Abstract
Molecular phylogenetic analysis of a naturally occurring microbial community in a deep-subsurface geothermal environment indicated that the phylogenetic diversity of the microbial population in the environment was extremely limited and that only hyperthermophilic archaeal members closely related to Pyrobaculum were present. All archaeal ribosomal DNA sequences contained intron-like sequences, some of which had open reading frames with repeated homing-endonuclease motifs. The sequence similarity analysis and the phylogenetic analysis of these homing endonucleases suggested the possible phylogenetic relationship among archaeal rRNA-encoded homing endonucleases.
Since sizable populations of viable microorganisms were found in terrestrial and ocean subsurface environments, there has been increasing interest in microbial communities and diversities in deep-subsurface environments (3, 7, 13, 17, 21, 27–29, 34). Subsurface microorganisms are often viewed as important because they could play a significant role in subsurface geochemical processes, and some of these strains may have novel metabolic properties potentially useful for industrial processes, bioremediation, or biotechnology (10, 11). Hyperthermophilic or thermophilic microorganisms are likely major and important members of deep-subsurface microbial communities, considering that the temperature of subsurface environments increases with increasing depth. In fact, a number of thermophiles and hyperthermophiles within the domains of Bacteria and Archaea have been isolated from deep continental or sea oil reservoirs (14, 21, 26, 27). However, thermophilic microbial diversity in deep-subsurface environments other than oil reservoirs is poorly understood.
Here, we sought to determine the microbial diversity in a hot-subsurface biosphere. Deep-subsurface geothermal water pools are found in active volcanic areas and are often tapped by geothermal electric power plants. As the first step in comprehending thermophilic microbial diversity in such environments, molecular phylogenetic analysis based on small-subunit rRNA gene (SSU rDNA) sequencing was undertaken. In this study, a sample from a deep-subsurface geothermal water pool was obtained 1,500 m down in a production well of the Hacchoubaru geothermal plant in Oita Prefecture, Japan. Approximately 100 liters of effluent hot water (96°C, but over 250°C in situ at a depth of 1,500 m) was collected from immediately beyond the end of the production well. The chemical composition of the water was described by Hirowatari et al. (15). Sixty liters of water was immediately filtered by 0.22-μm-pore-size 47-mm-diameter cellulose acetate filters (Advantec, Tokyo, Japan), and the microbial particles were collected on the filters. The filters were washed with NET buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 100 mM EDTA) twice. Nucleic acids were extracted from the microbial particles on the filters and purified according to the method of Takai and Sako (31). Approximately 10 ng of DNA was recovered from 60 liters of the water sample. Based on this DNA yield (0.17 pg/ml) and assuming an average cellular DNA content of 2 fg (2), the calculated microbial population density was 0.8 × 102 cells/ml. This was in good agreement with the population density determined by epifluorescence microscopy direct count (approximately 102 cells/ml) with 4′,6-diamidino-2-phenylindole (DAPI). No experimental contaminant (32) was detected throughout the procedure of DNA extraction and purification.
SSU rDNAs were amplified by PCR using LA Taq polymerase with GC buffer (TaKaRa, Kyoto, Japan). Reaction mixtures in which the concentration of each oligonucleotide primer was 0.1 μM and that of DNA template was 0.1 ng/μl were prepared. Thermal cycling was performed with the GeneAmp PCR system 9600 (Perkin-Elmer, Foster City, Calif.), and the conditions were as follows: denaturation at 96°C for 20 s, annealing at 50°C for 45 s, and extension at 72°C for 120 s for a total of 30 cycles. The oligonucleotide primers Arch21F (12) and 1492R (19) were used for archaeal rDNA, and Uni515F and Uni1408R (30, 31) were used for all microbial rDNAs. As usual, PCR products approximately 1.5 and 0.9 kb in size are expected with these archaeon-specific primers and universal primers, respectively. However, products 2.9 and 2.4 kb in size, respectively, were obtained from the reactions. These rDNA products of unusual length have often been reported in characterization of hyperthermophilic archaea such as Pyrobaculum, Thermoproteus, and Aeropyrum (8, 16, 25) that are isolated from various hot-water environments. In view of the occurrence of introns in rDNA of hyperthermophilic archaea, the cloning and sequencing of these PCR products were carried out as described by Takai and Horikoshi (30).
