Abstract
The impact of the COVID-19 pandemic has reinforced the need for rapid, cost-effective, and reliable point-of-care testing (POCT) devices for massive population screening. The co-circulation of SARS-CoV-2 with several seasonal respiratory viruses highlights the need for multiplexed biosensing approaches. Herein, we present a fast and robust all-in-one POCT device for parallel viral antigen and serological analysis. The biosensing approach consists of a functionalized polycarbonate disc-shaped surface with microfluidic structures, where specific bioreagents are immobilized in microarray format, and a portable optoelectronic analyzer. The biosensor quantifies the concentration of viral antigens and specific immunoglobulins G and M for SARS-CoV-2, influenza A/B, adenovirus, and respiratory syncytial virus, using 30 μL of a sample. The semi-automated analysis of 6 samples is performed in 30 min. Validation studies performed with 135 serum samples and 147 nasopharyngeal specimens reveal high diagnostic sensitivity (98–100%) and specificity (84–98%), achieving an excellent agreement (κ = 0.937) with commercial immunoassays, which complies with the World Health Organization criteria for POC COVID-19 diagnostic tests. The versatility of the POCT device paves the way for the detection of other pathogens and analytes in the incoming post-pandemic world, integrating specific bioreagents against different variants of concerns and interests.
Keywords: COVID-19, SARS-CoV-2, Influenza, Adenovirus, Respiratory syncytial virus, Microfluidics, Immunoassay
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late December 2019 (Wu et al., 2020; Zhu et al., 2020), causing the Coronavirus Disease 2019 (COVID-19) pandemic, which has been responsible for more than 515 million confirmed cases and 6.25 million deaths (World Health Organization, 2021). Since the beginning of the pandemic, the need to contain the disease brought the development of molecular and serological assays to identify COVID-19 infections (Afzal, 2020).
Several analytical methods have been developed to monitor the status of the coronavirus disease. The concept of the molecular techniques for COVID-19 diagnosis relies on identifying the viral RNA (Liu et al., 2020; Shetti et al., 2021). While RNA detection via quantitative reverse-transcription polymerase chain reaction (q-PCR) has proved to be highly specific and sensitive, only a few amplicons can be detected per reaction, specific thermal cyclers are needed, and complex, time-consuming operations are required (Dramé et al., 2020). However, the scalability of such a method is limited by cost and equipment availability (Yelagandula et al., 2021), which is not compatible with the POCT requirements.
Other approaches have also focused on developing methods for detecting viral antigens to facilitate some aspects of the logistics of mass testing that have been part of the first-line surveillance strategy during the pandemic (see Supplementary Table S1). These tests comprise lateral flow immunoassays and the qualitative enzyme-linked immunosorbent assays (ELISA), being the first of them more appropriate for point-of-care testing (Parolo et al., 2020; Weiss et al., 2020). However, the reliability of the lateral flow immunoassays for the detection of virus infection is still questioned by a continuous report of a lack of both sensitivity and quantitative measurements (Deeks and Raffle, 2020; Surkova et al., 2020). Although ELISA tests are more sensitive and widely used by clinical laboratories worldwide, they would require more time, would need expertise in procedures, and would have bulky benchtop analytical instruments (Dysinger et al., 2017; Elshal and McCoy, 2006; Lewis et al., 2015), and therefore, of limited use for medical testing done at or near the point of care.
Serological testing has also been used to understand viral circulation, complementing virus detection by indicating past infection, which could be exploited for therapeutic advances. These tests detect antiviral IgA, IgG, and IgM antibodies in serum and are mainly used for epidemiological studies to explore the protective value of the neutralizing antibodies (Chen et al., 2020; Krammer and Simon, 2020; Lin et al., 2020). In addition, most efforts are still focused on strengthening the accuracy and reliability of serological tests (Abid et al., 2021; Shen et al., 2021). Even though large-scale serological testing is desirable to approach the challenge of vaccinating the entire population, the lack of portable devices to afford the demanding logistical requirements makes the challenge harrowing.
