ABSTRACT
Natural Killer (NK) cells are known for their high intrinsic cytotoxic capacity, and the possibility to be applied as ‘off-the-shelf’ product makes them highly attractive for cell-based immunotherapies. In patients with multiple myeloma (MM), an elevated number of NK cells has been correlated with higher overall-survival rate. However, NK cell function can be impaired by upregulation of inhibitory receptors, such as the immune checkpoint NKG2A. Here, we developed a CRISPR-Cas9-based gene editing protocol that allowed us to knockout about 80% of the NKG2A-encoding killer cell lectin like receptor C1 (KLRC1) locus in primary NK cells. In-depth phenotypic analysis confirmed significant reduction in NKG2A protein expression. Importantly, the KLRC1-edited NK cells showed significantly increased cytotoxicity against primary MM cells isolated from a small cohort of patients, and maintained the NK cell-specific cytokine production. In conclusion, KLRC1-editing in primary NK cells has the prospect of overcoming immune checkpoint inhibition in clinical applications.
KEYWORDS: Genome editing, CRISPR-Cas, NK cells, inhibitory receptors, immunotherapy, NKG2A, HLA-E, KLRC1, multiple myeloma
Key points:
KLRC1-disruption in primary NK cells increases anti-tumor capacity against primary tumor isolates from multiple myeloma patients
Targeting the immune checkpoint NKG2A by CRISPR-Cas9 gene editing in NK cells offers a novel approach to overcome tumor immune evasion
Introduction
Multiple myeloma is a malignant hematological disease, which origins from an excess of monoclonal plasma cells, leading to elevated levels of dysfunctional monoclonal protein in the blood and/or urine. Standard treatment consists of induction therapy followed by high-dose chemotherapy and autologous stem cell transplantation (autoSCT) for eligible patients.1 Major advances in the treatment of MM have been made by the approval of novel proteasome inhibitors, monoclonal antibodies, and immunomodulatory drugs (IMiDs).2 However, MM patients are still prone to relapse, and there is an urgent medical need to improve their long-term survival rate.3 In this context, cellular therapies have gained in interest, especially since CAR-T cell therapeutic approaches have been successful in treatment-resistant or relapsed B cell malignancies.4 Beyond genetically engineered T cells, also NK cells have demonstrated encouraging results in early clinical trials.5 One reason is the high intrinsic cytotoxic capacity of NK cells. Furthermore, the possibility to apply NK cells as an ‘off-the-shelf’ third-party donor cell therapy product makes them highly attractive for clinical use. In cancer patients suffering from MM, an elevated number of NK cells has been correlated with a higher overall-survival rate.6 However, NK cell function can be impaired by upregulation of inhibitory receptors, such as the immune checkpoint NKG2A (natural killer group 2A), expressed on the surface of cytotoxic lymphocytes, including subsets of activated CD8 T and NK cells. Recently, blocking antibodies of the immune checkpoint HLA-E/NKG2A axis, such as monalizumab, have been reported to unleash the anti-tumor activity of T and NK effector cells in several preclinical mouse models as well as in early clinical trials.7,8 In a pre-clinical experimental setting, we have recently shown that the use of ex vivo cytokine-activated NK cells in combination with NKG2A blockade might be a valuable immune therapeutic approach to treat MM patients within an autoSCT setting.9 Here, we report a novel strategy to delete inhibitory immune checkpoint expression in NK cells by genome editing, resulting in a significant increase in NK cell-mediated cytotoxicity against allogenic primary bone marrow-derived MM cells in a small cohort of patients.
