Abstract
A toluene-degrading methanogenic consortium enriched from creosote-contaminated aquifer material was maintained on toluene as the sole carbon and energy source for 10 years. The species in the consortium were characterized by using a molecular approach. Total genomic DNA was isolated, and 16S rRNA genes were amplified by using PCR performed with kingdom-specific primers that were specific for 16S rRNA genes from either members of the kingdom Bacteria or members of the kingdom Archaea. A total of 90 eubacterial clones and 75 archaeal clones were grouped by performing a restriction fragment length polymorphism (RFLP) analysis. Six eubacterial sequences and two archaeal sequences were found in the greatest abundance (in six or more clones) based on the RFLP analysis. The relative abundance of each putative species was estimated by using fluorescent in situ hybridization (FISH), and the presence of putative species was determined qualitatively by performing slot blot hybridization with consortium DNA. Both archaeal species and two of the six eubacterial species were detected in the DNA and FISH hybridization experiments. A phylogenetic analysis of these four dominant organisms suggested that the two archaeal species are related to the genera Methanosaeta and Methanospirillum. One of the eubacterial species is related to the genus Desulfotomaculum, while the other is not related to any previously described genus. By elimination, we propose that the last organism probably initiates the attack on toluene.
Hydrocarbons, such as toluene, frequently contaminate soils, sediments, and groundwater as a result of inadvertent spills, leaks, or improper methods of disposal. Microbial degradation of toluene occurs readily under both aerobic and anaerobic conditions (15, 24, 28). While much is known about aerobic toluene degradation pathways and the many aerobic species that mineralize toluene, comparatively little is known about anaerobic degradation of toluene. Anaerobic conditions often prevail at contaminated sites, and therefore a better understanding of the fate of toluene and other contaminants under anaerobic conditions is required for proper site assessment and remediation.
Toluene degradation occurs under all of the major anaerobic electron-accepting conditions, including nitrate-reducing (1, 20, 24), sulfate-reducing (7, 18, 32), iron(III)-reducing (29), and methanogenic (17, 40) conditions, and pure cultures of nitrate-reducing, sulfate-reducing, and iron-reducing bacteria that degrade toluene have been isolated. In contrast, toluene degradation to methane and CO2 requires more than one species because of the limited substrate range of methanogenic bacteria. It is thought that fermentative or acetogenic bacteria first transform toluene to methanogenic precursors, such as acetate and hydrogen; methanogenic bacteria then convert these substrates to methane and CO2. Since transformation of toluene to acetate and hydrogen is energetically favorable only when the concentrations of hydrogen and acetate are kept low by the activity of methanogenic bacteria, toluene degradation is necessarily dependent on syntrophic relationships between species in a consortium. To date, no toluene-degrading organism has been isolated from such a consortium.
A methanogenic mixed culture that mineralizes toluene to carbon dioxide and methane was enriched from creosote-contaminated aquifer sediments (17). This stable culture (or consortium) has been maintained on toluene as the sole source of carbon and energy for the last 10 years. Our attempts to isolate toluene-degrading organisms from this consortium by conventional serial dilution or plating techniques have been unsuccessful, presumably because it is difficult to resolve syntrophic associations. The objectives of this research were to identify the different species in this consortium by using a molecular approach and to determine the role which each species plays in the degradative process. We identified the majority of the species in the consortium, and we propose a model which explains the roles of these species in toluene degradation. Identifying the different organisms in a consortium and determining their roles are important steps in improving our understanding of anaerobic biodegradation and, specifically, methanogenic biodegradation. Methanogenic transformation processes are independent of an external electron acceptor source, such as sulfate, nitrate, or oxygen, and therefore represent the ultimate degradation mechanism for organic compounds in the environment.
MATERIALS AND METHODS
Bacterial consortium.
A toluene-degrading methanogenic enrichment culture described previously was maintained in a series of 160-ml serum bottles in a reduced, defined, mineral medium (16, 17). Each bottle contained 100 ml of liquid culture and was fed 10 μl of neat toluene every 2 weeks. The bottles were incubated in a Coy anaerobic chamber. Prior to each feeding, the accumulated methane (approximately 16 ml) was removed from each bottle. Occasionally (about twice a year), cultures were transferred (20 to 50% inoculum) into new bottles and topped up with fresh medium. The rate of degradation has been constant for the past 5 years, and there is no indication that any trace element or vitamin in the medium is actually essential. The cultures in several bottles have not been transferred for more than 2 years, and there has been no significant reduction in the rate of toluene degradation; furthermore, the rate of degradation does not increase appreciably when the cultures are transferred. Samples used for DNA extraction were taken in September 1996, March 1998, and July 1998 from stable cultures that had been receiving toluene every 2 weeks for at least 6 months and were actively degrading toluene at the time of sampling.
Chemicals.
All chemicals were purchased from Sigma and were more than 99.9% pure.
DNA extraction and purification.
DNA was extracted by using a modified version of the protocol developed by Zhou et al. (41). The cells from 100 ml of an active culture (ca. 109 cells/ml) were harvested by centrifugation under anaerobic conditions at 4,000 × g for 30 min. The cells were resuspended in 1 ml of anaerobic medium, transferred to a 1.5-ml Eppendorf tube, and centrifuged at 14,000 rpm for 15 min. The supernatant was decanted, and the pellet was transferred into a sterile mortar to which 5 g of sterile glass beads had been added. The pellet and glass beads were ground with a mortar and pestle under liquid nitrogen to physically disrupt the cells. DNA extraction buffer (13.5 ml) was added to the mortar, and the resulting mixture was transferred to a clean tube. DNA extraction buffer consisted of 100 mM Tris-HCl (pH 8.0), 100 mM sodium EDTA (pH 8.0), 100 mM sodium phosphate (pH 8.0), 1.5 M NaCl, and 1% hexadecylmethylammonium bromide. For the rest of the protocol we used the procedures described by Zhou et al. (41). The DNA pellet was resuspended in 500 μl of double-distilled water (ddH2O) and stored at −20°C.
Oligonucleotide synthesis.
Oligonucleotides were synthesized by Dalton Chemical Laboratories Ltd. (Mississauga, Ontario, Canada) or the MOBIX Facility at McMaster University (Hamilton, Ontario, Canada).
Amplification of 16S rRNA genes.
Eubacterial 16S rRNA genes were selectively amplified from purified genomic DNA by PCR by using forward primer 5′-AGAGTTTGATCCTGGCTCAG-3′ (corresponding to positions 21 to 41 of the Escherichia coli 16S rRNA gene [12, 39]) and reverse primer 5′-GGTTACC TTGTTACGACTT-3′ (corresponding to positions 1510 to 1492 [39]). Archaeal 16S rRNA genes were amplified by performing PCR with forward primer 5′-TTCCGGTTGATCCYGCCGGA-3′ (corresponding to positions 21 to 41 of the E. coli 16S rRNA gene [12, 13]) and the reverse primer described above.
