Abstract
Hepatitis C virus (HCV) is responsible for more than 180 million infections worldwide, and about 80 % of infections are reported in Low and Middle-income countries (LMICs). Therapy is based on the administration of interferon (INF), ribavirin (RBV) or more recently Direct-Acting Antivirals (DAAs). However, amino acid substitutions associated with resistance (RAS) have been extensively described and can contribute to treatment failure, and diagnosis of RAS requires considerable infrastructure, not always locally available. Dried serum spots (DSS) sampling is an alternative specimen collection method, which embeds drops of serum onto filter paper to be transported by posting to a centralized laboratory. Here, we assessed feasibility of genotypic analysis of HCV from DSS in a cohort of 80 patients from São Paulo state Brazil. HCV RNA was detected on DSS specimens in 83 % of samples of HCV infected patients. HCV genotypes 1a, 1b, 2a, 2c and 3a were determined using the sequence of the palm domain of NS5B region, and RAS C316N/Y, Q309R and V321I were identified in HCV 1b samples. Concerning therapy outcome, 75 % of the patients who used INF +RBV as a previous protocol of treatment did not respond to DAAs, and 25 % were end-of-treatment responders. It suggests that therapy with INF plus RBV may contribute for non-response to a second therapeutic protocol with DAAs. One patient that presented RAS (V321I) was classified as non-responder, and combination of RAS C316N and Q309R does not necessarily imply in resistance to treatment in this cohort of patients. Data presented herein highlights the relevance of studying circulating variants for a better understanding of HCV variability and resistance to the therapy. Furthermore, the feasibility of carrying out genotyping and RAS phenotyping analysis by using DSS card for the potential of informing future treatment interventions could be relevant to overcome the limitations of processing samples in several location worldwide, especially in LMICs.
Keywords: direct-action antivirals, dried serum spot, genotyping, Hepatitis C Virus, NS5B, resistance-associated substitutions
Introduction
Hepatitis C virus (HCV) is a major cause of chronic hepatitis and hepatocellular carcinoma globally [1]. This pathogen has infected nearly 180 million people worldwide [2, 3], and about 80 % of infections are reported in Low and Middle-income countries (LMICs) [4, 5]. Despite this high prevalence rate, data on molecular epidemiology of HCV seem relatively scarce in LMICs, which can be associated with a lack of adequate throughput facilities to study the virus [6, 7].
HCV is an enveloped, icosahedral, positive sense single-stranded RNA virus that belongs to the family Flaviviridae, genus Hepacivirus [8, 9]. The viral genome is composed of approximately 9600 nucleotides and represents an uninterrupted open reading frame (ORF) encoding a polyprotein precursor of approximately 3000 amino acids [10, 11]. The HCV polyprotein is co- and post-translationally processed by a combination of cellular and viral proteases into three structural proteins (Core, E1, and E2) and seven non-structural (NS) proteins (P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). Phylogenetic analysis of global strain diversity resulted in the distinction of eight HCV genotypes (1 to 8) and 86 subtypes (a, b, c etc.), based on the nucleotide sequences of the Core/E1 and NS5B regions [3, 10, 12–16].
The NS5B is the viral RNA-dependent RNA-polymerase (RdRp) responsible for HCV RNA replication [15], an enzyme that lacks proof-reading activity, leading to the emergence of intrahost diversity of viral populations circulating in the blood of an individual [10]. The variability and dynamic of viral infection in the host organism are related to the progression of antiviral treatments, which can result in selective pressure for the emergence and spread of drug-resistant viral strains [10, 17]. The initial therapies used against HCV were interferons-based (IFN), resulting in several collateral effects. In 2015, Food and Drug Administration (FDA) approved the therapy with IFN-free direct-acting antivirals (DAAs) [12, 18], classified as NS3/4A protease inhibitors, NS5A inhibitors and NS5B polymerase inhibitors (NI - nucleoside inhibitor or NNI - non-nucleoside inhibitor). Treatment with DAAs presents high levels of sustained virologic response (SVR) rates, typically >90 %, contributing to both the control of HCV epidemics and improvement of the quality of life of patients, with reduced side effects compared to IFN [12]. However, under treatment, resistance-associated substitutions (RAS) might occur into the genomic regions related to the viral proteins targeted by anti-HCV drugs, which may confer resistance to the DAA classes [12, 19].
The three-dimensional structure of the viral NS5B polymerase contains three domains: Palm, Fingers, and Thumb [20–22]. Structural analyses of this protein and its active site in the palm domain demonstrated that substitutions that occurred close to the catalytic triad of HCV NS5B polymerase interfered with the effectiveness of antivirals. The most common RAS that affects the activity of NS5B inhibitors are L159F, D244N, S282T/R, Q309R, D310N, and A333E [23–25].
Surveying the viral evolution and identifying RAS in the NS5B of HCV is a relevant approach. However, HCV genotyping and RAS diagnosis can be a challenge in many locations, as for instance in Brazil. In this context, the use of desiccated patient specimens such as dried serum spots (DSS) or dried blood spots (DBS) may facilitate the monitoring of HCV infections in regions with limited access to laboratory services, reaching vulnerable populations [26–29]. DBS samples blotted on filter paper have revolutionized the point of care management of patients living with HCV and the study of HCV in developed countries [30, 31]. DSS requires traditional phlebotomy prior to blotting but provides identical downstream storage and transport potential where primary diagnostic facilities and / or secondary reference laboratories have significant distance from point of care [32].
Regrettably, LMICs that should benefit more from using this technology are lagging far behind with relatively few studies utilising the technique published [33, 34]. Besides, the HCV ribonucleic acid degradation of the specimen (whole blood/serum/plasma) at the collection site poses a serious challenge in LMICs, especially when serology or molecular studies cannot be performed immediately, or additional analyses requires further shipment to a reference laboratory. The dried spot technique proffers solutions to this problem. More importantly, many studies have shown no significant variation in the assay performance between the DBS and direct sample methods [31, 35, 36]. Also, DBS been proven efficacious for screening high-risk patients such as intravenous drugs users [37, 38] and require no cold chain for onward transportation from the sample collection site to the point of final analysis [39, 40]. Although, to date there is no standard operating procedure for elution and recovery of HCV RNA from desiccated material [41], the World Health Organization (WHO) testing guidelines recommend the use of DBS specimens as an option for HCV testing and analysis, especially where there are no facilities for samples processing [42].
In this study, we utilised surplus archived serum taken for routine HCV diagnostics to prepare DSS cards to carry out additional genotyping and RAS identification by analysing the NS5B palm domain variability and treatment response data in a cohort of chronically infected patients from São Paulo state, Brazil. Considering that Brazil has limited facilities for genotyping and resistance testing, our data provides relevant information to overcome the limitations of processing samples in several location worldwide, especially in the LMICs [43].
Methods
Clinical samples
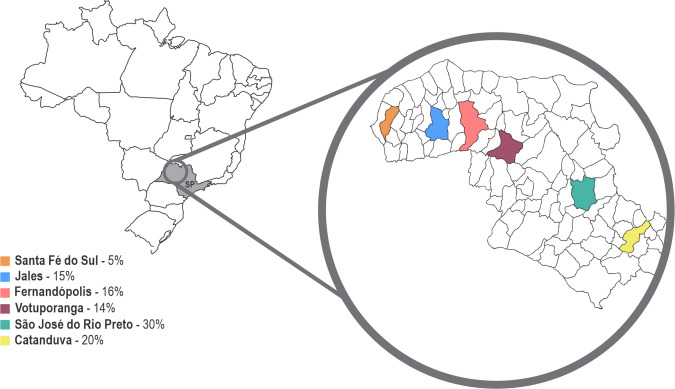
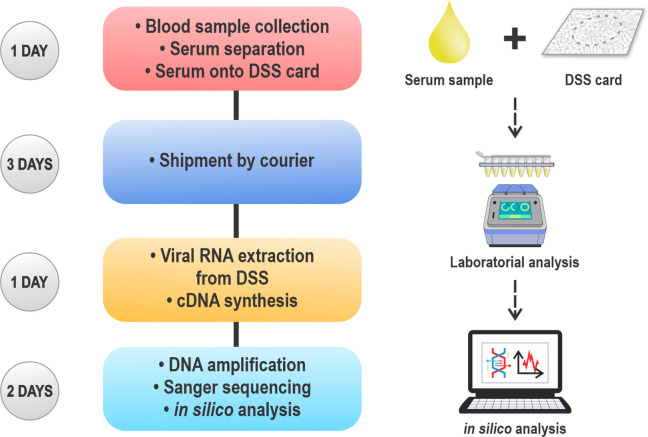
Eighty serum samples from patients previously diagnosed positive for HCV attending four specialized HCV screening and treatment centres from 2010 to 2017 were obtained after long-term storage at −20 °C or below. Samples were collected in specialized centres of the northwest region of the state of São Paulo, Brazil, serving the cities of São José do Rio Preto, Catanduva, Votuporanga, Fernandópolis, Jales, and Santa Fé do Sul (Fig. 1). Samples were taken for routine diagnostic investigation according to the varied methods and storage protocols of the four centres. The dried serum spot samples that were RNA positive presented a viral load ranging from 3.0×104 IU ml−1 to 5.3×107 IU ml−1. The core demographic data such as age, sex, viral loads, serum collection centres, and suspected route of infection were analysed. Additional retrospective data analysis as outlined in the following three sections of this study were completed within approximately 1 week of sample receipt (Fig. 2).
