Abstract
Salmonella Kentucky is commonly found in poultry and rarely associated with human disease. However, a multidrug-resistant (MDR) S. Kentucky clone [sequence type (ST)198] has been increasingly reported globally in humans and animals. Our aim here was to assess if the recently reported increase of S. Kentucky in poultry in Spain was associated with the ST198 clone and to characterize this MDR clone and its distribution in Spain. Sixty-six isolates retrieved from turkey, laying hen and broiler in 2011–2017 were subjected to whole-genome sequencing to assess their sequence type, genetic relatedness, and presence of antimicrobial resistance genes (ARGs), plasmid replicons and virulence factors. Thirteen strains were further analysed using long-read sequencing technologies to characterize the genetic background associated with ARGs. All isolates belonged to the ST198 clone and were grouped in three clades associated with the presence of a specific point mutation in the gyrA gene, their geographical origin and isolation year. All strains carried between one and 16 ARGs whose presence correlated with the resistance phenotype to between two and eight antimicrobials. The ARGs were located in the Salmonella genomic island (SGI-1) and in some cases (bla SHV-12 , catA1, cmlA1, dfrA and multiple aminoglycoside-resistance genes) in IncHI2/IncI1 plasmids, some of which were consistently detected in different years/farms in certain regions, suggesting they could persist over time. Our results indicate that the MDR S. Kentucky ST198 is present in all investigated poultry hosts in Spain, and that certain strains also carry additional plasmid-mediated ARGs, thus increasing its potential public health significance.
Keywords: Salmonella Kentucky, whole-genome sequencing, poultry, antimicrobial resistance, plasmid
Impact Statement
The emergence of a multidrug-resistant (MDR) Salmonella enterica Kentucky ST198 clone has been reported worldwide in the last decade, typically associated with cases in travellers. In addition, MDR S. Kentucky strains have been increasingly reported from poultry in several EU countries. In this article, we present a comprehensive analysis of the genomes from a large collection of isolates of this serotype obtained through the antimicrobial resistance surveillance programme in poultry flocks in Spain that confirms that this MDR clone is present in broiler, laying hen and turkey Spanish populations. Our results show that several of the strains analysed carry (in addition to the resistance traits characteristic of this MDR clone) a high number of antimicrobial resistance genes (ARGs) in mobile (plasmid and integron) structures. These MDR strains were retrieved from different locations and animal hosts over the years studied, suggesting their potential for persistence, and showed a high genetic similarity with sequences described in isolates from other countries and sources. The presence of ARGs in these mobile structures and the dynamics of establishment of this clone in the poultry population studied indicate that given its public health significance this serotype should be closely monitored in the future.
Data Summary
Short-read and long-read sequences along with complete genome assemblies generated in this study have been deposited in the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena) under BioProject ID PRJNA732654 (DDBJ/ENA/GenBank). SRA accession numbers can be found in Table S1 (available in the online version of this article). The phylogeny and associated metadata can be found in Fig. 2 and Table S1, respectively.
Introduction
Salmonella is one of the major causes of food-borne diseases worldwide and the is second most common zoonotic pathogen in humans in the European Union (EU) [1]. It usually causes a self-limiting disease, but in case of invasive infections in high-risk patients such as children, the elderly and immunocompromised patients, antibiotic treatment may be necessary. Besides its direct impact on human health, this pathogen causes considerable economic losses due to the need for healthcare measures and treatment implementation [2, 3].
Poultry is one of the main sources of Salmonella , and for this reason there are monitoring and control programmes worldwide with the aim of reducing its prevalence and identifying the main serovars present and their antimicrobial resistance phenotypes. Typically, predominant serovars in poultry differ depending on the commercial production type:laying hen, broiler and turkey. Therefore, control programmes may target different serovars depending on the host [4]. Predominant serovars may also change over time, possibly the product of serovar-specific control strategies (i.e. vaccination against S. Enteritidis, control programmes for S. Typhimurium and S. Enteritidis in poultry flocks) [5–7] but also the result of the expansion of emergent clones such as S. Typhimurium DT104 in the 1990s [8].
Since the 2000s, isolates belonging to the serovar Kentucky have been increasingly detected in travellers, although the proportion of S. Kentucky isolates among all human clinical Salmonella isolates reported in the EU has remained stable from 2010 to 2018 (~0.8 %) [6, 9]. These isolates, typically belonging to sequence type (ST)198, showed high levels of multidrug-resistant (MDR) profiles, including resistance to fluoroquinolones, a critically important antimicrobial class [10]. Furthermore, detection of resistance to third-generation cephalosporins has been recently reported in some isolates, and around 20 % of all human S. Kentucky isolates notified in the EU in 2017 carried extended-spectrum beta-lactamase (ESBL) genes [1].
While human cases in the EU are still typically associated with international travel (with up to 53.2 % of all travel-associated cases caused by this serotype in 2016) [6], broiler and turkey flocks have been identified as a potential reservoir of this MDR clone, and there has been an increase in the prevalence of serovar Kentucky in poultry in Europe since 2009 [11–13]. Recent reports have demonstrated the presence of isolates belonging to this serotype and showing high levels of resistance in poultry flocks from several EU Member States (MSs), suggesting a clonal expansion in poultry populations in Europe [1].
Accelerated dissemination of S. Kentucky in poultry flocks has been attributed to a better ability of this serovar to survive in low acid environments, such as the poultry caecum [14], and to the carriage of acquired virulence plasmids (pColV-like) [15], which may facilitate colonization of poultry hosts [7, 16]. However, the reasons why this serotype has been traditionally less associated with human clinical cases are not clear. It has been suggested that the decreased pathogenicity for humans could be due to the lack of certain virulence genes including gvra, sseI, sopE and sodCI [17] or sopD2, pipB2, sspH2 and srfH [18]
Previous studies suggest the S. Kentucky MDR ST198 clone has been emerging globally since the 1990s, and its origin has been traced back to Africa [12, 19], where it would have acquired a certain type of Salmonella genomic island (SGI-1) around 1989, probably in Egypt [11, 20]. The SGI harbours several genes conferring its characteristic MDR phenotype (including resistance to ampicillin, streptomycin, gentamicin, sulfamethoxazole and tetracycline). In addition, ST198 strains have accumulated various SNPs in the quinolone-resistance-determining regions (QRDRs) of genes coding for the DNA gyrase and DNA topoisomerase IV enzymes (gyrA and parC, respectively). These chromosomal mutations confer resistance to ciprofloxacin. Moreover, some strains have acquired genes encoding carbapenemases and ESBLs transferred by IncC and IncI1 plasmids [21, 22]
Due to the spread of the MDR ST198 clone in broiler production in the EU, France included this serotype among the target regulated serovars for poultry in 2016 [6, 23], and it has been proposed as a candidate target serovar for inclusion in national control programmes for breeding flocks in other EU MSs [6]. In Spain, the analysis of Salmonella isolates obtained in the antimicrobial resistance (AMR) surveillance programmes in poultry during 2011–2017 revealed that MDR S. Kentucky strains were recovered from all hosts considered (turkeys, broilers and laying hens) since 2011–2012, although their relative frequency was much higher in turkeys compared to broilers and especially to laying hens [24]. Moreover, MDR isolates belonging to this serotype have also been detected in wildlife in Spain, suggesting other species may be involved in its epidemiology [25]. However, the genetic diversity and potential carriage of ARGs among the S. Kentucky strains currently circulating in Spanish poultry flocks is currently unknown.
Thus, the aim of this study was to evaluate the genetic diversity among Spanish S. Kentucky strains circulating in poultry in the period 2011–2017 to establish if MDR strains recovered from the different poultry hosts belong to the ST198 emergent clone, along with their genetic relatedness and the presence of possible molecular mechanisms conferring AMR or increased virulence. This study can help to understand the transmission dynamics of MDR Salmonella in poultry and to inform enhanced prevention and control strategies to mitigate their potential impact.