The conditions used for PCR amplification with the eubacterial primers were as follows: denaturation at 95°C for 1 min, primer annealing at 52°C for 1.5 min, and chain extension for 1.5 min at 72°C for 30 cycles, followed by a final extension step consisting of 72°C for 10 min. The same conditions were used with the archaeal primers, except that the annealing temperature was 55°C. The amplified products were separated on a 1% agarose gel that was stained with ethidium bromide and were visualized with UV excitation.
Cloning of 16S rRNA genes and restriction fragment length polymorphism (RFLP) analysis.
PCR products were purified by using a QIAEX gel purification kit (Qiagen Inc., Chatsworth, Calif.) and were cloned into pCR 2.1 by using a TA cloning kit (Invitrogen, San Diego, Calif.). Plasmid DNA was purified by the alkaline lysis method (35), the insert was excised with restriction enzymes BamHI and NotI (New England Biomedical, Mississauga, Ontario, Canada), and the products were resolved on a 1% agarose gel and stained with ethidium bromide. The inserts were excised from the gel, purified with a QIAEX gel purification kit, and then digested at 37°C for 1 to 2 h with either HhaI or RsaI (New England Biomedical) in parallel digestion reactions. Each reaction mixture (total volume, 30 μl) contained 300 ng of insert DNA, the appropriate buffer, bovine serum albumin (if required), 1 μl of RsaI or 1 μl of HhaI, and ddH2O. The digested DNA fragments were separated by agarose gel electrophoresis, stained with ethidium bromide, and visualized with UV excitation.
Sequencing of cloned 16S rRNA gene fragments.
The flanking regions of each unique clone were sequenced with forward primer M13 and reverse primer T7 located on plasmid pCR 2.1. The complete 16S rRNA genes were sequenced by using internal primers designed for this study. The internal eubacterial primers used were 5′-CCTAGGG(A/T)GCAGCAG-3′ (complementary to E. coli positions 341 to 357) and 5′-CACCAGT(C/G)GCGAAGGCGG-3′ (complementary to positions 718 to 735). The internal archaeal primers used were 5′-GAGACACGAATCCAGGC-3′ (complementary to positions 324 to 340) and 5′-AAGCGTCTCACCAGAACG-3′ (complementary to positions 727 to 745).
Phylogenetic analyses of sequences.
Unaligned sequences were entered into the GenBank BLAST search program (2) and the Ribosomal Database Project SIMILARITY_RANK program (30) in order to obtain closely related phylogenetic sequences. Sequences were aligned by using ClustalW (26). Maximum-likelihood phylogenetic trees were created for eubacterial and archaeal sequences by using the DNAml program of PHYLIP, version 3.5 (21). The trees were rooted by including an archaeal sequence in the eubacterial tree and a eubacterial sequence in the archaeal tree. We assessed the significance of the maximum-likelihood branch points by performing a bootstrap analysis with 100 replicates in order to generate a consensus tree (19).
Oligonucleotide probe sequences.
Oligonucleotide probe sequences complementary to amplified rRNA sequences either were obtained from previously published papers or were designed de novo for the new organisms identified in our culture. The sequences of the following four probes were obtained from previously published papers: eubacterial probe EUB338 (4), archaebacterial probe ARCH915 (34), probe MX825 specific for Methanosaeta sp. (34), and probe SRB385 specific for Desulfovibrio sp. (4). A nonsense probe complementary to EUB338 was used as a negative control (4). The probes used for the cloned 16S rRNA genes were designed to be complementary to regions unique to each sequence by using a primer design program (39a). The sequences of the specific primers differed from all other sequences by at least 2 bp. Potential probe sequences were analyzed with the CHECK_PROBE program of the Ribosomal Database Project (30).
Probe labeling.
The oligonucleotides used for fluorescent in situ hybridization (FISH) analysis were purchased from Dalton Chemicals already conjugated with a fluorescent label at the 5′ end. Probes ARCH915, Eub-1, and MX825 were conjugated with rhodamine. Probes EUB338, Eub-2, Eub-3, Eub-4, SRB325, Eub-6, nonsense, and Arch-2 were conjugated with fluorescein. The oligonucleotides used for slot blot analysis were 5′ end labeled with 5 μl of [γ-32P]ATP (6,000 Ci/mmol; 10 mCi/ml; Amersham) by using 10 pmol of oligonucleotide and 10 U of T4 polynucleotide kinase (New England Biolabs) in a 25-μl reaction mixture that was incubated for 45 min at 37°C. Labeled oligonucleotides were purified by using Qiaquick Nucleotide Removal spin columns (Qiagen), and the level of incorporation was quantified (35).
Preparation of slides for FISH.
HTC supercured Teflon-coated slides with 10 or 12 wells per slide (Cel-Line, Newfield, N.J.) were used for FISH analysis. Prior to probing, the slides were soaked for 1 h in ethanolic KOH (10% [wt/vol] potassium hydroxide), rinsed with ddH2O, and air dried. To enhance cell adhesion, all of the slides were dipped in a gelatin solution (0.1% gelatin, 0.01% chromium potassium sulfate; 70°C) and dried in a vertical position (4).
Cell fixation and whole-cell hybridization.
