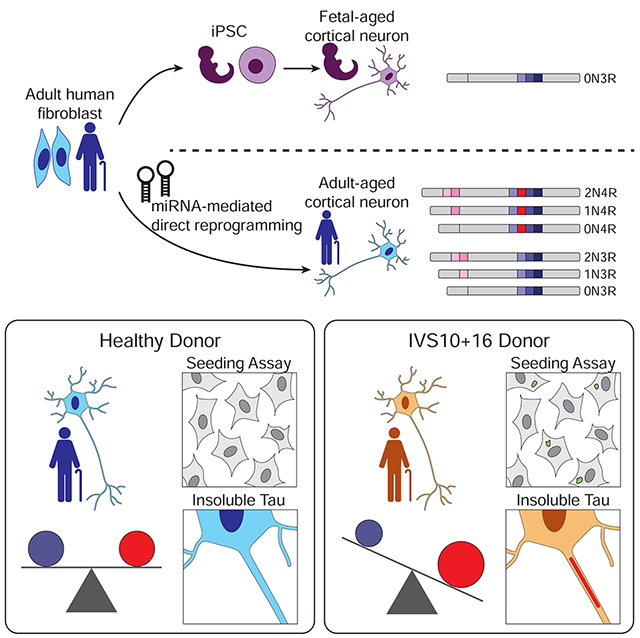
SUMMARY
Tau is a microtubule-binding protein expressed in neurons and the equal ratio between 4-repeat (4R) and 3-repeat (3R) isoforms are maintained in normal adult brain function. Dysregulation of 3R:4R ratio causes tauopathy and human neurons that recapitulate tau isoforms in health and disease will provide a platform for elucidating pathogenic processes involving tau pathology. We carried out extensive characterizations of tau isoforms expressed in human neurons derived by microRNA-induced neuronal reprogramming of adult fibroblasts. Transcript and protein analyses showed miRNA-induced neurons expressed all six isoforms with the 3R:4R isoform ratio equivalent to that detected in human adult brains. Also, miRNA-induced neurons derived from familial tauopathy patients with a 3R:4R ratio-altering mutation showed increased 4R tau and the formation of insoluble tau with seeding activity. Our results collectively demonstrate the utility of miRNA-induced neuronal reprogramming to recapitulate endogenous tau regulation comparable to the adult brain in health and disease.
Graphical Abstract
eTOC
Capano et al. show that human neurons directly reprogrammed by microRNAs from adult fibroblasts capture the endogenous expression of all tau isoforms detected in adult brains, including the vital 1:1 ratio of 3R:4R tau. Also, reprogrammed neurons derived from fibroblasts of tauopathy patients show formation of seed-capable and insoluble tau.
INTRODUCTION
Tauopathies are adult-onset, neurodegenerative disorders whose shared pathology is intracellular tau protein aggregates (Gao et al., 2018; Götz et al., 2019). Tau, encoded by the Microtubule Associated Protein Tau (MAPT) gene, produces six protein isoforms whose expression is under strict developmental regulation, with only one isoform (0N3R) present during fetal development but all six present perinatally and maintained throughout lifespan (Goedert and Jakes, 1990b; Goedert et al., 1989b; Hefti et al., 2018; Himmler et al., 1989). Tau isoforms are defined by the inclusion or exclusion of three alternatively spliced exons: 2, 3, and 10. Exon 2 and 3 inclusion produces two N-terminal domains (N1 and N2) whereas exon 10 encodes the second imperfect-repeat microtubule-binding domain (R2) among three other repeat domains (R1, R3, R4) (Goedert et al., 1989a). The inclusion or exclusion of exon 10 gives rise to 4-repeat (4R) and 3-repeat (3R) tau, respectively, and these two isoform families are expressed at an approximately 1:1 ratio in the healthy adult human brain (Goedert et al., 1989a; Kosik et al., 1989). Perturbation of the 3R:4R tau ratio contributes to the pathogenesis of various tauopathies, and out of the 53 known familial pathogenic tau mutations, almost 50% are located within or related to the expression of exon 10 (Ghetti et al., 2015). Thus, it is critical to establish a human neuron-based system that recapitulates the endogenous tau isoforms seen in the adult brain and tau pathology resulting from 3R:4R dysregulation in patient-derived neurons. The significance of human neuron-based approaches is also highlighted by the fact that humanized mouse tau models lack the 1:1 3R:4R ratio and requires genetic perturbation to achieve 3R:4R tau level comparable to the adult human brain (Duff et al., 2000; He et al., 2020).
The past decade has primarily used neuronal differentiation of induced pluripotent stem cells (iPSCs) as the primary methodology for generating human neurons (iPSC-Ns). While these cells offer a robust means to generate neurons at a large quantity, their utility in studying processes that occur in aged neurons has been limited due to the reversion of the cellular age, and previous studies revealed iPSC-Ns represent fetal stages of development (Lapasset et al., 2011; Patterson et al., 2012) and express only marginal levels of 1N, 2N, and 4R tau expression (Ehrlich et al., 2015; Sato et al., 2018; Sposito et al., 2015). To increase 4R tau levels in iPSC-Ns, several experimental perturbations have been used including transgene overexpression (Verheyen et al., 2015), splicing mutations (Verheyen et al., 2018), or extended culturing times (188-365 days) (Beevers et al., 2017; Sposito et al., 2015). However, these experimental manipulations still fall short of generating neurons with the endogenous ratio of 3R:4R in the adult human brain. Only the immortalized human neural progenitor ReNcells, which, after differentiation into a neuronal population, have expressed 4R tau mRNA along with 3R tau (Choi et al., 2014b; Kwak et al., 2020). ReNcells serve as a model for human neurons, but will have to be engineered to carry known human tau mutations. Therefore, an alternative strategy to generate human neurons that mirror the endogenous tau expression and harbor familial mutations will greatly increase the ability to study tauopathies.
Brain-enriched microRNAs (miRNAs), miR-9/9* and miR-124 (miR-9/9*-124) are potent neurogenic miRNAs whose expression is present in fetal and adult brain. During neural development, these miRNAs regulate cell proliferation and neuronal differentiation (Lim et al., 2005; Tan et al., 2012; Zhao et al., 2009). When ectopically expressed in human adult fibroblasts, miR-9/9*-124 can directly convert fibroblasts to neurons (miNs) (Lu and Yoo, 2018; Yoo et al., 2011). The miRNA-induced neuronal state can be synergized with subtype-defining transcription factors (TFs) that guide conversion to specific neuronal subtypes (Abernathy et al., 2017; Victor et al., 2014). During reprogramming, miRNAs first instruct cell cycle exit and fibroblast identity erasure, followed by the neuronal program activation in sequence (Cates et al., 2020). The fate conversion relies on miRNAs targeting components of chromatin modifiers and transcription factors, coordinating extensive reconfiguration of chromatin landscape without passing through pluripotent and multipotent stem cell stages (Abernathy et al., 2017; Cates et al., 2020; Huh et al., 2016; Richner et al., 2015; Victor et al., 2018; Yoo et al., 2009). This property in turn allows for the maintenance of the epigenetic age signature of fibroblast donors (Huh et al., 2016). Additionally, modeling adult-onset disorders using miNs derived from symptomatic Huntington’s disease patients exhibited hallmark adult-onset pathologies such as Huntingtin inclusion bodies, while their iPSC-N counterparts failed to do so (Victor et al., 2018).
Here, we reveal that miRNA-mediated direct reprogramming generates human neurons that express both 3R and 4R tau in the same ratio detected in adult human brains. RNA profiling by transcript assays and protein profiling by mass spectrometry demonstrates that adult human brain and miNs from adult fibroblasts have indistinguishable 4R tau profiles, which starkly contrasts with the low 4R tau expression in primary fetal human neurons and iPSC-Ns. The endogenous tau isoform regulation in miNs is sensitive to a point mutation within the splice site proximal to exon 10 in tauopathy patients resulting in reprogrammed neurons that showed increased 4R:3R tau ratio. Significantly, the increased level of 4R tau correlated with the formation of insoluble tau and seed-competent tau, reminiscent of that seen in human 4R tauopathies, and contrasting the lack of seed-competent tau seen in iPSC-Ns harboring the same patient mutation. Therefore, miRNA-mediated neuronal reprogramming provides a robust model for studying both the normal and abnormal biology of tau and exon 10 pathological mutations.
RESULTS
MiNs display neuronal markers similar to iPSC-Ns and fibroblast fate erasure
In this study, we adapted the neuronal conversion protocol based on miR-9/9*-124, Neuronal Differentiation 2 (NEUROD2), and Myelin Transcription Factor 1 Like (MYT1L) transcription factors (TFs) to generate neurons of cortical lineage (Yoo et al., 2011) (Fig 1). To minimize the number of lentiviruses used for transduction in fibroblasts, we replaced NEUROD2 with ISX9, a neurogenic compound that has been previously shown to activate endogenous NEUROD-family TFs (Li et al., 2015).
Figure 1 |. Directly-reprogrammed neurons display neuronal morphology and transcriptome.
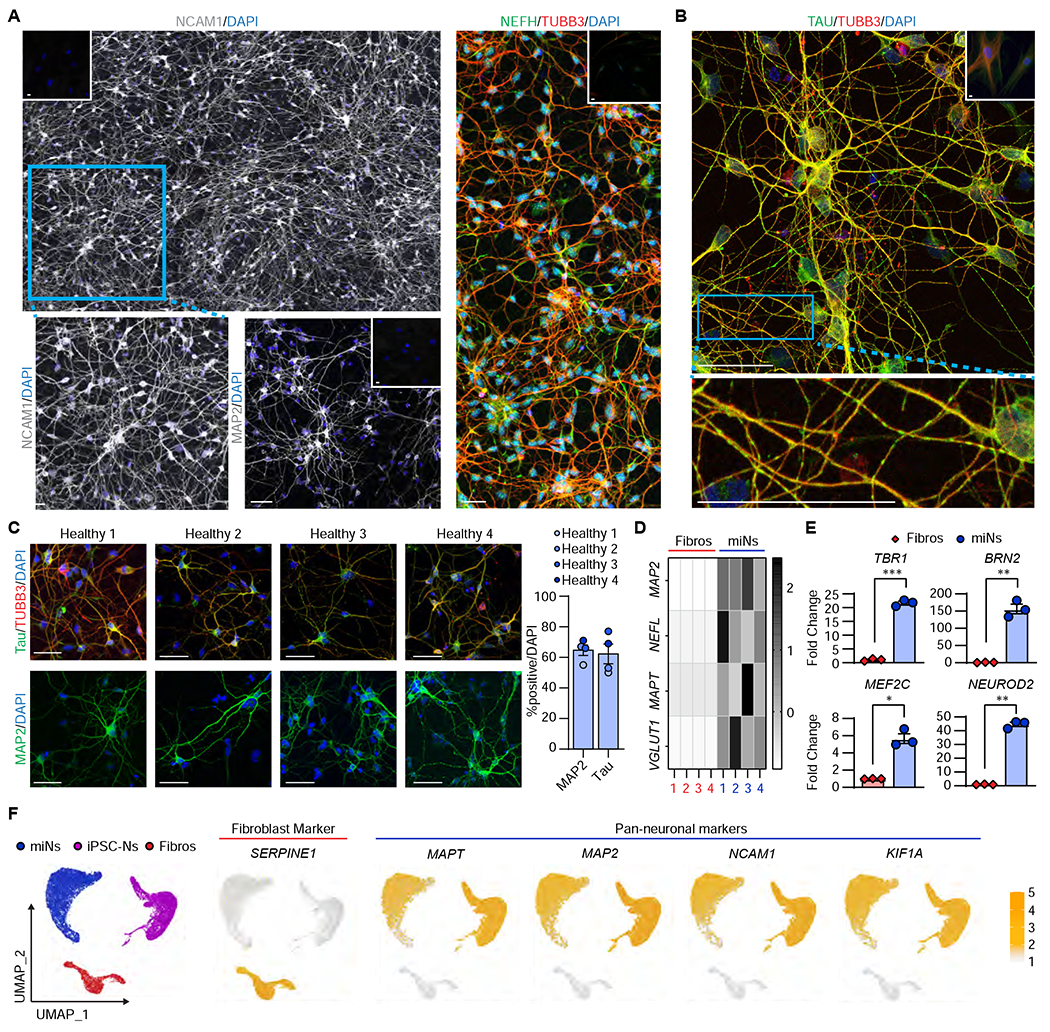
(A) Representative images of miNs from healthy adult individuals (Healthy 3+4) at post-induction day (PID) 30 immunostained with NCAM (composite of 3x2 40x images stitched together), MAP2, NEFH, and TUBB3. Insets at the corners are immunostaining images performed in starting fibroblasts. Blue rectangular outline depicts the region shown in magnified images. Scale bars = 50 μm. (B) Representative image of Healthy 4 immunostained for total tau and TUBB3. Inset is starting fibroblasts. Blue rectangular outline depicts the magnified region showing tau islands. Scale bars = 50 μm. (C) Left, representative images of healthy miNs stained for tau and TUBB3. Right, quantification of % positive over DAPI in reprogrammed cells from four independent fibroblast samples (MAP2 Number of cells counted: miN 1=140, miN 2=82, miN 3=112, miN 4=169; MAPT Ns: miN 1=113, miN 2=118, miN 3=78, miN 4=87). Scale bars = 50 μm. (D) Heatmap plots Z-scores comparing qPCR of neuronal genes between miNs and starting fibroblasts from four independent samples. (E) Fold change of cortical genes between starting fibroblasts and day 30 miNs. n=3 replicates from miNs grown in three independent wells. Mean±SEM.; two-tailed Student’s t-test; *p ≤ 0.05, **p < 0.01, ***p ≤ 0.001. (F) Left, UMAP projection of cells colored by starting cell type (Fibroblast: n=2533 cells; iPSC-N: n=7212 cells; miN: n=9305 cells). Right, UMAP projection of cells colored by non-neuronal (SERPINE1) and pan-neuronal markers (MAPT, MAP2, NCAM1, KIF1A). See also Figure S1.
At thirty days post-induction (PID 30), immunostaining for pan-neuronal marker Neuronal Cell Adhesion Molecule 1 (NCAM1) showed a robust reprogramming output with miNs displaying extensive process outgrowths (Figure 1A left, bottom-left: zoomed in view of NCAM1 expression). The reprogrammed cells were also enriched for other pan-neuronal markers, MAP2 (Microtubule Associated Protein 2) (Figure 1A, bottom-right) and Neurofilament Heavy Chain (Figure 1A, right). Beta III tubulin (TUBB3) was used to monitor cell morphologies. Importantly, immunostaining against tau revealed the miN population highly enriched with tau expression showing prototypical patterns of tau islands in TUBB3-positive outer neurites (Figure 1B), an endogenous tau feature suggested to be reflective of tau-tau interactions (Siahaan et al., 2019; Tan et al., 2019).