Fig. 1.
Location of the samples collection distributed by percentage according to the city in the highlighted northwest region of the state of São Paulo (SP).
Fig. 2.
Remote genotyping workflow: duration between conveyed samples by dried serum spots (DSS) card until final results.
Dried serum spot preparation and HCV RNA elution
Thirty microlitres of each serum sample was spotted onto a Protein Saver 903 Card (GE Healthcare), filling the demarcated 12 mm diameter circles and allowed to air dry at room temperature (25 °C) for approximately 1 h, then stored at 4 °C prior to shipment. Ambient shipment of the samples took approximately 1 week by international courier (DHL). Each entire serum spot was recovered from the DSS Card, by using a 5 mm puncher to take multiple discs from the sample area. The discs were suspended in a 2 ml tube containing 300 µl Phosphate Buffered Saline (PBS). The tubes were incubated with agitation at room temperature for 10 min, and then 140 µl of supernatant virus suspension/eluate were obtained for RNA extraction. Single punches of clean card were taken between samples and negative extraction controls undertaken to counteract and monitor for carryover contamination.
NS5B amplification and sequencing
RNA extraction was carried out using QIAamp MinElute virus spin RNA extraction kit (QIAGEN) according to the manufacturer’s instructions. Then, 50 µl HCV RNA eluate from each sample were stored at −70 °C and used for cDNA synthesis. A 20 µl RNA extract (a template) was added to a lyophilized EcoDry Premix (random hexamers) (Takara-Clontech), incubated at 42 °C for 60 min to generate complementary DNA, in a reverse transcriptase-polymerase chain reaction.
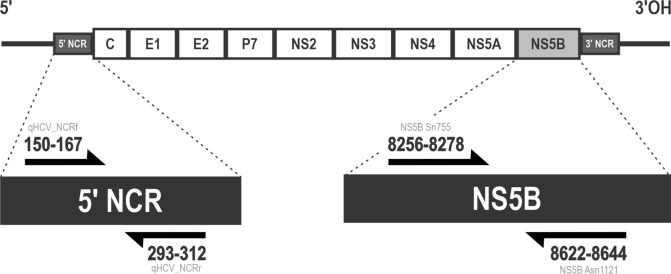
PCR reactions were performed using HotStarTaq (QIAGEN), according to manufacturer's instruction. Primers qHCV_NCRf (5′-GCGCAACCGGTGAGTACA-3′) and qHCV_NCRr (5′-ACTCGCAAGCACCCTATCAG-3′) [44] were used to amplify the 5′ untranslated region; and Sn755 (5′-TATGAYACCCGCTGYTTTGACTC-3′) and Asn1121 (5′-GCNGARTAYCTVGTCATAGCCTC-3′) [45] for NS5B (Fig. 3). Reactions were performed using the following conditions: 94 °C for 15 min for enzyme activation; 55 cycles of denaturation at 95 °C for 20 s, annealing at 55 °C for 20 s and extension at 72 °C for 20 s; and final extension at 72 °C for 10 min. Expected amplicon was 388 bp, confirmed by electrophoresis. PCR products were diluted 1:10 with water and ambient shipped direct for Sanger sequencing (Source BioScience, Nottingham, UK).
Fig. 3.
Schematic representation of the HCV genome, its polyprotein and the primers used in the fragment amplification.
Sequence analysis, RAS identification and phylogeny reconstruction
Putative Sanger sequences were visualized and manually edited by removing the terminal primer sequences using Finch TV software. Sequence assembly, alignment, and phylogenetic analysis were performed using mega six software [46]. Sequences were trimmed to 288 bases in length corresponding to 8311–8598 of H77 reference, accession NC_004102. The curated NS5B sequences were uploaded to seven different online HCV subtyping tools to compare outcomes: [1] Los Alamos database (LANL) [47]; [2] Virus Pathogen Database and Analysis (VIRP) [48]; [3] Oxford HCV automated subtyping tool (version 2.0) [49]; [4] NCBI genotyping tool [50]; [5] Geno2Pheno HCV Subtyping tool [51]; [6] HCV Comet [52]; and [7] HCV Glue Subtyping tool [53]. Further characterization of the sequences was performed by phylogenetic analysis using each confirmed HCV genotypes/subtypes for easy visualization/visibility from reference datasets in the International Committee on Taxonomy of Viruses (ICTV) to determine HCV subtype [15].
Maximum-likelihood phylogeny reconstruction with bootstrap evaluation was conducted in mega X programme, using the GTR nucleotide substitution model and gamma (Γ) distribution of rate variability among sites (GTR + gamma) [54]. The bootstrap (1000 replicates) cut-off greater than or equal to 70 % of each cluster clades was considered a reliable confidence threshold [55].
Amino acid sequence data corresponding to amino acid positions −223 to −339 were aligned using the Geno2Pheno software platform and NS5B RAS was thereby determined. The investigated RAS were selected based on the substitutions described in the literature in the NS5B palm domain genomic region (Table 1). Study sequences were submitted to GenBank, with accession numbers OL752714 - OL752779.
Table 1.
Substitutions in the NS5B polymerase palm region related to resistance to direct-acting antivirals
|
RAS |
Genotype |
Reference |
|---|---|---|
|
L 159 F |
1a, 1b, 3a |
Donaldson et al. (2014); Svarovskaia et al. (2014); Tong et al. (2014); Lontok et al. (2015); Noble et al. (2017) [23, 25, 103–105] |
|
D 244 N |
3a |
Asahina et al. (2005); Castilho et al. (2011) [24, 96] |
|
S 282 T/R |
1a, 1b |
Donaldson et al. (2014); Tong et al. (2014); Svarovskaia et al. (2016) [23, 104, 106] |
|
Q 309 R |
1a, 1b, 3a |
Hamano et al. (2005); Castilho et al. (2011) [24, 107] |
|
D 310 N |
1b, 3a |
Asahina et al. (2005); Castilho et al. (2011) [24, 96] |
|
C 316 H/N/Y |
1a, 1b |
Shi et al. (2008), McCown et al. (2009); Castilho et al. (2011); Donaldson et al. (2014); Lontok et al. (2015); Noble et al. (2017) [23–25, 105, 108, 109] |
|
L 320 F |
1a, 1b, 3a |
Tong et al. (2014); Svarovskaia et al. (2016) [104, 106] |
|
V 321 A/I |
1b, 3a |
Donaldson et al. (2014); Svarovskaia et al. (2014); Lontok et al. (2015); Noble et al. (2017); Hezode et al. (2018) [25, 103, 105] |
|
A 333 E |
1a |
Hamano et al. (2005); Castilho et al. (2011) [24, 107] |
Statistical analysis
All the data were analysed appropriately by GraphPad Prism version 8.0.1 for Windows, GraphPad Software, San Diego, California USA (www.graphpad.com). The descriptive data are presented as mean±standard deviation. The data were categorized in groups and analysed according to log normality test. Non-parametric data were compared by the Mann Whitney test, and the parametric data, by the t-test. Statistical inferences were based on a p-value of less than 0.05 as significant.
Results
Clinical characteristics of the patients
The study included 20 % of samples from Catanduva, 16 % from Fernandópolis, 15 % from Jales, 30 % from São José Do Rio Preto, 14 % from Votuporanga and 5 % from Santa Fé Sul (Fig. 1). The participants were within the age ranging from 30 to 80 years. Males were predominant, accounting for 53 %. The frequency of suspected route of infections was 33 % parenteral /drug use, 12 % of blood transfusion, 12 % surgical/tattoo, 11 % sexual contact, whilst 32 % did not disclose any known route of infection. Two patients were HIV/HBV co-infected, while ten tested positive for HIV infection (Table 2).