Methods
Study population
The study population was formed by all (n=228) S. Kentucky isolates included in the Salmonella AMR surveillance programme in poultry flocks (laying hen, broiler and turkey) in Spain between 2011 and 2017 and and were part of a larger panel of 3047 Salmonella isolates analysed in a previous study [24]. Isolates included in the AMR surveillance programme are selected at random (laying hen) and occasionally by convenience sampling (broiler and turkey) among those retrieved through official and auto-control checks performed in poultry flocks according to national and European legislation [26–28]. Therefore, the proportion of isolates of each serotype may not be representative of their actual distribution in the sampled populations (and hence of the Salmonella -positive farms) as previously discussed [24], but because no information on their resistance phenotype is available before their inclusion in the AMR surveillance programme, their phenotypes can be considered representative of the underlying serotype-specific population.
Information on their antimicrobial susceptibility profile for eight antibiotics (ciprofloxacin, nalidixic acid, gentamycin, ampicillin, sulfamethoxazole, tetracycline, chloramphenicol, colistin) determined using the two-fold broth microdilution method (ISO 20776-1 : 2006) was transformed into dichotomous variables (wild type, here referred to as susceptible, and non-wild type or resistant from here on) using the epidemiologic cut-offs (ECOFFs) indicated by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (Table S2). Isolates were classified as presenting or not presenting a certain MDR profile, including resistance to quinolones plus ampicillin, gentamycin, sulfamethoxazole and tetracycline (AGSuT), similar to the ASSuT profile [29–31] but replacing the aminoglycoside streptomycin (not available in our panel) by another aminoglycoside for which results were available, gentamicin (Table 1).
Table 1.
AMR profile of the 66 selected isolates depending on the host
|
Resistotype |
Laying hen |
Broiler |
Turkey |
% Isolates (n) |
|
|---|---|---|---|---|---|
|
2 |
Cip, Nal |
1 |
2 |
2 |
7.6 % (5) |
|
3 |
Cip, Nal, Amp |
– |
2 |
1 |
4.5 % (3) |
|
Cip, Nal, Tet |
1 |
– |
– |
1.5 % (1) |
|
|
Cip, Nal, Gen |
– |
2 |
1 |
4.5 % (3) |
|
|
Cip, Amp, Chl |
– |
1 |
– |
1.5 % (1) |
|
|
4 |
Cip, Nal, Gen, Smx |
1 |
2 |
1 |
6.1 % (4) |
|
5 |
Cip, Nal, Gen, Amp, Smx |
1 |
5 |
3 |
13.6 % (9) |
|
Cip, Nal, Gen, Smx, Tet |
2 |
2 |
3 |
10.6 % (7) |
|
|
6* |
Cip, Nal, Gen, Amp, Smx, Tet |
4 |
8 |
9 |
31.8 % (21) |
|
7* |
Cip, Nal, Gen, Amp, Smx, Tet, Chl |
– |
– |
2 |
3.0 % (2) |
|
Cip, Nal, Gen, Amp, Smx, Tet, Tmp |
– |
2 |
2 |
6.1 % (4) |
|
|
8* |
Cip, Nal, Gen, Amp, Smx, Tet, Tmp, Chl |
1 |
1 |
4 |
9.1 % (6) |
|
Total |
11 |
27 |
28 |
66 |
*Isolates presenting the AGSuT phenotype: resistance to ampicillin, gentamicin, sulfamethoxazole and tetracycline.
Ampicillin (Amp), chloramphenicol (Chl), ciprofloxacin (Cip), gentamicin (Gen), nalidixic acid (Nal), sulfamethoxazole (Smx), tetracycline (Tet) and trimethoprim (Tmp).
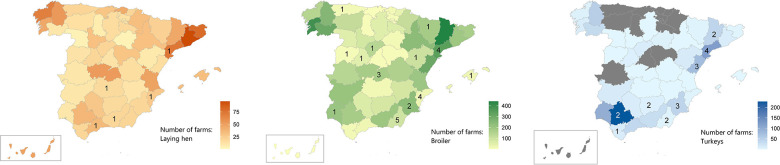
Sixty-six out of the total 228 S. Kentucky isolates were included in the study (Table S3) so that all hosts (laying hen, broiler or turkey), years (2011–2017) and AMR profiles (AGSuT/non-AGSuT) were represented (Table S3). Eleven (16.7 %), 27 (40.9 %) and 28 (42.4 %) isolates were retrieved from laying hen, broiler and turkey, respectively, roughly representing the proportions of isolates from each host among the 228 isolates [24]. The farms of origin of 56 isolates (no information was available for ten isolates) were in 21 provinces of Spain, with most laying hen and turkey farms being in the South and East of the country while broiler farms were more scattered (Fig. 1), in agreement with the overall distribution of the poultry farms from each production type in Spain based on farms sampled in the frame of the national Salmonella control programme (Fig. 1). Most isolates (n=44) originated from flocks coming from different farms, while six farms contributed two to four isolates collected from different flocks housed in different years (only two out of these six farms contributed two isolates in the same year, which originated from different flocks).
Fig. 1.
Maps indicating the total number of farms sampled in the frame of the Salmonella national control programmes during 2011–2017 per province in Spain (source: Ministry of Agriculture, Food and Fisheries). Colour indicates poultry production (orange: laying hen; green: broiler; blue: turkey) and colour intensity the number of farms. The number of farms from which isolates used in this study originated are indicated inside the relevant provinces.
Among the selected isolates, 5/11, 11/27 and 17/28 presented the AGSuT phenotype (representing 45.5, 40.7 and 60.7 % of the AGSuT S. Kentucky isolates from each host in the original collection). The remaining 33 isolates included in the selection were resistant to two to five antimicrobials and were also included to assess their genetic relatedness with the AGSuT isolates (Table 1, Table S3).
The strains were recovered from frozen stocks (kept at −80 °C) by culture in Columbia 5 % sheep blood agar (bioMérieux) for 24±3 h at 37 ± 1°C and submitted to the Laboratorio Tecnológico Agrario de Castilla y León (ITACYL) for whole-genome sequencing analysis.
Short-read sequencing
DNA from the 66 isolates was extracted and purified using commercial kits [32]. Libraries were prepared using the Nextera XT kit by standard protocols [33]. Extracted DNA was sequenced in the MiSeq Illumina platform using v3 reagents with 2×300 cycles.
The bioinformatics analysis was based on the Tormes pipeline (v 1.2) [34]. This included several steps (details and versions/parameters provided in Table S4): first, adaptors were removed and low-quality raw reads were filtered out with Trimmomatic [35]. Reads that passed the quality control were classified taxonomically using Kraken2 [36]. Genomes were assembled with SPAdes [37], and the quality of the assemblies was evaluated with QUAST [38] (contigs <200 bp long were discarded). Then, assembled contigs were annotated using PROKKA. Multilocus sequence typing (MLST) was performed with the mlst software (T. Seemann, https://github.com/tseemann/mlst). Assemblies were screened for the presence of antimicrobial resistance genes (ARGs) using the blast-based ResFinder software [39, 40] and for virulence genes using ABRicate (T. Seemann, Abricate Github, https://github.com/tseemann/abricate) and the Virulence Factors Database (VFDB) [41]. The identification of plasmid replicons was carried out using PlasmidFinder [42] with a coverage threshold of >90 %, and point mutations associated with AMR were identified using PointFinder [43]. Serotyping analysis was performed using SISTR [44] for Salmonella . The raw reads generated in this study have been deposited in the European Nucleotide Archive (ENA: PRJNA732654).