Using immunocytochemistry on four independent fibroblast samples reprogrammed into miNs (Figure 1C), approximately 60-70% of DAPI-positive cells showed MAP2- and tau-positivity, which is consistent with the reprogramming efficiency previously reported (Abernathy and Yoo, 2015; Cates et al., 2020; Huh et al., 2016; Victor et al., 2014; Victor et al., 2018). qPCR analyses of neuronal genes, MAP2, NEFL (Neurofilament Light Chain), MAPT, and the glutamatergic marker, SLC17A7(also known as VGLUT1), showed enrichment over starting fibroblasts (Figure 1D). Also, qPCR analyses of the cortical markers TBR1 (T-Box Brain Transcription Factor 1), MEF2C (Myocyte Enhancer Factor 2C), BRN2 (POU Class 3 Homeobox 2), and NEUROD2 demonstrated that the combination of MYT1L and ISX9 are sufficient to drive the cortical fate (Figure 1E).
Using single-cell RNA-sequencing (scRNA-seq) to stratify starting fibroblasts (n=2533), reprogrammed miNs (n=9305), and iPSC-Ns (n=7212) by their transcriptome, we identified three distinct cell populations corresponding to fibroblasts, miNs, and iPSC-Ns (Figure 1F, left). For instance, the non-neuronal marker SERPINE1 (Docagne et al., 1999; Mehra et al., 2016) was enriched only in starting fibroblasts, whereas the miNs and iPSC-Ns demonstrated thorough expression of pan-neuronal markers, MAPT, MAP2, NCAM1, and KIF1A (Figure 1F, right). Additionally, miNs were characterized by high expression of SLC17A7 (VGLUT1) whereas iPSC-Ns were positive for SLC17A6 (VGLUT2) (Figure S1A–C). MiNs and iPSC-Ns expressed different glutamate receptor subunits, but both were highly enriched for the glutamine synthetase, GLUL.GAD1 and GAD2 were present in a subset of miNs and iPSC-Ns, respectively. However, the expression of GABA transporter, SLC6A1, was only marginal. MiNs showed a low number of PVALB-positive cells, whereas iPSC-Ns had small populations of SST and TH positive cells.
Together, these results support the notion that miNs robustly acquire a neuronal fate due to miRNA-mediated reprogramming.
Age-maintained miNs endogenously express a 1:1 3R:4R tau isoform ratio equivalent to adult brain
scRNA-seq revealed that age-associated markers as main drivers for distinct segregation of miNs and iPSC-Ns. For instance, miNs were positive for the advanced age markers CDKN1A (p21), CDKN2A (p16), BAG3, and CAV1 (Figure 2A). BAG3 expression dramatically increases with aging in mouse neurons (Gamerdinger et al., 2009) and the human brain (brainspan.org), and has been implicated in tau clearance (Lei et al., 2015). Moreover, the expression of CAV1 has been shown to be increased in aged human brains (Kang et al., 2006) and its abnormal expression has been implicated in Parkinson’s Disease (Ha et al., 2021). In contrast, fetal-associated markers such as EPH3A (brainspan.org) and BCL2 (Merry et al., 1994) were enriched in iPSC-Ns. These results conform to previous studies indicating the maintenance of cellular age in direct neuronal reprogramming (Huh et al., 2016; Mertens et al., 2015).
Figure 2 |. MiNs express exon 10 inclusion at equivalent ratio to the adult human brain.
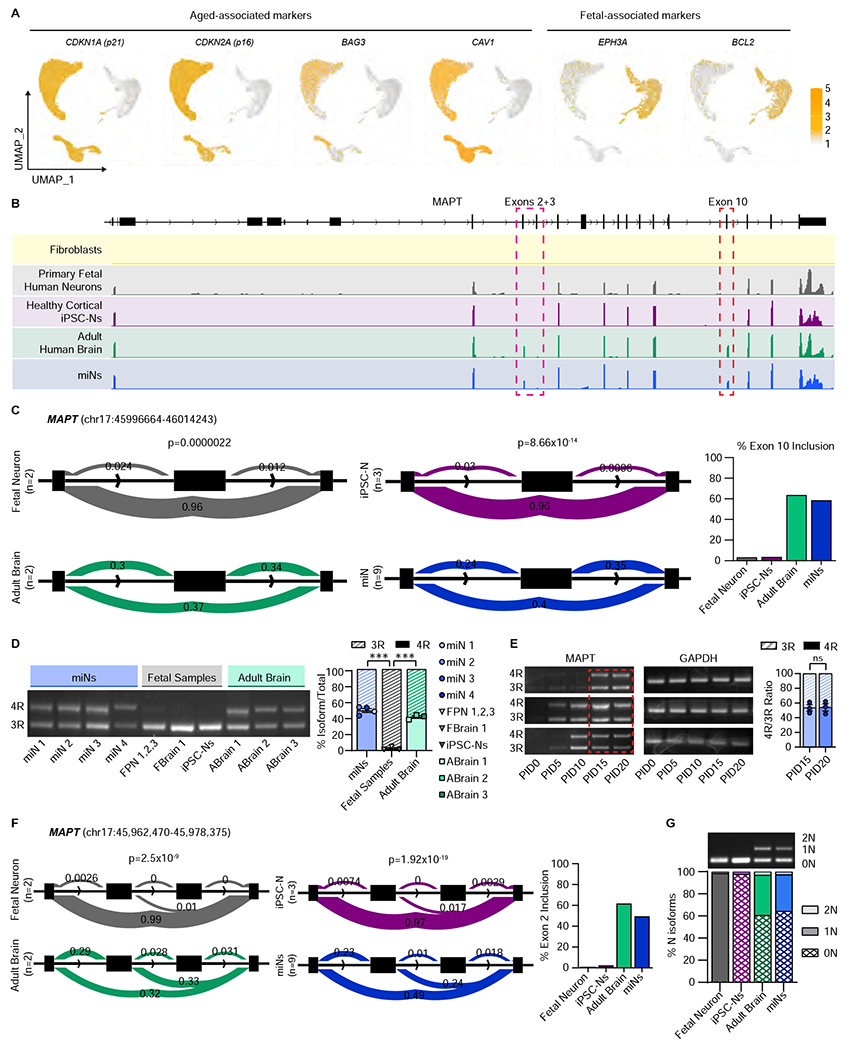
(A) UMAP projection of cells colored by age-associated markers (CDKN1A, CDKN2A, BAG3, and CAV1) and fetal-associated markers (EPHA3 and BCL2). Cell numbers as indicated in Figure 1F. (B) Representative RNA-seq tracks at the MAPT locus of fibroblasts, primary human neurons, cortical iPSC-Ns, adult human brain, and miNs at PID21. (C) Leafcutter analysis of MAPT exon 10 inclusion between fetal neurons and adult brain, and healthy iPSC-Ns and healthy miNs. Percent exon 10 inclusion is quantified on the right. For primary fetal neurons and adult brain, n= sequencing replicates performed from purchased purified RNA (see STAR Methods). For iPSC-Ns, n=replicates grown in three independent wells. For miNs, n=three independent well replicates from three independent individuals. (D) Left, semi-quantitative PCR of 3R and 4R tau isoforms. Right: Quantification of 3R and 4R percent ratio of samples grouped by miNs, fetal samples, and adult brain from independent individuals as indicated in the graph legend. Mean±SEM; One-way ANOVA with multiple comparisons with post hoc Tukey’s test; ***p ≤ 0.001. (E) Semi-quantitative PCR of 3R and 4R tau expression. Red rectangular outline depicts the time point when 4R tau starts being expressed consistently in multiple samples (n=three independent individuals). Right, Two-tailed Student’s T-test of PID15 vs PID20. Mean±SEM; ns p = 0.92. See also Figure S2 and Tables S1 and S2.
Tau isoform expression is developmentally regulated in which 4R isoforms begin to be expressed perinatally and are maintained throughout adulthood (Hefti et al., 2018). Extending the notion of age maintenance, we tested if miNs expressed 4R tau by bulk RNA-sequencing (RNAseq). Strikingly, miNs and adult brain showed robust signal of exon 10 in the RNAseq tracks (Figure 2B, red box: blue and green tracks, respectively), peaks that were distinctly missing in primary fetal human neurons and iPSC-Ns (Figure 2B, red dashed box: grey and purple tracks, respectively). The alternatively spliced exons 2 and 3 are also clearly expressed in miNs and adult human brain (Figure 2B, pink dashed box). We also used the Leafcutter analysis, which calculates splicing events (Li et al., 2018), to quantify inclusion of exon 10 and found significant differences between fetal and adult brain (p=2.2x10−6) and iPSC-Ns and miNs (p=8.66x10−14) (Figure 2C). We also identified 630 genes which demonstrate similar and significant differences in alternative splicing between adult brain versus fetal neurons, also manifested in miNs versus iPSC-Ns (Table S1). Some known age-associated splicing events captured between our aged and fetal samples include SCN2A (Kasai et al., 2001; Thompson et al., 2020), DCTN4 (Tanner et al., 2021), MAPRE3 (Mazin et al., 2013), and NEO1 (Mazin et al., 2013) (Figure S2A).
To specifically look at the 3R to 4R tau mRNA ratio, we used semi-quantitative PCR (sq-PCR) (Marone et al., 2001) in which PCR is performed in a cDNA library with primers in exons 9 and 11, flanking exon 10, creating a long (4R) and short (3R) PCR product corresponding to the inclusion and exclusion of exon 10, respectively. We found consistent expression of 4R tau mRNA in miNs comparable to adult human brain samples, whereas all three types of fetal samples primarily expressed 3R tau (Figure 2D). MiNs were found to express 50.3% of tau mRNA as 4R tau (Figure 2D, right). We asked at what time point during the neuronal reprogramming tau expression begins, and in multiple fibroblast samples we found consistent, robust expression of both 3R and 4R tau transcripts starting at PID15 (Figure 2E). Of note, this two-week point corresponds to the time that miNs start activating the neuronal program required for neuronal identity (Cates et al., 2020).
In addition to exon 10, MAPT exons 2 and 3 are also spliced under developmental control, with fetal samples only expressing a singular 0N isoform, whereas adult human brain expresses around 40% 0N, 50% 1N, and 10% 2N isoforms (Goedert and Jakes, 1990a). Leafcutter determined that the inclusion of exon 2 was also differentially spliced between adult versus fetal samples, also reflected in miNs versus iPSC-Ns, where the fetal-aged samples showed no inclusion, but adult-aged samples expressed ~50% 1N isoforms (Figure 2F). To validate, we designed sqPCR primers which flank exons 2 and 3, generating long, medium, and short PCR products depending on mRNA expression of 2N, 1N, and 0N isoforms, respectively. Fetal-aged samples only expressed 0N isoforms, whereas adult brain and miNs had equivalent expression of all three N isoforms (Figure 2G).
To determine if the miRNAs were controlling the inclusion of exon 10, we overexpressed either non-specific miRNAs or miR-9/9*-124 in neural progenitor cells (NPCs) then differentiated them to iPSC-Ns through small molecule differentiation (Mahali and Karch, 2021). MiR-9/9*-124 had no effect on exon 10 inclusion and all iPSC-N samples expressed only 3R tau (Figure S2B). This data supports that miNs maintain the age of the starting cells (Huh et al., 2016), and this age drives exon 10 inclusion, and miNs recapitulate the endogenous 4R expression, generating the 1:1 3R:4R ratio seen in the adult brain.
What controls exon 10 inclusion has not been identified in the adult human brain context. It is possible that age-associated, differentially-expressed RNA binding proteins and splicing factors direct the inclusion of exon 10 in adult-aged samples. We compared pairwise, bulk RNAseq between adult brain versus fetal neurons and miNs versus iPSC-Ns and identified 88 differentially expressed genes identified by Ingenuity Pathway Analysis as splicing regulators (Table S2). This gene list will have to be validated for potential age-associated splicing regulators in future studies.
MiNs produce endogenous 4R tau protein similar to adult brain
To validate tau isoform expression at the protein level, we performed mass spectrometry that can quantitatively and sensitively measure the tau isoforms in both cell culture and brain (Barthélemy et al., 2020a; Barthélemy et al., 2020b; Horie et al., 2020; Sato et al., 2018) on miNs from four independent adult individuals, four independent samples of fetal primary neurons or brain, adult brains from five individuals, and three iPSC-N replicates (Figure 3A, solid line = mean). We quantified tau peptides across full length intracellular tau protein, including peptides specific for 4R (residues 275-280, 282-290, and 299-317). In iPSC-Ns and fetal samples, the 1N, 2N and 4R peptides were expressed at less than 3% of the sum of all tau isoforms (i.e. 0N+1N+2N, and 3R+4R, respectively) (Figure 3A–D), consistent with previous literature showing iPSC-Ns recapitulate only fetal neuronal tau isoforms (Ehrlich et al., 2015; Sato et al., 2018; Sposito et al., 2015). Importantly, the 1N, 2N, and 4R peptide profiles of miNs showed significant enrichment over that of fetal samples, mirroring the tau profile of the adult brain samples (Figure 3A–D). To specifically assay 3R:4R in miNs, we analyzed the ratio of all three 4R-specific peptides to the adjacent, constitutive R1 domain peptide (260-267). Remarkably, miNs display an approximately 1:1 3R to 4R ratio equivalent to that of the adult human brain, which drastically contrasted iPSC-Ns and fetal neurons (Figure 3A–B). The presence of multiple tau isoforms in miNs and adult brain was confirmed via western blot, which differed with fetal brain expressing only 0N3R isoform (Figure 3D). Further, a time course of western blot for tau in miNs at days 15-24 into reprogramming showed clear expression of both 3R and 4R isoforms at 1:1 3R:4R ratio throughout the immunoblots (Figure 3E).
Figure 3 |. MiN tau protein profile mirrors that of the adult human brain.