Table 2.
Demographic characteristics of the study samples
|
Characteristics |
no. (%) |
GT 1a (N=34) |
GT 1b (N=18) |
GT 3a (N=12) |
GT 2a (N=1) |
GT 2c (N=1) |
P-value |
|---|---|---|---|---|---|---|---|
|
Gender |
|||||||
|
Male |
35 (53) |
18 |
9 |
6 |
1 |
1 |
0.757 |
|
Female |
31 (47) |
16 |
9 |
6 |
– |
– |
|
|
Age Group |
|||||||
|
<39 |
10 (15) |
5 |
3 |
2 |
– |
– |
0.042 |
|
40–49 |
21 (32) |
14 |
3 |
4 |
– |
– |
|
|
50–59 |
15 (3) |
8 |
4 |
3 |
– |
– |
|
|
60–69 |
14 (21) |
5 |
7 |
2 |
– |
– |
|
|
70–79 |
5 (8) |
1 |
1 |
1 |
1 |
1 |
|
|
80–89 |
1 (1) |
1 |
– |
– |
– |
– |
|
|
Suspected route of infection |
|||||||
|
Blood products |
8 (12) |
4 |
3 |
1 |
– |
– |
0.657 |
|
Drug use/parenteral |
22 (33) |
12 |
5 |
4 |
– |
1 |
|
|
Sexual contact |
7 (11) |
3 |
2 |
2 |
– |
– |
|
|
Surgical/tattoo |
8 (12) |
3 |
1 |
4 |
– |
– |
|
|
Unknown |
21 (32) |
12 |
7 |
1 |
– |
– |
|
|
Viral Load |
|||||||
|
<6x104 |
1 (1) |
– |
1 |
– |
– |
– |
0.801 |
|
6×104 - 8×104 |
2 (3) |
2 |
– |
– |
– |
– |
|
|
>8x104 |
63 (96) |
32 |
17 |
12 |
1 |
1 |
|
|
Co-infection |
|||||||
|
HIV |
10 (15) |
6 |
3 |
1 |
– |
– |
0.977 |
|
HIV/HBV |
2 (3) |
2 |
– |
– |
– |
– |
|
|
Unknown |
6 (9) |
23 |
14 |
9 |
1 |
1 |
Concerning the treatment profile of the patients, 23 % of the analysed samples were from naive patients, which means they had not undergone previous treatment against HCV. Patients who undergone therapy with direct-action antivirals (DAAs) presented three distinct patterns of response to treatment, classified as: i) the sustained virological response (SVR), meaning that blood tests of the patient continue to show no detectable RNA 12 weeks or more after treatment; ii) end-of-treatment responders (ETR), when there is no detectable HCV RNA in the blood at the completion of treatment but it was detected after the end of the treatment; iii) or non-responders (NR), when HCV RNA is detectable during and after the treatment. In total, 90 % of the patients were previously treated with interferon and ribavirin, and 58 % of them submitted to a second protocol of treatment with DAAs. Among patients who used sofosbuvir and daclatasvir, or simeprevir, as the second therapeutic approach, 75 % did not respond to treatment and 25 % were ETR, representing 0 % of SVR. Therefore, therapy with DAAs followed by a previous IFN-based protocol demonstrated reduced SVR. Patients who received DAAs as the first therapeutic approach represented 39 % of the patients, with an SVR rate of 96 % (Table S1, available in the online version of this article).
DSS samples processing and genotype assignment
From the 80 DSS samples investigated in this study, 66 (83 %) partial sequences of the NS5B coding region corresponding to the NS5B palm domain were successfully amplified and sequenced for analysis. A possible reason for the failure of 17 % of unamplified samples despite medium to high viral loads (3.04×104 IU ml−1 to 5.39×107 IU ml−1) in the initial diagnostic assay from the same specimen might be related to inconsistent handling over a broad collection period at the various collection sites. Each diagnostic and long-term storage pathway were prior to our retrospective investigation and thus may have been sub-optimal in some instances, but no further information was available. There was statistical difference in the diagnostic lab-tested HCV RNA viral load between the DSS PCR positive and negative samples (P=0.0042) (Fig. 4).
Fig. 4.
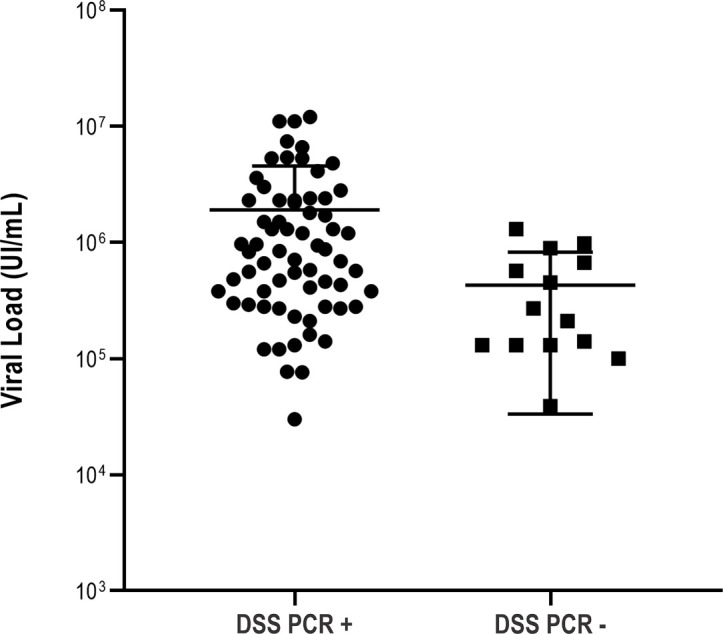
RNA viral load in PCR positive and PCR-negative Brazilian HCV patients; displaying the impact of viral load on PCR outcome
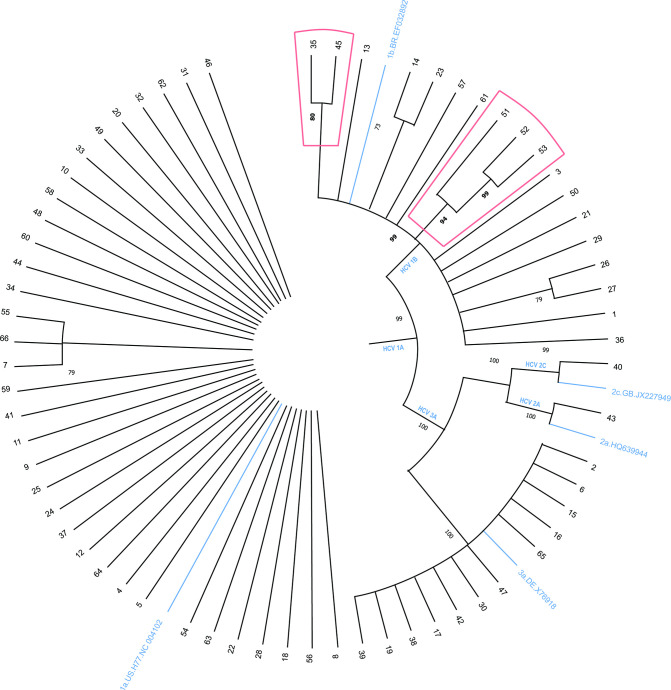
Genotype assignment demonstrated 100 % concordance using all of the seven online subtyping tools. The online tool prediction was also consistent with the phylogenetic analysis (Table S2). More importantly, all the sequences could be assigned at the subtype level (Fig. 5). The most frequent genotypes were 1a (34/66, 52%), 1b (18/66, 27%), and 3a (12/66, 18%), with the remainder being subtypes 2a (1/66, 1.5%) and 2c (1/66, 1.5%). There were no significant associations between subtypes/gender (P=0.757) and subtypes/suspected route of infections (P=0.765). However, the association between subtypes and age groups was significant (P=0.0442). Genotype 1a has the highest frequency in the age groups of 40–49 (41 %, 14/34) and 50–59 (24 %, 8/34). Genotype 1b was predominant in the age group 60–69 years, with a frequency of 39 % (7/18). Genotype 3a has the lowest incidence when compared to the others, yet was the second predominant in 40–49 group, with 33 % (4/12) frequency (Table 2).
Fig. 5.