Phylogenetic analysis
Thirteen S. Kentucky already sequenced and described in previous studies [11] were selected in order to include strains from different world regions [Europe (n=3), Africa (n=7) and Asia (n=2)], hosts (animal and human) and tree clades (linked to point mutations in the gyrA and parC genes) (Table S1). Reads from the isolates sequenced in this study and the above-mentioned downloaded sequences were mapped and aligned to the reference genome sequence 201001922 (CP028357.1) using bwa [45] with default parameters. The resulting SAM files were sorted and compressed into bam files using SAMtools [46]. SNPs were called using the ‘mpileup’ and ‘call’ commands from BCFtoools, excluding those with a coverage threshold lower than 10 and an average base quality (Phred score) lower than 30. SNPs selected were output in vcf (variant calling format) files to be parsed and examined.
Phage regions for the reference genome were found using the PHAST web server tool [47] and SNPs within such regions were masked using a custom python script. Consensus genome sequences from each strain were created by BCFtools from the corresponding vcf file and the reference sequence. All consensus sequences were aligned with MAFFT [48], and then Gubbins [49] was used to filter out polymorphic sites from the resulting multiple alignment file.
A maximum-likelihood phylogenetic tree was built in RAxML [50] using the general time-reversible substitution evolutionary model with gamma correction (GTR+Γ) and 10 000 rapid bootstrap replicates to correct for variability between positions in the genome. The tree was then rooted using one of the sequences downloaded from GenBank (SRR2124413) and visualized using FigTree (FigTree https://github.com/rambaut/figtree/releases) and iTol editor [51]. The presence of clusters of isolates in the phylogeny was evaluated by analysing the SNP alignment with RhierBAPs [52] in R.
Identification of SGI sequences
To characterize the sequence of SGI-1 in the 66 sequenced strains, assembled contigs were ordered against the S. Kentucky reference genome CP028357 using Abacas [53] and queried for the presence of SGI-l sequences with blast using the SGI1-K sequence (reference AY463797.8) as the target. Contigs in which alignments with an e-value lower than 0.01 and alignment length greater than 500 bp were found were extracted. Artemis ACT [54] was used to visualize the results.
Statistical analysis
The correlation between the number of antimicrobials against which the strains were resistant (phenotypical resistance) and the number of ARGs present in the strains was assessed using the Kendall rank correlation tau.b, and a Kruskal–Wallis test was applied to assess differences in the number of AMR genes of isolates depending on their BAPS clade (A, B, C; see below). Fisher’s exact test was used to compare frequencies of isolates presenting a certain minimum inhibitory concentration (MIC) to fluoroquinolones (<256 or ≥256 mg l−1 for nalidixic acid, <16 or ≥16 for ciprofloxacin) and specific quinolone-resistance point mutations in gyrA (D87N/D87Y) (Table S5). Fisher’s exact test was also used to compare frequencies of isolates from certain hosts, regions and period of isolation depending on the BAPS informed clades. A P-value cut-off of 0.05 was used to determine significance in all statistical analyses. All statistical tests were performed in R [55]
Long-read sequencing analysis
Following short-read analysis, a selection of isolates presenting plasmid replicons belonging to the IncHI2 and IncI1 groups and ARGs for more than four antimicrobial classes or harbouring mcr genes were further studied using MinION (Oxford Nanopore Technology) long-read sequencing. These included 13 isolates harbouring more than five aminoglycoside resistance genes, other specific resistance genes (mcr-9, dfrA1, blaSHV-12, catA1, cmlA), and/or certain plasmid replicons (IncHI2, IncI1) (see Results). The selected strains were recovered from frozen stocks (kept at −80 °C) and cultured in Columbia 5 % sheep blood agar (bioMérieux) for 24±3 h at 37±1 °C, followed by liquid cultures in LB broth at 37 °C. DNA extraction and purification were performed using commercial kits (DNeasy Blood and Tissue Kit; Qiagen). The sequencing library was prepared with the Ligation Sequencing Kit (SQK-LSK109) and the Native Barcoding Expansion (EXP-NBD104 and EXP-NBD114) protocols from Oxford Nanopore Technologies (ONT). Sequencing was performed in a 9.4 MinION flowcell (FLO-MIN106) for 24 h. Local base calling and demultiplexing were performed with Guppy, available via the ONT community site (https://community.nanoporetech.com). Porechop (https://github.com/rrwick/Porechop) was used to remove the adapters from the reads. Reads with an average read quality score lower than 10 or a read length lower than 500 bases were filtered out with NanoFilt [56]. A hybrid assembly of combined MinION and Illumina Hi Seq reads was performed with Unicycler [57]. The hybrid assemblies were again screened with ResFinder and then explored to verify the genomic context (chromosomal or plasmid) of the identified ARGs (Table S6).
Once the genomes were assembled, plasmid and chromosome sequences were examined separately. To characterize the plasmid sequences, plasmid type was confirmed by PlasmidFinder, and MOB suite [58] was used to provide relaxase typing and prediction conjugation potential (Table S7). Complete plasmid sequences were compared among them using blast n and then blasted against the NCBI database (only hits with at least 90 % sequence identity and 90 % coverage were selected) (Table S7). Core genes of the plasmids were identified with Roary using PRANK [59] and after exclusion of hypothetical proteins with an in-home pipeline a multiple alignment file was created with MAFFT to determine sequence variation between plasmids. The same methodology was followed to align and compare the chromosomal content of the strains.
The content of integrons in the consensus sequences of hybrid assemblies was determined using a sequence identity approach. Additionally, plasmid visualization was performed using blast Ring Image Generator (BRIG) [60]
Results
One of the 66 isolates (retrieved from a turkey) was excluded from the analysis due to the low quality of the reads (<20×) (Table S1). In silico serotyping and MLST confirmed the identity of the remaining 65 isolates as S. Kentucky ST198 (Table S1).
According to the phylogenetic analysis based on the SNP multi-alignment (812 bp length) of the 79 selected isolates (65 from this study and 13 external), four BAPS clades were identified, three of which included all the strains from this study (A, B and C), while the fourth (‘external’) included three strains whose genome was downloaded from GenBank (SRR2124413, SRR6898552, SRR6898566) (Fig. 2). All three hosts were represented in clades A, B and C, but isolates from farms in provinces in the West and Southeast of Spain were significantly (P<0.001) associated with clade C, while those coming from provinces in the North belonged more often to clades A and B (Table 2). Isolates from the initial years (2011–2015) mostly belonged to clade C, while those retrieved in 2016–2017 were more evenly distributed between all three clades.
Fig. 2.
Maximum-likelihood phylogenetic tree of 78 S. Kentucky ST198 isolates. Sequences were aligned to genome CP028357.1 (id in red). The coloured labels (yellow, blue and purple) indicate the three identified clades (a, b, c). Major internal nodes are labelled with triangles (grey, orange, blue, pink) indicating amino acid mutation change in the gyrA gene in each strain. Isolates identified with a star were selected for hybrid assembly analysis (n=8). The following information is presented to the left of the isolate IDs: host and year of origin, presence/absence of ARGs, plasmid replicons, and region of origin and plasmid group. Bootstrap values (>70 support) are shown in the tree.
Table 2.
Number of isolates allocated to each of the clades identified in the phylogenetic analysis depending on host, region and year of collection, as well as median number of ARGs
|
Variable |
Clade A (n=10), 14% |
Clade B (n=12), 17% |
Clade C (n=43), 59% |
Total (n=65), 100% |
|
|---|---|---|---|---|---|
|
Host |
Broiler |
5 (19 %) |
4 (15 %) |
18 (67 %) |
27 |
|
Laying hen |
2 (18 %) |
1 (9 %) |
8 (73 %) |
11 |
|
|
Turkey |
3 (11 %) |
7 (26 %) |
17 (63 %) |
27 |
|
|
Year range |
2011–2013 |
0 (0 %) |
0 (0 %) |
9 (100 %) |
9 |
|
2014–2015 |
4 (14 %) |
3 (10 %) |
22 (76 %) |
29 |
|
|
2016–2017 |
6 (22 %) |
9 (33 %) |
12 (44 %) |
27 |
|
|
Region* |
North a |
7 (41 %) |
7 (41 %) |
3 (18 %) |
17 |
|
South-East b |
3 (13 %) |
4 (17 %) |
17 (71 %) |
24 |
|
|
West c |
0 (0 %) |
0 (0 %) |
13 (100 %) |
13 |
|
|
ARGs |
Median (range) |
6 (2–11) |
8 (1–8) |
5 (1–16) |
6 (1–16) |
*No information of origin in 11 isolates (one from clade B and 10 from clade C).
a, Huesca, Teruel, Lleida, Tarragona, Baleares; b, Castellon, Alicante, Murcia, Almeria; c, Asturias, Salamanca, Avila, Segovia, Toledo, Ciudad Real, Huelva, Sevilla, Cadiz, Málaga, Jaen, Granada.