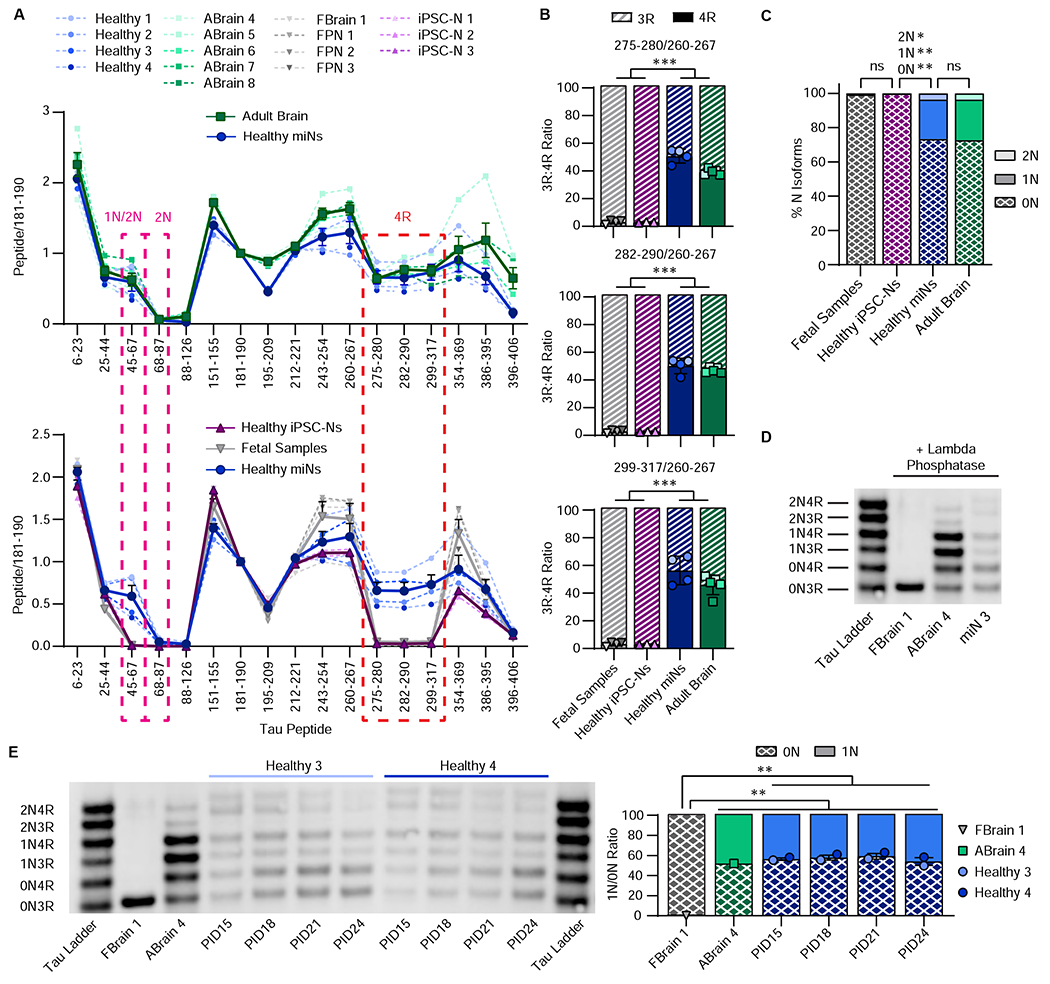
(A) Quantitation of relative tau peptides from multiple independent samples as indicated in the plot legend. Mean(thick line)±SEM. Pink and red dashed outline depicts1N, 1N/2N, and 4R-specific tau peptides, respectively. (B) Quantification of 3R:4R isoform ratio of all samples used in Figure 3A. Histogram colors are matched to sample colors shown in the plot legend of Figure 3A. Mean±SEM; One-Way ANOVA with multiple comparisons and post hoc Tukey’s test: Between any fetal vs adult sample for all three 4R peptides: ***p ≤ 0.001. (C) Percent of N isoforms (calculation in methods) using all samples as indicated in the plot legend in Figure 3A. Mean±SEM; One-Way ANOVA with multiple comparisons and post hoc Tukey’s test. All N isoforms between fetal vs healthy iPSC-Ns and between adult vs healthy miNs: ns p ¾ 0.999; between healthy iPSC-Ns vs healthy miNs: 0N: **p = 0.002, 1N: **p = 0.002, 2N: *p = 0.015. (D) Western blot of tau isoforms. (E) Left, western blot of tau isoforms over reprogramming timecourse. Right, quantification of 3R:4R ratio of 0N and 1N isoforms during the time course of reprogramming of two independent samples indicated in the plot legend. Mean±SEM; One-Way ANOVA with multiple comparisons and post hoc Tukey’s test; Between fetal vs any miN: **p = 0.002; between fetal and adult: **p = 0.004.
Directly reprogrammed miNs from tauopathy patient fibroblasts recapitulate abnormal 4R tau mRNA expression
We next asked if the neuronal reprogramming would be sensitive to capture the increased 4R tau resulting from a point mutation at an intronic splice site known as IVS10+16 C>T (IVS10+16) (Hutton et al., 1998), which disrupts a stem-loop structure and biases splicing towards exon 10 inclusion (Grover et al., 1999). This familial mutation was shown to cause an approximately 2- to 4-fold increase in 4R tau mRNA (Connell et al., 2005) and an imbalanced 3R:4R protein ratio, skewing towards increased 4R. First, we directly converted fibroblast samples with confirmed mutation status (Figure S3A–B) from four symptomatic patients, quantifying successful neuronal conversion, with 75% of DAPI-positive cells expressing MAP2 and tau (Figure 4A). IVS10+16 miNs showed enrichment of neuronal genes over their starting fibroblasts (Figure 4B). Neurons are different from most other cell types in their robust expression of long genes (> 100kb) (Gabel et al., 2015; King et al., 2013; Sugino et al., 2014). Using LONGO, a gene length analysis R package (McCoy et al., 2018), we found miNs from both healthy control and IVS10+16 fibroblasts were successfully reprogrammed to neurons as seen by the forked trajectory between miNs and fibroblasts starting around 200kb (Figure 4C). Reprogrammed miNs had significantly higher long gene expression over the starting fibroblasts (P<0.001), but there was no significant difference in long gene expression between healthy and IVS10+16 miNs (p=0.177) indicating a similar conversion efficiency and no effect of the IVS10+16 mutation on neuronal reprogramming.
Figure 4 |. IVS10+16 patient-derived miNs show increased 4R tau mRNA.

(A) Left, representative images of reprogrammed IVS10+16 miNs from four independent, symptomatic IVS10+16 patients, immunostained for tau, MAP2 and TUBB3. Right, quantification of % positive over DAPI (MAP2 Number of cells counted: IVS10+16 1=118, IVS10+16 2=173, IVS10+16 3=105, IVS10+16 4=152; MAPT Ns: IVS10+16 miN 1=98, IVS10+16 miN 2=125, IVS10+16 miN 3=89, IVS10+16 miN 4=88). Scale bars = 50 μm. (B) Z score heatmap (from qPCR) representation of neuronal marker gene expression MAP2, NEFL, MAPT, and VGLUT1 in IVS10+16 miNs and starting fibroblasts from four independent lines. (C) LONGO long gene analysis between starting fibroblasts (red) and reprogrammed healthy and IVS10+16 miNs (blue and orange, respectively). (D) Leafcutter analysis of MAPT exon 10 inclusion between healthy and IVS10+16 miNs. Percent exon 10 inclusion is quantified on the right. For miN groups, n=three independent well replicates from three independent individuals. (E) Left, sqPCR for detecting 4R and 3R tau ratio. Right: sqPCR intensity values grouped by healthy miNs, IVS10+16 miNs, and healthy adult brain for 3R and 4R. Mean±SEM. One-Way ANOVA with multiple comparisons and post hoc Tukey’s test; Between healthy miNs vs IVS10+16 miNs and IVS10+16 miNs vs adult brain: ***p ≤ 0.001. (F) sqPCR of both healthy and IVS10+16 human brain and miNs. See also Figure S3.
We next used Leafcutter on bulk RNAseq to dissect the amount of exon 10 inclusion between healthy and IVS10+16 miNs, and found there was a significant increase in 4R tau in IVS10+16 samples (p=0.012) (Figure 4D). This was validated using sq-PCR in which IVS10+16 miNs had a significant increase in exon 10 inclusion over that of healthy miNs and adult brain, with a 1.46-fold increase in their 4R:3R mRNA ratio (Figure 4E). As this fold change differed from those published when this familial mutation was discovered (Hutton et al., 1998), we performed sqPCR on IVS10+16 human brain and found the same fold change between IVS10+16 miNs and human brain over their corresponding healthy controls (Figure 4F). As expected, N isoform expression ratios were not altered in IVS10+16 miNs (Figure S3C).
Increase in 4R tau protein in IVS10+16 miNs
To validate the increase in 4R mRNA on the protein level, we performed mass spectrometry analyses on four independent patient-derived IVS10+16 miNs and isogenic IVS10+16 iPSC-Ns, both of which demonstrated an increase in the 4R tau peptides over the age-matched, healthy counterparts (Figure 5A, solid line = mean). IVS10+16 miNs had a significant increase in all three 4R-specific peptides over that of healthy miNs (Figure 5B) IVS10+16 iPSC-Ns also showed significant increase in 4R tau over healthy iPSC-Ns but their final 4R tau expression is still far below that of the 3R tau level (Figure 5A bottom: purple line, Figure 5B: pink bars). The ratio of all three 4R-specific peptides matched the increase seen in mRNA, with a 1.31-1.41-fold increase in the IVS10+16 patient miNs over the healthy miNs (Figure 5C). As expected, there was no significant difference in the profile of any other regions of tau including the ratio of 0N, 1N, or 2N isoforms between WT and IVS10+16 miNs (Figure 5D).
Figure 5 |. IVS10+16 patient-derived miNs demonstrate increased 4R protein corresponding to increased 4R mRNA.
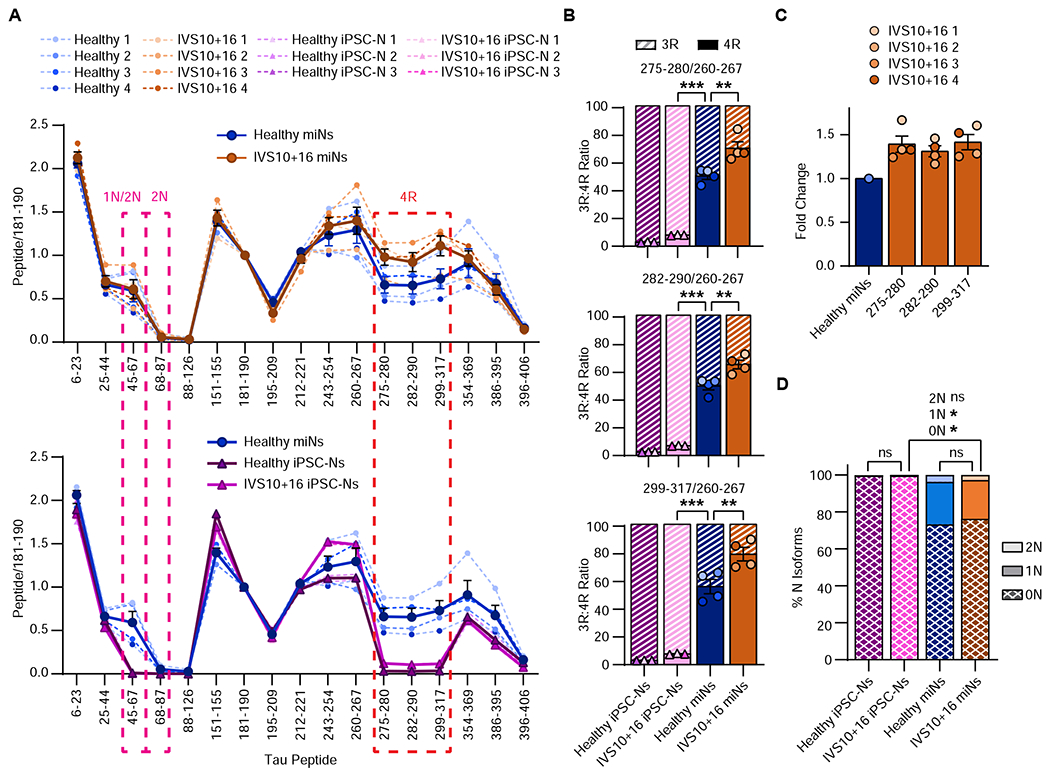
(A) Quantitation of relative tau peptides from multiple independent samples as indicated in the plot legend. Mean(thick line)±SEM. (B) Quantification of 3R:4R isoform ratio of all samples used in Figure 5A. Histogram colors are matched to sample colors shown in the plot legend of Figure 5A. Mean±SEM. One-Way ANOVA with multiple comparisons and post hoc Tukey’s test; healthy miNs vs IVS10+16 miNs: 275-280 **p = 0.004, 282-290 **p = 0.004, 299-317 **p = 0.008. Between any miN and iPSC-N: all peptides ***p ≤ 0.001. C) Fold change of 4R isoform expression for all three 4R-specific peptides over healthy miNs. (D) Percentage of N isoforms using all samples as indicated in the plot legend in Figure 5A. Mean±SEM; One-Way ANOVA with multiple comparisons and post hoc Tukey’s test. For all N isoforms: between healthy iPSC-Ns and IVS10+16 iPSC-Ns: ns p > 0.999; between healthy miNs vs IVS10+16 miNs: 0N: ns p = 0.97; 1N: ns p = 0.983; 2N: ns p = 0.887; between IVS10+15 miNs vs IVS10+16 iPSC-Ns: 0N: *p = 0.038; 1N: *p = 0.027; 2N: ns p = 0.285.
Increased insoluble tau and seeding capacity in IVS10+16 miNs
As a model system to endogenously express both the altered 3R:4R tau ratio seen in patient samples and have the advanced-age required for tau-associated phenotypes, we assessed IVS10+16 miNs for human tauopathy-specific pathologies. Previous studies have indicated that pathological tau has seeding capacity that precedes the formation of insoluble tau (Holmes et al., 2014) and which can be assayed using a tau-FRET Biosensor Assay (Clavaguera et al., 2009; Frost et al., 2009; Guo and Lee, 2011; Liu et al., 2012). We found the patient-derived IVS10+16 miNs produced seeding-competent tau, with a 50% increase in FRET-positive signal from IVS10+16 miN lysate than that of healthy miN lysate (Figure 6A) indicating the presence of tau seeds. Importantly, control and isogenic IVS10+16 iPSC-Ns did not show any seeding capacity (Figure 6A).
Figure 6 |. Formation of seed-capable and insoluble tau in IVS10+16 patient-derived miNs.