Phylogenetic tree not rooted with 66 sequences of 308 nucleotides from the NS5B region from samples from patients infected with HCV genotypes 1a, 1b, 2a, 2c and 3a. Red highlights indicate the clade of sequences 51, 52, and 53, which contains RAS C316N and Q309R, and the clade of sequences 35 and 45, with V321I. Bootstrap values obtained with 1000 replicates. The figure shows the bootstraps with values above 60.
Resistance association prediction
The palm domain of NS5B (aa 188 to 227 and aa 287 to 370) [56] was the region evaluated for RAS investigation by subjecting the sequences to the Geno2Pheno software tool. The amino acid sequence in this analysis overlaps 240–342 fragment. This segment is well characterized and was used for the analysis (Table 1). Either RAS C316Y and Q309R, were observed in the sequences of patients infected with HCV 1a (Table 3). Among genotype 1b sequences, samples of patients 51, 52 and 53 presented RAS Q309R and C316N, previously described for the amplified region (Table 1); and samples of patients 35 and 45 presented RAS V321I (Table 3).
Table 3.
NS5B Resistance associated substitution (RAS) analysis. NS5B sequences generated for 66 samples were analysed with Geno2Pheno software for the presence of described RAS by genotype to DAAs
|
Genotype |
No. of patients |
Patients with RAS |
Amino acid position associated with NS5B RAS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
L 159 F |
D 244 N |
S 282 T/R |
Q 309 R |
D 310 N |
C 316 H/N/Y |
L 320 F |
V 321 A/I |
A 333 E |
|||
|
1a |
35 |
16 |
15 |
1 C 316 Y |
|||||||
|
1b |
17 |
5 |
3 |
3 C 316 N |
2 – V 321 I |
||||||
|
2a |
1 |
0 |
|||||||||
|
2c |
1 |
0 |
|||||||||
|
3a |
12 |
0 |
|||||||||
Phylogenetic analysis
The tree generated from the phylogenetic reconstruction with the 66 analysed sequences is shown in Fig. 5. Genotypes 1a, 1b, 2a, 2c and 3a were grouped in different monophyletic branches, strongly sustained by the bootstrap values of 99, 99, 100, 99, and 100%, respectively, confirming previous genotyping analyses (Fig. 5). HCV 1b sequences 51, 52 and 53, which presented RAS C316N and Q309R, grouped in the same monophyletic branch, with 94 % of bootstrap. Additionally, sequences 52 and 53 grouped in a clade strongly sustained by a bootstrap of 99 % (Fig. 5). Furthermore, HCV 1b sequences 35 and 45 presented RAS V321I, and grouped in a clade sustained by the bootstrap value of 80 %.
Discussion
In this study, we reported an 83 % (66/80) success rate using dried serum spots (DSS) cards to genotype samples of HCV patients from the northeast region of São Paulo, Brazil. This is comparable to the 85 % success rate reported by Soulier and colleagues, in which they determined the HCV genotype using a similar DBS technique [57]. Available evidence showed that HCV RNA in DBS can be detected at levels >1000 IU ml−1 and detection limit range between 178 to 1779 IU ml−1 [58]. In accordance to the recent WHO guidelines on Hepatitis B and C testing, a detection limit of 3000 IU ml−1 or lower is acceptable and would identify 95 % of those with viraemic infection [59, 60]. Also, this limit is higher than the 45 IU ml−1 detection limit for the plasma sample [61]. This method has shown immense benefit in screening, surveillance, diagnosis, and monitoring of HCV treatment [57, 62], aside from success rates of about 90–100 % reported by Mahajan and co-workers and Vázquez-Morón and colleagues [41, 63].
Whilst we utilised Sanger sequencing, Next-generation Sequencing (NGS) technology could equally be applied to nucleic acid extracted from DSS cards. We have previously shown comparable results between Sanger and Nanopore sequencing technologies with Hepatitis B amplicons from DSS-derived material, but improved depth of coverage of minority variants from the NGS reads [32]. Elsewhere, Alidjinou and co-workers have evaluated Sanger sequencing with alternative NGS technology for HIV-1 drug resistance testing in treatment-naive patients, with NGS again facilitating detection of additional minority drug-resistant mutations [64].
According to our data, genotypes 1a, 1b, and 3a were the most prevalent, followed by 2a and 2c, respectively. It confirms the worldwide panorama since the widespread HCV genotypes are 1 (44 %) and genotype 3 (25 %) [65]. The prevalence of HCV genotype distribution in this study is also in accordance with previous studies performed in Brazil [66–68]. Recently, Mutini and colleagues described a high prevalence of genotypes 1a (41%), 1b (30%), 3a (24%), and 2 (4 %) in about 29071 of either healthy people or blood donors in Brazil. Our data is in agreement with most of previous studies in São Paulo state; the predominant genotypes being 1a, 1b, and 3a [69, 70]. However, genotypes 3 and 1b have been reported as the most prevalent genotypes in Salvador [71] and Pernambuco [67], respectively; both in north-eastern Brazil.
Herein, the suspected acquisition of HCV infection was 33%, similar to previous studies that observed 36 % of risk of transmission by parenteral/drug use in the North and Northeast regions in Brazil [72, 73]. Coutinho and co-workers, however, emphasize this transmission route as the most significant in the country. According to their data, the odds of HCV infection are 85 % higher among multidrug users and almost eight times higher among injecting drug users [74].
No significant association was observed between genotypes and suspected route of infection, despite the majority of patients being infected with subtypes 1a through parenteral/injection drug use. This is in agreement with some studies which have associated genotype 1a with injectable drugs users [75, 76]. However, some studies in developed countries have shown that genotype 3a is more prevalent in this group of HCV infected patients [77]. These different profiles may likely be due to some particular variables [78]. There was a significant association between subtypes and age group, being genotype 1a the most prevalent in the age group 40–49 years. This data corroborates with the study of Guimarães and co-workers that underpins the idea of late diagnosis of chronic HCV infection and thus a historical predominance of genotype 1a [79]. The risk of HCV transmission via blood transfusion (12 %) in this study is low compared to previous study (38%) [80]. This is expected because of high transfusion-transmitted disease awareness and the level of blood donor screening [81–83]. Also, the risk of surgical/tattooing agrees with previous reports of negligence to the sterilization of body piercing instruments in Brazil [84]. Sexual exposure risk is 11 % in this study; the transmission of HCV via sex is controversial in HCV infection [85, 86]. However, studies have shown high risk in people that trade sexual services, injecting drugs, STI – coinfection (Sexually Transmitted Co-Infections) and, MSM (men who have sex with men) [74, 87, 88]. The HCV co-infection prevalence observed herein is consistent with the previous reports in Brazil [89].
The new findings on HCV viral replication allowed a better understanding of non-structural 5B (NS5B) activity, resulting in the polymerase inhibitors implementation. The DAAs of this class act mainly in the NS5B palm domain, between the amino acids at positions 287 and 371, which is the most well-conserved domain and contains the catalytic residues. Therefore, RAS to these antivirals are concentrated in this HCV region [23]. Until now, there are few studies in Brazil associating the effectiveness of DAA therapeutic regimens and the role of RAS in a flaw treatment outcome [90]. Monitoring the identification and circulation of the variants in Brazilian population, and worldwide, is relevant to evaluate the possible impacts in therapeutic failure with DAAs [91]. One reason for the lack of data on the circulation of RAS in several countries is the difficulty in testing HCV infected patients [92, 93]. The DSS sampling is, however, a reliable alternative specimen collection method that may facilitate the monitoring of ongoing RAS into vulnerable populations [26–29].