The partial rDNA sequences (400 bp corresponding to positions 536 to 935 of Escherichia coli rDNA numbering) were analyzed with the similarity_rank and align_sequence from the Ribosomal Database Project (20) and the gapped-BLAST search algorithm (1, 6) to examine the microbial and archaeal populations in a deep-subsurface geothermal water pool. Sixty-eight and fifty-two clones were analyzed from the universal and archaeal primer PCR libraries, respectively. Sequence similarity analysis among the partially sequenced clones indicated that two distinct clone types (pHGPU6 and pHGPU21, 37 and 31 clones of 68 clones, respectively) were recovered from the universal primer PCR library and that these clone types almost completely matched two distinct clone types (pHGPA1 and pHGPA13, 24 and 28 clones of 52 clones, respectively) observed in the archaeal primer PCR library. Low sequence similarity (below 65%) between the partial sequences of pHGPA1 and pHGPA13 was found. Gapped-BLAST analysis of the partial sequences of pHGPA1 and pHGPA13 revealed coexistence of highly and poorly conserved regions in their sequences. When analysis was performed on these regions, the highly conserved regions of pHGPA1 and pHGPA13 were found to be quite similar to the rDNA sequences of Pyrobaculum species (over 99.0%). In contrast, the poorly conserved regions had certain similarities to the sequences of rRNA introns (033-II and 061-II) of Thermoproteus sp. strains IC-033 and IC-061 (16). These results suggested that the clones in the universal and archaeal primer PCR libraries represented the microbial rDNAs containing intron-like sequences and that all of the rDNA sequences recovered were derived from hyperthermophilic archaeal members. In order to characterize the gene structure of rDNA clones, almost complete sequences of the representative types of rDNA clones (2,930 bp for pHGPA1 and 2,909 bp for pHGPA13) were determined.
Based on the multiple alignments with the rDNA sequences of Pyrobaculum members, the insertion sites of intron-like sequences were roughly assumed, and then the secondary structures around exon-intron junction sites were manually constructed according to the convention proposed by Thompson and Daniels (33) and Lykke-Andersen et al. (23). As a result, three (pHGPA1-a, -b, and -c) and four (pHGPA13-a, -b, -c, and -d) possible introns were found in the rDNA sequences of pHGPA1 and pHGPA13, respectively. All the introns except for the second intron of pHGPA13 (pHGPA13-b) had a secondary structure specific to archaeal rRNA introns, having a core structure consisting of a bulge-helix-bulge structure, a long stable stem, and a terminal loop. In terms of their core structures, these putative introns were quite similar to the rRNA introns of Thermoproteus sp. strains IC-033 and IC-061 (16). These results strongly suggested that the unusually long rDNA clones obtained from the deep-subsurface geothermal pool contained several intervening sequences that were closely related to archaeal rRNA introns.
Several putative introns having relatively long terminal inserts (pHGPA1-a, pHGPA1-c, pHGPA13-b, and pHGPA13-d) were found to contain a single open reading frame (ORF) each. The ORFs encoded proteins consisting of 234, 69, 160, and 108 residues in the case of pHGPA1-a, pHGPA1-c, pHGPA13-b, and pHGPA13-d, respectively. These ORFs were found to contain amino acid sequences similar to LAGLI-DADG-like stretches, which are considered to be motifs shared by intron- and intein-encoded homing endonucleases (24). In the archaeal rRNA intron-encoded homing endonucleases found so far, the LAGLI-DADG-like stretches appear to be repeated with an interval of about 60 to 90 amino acid residues (8, 9, 16, 18, 22, 25). It seems likely, therefore, that the repeated LAGLI-DADG-like stretch is a feature of a proper ORF incurring no lethal mutation or frameshift. Although the products of the ORFs in pHGPA1-a and pHGPA13-b had the repeated LAGLI-DADG-like stretches with intervals of 88 and 73 residues, respectively, pHGPA1-c and pHGPA13-d products had one LAGLI-DADG-like stretch each. A second LAGLI-DADG-like stretch was found in the products of other reading frames in the case of pHGPA1-c and pHGPA13-d. When the amino acid sequences encoded by the ORFs in pHGPA1-c and pHGPA13-d were modified by a single nucleotide deletion at the nucleotide position 206 of pHGPA1-c and in the nucleotide position 224 of pHGPA13-d, the highest sequence similarity was obtained with other archaeal homing endonucleases. These results indicated that both ORFs of pHGPA1-c and pHGPA13-d incurred mutations that shifted the reading frames, resulting in smaller size and loss of the second LAGLI-DADG-like stretch. The putative junction sites and lengths of intron-like sequences and the initiation and terminal sites and amino acid sequences of ORF products were described in the DDBJ database under accession no. AB027539 for pHGPA1 and AB027540 for pHGPA13.