AMR determinants
Different determinants conferring resistance against quinolones, aminoglycosides, beta-lactams, colistin, phenicols, sulphonamides, tetracyclines and trimethoprim were found in the sequenced strains (Tables S1 and S5). Each isolate harboured between one and 16 ARGs (median=6), and there were no significant differences (P<0.0001) in the number of ARGs depending on the clade (Table 2). The number of resistance genes found was highly correlated with the number of phenotypical resistances per isolate, as expected (tau=0.92, P<0.001).
Regarding fluoroquinolone resistance, all isolates were resistant to ciprofloxacin and nalidixic acid, but no plasmid-mediated quinolone resistance genes [qnr, aac(6′)-Ib-cr, qepA or oqxAB] were detected in their genomes. However, we found several mutations in the QRDRs previously reported in ST198 MDR S. Kentucky strains: all the isolates had non-synonymous mutations in the parC (codon S80I and codon T57S) and gyrA (codon 83 S83F) genes. In addition, the sequenced strains presented one of two mutations in codon 87 of the gyrA gene: D87Y was found in 10 isolates (15.4%) and D87N in the remaining 55 isolates (84.6%). The D87Y/D87N genotype was associated with the grouping based on the phylogenetic tree: the 11 isolates presenting the D87Y substitution were included in clade A, while the D87N substitution was present in the remaining strains in clades B and C. The sequences external to this study clustering close to clade C also presented the D87N genotype (Fig. 2).
Similar MICs for nalidixic acid were observed irrespective of the mutation detected (P>0.05), while higher values for ciprofloxacin were observed in isolates presenting the D87N mutation (P<0.05) (Table S5).
A total of 10 determinants [aac(6)-Iaa, aac(3)-IIa, aac(3)-VIa, aacC5, aadA1, aadA2, aadA7, aph(3)-Ia, aph(3)-Ib, aph(6)-Id] conferring resistance to aminoglycosides were found in the collection, with between one and 10 genes per isolate (Table S1). The 10 strains susceptible to the only aminoglycoside included in the panel (gentamycin) carried only the aac(6)-Iaa gene, present in all 65 isolates. The 55 gentamycin-resistant isolates presented different combinations of genes, with the most frequent one being aac(6')-Iaa, aacC5 and aadA7 (34/55).
Up to three beta-lactamase-encoding genes (bla SHV-12 , blaTEM-1A, blaTEM-1B ) were found in 44 isolates (all resistant to ampicillin), of which nine carried blaTEM-1A , 33 blaTEM-1B and two blaSHV-12 (in addition to blaTEM-1B ). The two blaSHV-12 positive isolates were also resistant to cefotaxime (data not shown) and were recovered from a laying hen and a turkey flock in 2015. One ampicillin-resistant isolate did not harbour any beta-lactam resistance determinant.
Fifty out of the 52 phenotypically resistant isolates carried resistance genes against sulphonamides: 39 isolates carried only sul1, and 11 carried sul1 and sul3 genes. Two isolates were phenotypically resistant in the absence of sulfamethoxazole-resistant genes, and one out of the 13 isolates susceptible to sulphonamides carried sul1.
The tet(A) gene was found in 40 isolates, all resistant to tetracycline, while no resistance gene was found in the remaining 25 tetracycline-susceptible isolates.
Among the less frequently reported resistance genes in ST198 S. Kentucky isolates, only one gene conferring resistance to trimethoprim (dfrA1) was found in eight isolates, all of which were phenotypically resistant to trimethoprim. Also, two phenicol-resistant genes (catA1, cmlA1) were present in nine isolates, all of which were resistant to chloramphenicol: seven carried only cmlA1 and two carried both genes. In addition, two other phenotypically chloramphenicol-resistant isolates did not carry any phenicol resistance gene. Finally, the mcr-9 gene was found in two isolates recovered from broiler samples collected in the same province of Spain in 2013 and 2015. Both isolates were only resistant to ciprofloxacin, gentamicin and nalidixic acid, and were susceptible to colistin (MICs of 1 and 2 mg l−1).
Different SGI-K, Q and P-like structures were detected with the short-read assembly (Table S1). Information on the presence of virulence genes is presented in (Fig S1). Most of the 92 virulence genes searched were identified in all isolates, and for the 17 genes absent in one or more isolates no clear pattern was observed (i.e. they were not missing specifically in certain regions of the tree).
Plasmid replicons
Plasmid replicons belonging to five different plasmid types were detected in 38 strains of the collection. Replicons from long plasmids (>50000 bp, IncI1 and IncHI2) were present in 17 isolates, while short plasmid replicons [<7000 bp, ColpVC, Col156, Col(MG828)] were found in 27 isolates. (Table S1). The presence of long plasmids [IncI1 (n=3) or IncHI2 (n=10) replicon] was associated with specific resistance genes: isolates harbouring the mcr-9 gene or isolates with more than five aminoglycoside resistance genes plus resistance genes against phenicols, trimethoprim and/or the bla SHV-12 gene.
Hybrid assembly analysis
Eight isolates carrying a particularly high number of aminoglycoside genes (eight or nine), blaTEM-1A, sul1, sul3, tetA, dfrA and cmlA1 (in six out of the eight isolates), along with IncHI2 plasmid replicons were selected for long-sequencing analysis (n=8). In addition, two isolates carrying the mcr-9 gene and IncHI2 replicons and three harbouring five aminoglycoside genes, blaTEM-1B, sul1, sul3 dfrA1, tetA and blaSHV-12 (in two out of the three) along with IncI1 plasmid replicon were also included. High-quality reads and subsequent hybrid assembly were obtained and completed for 12 out of the 13 selected isolates (all but one carrying IncHI2, isolate ZTA19/00842). In all cases, at least two contigs were differentiated after hybrid assembly, one of ~4 800 000 bp representing the bacterial chromosome, and another of between 111 000 and 390 000 bp representing the plasmid sequence.
The nine IncHI2 plasmids that characterized sequences retrieved from the selected isolates were clustered into four groups (IncHI2a, IncHI2b, IncHI2c and IncHI2d) based on presence/absence of shared genes and sequence similarity (identity and coverage). All nine plasmids shared 23 of the total 139 genes identified in their sequences, with plasmids in the same groups sharing the same genes (Table S6, Fig. 3a, b), and having very similar sequences (>99.98 % identity and 100 % coverage) (Table S8). The IncHI2a group included three plasmids (isolates ZTA19/00814, ZTA19/00831 and ZTA19/00847) with 11 resistance genes (six aminoglycoside resistance genes plus cmlA1, sul1, sul3, tetA and dfrA). These plasmids were found in isolates from different flocks (two turkey flocks and one broiler flock) housed in three different years (2015, 2016 and 2017) in the same farm in the southeast of Spain. The IncHI2b group contained two plasmids (isolates ZTA19/00816 and ZTA19/00830) retrieved from different turkey farms in the same region in 2015 and 2016 and with the same resistant genes identified in the IncHI2a group (except one aminoglycoside gene, aadA2). The IncHI2c group included two plasmids found in isolates (ZTA19/00785 and ZTA19/00789) from turkey and laying hen samples collected in 2015 and 2016 (no geographical location information available, same ARGs as IncHI2a, IncHI2b groups plus catA1). Finally, the IncHI2d group contained the two plasmids carrying the mcr-9 gene. Plasmid sequences from this subgroup presented 69 unique genes and were very different (<53 % coverage) from plasmids of the IncHI2a, IncHI2b and IncHI2c groups. These two plasmids also contained aadA1 and aac(3)-VIa resistance genes (Fig. 3b).