As stated in our data, patients who previously undergone therapy with interferon (INF) and ribavirin (RBV) were submitted to a second therapeutic approach, using DAAs (sofosbuvir and daclatasvir, or simeprevir). These patients did not achieve SVR, suggesting that previous therapy with INF and RBV could interfere in the effectiveness of subsequent therapy with DAAs. Additionally, patients who received DAAs as the first therapeutic approach presented higher SVR rates. Treatments with INF and RBV present many side effects and, the RBV mechanism action is still unclear. It is known that RBV is a purine analogue, and it is involved in several cellular pathways, acting synergistically with INF [94]. An important antiviral mechanism of action of RBV is the inhibition of HCV replication, which interferes with both the NS5B polymerase and the NS5A multifunctional protein [95]. The action of RBV as a purine analogue, associated with the lack of corrective activity of NS5B, results in high rates of mutations in the viral variants of chronically infected patients undergoing this therapeutic protocol [96]. It could potentially cause the failure of the first treatment and result in a bottleneck effect in the viral population. In that way, previous treatment with INF and RBV could select resistant variants, which could explain why patients did not respond to the treatment with DAAs as a second therapeutic approach. Jardim and co-workers evaluated intrahost viral diversity in HCV chronically infected patients and suggested that the composition of quasispecies population at the beginning of the treatment, followed by an increase in some predominant quasispecies after treatment of NR and ETR in their patients, represented an advantage for the virus as it remains in the organism [97]. Although the heterogeneous population of HCV sequences in vivo is the main cause of DAAs treatment failures, it seems to have no differences between the hepatic and plasma viral quasispecies, as demonstrated by Hedegaard and colleagues [98]. In this context, based in preclinical in vivo evidence, an alternative presented by Vercauteren and colleagues was that the addition of an entry inhibitor to an anti-HCV DAA regimen restricts the breakthrough of DAA-resistant viruses. According to their work, this strategy may prevent therapeutic failure caused by RAS developing, and this combination can increase response rates, especially in difficult-to-treat patient populations [99].
Treatment resistance is associated with several factors. The resistance mechanism associated with RAS is not completely understood; therefore the presence of an isolated substitution may not confer resistance to the treatment. It is known that the combination of RAS L159F and L320F or C316N, for example, directly interferes in the action of sofosbuvir, targeting the palm domain of NS5B [23, 100]. Furthermore, a study by Uchida and colleagues demonstrated that patients with previous treatment with RBV had a lower frequency of C316N replacement than those who did not previously treat with this drug [101]. The RAS C316N / Y, Q309R and V321I are also associated with DAA resistant variants. The spread of these mutations in the northwest region of São Paulo, Brazil, is worrisome, as it may be associated with an increase in the number of individuals resistant to the available treatment. Therefore, surveillance of circulating variants in the infected population should be performed regularly. A recent study evaluated more than 100 HCV-infected patients in Rio de Janeiro, Brazil, identifying amino acid substitutions in positions 159 and 316 of the NS5B of genotype 1b [91]. Although the majority of Brazilian patients with HCV are susceptible to therapeutic regimens with DAAs, the presence of these RAS in the variants of this region opens avenues for the implementation of resistant viral variants [91].
Here both RAS C316N and Q309R located in the NS5B palm region were observed in the sequences from patients 51, 52, and 53. These sequences were grouped in the same monophyletic branch, showing high similarity among the variants of these patients. Despite these patients presented SVR after therapy with DAAs, the identification of the RAS in these samples is a relevant indicator that these substitutions are circulating in this location. Mutations in the amino acid C316 have been frequent in Asian variants, and have also been reported in many analyses in Brazil [102]. Moreover, a second monophyletic branch was presented in the reconstruction of the topology of the phylogenetic tree, grouping sequences 52 and 53, demonstrating that these sequencers are genetically closer than to sequence 51. Interestingly, patients 52 and 53 were assessed in Votuporanga, unlike patient 51 who was examined in São José do Rio Preto. Besides that, the RAS V321I was observed in sequences 35 and 45, which grouped in a separated clade. One of the sequences is from a patient from Jales, the only non-responder patient, and presented one of the targets RAS we listed in the studied population.
To the best of our knowledge, this study has shown for the first time that cascade of HCV care such as diagnosis, genotyping, and drug resistance analysis is achievable using DSS. This can be implemented in LMICs where facilities are limited. Finally, it plays an important role in the accomplishment of the WHO mandate towards global eradication of HCV by the year 2023.
Supplementary Data
Funding information
Funding was provided by the Royal Society – Newton Advanced Fellowship, and CAPES (Coordination for the Improvement of Higher Education Personnel – Code 001). VRG received fellowship from the pro-rectory of undergraduate studies program of the Federal University of Uberlândia. KA was supported by the Tertiary Education Trust Fund (TETFUND) and the NIHR Nottingham Biomedical Research Centre.
Acknowledgements
We would like to thank the support of fellowships and the samples provided by the Adolfo Lutz Institute.
Author contributions
K.A carried out the experimental work, performed genetic and phylogenetic analysis and drafted the manuscript; V.R.G. performed phylogenetic analysis, drafted the manuscript and illustrations; G.M.F. drafted the manuscript; M.C.N.S. provided samples and associated clinical data; J.F.S. prepared D.S.S. cards; C.P.M., D.P.J. and A.C.G.J. designed the experiments, analysed data and critically revised the manuscript; B.J.K., A.W.R. and J.K.B. analysed the data and drafted the manuscript. All the authors read and approved the final manuscript.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical statement
Approval for the extended diagnostic investigation in this study was granted by the Research Ethics Committee of the Adolfo Lutz Institute, Brazil, in accordance with resolution 196/96 of the National Health Council, with study number 290.504.
Footnotes
Abbreviations: DAAs, direct-acting antivirals; DBS, dried blood spots; DSS, dried serum spots; ETR, end-of-treatment responders; HCV, Hepatitis C virus; INF, interferon; LMICs, low and middle-income countries; MSM, men who have sex with men; NGS, next-generation sequencing; NR, non-responders; NS, non-structural proteins; ORF, open-reading frame; RAS, resistance-associated substitution; RBV, ribavirin; RdRp, RNA-dependent RNA-polymerase; STI, sexually transmitted infections; SVR, sustained virologic response; WHO, World Health Organization.
Two supplementary tables are available with the online version of this article.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29 Suppl 1:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Paraskevis D, Stylianou DC, Hezka J, Stern Z, Oikonomopoulou M, et al. HCV phylogeography of the general population and high-risk groups in cyprus identifies the island as a global sink for and source of infection. Sci Rep. 2019;9:10077. doi: 10.1038/s41598-019-46552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, et al. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology. 2014;59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Geneva, Switzerland: WHO; 2018. Progress report on access to hepatitisC treatment.https://www.who.int/hepatitis/publications/hep-c-access-report-2018/en [Google Scholar]
- 5.Graham CS, Swan T. A path to eradication of hepatitis C in low- and middle-income countries. Antiviral Res. 2015;119:89–96. doi: 10.1016/j.antiviral.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J Gastroenterol. 2016;22:7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishizaki A, Bouscaillou J, Luhmann N, Liu S, Chua R, et al. Survey of programmatic experiences and challenges in delivery of hepatitis B and C testing in low- and middle-income countries. BMC Infect Dis. 2017;17:696. doi: 10.1186/s12879-017-2767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaito M, Watanabe S, Tsukiyama-Kohara K, Yamaguchi K, Kobayashi Y, et al. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994;75:1755–1760. doi: 10.1099/0022-1317-75-7-1755. [DOI] [PubMed] [Google Scholar]
- 9.Pol S, Lagaye S. The remarkable history of the hepatitis C virus. Genes Immun. 2019;20:436–446. doi: 10.1038/s41435-019-0066-z. [DOI] [PubMed] [Google Scholar]
- 10.Tsukiyama-Kohara K, Kohara M. Hepatitis C Virus: viral quasispecies and genotypesViral Quasispecies and Genotypes. Int J Mol Sci. 2017;19:1–8. doi: 10.3390/ijms19010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 12.Manns MP, Buti M, Gane E, Pawlotsky J-M, Razavi H, et al. Hepatitis C virus infection. Nat Rev Dis Primers. 2017;3:17006. doi: 10.1038/nrdp.2017.6. [DOI] [PubMed] [Google Scholar]
- 13.Hedskog C, Parhy B, Chang S, Zeuzem S, Moreno C, et al. Identification of 19 novel hepatitisnovel hepatitis c virus subtypes-further expandingvirus subtypes-further expanding HCV cclassification. Open Forum Infect Dis. 2019;6:fz076. doi: 10.1093/ofid/ofz076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borgia SM, Hedskog C, Parhy B, Hyland RH, Stamm LM, et al. Identification of a novel hepatitis C virus genotype from Punjab, India: expanding classification of hepatitis C virus into 8 genotypes. J Infect Dis. 2018;218:1722–1729. doi: 10.1093/infdis/jiy401. [DOI] [PubMed] [Google Scholar]
- 15.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, et al. International Committee on Taxonomy of Viruses (ICTV). HCV Classification. 2019.