Methods
Primary NK cells were isolated and expanded according to previously described protocols using only IL-15 under feeder-cell free conditions.10 Upon transfer of CRISPR-Cas9 nucleases targeting KLRC1, NK cells were cultured for two to three weeks and the extent of KLRC1 KO was determined by TIDE11 and T7E1 analyses.12 In-depth evaluation of disrupted alleles was performed by targeted amplicon next-generation sequencing (NGS).13 The NKG2A expression of gene-edited, IL-15 expanded bulk NK cells was compared with non-edited bulk NK cells in terms of cell viability and phenotypic changes by flow cytometry prior to functional analyses. Cytotoxicity of IL-15 expanded, gene-edited NK cells against different tumor cell lines and primary patient bone marrow-derived MM cells was analyzed after 24 h co-culture experiments, comparing NKG2A-KO (KO) NK cells with non-treated (NT) NK cells. Indicated cytokines in the supernatants from these functional analyses were measured using BD CBA Flex Sets (BD Biosciences).10 Detailed protocols, statistical analysis, and materials were described in supplemental material and methods.
Results and discussion
With the aim to overcome HLA-E/NKG2A-mediated suppression of anti-tumor NK cell function, we used the CRISPR-Cas9 system to knockout (KO) the KLRC1 locus encoding NKG2A in primary NK cells. In preliminary experiments, we identified CRISPR-Cas9 #3 nuclease (Figure S1A-E) as the best performing nuclease to disrupt KLRC1. Genotypic analysis indicated 70% to 80% gene disruption as assessed by T7E1 assay or TIDE analysis, respectively (Figure S1B-E). Next, we validated the developed protocol in NK cells derived from several donors. High gene disruption activity was confirmed by genotyping using T7E1 (70%) and TIDE (75%) analyses (Figure 1(a,b)), as well as next-generation targeted amplicon sequencing (85.6%) (Figure 1(c)) with a predominant +1 insertion at the target site (Figure 1(d)) across several donors (Figures 1(b) and S1D). Moreover, we addressed the specificity of the developed nuclease by targeted amplicon NGS of the top 10 predicted off-target (OT) sites (Figure S1H). Our data indicated only minor activity of the CRISPR-Cas9 nuclease at OT1 (~0.1%) which is located in an intergenic region (Chr2:63612892–63612914) (Figure S1H). KLRC1 disruption reduced the NKG2A expression on feeder-cell free, IL-15 expanded primary NK cells after two to three weeks of cultivation from 90% for non-edited (non-treated, NT) NK cells to 43.5% for the NKG2A-KO NK cell population (Figure 1(e–g)). This significant reduction in NKG2A expression did not affect NK cell viability (Figure 1(h)). Moreover, gene-edited bulk NK cells showed a significant expansion of up to 10-folds, 18 days following CRISPR-Cas9 nuclease nucleofection (Figure S1F) under a feeder-cell free, IL-15-based cultivation system. Phenotyping analysis of NKG2A-KO NK cells revealed an equally high surface expression of CD56 (Figure 1(g)) in both NK cell preparations, NT and NKG2A-KO, which is related to the IL-15-based expansion of the NK cells.14 In addition, the surface expression of CD16 and CD57 was significantly reduced in NKG2A-KO NK cells compared to NT NK cells (Figure 1(i) and S1G), while the expression of additionally analyzed NK cell-specific activation, inhibition, and migration markers were not significantly altered in the gene-edited NK cell population (Figure 1(i)).
Figure 1.