The spread of SARS-CoV-2 is significantly affected due to its concurrence with seasonal influenza (Bordi et al., 2020) and other viruses, which can cause similar symptoms to those produced by the COVID-19. Thus, another significant research challenge deals with the distinction between the different co-circulating viruses. In addition, detecting several targets in a row may require a time-consuming optimization process or even unique instrumentation (Trivedi et al., 2019; Mas et al., 2020). In this scenario, multiplexed POCT for the simultaneous detection of common respiratory viruses concomitantly with the mass population screening opens potential venues to materialize a milestone in the fight against the COVID-19 pandemic. Indeed, multiplexing has become more critical for point-of-care testing in the last decade (Dincer et al., 2017). In this line, POCT devices with analytical performance comparable to or beyond that of laboratory testing technologies are needed to ensure the requirements of in vitro diagnostics, paving the way for novel home health-monitoring systems. Despite numerous efforts to develop sensitive and selective assays, POCT devices for the multiplex detection of respiratory infections have not yet been established (Lu et al., 2021).
Motivated by finding a reliable alternative to existing single-based detection antigen and serological assays, we present an all-in-one multiplexed and cost-effective POCT device to detect and quantify several viral antigens or specific antibodies (IgG and IgM) against the respiratory viruses SARS-CoV-2, Influenza A/B, adenovirus, and respiratory syncytial virus, simultaneously. The immunochemical solution comprises a DVD drive as an optoelectronic analyzer and a transparent microfluidic polycarbonate disc-shaped platform. The analytical and clinical performances of the POCT device are evaluated with the analysis of a cohort of 282 human samples, offering reliable results within 30 min. To our best knowledge, this is the first POCT technology using a consumer electronics device, coupling serological and viral antigen testing in parallel to detect respiratory infections.
2. Materials and methods
2.1. Reagents and materials
Influenza A and B recombinant nucleoproteins, Adenovirus HEXON protein, Respiratory Syncytial Virus (RSV) recombinant fusion protein, SARS-CoV-2 recombinant Receptor Binding Domain (RBD), SARS-CoV-2 recombinant nucleoprotein (N), anti-Influenza A and B monoclonal antibodies (Mab), anti-Adenovirus Mab, and anti-RSV Mab, were purchased from Certest Biotec (Zaragoza, Spain). SARS-CoV-2 spike protein (S-ECD/RBD) monoclonal antibody (Ab2) was purchased from Thermo Fisher Scientific (Massachusetts, USA). Goat anti-mouse (GAM) antibody, Bovine serum albumin (BSA), Tween-20, phosphate-buffered saline (PBS) tablets, and anti-human IgG (anti-hIgG) were obtained from Sigma-Aldrich (Madrid, Spain). Horseradish peroxidase (HRP) kit and goat anti-Mouse-HRP labeled antibody were purchased from Abcam (Cambridge, UK). 3,3′,5,5′-Tetramethylbenzidine (TMB) was provided by Stereospecific Detection Technologies (Baesweiler, Germany). The information about the commercial SARS-CoV-2 IgG immunoassays is available in the Supplementary Material (Supplementary Information 2).
2.2. Point-of care-testing device
The POCT device comprises a portable optoelectronic analyzer, referred to as a reader, and a transparent microfluidic disc-shaped platform (Fig. 1 ). The reader is based on a standard DVD drive's mechanical and electrical components, structures, and configurations (see Supplementary Information 1 and Fig. S1).
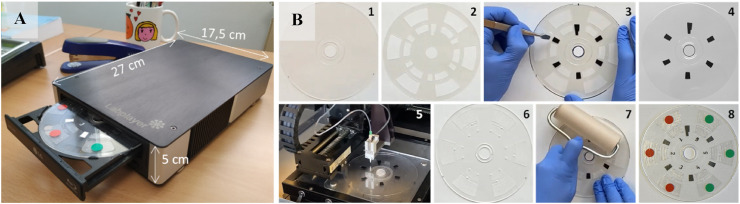
Fig. 1.
(A) Image of the point-of-care all-in-one testing device, composed of a portable optoelectronic analyzer and a microfluidic transparent disc. (B) Microfluidic disc assembling: (1) Bottom disc; (2) Pressure-sensitive adhesive film (PSA); (3) Placing the black patterns on the bottom disc, using the PSA as a template; (4) Bottom disc with the black patterns; (5) Printing the microarrays onto the disc (4); (6) Top disc with microfluidic structures; (7) Assembling the bottom (5) and top (6) discs using the PSA film; (8) Ready-for-use disc. The red and green dot stickers indicate the microfluidic structure intended to perform serological and antigen assays, respectively.