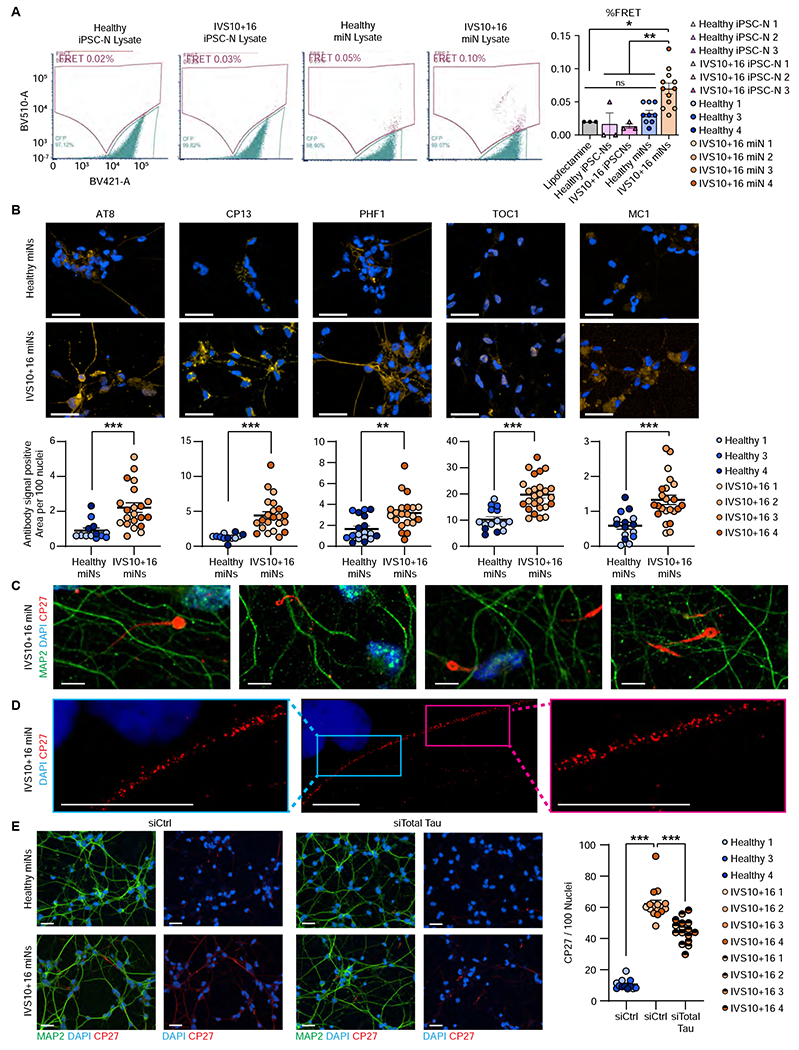
(A) Left, Representative FRET signals for healthy and IVS10+16 iPSC-N and miN lysate. Right, quantification of % FRET. n=20,000 cells per FRET run. For iPSC-N samples, we performed three independent FRET runs per cell pellet. For miNs, we used three healthy and four IVS10+16 independent, individual samples, with three replicate FRET runs per sample. One-way ANOVA with multiple comparisons. *p ≤ 0.05, **p ≤ 0.01. (B) Top, representative images of methanol-fixed healthy and IVS10+16 miNs. Cells were co-labeled with MAP2 antibody (green) and one of five anti-tau antibodies: AT8, CP13, MC1, PHF1, and TOC1 (yellow). Below, quantification of anti-tau antibody signal positive area per 100 nuclei from three healthy and four IVS10+16 independent, individual samples as indicated in plot legend. Nuclei counted per condition: AT8: healthy miN=70939, IVS10+16 miN=84046; CP13: healthy miN=69708, IVS10+16 miN=88775; PHF1: healthy miN=93443, IVS10+16 miN=71070; TOC1: healthy miN=47280, IVS10+16 miN=91376; MC1: healthy miN=94171, IVS10+16 miN=97931. Scale bars = 50 μm. (C) High magnification of IVS10+16 miNs and insoluble tau threads and tendrils stained with total tau antibody, CP27. Scale bars = 50 μm. (D) Stimulated emission depletion (STED) microscopy of insoluble tau in IVS10+16 miNs. Scale bars =10 μm. (E) Left, arrowheads indicate the location of insoluble tau signals in healthy miNs (top) and IVS10+16 miNs (bottom). Left images: cells treated with control siRNA (siCtrl). Right images: cell treated with siRNA against total tau (siTotal tau). Right plot: Quantification of tau signals detected per 100 nuclei (DAPI) in healthy miNs (healthy miNs treated with siCtrl) in comparison to IVS10+16 miNs treated with siCtrl and IVS10+16 miNs treated with siTotal tau from three healthy and four IVS10+16 independent, individual samples. Nuclei counted per condition: siCTRL on healthy miNs =140,283; siCTRL on IVS10+16 miNs=117,781; siTau on IVS10+16 miNs=114,050. One-way ANOVA with multiple comparisons. ***p ≤ 0.001. Scale bars = 50 μm. See also Figure S4.
We next used methanol fixation method to remove soluble proteins (Guo et al., 2016; Katsikoudi et al., 2020) and five tau-specific antibodies: AT8, (phospho-Ser202/Thr205), CP13 (pS202 tau), PHF1 (phosphor-Ser396/Ser404), TOC1 (oligomeric tau), and MC1 (conformationally abnormal tau). All five antibodies showed a significant increase in signal in IVS10+16 miNs over that of healthy miNs, indicating the increased presence of insoluble tau in patient-derived miNs (Figure 6B). No exogenous perturbation was applied to these cells; the insoluble tau developed through endogenous methods. This is in striking contrast to previously published data from IVS10+16 iPSC-Ns which were devoid of insoluble tau (Verheyen et al., 2018). We also observed thread-like tau staining (Braak and Del Tredici, 2010; Guo et al., 2016; Katsikoudi et al., 2020) in IVS10+16 miNs (Figure 6C). Using stimulated emission depletion (STED) confocal microscope, we obtained higher resolution images of CP27-positive insoluble tau which demonstrated speckles of varying sizes along the neurites imaged (Figure 6D). We further confirmed that the insoluble tau signal was specific and caused by the tau protein by applying tau siRNA to IVS10+16 miNs. Total tau was reduced by 30-40% (Figure S4), which led to significant reduction in the amount of insoluble tau (Figure 6E).
DISCUSSION
The ability to study tau expression, isoform control, and age-associated disease pathologies will rely on a model system which endogenously recapitulates tau expression of the adult human brain. ReNcell-derived neurons display a dramatic increase in 4R tau transcript whose expression becomes enhanced in 3D culturing condition (Choi et al., 2014b; Kwak et al., 2020). Results shown here demonstrate miRNA-mediated neuronal conversion as a means to generate adult human neurons that mimic endogenous tau isoforms at both transcript and protein levels mirroring the 1-to-1 ratio of 3R:4R in the adult human brain. This ratio was strictly regulated in multiple independent donors. Using patient fibroblasts which harbor the IVS10+16 intronic mutation, we found that miRNA-reprogramming were sensitive to capture the difference in mRNA levels caused by the genetic mutation and the subsequent increase in 4R protein levels that reflect fold increase at mRNA level.
The directly converted human neuron model with robust 4R tau expression raises interesting future questions, for instance, which age-associated splicing factors drive exon 10 inclusion. Engineered mouse models expressing the entire human MAPT genomic sequence deviated from the expected 1:1 ratio of 3R:4R, and showed a bias towards 3R (Duff et al., 2000), indicating there may exist splicing events and factors unique for 4R tau regulation in adult human neurons. Additionally, previous studies using tau minigenes to assay the regulation of tau exon 10 splicing in immortalized cell lines, has implicated over 100 putative splicing effectors including SRSF7 (Cavaloc et al., 1994), SFPQ (Ishigaki et al., 2017), NOVA1 (Wang et al., 2004), and DYRK1A (Ding et al., 2012), but these remain to be validated in an adult human neuronal context which could be addressed using miNs.
The maintenance of age is required for modeling adult-onset disorders supported by recently published recent study of age-associated phenotypes in patient-derived neurons (Mertens et al., 2021; Victor et al., 2018). IVS10+16 patient-derived miNs develop increased insoluble tau inclusions and seed-competent tau, two defining characteristics of pathological tau from adult-onset tauopathy brains (Frost et al., 2009; Guo and Lee, 2011; Guo et al., 2016). The ability for a model to endogenously produce pathogenic tau will provide a system for assaying the prevention and clearing of insoluble tau. Of note, the model responded to the genetic knock-down of tau using siRNA which has therapeutic implications as anti-sense oligomer directed reduction of tau is in human clinical trials (Safety, Tolerability and Pharmacokinetics of Multiple Ascending Doses of NIO752 in Progressive Supranuclear Palsy).
Given the global increase in the aging population and the correlating increase in tauopathies, a model system that mirrors the adult human brain and its tau expression will provide an alternative avenue for studying these devastating diseases. The generation of human neurons that regulate tau splicing in the same way as the human adult brain and can replicate human tau-associated phenotypes represents a significant experimental advancement towards the investigation of tau-based pathology.
LIMITATIONS OF STUDY
While the data show that miNs provide an excellent model for studying tau isoform regulation and that they have the potential for testing pathological interventions in patient neurons, there is a limitation to direct reprogramming that should be noted. Scalability is limited due to tauopathy patient-derived fibroblasts being a rare commodity, especially those collected after symptom onset. This limitation highlights the need for more patient sample collection and availability to increase the scientific community’s ability to leverage the benefit of direct neuronal conversion as an experimental system for adult-onset tauopathies.
STAR METHODS
Resource Availability
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Andrew S. Yoo (yooa@wustl.edu).
Materials Availability
All unique and stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Single-cell and bulk RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper available from the Lead Contact upon request
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-MAP2 | Cell Signaling Technology | Cat# 4542S; RRID: AB_ 10693782 |
| Rabbit polyclonal anti-MAP2 | Millipore | Cat# AB5622; RRID: AB_91939 |
| Rabbit polyclonal anti-Tau | Aligent/Dako | Cat# 002401-2; RRID: AB_10013724 |
| Mouse monoclonal anti-TUBB3 | BioLegend | Cat# 801202; RRID: AB_ 10063408 |
| Rabbit polyclonal anti-TUBB3 | BioLegend | Cat# 802001, RRID: AB_2564645 |
| Chicken polyclonal anti-TUBB3 | Novus | Cat# NB100-1612, RRID: AB_10000548 |
| Mouse monoclonal anti-Tau (CP-27) | Peter Davies, Albert Einstein College of Medicine | Cat# CP-27; RRID: AB_2716722 |
| Mouse monoclonal anti-Tau (Tau1) | Nicholas M. Kanaan, Michigan State University | Cat# Tau1; RRID: AB_2721197 |
| Mouse monoclonal anti-Tau (HJ8.5) | David Holtzman, Washington University in St. Louis | Cat# HJ8.5; RRID: AB_2721237 |
| Mouse monoclonal anti-NCAM1 | Santa Cruz Biotechnology | Cat#SC-106; RRID: AB_627128 |
| Mouse monoclonal anti-pTau (AT8) | Thermo Fisher Scientific | Cat# MN1020; RRID: AB_223647 |
| Mouse monoclonal anti-pTau (CP13) | Peter Davies, Albert Einstein College of Medicine | Cat# CP13; RRID: AB_2314223 |
| Mouse monoclonal anti-conformationally abnormal Tau (MC1) | Peter Davies, Albert Einstein College of Medicine | Cat# MC1; RRID: AB_2314773 |
| Mouse monoclonal anti-pTau (PHF1) | Peter Davies, Albert Einstein College of Medicine | Cat# PHF1; RRID: AB_2315150 |
| Mouse monoclonal anti-oligomeric Tau (TOC1) | Nicholas M. Kanaan, Michigan State University | Cat# TOC1; RRID: AB_2832939 |
| Rabbit polyclonal anti-Neurofilament Heavy Chain | Abcam | Cat# ab8135; RRID: AB_306298 |
| Abberior StarRed goat anti-mouse | Abberior | Cat# STRED-1001-500UG |
| Biological samples | ||
| Fetal Brain RNA– Fbrain 1 | Tebu Bio | 1F01-50 |
| Fetal Primary Neurons – FPN 1 | ScienCell | 1520 |
| Fetal Primary Neurons – FPN 2 | ScienCell | 1520 |
| Fetal Primary Neurons – FPN 3 | ScienCell | 1520 |
| iPSC-derived cortical neurons – iPSC-CN 1 | BrainXell | BX-0300 |
| Fetal neuron lysate – FNL 1 | ScienCell | 1526 |
| Adult Brain RNA, Male 29yr - ABrain 1 | BioChain | R1234066-50 |
| Adult Brain RNA, Male 66yr - ABrain 2 | BioChain | R1234066-50 |
| Adult Brain RNA, Female 78yr - ABrain 3 | BioChain | R1234066-50 |
| Adult Brain RNA, Male 82yr - ABrain 4 | NovusBio | NB820-59177 |
| Adult Brain Lysate, Male 90yrs - ABrain 5 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| Adult Brain Lysate, Male 80yrs - ABrain 6 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| Adult Brain Lysate, Male 87yrs - ABrain 7 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| Adult Brain Lysate, Male 72yrs - ABrain 8 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| IVS10+16 Adult Brain RNA, Male 66rs – IVS10+16 ABrain | Queen Square House Brain Bank | P2/08 |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum (FBS), qualified, US Department of Agriculture (USDA)-approved regions | Life Technologies | Cat# 10437028 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Neurobasal-A | Gibco | Cat# 10888022 |
| GlutaMAX | Gibco | Cat# 35050061 |
| B27 Plus Supplement | Gibco | Cat# A3582801 |
| BrainPhys Neuronal Medium | StemCell | Cat# 05790 |
| NeuroCult SM1 Neuronal Supplement | StemCell | Cat# 05711 |
| N2 Supplement-A | StemCell | Cat# 07152 |
| Doxycycline hyclate (Dox) | Sigma-Aldrich | Cat# D9891 |
| Poly-L-ornithine solution | Sigma-Aldrich | Cat# P4957 |
| Laminin | Sigma-Aldrich | Cat# L2020 |
| Fibronectin | Sigma-Aldrich | Cat# F4759 |
| Valproic acid (VPA), sodium salt | Sigma-Aldrich | Cat# 676380 |
| Dibutyryl-cAMP sodium salt | Sigma-Aldrich | Cat# D0627 |
| Retinoic acid (RA) | Sigma-Aldrich | Cat# R2625 |
| Neurotrophin-3 (NT-3) | PeproTech | Cat# 450-03 |
| Brain-derived neurotrophic factor (BDNF) | PeproTech | Cat# 450-02 |
| Glial-derived Neurotrophic Factor (GDNF) | PeproTech | Cat# 450-10 |
| RevitaCell Supplement | Thermo Fisher Scientific | Cat# A2644501 |
| ISX-9 | StemCell Technologies | Cat# 73202 |
| DAPT | Millipore Sigma | Cat# D5942 |
| Critical commercial assays | ||
| Go Taq DNA Polymerase | Promega | Cat# M3001 |
| RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 |
| SMARTer Ultra Low RNA Kit for Illumina sequencing | Clontech | Cat# 635029 |
| TruSeq Small RNA Library Preparation Kit | Illumina | Cat# RS-200-0012 |
| ReliaPrep™ RNA Miniprep Systems | Promega | Cat# Z6011 |
| QIAamp DNA Mini Kit | QIAGEN | Cat# 51304 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat# 28706 |
| Deposited data | ||
| Raw and processed data (Single-cell RNA-sequencing, bulk RNA-sequencing) | NCBI Gene Expression Omnibus | Accession #: GSE201084 |
| Western Blot | Mendeley Data | DOI: 10.17632/t93byrznm6.1 |
| Experimental models: Cell lines | ||
| Lenti-X 293LE cell line | Clontech | Cat# 632180 |
| AG04148, Male 56yrs – Control 1 | Coriell | Cat# AG04148 |
| AG08260, Male 61yrs – Control 2 | Coriell | Cat# AG08260 |
| AG08379, Female 60yrs – Control 3 | Coriell | Cat# AG08379 |
| AG13369, Male 68yrs – Control 4 | Coriell | Cat# AG13369 |
| UCL455, Male 53yrs – IVS10+16 1 | Dr.Henry Houlden, University College London | n/a |
| UCL457, Male 52yrs – IVS10+16 2 | Dr.Henry Houlden, University College London | n/a |
| UCL497, Female 54yrs – IVS10+16 3 | Dr.Henry Houlden, University College London | n/a |
| ES046, Male 57yrs – IVS10+16 4 | Dr. Barbara Corneo, Columbia University | n/a |
| Oligonucleotides | ||
| Primers for qPCR; see Table S3 | This paper | n/a |
| Primers for sqPCR; see Table S3 | This paper | n/a |
| Primers for sequencing; see Table S3 | This paper | n/a |
| Recombinant DNA | ||
| pMD2.G | Didier Trono | 12259 |
| psPAX2 | Didier Trono | 12260 |
| rtTA-N144 | Richner et al., 2015 | 66810 |
| pTight-9/9*-124-Bclxl | Victor et al., 2014 | 60857 |
| MYT1L-N174 | Richner et al., 2015 | 66809 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| LAS X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Harmony v4.9 | Perkin Elmer | https://www.perkinelmer.com/product/harmony-4-8-office-hh17000001 |
| Graphpad Prism 9 | GraphPad Software Inc | http://www.graphpad.com/ |
| FCS Express 7 Research software | De Novo Software | https://denovosoftware.com/ |
| Ingenuity Pathway Analysis | QIAGEN | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ |
| Skyline | MacCoss Lab Software, MacLean et al., 2010 | https://skyline.ms/project/home/software/Skyline/begin.view |
| Cell Ranger (v3.1.0) | 10x Genomics, Zheng et al., 2017 | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat (v3.2.3) | Santija Lab, Satija et al., 2015 | https://satijalab.org/seurat/ |
| Cutadapt v2.4 | Martin, 2011; National Bioinformatics Infrastructure Sweden | https://github.com/marcelm/cutadapt/ |
| DESeq2 | Love, Huber, and Anders, 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| FastQC v0.11.4 | Simon Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Deeptools (v3.5.1) | Ramirez et al., 2014 | http://deeptools.ie-freiburg.mpg.de |
| Wiggletools (v1.2) | Zerbino et al., 2014 | www.github.com/Ensembl/Wiggletools |
| Samtools v1.9 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Picard v2.18 | Broad Institute | https://broadinstitute.github.io/picard/ |
| IGV | Thorvaldsdóttir et al., 2013 | https://software.broadinstitute.org/software/igv/download |
| STAR | Dobin et al., 2015 | https://github.com/alexdobin/STAR |
| LONGO | McCoy et al., 2018 | https://github.com/biohpc/longo |
| Leafcutter | Li et al., 2018 | https://davidaknowles.github.io/leafcutter/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fibroblast cell lines
Healthy healthy human fibroblasts were obtained from Coriell NINDS (AG04148 – Male 56yrs, AG08260 – Male 61yrs, AG08379 – Female 60yrs, and AG13369 – Male 68yrs) while IVS10+16 patient fibroblasts were acquired from Columbia University (ES046 – Male 57yrs) and University College London (UCL455 – Male 53yrs, UCL457 – Male 52yrs, and UCL497 – Female 54yrs). All fibroblasts were maintained in DMEM with 15% FBS for growth. We do not have access to original patient data, and therefore cannot identify the donor. The research falls outside the federal definition under the jurisdiction of an institutional review board and is exempt from human subject studies. Samples were grouped by MAPT genotype.