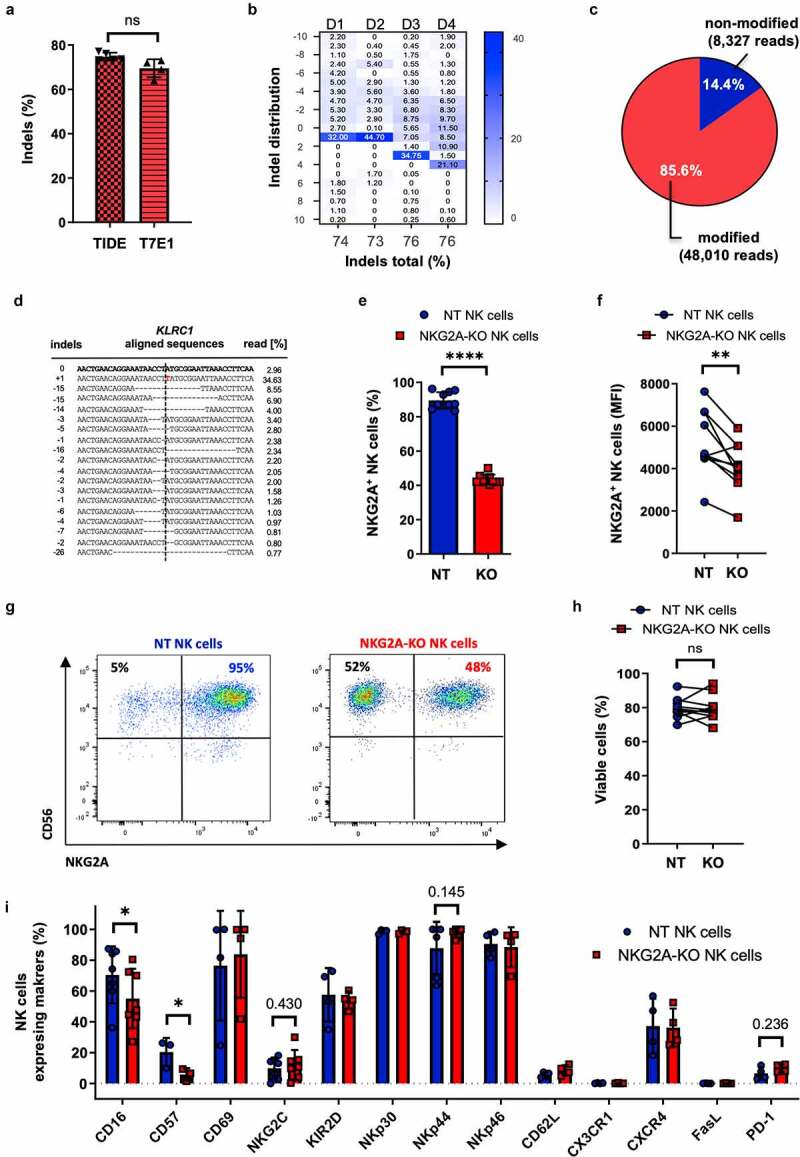
Characterization of KLRC1-edited primary NK cells. Primary NK cells were nucleofected with CRISPR-Cas9 ribonucleoprotein (RNP) complex targeting KLRC1 locus. (a-d) Genotyping. Frequency of KLRC1 disruption was evaluated by T7E1 assay (a), TIDE analysis (a, b), and targeted amplicon next-generation sequencing (NGS) (c-d). Insertion/deletion (indel) distribution profiles were analyzed by TIDE for four different donors (D1-4) while NGS was performed for a single donor. (e-i) Phenotyping. KLRC1-edited NKG2A knock-out NK cells (NKG2A-KO NK cells) were compared to non-treated NK cells (NT NK cells) by flow cytometry (n = 10 donors). Percentages of NKG2A-positive NK cells (e), Mean Fluorescence Intensities (MFI) (f), FACS plots of a representative donor (g) and percentages of viabilities (h) are indicated. Analyses of NK cell receptor expression was measured by flow cytometry (n = 4 donors) (i). Paired t-test. **** p ≤ .0001; ** p ≤ .01; * p ≤ .05; ns p > .05; if not mentioned, no significant difference was observed. T7E1, T7 endonuclease I; TIDE, tracking of indels by decomposition.
To further evaluate the anti-tumor activity of NKG2A-KO NK cells, we tested cytotoxicity against different human MM cells. It is well known that inhibition of NKG2A-expressing NK cells is mainly related to HLA-E ligand expression, which is often overexpressed in anti-tumor response and can be particularly upregulated by IFN-γ.9,15 In line with this, we confirmed an increase in HLA-E expression following IFN-γ stimulation on the MM tumor cell lines MM1.S, U266, as well as on patient-derived primary MM cells (Figure 2(b)). In addition, for a more detailed evaluation of the role of HLA-E/NKG2A interaction in tumor/NK cells context, we established HLA-E deficient MM1.S cells (MM1.S HLA-Eneg) as described in the supplemental methods (Figure 2(a) and 2SA).