The optical performance of the optoelectronic analyzer was evaluated using a calibration disc (Fig. S2) comprised of a transparent polyester adhesive film (0.1 mm thick) enclosed between two dummy polycarbonate top (0.6 mm thick) and bottom (1.2 mm thick) discs. Matrices of shaded black (RGB 0, 0, 0) and blue (RGB 0, 0, 250) dots and black patterns for synchronization of the readouts were previously printed on the transparent film using a laser printer (Develop ineo +3080, Konika Minolta, Tokyo, Japan). Each chamber of the calibration disc contained a matrix of dots (3 × 11) printed with one color and one diameter (see Fig. S2). Shaded dots were achieved by varying color intensity (0, 10, 20, 30, 50, and 70%), covering the whole measurement range of the optoelectrical biosensor. The printed adhesive polyester transparent film was taped on the bottom disc, and then the top disc was stuck to the bottom using double-sided adhesive tape.
The microfluidic disc is comprised of two top and bottom polycarbonate disc-shaped surfaces (120 mm in diameter) assembled by a pressure-sensitive adhesive film (Fig. 1B). The bottom surface is 0.6 mm thick and includes a black pattern to synchronize the readouts. The top disc has a thickness of 1.2 mm and contains six fluidic structures manufactured by micro-milling. Each microfluidic structure includes detection and waste chambers (see Supplementary Information 3 and Fig. S3). Before assembling, the bottom disc was functionalized with specific bioreceptors (antigens and antibodies), which were immobilized by passive adsorption in microarray format (6 arrays per disc of 3 x 11 spots), dispensing 50 nL of viral antigens, antibodies, and positive and negative control solutions, using a noncontact printing liquid dispenser (AD1500 BioDot, Inc., Irvine, CA). The spots had a diameter of 500 μm, with a center-to-center distance of 1.2 mm, achieving an array density of 4.0 spot mm−2.
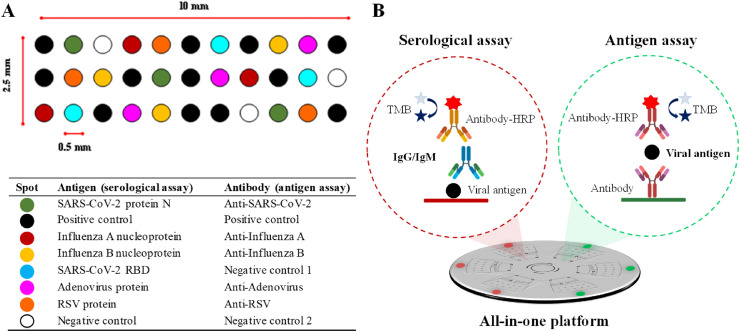
Three microfluidic structures were designated for the serological assay (identified with a red dot sticker), and the other three for detecting viral antigens (green dot sticker). Fig. 2 A shows the microarray layout and the list of bioreceptors used for each type of immunoassay. The concentrations of specific viral antigens, capture antibodies, and positive and negative controls are shown in the Supplementary Material (Table S2). After printing the microarrays, the disc was assembled and then incubated overnight at 37 °C before use.
Fig. 2.
(A) Layout of the microarrays, and (B) schemes of the immunoassays developed on the all-in-one platform.
2.3. Human serum and nasopharyngeal samples
Human blood samples were drawn through venipuncture of forearm veins from 78 cases hospitalized at the Hospital Clínico Universitario de València. Sera were obtained by centrifuging the blood at 2000 rpm for 10 min. Samples of 40 patients (52%) were received within the first two weeks after the onset of symptoms, 26 (33%) were between the third and the fourth weeks, and 12 (15%) after the fifth week. Besides, sera from 57 individuals collected before the pandemic (2016–2017) were used as negative controls. Serum samples were analyzed using four commercial enzyme immunoassays (see Supplementary Information 2).
A cohort of 147 nasopharyngeal samples was collected from 143 COVID-19 suspicious subjects and 4 PCR negative individuals who were controls for the SARS-CoV-2 antigen detection assay. The samples were taken by the medical staff of the Health Centre of the Universitat Politècnica de València and analyzed following the instructions of the Panbio™ COVID-19 Ag Rapid Test (Abbott Laboratories, IL, USA).