Fig. 3.
Alignment of the identified plasmids in strains selected for hybrid analysis. Incompatibility group and number of plasmids per group are indicated in the top-middle of each panel. The inner ring in each group is used as a reference for the alignment (size indicated in the bottom-middle of each panel). Plasmid names (coloured key) refer to the strain of origin. Annotation of ARGs is shown on the outermost ring. ARGs in red are detected in all the plasmids of the group. ARGs in dark/soft grey are found in one/some of the plasmids, respectively. (a) IncHI2 plasmids (n=7, groups IncHI2a, IncHI2b and IncHI2c). (b) IncHI2 plasmids (n=2, group IncHI2d). (c) IncI1 plasmids (n=3, group IncI1).
The three IncI1 plasmid sequences only shared 83–95 % of their sequences, but all harboured the same resistance aminoglycosides and sulphonamide resistance genes, plus the beta-lactamase gene blaSHV-12 and the tetA gene in two of them (Fig. 3c).
The chromosomal structure of isolates with IncHI2 plasmids was very similar. All strains in group 1 carried four aminoglycoside resistance genes [aacC5, aac(6)-Iaa, aadA7 and sul1] in the chromosome, while all the remaining genes, aforementioned, were found in the plasmids. The chromosomal resistance genes were located in the SGI-1 sequence, which presented a similar structure and content for all isolates, with an In4‐type complex integron on which aacC5, aadA7 and sul1 genes were encoded, and transposon Tn21 on which the mercury resistance operon (merEDACPTR) was encoded. Core genomes of the chromosomic contig of these seven isolates differed by 20–76 SNPs. Chromosomal structures of isolates carrying the IncHI2d plasmid group were also similar, differing only in 53 SNPs. In contrast, and similar to what was observed in the plasmid analysis, the three isolates carrying IncI1 plasmids were more different, presenting between 115 and 582 SNPs (Table S8).
Plasmids from the 12 strains were predicted as being conjugative according to MOBtyper. Seven IncHI2 plasmids (IncHI2a, IncHI2b and IncHI2c groups) presented MOBH relaxase type, two carrying the mcr-9 gene (IncHI2d) had both MOBH and MOBP relaxases, and all three IncI1 plasmids presented MOBP type (Table S6).
In addition, we characterized the integron content of the 12 strains sequenced with both Illumina and MinION technologies (Fig. 4). We identified four types of class 1 integrons in the 12 closed genomes, with seven strains containing three of them simultaneously (Fig. 4a). Together, these integrons contained eight resistance genes against seven different antimicrobials, namely chloramphenicol, trimethoprim, sulphonamides, quaternary ammonium compounds, and the aminoglycosides gentamycin, streptomycin and tobramycin (Fig. 4b). As mentioned previously, one of them (IC1) is in the chromosomal genomic island SGI-1. The other three (IP1, IP2 and IP3) are located in the IncHI2 and IncI1-type plasmid
Fig. 4.
(a) Integron content, strain distribution and variation among strains. (b) Schematic representation of the structure and content of integrons (IC: integron on the chromosome; IP: integron on a plasmid).
Discussion
Historically, Salmonella isolates belonging to serotype Kentucky have been rarely associated with human illness [61]. However, the recent emergence of the ciprofloxacin-resistant S. Kentucky ST198 clone has highlighted the potential of this serotype to become a public health concern [6]. The recent reports describing MDR S. Kentucky isolates circulating in livestock in several EU MSs [1] suggest the possible establishment of this clone in the European food animal production systems. However, the usefulness of serotype-based identification for epidemiological purposes is limited because it may lead to the clustering of highly unrelated strains [62]. For this reason, we performed the present study based on whole genome sequencing (WGS) analysis in an attempt to assess the genetic relatedness between S. Kentucky strains circulating in poultry species in Spain, given the recent reports of the presence of MDR isolates belonging to this serotype [24].
The WGS results confirmed that all the sequenced isolates belonged to ST198 and were, in fact, related to the described emerging clone, including those resistant phenotypically to a low (two or three) number of antimicrobials (non-AGSuT). Although the limited variability observed within the isolates analysed in this study hampers the assessment of their genetic relationship (as evidenced by the low bootstrapping values obtained in the internal branches of the phylogenetic tree, Fig. 2), at least three well-separated clades were identified (A, B, C). If only the mutations in the QRDRs in the parC and gyrA genes are considered, two groups were differentiated by the mutation in codon 87 of the gyrA gene, in agreement with previous studies describing two distinct phylogenetic groups based on that SNP [20]. Divergence of these two groups was dated to around the 2000s and different geographical distributions for each of them have been described, although all have been detected in European countries [11]. In our study, both genotypes were found in isolates coming from the three sampled hosts, but strains with the least represented genotype (D87Y) (classified in clade A) were predominantly retrieved from farms located in the North and to a lower extent in the South-east of Spain and during the last few years (2016–2017), while strains with the alternative D87N genotype originated from all sampled regions and were retrieved throughout the whole study period, being the only ones found in the first few years (Table 2). This may indicate a different spatial and temporal distribution of the D87N/D87Y genotypes in Spain, as previously described at a larger scale [11], suggesting more than one successful introduction of different ST198 S. Kentucky strains in the Spanish poultry population, as was also hypothesized in China based on the genetic diversity of isolates from human and food sources, indicating that multiple introductions of strains with different origins could have occurred [63].
The high number of resistance genes found in most isolates was expected since they were selected because of their MDR phenotype, and is largely in agreement with previous reports describing several resistance genes against multiple antimicrobial classes in this clone [11, 63, 64]. Although we had no information on phenotypic resistance to streptomycin, 16 isolates carried several genes conferring resistance to this antimicrobial [e.g. aph(3)-Ib and aph(6)-Id], suggesting a resistant phenotype as previously described for ST198 S. Kentucky isolates. However, several strains presented in addition an unusually high number of aminoglycoside resistance genes and/or genes conferring resistance to other antimicrobial classes less commonly reported in S. Kentucky ST198, such as phenicols (catA, cmlA1), trimethoprim (dfrA1) or polymyxins (mcr-9) (Fig. 1). Hybrid assembly analysis demonstrated that these genes were primarily associated with IncHI2 and IncI1 plasmids. Among the phenicol resistance genes, cmlA1 and catA1 had been occasionally reported before in S. Kentucky ST198 in plasmids [11, 63] but always at frequencies below 5 % compared with 9/66 in this study, and they had not been reported together. The trimethoprim dfrA1 gene, present in 8/66 of our strains (along with phenicol genes), had been reported only in one ST198 strain before together with an IncI1 plasmid replicon [64]. In contrast, other resistance genes to these antimicrobial classes previously reported (always at low frequencies) such as floR or dfrA12 were not found in our collection.
The high content of antimicrobial resistance genes in our strains was also associated with the presence of integrons. Integrons are major elements in AMR because they mobilize and stockpile more than 130 different resistance genes [65] and provide their bacterial host with adaptation on demand [66, 67]. All strains selected for MinION sequencing for their high resistance profile (12) possessed at least one integron, and the majority (seven) had three (Fig. 4). Our analysis illustrates the relevance of horizontal transfer of integrons, as their presence and content did not correlate with the genetic relationship of the isolates based on the phylogenetic analysis (Fig. 2). Even for the same integron we found variations between strains (Fig. 4a). Rearrangements mediated by insertion sequences (IS6 family) are known to disrupt integrons in clinical isolates [68] and seem to be at the basis of major rearrangements in the IncHI2 plasmid groups (Fig. 4a). It is important to note that AMR genes against almost all clinically relevant antimicrobial families have been identified in these genetic structures [65], emphasizing the importance of the correct identification and characterization of these platforms.
