorded with ~760 containing hits (1.77% hit rate) of S816–4 ApoA1 microcrystals on the bare chip. 40,500 shots were recorded with 250 containing hits for S1022 ApoE4 and 41,100 shots with 176 containing hits for M1328 ApoE4, resulting in 0.6 % and 0.4 % hit rates, respectively. Differences in hit rates are likely due to both differences in crystal density on the chip as deposited and variations in the strength of diffraction due to crystal size and quality. For each apolipoprotein and crystallization condition, diffraction was highly anisotropic but displayed distinct Bragg peaks within layer lines (Figure 4). For S816–4 ApoA1, reflections out to ~7 Å were observed for some hits although 11 Å was typical. It is likely some weak or diffuse diffraction was obscured by the low-angle attenuator. Diffraction from S1022 and M1328 ApoE4 crystals have similar characteristics in that Bragg peaks within layer lines are generally more diffuse than for ApoA1 and reflections along the direction of most diffraction did not exceed 9 Å (Figure 5).
Figure 4.
An example of hits collected at MFX for S816–4 ApoA1 at MFX showing A) anisotropic diffraction and diffuse features at 4.2 Å, and averaging of signal intensities along B) layer lines and D) an orthogonal crystallographic axis. The equivalent histograms of the reflection spacing measured from B) and D) are shown in C) and E) respectively.
Figure 5.
An example of hits collected at MFX for A) S816–4 ApoA1 and B) S1022 ApoE4 showing differences in diffusivity of peaks within layer lines at low resolution.
The hits appear to have a very limited range of orientations about at least one crystallographic axis, which is consistent with a rod or needle-like morphology. In general, the crystals were much larger in one dimension than the chip pore diameter, as evident from optical microscopy of crystals deposited onto micropatterned chips prior to beamtime. Attempts at indexing the patterns failed likely due to the sparsity, low-resolution, and diffuseness of the reflections. Thus, two unit cell dimensions were approximated by measuring the reflection spacing along putative zero-level axes and parallel axes over many of the recorded hits. For approximately 30% of the available hits for S816–4 ApoA1, signal intensities along layer lines and another clear axis were integrated over a width of 3 pixels and inter-peak distances were determined. These spacings of the reciprocal lattice points were translated into real space and histograms of the resulting unit cell values were fit to determine average values, in this case 73.0 ± 7.6 Å and 38.5 ± 4.8 Å . Fewer hits with less well-defined peak positions were available for S1022 and M1328 ApoE4, thus approximately 15 individual measurements of reflection spacing were simply averaged. Analogous values obtained were 90.2 ± 10.2 Å and 41.6 ± 5.5 Å for S1022 ApoE4 and for 111.4 ± 12.1 Å and 40.8 ± 6.7 Å M1328 ApoE4. During SFX experiments at CXI in vacuum, approximately 60,000 shots were recorded for S816–4 ApoA1 microcrystals. While hit finding was somewhat hampered by the prevalence of diffraction from what appear to be partially dried buffer components, at least 100 frames show hits with identifiable NLP diffraction similar to data collected at MFX. Using similar methods and assumptions for hits analyzed from the MFX experiment for the CXI experiment, unit cell parameters of 68.4 ± 6.9Å and 36.9 ± 5.3Å were determined from 21 individual measurements of reflection spacing.