Culture samples were removed 2 days after feeding with toluene. The cells were harvested by centrifugation at 13,000 × g for 5 min, and each cell pellet was resuspended in 750 μl of a solution containing freshly prepared 4% paraformaldehyde (PFA) in water (33). The PFA fixation solution was prepared by mixing 1 drop of 10 M NaOH, 2 g of PFA, and 16.5 ml of 3× phosphate-buffered saline with 33 ml of ddH2O. The PFA solution was heated to 60°C and cooled on ice, the pH was adjusted to 7.2, and finally the solution was filtered through a 0.45-μm-pore-size filter. The cells were resuspended in PFA by vortexing the preparation for approximately 60 s and then were incubated at room temperature for 1 min or 2 h or 12 h. The cells were recovered by centrifugation and washed in a solution containing 900 μl of phosphate-buffered saline (130 mM NaCl plus 10 mM sodium phosphate, pH 7.2) and 100 μl of 0.1% Igepal CA-630 (Sigma) (a nonionic detergent). Then the cells were recovered by centrifugation and washed with 500 μl of 0.1% Igepal CA-630. The final cell pellet was resuspended in a solution containing 200 μl of storage buffer (20 mM Tris-HCl [pH 7.2], 0.1% Igepal CA-630) and an equal amount of absolute ethanol (33) and stored at 4°C. One microliter of the fixed cell suspension was added to each well of a gelatin-treated, Teflon-coated slide. The cell smear was allowed to air dry. The slides were then dehydrated by immersing them in 50% ethanol for 3 min, in 80% ethanol for 3 min, and then in 100% ethanol for 3 min and finally were air dried. Ten microliters of proteinase K (10 mg/ml in 20 mM Tris-HCl–2 mM CaCl2 [pH 7.4]) was added to each well. The slides were incubated at 37°C for 10 min, washed with ddH2O, and air dried. Nine microliters of hybridization buffer, 1 μl of 4′,6′-diamidino-2-phenylindole (DAPI) stain (0.2 μg/μl), and 25 to 50 ng of probe were added to each well. The hybridization solutions contained 0.9 M NaCl, 0.1% sodium dodecyl sulfate (SDS), 20 mM Tris-HCl (pH 7.2), and different concentrations of formamide (0 to 60%). The optimum formamide concentration was different for each probe but ranged from 20 to 40%. The concentration of SDS (0.01, 0.1, or 1%) had no effect on the success of hybridization; therefore, 0.01% SDS was used throughout the study. The slides were incubated at 34 to 52°C for 4 h in a tissue culture dish sealed with Parafilm in a humid atmosphere. After incubation, the wells were washed once with 20 μl of prewarmed hybridization buffer and then incubated at 34 to 52°C for 20 min. The slides were briefly rinsed with ddH2O, air dried in the dark, and mounted in Dako fluorescent mounting medium (Dako Corporation, Carpinteria, Calif.). Fluorescence was detected with an Axioskop microscope (Zeiss, Oberkochen, Germany) fitted for epifluorescence microscopy with a type HBO 100-W AttoArc bulb and Zeiss filter sets I, III, and IV. The magnification used was ×1,000. Color photographs were taken with Kodak Gold 400 ASA film; the exposure times were 8 s for DAPI photographs and 8 to 15 s for epifluorescence photographs.
Slot blot hybridization.
Nucleic acids were diluted with 200 μl of 6× SSPE (20× SSPE is 3.6 M sodium chloride, 0.2 M sodium phosphate [monobasic], and 20 mM EDTA [pH 7.4]) to give final concentrations of 0.1, 1, and 2 μg of nucleic acids per 200-μl sample. Each diluted sample was boiled for 10 min and then kept on ice. Samples were applied to Hybond N membrane filters (Amersham) by using a Minifold II slot blot apparatus (Schleicher & Schuell). The membrane filter preparations were denatured for 5 min with 1.5 M sodium hydroxide–0.5 M sodium chloride and were neutralized for 5 min with 1.5 M sodium chloride–0.5 M Tris (pH 7.4). The DNA was cross-linked by treatment with UV light (1,200 μJ; Stratagene 1800). The membrane filters were prehybridized for at least 2 h at 40°C in heat-sealed bags by using 0.2 ml of hybridization solution (6× SSPE, 5× Denhardt’s reagent, 1% SDS, 50 μg of denatured single-stranded DNA per ml) per cm2. Labeled oligonucleotides were added to the appropriate membrane filters along with fresh hybridization solution, and the preparations were incubated at 40°C overnight. The membrane filters were washed three times (10 min each) in low-stringency wash solution (6× SSPE, 1% SDS) at room temperature and once for 45 min in high-stringency wash solution (1× SSPE, 1% SDS) at the appropriate temperature. The high-stringency wash temperatures used were as follows: UNIV522, 45°C; Eub-1, 50°C; Eub-2, 50°C; Eub-3, 40°C; Eub-4, 55°C; SRB385, 50°C; Eub-6, 50°C; MX825, 65°C; and Arch-2, 40°C. The membranes filters were wrapped in plastic wrap and exposed to Biomax MR film (Kodak) either overnight or for up to 2 days by using an intensifying screen.
RESULTS
DNA extraction and amplification.
Approximately 300 μg of genomic DNA was isolated from 100 ml of bacterial culture. The extracted DNA was large (>12 kb). Both eubacterial and archaeal 16S rRNA genes were readily amplified with the primers used. No amplification occurred with the negative PCR controls (i.e., samples without a DNA template), indicating that there was no contamination with other sources of DNA.
Clones and RFLP analysis.
The amplified 16S rRNA gene sequences were cloned. A total of 90 eubacterial clones and 75 archaeal clones were grouped by performing an RFLP analysis. While it has been recommended that three restriction enzymes should be used in order to ensure that all organisms are differentiated properly, we used only two enzymes, RsaI and HhaI, in separate digestions, because these enzymes are reported to have a high combined efficacy for distinguishing known bacterial taxa (31). Using these restriction enzymes resulted in between two and seven bands per digest for both eubacterial and archaeal clones.
Six dominant eubacterial operational taxonomic units (OTUs), designated OTUs Eub-1 though Eub-6, were identified based on the RFLP patterns. Fourteen percent of the clones grouped with OTU Eub-1, 18% grouped with OTU Eub-2, 7% grouped with OTU Eub-3, 11% group with OTU Eub-4, 15% grouped with OTU Eub-5, and 8% grouped with OTU Eub-6. The remaining 27% of the eubacterial clones produced RFLP patterns that differed from the six dominant patterns found. RFLP patterns that were obtained for only one clone were not examined further, as they were probably the result of an operational artifact (e.g., incomplete digestion). In four cases, a unique RFLP pattern was obtained for two clones. To determine if these patterns were distinct from the six patterns described above, we sequenced the first 500 bp of the inserts of the clones. We found that the sequences were essentially the same as sequences obtained for the dominant six clones and therefore did not represent additional OTUs.
Two archaeal OTUs, designated OTUs Arch-1 and Arch-2, were identified; 84% of the clones belonged to the Arch-1 group, and 13% belonged to the Arch-2 group. The remaining 3% of the clones each produced a unique RFLP pattern and were not considered further.
Phylogenetic analysis.
The 16S rRNA gene insert sequences were determined for two representative clones belonging to each of the six eubacterial OTUs and two archaeal OTUs. The CHECK_CHIMERA program of the Ribosomal Database Project (30) did not detect chimeric sequences in any of the eight cloned sequences. Phylogenetic trees were generated for the eubacterial and methanogenic OTUs by using maximum-likelihood methods at two separate points in the sequencing process; the first was when approximately 800 bp of the sequence was known (data not shown), and the second was when the entire amplified fragment had been sequenced (Fig. 1 and 2). The groups on the trees did not differ, an observation that supports the results of previous work in which the results of analyses based on the first 500 bp of the 16S rRNA gene were consistent with the results of analyses based on the entire sequence (9, 11). It is generally accepted that bootstrap values greater than 95% are statistically significant (22).