Figure 2.
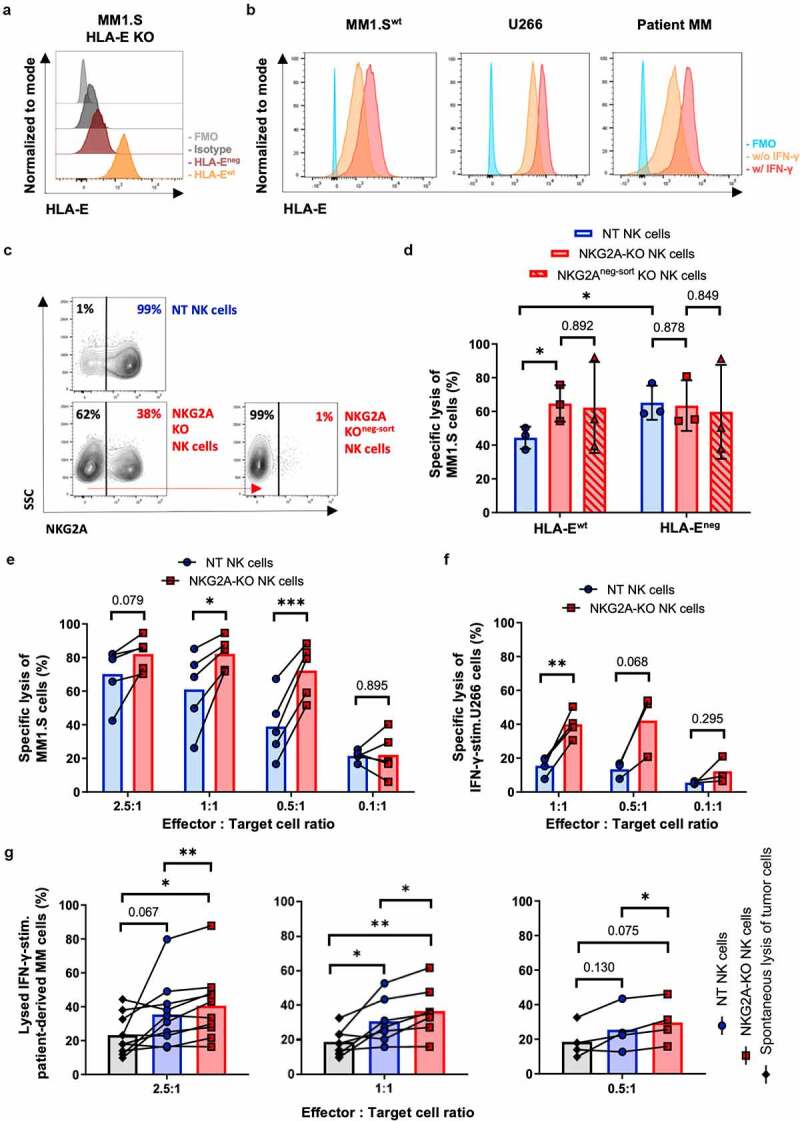
Cytotoxicity of KLRC1-edited NK cells. MM tumor cells were with pre-stimulated with (w/IFN-γ) or without (w/o) IFN-γ to induce HLA-E expression and then co-cultured with different NK effector cells (non-treated (NT) NK cells, NKG2A-KO NK cells, NKG2Aneg-sort KO NK cells). (a-b) HLA-E expression of tumor cells measured by flow cytometry. Indicated is the HLA-E expression of HLA-E KO (HLA-Eneg) MM1.S cells as compared to HLA-Ewt MM1.S cells (a). MM1.Swt, U266 and primary patient-derived bone marrow MM tumor cells after IFN-γ stimulation (b). (c) Representative FACS plots of NT NK cells, NKG2A-KO NK cells and NKG2A-KO NK cells that were negatively selected (NKG2Aneg-sort NK cells) and expanded for one additional week of cultivation. (d-g) MM tumor cells were co-incubated with indicated effector NK cells for 24 h and the extent of lysed tumor cells measured by flow cytometry. Percentages of lysed MM tumor cells were determined for different effector (NK cells) to target (MM tumor cells) (E:T) ratios. (d) MM1.S HLA-Ewt and HLA-Eneg tumor cells (E:T 0.5:1, n = 3 donors). (e) MM1.Swt tumor cells (n = 5 donors); (f) IFN-γ pre-stimulated U266 tumor cells (E:T 1:1, n = 4 donors; E:T 0.5:1 or 0.1:1, n = 3 donors); (g) IFN-γ pre-stimulated patient-derived MM tumor cells. Indicated are results for NKG2A-KO NK cells, NT NK cells, and tumor cells only (spontaneous lysis of MM cells) for 10 patients (E:T 2.5:1), seven patients (E:T 1:1) and four patients (0.5:1). FMO, Fluorescence minus one control. Paired t-test and (D) unpaired t-test. ** p ≤ .01; * p ≤ .05; ns p > .05; if not mentioned no significant difference was observed.