All crystallization conditions measured at MFX have strong diffuse scattering features at 4.2 Å in the direction perpendicular to the direction of strongest diffraction. These features are similar to those exhibited by previous studies of stacked lamellar membranes of DPPC at room temperature70, 71 and are attributed to partial ordering of the phospholipids in the form of packing distances between the hydrocarbon tails. This is a strong indication of the presence of a DMPC lipid bilayer in the NLP crystals. One of the measured unit cell dimensions that is fairly consistent across apolipoprotein and crystallization conditions is also consistent with the thickness of individual NLPs as measured by AFM64 as well as the thickness of a DMPC bilayer at room temperature (3.6 nm). The other unit cell dimension increases for both ApoE4 conditions compared to ApoA1, which is consistent with a larger-diameter disc as previously observed.27
The anisotropy of the measured diffraction and the diffuse nature of some reflections within layer lines is suggestive of fiber-like diffraction. NLPs containing neutral zwitterionic phosphocholine lipids as well as discoidal HDL and LDL are known to form “rouleaux”, where disc bilayers associate to form lamellar-like columnar structures, under certain staining and deposition conditions for negative-stain TEM, where the negatively charged stain is thought to associate with the positive surface charge of the choline head group and allow the bilayer surfaces to come together.72 This effect is enhanced in the presence of monovalent salt conditions that are reproduced in many of our crystallization conditions and may contribute to fibril formation. Diffraction with very similar characteristics was reported for synchrotron crystallography of ApoE4 single crystals, but with slightly more well-defined Bragg spots particularly compared to SFX results for the same apolipoprotein, possibly due in part to cryo cooling.33, 72
While diffraction observed in the SFX experiment is similar to results obtained from previous synchrotron cryo-crystallography experiments, SFX enables collection of effectively damage-free data36, 37, 73 that would not otherwise be achievable at room temperature, especially for radiation damage prone proteins as lipid-bound lipoproteins have been noted to be.34 Radiation damage makes room temperature data collection particularly difficult for small or needle-like crystals (with two small size dimensions), which is circumvented by both the small XFEL beam focus and short pulse duration. Moreover, room temperature data collection facilitates dynamic studies of membrane protein function, maturation processes associated with lipid and cholesterol binding to apolipoproteins, and functional processes of crystallized NLP-encapsulated membrane proteins. The capability for room temperature measurement also represents the major advantage of this approach over cryoEM for nanodisc-encapsulated membrane protein structure determination, which has recently seen great success.2–8 Cell-free expression and assembly of membrane proteins into NLPs and other nanodiscs is equally amenable to detergent-free structural studies with cryoEM, though this is not currently widely implemented, but in that case all dynamics must be inferred from an equilibrium conformational ensemble. Some time resolved capabilities are being developed (ref) but the time resolution is still restricted to the ~10s-100s of millisecond freezing time,74–78 where the SFX time resolution is only limited by the XFEL pulse duration and the duration of the trigger (substrate mixing, laser pulse duration, etc).
4. Summary
We present robust crystallization conditions for multiple types of lipid-bound apolipoproteins produced via cell-free expression. This study represents a broad search of the crystallization chemical space that will inform further optimization for membrane proteins supported in NLP scaffolds produced via cell-free expression. Our unit cell calculations are consistent with the presence of a discoidal structure containing a centralized DMPC lipid bilayer surrounded by apolipoproteins for these self-assembled NLPs produced with cell-free expression in a detergent-free process. Importantly, the diffraction pattens from cell-free lipid-bound apolipoproteins on fixed stages at CXI and MFX produced similar diffraction characteristics to previously published studies using more conventional NLP assembly procedures,33, 34 validating our approach for structural studies of detergent-sensitive membrane protein-nanodisc complexes. This is the first study to take advantage of SFX at the LCLS with low-background fixed-target sample delivery with high rep rate scanning for studying these types of nanoparticles, containing both forms of lipid bound apolipoproteins. The combined low sample consumption, low background, minimal crystal size requirements, and lack of radiation damage effects on measurement make fixed target SFX an ideal approach for characterization of the diffuse NLP diffraction, while room temperature measurement enables future dynamic studies. The crystallization protocols outlined will enable investigation of the maturation processes associated with lipid and cholesterol binding, while also potentially enabling the dynamic study of membrane bound proteins within NLPs.
Supplementary Material
Acknowledgments:
The authors would like to thank the scientific staff at the Linac Coherent Light Source for operational support at the MFX (proposals P088, P117) and CXI (proposal LS18) end stations. Petra Fromme, Carolin Seuring, and their teams were extremely helpful during preparation for beamtime.
Funding: The LCLS is a national user facility operated by Stanford University on behalf of the US Department of Energy and supported by the US Department of Energy Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. This work was performed, in part, under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. M.L.S., D.G., M.A.C. and M.F. were supported by the NIH grant 1R01GM117342. Partial support was also provided by NIH grant U19AI144184. Support was also provided through the NSF-STC “BioXFEL” through award STC-1231306.
Footnotes
Supplementary Materials: The following are available online at www.mdpi.com/xxx/s1, Figure S1: Layout of chips used in experiments at MFX and CXI, Figure S2: Optical microscopy of graphene/PMMA enclosed crystals after vacuum exposure, Figure S3: Size distributions of ApoA1 and ApoE4 NLPs measured with DLS.
Conflicts of Interest: The authors declare no conflict of interest.