Adult human brain samples
Three biological replicate lots of adult human brain RNA (R1234066-50) (Lot C.210018 – ABrain1 Male 29yrs, Lot B.210080 – ABrain2 Male 66yrs, Lot B.811107 – ABrain3 Female 78yrs) was purchased from BioChain. Adult human brain protein samples include Novus purchased lysate (NB820-59177) (Lot C.111050 – ABrain4 Male 82yrs) and four previously published healthy healthy samples (Lot 61732 – ABrain5 Male 90yrs, Lot 65241 – ABrain6 Male 80yrs, Lot 65318– ABrain7 Male 87yrs, Lot 84868 – ABrain8 Male 72yrs) obtained from Washington University Alzheimer’s Disease Research Center (ADRC) (Horie et al., 2020). All participants were between A0B1 and A3B1 on the ABC Alzheimer’s Disease lesion scoring system (Kovacs and Gelpi, 2012) (none had clinical symptoms and they were not diagnosed with Alzheimer’s Disease). The study was approved by the Washington University Institutional Review Board, and all participants were purchased from BrainXell (#BX-0300). IVS10+16 human brain (P2/08) from Queen Square House Brain Bank. Samples were grouped by MAPT genotype.
Fetal-aged human brain samples
Fetal human brain RNA (#1F01-50) (Lot 1333 – Fbrain1 Male 21weeks) was purchased from Cell Applications Inc. Fetal human neuron lysate (#1526) (Lot 2219 – FNL1 sex and age not available) was purchased from ScienCell. Primary fetal neurons (#1520) (Lot 29207 – FPN1 sex unknown 19wks post-conception, Lot 28630 – FPN2 Male 22wks post conception, Lot 29390 – FPN3 Male 18wks post-conception) were purchased from ScienCell and were thawed and cultured in Neuronal Medium (ScienCell #1521). Samples were grouped by cellular age.
Purchased iPSC-derived neuron samples
For bulk and single cell RNA-sequencing and sqPCR, differentiated iPSC-derived Cortical Glutamatergic Neurons (Lot 200107 – iPSC-CN1 Female) were purchased from BrainXell (#BX-0300) and cultured according to BrainXell protocol in DMEM with F12 and additional neuronal supplements. Samples were grouped by cellular age.
METHOD DETAILS
Lentiviral Production
Lentiviruses were generated as previously described (Church et al., 2021) with minor changes. To make supernatant virus, viral supernatant was collected, spun at 1200g for 5min at 4°C, then passed through a 0.45uM filter. This supernatant was then aliquoted and directly frozen at −80°C until used (less than 1 year after production date).
Direct Neuronal Reprogramming
Human fibroblasts were directly reprogrammed to miNs as previously described (Church et al., 2021; Richner et al., 2015; Yoo et al., 2011) with slight modification. Briefly, fibroblasts were transduced with supernatant lentivirus mix comprised of dox-inducible miR-9/9*-124, rtTA, and the transcription factor MYT1L. From PID1 to PID14 cells were treated with DAPT (2uM) to increase neurite outgrowth and neuronal differentiation, and on PID 3, 6, 10, and 14, cells were also treated with the NEUROD1-activator ISX9 (10uM) to push cortical fate. Fibroblasts and reprogramming cells were cultured in DMEM+10%FBS through replating at PID5. On PID6, cells were switched to Neurobasal-A with B27+ (1000x) and Glutamax (500x), containing 1 μg/mL doxycycline, 200 μΜ dibutyl cyclic AMP, 1 mM valproic acid, 2uM DAPT, 200nM Ascorbic Acid, 10 ng/mL BDNF, 10 ng/mL NT-3, 1 μM retinoic acid, 10uM ISX9, 100x RVC, and 3 μg/mL puromycin. Cells were half fed every 4 days and doxed every 4 days on an offsetting 2 day scheduled. On PID14, miNs were half fed using BrainPhys containing N2A and SM1 (StemCell) with the following goodies: 1 μg/mL doxycycline, 200 μM dibutyl cyclic AMP, 1 mM valproic acid, 2uM DAPT, 200nM Ascorbic Acid, 10 ng/mL BDNF, 10 ng/mL NT-3, and 1 μM retinoic acid.
iPSC Generation and Genome Engineering
Dermal fibroblasts from MAPT IVS10+16 carriers (GIH36) were transduced with non-integrating Sendai virus carrying OCT3/4, SOX2, KLF4, and cMYC (Life Technologies) as previously described (Karch et al., 2019). iPSC that were heterozygous for MAPT IVS10+16 were edited to WT (GIH36.2Δ1D01) using CRISPR/Cas9 as previously reported (Karch et al., 2019). Mutation status was confirmed by Sanger sequencing. Cell lines were maintained in mTesR medium (StemCell Technologies) on Matrigel. Cell lines were confirmed to be free of mycoplasma.
iPSC Differentiation
MAPT IVS10+16 iPSC (n=1) and isogenic controls (n=1) were differentiated into neural progenitor cells (NPCs) as previously described (Jiang et al., 2018; Karch et al., 2019). Briefly, iPSC were dissociated with Accutase (Life Technologies). iPSCs were then plated at 65,000 cells per well in Neural Induction Media (NIM; Stem Cell Technologies) in a 96-well v-bottom plate to form neural aggregates. After 5 days, neural aggregates were plated on Poly-L-Ornithine (PLO) and laminin-coated plates to form neural rosettes. After 5 to 7 days, neural rosettes were isolated by enzymatic selection and cultured as NPCs. NPCs were cultured on PLO and laminin-coated plates and terminal differentiation was initiated with the addition of cortical maturation medium (Neurobasal-A (Life Technologies) supplemented with 1x B27 (Gibco), 20ng/mL BDNF (Peprotech), 20ng/mL GDNF (Peprotech), 0.5mM cAMP (Sigma) and 1% L-glutamate (Sigma)). Neural cultures were maintained for six weeks.
Immunocytochemistry
Cells are fixed with either 4% PFA for 20min and washed 3x PBS after fixation. Fixed cells were permeabilized for 10min at RT in permeabilization buffer, blocked for 1hr RT in 5% BSA/1% NGS, then stained in primary antibody in blocking buffer overnight 4°C. The next day, cells were washed 3x in PBS and placed in secondary antibody 1:1000 in blocking buffer for 1 hr room temp. Cells were washed 3x in PBS and stained with DAPI for 10min RT, washed once with PBS, then mounted in ProLong Gold antifade Mountant. Primary antibodies used for the immunofluorescence imaging: mouse anti-NCAM (Santa Cruz, SC-106 1:50), rab anti-Tau (Aligent/DAKO, A002401-2 1:200), rab anti-TUBB3 (Biolegend, 802001 1:2000), mouse anti-TUBB3 (Biolegend, 801202 1:2000), chicken anti-TUBB3 (Novus, NB100-1612 1:1000), mouse anti-tau (CP27) (generously provided by Dr. Peter Davies, 1:400), mouse anti-pTau (PHF1) (generously provided by Dr. Peter Davies, 1:1000), mouse anti-pTau (AT8) (Invitrogen, MN1020, 1:1000), mouse anti-pTau (CP13) (generously provided by Dr. Peter Davies, 1:500), mouse anti-Tau (MC1) (generously provided by Dr. Peter Davies, 1:500), mouse anti-Tau (TOC1) (generously provided by Dr. Nicholas Kanaan, 1:500), rabbit anti-MAP2 (Cell Signaling, #4542 1:200), rabbit anti-MAP2 (Millipore, AB5622 1:1000), The secondary antibodies were goat anti-mouse, -rabbit, or -chicked IgG conjugated with Alexa-488, Alexa-594, or Alexa-647 (Invitrogen).
Immunostained images were taken using a Leica SP5X white light laser confocal system with Leica Application Suite (LAS) Advanced Fluorescence 2.7.3.9723. All antibodies were validated for functionality through negative control screening of fibroblasts. Composite images were stitched during acquisition in LAS software.
Quantification of cell fate immunocytochemistry was performed using imageJ multi tool counter. Intact nuclei were used to count total cells, and MAP2 and tau positive cells were defined by the presence of two or more fluorescent-positive neurites whose length is twice the size of the soma. N represents the total number of live cells counted per cell line, as outlined in figure legends. Percentages were calculated in Excel.
Quantitative PCR (qPCR)
Total RNA was extracted from miNs using TRIzol (Invitrogen, USA) following manufacturer protocol. Reverse-transcription was performed with 150-200ng of RNA with Superscript IV First Strand Synthesis SuperMix (Invitrogen, USA). qPCR assay was run with SYBR Green PCR Master Mix and plate was run and analyzed on StepOnePlus Real-Time PCR System (AB Applied Biosystems, Germany). Each sample was run in triplicate and the mean of each sample is represented as a single data point. Expression values were calculated in Excel using the delta Ct method and Z-score calculation.
Single-cell RNAseq sample preparation
Reprogrammed cells, fibroblasts, and cultured iPSC-Ns were collected as previously published (Cates et al., 2020). Briefly, cells were washed once with IxDPBS, 200μL of 0.25% trypsin was added to the wells and plates were placed at 37°C for 5 minutes. Wells were flooded with 500μL warm 10% DMEM. Cells were collected in a 5mL Eppendorf and centrifuged at 300g for 5 minutes at 37°C. Supernatant was removed and the pellet was resuspended in 0.04% BSA in PBS and spun again. Pellet was resuspended in 0.04% BSA in PBS, cells were counted on a hemocytometer, and volume was adjusted to achieve 1,000 cells/μL. All samples were placed on ice and immediately brought to the Genome Technology Access Center at Washington University in Saint Louis (https://gtac.wustl.edu/).
10x
Single-cell RNAseq was performed on the 10x Genomics platform, using the Chromium Single Cell 3’ kits: Library & Gel Bead Kit v2 (PN-120237), Chip kit v2 (PN-120236), and i7 Multiplex Kit (PN-120262), following manufacturer-provided user guide. Agilent Bioanalyzer was used for cDNA library quantification.
Single-cell RNAseq data processing and analysis
The raw 10x reads were processed with the Cell Ranger count pipeline using default parameters (Cell Ranger v3.1.0, 10x Genomics. Reads were aligned to the hg38 reference index provided by 10x Genomics (refdata-cellranger-GRCh38 v3.0.0). Cell barcode and unique molecular identifier (UMI) were extracted and corrected from the feature library using the same methods as gene expression read processing. Feature-barcode matrices were generated by counting distinct UMIs of each gene within a given individual cell (Zheng et al., 2017).
The R package, Seurat (v3.2.3) was used for quality control, analysis, and exploration of scRNA-seq data (Satija et al., 2015). We first removed cells where features less than 200 were detected and mitochondrial counts were high, then filtered features detected in cells less than 10. Seurat was used to remove unwanted variation from the gene expression by regressing out proportion of mitochondrial UMIs and overall UMIs. Highly variable genes were identified and used as input for dimensionality reduction via Principal Component Analysis (PCA). The resulting PCs and the correlated genes were examined to determine the number of components to be included in downstream analysis. These principal components were then used as inputs to cluster individual cells, using a K-nearest neighbor graph and the Louvain algorithm. The resulting cell clusters were visualized and explored using UMAP as a non-linear dimensional reduction technique.
Genotyping
Genomic DNA was extracted from fibroblasts using QIAamp DNA Mini Kit (Qiagen). PCR was performed using Phusion DNA polymerase (NEB) following the published protocol, with amplification primers (see Table S3) at the annealing temperature of 63°C for 35 cycles. PCR product was run on 1% agarose gel to confirm size and product was extracted using QIAquick Gel Extraction Kit (Qiagen). For restriction digest test, 500ng of PCR product was then digested with Nspl (NEB) then run on a 1% gel and imaged. For sequencing, purified PCR product was submitted to GeneWiz with sequencing primer (see Table S3) and resulting tracks were visualized compared to control MAPT sequence.