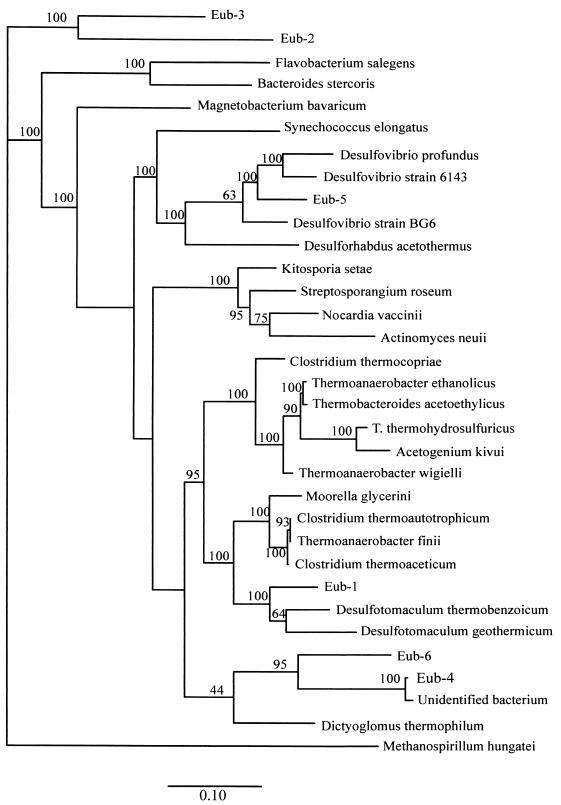
FIG. 1.
Phylogenetic tree for eubacteria, showing the positions of the six OTUs obtained by RFLP analysis. Scale bar = 10 substitutions/100 nucleotide positions.
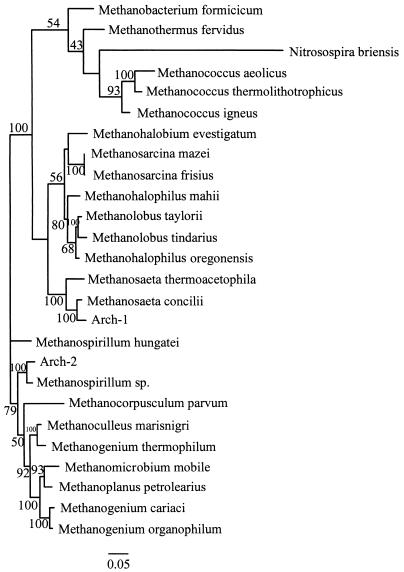
FIG. 2.
Phylogenetic tree for archaebacteria, showing the positions of the two OTUs obtained by RFLP analysis. Scale bar = 5 substitutions/100 nucleotide positions.
Some of the OTUs grouped with previously described genera in the phylogenetic analysis when 16S rRNA genes were used. In general, species are defined as organisms that exhibit 95 to 99% sequence similarity (3, 8, 9), whereas similarity values of 85 to 95% are used to group organisms belonging to families or genera (11, 14, 38). The exact criteria used for grouping organisms vary depending on the author and the taxonomic groups. When the guidelines described above were used, three of the eubacterial OTUs grouped with previously described organisms; OTU Eub-5 grouped with Desulfovibrio sp., OTU Eub-1 grouped with Desulfotomaculum sp. (both groups of organisms are sulfate-reducing bacteria), and OTU Eub-4 grouped very closely with an unidentified organism isolated from the aeration basin of a municipal sewage plant in Munich-Grosslappen, Germany (36). Both archaeal OTUs grouped with previously described taxa; OTU Arch-1 grouped with Methanosaeta sp., and OTU Arch-2 grouped with Methanospirillum sp. OTUs Eub-2, Eub-3, and Eub-6 did not group closely with any previously described organism in the database.
Oligonucleotide probe sequences.
PCR-RFLP analysis is a crude first step for examining community composition, especially in slowly growing, batch cultures with very low dilution rates. Confirmation is required to establish that sequences which are found actually correspond to dominant organisms in a consortium. To do this, we designed species-specific probes for the six eubacterial OTUs and the two methanogenic OTUs or used previously described species-specific probes. Kingdom-specific probes were obtained from previously published papers. The sequences of 16S rRNA probes used and their target groups are shown in Table 1.
TABLE 1.
Oligonucleotide probe sequences used in slot blot and fluorescent hybridization experiments
| Probe | Sequence (5′ to 3′) | Target (E. coli positions) | Specificity | Reference |
|---|---|---|---|---|
| UNIV522 | GWATTACCGCGGCKGCTG | 522–540 | All bacteria | 25 |
| EUB338 | GCTGCCTCCCGTAGGAGT | 338–355 | Eubacteria | 4 |
| ARCH915 | GTGCTCCCCCGCCAATTCCT | 915–935 | Archaebacteria | 34 |
| Nonsense | ACTCCTACGGGAGGCAGC | 338–355 | Control | 4 |
| Eub-1 | GGGAACTCCACTCCCCTG | 664–681 | OTU Eub-1 | This study |
| Eub-2 | ACTGGTTGTGCGCCTCC | 927–942 | OTU Eub-2 | This study |
| Eub-3 | AATCTTTACTCTAAAACACAC | 197–218 | OTU Eub-3 | This study |
| Eub-4 | CCACCTCCCTCTACCATCC | 656–674 | OTU Eub-4 | This study |
| Eub-5 (SRB385) | CGGCGTCGCTGCGTCAGG | 385–402 | Desulfovibrio | 4 |
| Eub-6 | CCACTCCCCTCTGTCACC | 657–674 | OTU Eub-6 | This study |
| Arch-1 (MX825) | TCGCACCGTGGCCGACACCTAGC | 825–847 | Methanosaeta | 34 |
| Arch-2 | ATAGTCTATGGGGTATTATC | 171–190 | OTU Arch-2 | This study |
FISH.
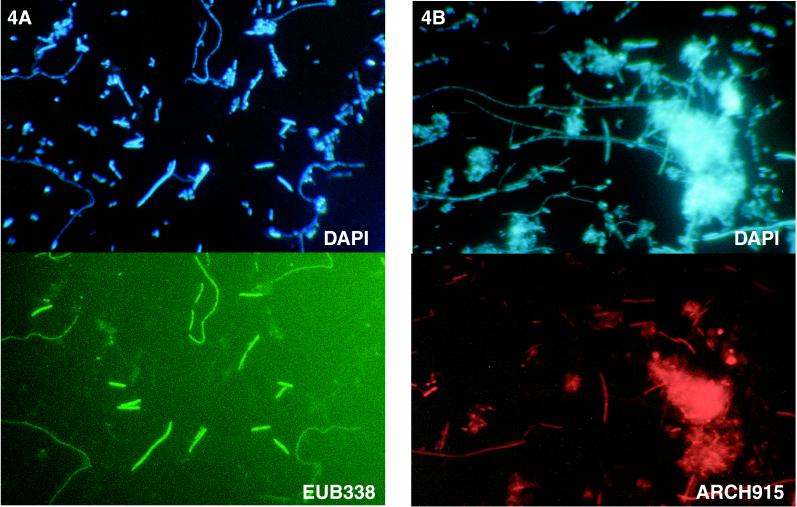
FISH was used to determine if the 16S rRNA sequences that were obtained in the PCR-RFLP analysis actually represented major species in the toluene-degrading culture. The FISH technique provides definitive confirmation of the presence of active species in a consortium, since the probes target labile rRNA and not DNA, which is targeted in slot blot experiments. Furthermore, FISH data provide information that links morphology to identity. All of the probes listed in Table 1 except the universal probe were used in FISH experiments. Preliminary experiments were conducted to validate the FISH analysis procedure; known eubacterial and archaeal populations were probed with the kingdom-specific probes (EUB338 and ARCH915 at 45°C and in the presence of 30% formamide, respectively) and with the nonsense probe. The kingdom-specific probes hybridized only with expected targets. The nonsense probe (same strand as the target sequence) highlighted only clumps of cells and not individual cells, and the fluorescence was thought to result from trapping of the probe between cells in the clumps rather than from specific hybridization.