References
- 1.Feher VA; Randall A; Baldi P; Bush RM; de la Maza LM; Amaro RE, A 3-dimensional trimeric beta-barrel model for Chlamydia MOMP contains conserved and novel elements of Gram-negative bacterial porins. PLoS One 2013, 8 (7), e68934. [Google Scholar]
- 2.Autzen HE; Myasnikov AG; Campbell MG; Asarnow D; Julius D; Cheng Y, Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 2018, 359 (6372), 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roh S-H; Stam NJ; Hryc CF; Couoh-Cardel S; Pintilie G; Chiu W; Wilkens S, The 3.5-Å CryoEM Structure of Nanodisc-Reconstituted Yeast Vacuolar ATPase Vo Proton Channel. Molecular Cell 2018, 69 (6), 993–1004.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machen AJ; Akkaladevi N; Trecazzi C; O’Neil PT; Mukherjee S; Qi Y; Dillard R; Im W; Gogol EP; White TA; Fisher MT, Asymmetric Cryo-EM Structure of Anthrax Toxin Protective Antigen Pore with Lethal Factor N-Terminal Domain. Toxins 2017, 9 (10), 298. [Google Scholar]
- 5.Gao Y; Cao E; Julius D; Cheng Y, TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 2016, 534, 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedrov A; Sustarsic M; de Keyzer J; Caumanns JJ; Wu ZC; Driessen AJM, Elucidating the Native Architecture of the YidC: Ribosome Complex. J. Mol. Biol. 2013, 425 (22), 4112–4124. [DOI] [PubMed] [Google Scholar]
- 7.Akkaladevi N; Hinton-Chollet L; Katayama H; Mitchell J; Szerszen L; Mukherjee S; Gogol EP; Pentelute BL; Collier RJ; Fisher MT, Assembly of anthrax toxin pore: lethal-factor complexes into lipid nanodiscs. Protein science : a publication of the Protein Society 2013, 22 (4), 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gogol EP; Akkaladevi N; Szerszen L; Mukherjee S; Chollet-Hinton L; Katayama H; Pentelute BL; Collier RJ; Fisher MT, Three dimensional structure of the anthrax toxin translocon-lethal factor complex by cryo-electron microscopy. Protein science : a publication of the Protein Society 2013, 22 (5), 586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frauenfeld J; Gumbart J; Sluis EO; Funes S; Gartmann M; Beatrix B; Mielke T; Berninghausen O; Becker T; Schulten K; Beckmann R, Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol 2011, 18 (5), 614–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Midtgaard Søren R.; Pedersen Martin C.; Arleth L, Small-Angle X-Ray Scattering of the Cholesterol Incorporation into Human ApoA1-POPC Discoidal Particles. Biophysical Journal 2015, 109 (2), 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayburt TH; Grinkova YV; Sligar SG, Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys 2006, 450 (2), 215–22. [DOI] [PubMed] [Google Scholar]
- 12.Raschle T; Hiller S; Yu T-Y; Rice AJ; Walz T; Wagner G, Structural and Functional Characterization of the Integral Membrane Protein VDAC-1 in Lipid Bilayer Nanodiscs. Journal of the American Chemical Society 2009, 131 (49), 17777–17779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kijac AZ; Li Y; Sligar SG; Rienstra CM, Magic-Angle Spinning Solid-State NMR Spectroscopy of Nanodisc-Embedded Human CYP3A4. Biochemistry 2007, 46 (48), 13696–13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama H; Wang J; Tama F; Chollet L; Gogol EP; Collier RJ; Fisher MT, Three-dimensional structure of the anthrax toxin pore inserted into lipid nanodiscs and lipid vesicles. PNAS 2010, 107 (8), 3453–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayburt TH; Sligar SG, Membrane protein assembly into Nanodiscs. FEBS Letters 2010, 584 (9), 1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glomset JA, The plasma lecithins:cholesterol acyltransferase reaction. Journal of lipid research 1968, 9 (2), 155–167. [PubMed] [Google Scholar]
- 17.Glomset JA; Janssen ET; Kennedy R; Dobbins J, Role of plasma lecithin:cholesterol acyltransferase in the metabolism of high density lipoproteins. Journal of lipid research 1966, 7 (5), 638–648. [PubMed] [Google Scholar]