RNAseq cell collection, sequencing, data processing, and analysis
MiNs were collected on PID21 in triplicate for RNA-sequencing using RNeasy Plus Micro Kit (Qiagen) according to the manufacturer’s instructions. Cortical glutamatergic iPSC-Ns were purchased from BrainXell (BX-0300e), human adult whole brain RNA was purchased from Biochain (R123403-50), and human fetal brain RNA sample was purchased from Cell Applications (1F01-50). Adult human brain and fetal brain RNA samples were run by GTAC through the NovaSeq6000 using SMARTer 150PE with 30 million reads per sample input. MiNs and iPSC-Ns were sequenced by DNALink (www.dnalink.com) on NovaSeq6000 using SMARTer 100PE and 40-50 million reads per sample input.
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used to determine sequencing quality and identify adapter contamination and FastQ files were trimmed using cutadapt for adapter contamination (if needed). Trimmed sequences were mapped to hg38 using STAR (Dobin and Gingeras, 2015). Raw gene counts were extracted using deepTools multiBamSummary option –outRawCounts ((Ramírez et al., 2016). Differential gene expression analysis was performed using DESeq2 normalizing to sequencing depth (Love et al., 2014). Genes defined as differentially expressed had an adjusted p-value < 0.05. Genes were categorized as RNA binding or splicing regulators by Ingenuity Pathway Analysis (QIAGEN).
Leafcutter data processing and analysis
New BAM files were generated by STAR using the --outSAMstrandField intronMotif option were used for Leafcutter (Li et al., 2018), according to the published and recommended protocol (https://davidaknowles.github.io/leafcutter/). Samples were grouped by both cell-type and age and comparisons were performed pairwise, as required for Leafcutter analysis. Differentially spliced clusters and introns were defined by FDR < 0.05, were exported using LeafViz by Jack Humphry (https://github.com/jackhump/leafviz), and loci colors were edited using illustrator (Adobe). Splicing line weights were linearly-scaled so all clusters had similar line weights for easier viewing. Per cluster, cryptic splicing events with less than 5% inclusion were removed.
Semi-quantitative PCR (sq-PCR)
Semi-quantitative PCR was performed to quantify the ratio of 3R:4R mRNA. To perform sqPCR, cDNA was generated from RNA (Superscript IV Reverse Transcriptase, ThermoFisher 18090010) then amplified using primers (Choi et al., 2014a) flanking exon 10 (forward 5′-AAGTCGCCGTCTTCCGCCAAG-3′; reverse 5′-GTCCAGGGACCCAATCTTCGA-3′). The PCR product was then run on a 2% agarose gel with 381bp and 288bp fragments indicating 4R and 3R, respectively. Ratios were then calculated as previously described (Antiabong et al., 2016), via imageJ box plots and measure plots, summed pixel intensity values were exported to Excel, where each isoform value was divided by the summed total to generate percentage of each isoform. N isoform primers for sqPCR: forward 5’-TACGGGTTGGGGGACAGGAAACAT-3’; reverse 5’- GGGGTGTCTCCAATGCCTGCTTCT-3’.
Lysate Preparation for mass spectrometry analysis
MiNs were collected on PID 30. iPSC-Ns were collected after six weeks. Both were collected in the same manner. Briefly, media was removed and cells were washed once with DPBS. Fresh DPBS was added to the wells and cells were scraped off. Resuspended cells were pelleted at 1000g for 5min at RT, supernatant was removed, and pellets were frozen at −80°C until use.
Three different lots of Human Primary Neurons were purchased from ScienCell and plated according to manufacturer’s instructions. Cells were cultured in Neuronal Medium (ScienCell #1521) for one week and collected same as the miNs and iPSC-Ns.
Adult brain frozen tissues was sliced via a cryostat at −20°C, from which 300-400mg was sonicated in 4°C buffer containing 25mM tris-hydrochloride (pH 7.4), 150 mM sodium chloride, 10 mM ethylenediaminetetraacetic acid, 10 mM ethylene glycol tetraacedic acid, phosphatase inhibitor cocktail, and protease inhibitor cocktail, with final brain suspension at 0.3 mg/μL buffer. Suspension was cleared using centrifugation for 20 minutes at 11,000 g at4°C and the resulting supernatant was defined as “brain homogenate” (Horie et al., 2020).
Immunoprecipitation and mass spectrometry of tau isoforms
Mass spectrometry analyses of tau proteins were performed as previously described with some modifications (Barthélemy et al., 2020a; Barthélemy et al., 2020b; Horie et al., 2020; Sato et al., 2018). Cells and brain homogenates were diluted with PBS and 2% human serum albumin (Sigma D4197) respectively, and lysed with final concentration of 0.5% NP-40 and 2.5mM guanidine. Tau protein was immunoprecipitated with mouse anti-Tau (Tau1) (provided by Dr. Nicholas Kanaan, 1.125μg/sample), and mouse anti-human tau (HJ8.5) (provided by Dr. David Holtzman, 2.25μg/sample), digested with trypsin, oxidized, desalted and subjected to nano-Acquity LC and MS analyses using Orbitrap Eclipse Tribrid Mass Spectrometer (Thermo Scientific). Mass spectrometry data were extracted using Skyline software (Maclean et al., 2010).
All samples were spiked with full length 15N-labeled 2N4R recombinant tau as an internal standard and ratios of isoforms were calculated by using 2N and 4R isoform-specific peptides compared to constitutive peptides (all peptide numbers are in reference to 2N4R isoform). For 2N, the 2N-specific tryptic peptide 68-87 was divided by the common peptide 151-155 to obtain percentage of 2N. 1N isoform percentage was calculated by dividing a shared 1N/2N peptide, 45-67, by the common peptide 151-155, to obtain the cumulative percentage of 1N+2N isoforms. The previously calculated 2N percentage was subtracted from this shared 1N/2N percentage to calculate the 1N percentage. 0N percentage was calculated by subtracting both 1N and 2N percentages from 100. Percent 4R was calculated three ways by dividing each of the R2 region, 4R-specific peptides, 275-280, 282-290, and 299-317, by the constitutive adjacent R1 peptide, 260-267. 3R was calculated by subtracting the 4R percentage from 100.
Immunoblot analysis
Media was aspirated from wells, cells were washed with DPBS, and lysed in the plate with RIPA (Sigma #R0278) supplemented with cOmplete Mini Protease Inhibitor Cocktail (Roche #11836153001). Lysate was collected and centrifuged at 13,000g for 10 minutes at 4°C. BCA protein assay determined protein content and samples were normalized with RIPA/protease inhibitor. Samples were treated with Lambda Phosphatase (NEB #P0753L) for 3 hours at 30°C. LDS NuPAGE sample buffer with 5% β-mercaptoethanol was added and samples were heated to 95°C for 3 minutes then loaded onto a NuPAGE 8% 20-well Midi gel (Invitrogen WG1002), with Tau Peptide Ladder (rPeptideT-1007–2). After running, the gel was transferred onto either 0.45um PVDF membrane for 2hr at 400mA. Membrane was both blocked and treated with primary antibodies in TBS/0.1% Tween20 overnight at 4°C: rabbit anti-Tau antibody (Aligent/DAKO, A002401-2 1:1000-10,000). The next day, membrane was washed 3 times with TBST, then treated with peroxidase-conjugated goat anti-rabbit secondary antibodies in 5% Milk/TBS/0.1% Tween 20 for up to 3hrs at RT. Membrane was washed 3 times with TBS/0.1% Tween20 and developed with the ECL system (Thermo Scientific, #34076) and imaged on a Sapphire Imager (Azure Biosystems).
Biosensor cell culture methods
MiNs were harvested on PID 26 in 50 mM Tris-HCl, pH 7.5 150 mM NaCl. The lysates were sonicated for 4.5 minutes at 50% amplitude (QSonica, Q800R3 Sonicator). BCA assay was used determine protein concentration of protein. HEK biosensor Tau RD-CFP/YFP cell line, kindly provided by Dr. Marc Diamond (Furman et al., 2015), were cultured in 10 cm dish in DMEM (+Pyruvate, + D-glucose/D-glutamine) (Thermofisher) with 10%FBS and 1% Pen-Strep. The day before the seeding experiment, cells were replated in a 96-well plate at a density of 40,000 cells/well in 130uL of media and left adhere over-night. The following day, cells were seeded with miN lysates; the seeding mixes were made by combining 17μg of total protein lysate per well to 3.75μL of Opti-MEM (Gibco) and 1.25uL Lipofectamine 2000 (Invitrogen) for a total volume of 20 μL per well. Liposome preparations were incubated at room temperature for 30 min before adding to cells, and each condition was done in triplicate. Cells were incubated with seeding mixes for 72 h before the analysis.
FRET Flow Cytometry
After 72 h from the seeding, cells were washed in PBS (Gibco), harvested with 0.25% trypsin and fixed in 4% paraformaldehyde for 10 min, then resuspended in flow cytometry buffer (1 mM EDTA in PBS). The BD LSRFortessa™ Flow Cytometer was used to perform FRET flow cytometry as previously described (Furman et al., 2015). Briefly, to measure CFP and FRET, cells were excited with the 405 nm laser, and to measure YFP cells were excited with a 488 laser. Fluorescence was captured with a 405/50 nm, 525/50 nm and 525/50 nm filters respectively.
To quantify FRET, a gating strategy similar to that previously described was used (Furman et al., 2015): first the CFP bleed-through into the YFP and FRET channels was compensated, and cells were gated in order to exclude YFP-positive only cells emitting in the FRET signal. A bivariate plot of FRET vs. CFP was made to assess the number of FRET-positive cells. The percentage of FRET (i.e., the number of FRET-positive cells per total cell count) and the Integrated FRET density (i.e. product of percent positivity and median fluorescence intensity) were used for the analyses. Data analysis was performed using FCS Express 7 Research software (De Novo Software).
Immunocytochemistry, imaging and analysis
Methanol fixed cells in DPBS had the liquid tipped off, and the cells were blocked with 200 μL/well of Intercept Blocking Buffer (LI-COR) containing 0.1% Triton for one hour at room temperature. The primary antibodies (see STAR methods) were prepared in Blocking Buffer and incubated overnight at 4°C with gentle agitation.
On the following day, the primary antibody was tipped off, and the cells were washed three times with 200 μL/well of DPBS. The secondary antibodies Goat anti-mouse IgG2b 647 Alexa Flour-conjugated antibody (A-21242) and Goat anti-rabbit 488 Alexa Flour-conjugated antibody (A-11008), as well as Hoechst 33342 (Invitrogen) were prepared at 1:1000 in Blocking Buffer and left for 1 hour at room temperature.
The plates were then washed three times with 200 μL/well of DPBS, before being left in DPBS at 4°C until high-content imaging using the Opera Phenix (Perkin Elmer). Images were taken using a 20X water objective, with 75 fields of view per well, and a Z-stack of 6 slices. The images were analysed using Perkin Elmer software Harmony as previously described (Katsikoudi et al., 2020). Briefly, stacks of each image were maximum projected and filtered using the sliding parabola to smoothen the background and get rid of the fluorescent noise. Subsequently, machine learning was used to define the tau positive area used as region in which the software was trained to identify the tau positive threads (for TOC1, only cytoplasmic signal was included in the analysis). Total nuclei were also detected and analyzed. At the end of the analysis results were exported in an Excel file divided by object (total nuclei and tau count) and well. The final readout was “Tau count normalized to the total nuclei count”. Any well with fewer than 800 nuclei was not included in analysis. n=total number of nuclei per line. Data was plotted in GraphPad.
STED Microscopy
STED microscopy was performed on a Leica STELLARIS 8 STED. Cells were methanol fixed as previously described. Primary antibodies were used at 1:1000. Secondary antibody was purchased from Abberior and used at 1:250.
SiRNA treatment of miNs
MiNs were replated, as per the above protocol, at PID 5 on 24 well Sensoplate (Greiner, 662892) previously coated with poly-ornithine, laminin, and fibronectin.
At PID 18, cells were treated with 1uM of Dharmacon™ Accell™siRNAs: Accell Human MAPT (4137) siRNA-SMARTpool (E-012488-00-0050) and Accell Non-targeting Pool (D-001910-10-05). Treatment was left on until PID 26 when cells were fixed with 100% methanol to extract soluble proteins, as described previously (Guo et al., 2016; Katsikoudi et al., 2020). In brief, the growth media was completely aspirated, and cells were washed twice with 200 μL/well of DPBS. Ice-cold 100% methanol was added 200 μL/well for 15 minutes at room temperature. Subsequently, the cells were washed three times with 200 μL/well of DPBS, with the final wash being left on for storage. Plates were kept at 4C° until immunocytochemistry (ICC) was performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Two-tailed Student’s t-test were performed for datasets containing two groups. ANOVA analyses were used for datasets with more than two groups. Specific statistical details and n information for each assay can be found in the corresponding figure legends and methods sections. All data is presented as mean±SEM. Differences were considered statistical at *p<0.05, **p<0.01, ***p<0.001.
Supplementary Material
Figure S1, related to Figure 1 – Single cell RNAseq subtyping
Figure S2, related to Figure 2 – Age-associated splicing events in miNs and 4R expression independent of miRNA over-expression
Figure S3, related to Figure 4 – Confirmation of mutational status in healthy and IVS10+16 fibroblasts
Figure S4, related to Figure 6 – siRNA against total tau shows reduction in both mRNA and protein levels
Table S1, related to Figure 2 – Differential age-associated splicing events overlapping between adult and fetal and miNs and iPSC-Ns
Table S2, related to Figure 2 – Age-associated expression of RNA binding proteins
Table S3, related to STAR methods – Oligonucleotide Primers.
Highlights.