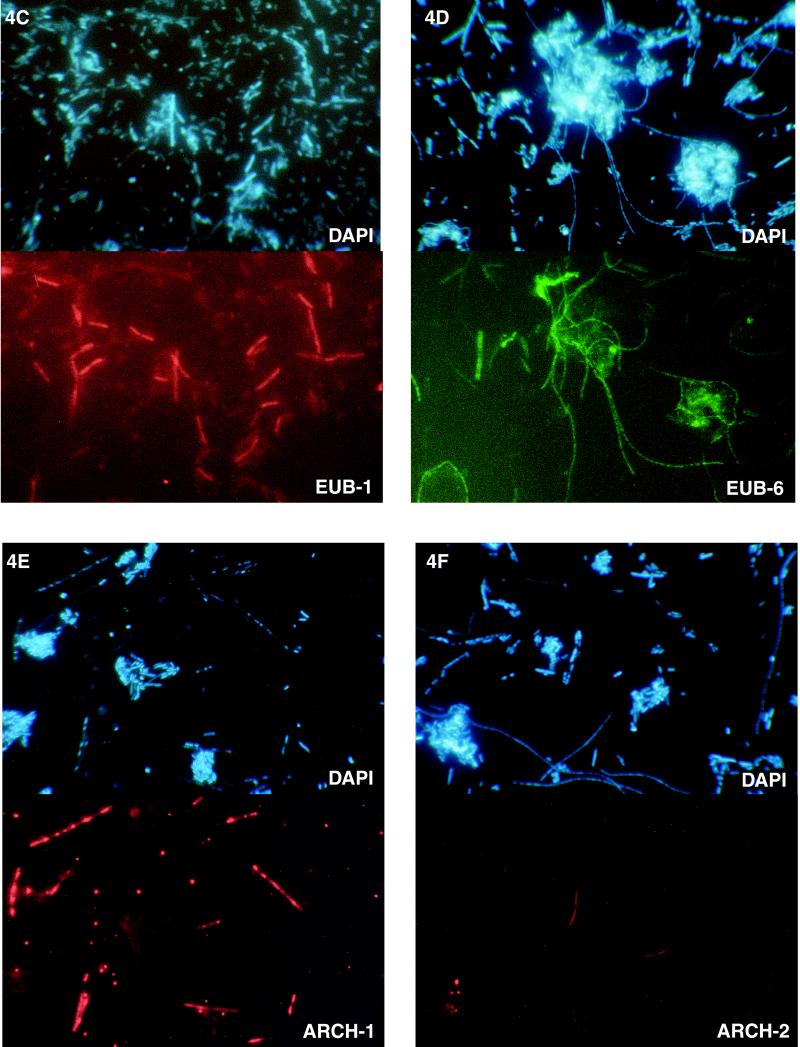
The probes were then tested with cell preparations obtained from the toluene-degrading culture. The kingdom-specific eubacterial probe, EUB338, hybridized to the rRNA of cells having two different morphologies (thick rods that formed small straight chains and extremely long curving chains) (Fig. 4A). The archaeal probe, ARCH915, highlighted short rods and filamentous chains (Fig. 4B). Next, the species-specific probes were tested. The probe for OTU Eub-1 hybridized to short to medium-length rods that occurred in chains (Fig. 4C). The probe for OTU Eub-6 hybridized to long slender chains similar to the chains highlighted by EUB338 (Fig. 4D). The fluorescent probes for OTUs Eub-2, Eub-3, Eub-4, and Eub-5 (SRB385, specific for Desulfovibrio sp.) did not hybridize to any cells (data not shown). The probe for OTU Arch-1 (MX825), which was specific for Methanosaeta sp., hybridized to distinctive chain-forming rods which appeared to be encased in a sheath (Fig. 4E). In this preparation, the cells in a chain appeared to have been stained unevenly with both DAPI and the fluorescent probe. The small coccoidlike cells in Fig. 4E are just single cells of the same species as the species whose cells occurred in chains. This observation is consistent with the unique morphology of Methanosaeta sp. The probe for OTU Arch-2 hybridized to rods that characteristically formed short chains, although the number of cells of this type was apparently low (Fig. 4F). The ratio of fluorescent cells to total (DAPI-stained) cells was determined by directly counting the visible cells in at least two fields for each probe (Table 2). Together, the two kingdom-specific probes, EUB338 and ARCH915, hybridized to all of the cells in the culture since the cells detected with these two probes accounted for 104% of the DAPI-stained cells. About two-thirds of the cells were methanogenic bacteria, and one-third were eubacteria. Probes Eub-1 and Eub-6 hybridized to all of the eubacterial cells since these two probes accounted for 34% of the total counts, which compares well with the value obtained with the eubacterial probe (39%) (Table 2). In contrast, the two probes for methanogenic species, MX825 and Arch-2, hybridized to only about 19% of the total cells, compared to the value of 65% obtained with the archaeal probe; this indicated that not all of the archaeal cells were detected with probes Arch-2 and MX825 (Table 2). The FISH results obtained with Arch-2 were very poor, although this probe hybridized quite well in slot blot experiments (see below); these findings suggest that probe Arch-2 may not be suitable for detecting cells by FISH and should be redesigned.
FIG. 4.
Photomicrographs of DAPI-stained cells (upper panels) and FISH results (lower panels) for the same fields of view for a toluene-degrading methanogenic consortium. The fluorescent probes used were EUB338 (A), ARCH915 (B), Eub-1 (C), Eub-6 (D), Arch-1/MX825 (E), and Arch-2 (F).
TABLE 2.