- 18.Fielding CJ; Shore VG; Fielding PE, A protein cofactor of lecithin:Cholesterol acyltransferase. Biochemical and Biophysical Research Communications 1972, 46 (4), 1493–1498. [DOI] [PubMed] [Google Scholar]
- 19.Jonas A; Sweeny SA; Herbert PN, Discoidal complexes of A and C apolipoproteins with lipids and their reactions with lecithin: cholesterol acyltransferase. J Biol Chem 1984, 259 (10), 6369–75. [PubMed] [Google Scholar]
- 20.Jonas A, Lecithin-cholesterol acyltransferase in the metabolism of high-density lipoproteins. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1991, 1084 (3), 205–220. [DOI] [PubMed] [Google Scholar]
- 21.Rajaram OV; Barter PJ, Reactivity of human lipoproteins with purified lecithin: cholesterol acyltransferase during incubations in vitro. Biochimica et biophysica acta 1985, 835 (1), 41–49. [DOI] [PubMed] [Google Scholar]
- 22.Remaley AT; Amar M; Sviridov D, HDL-replacement therapy: mechanism of action, types of agents and potential clinical indications. Expert Rev Cardiovasc Ther 2008, 6 (9), 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorci-Thomas MG; Bhat S; Thomas MJ, Activation of lecithin: cholesterol acyltransferase by HDL ApoA-I central helices. Clinical Lipidology 2009, 4 (1), 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chromy BA; Arroyo E; Blanchette CD; Bench G; Benner H; Cappuccio JA; Coleman MA; Henderson PT; Hinz AK; Kuhn EA; Pesavento JB; Segelke BW; Sulchek TA; Tarasow T; Walsworth VL; Hoeprich PD, Different apolipoproteins impact nanolipoprotein particle formation. J Am Chem Soc 2007, 129 (46), 14348–54. [DOI] [PubMed] [Google Scholar]
- 25.Marcink TC; Simoncic JA; An B; Knapinska AM; Fulcher YG; Akkaladevi N; Fields GB; Van Doren SR, MT1-MMP Binds Membranes by Opposite Tips of Its β Propeller to Position It for Pericellular Proteolysis. Structure (London, England: 1993) 2019, 27 (2), 281–292.e6. [Google Scholar]
- 26.Bibow S; Polyhach Y; Eichmann C; Chi CN; Kowal J; Albiez S; McLeod RA; Stahlberg H; Jeschke G; Güntert P; Riek R, Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nature Structural & Molecular Biology 2017, 24 (2), 187–193. [Google Scholar]
- 27.Chromy BA; Arroyo E; Blanchette CD; Bench G; Benner H; Cappuccio JA; Coleman MA; Henderson PT; Hinz AK; Kuhn EA; Pesavento JB; Segelke BW; Sulchek TA; Tarasow T; Walsworth VL; Hoeprich PD, Different Apolipoproteins Impact Nanolipoprotein Particle Formation. J. Am. Chem. Soc. 2007, 129 (46), 14348–14354. [DOI] [PubMed] [Google Scholar]
- 28.Mackey RH; Greenland P; Goff DC; Lloyd-Jones D; Sibley CT; Mora S, High-Density Lipoprotein Cholesterol and Particle Concentrations, Carotid Atherosclerosis, and Coronary Events: MESA (Multi-Ethnic Study of Atherosclerosis). J Am Coll Cardiol 2012, 60 (6), 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson WS, HDL-C vs HDL-P: How Changing One Letter Could Make a Difference in Understanding the Role of High-Density Lipoprotein in Disease. Clinical Chemistry 2014, 60 (11), e1–e3. [DOI] [PubMed] [Google Scholar]
- 30.He W; Felderman M; Evans AC; Geng J; Homan D; Bourguet F; Fischer NO; Li Y; Lam KS; Noy A; Xing L; Cheng RH; Rasley A; Blanchette CD; Kamrud K; Wang N; Gouvis H; Peterson TC; Hubby B; Coleman MA, Cell-free production of a functional oligomeric form of a Chlamydia major outer-membrane protein (MOMP) for vaccine development. J Biol Chem 2017, 292 (36), 15121–15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pourmousa M; Song HD; He Y; Heinecke JW; Segrest JP; Pastor RW, Tertiary structure of apolipoprotein A-I in nascent high-density lipoproteins. Proceedings of the National Academy of Sciences 2018, 115 (20), 5163–5168. [Google Scholar]