Recapitulation of all adult human tau isoforms in directly reprogrammed neurons
miNs capture 4R increase caused by familial IVS10+16 splice site mutation
Patient-derived miNs endogenously generate seed-competent and insoluble tau
Reduction of tau decreases pathogenic tau species
ACKNOWLEDGEMENTS
The authors thank Edward D. Huey and Barbara Corneo (Columbia University) for providing one IVS10+16 fibroblast line, the Genome Technology Access Center and the fund from the Institute of Clinical and Translational Sciences (ICTS) (UL1TR002345) for their assistance in generating transcriptome dataset and iPSC lines, Mariana Beltcheva for iPSC to neuron differentiation, Kuo-Chan Weng for NPC expansion, Kitra Cates and Dr. Seongwon Lee for qPCR primers, Dr. Selina Wray for control and IVS10+16 iPSC-N lysate, Dr. Victoria Church for human brain miRNA, and Dr. Chloe He and Faris Shaikh for assisting with IP/MS experiments. This work was supported the following grants and awards. MCB training grant (T32 GM007067) (L.S.C.), ICTS JIT (JIT659H) (L.S.C and A.S.Y.), Farrell Family Fund for Alzheimer’s Disease (A.S.Y, C.M.K., and D.M.H), Cure Alzheimer’s Fund (A.S.Y.), NIH/NIA RF1AG056296 (A.S.Y.), NIH/NINDS R01NS107488 (A.S.Y.), Mallinckrodt Scholar Award (A.S.Y.), NIH/NIA K01AG062796 (C.S.), SILQ Center (R.J.B.), NIH/NINDS R01 NS095773 (R.J.B.), NIH/NIA AG063521 (K.E.D and E.F) and the UK DRI funds from DRI Ltd, from the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK (K.E.D and E.F), MRC and Wellcome Trust (H.H.), NIH R56 NS110890 (C.M.K.), Rainwater Charitable Organization (C.M.K.). ADRC Resources provided by Washington University Neuropathology core was supported by Healthy Aging and Senile Dementia [P01 AG03991], Alzheimer’s Disease Research Center [P30 AG066444], Healthy Aging and Senile Dementia [P01 AG03991], Alzheimer Disease Research Center [P50 AG05691], and the Adult Children Study [P01 AG026276].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
D.M.H. is as an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. C2N Diagnostics has licensed certain anti-tau antibodies to AbbVie for therapeutic development. D.M.H. is on the scientific advisory board of Denali, Genentech, and Cajal Neurosciences and consults for Genentech, Takeda, Casma, and Eli Lilly. A.S.Y. consults for Roche. K.H. is an Eisai-sponsored visiting researcher at Washington University and has received salary from Eisai. K.E.D is a board member and advisor for Ceracuity LLC. All other authors declare no competing interests.
REFERENCES
- Abernathy DG, Kim WK, McCoy MJ, Lake AM, Ouwenga R, Lee SW, Xing X, Li D, Lee HJ, Heuckeroth RO, et al. (2017). MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell 21, 332–348.e339. 10.1016/j.stem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy DG, and Yoo AS (2015). MicroRNA-dependent genetic networks during neural development. Cell Tissue Res. 359, 179–185. 10.1007/s00441-014-1899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiabong JF, Ngoepe MG, and Abechi AS (2016). Semi-quantitative digital analysis of polymerase chain reaction-electrophoresis gel: Potential applications in low-income veterinary laboratories. Vet World 9, 935–939. 10.14202/vetworld.2016.935-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélemy NR, Bateman RJ, Hirtz C, Marin P, Becher F, Sato C, Gabelle A, and Lehmann S (2020a). Cerebrospinal fluid phospho-tau T217 outperforms T181 as a biomarker for the differential diagnosis of Alzheimer’s disease and PET amyloid-positive patient identification. Alzheimer’s Research & Therapy 12. 10.1186/s13195-020-00596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélemy NR, Li Y, Joseph-Mathurin N, Gordon BA, Hassenstab J, Benzinger TLS, Buckles V, Fagan AM, Perrin RJ, Goate AM, et al. (2020b). A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nature Medicine 26, 398–407. 10.1038/s41591-020-0781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers JE, Lai MC, Collins E, Booth HDE, Zambon F, Parkkinen L, Vowles J, Cowley SA, Wade-Martins R, and Caffrey TM (2017). MAPT Genetic Variation and Neuronal Maturity Alter Isoform Expression Affecting Axonal Transport in iPSC-Derived Dopamine Neurons. Stem Cell Reports 9, 587–599. 10.1016/j.stemcr.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, and Del Tredici K (2010). Neurofibrillary Tangles. In Encyclopedia of Movement Disorders, Kompoliti K, and Metman LV, eds. (Academic Press; ), pp. 265–269. 10.1016/B978-0-12-374105-9.00269-0. [DOI] [Google Scholar]
- Cates K, Mccoy MJ, Kwon J-S, Liu Y, Abernathy DG, Zhang B, Liu S, Gontarz P, Kim WK, Chen S, et al. (2020). Deconstructing Stepwise Fate Conversion of Human Fibroblasts to Neurons by MicroRNAs. Cell Stem Cell. 10.1016/j.stem.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs JP, Gattoni R, and Stévenin J (1994). Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. The EMBO Journal 13, 2639–2649. 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, et al. (2014a). A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278. 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, et al. (2014b). A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278. 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church VA, Cates K, Capano L, Aryal S, Kim WK, and Yoo AS (2021). Generation of Human Neurons by microRNA-Mediated Direct Conversion of Dermal Fibroblasts. In (Springer US; ), pp. 77–100. 10.1007/978-1-0716-1084-8_6. [DOI] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, et al. (2009). Transmission and spreading of tauopathy in transgenic mouse brain. Nature Cell Biology 11, 909–913. 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell JW, Rodriguez-Martin T, Gibb GM, Kahn NM, Grierson AJ, Hanger DP, Revesz T, Lantos PL, Anderton BH, and Gallo JM (2005). Quantitative analysis of tau isoform transcripts in sporadic tauopathies. Molecular Brain Research 137, 104–109. 10.1016/j.molbrainres.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Ding S, Shi J, Qian W, Iqbal K, Grundke-Iqbal I, Gong C-X, and Liu F (2012). Regulation of alternative splicing of tau exon 10 by 9G8 and Dyrk1A. Neurobiology of Aging 33, 1389–1399. 10.1016/j.neurobiolaging.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, and Gingeras TR (2015). Mapping RNA-seq Reads with STAR. Current Protocols in Bioinformatics 51. 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docagne F, Nicole O, Marti HH, Mackenzie ET, Buisson A, and Vivien D (1999). Transforming growth factor-βI as a regulator of the serpins/t-PA axis in cerebral ischemia. The FASEB Journal 13, 1315–1324. 10.1096/fasebj.13.11.1315. [DOI] [PubMed] [Google Scholar]
- Duff K, Knight H, Refolo LM, Sanders S, Yu X, Picciano M, Malester B, Hutton M, Adamson J, Goedert M, et al. (2000). Characterization of Pathology in Transgenic Mice Over-Expressing Human Genomic and cDNA Tau Transgenes. Neurobiology of Disease 7, 87–98. 10.1006/nbdi.1999.0279. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210. 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Hallmann A-L, Reinhardt P, Araúzo-Bravo J, Marcos, Korr S, Röpke A, Psathaki E, Olympia, Ehling P, Meuth G, Sven, Oblak L, Adrian, et al. (2015). Distinct Neurodegenerative Changes in an Induced Pluripotent Stem Cell Model of Frontotemporal Dementia Linked to Mutant TAU Protein. Stem Cell Reports 5, 83–96. 10.1016/j.stemcr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, and Diamond MI (2009). Propagation of Tau Misfolding from the Outside to the Inside of a Cell. Journal of Biological Chemistry 284, 12845–12852. 10.1074/jbc.m808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman JL, Holmes BB, and Diamond MI (2015). Sensitive Detection of Proteopathic Seeding Activity with FRET Flow Cytometry. Journal of Visualized Experiments. 10.3791/53205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, Hemberg M, Ebert DH, and Greenberg ME (2015). Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93. 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, and Behl C (2009). Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. The EMBO Journal 28, 889–901. 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-L, Wang N, Sun F-R, Cao X-P, Zhang W, and Yu J-T (2018). Tau in neurodegenerative disease. Ann Transl Med 6. 10.21037/atm.2018.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B, Oblak AL, Boeve BF, Johnson KA, Dickerson BC, and Goedert M (2015). Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging: MAPT mutations and FTD. Neuropathology and Applied Neurobiology 41, 24–46. 10.1111/nan.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, and Jakes R (1990a). Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. The EMBO Journal 9, 4225–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, and Jakes R (1990b). Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. The EMBO Journal 9, 4225–4230. 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Jakes R, Rutherford D, and Crowther RA (1989a). Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3, 519–526. 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Potier MC, Ulrich J, and Crowther RA (1989b). Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. The EMBO Journal 8, 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz J, Halliday G, and Nisbet RM (2019). Molecular Pathogenesis of the Tauopathies. Annual Review of Pathology: Mechanisms of Disease 14, 239–261. 10.1146/annurev-pathmechdis-012418-012936. [DOI] [PubMed] [Google Scholar]
- Grover A, Houlden H, Baker M, Adamson J, Lewis J, Prihar G, Pickering-Brown S, Duff K, and Hutton M (1999). 5’ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. The Journal of Biological Chemistry 214, 15134–15143. [DOI] [PubMed] [Google Scholar]
- Guo JL, and Lee VM-Y (2011). Seeding of Normal Tau by Pathological Tau Conformers Drives Pathogenesis of Alzheimer-like Tangles. Journal of Biological Chemistry 286, 15317–15331. 10.1074/jbc.m110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Narasimhan S, Changolkar L, He Z, Stieber A, Zhang B, Gathagan RJ, Iba M, McBride JD, Trojanowski JQ, and Lee VMY (2016). Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. Journal of Experimental Medicine 213, 2635–2654. 10.1084/jem.20160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T-Y, Choi YR, Noh HR, Cha S-H, Kim J-B, and Park SM (2021). Age-related increase in caveolin-1 expression facilitates cell-to-cell transmission of α-synuclein in neurons. Molecular Brain 14. 10.1186/s13041-021-00834-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Mcbride JD, Xu H, Changolkar L, Kim S-J, Zhang B, Narasimhan S, Gibbons GS, Guo JL, Kozak M, et al. (2020). Transmission of tauopathy strains is independent of their isoform composition. Nature Communications 11. 10.1038/s41467-019-13787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefti MM, Farrell K, Kim S, Bowles KR, Fowkes ME, Raj T, and Crary JF (2018). High-resolution temporal and regional mapping of MAPT expression and splicing in human brain development. PLOS ONE 13, e0195771. 10.1371/journal.pone.0195771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmler A, Drechsel D, Kirschner MW, and Martin DW (1989). Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Molecular and Cellular Biology 9, 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, and Diamond MI (2014). Proteopathic tau seeding predicts tauopathy in vivo. Proceedings of the National Academy of Sciences of the United States of America 111, E4376–E4385. 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Barthélemy NR, Mallipeddi N, Li Y, Franklin EE, Perrin RJ, Bateman RJ, and Sato C (2020). Regional correlation of biochemical measures of amyloid and tau phosphorylation in the brain. Acta Neuropathologica Communications 8. 10.1186/s40478-020-01019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh CJ, Zhang B, Victor MB, Dahiya S, Batista LF, Horvath S, and Yoo AS (2016). Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. eLife 5. 10.7554/eLife.18648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. (1998). Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705. 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- Ishigaki S, Fujioka Y, Okada Y, Riku Y, Udagawa T, Honda D, Yokoi S, Endo K, Ikenaka K, Takagi S, et al. (2017). Altered Tau Isoform Ratio Caused by Loss of FUS and SFPQ Function Leads to FTLD-like Phenotypes. Cell Reports 18, 1118–1131. 10.1016/j.celrep.2017.01.013. [DOI] [PubMed] [Google Scholar]
- Jiang S, Wen N, Li Z, Dube U, Del Aguila J, Budde J, Martinez R, Hsu S, Fernandez MV, Cairns NJ, et al. (2018). Integrative system biology analyses of CRISPR-edited iPSC-derived neurons and human brains reveal deficiencies of presynaptic signaling in FTLD and PSP. Transl Psychiatry 8. 10.1038/s41398-018-0319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M-J, Chung YH, Hwang C-I, Murata M, Fujimoto T, Mook-Jung I-H, Cha CI, and Park W-Y. (2006). Caveolin-1 upregulation in senescent neurons alters amyloid precursor protein processing. Experimental & Molecular Medicine 38, 126–133. 10.1038/emm.2006.16 [DOI] [PubMed] [Google Scholar]
- Karch CM, Kao AW, Karydas A, Onanuga K, Martinez R, Argouarch A, Wang C, Huang C, Sohn PD, Bowles KR, et al. (2019). A Comprehensive Resource for Induced Pluripotent Stem Cells from Patients with Primary Tauopathies. Stem Cell Reports 13, 939–955. 10.1016/j.stemcr.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai N, Fukushima K, Ueki Y, Prasad S, Nosakowski J, Sugata KI, Sugata A, Nishizaki K, Meyer NC, and Smith RJH (2001). Genomic structures of SCN2A and SCN3A – candidate genes for deafness at the DFNA16 locus. Gene 264, 113–122. 10.1016/s0378-1119(00)00594-1. [DOI] [PubMed] [Google Scholar]
- Katsikoudi A, Ficulle E, Cavallini A, Sharman G, Guyot A, Zagnoni M, Eastwood BJ, Hutton M, and Bose S (2020). Quantitative propagation of assembled human Tau from Alzheimer’s disease brain in microfluidic neuronal cultures. Journal of Biological Chemistry 295, 13079–13093. 10.1074/jbc.ra120.013325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Yandava CN, Mabb AM, Hsiao JS, Huang H-S, Pearson BL, Calabrese JM, Starmer J, Parker JS, Magnuson T, et al. (2013). Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62. 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS, Orecchio LD, Bakalis S, and Neve RL (1989). Developmentally regulated expression of specific tau sequences. Neuron 2, 1389–1397. 10.1016/0896-6273(89)90077-9 [DOI] [PubMed] [Google Scholar]
- Kwak SS, Washicosky KJ, Brand E, Von Maydell D, Aronson J, Kim S, Capen DE, Cetinbas M, Sadreyev R, Ning S, et al. (2020). Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer’s disease. Nature Communications 11. 10.1038/s41467-020-15120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez J-M, De Vos J, et al. (2011). Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes & Development 25, 2248–2253. 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Brizzee C, and Johnson GVW (2015). BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiology of Aging 36, 241–248. 10.1016/j.neurobiolaging.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zuo X, Jing J, Ma Y, Wang J, Liu D, Zhu J, Du X, Xiong L, Du Y, et al. (2015). Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 17, 195–203. 10.1016/j.stem.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, and Pritchard JK (2018). Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158. 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, and Johnson JM (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, and Duff K (2012). Trans-Synaptic Spread of Tau Pathology In Vivo. PLOS ONE 7, e31302. 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-L, and Yoo AS (2018). Mechanistic Insights Into MicroRNA-Induced Neuronal Reprogramming of Human Adult Fibroblasts. Frontiers in Neuroscience 12, 522. 10.3389/fnins.2018.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and Maccoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahali S, and Karch C (2021). Defective proteostasis in patient-derived iPSC-astrocytes and neurons carrying a MAPT IVS10+ 16 mutation. Alzheimer’s & Dementia 17, e058727. [Google Scholar]
- Marone M, Mozzetti S, De Ritis D, Pierelli L, and Scambia G (2001). Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 3, 19–25. 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen YP, et al. (2013). Widespread splicing changes in human brain development and aging. Molecular Systems Biology 9, 633. 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MJ, Paul AJ, Victor MB, Richner M, Gabel HW, Gong H, Yoo AS, and Ahn T-H (2018). LONGO: an R package for interactive gene length dependent analysis for neuronal identity. Bioinformatics 34, i422–i428. 10.1093/bioinformatics/bty243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Ali C, Parcq J, Vivien D, and Docagne F (2016). The plasminogen activation system in neuroinflammation. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1862, 395–402. 10.1016/j.bbadis.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Merry DE, Veis DJ, Hickey WF, and Korsmeyer SJ (1994). bcl-2 protein expression is widespread in the developing nervous system and retained in the adult PNS. Development 120, 301–311. [DOI] [PubMed] [Google Scholar]
- Mertens J, Herdy JR, Traxler L, Schafer ST, Schlachetzki JCM, Böhnke L, Reid DA, Lee H, Zangwill D, Fernandes DP, et al. (2021). Age-dependent instability of mature neuronal fate in induced neurons from Alzheimer’s patients. Cell Stem Cell. 10.1016/j.stem.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. (2015). Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 17, 705–718. 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Chan DN, Ha I, Case D, Cui Y, Van Handel B, Mikkola HK, and Lowry WE (2012). Defining the nature of human pluripotent stem cell progeny. Cell Res. 22, 178–193. 10.1038/cr.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research 44, W160–W165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner M, Victor MB, Liu Y, Abernathy D, and Yoo AS (2015). MicroRNA-based conversion of human fibroblasts into striatal medium spiny neurons. Nat Protoc 10, 1543–1555. 10.1038/nprot.2015.102. [DOI] [PubMed] [Google Scholar]
- Safety, Tolerability and Pharmacokinetics of Multiple Ascending Doses of NIO752 in Progressive Supranuclear Palsy. https://ClinicalTrials.gov/show/NCT04539041.