- 16.Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, et al. Resistance analysis in patients with genotype 1–6 HCV infection treated with sofosbuvir/velpatasvir in the phase iii studies. J Hepatol. 2017;89:6105–6116. doi: 10.1007/s00705-018-3920-9. [DOI] [Google Scholar]
- 17.Perales C, Quer J, Gregori J, Esteban JI, Domingo E. Resistance of hepatitis C virus to inhibitors: complexity and clinical implications. Viruses. 2015;7:5746–5766. doi: 10.3390/v7112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manns MP, von Hahn T. Novel therapies for hepatitis C - one pill fits all? Nat Rev Drug Discov. 2013;12:595–610. doi: 10.1038/nrd4050. [DOI] [PubMed] [Google Scholar]
- 19.Dietz J, Susser S, Vermehren J, Peiffer K-H, Grammatikos G, et al. Patterns of resistance-associated substitutions in patientswith chronic HCV infection following treatment with direct-actingantivirals. Gastroenterology. 2018;154:976–988. doi: 10.1053/j.gastro.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Eltahla AA, Luciani F, White PA, Lloyd AR, Bull RA. Inhibitors ofthe hepatitis C virus polymerase; mode of action and resistance. Viruses. 2015;7:5206–5224. doi: 10.3390/v7102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Membreno FE, Lawitz EJ. The HCV NS5B nucleoside and non-nucleoside inhibitors. Clin Liver Dis. 2011;15:611–626. doi: 10.1016/j.cld.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Choi KH. In: Viral Molecular Machines. Advances in Experimental Medicine and Biology. Rossmann M, Rao V, editors. Vol. 726. Viral Polymerases; pp. 267–304. vol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaldson EF, Harrington PR, Rear JJO, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2014;61:56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 24.Castilho MCB, Martins AN, Horbach IS, Perez RM, Figueiredo FAF, et al. Association of hepatitis C virus NS5B variants with resistance to new antiviral drugs among untreated patients. Mem Inst Oswaldo Cruz. 2011;106:968–975. doi: 10.1590/s0074-02762011000800011. [DOI] [PubMed] [Google Scholar]
- 25.Noble CF, Malta F, Lisboa-Neto G, Gomes-Gouvêa MS, Leite AGB, et al. Natural occurrence of NS5B inhibitor resistance-associated variants in Brazilian patients infected with HCV or HCV and HIV. Arch Virol. 2017;162:165–169. doi: 10.1007/s00705-016-3094-2. [DOI] [PubMed] [Google Scholar]
- 26.Lange B, Roberts T, Cohn J, Greenman J, Camp J, et al. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples - a systematic review and meta-analysis. BMC Infect Dis. 2017;17:693. doi: 10.1186/s12879-017-2776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komas NP, Vickos U, Hübschen JM, Béré A, Manirakiza A, et al. Cross-sectional study of hepatitis B virus infection in rural communities, Central African Republic. BMC Infect Dis. 2013;13:286. doi: 10.1186/1471-2334-13-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobari RF, Meshkati M, Ataei B, Yazdani MR, Heidari K, et al. Identification of patients with hepatitispatients with hepatitis C virus infection in persons with background of intravenous drug use: the first community announcement-based study fromvirus infection in persons with background of intravenous drug use: the first community announcement-based study from iran. Int J Prev Med. 2012;3:S170–5. [PMC free article] [PubMed] [Google Scholar]
- 29.Lopes CLR, Teles SA, Espírito-Santo MP, Lampe E, Rodrigues FP, et al. Prevalence, risk factors and genotypes of hepatitis C virus infection among drug users, Central-Western Brazil. Rev Saude Publica. 2009;43 Suppl 1:43–50. doi: 10.1590/s0034-89102009000800008. [DOI] [PubMed] [Google Scholar]
- 30.Saludes V, Folch C, Morales-Carmona A, Ferrer L, Fernàndez-López L, et al. Community-based screening of hepatitis C with a one-step RNA detection algorithm from dried-blood spots: analysis of key populations in Barcelona, Spain. J Viral Hepat. 2018;25:236–244. doi: 10.1111/jvh.12809. [DOI] [PubMed] [Google Scholar]
- 31.Tuaillon E, Mondain A-M, Meroueh F, Ottomani L, Picot M-C, et al. Dried blood spot for hepatitis C virus serology and molecular testing. Hepatology. 2010;51:752–758. doi: 10.1002/hep.23407. [DOI] [PubMed] [Google Scholar]
- 32.Astbury S, Costa Nunes Soares MM, Peprah E, King B, Jardim ACG, et al. Nanopore sequencing from extraction-free direct PCR of dried serum spots for portable hepatitis B virus drug-resistance typing. J Clin Virol. 2020;129:104483. doi: 10.1016/j.jcv.2020.104483. [DOI] [PubMed] [Google Scholar]
- 33.Ndiaye O, Gozlan J, Diop-Ndiaye H, Sall AS, Chapelain S, et al. Usefulness of Dried Blood Spots (DBS) to perform hepatitis C virus genotyping in drug users in Senegal. J Med Virol. 2017;89:484–488. doi: 10.1002/jmv.24460. [DOI] [PubMed] [Google Scholar]
- 34.Kania D, Bekalé AM, Nagot N, Mondain A-M, Ottomani L, et al. Combining rapid diagnostic tests and dried blood spot assays for point-of-care testing of human immunodeficiency virus, hepatitis B and hepatitis C infections in Burkina Faso, West Africa. Clin Microbiol Infect. 2013;19:E533–41. doi: 10.1111/1469-0691.12292. [DOI] [PubMed] [Google Scholar]
- 35.Dokubo EK, Evans J, Winkelman V, Cyrus S, Tobler LH, et al. Comparison of Hepatitis C Virus RNA and antibody detection in dried blood spots and plasma specimens. J Clin Virol. 2014;59:223–227. doi: 10.1016/j.jcv.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marques BLC, do Espírito-Santo MP, Marques VA, Miguel JC, da Silva EF, et al. Evaluation of dried blood spot samples for hepatitis C virus detection and quantification. J Clin Virol. 2016;82:139–144. doi: 10.1016/j.jcv.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 37.De Crignis E, Re MC, Cimatti L, Zecchi L, Gibellini D. HIV-1 and HCV detection in dried blood spots by SYBR Green multiplex real-time RT-PCR. J Virol Methods. 2010;165:51–56. doi: 10.1016/j.jviromet.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Naidoo A, Parboosing R, Moodley P. Real-time polymerase chain reaction optimised for hepatitis C virus detection in dried blood spots from HIV-exposed infants, KwaZulu-Natal, South Africa. Afr J Lab Med. 2016;5:269. doi: 10.4102/ajlm.v5i1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenman J, Roberts T, Cohn J, Messac L. Dried blood spot in the genotyping, quantification and storage of HCV RNA: a systematic literature review. J Viral Hepat. 2015;22:353–361. doi: 10.1111/jvh.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe K, Konomi N. Hepatitis C virus RNA in dried serum spotted onto filter paper is stable at room temperature. J Clin Microbiol. 1998;36:3070–3072. doi: 10.1128/JCM.36.10.3070-3072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahajan S, Choudhary MC, Kumar G, Gupta E. Evaluation of dried blood spot as an alternative sample collection method for hepatitis C virus RNA quantitation and genotyping using a commercial system. Virusdisease. 2018;29:141–146. doi: 10.1007/s13337-018-0441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WHO Guidelines on Hepatitis B and C Testing. 2017
- 43.WDI Classifying countries by income [Internet] 2019. [ November 19; 2020 ]. https://datatopics.worldbank.org/world-development-indicators/stories/the-classification-of-countries-by-income.html accessed.