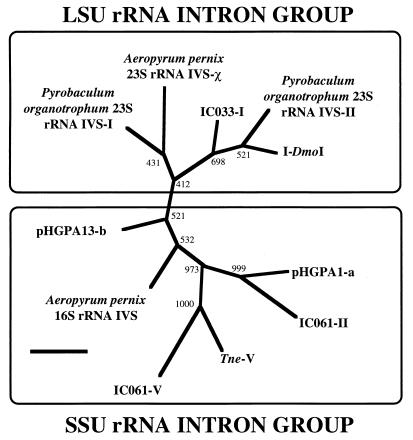
Although an increasing number of homing-endonuclease-like proteins have been found to be encoded in archaeal rRNA genes, the amino acid sequence-based relatedness and the phylogenetic relationship among such homing-endonuclease-like proteins are little understood. The amino acid sequences of archaeal rRNA intron-encoded proteins, including those of pHGPA1-a and pHGPA13-b, were aligned to focus on the repeated LAGLI-DADG-like stretch. Except for the ORF in the Pyrobaculum aerophilum 16S rRNA intron, all of the amino acid sequences reported so far were potentially alignable. Although no identical amino acid residue was found in the multiple alignment, 16 similar amino acid residues were identified in the LAGLI-DADG-like stretches and in other parts. The result indicated that most of the archaeal rRNA intron-encoded homing endonucleases were related to each other on the primary structure level. The phylogenetic analysis also supported the relationship among these archaeal rRNA intron-encoded homing endonucleases (Fig. 1). Although the general tree topology was not significantly supported by bootstrap examination, it revealed the presence of two distinct lineages: one group contained most of the SSU rRNA intron-encoded endonucleases, and the other group consisted of the large subunit rRNA intron-encoded entities and several SSU rRNA intron types (Fig. 1).
FIG. 1.
Unrooted phylogenetic tree of archaeal rRNA intron-encoded homing endonucleases. The tree was inferred by applying the neighbor-joining method to 81 homologous positions of the amino acid sequence. Each number shows the bootstrap value on the branching (1,000 replicates). The scale bar indicates 0.1 substitution per residue. The amino acid sequences in this figure are based on the nucleotide sequences of archaeal rRNA introns under the following GenBank accession numbers: Aeropyrum pernix 16S and 23S rRNA IVSs, AB008745; IC 033-I, AB009616; IC 061-II and -V, AB009617; TneV, AB009618; I-DmoI, X03263; pHGPA1-a, AB027539; and pHGPA13-b, AB027540. The amino acid sequences of Pyrobaculum organotrophum 23S rRNA IVS-I and -II are from GenPept, under accession no. JC1382 and JC1383, respectively. LSU, large subunit.
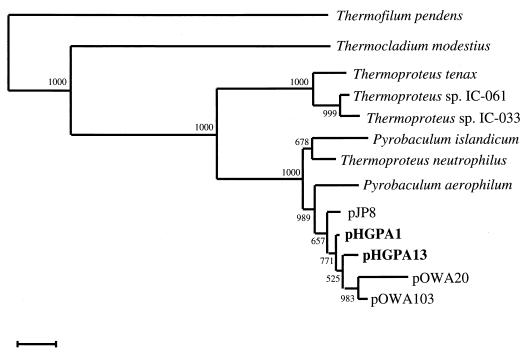
Using the exon regions of sequences, the phylogenetic analysis of rDNA clones obtained from a deep-subsurface geothermal environment was carried out by the neighbor-joining and maximum-likelihood methods with the ODEN software package (version 1.1; National Institute of Genetics, Mishima, Japan) and the PHYLIP package (version 3.5; obtained from J. Felsenstein, University of Washington, Seattle). Phylogenetic analysis by both methods resulted in trees with similar topologies (Fig. 2). As indicated by the extent of sequence similarity, both archaeal rDNA clones were closely related to the members of Thermoproteales, especially to Pyrobaculum species (Fig. 2). However, both rDNA clones had closer relationships with rDNA clones pJP8, obtained from the sediment of a Yellowstone National Park hot spring, and pOWA20 and pOWA103, obtained from the vent water of a shallow marine hydrothermal vent, than with the cultivated Pyrobaculum species (4, 5, 31). This implied that uncultivated and unidentified Thermoproteales members were widely distributed in terrestrial, shallow marine, and deep-subsurface hot-water environments.
FIG. 2.