All subjects participated after giving written informed consent according to protocols approved by the Research Ethics Committee of Hospital Clínico Universitario INCLIVA (March 2020, Valencia, Spain).
2.4. Assay procedure
Fig. 2B illustrates the procedure for the serological and viral antigen assays. Briefly, 30 μL of a sample (serum previously diluted (1:5, v/v) in PBST (PBS solution with 0.05% Tween 20), or nasopharyngeal swab specimens collected in 300 μL of lysis buffer solution) is loaded into the detection chamber. The disc is placed on the tray, and the tray slides back when clicking the insert button. After 10 min, the disc is centrifuged at 4000 rpm to deliver the non-reacted sample to the waste chamber. Then, the disc is ejected, and 30 μL of detector antibody solution (HRP-labeled anti-hIgG and specific antiviral antibody for serological and antigen assays, respectively) is loaded into the detection chamber. After 10 min, the disc is centrifuged as before. Next, the disc is ejected to load 30 μL of washing solution (PBST) into the detection chamber. The disc is centrifuged as described before to deliver the PBST to the waste chamber. Finally, the immunoreaction is developed by loading 30 μL of TMB into the detection chamber. After 5 min, the disc is centrifuged at 4000 rpm to deliver the developer to the waste chamber. Then, the reader scans the detection chamber in 5 min and quantifies the optical density of the colored spots, which is proportional to the concentration of the analytes (viral antigens and specific IgG and IgM). The total assay time takes 30 min. An “Assay protocol” video clip is attached to the Supporting Material (Supplementary Information 6).
3. Results and discussion
3.1. Performances of the optical POCT device
The optical resolution of the POCT instrument was tested by scanning the calibration disc, which contains arrays of dots of several diameters (150, 280, and 525 μm). The calibration disc was scanned three times, and the mean signal was calculated for the same color and diameter. Figs. S2A and S2B show the images obtained and the calibration curves for black and blue dots, respectively. Linear mathematical models for each calibration curve were obtained with goodness of adjustment greater than 99%. Through an ANOVA analysis for each color intensity (0–70%), no statistically significant differences were observed between the three performed measurements for both black and blue calibration curves (p-value > 0.05 in all cases), revealing the excellent precision of the readouts.
As far as the optical resolution is concerned, the POCT instrument can detect dots of 150 μm in diameter, achieving a good reproducibility within the different measurements. Compared to other sizes of spots (150, 280, and 525 μm), the ranges of signal intensity are similar for the three tested diameters, as shown in Fig. S2C, and the 95% confidence bands reveal no statistically significant differences between the calibration curves obtained for the three analyzed diameters. These results indicate the excellent optical performance of the POCT device, capable of detecting spots of different colors (blue and black) and sizes as small as 150 μm. In addition, signals of other colored dots (black, blue, green, red, and yellow) were evaluated, as shown in Fig. S2D, using a spot size of 525 μm. Differences between the signals obtained from blue, black, or green spots are not statistically significant in any case for a 95% confidence interval. As expected, red and yellow spots give lower signal intensities than blue, black, or green spots since the reader uses the laser emitting at 650 nm (red). These performances make it a versatile analytical instrument for clinical diagnostics and with the potential capability to read high-density microarrays using various different-colored immunoreagents.
3.2. Analytical performances
All the experimental variables (concentration of immobilized bioreceptors, dilutions of HRP-labeled detector antibodies, and incubation time) involved in the POCT functioning were studied using sensitivity and the linear dynamic range as the selection criterion. Table S2 and Fig. S4 summarize the selected values of such parameters.