Following CRISPR-Cas9-based gene editing, we increased the fraction of NKG2A–KO NK cells by fluorescence-activated cell sorting and reached a stable 99% NKG2A-negative NK cell population one-week after selection, hereafter called NKG2Aneg-sort NK cells (Figure 2(c)). After co-culture of long-term IL-15 expanded NK cells, we compared NT, NKG2A-KO, and NKG2Aneg-sort NK cells regarding their killing capacity of the HLA-E-expressing wild-type MM1.Swt cell line. The resulting data confirmed a significantly increased killing capacity of the NKG2A-KO NK cells compared to NT NK cells (65% vs 44%), but no relevant enhancement of tumor cell lyses for NKG2Aneg-sort NK cells compared to the bulk NKG2A-KO NK cell preparation (62% vs 65%) (Figure 2(d)).
In parallel, we addressed the NK cell-mediated anti-tumor capacity against HLA-Eneg compared to HLA-Ewt MM1.S cells. As expected, NT NK cells presented a significantly increased killing capacity against HLA-Eneg MM1.S compared to HLA-Ewt MM1.S cells (Figure 2(d)). Importantly, there was no additional increase in tumor lyses of HLA-Eneg MM1.S compared to HLA-Ewt MM1.S cells by either NKG2A-KO or NKG2Aneg-sort NK cells compared to NT NK cells (63% vs 60% vs 63%) (Figure 2(d)). These data underline the biological relevance of HLA-E as ligand for the inhibitory NKG2A-receptor on NK cells. Remarkably, extinction of NKG2A in approximately 50% of NK cells was sufficient to bypass HLA-E-mediated immune checkpoint inhibition by different tumor cell lines and primary tumor cells (Figure 2(e–g)). Notably, a significantly increased cytotoxicity of NKG2A-KO NK cells compared to the already high intrinsic NT NK cell lysis against MM1.Swt cells was confirmed at different E:T ratios (Figure 2(e)). Remarkably, the NKG2A-KO NK cells mediated a significantly increased cytotoxicity, with more than 80% of MM cells being lysed at E:T ratios of 1:1 or 0.5:1, compared to NT NK cells (Figure 2(e)).
To mimic a more patient-related situation ex-vivo, we performed killing assays with IFN-γ pre-stimulated U266 tumor cells. U266 is known for a higher HLA-E expression and immune resistance in vivo than in vitro.9,15 Therefore, we used pre-stimulated IFN-γ to enhance HLA-E expression (Figure 2(b)) and observed low cytotoxic capacity of NT NK cells (Figure S2C), but significantly enhanced killing by the NKG2A-KO NK cell preparations (Figure 2(f)).