- 32.Cleveland TE; He W; Evans AC; Fischer NO; Lau EY; Coleman MA; Butler P, Small-angle X-ray and neutron scattering demonstrates that cell-free expression produces properly formed disc-shaped nanolipoprotein particles. Protein Science 2018, 27 (3), 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters-Libeu CA; Newhouse Y; Hatters DM; Weisgraber KH, Model of Biologically Active Apolipoprotein E Bound to Dipalmitoylphosphatidylcholine. J Biol Chem 2006, 281 (2), 1073–1079. [DOI] [PubMed] [Google Scholar]
- 34.Newhouse Y; Peters-Libeu C; Weisgraber KH, Crystallization and preliminary X-ray diffraction analysis of apolipoprotein E-containing lipoprotein particles. Acta Cryst F, Acta Cryst Sect F, Acta Crystallogr F, Acta Crystallogr Sect F, Acta Cryst F Struct Biol Cryst Commun, Acta Cryst Sect F Struct Biol Cryst Commun, Acta 2005, 61 (11), 981–984. [Google Scholar]
- 35.Neutze R; Wouts R; Spoel D. v. d.; Weckert E; Hajdu J, Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406 (6797), 752. [DOI] [PubMed] [Google Scholar]
- 36.Chapman HN; Fromme P; Barty A; White TA; Kirian RA; Aquila A; Hunter MS; Schulz J; DePonte DP; Weierstall U; Doak RB; Maia FRNC; Martin AV; Schlichting I; Lomb L; Coppola N; Shoeman RL; Epp SW; Hartmann R; Rolles D; Rudenko A; Foucar L; Kimmel N; Weidenspointner G; Holl P; Liang M; Barthelmess M; Caleman C; Boutet S; Bogan MJ; Krzywinski J; Bostedt C; Bajt S; Gumprecht L; Rudek B; Erk B; Schmidt C; Hömke A; Reich C; Pietschner D; Strüder L; Hauser G; Gorke H; Ullrich J; Herrmann S; Schaller G; Schopper F; Soltau H; Kühnel K-U; Messerschmidt M; Bozek JD; Hau-Riege SP; Frank M; Hampton CY; Sierra RG; Starodub D; Williams GJ; Hajdu J; Timneanu N; Seibert MM; Andreasson J; Rocker A; Jönsson O; Svenda M; Stern S; Nass K; Andritschke R; Schröter C-D; Krasniqi F; Bott M; Schmidt KE; Wang X; Grotjohann I; Holton JM; Barends TRM; Neutze R; Marchesini S; Fromme R; Schorb S; Rupp D; Adolph M; Gorkhover T; Andersson I; Hirsemann H; Potdevin G; Graafsma H; Nilsson B; Spence JCH, Femtosecond X-ray protein nanocrystallography. Nature 2011, 470 (7332), 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutet S; Lomb L; Williams GJ; Barends TRM; Aquila A; Doak RB; Weierstall U; DePonte DP; Steinbrener J; Shoeman RL; Messerschmidt M; Barty A; White TA; Kassemeyer S; Kirian RA; Seibert MM; Montanez PA; Kenney C; Herbst R; Hart P; Pines J; Haller G; Gruner SM; Philipp HT; Tate MW; Hromalik M; Koerner LJ; Bakel N. v.; Morse J; Ghonsalves W; Arnlund D; Bogan MJ; Caleman C; Fromme R; Hampton CY; Hunter MS; Johansson LC; Katona G; Kupitz C; Liang M; Martin AV; Nass K; Redecke L; Stellato F; Timneanu N; Wang D; Zatsepin NA; Schafer D; Defever J; Neutze R; Fromme P; Spence JCH; Chapman HN; Schlichting I, High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science 2012, 337 (6092), 362–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seibert MM; Ekeberg T; Maia FRNC; Svenda M; Andreasson J; Jönsson O; Odić D; Iwan B; Rocker A; Westphal D; Hantke M; DePonte DP; Barty A; Schulz J; Gumprecht L; Coppola N; Aquila A; Liang M; White TA; Martin A; Caleman C; Stern S; Abergel C; Seltzer V; Claverie J-M; Bostedt C; Bozek JD; Boutet S; Miahnahri AA; Messerschmidt M; Krzywinski J; Williams G; Hodgson KO; Bogan MJ; Hampton CY; Sierra RG; Starodub D; Andersson I; Bajt S; Barthelmess M; Spence JCH; Fromme P; Weierstall U; Kirian R; Hunter M; Doak RB; Marchesini S; Hau-Riege SP; Frank M; Shoeman RL; Lomb L; Epp SW; Hartmann R; Rolles D; Rudenko A; Schmidt C; Foucar L; Kimmel N; Holl P; Rudek B; Erk B; Hömke A; Reich C; Pietschner D; Weidenspointner G; Strüder L; Hauser G; Gorke H; Ullrich J; Schlichting I; Herrmann S; Schaller G; Schopper F; Soltau H; Kühnel K-U; Andritschke R; Schröter C-D; Krasniqi F; Bott M; Schorb S; Rupp D; Adolph M; Gorkhover T; Hirsemann H; Potdevin G; Graafsma H; Nilsson B; Chapman HN; Hajdu J, Single mimivirus particles intercepted and imaged with an X-ray laser. Nature 2011, 470 (7332), 78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Z; Fan J; Li H; Jiang H, Current Status of Single Particle Imaging with X-ray Lasers. Applied Sciences 2018, 8 (1), 132. [Google Scholar]