- 44.Vercauteren K, Brown RJP, Mesalam AA, Doerrbecker J, Bhuju S, et al. Targeting a host-cell entry factor barricades antiviral-resistant HCV variants from on-therapy breakthrough in human-liver mice. Gut. 2016;65:2029–2034. doi: 10.1136/gutjnl-2014-309045. [DOI] [PubMed] [Google Scholar]
- 45.Morice Y, Roulot D, Grando V, Stirnemann J, Gault E, et al. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J Gen Virol. 2001;82:1001–1012. doi: 10.1099/0022-1317-82-5-1001. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuiken C, Thurmond J, Dimitrijevic M, Yoon H. The LANL hemorrhagic fever virus database, a new platform for analyzing biothreat viruses. Nucleic Acids Res. 2012;40:D587–92. doi: 10.1093/nar/gkr898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–8. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 50.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Döring M, Büch J, Friedrich G, Pironti A, Kalaghatgi P, et al. geno2pheno[ngs-freq]: a genotypic interpretation system for identifying viral drug resistance using next-generation sequencing data. Nucleic Acids Res. 2018;46:W271–W277. doi: 10.1093/nar/gky349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Struck D, Lawyer G, Ternes A-M, Schmit J-C, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014;42:e144. doi: 10.1093/nar/gku739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singer JB, Thomson EC, McLauchlan J, Hughes J, Gifford RJ. GLUE: a flexible software system for virus sequence data. BMC Bioinformatics. 2018;19:532. doi: 10.1186/s12859-018-2459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platformsMolecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182. doi: 10.2307/2992540. [DOI] [Google Scholar]
- 56.Di Maio VC, Cento V, Mirabelli C, Artese A, Costa G, et al. Hepatitis C virus genetic variability and the presence of NS5B resistance-associated mutations as natural polymorphisms in selected genotypes could affect the response to NS5B inhibitors. Antimicrob Agents Chemother. 2014;58:2781–2797. doi: 10.1128/AAC.02386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soulier A, Poiteau L, Rosa I, Hézode C, Roudot-Thoraval F, et al. Dried blood spotsBlood Spots: a tool to ensure broad access to hepatitisA Tool to Ensure Broad Access to Hepatitis C screening, diagnosis, and treatment monitoringScreening, Diagnosis, and Treatment Monitoring. J Infect Dis. 2016;213:1087–1095. doi: 10.1093/infdis/jiv423. [DOI] [PubMed] [Google Scholar]
- 58.Shepherd SJ, Baxter RE, Gunson RN. Evaluation of the Abbott m2000 system for dried blood spot detection of hepatitis C virus RNA. J Clin Virol. 2019;110:7–10. doi: 10.1016/j.jcv.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 59.Hezode C, Reau N, Svarovskaia ES, Doehle BP, Shanmugam R, et al. Resistance analysis in patients with genotype 1-6 HCV infection treated with sofosbuvir/velpatasvir in the phase III studies. J Hepatol. 2018;68:895–903. doi: 10.1016/j.jhep.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 60.Llibre A, Shimakawa Y, Mottez E, Ainsworth S, Buivan T-P, et al. Development and clinical validation of the Genedrive point-of-care test for qualitative detection of hepatitis C virus. Gut. 2018;67:2017–2024. doi: 10.1136/gutjnl-2017-315783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krajden M, Ziermann R, Khan A, Mak A, Leung K, et al. Qualitative detection of hepatitis C virus RNA: comparison of analytical sensitivity, clinical performance, and workflow of the Cobas Amplicor HCV test version 2.0 and the HCV RNA transcription-mediated amplification qualitative assay. J Clin Microbiol. 2002;40:2903–2907. doi: 10.1128/JCM.40.8.2903-2907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandão CPU, Marques BLC, Marques VA, Villela-Nogueira CA, Do Ó KMR, et al. Simultaneous detection of hepatitis C virus antigen and antibodies in dried blood spots. J Clin Virol. 2013;57:98–102. doi: 10.1016/j.jcv.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 63.Vázquez-Morón S, Ardizone Jiménez B, Jiménez-Sousa MA, Bellón JM, Ryan P, et al. Evaluation of the diagnostic accuracy of laboratory-based screening for hepatitis C in dried blood spot samples: a systematic review and meta-analysis. Sci Rep. 2019;9:7316. doi: 10.1038/s41598-019-41139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alidjinou EK, Deldalle J, Hallaert C, Robineau O, Ajana F, et al. RNA and DNA Sanger sequencing versus next-generation sequencing for HIV-1 drug resistance testing in treatment-naive patients. J Antimicrob Chemother. 2017;72:2823–2830. doi: 10.1093/jac/dkx232. [DOI] [PubMed] [Google Scholar]
- 65.Blach S, Zeuzem S, Manns M, Altraif I, Duberg A-S, et al. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol. 2017;2:161–176. doi: 10.1016/S2468-1253(16)30181-9. [DOI] [PubMed] [Google Scholar]
- 66.Campiotto S, Pinho JRR, Carrilho FJ, Da Silva LC, Souto FJD, et al. Geographic distribution of hepatitis C virus genotypes in Brazil. Braz J Med Biol Res. 2005;38:41–49. doi: 10.1590/s0100-879x2005000100007. [DOI] [PubMed] [Google Scholar]
- 67.Alvarado-Mora MV, Moura IM, Botelho-Lima LS, Azevedo RS, Lopes E, et al. Distribution and molecular characterization of hepatitis C virus (HCV) genotypes in patients with chronic infection from Pernambuco State, Brazil. Virus Res. 2012;169:8–12. doi: 10.1016/j.virusres.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 68.Vieira DS, Alvarado-Mora MV, Botelho L, Carrilho FJ, Pinho JR, et al. Distribution of hepatitis c virus (HCV) genotypes in patients with chronic infection from Rondônia, Brazil. Virol J. 2011;8:165. doi: 10.1186/1743-422X-8-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishiya AS, de Almeida-Neto C, Ferreira SC, Alencar CS, Di-Lorenzo-Oliveira C, et al. HCV genotypes, characterization of mutations conferring drug resistance to protease inhibitors, and risk factors among blood donors in São Paulo, Brazil. PLoS One. 2014;9:e86413. doi: 10.1371/journal.pone.0086413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romano CM, de Carvalho-Mello IMVG, Jamal LF, de Melo FL, Iamarino A, et al. Social networks shape the transmission dynamics of hepatitis C virus. PLoS One. 2010;5:e11170. doi: 10.1371/journal.pone.0011170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zarife MAS, Silva LK, Silva MBS, Lopes GB, Barreto ML, et al. Prevalence of hepatitis C virus infection in north-eastern Brazil: a population-based study. Trans R Soc Trop Med Hyg. 2006;100:663–668. doi: 10.1016/j.trstmh.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 72.Pacheco SDB, Silva-Oliveira GC, Maradei-Pereira LMC, Crescente JÂB, Lemos JAR de, et al. Prevalence of HCV infection and associated factors among illicit drug users in Breves, State of Pará, northern Brazil. Rev Soc Bras Med Trop. 2014;47:367–370. doi: 10.1590/0037-8682-0153-2013. [DOI] [PubMed] [Google Scholar]
- 73.Silva MBS, Andrade TM, Silva LK, Rodart IF, Lopes GB, et al. Prevalence and genotypes of hepatitis C virus among injecting drug users from Salvador-BA, Brazil. Mem Inst Oswaldo Cruz. 2010;105:299–303. doi: 10.1590/s0074-02762010000300009. [DOI] [PubMed] [Google Scholar]
- 74.Coutinho C, Bastos LS, da Mota JC, Toledo L, Costa K, et al. The risks of HCV infection among Brazilian crack cocaine users: incorporating diagnostic test uncertainty. Sci Rep. 2019;9:443. doi: 10.1038/s41598-018-35657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SYW, Shapiro B, et al. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 2009;6:e1000198. doi: 10.1371/journal.pmed.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakano T, Lu L, Liu P, Pybus OG. Viral gene sequences reveal the variable history of hepatitis C virus infection among countries. J Infect Dis. 2004;190:1098–1108. doi: 10.1086/422606. [DOI] [PubMed] [Google Scholar]
- 77.Morice Y, Cantaloube J-F, Beaucourt S, Barbotte L, De Gendt S, et al. Molecular epidemiology of hepatitis C virus subtype 3a in injecting drug users. J Med Virol. 2006;78:1296–1303. doi: 10.1002/jmv.20692. [DOI] [PubMed] [Google Scholar]
- 78.Pereira LMMB, Martelli CMT, Moreira RC, Merchan-Hamman E, Stein AT, et al. Prevalence and risk factors of Hepatitis C virus infection in Brazil, 2005 through 2009: a cross-sectional study. BMC Infect Dis. 2013;13:60. doi: 10.1186/1471-2334-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guimarães V de S, Melo T de, Ferreira R de C, Almeida S de, Martins LC. Prevalence of hepatitis C virus genotypes in the State of Pará, Brazil. Rev Soc Bras Med Trop. 2018;51:508–512. doi: 10.1590/0037-8682-0457-2017. [DOI] [PubMed] [Google Scholar]