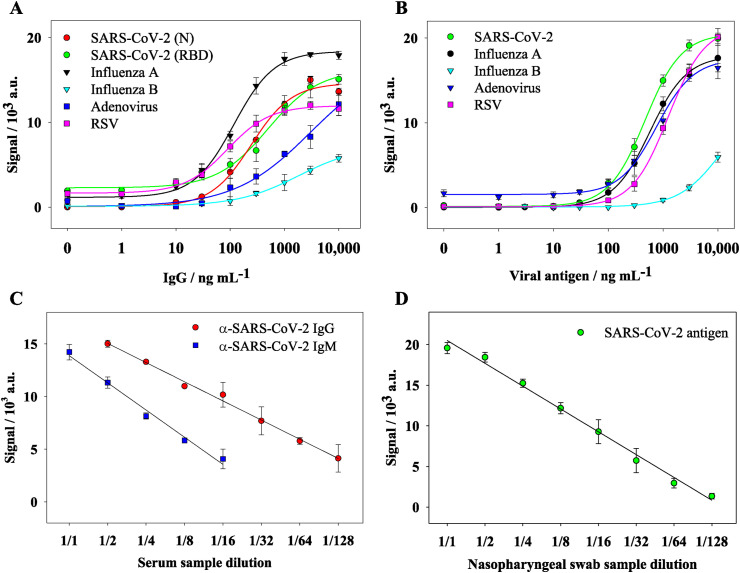
Since a quantitative micro-immunoassay should provide results in units related to a standard, calibration curves for the serological and antigen assays were performed by evaluating serial dilutions (from 0.1 ng mL−1 to 10,000 ng mL−1 in PBST) of specific antibodies and viral antigens, respectively. The limits of detection (LOD) and quantification (LOQ) were determined by measuring the signal of 10 blank samples and calculating the mean value of the signal plus 3 and 10 times its standard deviation, respectively. As observed in Fig. 3 A and B, the signals fit well to a four-parameter logistic curve (R2 > 0,997). Table 1 shows the figures of merit of the multiplexed serological and antigen assays.
Fig. 3.
Calibration curves for the detection of respiratory viruses through (A) serological and (B) antigen assays. Dilution linearity studies for (C) serological and (D) antigen detection of SARS-CoV-2 from human serum and nasopharyngeal swabs samples, respectively.
Table 1.
Limit of detection and working range for the serological and antigen assays.
| Serological assayb |
Antigen assay |
|||
|---|---|---|---|---|
| LODa | Working range | LOD | Working range | |
| SARS-CoV-2c | 17 | 34–3450 | 18 | 100–6500 |
| Influenza A | 30 | 62–850 | 16 | 125–6500 |
| Influenza B | 280 | 520–10,000 | 635 | 850–10,000 |
| Adenovirus | 110 | 250–10,000 | 33 | 125–6500 |
| RSV | 12 | 25–325 | 41 | 220–8000 |
LOD: Limit of detection.
Inmunglobulin type G.
Nucleoprotein (N). All values are expressed in ng mL−1.
Though the assays for detecting Influenza B showed a lower sensitivity, Influenza A accounts for approximately 75% of total flu virus infections (Hayward et al., 2014), making the POCT device very sensitive to detecting flu infections. Under the selected conditions, cross-reactivity studies were performed, and the results show that the assays are selective to discriminate against the tested respiratory viruses (see Supplementary Information 5).
The linearity of dilution study was performed using a pool of serum samples with known concentrations of specific IgM and IgG against SARS-CoV-2, submitted to 2-fold serial dilutions (1:2–1:128). Fig. 3C shows the results of the linearity of the dilution assay. The solid lines represent the corresponding linearity of dilution plots for the experimental concentration. The concentrations of specific IgG and IgM s were calculated using the standard calibration curve and the estimated concentration measured by the known dilution factors. As shown in Fig. 3C, the linearity was good over a wide range of dilutions, revealing that the methodology provided flexibility to test serum samples with different levels of specific IgG and IgM antibodies. As can be seen in Fig. 3 C, the determined specific IgG and IgM concentrations differed slightly from the estimated concentration, ranging from 82 to 120%. The high ratio values observed for IgG and IgM (>120% for 1:128 dilutions) are likely due to the initial low IgG and IgM concentrations. Interestingly, the relative standard deviation values were below 20% for all dilutions.
Similarly, the linearity of dilution was also performed for the antigen assay using a representative positive nasopharyngeal swab sample with a known concentration of SARS-CoV-2 nucleoprotein, submitted to 2-fold serial dilutions (1:2–1:512). Fig. 3D shows the results of the linearity of the dilution assay. The solid lines represent the corresponding linearity of dilution plots for the experimental concentration.