- 40.Pedrini B; Tsai C-J; Capitani G; Padeste C; Hunter MS; Zatsepin NA; Barty A; Benner WH; Boutet S; Feld GK; Hau-Riege SP; Kirian RA; Kupitz C; Messerschmitt M; Ogren JI; Pardini T; Segelke B; Williams GJ; Spence JCH; Abela R; Coleman M; Evans JE; Schertler GFX; Frank M; Li X-D, 7 Å resolution in protein two-dimensional-crystal X-ray diffraction at Linac Coherent Light Source. Phil. Trans. R. Soc. B 2014, 369 (1647), 20130500. [Google Scholar]
- 41.Frank M; Carlson DB; Hunter MS; Williams GJ; Messerschmidt M; Zatsepin NA; Barty A; Benner WH; Chu K; Graf AT; Hau-Riege SP; Kirian RA; Padeste C; Pardini T; Pedrini B; Segelke B; Seibert MM; Spence JCH; Tsai C-J; Lane SM; Li X-D; Schertler G; Boutet S; Coleman M; Evans JE, Femtosecond X-ray diffraction from two-dimensional protein crystals. IUCrJ 2014, 1 (2), 95–100. [Google Scholar]
- 42.Casadei CM; Tsai CJ; Barty A; Hunter MS; Zatsepin NA; Padeste C; Capitani G; Benner WH; Boutet S; Hau-Riege SP; Kupitz C; Messerschmidt M; Ogren JI; Pardini T; Rothschild KJ; Sala L; Segelke B; Williams GJ; Evans JE; Li XD; Coleman M; Pedrini B; Frank M, Resolution extension by image summing in serial femtosecond crystallography of two-dimensional membrane-protein crystals. IUCrJ 2018, 5 (1), 103–117. [Google Scholar]
- 43.Seuring C; Ayyer K; Filippaki E; Barthelmess M; Longchamp J-N; Ringler P; Pardini T; Wojtas DH; Coleman MA; Dörner K, Femtosecond X-ray coherent diffraction of aligned amyloid fibrils on low background graphene. Nature communications 2018, 9 (1), 1836. [Google Scholar]
- 44.Kirian RA; Wang X; Weierstall U; Schmidt KE; Spence JCH; Hunter M; Fromme P; White T; Chapman HN; Holton J, Femtosecond protein nanocrystallography—data analysis methods. Optics Express 2010, 18 (6), 5713–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chavas LMG; Gumprecht L; Chapman HN, Possibilities for serial femtosecond crystallography sample delivery at future light sources. Structural Dynamics 2015, 2 (4), 041709. [Google Scholar]
- 46.White TA; Mariani V; Brehm W; Yefanov O; Barty A; Beyerlein KR; Chervinskii F; Galli L; Gati C; Nakane T; Tolstikova A; Yamashita K; Yoon CH; Diederichs K; Chapman HN, Recent developments in CrystFEL. Journal of Applied Crystallography 2016, 49 (2), 680–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roedig P; Ginn HM; Pakendorf T; Sutton G; Harlos K; Walter TS; Meyer J; Fischer P; Duman R; Vartiainen I; Reime B; Warmer M; Brewster AS; Young ID; Michels-Clark T; Sauter NK; Kotecha A; Kelly J; Rowlands DJ; Sikorsky M; Nelson S; Damiani DS; Alonso-Mori R; Ren J; Fry EE; David C; Stuart DI; Wagner A; Meents A, High-speed fixed-target serial virus crystallography. Nature Methods 2017, advance online publication. [Google Scholar]
- 48.Roedig P; Vartiainen I; Duman R; Panneerselvam S; Stübe N; Lorbeer O; Warmer M; Sutton G; Stuart DI; Weckert E; David C; Wagner A; Meents A, A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering. Scientific Reports 2015, 5, 10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shelby ML; Gilbile D; Grant TD; Seuring C; Segelke BW; He W; Evans AC; Pakendorf T; Fischer P; Hunter MS; Batyuk A; Barthelmess M; Meents A; Coleman MA; Kuhl TL; Frank M, A fixed-target platform for serial femtosecond crystallography in a hydrated environment. IUCrJ 2020, 7 (1). [Google Scholar]
- 50.Cappuccio JA; Blanchette CD; Sulchek TA; Arroyo ES; Kralj JM; Hinz AK; Kuhn EA; Chromy BA; Segelke BW; Rothschild KJ; Fletcher JE; Katzen F; Peterson TC; Kudlicki WA; Bench G; Hoeprich PD; Coleman MA, Cell-free Co-expression of Functional Membrane Proteins and Apolipoprotein, Forming Soluble Nanolipoprotein Particles. Molecular & Cellular Proteomics 2008, 7 (11), 2246–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klammt C; Schwarz D; Lohr F; Schneider B; Dotsch V; Bernhard F, Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J 2006, 273 (18), 4141–53. [DOI] [PubMed] [Google Scholar]