- 80.Bassit L, Da Silva LC, Ribeiro-Dos-Santos G, Maertens G, Carrilho FJ, et al. Chronic hepatitis C virus infections in brazilian patients: association with genotypes, clinical parameters and response to long term alpha interferon therapy. Rev Inst Med Trop Sao Paulo. 1999;41:183–189. doi: 10.1590/s0036-46651999000300010. [DOI] [PubMed] [Google Scholar]
- 81.Stramer SL, Glynn SA, Kleinman SH, Strong DM, Caglioti S, et al. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid–amplification testing. N Engl J Med. 2004;351:760–768. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- 82.Tobler LH, Stramer SL, Lee SR, Masecar BL, Peterson JE, et al. Impact of HCV 3.0 EIA relative to HCV 2.0 EIA on blood-donor screening. Transfusion. 2003;43:1452–1459. doi: 10.1046/j.1537-2995.2003.00521.x. [DOI] [PubMed] [Google Scholar]
- 83.Jaques B, Saldanha PC de A, Moraes ACR de. Profile of blood donations with a positive serology in Southern Brazil. Hematol Transfus Cell Ther. 2020;42:129–133. doi: 10.1016/j.htct.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kretzer IF, do Livramento A, da Cunha J, Gonçalves S, Tosin I, et al. Hepatitis C worldwide and in Brazil: silent epidemic--data on disease including incidence, transmission, prevention, and treatment. Sci World J. 2014;2014:1–10. doi: 10.1155/2014/827849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terrault NA, Dodge JL, Murphy EL, Tavis JE, Kiss A, et al. Sexual transmission of hepatitis C virus among monogamous heterosexual couples: the HCV partners study. Hepatology. 2013;57:881–889. doi: 10.1002/hep.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neumayr G, Propst A, Schwaighofer H, Judmaier G, Vogel W. Lack of evidence for the heterosexual transmission of hepatitis C. QJM. 1999;92:505–508. doi: 10.1093/qjmed/92.9.505. [DOI] [PubMed] [Google Scholar]
- 87.Danta M, Brown D, Bhagani S, Pybus OG, Sabin CA, et al. Recent epidemic of acute hepatitis C virus in HIV-positive men who have sex with men linked to high-risk sexual behaviours. AIDS. 2007;21:983–991. doi: 10.1097/QAD.0b013e3281053a0c. [DOI] [PubMed] [Google Scholar]
- 88.de Bruijne J, Schinkel J, Prins M, Koekkoek SM, Aronson SJ, et al. Emergence of hepatitis C virus genotype 4: phylogenetic analysis reveals three distinct epidemiological profiles. J Clin Microbiol. 2009;47:3832–3838. doi: 10.1128/JCM.01146-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuehlkamp VM, Schuelter-Trevisol F. Prevalence of human immunodeficiency virus/hepatitis C virus co-infection in Brazil and associated factors: a review. Braz J Infect Dis. 2013;17:455–463. doi: 10.1016/j.bjid.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheinquer H, Sette H, Wolff FH, de Araujo A, Coelho-Borges S, et al. Treatment of cChronic HCV infection with the new direct acting antiviralsInfection with the New Direct Acting Antivirals (DAA): first report of a real world experienceFirst Report of a Real World Experience in Southern Brazil. Ann Hepatol. 2017;16:727–733. doi: 10.5604/01.3001.0010.2717. [DOI] [PubMed] [Google Scholar]
- 91.Costa VD, Brandão-Mello CE, Nunes EP, Dos Santos Silva PGC, de Souza Rodrigues L, et al. Treatment of chronic HCV infection with DAAs in Rio de Janeiro/Brazil: SVR rates and baseline resistance analyses in NS5A and NS5B genes. PLoS One. 2019;14:e0216327. doi: 10.1371/journal.pone.0216327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ministério da Saúde Boletim Epidemiológico Hepatites Virais 2020. 2020 http://www.aids.gov.br/pt-br/pub/2020/boletim-epidemiologico-hepatites-virais-2020
- 93.Portari-Filho LH, Álvares-da-Silva MR, Gonzalez A, Ferreira AP, Villela-Nogueira CA, et al. How are HCV-infected patients being identified in Brazil: a multicenter study. Braz J Infect Dis. 2019;23:34–39. doi: 10.1016/j.bjid.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Puchades Renau L, Berenguer M. Introduction to hepatitis C virus infection: overview and history of hepatitis C virus therapies. Hemodial Int. 2018;22 Suppl 1:S8–S21. doi: 10.1111/hdi.12647. [DOI] [PubMed] [Google Scholar]
- 95.Te HS, Randall G, Jensen DM. Mechanism of action of ribavirin in the treatment of chronic hepatitis C. Gastroenterol Hepatol. 2007;3:218–225. [PMC free article] [PubMed] [Google Scholar]
- 96.Asahina Y, Izumi N, Enomoto N, Uchihara M, Kurosaki M, et al. Mutagenic effects of ribavirin and response to interferon/ribavirin combination therapy in chronic hepatitis C. J Hepatol. 2005;43:623–629. doi: 10.1016/j.jhep.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 97.Jardim ACG, Yamasaki LHT, de Queiróz ATL, Bittar C, Pinho JRR, et al. Quasispecies of hepatitis C virus genotype 1 and treatment outcome with peginterferon and ribavirin. Infect Genet Evol. 2009;9:689–698. doi: 10.1016/j.meegid.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 98.Hedegaard DL, Tully DC, Rowe IA, Reynolds GM, Bean DJ, et al. High resolution sequencing of hepatitis C virus reveals limited intra-hepatic compartmentalization in end-stage liver disease. J Hepatol. 2017;66:28–38. doi: 10.1016/j.jhep.2016.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vercauteren K, Brown RJP, Mesalam AA, Doerrbecker J, Bhuju S, et al. Targeting a host-cell entry factor barricades antiviral-resistant HCV variants from on-therapy breakthrough in human-liver mice. Gut. 2016;65:2029–2034. doi: 10.1136/gutjnl-2014-309045. [DOI] [PubMed] [Google Scholar]
- 100.Hang JQ, Yang Y, Harris SF, Leveque V, Whittington HJ, et al. Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus. J Biol Chem. 2009;284:15517–15529. doi: 10.1074/jbc.M808889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Uchida Y, Nakamura S, Kouyama J-I, Naiki K, Motoya D, et al. Significance of NS5B substitutions in genotypeSubstitutions in Genotype 1b hHepatitis C virus evaluated by bioinformatics analysisVirus Evaluated by Bioinformatics Analysis. Sci Rep. 2018;8:8818. doi: 10.1038/s41598-018-27291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peres-da-Silva A, Brandão-Mello CE, Lampe E. Prevalence of sofosbuvir resistance-associated variants in Brazilian and worldwide NS5B sequences of genotype-1 HCV. Antivir Ther. 2017;22:447–451. doi: 10.3851/IMP3131. [DOI] [PubMed] [Google Scholar]
- 103.Svarovskaia ES, Dvory-Sobol H, Parkin N, Hebner C, Gontcharova V, et al. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis. 2014;59:1666–1674. doi: 10.1093/cid/ciu697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tong X, Le Pogam S, Li L, Haines K, Piso K, et al. In vivo emergence of a novel mutant L159F/L320F in the NS5B polymerase confers low-level resistance to the HCV polymerase inhibitors mericitabine and sofosbuvir. J Infect Dis. 2014;209:668–675. doi: 10.1093/infdis/jit562. [DOI] [PubMed] [Google Scholar]
- 105.Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, et al. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology. 2015;62:1623–1632. doi: 10.1002/hep.27934. [DOI] [PubMed] [Google Scholar]
- 106.Svarovskaia ES, Gane E, Dvory-Sobol H, Martin R, Doehle B, et al. L159F and V321A sofosbuvir-associated hepatitis C virus NS5B substitutions. J Infect Dis. 2016;213:1240–1247. doi: 10.1093/infdis/jiv564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hamano K, Sakamoto N, Enomoto N, Izumi N, Asahina Y, et al. Mutations in the NS5B region of the hepatitis C virus genome correlate with clinical outcomes of interferon-alpha plus ribavirin combination therapy. J Gastroenterol Hepatol. 2005;20:1401–1409. doi: 10.1111/j.1440-1746.2005.04024.x. [DOI] [PubMed] [Google Scholar]
- 108.Shi ST, Herlihy KJ, Graham JP, Fuhrman SA, Doan C, et al. In vitro resistance study of AG-021541, a novel nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob Agents Chemother. 2008;52:675–683. doi: 10.1128/AAC.00834-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McCown MF, Rajyaguru S, Kular S, Cammack N, Nájera I. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob Agents Chemother. 2009;53:2129–2132. doi: 10.1128/AAC.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.