Proportions of the total population hybridizing in FISH experiments
| Probe | Type or target | % of population hybridizinga with:
|
|
|---|---|---|---|
| Kingdom-specific probes | Species-specific probes | ||
| Eub-1 | Desulfotomaculum | 16 | |
| Eub-2 | Species specific | 0 | |
| Eub-3 | Species specific | 0 | |
| Eub-4 | Species specific | 0 | |
| Eub-5 (SRB385) | Desulfovibrio | 0 | |
| Eub-6 | Species specific | 18 | |
| EUB338 | All eubacteria | 39 | |
| Arch-1 (MX825) | Methanosaeta | 17 | |
| Arch-2 | Methanospirillum | 2 | |
| ARCH915 | All archaeabacteria | 65 | |
| Total recovery | 104 | 53 | |
Data are expressed as the percentages of fluorescent cells that hybridized to a probe; the values were obtained by using the number of fluorescent cells and the total number of DAPI-stained cells in the same field of view.
When observing cells under the microscope with the filter set used to view fluorescein-labeled probes, we noticed that sometimes cells fluoresced even though no probe was applied to the preparation. Many methanogenic bacteria contain high levels of the electron carrier F420 and thus autofluoresce blue-green when they are illuminated with light at wavelengths near 420 nm (blue-violet) (42). To avoid interference with autofluorescence, we recommend that probes be synthesized with rhodamine as a fluorescent marker instead of fluorescein. For example, the autofluorescent cells are the fatter, diffuse cells in Fig. 4D. These cells were not included in the cell counts obtained for OTU Eub-6.
Only four of the eight species identified by PCR-RFLP analysis were detected in the consortium by FISH. However, a lack of hybridization in the FISH experiments did not necessarily mean that the species were not present. Since we had no way of testing the probes with known cultures, it is possible that the probes did not hybridize because the targeted region of the 16S rRNA was inaccessible or the specific cells were not sufficiently permeable for the probe to enter. To rule out these access problems, we used all of the probes in slot blot experiments to determine if they hybridized to purified nucleic acid preparations.
Slot blot hybridizations.
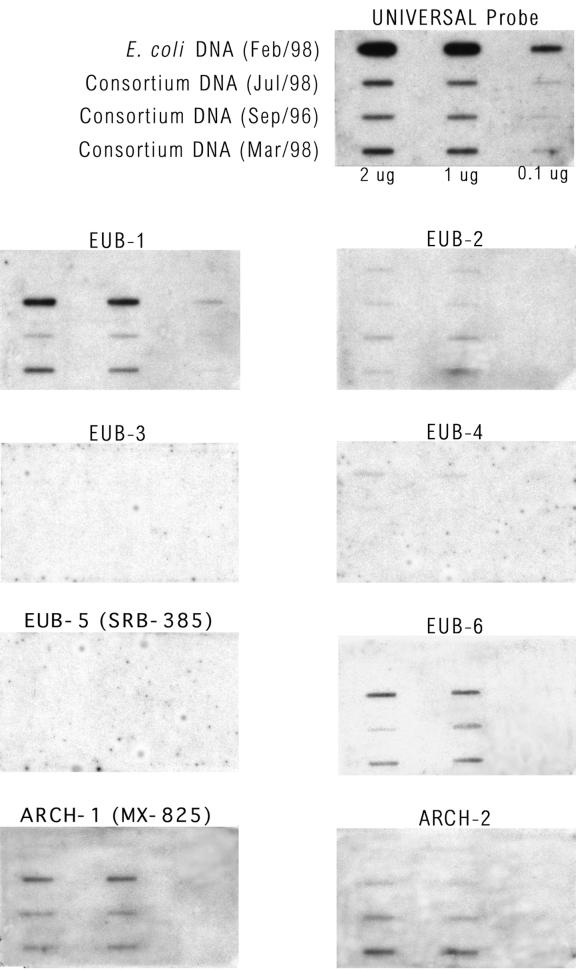
Aliquots of DNA extracted from the toluene-degrading consortium on three different dates (in September 1996, March 1998, and July 1998), as well as control DNA from E. coli, were adsorbed onto membranes at three different concentrations. The DNA extracted in September 1996 was the DNA used to generate the clone library described in this paper. The DNA samples were probed with a universal probe (as a control for the relative amount of DNA in each preparation) and with each of the species-specific probes (Fig. 3). The probes were also tested with plasmid DNA extracted from clones that belonged to each group; all of the probes used hybridized with the intended target sequences (data not shown). Only two of the six eubacterial probes tested, the probes for OTUs Eub-1 and Eub-6, exhibited significant and specific hybridization to the consortium DNA. The probes for OTUs Eub-3, Eub-4, and Eub-5 (SRB385) did not hybridize at all with DNA from the consortium. The probe for OTU Eub-2 hybridized slightly with all of the DNA samples, including the E. coli sample, suggesting that the probe was not specific. In Fig. 3, the hybridization signal in the bottom slot of the center column for OTU Eub-2 is artificially dark as a result of a spot on the film. The relative intensity of hybridization to probe Eub-2 was greatest with the DNA obtained in September 1996 compared to DNA samples obtained later (this was especially noticeable with the data obtained with 2 μg of DNA), suggesting that if OTU Eub-2 was present, its relative abundance decreased with time. In contrast, OTUs Eub-1 and Eub-6 became relatively more abundant with time. Both archaeal probes hybridized significantly and specifically with the consortium DNA.
FIG. 3.
Autoradiographs showing the results of slot blot hybridization experiments. In all nine panels, the first row contained E. coli DNA, and the next three rows contained different DNA samples from the consortium that were extracted on three separate dates (as indicated). Each DNA sample was applied at three concentrations (2, 1, and 0.1 μg in 200 μl). The DNA extracted from the consortium in September 1996 (third row) was the sample used for molecular characterization.
In summary, the slot blot experiments qualitatively confirmed the results of the FISH experiments, perhaps with the exception of the results for OTU Eub-2. Of the six eubacterial sequences obtained by PCR amplification of 16S rRNA genes, only two were clearly detected in the FISH experiments; the same two were clearly detected in slot blot hybridization experiments. Probe Eub-2 was the only other eubacterial probe which hybridized in the slot blot experiments, suggesting that OTU Eub-2 may have been missed by FISH; however, this probe may not be specific as it also hybridized to E. coli DNA. It is also possible that OTU Eub-2 was not sufficiently abundant to be detected by FISH. The two archaeal sequences obtained by PCR amplification of 16S rRNA genes were detected by both FISH and slot blot hybridization. However, the FISH results suggest that OTU Arch-2 may not have been very abundant and that other archaeal species may have been missed.
DISCUSSION
Complete anaerobic degradation of relatively complex organic molecules to carbon dioxide and methane requires the concerted effort of three major metabolic groups of bacteria, the hydrolytic fermentative bacteria, the syntrophic acetogenic bacteria, and the methanogenic bacteria (5). Because of the various syntrophic associations in methanogenic consortia, isolation of pure cultures is difficult, and few consortia have been thoroughly described.