- 52.Koglin A; Klammt C; Trbovic N; Schwarz D; Schneider B; Schafer B; Lohr F; Bernhard F; Dotsch V, Combination of cell-free expression and NMR spectroscopy as a new approach for structural investigation of membrane proteins. Magn Reson Chem 2006, 44 Spec No, S17–23. [DOI] [PubMed] [Google Scholar]
- 53.Cappuccio JA; Blanchette CD; Sulchek TA; Arroyo ES; Kralj JM; Hinz AK; Kuhn EA; Chromy BA; Segelke BW; Rothschild KJ; Fletcher JE; Katzen F; Peterson TC; Kudlicki WA; Bench G; Hoeprich PD; Coleman MA, Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Molecular & cellular proteomics : MCP 2008, 7 (11), 2246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieske J; Cerv M; Kreida S; Komadina D; Fischer J; Barthelmess M; Fischer P; Pakendorf T; Yefanov O; Mariani V; Seine T; Ross BH; Crosas E; Lorbeer O; Burkhardt A; Lane TJ; Guenther S; Bergtholdt J; Schoen S; Törnroth-Horsefield S; Chapman HN; Meents A, On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies. IUCrJ 2019, 6 (4), 714–728. [Google Scholar]
- 55.Schropp A; Hoppe R; Meier V; Patommel J; Seiboth F; Lee HJ; Nagler B; Galtier EC; Arnold B; Zastrau U; Hastings JB; Nilsson D; Uhlén F; Vogt U; Hertz HM; Schroer CG, Full spatial characterization of a nanofocused x-ray free-electron laser beam by ptychographic imaging. Scientific Reports 2013, 3. [Google Scholar]
- 56.Blaj G; Caragiulo P; Carini G; Carron S; Dragone A; Freytag D; Haller G; Hart P; Hasi J; Herbst R; Herrmann S; Kenney C; Markovic B; Nishimura K; Osier S; Pines J; Reese B; Segal J; Tomada A; Weaver M, X-ray detectors at the Linac Coherent Light Source. Journal of Synchrotron Radiation 2015, 22 (Pt 3), 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roedig P; Duman R; Sanchez-Weatherby J; Vartiainen I; Burkhardt A; Warmer M; David C; Wagner A; Meents A, Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering. Journal of Applied Crystallography 2016, 49 (3), 968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolstikova A; Levantino M; Yefanov O; Hennicke V; Fischer P; Meyer J; Mozzanica A; Redford S; Crosas E; Opara NL; Barthelmess M; Lieske J; Oberthuer D; Wator E; Mohacsi I; Wulff M; Schmitt B; Chapman HN; Meents A, 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector. IUCrJ 2019, 6 (5), 927–937. [Google Scholar]
- 59.Mariani V; Morgan A; Yoon CH; Lane TJ; White TA; O’Grady C; Kuhn M; Aplin S; Koglin J; Barty A; Chapman HN, OnDA: online data analysis and feedback for serial X-ray imaging. Journal of Applied Crystallography 2016, 49 (3), 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barty A; Kirian RA; Maia FRNC; Hantke M; Yoon CH; White TA; Chapman H, Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. Journal of Applied Crystallography 2014, 47 (3), 1118–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arvai A, ADXV-a program to display X-ray diffraction images. Scripps Research Institute, La Jolla, CA 2012. [Google Scholar]
- 62.Cappuccio J; Hinz A; Kuhn E; Fletcher J; Arroyo E; Henderson P; Blanchette C; Walsworth V; Corzett M; Law R; Pesavento J; Segelke B; Sulchek T; Chromy B; Katzen F; Peterson T; Bench G; Kudlicki W; Hoeprich P Jr.; Coleman M, Cell-Free Expression for Nanolipoprotein Particles: Building a High-Throughput Membrane Protein Solubility Platform. In High Throughput Protein Expression and Purification, Doyle S, Ed. Humana Press: 2009; pp 273–295. [Google Scholar]