To address the increased killing capacity of NKG2A-KO NK cells in a more clinical setting, we isolated primary MM tumor cells from bone marrow aspirates of a small cohort of differently treated MM-patients (Figure 2(g); n=10 for E:T 2.5:1, for detailed patient characteristics see supplemental Table S4). In order to simulate the pro-inflammatory in vivo patient setting, we used IFN-γ pre-stimulation of patient-derived MM tumor cells and observed increased resistance against killing by normal allogenic NK cells but not by NKG2A-KO NK cells (Figure S2D, data of one representative patient and allogenic NK cell donor shown). In fact, the cytolytic activities of NKG2A-KO NK cells to kill patient-derived tumor cells was significantly higher than cell lysis induced by NT NK cells at all tested E:T ratios (Figure 2(g)). There was no indication of gender- or age-related impact to NKG2A-KO NK cell-based tumor lysis in this patient cohort (age median: 70, range: 56–81 years; data not shown). Importantly, the secretion of GM-CSF, IFN-γ, MIP-1α, and TNF-α was not significantly different between NKG2A-KO NK and NT NK cells following co-cultivation with different tumor cell lines and primary MM tumor cells (Figure S3). In this regard, an increased cytokine concentration could be detected for the NKG2A-KO NK cells already after long-term cultivation without tumor cells compared to equally cultivated NT NK cells (Figure S3). Notably, IFN-γ and TNF-α are known mediators of NK cell activation16 and might support the higher activation level of all CRISPR-modified NK cells. This might be a possible explanation for the similar increase in cytotoxic activity of the 50% NKG2A-KO NK cells compared to 100% NKG2A negative selected NKG2Aneg-sort NK cells (Figure 2(d)).
In conclusion, we believe that KLRC1-editing in primary NK cells might be a valuable strategy to design cell therapeutic approaches, which will overcome immune checkpoint inhibition for treatment of multiple myeloma. In addition, our established protocol to edit the genome of primary NK cells provides a robust platform to perform a variety of further modulations to release the brakes and enhance the anti-tumor potential of NK cells against a variety of tumor entities.
Supplementary Material
Acknowledgments
We thank all patients who contributed to the study by their donation of bone marrow aspirates, and the German Red Cross Blood Donation Center Frankfurt and Blood Donation Center of the Medical Center University of Freiburg for support with buffy coats. The authors thank Katja Stein, Franziska Kalensee, Petra Schoen and Beate vom Hövel for their excellent technical assistance, Kay Ole Chmielewski and Simone Haas for support with bioinformatics, the Lighthouse Core Facility (Medical Center - University of Freiburg), Julia Campe for support concerning flow cytometry, and Michael Hudecek for kindly providing MM1.S tumor cells.
Funding Statement
This work was supported by the German Research Foundation /Deutsche Forschungsgemeinschaft DFG, project number 318346496, SFB1292/2 TP12 (to E.U.) and CRC /IRTG 1292 (to E.U. and T.B.) and CRC 1160 (to T.C.), by the German Cancer Aid / Deutsche Krebshilfe (MSNZ stipend to T.B.), and by the Alfred and Angelika Gutermuth Foundation.
Authors contribution
T.B., J.A., L.M.R., P.W. performed experiments, T.B., J.A., L.M.R., P.W., R.S. analyzed data. E.U., T.C. designed and directed the study; E.S.M. performed statistical analysis; E.U., T.C., R.S., E.S.M., I.v.M. discussed the results and interpreted the data together with all co-authors. T.B., J.A., E.U., T.C. wrote the manuscript with contributions of all authors.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2022.2081415
References