The concentration of SARS-CoV-2 nucleoprotein was calculated using the standard calibration curve and the estimated concentration measured by the known dilution factors. As is shown in Fig. 3D, the linearity found over a wide range of dilutions reveals that the methodology provided flexibility to test nasopharyngeal swab samples with different levels of SARS-CoV-2 nucleoprotein. Indeed, the determined SARS-CoV-2 nucleoprotein concentrations differed slightly from the estimated concentration, ranging from 75 to 125%. Finally, to test the robustness of the POCT device, the reproducibility of the quantitative results was determined. Tests corresponding to 60 replicas were carried out using different concentrations (0–10,000 ng mL−1) of monoclonal anti-SARS-CoV-2 antibody and nucleocapsid protein for serological and antigen assays, respectively. The POCT showed good precision as the relative standard deviation was below 11% (RSD 10.8% and 5.5% for intra-disc and inter-disc, respectively).
3.3. Analysis of human samples
A cohort of 135 human serum samples previously diluted in PBST (1:5, v/v) were tested by the POCT device. As described in the supplementary material, four commercial ELISA methods (Diasorin, Euroimmun, Maglumi, and Vircell) were also used to qualitatively detect IgG class antibodies to the S protein of SARS-CoV-2 in human serum. The results are shown in Table S3 and Table S4 for the positive cases and the negative controls, respectively. Results of the immunoassays were evaluated by calculating a ratio of the optical density of the control or patient sample over that of the calibrator. The ratio was used as a relative measure for the concentration of IgG antibodies in serum. The individual statistical correlation (Rho-parameter) between the 4 ELISA methods and the quantitative POCT device were 0.43, 0.34, 0.42, and 0.37 (P = 0.0002, 0.0043, 0.0003, and 0.0048), respectively. It reveals a good, positive relationship, considering that the commercial methods are qualitative and use different protocols, calibrators, reagents, and ratio-based analyses. In addition, it is worth mentioning that the correlation between the methods showed a much stronger relationship (Rho = 0.74; P = 0.0002) when the results are globally interpreted, based on a binary qualitative response, 1 or 0, assigning the value of 1 when two or more ELISA methods deemed positive (see Table S3 and Table S4). This is probably because the multiplex configuration allows detecting IgG and IgM antibodies against both S and N proteins simultaneously, obtaining a complete view and reliable information.
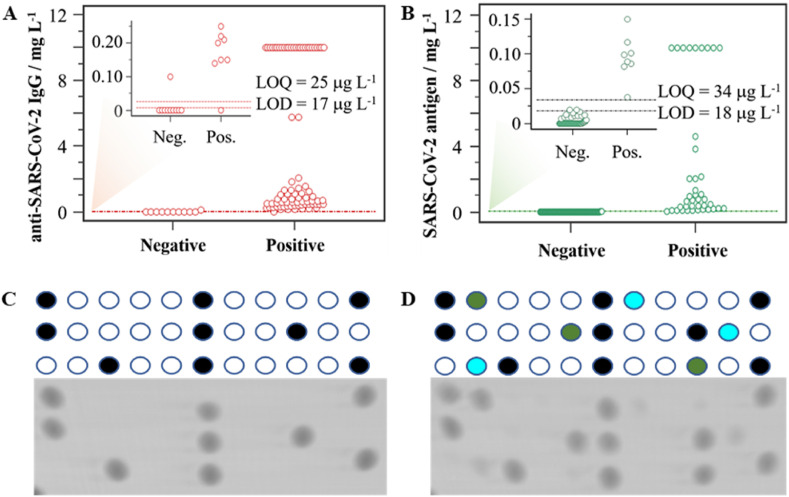
Furthermore, the interactive dot diagram illustrated in Fig. 4A reveals that the POCT device reaches a sensitivity and specificity of 98 and 84%, respectively, using 17 ng mL−1 as the cut-off threshold. As shown in Table S5, a positive predictive value of 88% and a negative predictive value of 98% were achieved. These results confirm the suitability of the developed micro-immunoassay for serological testing of SARS-CoV-2, which complies with the World Health Organization criteria for POC COVID-19 diagnostic tests. Furthermore, Cohen's kappa coefficient quantified the degree of agreement to assess the inter-rater reliability, revealing an excellent agreement (κ = 0.937) with the global response of the ELISA methods. According to the LOD obtained for the rest of the respiratory viruses in the micro-immunoassays (Table 1), the diagnosis performances might be at the same level of sensitivity and specificity achieved for SARS-CoV-2.
Fig. 4.
Case-control study: interactive-dot diagram for (A) serological (n = 135) and (B) viral antigen (n = 147) assays for SARS-CoV-2 detection. Representative results of a (C) negative control and (D) a COVID-19 positive case after scanning the disc (see Fig. 2A).