- 63.He W; Luo J; Bourguet F; Xing L; Yi SK; Gao T; Blanchette C; Henderson PT; Kuhn E; Malfatti M; Murphy WJ; Cheng RH; Lam KS; Coleman MA, Controlling the diameter, monodispersity, and solubility of ApoA1 nanolipoprotein particles using telodendrimer chemistry. Protein Science 2013, 22 (8), 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blanchette CD; Cappuccio JA; Kuhn EA; Segelke BW; Benner WH; Chromy BA; Coleman MA; Bench G; Hoeprich PD; Sulchek TA, Atomic force microscopy differentiates discrete size distributions between membrane protein containing and empty nanolipoprotein particles. Biochimica et Biophysica Acta (BBA) - Biomembranes 2009, 1788 (3), 724–731. [DOI] [PubMed] [Google Scholar]
- 65.Blanchette C; Segelke B; Fischer N; Corzett M; Kuhn E; Cappuccio J; Benner WH; Coleman M; Chromy B; Bench G; Hoeprich P; Sulchek T; Blanchette CD; Segelke BW; Corzett MH; Kuhn EA; Cappuccio JA; Coleman MA; Chromy BA; Hoeprich PD; Sulchek TA, Characterization and Purification of Polydisperse Reconstituted Lipoproteins and Nanolipoprotein Particles. International Journal of Molecular Sciences 2009, 10 (7), 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blanchette CD; Law R; Benner WH; Pesavento JB; Cappuccio JA; Walsworth V; Kuhn EA; Corzett M; Chromy BA; Segelke BW; Coleman MA; Bench G; Hoeprich PD; Sulchek TA, Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations. Journal of lipid research 2008, 49 (7), 1420–30. [DOI] [PubMed] [Google Scholar]
- 67.Luft JR; Collins RJ; Fehrman NA; Lauricella AM; Veatch CK; DeTitta GT, A deliberate approach to screening for initial crystallization conditions of biological macromolecules. Journal of Structural Biology 2003, 142 (1), 170–179. [DOI] [PubMed] [Google Scholar]
- 68.Koszelak-Rosenblum M; Krol A; Mozumdar N; Wunsch K; Ferin A; Cook E; Veatch CK; Nagel R; Luft JR; DeTitta GT; Malkowski MG, Determination and application of empirically derived detergent phase boundaries to effectively crystallize membrane proteins. Protein Science 2009, 18 (9), 1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alzate O; Osorio C; DeKroon RM; Corcimaru A; Gunawardena HP, Differentially charged isoforms of apolipoprotein E from human blood are potential biomarkers of Alzheimer’s disease. Alzheimers Res Ther 2014, 6 (4), 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McIntosh TJ; Simon SA, Area per molecule and distribution of water in fully hydrated dilauroylphosphatidylethanolamine bilayers. Biochemistry 1986, 25 (17), 4948–4952. [DOI] [PubMed] [Google Scholar]
- 71.McIntosh TJ; Simon SA, Hydration force and bilayer deformation: a reevaluation. Biochemistry 1986, 25 (14), 4058–4066. [DOI] [PubMed] [Google Scholar]
- 72.Zhang L; Song J; Cavigiolio G; Ishida BY; Zhang S; Kane JP; Weisgraber KH; Oda MN; Rye K-A; Pownall HJ; Ren G, Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. Journal of lipid research 2011, 52 (1), 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spence JCH, XFELs for structure and dynamics in biology. IUCrJ 2017, 4 (4), 322–339. [Google Scholar]
- 74.Dandey VP; Budell WC; Wei H; Bobe D; Maruthi K; Kopylov M; Eng ET; Kahn PA; Hinshaw JE; Kundu N; Nimigean CM; Fan C; Sukomon N; Darst SA; Saecker RM; Chen J; Malone B; Potter CS; Carragher B, Time-resolved cryo-EM using Spotiton. Nat Meth 2020, 17 (9), 897–900. [Google Scholar]
- 75.Mäeots M-E; Lee B; Nans A; Jeong S-G; Esfahani MMN; Ding S; Smith DJ; Lee C-S; Lee SS; Peter M; Enchev RI, Modular microfluidics enables kinetic insight from time-resolved cryo-EM. Nat Commun 2020, 11 (1), 3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaledhonkar S; Fu Z; Caban K; Li W; Chen B; Sun M; Gonzalez RL; Frank J, Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature 2019, 570 (7761), 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frank J, Time-resolved cryo-electron microscopy: Recent progress. Journal of Structural Biology 2017, 200 (3), 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fischer N; Konevega AL; Wintermeyer W; Rodnina MV; Stark H, Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 2010, 466 (7304), 329–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.