Most of the isolates carrying IncHI2 plasmids and integrons, and the catA1, cmlA1 and/or dfrA1 genes were also those with the highest number of aminoglycoside resistance genes (7/9). These isolates were retrieved from all hosts but predominantly from turkey (5/7), a species often associated with higher levels of MDR Salmonella infection [6, 24]. Interestingly, very similar plasmid sequences (100 % of coverage and >99.9 % of identity) were retrieved from isolates coming from flocks housed in the same farm over different years, and from flocks housed in different farms and sampled in different years (Table S1). Given that the chromosome sequences from the isolates harbouring these highly similar plasmids differed in 20–40 SNPs, it could be hypothesized that these S. Kentucky ST198 plasmids themselves were able to persist in the farm from which they were recovered over three different years. An alternative hypothesis would be that multiple introductions of strains carrying the plasmid from a contaminated source occurred, which could explain its presence in other farms in different years. The IncHI2 plasmids retrieved here shared ~99 % identity and between 96 and 99 % coverage with plasmid sequences harboured by Escherichia coli O157:H7 strains retrieved during a foodborne outbreak in the UK in 2012 [69] and from E. coli isolated from poultry meat in Italy [70]. These plasmids also carried a set of resistance genes against aminoglycosides, phenicol (cmlA1), trimethoprim (dfrA1) and other antimicrobial classes (Table S7), and were associated with increased fitness under certain environmental conditions in the O157:H7 strains and potentially linked to increased transmissibility/persistence of the carrier bacterial isolates [69], thus highlighting their clinical relevance. Given the role that IncHI2 plasmids can have in the acquisition and dissemination of ARGs in Salmonella and other Enterobacteriaceae [71], their detection in an already highly resistant Salmonella clone such as ST198 is a worrying finding since it could lead to decreased susceptibility to other antimicrobial classes, as hypothesized in China, where the circulation of ST198 S. Kentucky isolates harbouring IncHI2 plasmids that carried additional ARGs has also been described [63].
Despite the high frequency of resistance genes found in our collection, we did not find any of the carbapenem-resistance genes (bla OXA48 and bla NDM-1) previously reported in S. Kentucky clinical isolates from African countries [72, 73]. Similarly, only two out of 65 isolates carried ESBL genes (blaSHV-12 ), in contrast to what has been described for S. Kentucky isolates from turkey in Poland (carrying bla CTX-M-25) [21]. Similarly blaCTX-M-14b , found in a clone increasingly reported in humans in Europe [64] and in isolates from animal sources in Europe, Africa and Asia [64, 74, 75], was not detected in our study. The two isolates carrying the blaSHV-12 gene (in a very similar IncI1 plasmid) were genetically distinct (>600 SNPs), phenotypically resistant to cefotaxime and originated from turkey farms located in different provinces sampled in 2015. Plasmid sequences harbouring the blaSHV-12 gene were similar (>97 % coverage and >99 % identity) to other plasmids described in E. coli isolates from a German poultry farm in 2011 [76] and a Belgian broiler farm in 2013 [77, 78] (Table S7), thus suggesting its circulation in poultry populations in Europe and occasional carriage of ESBL genes [76].
The mcr9 gene found in two isolates in IncHI2 plasmids did not seem to confer phenotypic resistance to colistin even when exposed to increasing concentrations of this antimicrobial (data not shown), in agreement with previous results that suggest that its presence is not necessarily associated with a resistant phenotype [79–81].
In our panel, multiple variants of SGI-K, Q and P were observed in agreement with previous studies [11, 82], with strains with a less resistant phenotype presenting large deletions in this segment of the bacterial chromosome. Reorganization of SGI elements in this serotype did not impose an increased fitness cost under nutrient-limited conditions in Salmonella infection studies in an animal model, suggesting an adaptative advantage of this MDR strain [83]. In contrast, limited variability in the presence of virulence genes was found in our collection (Fig. S1) and most of T3SS (Type 3 Secretion System)-associated genes [84] were detected in all our strains, as commonly reported in isolates from this serotype. The reduced virulence of this serotype has been partially attributed to the absence of determined virulence genes such as sspH2 [18, 85], missing in the isolates analysed here. However, we found other virulence genes related to human infection (sopE, sopD2, pipB2, sseK2) [17, 18]. Therefore, the existence of poultry isolates carrying these genes deserves attention due to its potential ability to cause human infections.
From these data it is not possible to directly quantify the proportion or the underlying spatial distribution of S. Kentucky-positive farms in Spain, since isolates included in this study were part of the panel of Salmonella isolates tested in the frame of the AMR surveillance programme, which in turn is formed by a selection of isolates recovered through the national Salmonella control programmes, of which approximately half are selected by convenience sampling. Nevertheless, a previous study found no significant differences between the relative distribution of serotypes in Salmonella -positive farms in the routine auto-control checks and those included in the AMR surveillance panel [24]. Given that we analysed through WGS here between 25 and 38 % of all S. Kentucky isolates available from laying hen, broiler and turkey, our results probably approximate well the genetic variability in circulating S. Kentucky strains in poultry in Spain.
Our results indicate that the MDR S. Kentucky ST198 is present in all the investigated hosts in Spain, and that certain strains also carry additional plasmid and integron-mediated AMR genes, thus increasing its potential public health significance. Several isolates from poultry originating from other countries as well as from human and wildlife sources were genetically similar to the isolates sequenced in this study, demonstrating the dynamic and global transmission cycle of the ST198 clone and the involvement of multiple species in its epidemiology. The presence of MDR strains/plasmids in different flocks housed in the same or different farms over the years suggests that these may persist in the environment and/or be reintroduced from contaminated sources, possibly facilitated by a high antimicrobial selective pressure. Additional studies to clarify the epidemiology of this important serovar are needed in order to design preventive and mitigation measures.
Supplementary Data
Funding information
This work was partially supported by the European Union’s Horizon 2020 Research and Innovation programme under Grant Agreement No. 773830 (One Health European Joint Program) and the Spanish Ministry of Agriculture, Fisheries and Food. C.S.C. is funded with a predoctoral grant (reference CT19/20) from the Universidad Complutense de Madrid and Banco Santander. J.A. is the recipient of a Ramon y Cajal contract from the Spanish Ministry of Economy, Industry and Competitiveness (MINECO, RYC‐2016‐20422). F.T.R. holds a Ph.D. fellowship (SFRH/BD/144108/2019) from the Foundation for Science and Technology, Portugal.
Acknowledgements
We would like to acknowledge the work performed by the laboratory technicians from the ‘Foodborne Zoonoses and Antibiotic Resistance Unit’ (ZTA) at the VISAVET centre, and the Spanish Ministry of Agriculture, Fisheries and Food officers tasked with the routine sampling in the frame of the AMR surveillance programmes.
Author contributions
Conceptualization: J.A., J.L.S., M.A.M. Resources and investigation: C.S.C., M.U.R., S.C., J.L.S., C.dF., M.A. Data curation: C.S.C., B.D.R., M.U.R., S.L. Formal analysis and validation: C.S.C., B.D.R., F.T.R., S.L., E.E., J.A. Methodology and software: B.D.R., S.L., M.H., D.A. Project administration, funding acquisition and supervision: J.A. Writing original draft and visualization: C.S.C., B.D.R., F.T.R., J.A.E., J.A. Writing – review and editing: all authors. All authors contributed to the article and approved the submitted version
Conflicts of interest
The authors declare that there are no conflicts of interest
Ethical statement
All isolates used in this study were generated through the routine AMR monitoring surveillance programme conducted in the frame of the EU and national legislation, and therefore ethical approval was not required.