For the antigen assay, a positive result was considered equal to or greater than a cut-off threshold of 18 ng mL−1. Using this criterion, from the 147 nasopharyngeal samples analyzed, 42 tested positive for the N protein using the POCT device. Similarly, 42 (28.5%) of the nasopharyngeal samples also tested positive for SARS-CoV-2 using the PANBIO COVID-19 Ag rapid test (Abbot, Illinois, USA). Interestingly, all the positive samples were also positively indicated by the POCT device, achieving an agreement of 100% and a specificity of 98.1% (see Table S6 and Fig. 4B). The main differences found in this study are that our POCT device allows us to investigate the simultaneous presence of different viral antigens in the sample and provides quantitative results for all the viral antigens. Indeed, 2% of the tested samples indicated a positive result for influenza A at the low ng mL−1 level, and no adenovirus or RSV was detected. These results are in good accordance with the epidemiology status of the Spanish population during this study.
The naked eye can also detect the colored spots for visual discrimination between positive and negative persons. Representative images of the analysis of case-control studies are shown in Fig. 4C and D. As can be observed, the developed POCT solution distinguishes very well between the control and actual cases, showing its analytical potential and multiplex capability for qualitative measurements and rapid interpretation of the results. In this context, it is also worth mentioning the versatility of the POCT device, providing quantitative, semi-quantitative, or qualitative information to offer reliable and evidence-based health responses and thus promote a cost-effective strategy for helping manage new outbreaks.
4. Conclusions
This work presents a novel POCT device based on consumer electronics as an alternative analytical system to determine in parallel specific antibodies and multiple viral antigens of the most typical respiratory viruses in real clinical scenarios and other settings. The presented empirical evidence demonstrates the analytical potential for the diagnosis and immunological tracing of COVID-19 patients, measuring the impact of the virus on public health, and supporting the development of effective vaccines and therapeutics.
Table S1 shows a comparison of antigen and serological immunochemical methods at the R&D stage produced in response to respiratory virus infections. Considering the global social and personal impact of respiratory infections worldwide, the availability of multiplexed systems that can provide results in a cost-effective way, with a single assay, has clear additional benefits for healthcare systems. Furthermore, we envision complete automation of the assay by designing more advanced and complex microfluidic platforms that could simplify the analytical protocol, including the sample treatment. Another significant advantage is the cost-effectiveness of the quantitative instrument, which makes it very affordable to every laboratory and promising for primary healthcare centers and doctors' offices. This investigation provides the basis for the prospective implementation of the presented POCT device in epidemiological research studies, and surveillance vaccine assessments to develop personalized therapies based on antibody drugs. The versatility of the POCT device permits expanding the solution for clinical diagnostics to determine on-demand target analytes such as other viruses, microorganisms, biomarkers, etc., including immunochemical and DNA-based approaches.
CRediT authorship contribution statement
William Teixeira: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Yeray Pallás-Tamarit: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Augusto Juste-Dolz: Conceptualization, Methodology, Formal analysis, Investigation, Writing – original draft. Amadeo Sena-Torralba: Methodology, Formal analysis, Validation. Roberto Gozalbo-Rovira: Resources, Validation. Jesús Rodríguez-Díaz: Resources, Validation. David Navarro: Resources, Validation. Javier Carrascosa: Formal analysis, Methodology, Validation, Data curation. David Gimenez-Romero: Formal analysis, Validation. Ángel Maquieira: Conceptualization, Writing – review & editing, Resources, Project administration, Funding acquisition. Sergi Morais: Conceptualization, Writing – review & editing, Visualization, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
W. T and A. J-D acknowledge the financial support for the Ph.D. studies (PAID-19-01-06 and FPI-UPV 2017 grants, respectively). S.M awards grant PID2019-110713RB-I00 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe.” A.M awards grant DOGV no. 8815, May 19, 2020, pp. 16712–16726. The research was also supported by the Generalitat Valenciana (PROMETEO/2020/094, Proyecto “88/2021”).