The use of a combination of molecular techniques (PCR-RFLP, FISH, and slot blot hybridization) enabled us to identify several species in a very well-established, stable, toluene-degrading methanogenic consortium. Using PCR-RFLP analysis, we identified six different eubacterial OTUs and two different archaeal OTUs. While both archaeal OTUs were subsequently also detected by slot blot hybridization and in situ hybridization, the same was not true for the eubacterial OTUs. Only three of the six eubacterial OTUs were detected by slot blot hybridization (OTUs Eub-1, Eub-6, and possibly Eub-2), and only two were detected by FISH (OTUs Eub-1 and Eub-6). Each of these methods suffers from biases and shortcomings that may have resulted in overestimation or underestimation of the number of species in the consortium. PCR amplification may have amplified DNA from very minor species or dormant or dead residual cells that were not abundant and therefore were not detected by slot blot hybridization or in situ hybridization, whose detection limits are much higher. For example, OTU Eub-5 detected by PCR grouped very closely with Desulfovibrio species, and the sequence of the previously described Desulfovibrio-specific probe (SRB385) matched exactly the sequence of our clone, suggesting that this OTU was not an artifact; however, the probe did not hybridize in either slot blot or FISH experiments. Perhaps if we had used less template DNA and fewer cycles in the initial PCR we would have obtained better representation of the community when this method was used. Also, we may have completely missed some species in the consortium whose DNA were not amplified by PCR. Despite these caveats, both slot blot and fluorescent hybridization experiments confirmed that the toluene-degrading methanogenic consortium which we studied consists of at least two methanogenic species and two eubacterial species. These four species accounted for more than one-half of the total population (as determined by FISH).
In FISH experiments, the fluorescent eubacterial kingdom-specific probe hybridized to cells with two distinct morphologies (long thin chains and shorter fatter chains) which were also specifically targeted by species-specific probes for OTU Eub-1 (short fat rods) and OTU Eub-6 (long thin chains). Thus, all of the eubacterial cells detected by the more general probe were accounted for by the two species-specific probes. This conclusion is supported by the cell count data (Table 2); the two species-specific probes accounted for about 87% of the cells that hybridized to the kingdom-specific probes. Therefore, it is reasonable to conclude that all of the dominant eubacterial species were identified. The FISH results were less conclusive for the archaea; in this case the two species-specific probes accounted for only about 29% of the cells that hybridized to the kingdom-specific probe. The archaeal kingdom-specific probe did not hybridize to cells with easily distinguishable morphologies; thus, it was not possible to clearly distinguish all of the archaeal cell types present. However, as only two OTUs were detected by RFLP analysis and since both methanogenic sequences were clearly detected by slot blot hybridization, it is reasonable to conclude that two methanogenic species are present in the consortium and that they are probably the dominant species. In addition, while many different types of methanogenic bacteria autofluoresce blue-green when they are illuminated with light with wavelengths near 420 nm (blue-violet), previously described Methanosaeta species do not typically contain high enough concentrations of F420 to autofluoresce (42), suggesting that the autofluorescence which we observed in FISH control experiments without probes came from methanogenic species other than Methanosaeta, such as Methanospirillum-like organisms or other methanogenic species not detected by PCR.
While some archaeal species in the consortium may have escaped detection, the identity, morphology, and function of each of the four species that were detected are consistent with previous experimental observations of this culture and with how methanogenic consortia are known to operate. Three of the four organisms identified grouped relatively closely with previously described organisms, and thus we can postulate functions for these organisms. The two methanogenic species found in the consortium play complementary roles. Methanosaeta species (OTU Arch-1) are aceticlastic methanogens that split acetate, oxidizing the carboxylic group to CO2 and reducing the methyl group to methane. No other substrate supports growth. Thus, these organisms presumably utilize the acetate produced from toluene by other species in the consortium. Acetate has been detected in the culture medium, and when acetate is added to a culture, it inhibits toluene degradation (17). Methanosaeta species grow in sheaths which confer a rod shape; often the organisms form in chains which are typically longer than 100 μm (42). This type of blunt-ended rod-shaped organism was observed previously in scanning electron micrographs of our consortium and was at the time tentatively identified as a Methanosaeta sp. (17). Methanospirillum species (OTU Arch-2) use formate and hydrogen as electron donors (10). Thus, these organisms presumably utilize hydrogen or formate produced by other organisms in the culture. Hydrogen-utilizing methanogens have been detected previously in subcultures of toluene-degrading cultures that were fed only hydrogen. Furthermore, hydrogen has been detected at very low concentrations (around 10−4 atm) in culture headspaces, and when hydrogen was added to culture bottles at a high concentration, toluene degradation was inhibited (17). While OTU Arch-2 certainly groups with Methanospirillum spp. on a phylogenetic tree, its morphology appears to be unlike the morphology of previously described Methanospirilla in culture.
One of the two eubacterial species (OTU Eub-1) is related to Desulfotomaculum spp. The cells of Desulfotomaculum species are gram-positive, endospore-forming, straight or curved rods that are found primarily in soil (14). Gram-positive endospore-forming cells with a morphology similar to that highlighted by fluorescent probe Eub-1 were observed previously in the culture when light microscopy and an endospore stain were used (data not shown). Desulfotomaculum species are sulfate-reducing bacteria but in the absence of sulfate grow acetogenically on substrates such as ethanol, propionate, butyrate, benzoate, and other reduced intermediary metabolites (37). Since sulfate inhibited degradation in the culture (17, 23), it seems unlikely that this organism initiated the attack on toluene; if it was the toluene-degrading organism, one would expect that sulfate would stimulate toluene degradation, not inhibit it. The other eubacterial species found in the consortium (OTU Eub-6) does not group with any previously described genus. The fluorescent probe for this organism hybridized with long thin chains of cells. At least in some sections, these chains appeared to wrap around or intertwine with other bacteria in the consortium (Fig. 4D). Close proximity of organisms is probably necessary for interspecies metabolite transfer in syntrophic cultures. If it is argued that the species that initiates the attack on toluene is most probably a eubacterium (it is unlikely to be a methanogen), then by elimination we postulate that OTU Eub-6 is the organism that initiates the degradation of toluene.
Benzoate is a central intermediate in anaerobic transformation and mineralization of many aromatic compounds and has been shown previously to be an early intermediate during degradation of toluene by the consortium which we studied (16). Several methanogenic consortia that degrade benzoate have been described (27), and pure cultures of syntrophic benzoate degraders have been isolated in some cases. Belaich et al. (6) described a stable benzoate-degrading consortium that closely resembles our consortium. This benzoate-degrading consortium was composed of four morphologically dominant species that were tentatively identified as two methanogens (resembling Methanospirillum sp. and Methanosaeta sp.) and two eubacteria (a spore-forming sulfate reducer and a syntrophic benzoate degrader). The authors proposed that the sulfate-reducing bacterium might play a homoacetogenic role in a process in which H2 and CO2 resulting from benzoate oxidation by the syntroph are used. This benzoate-degrading consortium is remarkably similar to our toluene-degrading consortium. By analogy, we can begin to formulate a hypothetical model (that needs to be verified) for the degradation of toluene by the latter consortium. OTU Eub-6 may initiate the attack on toluene, converting toluene to some as-yet-undetermined intermediate(s). The intermediate(s) is then converted by OTU Eub-1 (Desulfotomaculum sp.) to acetate and hydrogen, which in turn are consumed by OTU Arch-1 (Methanosaeta sp.) and OTU Arch-2 (Methanospirillum sp.), respectively.