Footnotes
Abbreviations: AGSuT, ampicillin, gentamycin, sulfamethoxazole, tetracycline; AMR, antimicrobial resistance; ARG, antimicrobial resistance gene; BRIG, BLAST Ring Image Generator; ECOFFs, epidemiologic cut-offs; ENA, European Nucleotide Archive; ESBL, extended-spectrum beta-lactamase; EU, European Union; EUCAST, European Committee on Antimicrobial Susceptibility Testing; IC, integron on the chromosome; IP, integron on a plasmid; MDR, multidrug-resistant; MIC, minimum inhibitory concentration; MLST, multilocus sequence typing; MS, Member State; QRDR, quinolone-resistance-determining region; SGI, Salmonella genomic island; SNP, Single Nucleotide Polymorphism; ST, sequence type; T3SS, Type 3 Secretion System; VFDB, Virulence Factors Database; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Eight supplementary tables and one supplementary figure are available with the online version of this article.
References
- 1.European Food Safety Authority. European Centre for Disease Prevention and Control The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020;18:e06007. doi: 10.2903/j.efsa.2020.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015;12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. The Lancet Infectious Diseases. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messens W, Vivas-Alegre L, Bashir S, Amore G, Romero-Barrios P, et al. Estimating the public health impact of setting targets at the European level for the reduction of zoonotic Salmonella in certain poultry populations. Int J Environ Res Public Health. 2013;10:4836–4850. doi: 10.3390/ijerph10104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow PA, Methner U. Salmonella in Domestic Animals. CABI; 2013. Vaccination against Salmonella infections in food animals: rationale, theoretical basis and practical application; pp. 455–475. [Google Scholar]
- 6.Koutsoumanis K, Allende A, Alvarez-Ordóñez A, Bolton D, Bover-Cid S, et al. Salmonella control in poultry flocks and its public health impact. EFSA J. 2019;17:e05596. doi: 10.2903/j.efsa.2019.5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari RG, Rosario DKA, Cunha-Neto A, Mano SB, Figueiredo EES, et al. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: a Meta-analysis. Appl Environ Microbiol. 2019;85:e00591-19. doi: 10.1128/AEM.00591-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Threlfall EJ. Epidemic Salmonella typhimurium DT 104--a truly international multiresistant clone. J Antimicrob Chemother. 2000;46:7–10. doi: 10.1093/jac/46.1.7. [DOI] [PubMed] [Google Scholar]
- 9.European Food Safety Authority. European Centre for Disease Prevention and Control The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018;16:e05182. doi: 10.2903/j.efsa.2018.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO Critically important antimicrobials for human medicine, 6th revision. 2019. https://www.who.int/publications/i/item/9789241515528
- 11.Hawkey J, Le Hello S, Doublet B, Granier SA, Hendriksen RS, et al. Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb Genom. 2019;5 doi: 10.1099/mgen.0.000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Hello S, Hendriksen RS, Doublet B, Fisher I, Nielsen EM, et al. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J Infect Dis. 2011;204:675–684. doi: 10.1093/infdis/jir409. [DOI] [PubMed] [Google Scholar]
- 13.Le Hello S, Weill F-X, Guibert V, Praud K, Cloeckaert A, et al. Early strains of multidrug-resistant Salmonella enterica serovar Kentucky sequence type 198 from Southeast Asia harbor Salmonella genomic island 1-J variants with a novel insertion sequence. Antimicrob Agents Chemother. 2012;56:5096–5102. doi: 10.1128/AAC.00732-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joerger RD, Sartori CA, Kniel KE. Comparison of genetic and physiological properties of Salmonella enterica isolates from chickens reveals one major difference between serovar Kentucky and other serovars: response to acid. Foodborne Pathog Dis. 2009;6:503–512. doi: 10.1089/fpd.2008.0144. [DOI] [PubMed] [Google Scholar]
- 15.Johnson TJ, Thorsness JL, Anderson CP, Lynne AM, Foley SL, et al. Horizontal gene transfer of a ColV plasmid has resulted in a dominant avian clonal type of Salmonella enterica serovar Kentucky. PLoS One. 2010;5:e15524. doi: 10.1371/journal.pone.0015524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley SL, Johnson TJ, Ricke SC, Nayak R, Danzeisen J. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol Mol Biol Rev. 2013;77:582–607. doi: 10.1128/MMBR.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, Pedroso AA, Porwollik S, McClelland M, Lee MD, et al. rpoS-Regulated core genes involved in the competitive fitness of Salmonella enterica Serovar Kentucky in the intestines of chickens. Appl Environ Microbiol. 2015;81:502–514. doi: 10.1128/AEM.03219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhanani AS, Block G, Dewar K, Forgetta V, Topp E, et al. Genomic Comparison of Non-Typhoidal Salmonella enterica Serovars Typhimurium, Enteritidis, Heidelberg, Hadar and Kentucky Isolates from Broiler Chickens. PLoS One. 2015;10:e0128773. doi: 10.1371/journal.pone.0128773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westrell T, Monnet DL, Gossner C, Heuer O, Takkinen J. Drug-resistant Salmonella enterica serotype Kentucky in Europe. Lancet Infect Dis. 2014;14:270–271. doi: 10.1016/S1473-3099(14)70703-0. [DOI] [PubMed] [Google Scholar]
- 20.Le Hello S, Bekhit A, Granier SA, Barua H, Beutlich J, et al. The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front Microbiol. 2013;4:395. doi: 10.3389/fmicb.2013.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasyl D, Kern-Zdanowicz I, Domańska-Blicharz K, Zając M, Hoszowski A. High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying bla (CTX-M-25) gene. Vet Microbiol. 2015;175:85–91. doi: 10.1016/j.vetmic.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Wasyl D, Hoszowski A. First isolation of ESBL-producing Salmonella and emergence of multiresistant Salmonella Kentucky in turkey in Poland. Food Research International. 2012;45:958–961. doi: 10.1016/j.foodres.2011.07.024. [DOI] [Google Scholar]
- 23.Légifrance Arrêté du 1er août 2018 relatif à la surveillance et à la lutte contre les infections à Salmonella dans les troupeaux de l’espèce Gallus gallus en filière ponte d’œufs de consommation - Légifrance. 2021. https://www.legifrance.gouv.fr/jorf/id/JORFTEXT000037330841?r=xG21WLRFAo
- 24.Alvarez J, Lopez G, Muellner P, de Frutos C, Ahlstrom C, et al. Identifying emerging trends in antimicrobial resistance using Salmonella surveillance data in poultry in Spain. Transbound Emerg Dis. 2020;67:250–262. doi: 10.1111/tbed.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antilles N, García-Bocanegra I, Alba-Casals A, López-Soria S, Pérez-Méndez N, et al. Occurrence and antimicrobial resistance of zoonotic enteropathogens in gulls from southern Europe. Sci Total Environ. 2021;763:143018. doi: 10.1016/j.scitotenv.2020.143018. [DOI] [PubMed] [Google Scholar]
- 26.Anon Comission Regulation 517/2011 implementing Regulation 2160/2003 of the European Parliament and of the Council as regards a Union target for the reduction of the prevalence of certain Salmonella serotypes in laying hens of Gallus gallus and amending Regula. Official Journal of the European Union, L138, 7. 2021. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32011R0517
- 27.Anon Commission regulation 200/2012 concerning a Union target for the reduction of Salmonella enteritidis and Salmonella typhimurium in flocks of broilers, as provided for in Regulation 2160/2003 of the European Paliament and of the Council. Official Journal of the European Union, L71, 6. 2012. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32012R0200
- 28.Anon Comission Regulation 517/2011 implementing Regulation 2160/2003 of the European Parliament and of the Council as regards a Union target for the reduction of the prevalence of certain Salmonella serotypes in laying hens of Gallus gallus and amending Regula. Official Journal of the European Union, L349, 6. 2021. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32012R1190
- 29.Douard G, Praud K, Cloeckaert A, Doublet B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One. 2010;5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doublet B, Praud K, Bertrand S, Collard J-M, Weill F-X, et al. Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob Agents Chemother. 2008;52:3745–3754. doi: 10.1128/AAC.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006;8:1915–1922. doi: 10.1016/j.micinf.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 32.Hernández M, Iglesias MR, Rodríguez-Lázaro D, Gallardo A, Quijada N, et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 2017;22:30586. doi: 10.2807/1560-7917.ES.2017.22.31.30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Illumina Nextera XT DNA Library Prep Kit Reference Guide For Research Use Only. Not for use in diagnostic procedures. 2018. www.illumina.com/company/legal.html
- 34.Quijada NM, Rodríguez-Lázaro D, Eiros JM, Hernández M. TORMES: an automated pipeline for whole bacterial genome analysis. Bioinformatics. 2019;35:4207–4212. doi: 10.1093/bioinformatics/btz220. [DOI] [PubMed] [Google Scholar]
- 35.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Yang J, Yu J, Yao Z, Sun L, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33:D325–8. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, et al. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, et al. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft salmonella genome assemblies. PLoS One. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. RhierBAPS: An R implementation of the population clustering algorithm hierBAPS. Version 1. Wellcome Open Res. 2018;3:93. doi: 10.12688/wellcomeopenres.14694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 55.R Core Team R: a language and environment for statistical computing. 2021.