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502. 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Barthélemy NR, Mawuenyega KG, Patterson BW, Gordon BA, Jockel-Balsarotti J, Sullivan M, Crisp MJ, Kasten T, Kirmess KM, et al. (2018). Tau Kinetics in Neurons and the Human Central Nervous System. Neuron 97, 1284–1298.e1287. 10.1016/j.neuron.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siahaan V, Krattenmacher J, Hyman AA, Diez S, Hernández-Vega A, Lansky Z, and Braun M (2019). Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nature Cell Biology 21, 1086–1092. 10.1038/s41556-019-0374-6. [DOI] [PubMed] [Google Scholar]
- Sposito T, Preza E, Mahoney CJ, Setó-Salvia N, Ryan NS, Morris HR, Arber C, Devine MJ, Houlden H, Warner TT, et al. (2015). Developmental regulation of tau splicing is disrupted in stem cell-derived neurons from frontotemporal dementia patients with the 10 + 16 splice-site mutation in MAPT. Human Molecular Genetics 24, 5260–5269. 10.1093/hmg/ddv246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Okaty BW, Arnson HA, Kato S, Dani VS, and Nelson SB (2014). Cell-Type-Specific Repression by Methyl-CpG-Binding Protein 2 Is Biased toward Long Genes. Journal of Neuroscience 34, 12877–12883. 10.1523/jneurosci.2674-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R, Lam AJ, Tan T, Han J, Nowakowski DW, Vershinin M, Simó S, Ori-McKenney KM, and McKenney RJ (2019). Microtubules gate tau condensation to spatially regulate microtubule functions. Nature Cell Biology 21, 1078–1085. 10.1038/s41556-019-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S-L, Ohtsuka T, González A, and Kageyama R (2012). MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes to Cells 17, 952–961. 10.1111/gtc.12009. [DOI] [PubMed] [Google Scholar]
- Tanner MK, Tang Z, and Thornton CA (2021). Targeted splice sequencing reveals RNA toxicity and therapeutic response in myotonic dystrophy. Nucleic Acids Research 49, 2240–2254. 10.1093/nar/gkab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CH, Ben-Shalom R, Bender KJ, and George AL (2020). Alternative splicing potentiates dysfunction of early-onset epileptic encephalopathy SCN2A variants. Journal of General Physiology 152. 10.1085/jgp.201912442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen A, Diels A, Dijkmans J, Oyelami T, Meneghello G, Mertens L, Versweyveld S, Borgers M, Buist A, Peeters P, and Cik M (2015). Using Human iPSC-Derived Neurons to Model TAU Aggregation. PLOS ONE 10, e0146127. 10.1371/journal.pone.0146127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen A, Diels A, Reumers J, Van Hoorde K, Van den Wyngaert I, van Outryve d’Ydewalle C, De Bondt A, Kuijlaars J, De Muynck L, De Hoogt R, et al. (2018). Genetically Engineered iPSC-Derived FTDP-17 MAPT Neurons Display Mutation-Specific Neurodegenerative and Neurodevelopmental Phenotypes. Stem Cell Reports 11, 363–379. 10.1016/j.stemcr.2018.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng P-Y, Klyachko VA, Nerbonne JM, and Yoo AS (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84, 311–323. 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, Huh CJ, Zhang B, Davidson BL, Yang XW, and Yoo AS (2018). Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nature Neuroscience 21, 341–352. 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao Q-S, Wang Y, Lafyatis R, Stamm S, and Andreadis A (2004). Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. Journal of Neurochemistry 88, 1078–1090. 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, and Crabtree GR (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646. 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, and Crabtree GR (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, and Shi Y (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nature Structural & Molecular Biology 16, 365–371. 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nature Communications 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1 – Single cell RNAseq subtyping
Figure S2, related to Figure 2 – Age-associated splicing events in miNs and 4R expression independent of miRNA over-expression
Figure S3, related to Figure 4 – Confirmation of mutational status in healthy and IVS10+16 fibroblasts
Figure S4, related to Figure 6 – siRNA against total tau shows reduction in both mRNA and protein levels
Table S1, related to Figure 2 – Differential age-associated splicing events overlapping between adult and fetal and miNs and iPSC-Ns
Table S2, related to Figure 2 – Age-associated expression of RNA binding proteins
Table S3, related to STAR methods – Oligonucleotide Primers.
Data Availability Statement
Single-cell and bulk RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper available from the Lead Contact upon request
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-MAP2 | Cell Signaling Technology | Cat# 4542S; RRID: AB_ 10693782 |
| Rabbit polyclonal anti-MAP2 | Millipore | Cat# AB5622; RRID: AB_91939 |
| Rabbit polyclonal anti-Tau | Aligent/Dako | Cat# 002401-2; RRID: AB_10013724 |
| Mouse monoclonal anti-TUBB3 | BioLegend | Cat# 801202; RRID: AB_ 10063408 |
| Rabbit polyclonal anti-TUBB3 | BioLegend | Cat# 802001, RRID: AB_2564645 |
| Chicken polyclonal anti-TUBB3 | Novus | Cat# NB100-1612, RRID: AB_10000548 |
| Mouse monoclonal anti-Tau (CP-27) | Peter Davies, Albert Einstein College of Medicine | Cat# CP-27; RRID: AB_2716722 |
| Mouse monoclonal anti-Tau (Tau1) | Nicholas M. Kanaan, Michigan State University | Cat# Tau1; RRID: AB_2721197 |
| Mouse monoclonal anti-Tau (HJ8.5) | David Holtzman, Washington University in St. Louis | Cat# HJ8.5; RRID: AB_2721237 |
| Mouse monoclonal anti-NCAM1 | Santa Cruz Biotechnology | Cat#SC-106; RRID: AB_627128 |
| Mouse monoclonal anti-pTau (AT8) | Thermo Fisher Scientific | Cat# MN1020; RRID: AB_223647 |
| Mouse monoclonal anti-pTau (CP13) | Peter Davies, Albert Einstein College of Medicine | Cat# CP13; RRID: AB_2314223 |
| Mouse monoclonal anti-conformationally abnormal Tau (MC1) | Peter Davies, Albert Einstein College of Medicine | Cat# MC1; RRID: AB_2314773 |
| Mouse monoclonal anti-pTau (PHF1) | Peter Davies, Albert Einstein College of Medicine | Cat# PHF1; RRID: AB_2315150 |
| Mouse monoclonal anti-oligomeric Tau (TOC1) | Nicholas M. Kanaan, Michigan State University | Cat# TOC1; RRID: AB_2832939 |
| Rabbit polyclonal anti-Neurofilament Heavy Chain | Abcam | Cat# ab8135; RRID: AB_306298 |
| Abberior StarRed goat anti-mouse | Abberior | Cat# STRED-1001-500UG |
| Biological samples | ||
| Fetal Brain RNA– Fbrain 1 | Tebu Bio | 1F01-50 |
| Fetal Primary Neurons – FPN 1 | ScienCell | 1520 |
| Fetal Primary Neurons – FPN 2 | ScienCell | 1520 |
| Fetal Primary Neurons – FPN 3 | ScienCell | 1520 |
| iPSC-derived cortical neurons – iPSC-CN 1 | BrainXell | BX-0300 |
| Fetal neuron lysate – FNL 1 | ScienCell | 1526 |
| Adult Brain RNA, Male 29yr - ABrain 1 | BioChain | R1234066-50 |
| Adult Brain RNA, Male 66yr - ABrain 2 | BioChain | R1234066-50 |
| Adult Brain RNA, Female 78yr - ABrain 3 | BioChain | R1234066-50 |
| Adult Brain RNA, Male 82yr - ABrain 4 | NovusBio | NB820-59177 |
| Adult Brain Lysate, Male 90yrs - ABrain 5 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| Adult Brain Lysate, Male 80yrs - ABrain 6 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| Adult Brain Lysate, Male 87yrs - ABrain 7 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| Adult Brain Lysate, Male 72yrs - ABrain 8 | Dr. Chihiro Sato, Washington University in Saint Louis | n/a |
| IVS10+16 Adult Brain RNA, Male 66rs – IVS10+16 ABrain | Queen Square House Brain Bank | P2/08 |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum (FBS), qualified, US Department of Agriculture (USDA)-approved regions | Life Technologies | Cat# 10437028 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Neurobasal-A | Gibco | Cat# 10888022 |
| GlutaMAX | Gibco | Cat# 35050061 |
| B27 Plus Supplement | Gibco | Cat# A3582801 |
| BrainPhys Neuronal Medium | StemCell | Cat# 05790 |
| NeuroCult SM1 Neuronal Supplement | StemCell | Cat# 05711 |
| N2 Supplement-A | StemCell | Cat# 07152 |
| Doxycycline hyclate (Dox) | Sigma-Aldrich | Cat# D9891 |
| Poly-L-ornithine solution | Sigma-Aldrich | Cat# P4957 |
| Laminin | Sigma-Aldrich | Cat# L2020 |
| Fibronectin | Sigma-Aldrich | Cat# F4759 |
| Valproic acid (VPA), sodium salt | Sigma-Aldrich | Cat# 676380 |
| Dibutyryl-cAMP sodium salt | Sigma-Aldrich | Cat# D0627 |
| Retinoic acid (RA) | Sigma-Aldrich | Cat# R2625 |
| Neurotrophin-3 (NT-3) | PeproTech | Cat# 450-03 |
| Brain-derived neurotrophic factor (BDNF) | PeproTech | Cat# 450-02 |
| Glial-derived Neurotrophic Factor (GDNF) | PeproTech | Cat# 450-10 |
| RevitaCell Supplement | Thermo Fisher Scientific | Cat# A2644501 |
| ISX-9 | StemCell Technologies | Cat# 73202 |
| DAPT | Millipore Sigma | Cat# D5942 |
| Critical commercial assays | ||
| Go Taq DNA Polymerase | Promega | Cat# M3001 |
| RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 |
| SMARTer Ultra Low RNA Kit for Illumina sequencing | Clontech | Cat# 635029 |
| TruSeq Small RNA Library Preparation Kit | Illumina | Cat# RS-200-0012 |
| ReliaPrep™ RNA Miniprep Systems | Promega | Cat# Z6011 |
| QIAamp DNA Mini Kit | QIAGEN | Cat# 51304 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat# 28706 |
| Deposited data | ||
| Raw and processed data (Single-cell RNA-sequencing, bulk RNA-sequencing) | NCBI Gene Expression Omnibus | Accession #: GSE201084 |
| Western Blot | Mendeley Data | DOI: 10.17632/t93byrznm6.1 |
| Experimental models: Cell lines | ||
| Lenti-X 293LE cell line | Clontech | Cat# 632180 |
| AG04148, Male 56yrs – Control 1 | Coriell | Cat# AG04148 |
| AG08260, Male 61yrs – Control 2 | Coriell | Cat# AG08260 |
| AG08379, Female 60yrs – Control 3 | Coriell | Cat# AG08379 |
| AG13369, Male 68yrs – Control 4 | Coriell | Cat# AG13369 |
| UCL455, Male 53yrs – IVS10+16 1 | Dr.Henry Houlden, University College London | n/a |
| UCL457, Male 52yrs – IVS10+16 2 | Dr.Henry Houlden, University College London | n/a |
| UCL497, Female 54yrs – IVS10+16 3 | Dr.Henry Houlden, University College London | n/a |
| ES046, Male 57yrs – IVS10+16 4 | Dr. Barbara Corneo, Columbia University | n/a |
| Oligonucleotides | ||
| Primers for qPCR; see Table S3 | This paper | n/a |
| Primers for sqPCR; see Table S3 | This paper | n/a |
| Primers for sequencing; see Table S3 | This paper | n/a |
| Recombinant DNA | ||
| pMD2.G | Didier Trono | 12259 |
| psPAX2 | Didier Trono | 12260 |
| rtTA-N144 | Richner et al., 2015 | 66810 |
| pTight-9/9*-124-Bclxl | Victor et al., 2014 | 60857 |
| MYT1L-N174 | Richner et al., 2015 | 66809 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| LAS X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Harmony v4.9 | Perkin Elmer | https://www.perkinelmer.com/product/harmony-4-8-office-hh17000001 |
| Graphpad Prism 9 | GraphPad Software Inc | http://www.graphpad.com/ |
| FCS Express 7 Research software | De Novo Software | https://denovosoftware.com/ |
| Ingenuity Pathway Analysis | QIAGEN | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ |
| Skyline | MacCoss Lab Software, MacLean et al., 2010 | https://skyline.ms/project/home/software/Skyline/begin.view |
| Cell Ranger (v3.1.0) | 10x Genomics, Zheng et al., 2017 | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger |
| Seurat (v3.2.3) | Santija Lab, Satija et al., 2015 | https://satijalab.org/seurat/ |
| Cutadapt v2.4 | Martin, 2011; National Bioinformatics Infrastructure Sweden | https://github.com/marcelm/cutadapt/ |
| DESeq2 | Love, Huber, and Anders, 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| FastQC v0.11.4 | Simon Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Deeptools (v3.5.1) | Ramirez et al., 2014 | http://deeptools.ie-freiburg.mpg.de |
| Wiggletools (v1.2) | Zerbino et al., 2014 | www.github.com/Ensembl/Wiggletools |
| Samtools v1.9 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Picard v2.18 | Broad Institute | https://broadinstitute.github.io/picard/ |
| IGV | Thorvaldsdóttir et al., 2013 | https://software.broadinstitute.org/software/igv/download |
| STAR | Dobin et al., 2015 | https://github.com/alexdobin/STAR |
| LONGO | McCoy et al., 2018 | https://github.com/biohpc/longo |
| Leafcutter | Li et al., 2018 | https://davidaknowles.github.io/leafcutter/ |