Phylogenetic tree of cultivated strains and rDNA clones within Thermoproteales members. The tree was inferred by a neighbor-joining analysis of 414 homologous positions of the rDNA sequence in the case of each organism or clone. rDNA clones were derived from a shallow marine hydrothermal vent (pOWA) (31) and from sediment in a Yellowstone National Park hot spring (pJP) (5). Bold letters indicate rDNA clones obtained from a deep-subsurface geothermal environment. The scale bar represents 0.02 nucleotide substitution per sequence position. The number of bootstrap resamplings out of 1,000 is indicated. The rDNA sequences in this figure are from GenBank under the following accession numbers: Thermofilum pendens, X14835; Thermocladium modestius, AB005296; Thermoproteus tenax, M35966; Thermoproteus sp. strain IC-061, AB009617; Thermoproteus sp. strain IC-033, AB009616; Thermoproteus neutrophilus, AB009618; Pyrobaculum islandicum, L07511; P. aerophilum, L07510; pJP8, L25309; pOWA20, AB007314; pOWA103, AB007311; pHGPA1, AB027539; and pHGPA13, AB027540.
The presence of a thermophilic microbial population was evident upon analysis of DNA recovered from a deep-subsurface geothermal vent water pool. All of the microbial rDNA sequences obtained from the water sample from an environment with a temperature over 250°C represented archaeal rDNA sequences closely related to those of hyperthermophilic Thermoproteales members. As determined by microscopic observation, all of these microbial cells were straight rods with an average length of 4 to 8 μm and a width of about 0.5 to 0.8 μm, which corresponded well with the morphological characteristics of Thermoproteales members. The chemical composition of the hot water from the production wells was frequently analyzed by the water quality department of the power plant, and it was proved that the hot water and vapor were derived from the deep geothermal water pool (15). In addition, no rDNA sequences of mesophilic microorganisms were recovered from the sample, and no experimental contamination was detected. These results excluded possible contamination from upper layers of sediments and other contamination throughout the experimental procedure. Hence, the archaeal rDNA clone population might represent the true microbial population in the deep-subsurface geothermal environment.
To date, a great phylogenetic diversity of archaea has been found in a variety of hot-water environments, such as a Yellowstone National Park hot spring (4, 5), a shallow marine hydrothermal vent (31), and various deep-sea hydrothermal vents (30). Lower microbial population density (102 cells/ml) and less phylogenetic diversity were observed in a deep-subsurface geothermal water pool than in other hot-water environments. Two phylotypes of archaeal members found in the deep-subsurface water pool were closely related to Pyrobaculum species, which are known as the most hyperthermophilic archaeal members isolated from geothermal freshwater systems (28). The extraordinarily high temperature and the confined hydrological system of the deep-subsurface geothermal pool are likely to allow only the most hyperthermophilic archaeal members and to preclude an increase in deep-subsurface microbial diversity. In addition, the geochemical condition of the deep-subsurface hot-water environment might have great impact on the deep-subsurface microbial populations. Further molecular phylogenetic analyses will be helpful in comprehending the magnitude, composition, and diversity of the microbial communities in deep-subsurface environments. The isolation and cultivation method should also help in obtaining further insight into the thermophilic microbial communities in the deep-subsurface environments. These subjects are the focus of future work.
Nucleotide sequence accession numbers.
The sequences of the 2,930-bp and the 2,909-bp rDNA clones (pHGPA1 and pHGPA13, respectively) determined in this study have been deposited in DDBJ under accession no. AB027539 and AB027540.
Acknowledgments
We thank Wayne R. Bellamy for editing English usage in the manuscript.