- 56.De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:1–10. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah DH, Paul NC, Guard J. Complete Genome Sequence of a Ciprofloxacin-Resistant Salmonella enterica subsp. enterica Serovar Kentucky Sequence Type 198 Strain, PU131, Isolated from a Human Patient in Washington State. Genome Announc. 2018;6:e00125-18. doi: 10.1128/genomeA.00125-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elnekave E, Hong SL, Lim S, Johnson TJ, Perez A, et al. Comparing serotyping with whole-genome sequencing for subtyping of non-typhoidal Salmonella enterica: a large-scale analysis of 37 serotypes with a public health impact in the USA. Microb Genom. 2020;6:1–13. doi: 10.1099/mgen.0.000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen H, Song J, Zeng X, Chen D, Chen R, et al. National prevalence of Salmonella enterica serotype Kentucky ST198 with high-level resistance to ciprofloxacin and extended-spectrum cephalosporins in China, 2013 to 2017. mSystems. 2021;6:e00935-20. doi: 10.1128/mSystems.00935-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coipan CE, Westrell T, Hoek A, Alm E, Kotila S, et al. Genomic epidemiology of emerging ESBL-producing Salmonella Kentucky bla CTX-M-14b in Europe. Emerg Microbes Infect. 2020;9:2124–2135. doi: 10.1080/22221751.2020.1821582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 66.Escudero JA, Loot C, Nivina A, Mazel D. Mobile DNA III. American Society for Microbiology; 2014. The Integron: Adaptation On Demand; pp. 139–161. [Google Scholar]
- 67.Souque C, Escudero JA, MacLean RC. Integron activity accelerates the evolution of antibiotic resistance. Elife. 2021;10:1–47. doi: 10.7554/eLife.62474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naas T, Aubert D, Lambert T, Nordmann P. Complex genetic structures with repeated elements, a sul-type class 1 integron, and the bla VEB extended-spectrum beta-lactamase gene. Antimicrob Agents Chemother. 2006;50:1745–1752. doi: 10.1128/AAC.50.5.1745-1752.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cowley LA, Dallman TJ, Fitzgerald S, Irvine N, Rooney PJ, et al. Short-term evolution of Shiga toxin-producing Escherichia coli O157:H7 between two food-borne outbreaks. Microb Genom. 2016;2:e000084. doi: 10.1099/mgen.0.000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zurfluh K, Klumpp J, Nüesch-Inderbinen M, Stephan R. Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-β-lactamase-producing Escherichia coli isolates of different origins. Antimicrob Agents Chemother. 2016;60:5589–5591. doi: 10.1128/AAC.00935-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.García Fernández A, Cloeckaert A, Bertini A, Praud K, Doublet B, et al. Comparative analysis of IncHI2 plasmids carrying bla CTX-M-2 or bla CTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob Agents Chemother. 2007;51:4177–4180. doi: 10.1128/AAC.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ktari S, Le Hello S, Ksibi B, Courdavault L, Mnif B, et al. Carbapenemase-producing Salmonella enterica serotype Kentucky ST198, North Africa. J Antimicrob Chemother. 2015;70:3405–3407. doi: 10.1093/jac/dkv276. [DOI] [PubMed] [Google Scholar]
- 73.Le Hello S, Harrois D, Bouchrif B, Sontag L, Elhani D, et al. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infect Dis. 2013;13:672–679. doi: 10.1016/S1473-3099(13)70124-5. [DOI] [PubMed] [Google Scholar]
- 74.Lei C-W, Zhang Y, Wang X-C, Gao Y-F, Wang H-N. Draft genome sequence of a multidrug-resistant Salmonella enterica serotype Kentucky ST198 with chromosomal integration of bla CTX-M-14b isolated from a poultry slaughterhouse in China. J Glob Antimicrob Resist. 2020;20:145–146. doi: 10.1016/j.jgar.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 75.European Food Safety Authority. European Centre for Disease Prevention and Control The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019;17:e05598. doi: 10.2903/j.efsa.2019.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alonso CA, Michael GB, Li J, Somalo S, Simón C, et al. Analysis of bla SHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J Antimicrob Chemother. 2017;72:1589–1596. doi: 10.1093/jac/dkx024. [DOI] [PubMed] [Google Scholar]
- 77.Lambrecht E, Van Meervenne E, Boon N, Van de Wiele T, Wattiau P, et al. Characterization of cefotaxime- and ciprofloxacin-resistant commensal Escherichia coli originating from Belgian farm animals indicates high antibiotic resistance transfer rates. Microb Drug Resist. 2018;24:707–717. doi: 10.1089/mdr.2017.0226. [DOI] [PubMed] [Google Scholar]
- 78.Lambrecht E, Coillie EV, Boon N, Heyndrickx M, Wiele TV de. Transfer of antibiotic resistance plasmid from commensal E. coli towards human intestinal microbiota in the M-SHIME: effect of E. coli dosis, human individual and antibiotic use. Life (Basel) 2021;11:192. doi: 10.3390/life11030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, et al. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio. 2019;10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Osei Sekyere J, Maningi NE, Modipane L, Mbelle NM. Emergence of mcr-9.1 in extended-spectrum-β-lactamase-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics, resistome, and mobilome. mSystems. 2020;5:e00148-20. doi: 10.1128/mSystems.00148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayer SS, Lim S, Hong S, Elnekave E, Johnson T, et al. Genetic determinants of resistance to extended-spectrum cephalosporin and fluoroquinolone in Escherichia coli isolated from diseased pigs in the United States. mSphere. 2020;5:e00990-20. doi: 10.1128/mSphere.00990-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Curraize C, Siebor E, Varin V, Neuwirth C, Hall RM. Two New SGI1-LK Variants Found in Proteus mirabilis and Evolution of the SGI1-HKL Group of Salmonella Genomic Islands. mSphere. 2020;5:e00875-19. doi: 10.1128/mSphere.00875-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cohen E, Davidovich M, Rokney A, Valinsky L, Rahav G, et al. Emergence of new variants of antibiotic resistance genomic islands among multidrug-resistant Salmonella enterica in poultry. Environ Microbiol. 2020;22:413–432. doi: 10.1111/1462-2920.14858. [DOI] [PubMed] [Google Scholar]
- 84.Deng W, Marshall NC, Rowland JL, McCoy JM, Worrall LJ, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 85.El Hage R, Losasso C, Longo A, Petrin S, Ricci A, et al. Whole-genome characterisation of TEM-1 and CMY-2 β-lactamase-producing Salmonella Kentucky ST198 in Lebanese broiler chain. J Glob Antimicrob Resist. 2020;23:408–416. doi: 10.1016/j.jgar.2020.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.