- 1.Landgren O, Rajkumar SV.. New developments in diagnosis, prognosis, and assessment of response in multiple myeloma. Clin Cancer Res. 2016;22:5428–6. doi: 10.1158/1078-0432.CCR-16-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasche L, Wasch R, Munder M, Goldschmidt H, Raab MS.. Novel immunotherapies in multiple myeloma - chances and challenges. Haematologica. 2021;106:2555–2565. doi: 10.3324/haematol.2020.266858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehners N, Becker N, Benner A, Pritsch M, Löpprich M, Mai EK, Hillengass J, Goldschmidt H, Raab M-S. Analysis of long-term survival in multiple myeloma after first-line autologous stem cell transplantation: impact of clinical risk factors and sustained response. Cancer Med. 2018;7:307–316. doi: 10.1002/cam4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sterner RC, Sterner RM. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 2021;11:69. doi: 10.1038/s41408-021-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reindl LM, Albinger N, Bexte T, Muller S, Hartmann J, Ullrich E. Immunotherapy with NK cells: recent developments in gene modification open up new avenues. Oncoimmunology. 2020;9:1777651. doi: 10.1080/2162402X.2020.1777651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osterborg A, Nilsson B, Bjorkholm M, Holm G, Mellstedt H. Natural killer cell activity in monoclonal gammopathies: relation to disease activity. Eur J Haematol. 1990;45:153–157. doi: 10.1111/j.1600-0609.1990.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 7.Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, Bléry M, Bonnafous C, Gauthier L, Morel A, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175:1731–43 e13. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hall T, Andre P, Horowitz A, van Hall T, Ruan DF, Borst L, Zerbib R, Narni-Mancinelli E, van der Burg SH, Vivier E. Monalizumab: inhibiting the novel immune checkpoint NKG2A. J ImmunoTher Cancer. 2019;7:263. doi: 10.1186/s40425-019-0761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tognarelli S, Wirsching S, von Metzler I, Rais B, Jacobs B, Serve H, Bader P, Ullrich E. Enhancing the activation and releasing the brakes: a double hit strategy to improve NK cell cytotoxicity against multiple myeloma. Front Immunol. 2018;9:2743. doi: 10.3389/fimmu.2018.02743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller S, Bexte T, Gebel V, Kalensee F, Stolzenberg E, Hartmann J, Koehl U, Schambach A, Wels WS, Modlich U, et al. High cytotoxic efficiency of lentivirally and alpharetrovirally engineered CD19-specific chimeric antigen receptor natural killer cells against acute lymphoblastic leukemia. Front Immunol. 2019;10:3123. doi: 10.3389/fimmu.2019.03123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patsali P, Turchiano G, Papasavva P, Romito M, Loucari CC, Stephanou C, Christou S, Sitarou M, Mussolino C, Cornu TI, et al. Correction of IVS I-110(G>A) β-thalassemia by CRISPR/Cas-and TALEN-mediated disruption of aberrant regulatory elements in human hematopoietic stem and progenitor cells. Haematologica. 2019;104:e497–e501. doi: 10.3324/haematol.2018.215178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alzubi J, Lock D, Rhiel M, Schmitz S, Wild S, Mussolino C, Hildenbeutel M, Brandes C, Rositzka J, Lennartz S, et al. Automated generation of gene-edited CAR T cells at clinical scale. Mol Ther Methods Clin Dev. 2021;20:379–388. doi: 10.1016/j.omtm.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turchiano G, Andrieux G, Klermund J, Blattner G, Pennucci V, El Gaz M, Monaco G, Poddar S, Mussolino C, Cornu TI, et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. 2021;28:1136–47 e5. doi: 10.1016/j.stem.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Heinze A, Grebe B, Bremm M, Huenecke S, Munir TA, Graafen L, Frueh JT, Merker M, Rettinger E, Soerensen J, et al. The synergistic use of IL-15 and IL-21 for the generation of NK cells from CD3/CD19-depleted grafts improves their ex vivo expansion and cytotoxic potential against neuroblastoma: perspective for optimized immunotherapy post haploidentical stem cell transplantation. Front Immunol. 2019;10:2816. doi: 10.3389/fimmu.2019.02816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar S, van Gelder M, Noort W, Xu Y, Rouschop KMA, Groen R, Schouten HC, Tilanus MGJ, Germeraad WTV, Martens ACM, et al. Optimal selection of natural killer cells to kill myeloma: the role of HLA-E and NKG2A. Cancer Immunol Immunother. 2015;64:951–963. doi: 10.1007/s00262-015-1694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Jaw JJ, Stutzman NC, Zou Z, Sun PD. Natural killer cell-produced IFN-gamma and TNF-alpha induce target cell cytolysis through up-regulation of ICAM-1. J Leukoc Biol. 2012;91:299–309. doi: 10.1189/jlb.0611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