In summary, we found that a methanogenic toluene-degrading consortium was composed of at least two eubacterial species and two archaeal (methanogenic) species. Only one of these four species, the putative toluene-degrading eubacterium, was not closely related to any previously described genus. Further experimentation is required to verify the proposed roles of the identified species in the consortium and to determine if there are any species in the consortium that have not been detected yet.
ACKNOWLEDGMENTS
We gratefully acknowledge Aled Edwards (University of Toronto), Brian Golding, and Michael Rudnicki (McMaster University) for their help.
This research was supported by the Natural Science and Engineering Research Council of Canada through an operating grant awarded to E.E. and by the Medical Research Council of Canada through a grant awarded to Aled Edwards.
REFERENCES
- 1.Altenschmidt U, Oswald B, Fuchs G. Anaerobic toluene oxidation to benzyl alcohol and benzaldehyde in a denitrifying Pseudomonas strain. J Bacteriol. 1992;174:4860–4862. doi: 10.1128/jb.174.14.4860-4862.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amann R I. Fluorescently labeled, ribosomal-RNA-targeted oligonucleotide probes in the study of microbial ecology. Mol Ecol. 1995;4:543–553. [Google Scholar]
- 4.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaty P S, McInerney M J, Wofford N Q. Energetics of H2-producing syntrophic bacteria. In: Antonopoulos A A, editor. Biotechnological advances in processing municipal wastes for fuels and chemicals. Chicago, Ill: Argonne National Laboratory; 1986. pp. 67–83. [Google Scholar]
- 6.Belaich J-P, Heitz P, Rousset M, Garcia J L. Energetics of the growth of a new syntrophic benzoate degrading bacterium. In: Belaich J-P, editor. Microbiology and biochemistry of strict anaerobes involved in interspecies transfer. New York, N.Y: Plenum Press; 1990. pp. 269–280. [Google Scholar]
- 7.Beller H R, Spormann A M, Sharma P K, Cole J R, Reinhard M. Isolation and characterization of a novel toluene-degrading, sulfate-reducing bacterium. Appl Environ Microbiol. 1996;62:1188–1196. doi: 10.1128/aem.62.4.1188-1196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boivin-Jahns V, Ruimy R, Bianchi A, Daumas A, Christen R. Bacterial diversity in a deep-subsurface clay environment. Appl Environ Microbiol. 1996;62:3405–3412. doi: 10.1128/aem.62.9.3405-3412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boone D R, Whitman W B, Rouviere P. Diversity and taxonomy of methanogens. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry and genetics. New York, N.Y: Chapman & Hall; 1990. pp. 35–80. [Google Scholar]
- 11.Borneman J, Skroch P W, Osullivan K M, Palus J A. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duetz W A, Dejong C, Williams P A, Vanandel J G. Competition in chemostat culture between Pseudomonas strains that use different pathways for the degradation of toluene. Appl Environ Microbiol. 1994;60:2858–2863. doi: 10.1128/aem.60.8.2858-2863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards E A, Edwards A M, Grbic-Galic D. A method for the detection of metabolites at very low concentration: application to the detection of metabolites of anaerobic toluene degradation. Appl Environ Microbiol. 1994;60:323–327. doi: 10.1128/aem.60.1.323-327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards E A, Grbic-Galic D. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl Environ Microbiol. 1994;60:313–322. doi: 10.1128/aem.60.1.313-322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards E A, Wills L E, Reinhard M, Grbic-Galic D. Anaerobic degradation of toluene and xylene by aquifer microorganisms under sulfate-reducing conditions. Appl Environ Microbiol. 1992;58:794–800. doi: 10.1128/aem.58.3.794-800.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efron B, Gong G. A leisurely look at the bootstrap, the jackknife and cross-validation. Am Stat. 1983;37:36–48. [Google Scholar]
- 20.Evans P J, Mang D T, Kim K S, Young L Y. Anaerobic degradation of toluene by a denitrifying bacterium. Appl Environ Microbiol. 1991;57:1139–1145. doi: 10.1128/aem.57.4.1139-1145.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Ficker M. M.Eng. thesis. Hamilton, Ontario, Canada: McMaster University; 1998. [Google Scholar]
- 24.Fries M R, Zhou J, Chee-Sandford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins D G, Bleasby A J, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 27.Hopkins B T, McInerney M J, Warikoo V. Evidence for an anaerobic syntrophic benzoate degradation threshold and isolation of the syntrophic benzoate degrader. Appl Environ Microbiol. 1995;61:526–530. doi: 10.1128/aem.61.2.526-530.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krumholz L R, Caldwell M E. Biodegradation of “BTEX” hydrocarbons under anaerobic conditions. In: Crawford R L, Crawford D L, editors. Bioremediation: principles and applications. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 61–99. [Google Scholar]
- 29.Lovley D R, Lonergan D J. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism GS-15. Appl Environ Microbiol. 1990;56:1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabus R, Nordhaus R, Ludwig W, Widdel F. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol. 1993;59:1444–1451. doi: 10.1128/aem.59.5.1444-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 36.Snaidr J, Amann R, Huber I, Ludwig W, Scheifer K. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiele J H, Zeikus J G. Control of interspecies electron flow during anaerobic digestion: significance of formate transfer versus hydrogen transfer during syntrophic methanogenesis in flocs. Appl Environ Microbiol. 1988;54:20–29. doi: 10.1128/aem.54.1.20-29.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weidner S, Arnold W, Puhler A. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl Environ Microbiol. 1996;62:766–771. doi: 10.1128/aem.62.3.766-771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Whitehead Institute for Biomedical Research. 1996, 1997, copyright dates. [Online.] Primer 3, old version. http://www.genome.wi.mit.edu/cgi-bin/primer/primer3.cgi. [18 October 1999, last date accessed.]
- 40.Wilson B H, Smith G B, Rees J F. Biotransformations of selected alkylbenzenes and halogenated aliphatic hydrocarbons in methanogenic aquifer material: a microcosm study. Environ Sci Technol. 1986;20:997–1002. doi: 10.1021/es00152a005. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zinder S H. Physiological ecology of methanogens. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry and genetics. New York, N.Y: Chapman & Hall; 1990. pp. 128–206. [Google Scholar]