This work was supported in part by a domestic research fellowship provided by the Japan Science and Technology Corporation.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakken L R, Olsen R A. DNA content of soil bacteria of different cell size. Soil Biol Biochem. 1989;21:789–793. [Google Scholar]
- 3.Bale S J, Goodman K, Rochelle P A, Marchesi J R, Fry J C, Weightman A J, Parkes R J. Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int J Syst Bacteriol. 1997;47:515–521. doi: 10.1099/00207713-47-2-515. [DOI] [PubMed] [Google Scholar]
- 4.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boone D R, Liu Y, Zhao Z-J, Balkwill D L, Drake G R, Stevens T O, Aldrich H C. Bacillus infernus sp. nov., an Fe(III)- and Mn(IV)-reducing anaerobe from the deep terrestrial subsurface. Int J Syst Bacteriol. 1995;45:441–448. doi: 10.1099/00207713-45-3-441. [DOI] [PubMed] [Google Scholar]
- 8.Burggraf S, Larsen N, Woese C R, Stetter K O. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc Natl Acad Sci USA. 1993;90:2547–2550. doi: 10.1073/pnas.90.6.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgaard J Z, Garrett R A. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene. 1992;121:103–110. doi: 10.1016/0378-1119(92)90167-n. [DOI] [PubMed] [Google Scholar]
- 10.Fredrickson J K. DOE explores subsurface biosphere. ASM News. 1992;58:183. [Google Scholar]
- 11.Fredrickson J K, Brockman F J, Workman D J, Li S W, Stevens T O. Isolation and characterization of a subsurface bacterium capable of growth on toluene, naphthalene, and other aromatic compounds. Appl Environ Microbiol. 1991;57:796–803. doi: 10.1128/aem.57.3.796-803.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 13.Gold T. The deep, hot biosphere. Proc Natl Acad Sci USA. 1992;89:6045–6049. doi: 10.1073/pnas.89.13.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greene A C, Patel B K C, Sheehy A J. Deferribacter thermophilus gen. nov., sp. nov., a novel thermophilic manganese- and iron-reducing bacterium isolated from a petroleum reservoir. Int J Syst Bacteriol. 1997;47:505–509. doi: 10.1099/00207713-47-2-505. [DOI] [PubMed] [Google Scholar]
- 15.Hirowatari K, Nishiyama E, Tanaka A. Research in chemical processing of water in geothermal electric power plants. Proc Res Kyusyu Electric Power Co. 1981;56:15–27. . (In Japanese.) [Google Scholar]
- 16.Itoh T, Suzuki K, Nakase T. Occurrence of introns in the 16S rRNA genes of members of the genus Thermoproteus. Arch Microbiol. 1998;170:155–161. doi: 10.1007/s002030050628. [DOI] [PubMed] [Google Scholar]
- 17.Kieft T L, Fredrickson J K, Onstott T C, Gorby Y A, Kostandarithes H M, Bailey T J, Kennedy D W, Li S W, Plymale A E, Spadoni C M, Gray M S. Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl Environ Microbiol. 1999;65:1214–1221. doi: 10.1128/aem.65.3.1214-1221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.Kjems J, Garrett R A. An intron in the 23S ribosomal RNA gene of the archaebacterium Desulfurococcus mobilis. Nature. 1985;318:675–677. doi: 10.1038/318675a0. [DOI] [PubMed] [Google Scholar]
- 20.Lane D J. 16S/23S sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1985. pp. 115–176. [Google Scholar]
- 21.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L’Haridon S, Reysenbach A-L, Glenat P, Prieur D, Jeanthon C. Hot subterranean biosphere in a continental oil reservoir. Nature. 1995;377:223–224. [PubMed] [Google Scholar]
- 23.Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett G A. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem Sci. 1997;22:326–331. doi: 10.1016/s0968-0004(97)01113-4. [DOI] [PubMed] [Google Scholar]
- 24.Lykke-Andersen J, Garrett R A. Structural characteristics of the stable RNA introns of archaeal hyperthermophiles and their splicing junctions. J Mol Biol. 1994;243:846–855. doi: 10.1006/jmbi.1994.1687. [DOI] [PubMed] [Google Scholar]
- 25.Mueller J E, Bryk M, Loizos N, Belfort M. Homing endonucleases. In: Linn S M, Lloyd R S, Roberts R J, editors. Nucleases. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 111–143. [Google Scholar]
- 26.Nomura N, Sako Y, Uchida A. Molecular characterization and postsplicing fate of three introns within the single rRNA operon of the hyperthermophilic archaeon Aeropyrum pernix K1. J Bacteriol. 1998;180:3635–3643. doi: 10.1128/jb.180.14.3635-3643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravot G, Magot M, Fardeau M-L, Patel B K C, Prensier G, Egan A, Garcia J-L, Ollivier B. Thermotoga elfii sp. nov., a novel thermophilic bacterium from an African oil-producing well. Int J Syst Bacteriol. 1995;45:308–314. doi: 10.1099/00207713-45-2-308. [DOI] [PubMed] [Google Scholar]
- 28.Stetter K O, Huber R, Blöchl E, Kurr M, Eden R D, Fielder M, Cash H, Vance I. Hyperthermophilic archaea are thriving in deep North Sea and Alaskan oil reservoirs. Nature. 1993;365:743–745. [Google Scholar]
- 29.Stevens T O, McKinley J P. Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science. 1995;270:450–454. [Google Scholar]
- 30.Takai K, Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takai K, Sako Y. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol Ecol. 1999;28:177–188. [Google Scholar]
- 32.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson L D, Daniels C J. A tRNATrp intron endonuclease from Halobacterium volcanii: unique substrate recognition properties. J Biol Chem. 1988;263:17951–17959. [PubMed] [Google Scholar]
- 34.Whitman W B, Coleman D C, Wiebe W J. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]