The authors gratefully acknowledge the outstanding contribution of Alicia Parra, Violeta Espada, Roberto Renuncio, and Jose A. Marzal for collecting the nasopharyngeal swab samples from the Health Centre of the Universitat Politècnica de València. We also gratefully thank Dr. L. Mira and G. Rios for carefully reading this manuscript and the four anonymous reviewers for their insightful comments and suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114454.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abid S.A., Ahmed Muneer A., Al-Kadmy I.M.S., Sattar A.A., Beshbishy A.M., Batiha G.E.S., Hetta H.F. Life Sci. 2021;273 doi: 10.1016/j.lfs.2021.119117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal A. J. Adv. Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L., Nicastri E., Scorzolini L., Di Caro A., Capobianchi M.R., Castilletti C., Lalle E. Euro Surveill. 2020;25:2–5. doi: 10.2807/1560-7917.ES.2020.25.8.2000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- Deeks J.J., Raffle A.E. BMJ. 2020;371:1–2. doi: 10.1136/bmj.m4787. [DOI] [PubMed] [Google Scholar]
- Dincer C., Bruch R., Kling A., Dittrich P.S., Urban G.A. Trends Biotechnol. 2017;35:728–742. doi: 10.1016/j.tibtech.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramé M., Tabue Teguo M., Proye E., Hequet F., Hentzien M., Kanagaratnam L., Godaert L. J. Med. Virol. 2020;92:2312–2313. doi: 10.1002/jmv.25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dysinger M., Marusov G., Fraser S. J. Immunol. Methods. 2017;450:1–10. doi: 10.1016/j.jim.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Elshal M.F., McCoy J.P. Methods. 2006;38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.C., Fragaszy E.B., Bermingham A., Wang L., Copas A., Edmunds W.J., Ferguson N., Goonetilleke N., Harvey G., Kovar J., Lim M.S., McMichael A., Millett E.R., Nguyen-Van-Tam J.S., Nazareth I., Pebody R., Tabassum F., Watson J.M., Wurie F.B., Johnson A.M., Zambon M. Lancet Respir. Med. 2014;2(6):445–454. doi: 10.1016/S2213-2600(14)70034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Simon V. Science. 2020;80:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Lewis J.M., Macpherson P., Adams E.R., Ochodo E., Sands A., Taegtmeyer M. AIDS. 2015;29:2465–2471. doi: 10.1097/QAD.0000000000000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Anal. Chem. 2020;92:9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.H. Emerg. Microb. Infect. 2020;9:1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Lin S., Zhang H., Liang L., Shen S. Micromachines. 2021;12:697–715. doi: 10.3390/mi12060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas S., Badran A.A., Juárez M.J., Fernández de Rojas D.H., Morais S., Maquieira A. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112438. [DOI] [PubMed] [Google Scholar]
- Parolo C., Sena-Torralba A., Bergua J.F., Calucho E., Fuentes-Chust C., Hu L., Rivas L., Álvarez-Diduk R., Nguyen E.P., Cinti S., Quesada-González D., Merkoçi A. Nat. Protoc. 2020;15:3788–3816. doi: 10.1038/s41596-020-0357-x. [DOI] [PubMed] [Google Scholar]
- Shen Y., Anwar T. Bin, Mulchandani A. Sensor. Actuator. Rep. 2021;3 doi: 10.1016/j.snr.2021.100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetti N.P., Mishra A., Bukkitgar S.D., Basu S., Narang J., Raghava Reddy K., Aminabhavi T.M. ACS Appl. Bio Mater. 2021;4:1178–1190. doi: 10.1021/acsabm.0c01545. [DOI] [PubMed] [Google Scholar]
- Surkova E., Nikolayevskyy V., Drobniewski F. Lancet Respir. Med. 2020;8:1167–1168. doi: 10.1016/S2213-2600(20)30453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi S.U., Miao C., Sanchez J.E., Caidi H., Tamin A., Haynes L., Thornburg N.J. Sci. Rep. 2019;9:1390. doi: 10.1038/s41598-018-37747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C., Carriere M., Fusco L., Fusco L., Capua I., Regla-Nava J.A., Pasquali M., Pasquali M., Pasquali M., Scott J.A., Vitale F., Vitale F., Unal M.A., Mattevi C., Bedognetti D., Merkoçi A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L.G. ACS Nano. 2020;14:6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- World Health Organization 2021. https://www.who.int)/publications/m/item/weekly-epidemiological-update-on-covid-19
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R., Bykov A., Vogt A., et al. Nat. Commun. 2021;12(3132):1–17. doi: 10.1038/s41467-021-22664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.