Abstract
Serratia marcescens is an important nosocomial pathogen causing various opportunistic infections, such as urinary tract infections, bacteremia and sometimes even hospital outbreaks. The recent emergence and spread of multidrug-resistant (MDR) strains further pose serious threats to global public health. This bacterium is also ubiquitously found in natural environments, but the genomic differences between clinical and environmental isolates are not clear, including those between S. marcescens and its close relatives. In this study, we performed a large-scale genome analysis of S. marcescens and closely related species (referred to as the ‘ S. marcescens complex’), including more than 200 clinical and environmental strains newly sequenced here. Our analysis revealed their phylogenetic relationships and complex global population structure, comprising 14 clades, which were defined based on whole-genome average nucleotide identity. Clades 10, 11, 12 and 13 corresponded to S. nematodiphila , S. marcescens sensu stricto, S. ureilytica and S. surfactantfaciens, respectively. Several clades exhibited distinct genome sizes and GC contents and a negative correlation of these genomic parameters was observed in each clade, which was associated with the acquisition of mobile genetic elements (MGEs), but different types of MGEs, plasmids or prophages (and other integrative elements), were found to contribute to the generation of these genomic variations. Importantly, clades 1 and 2 mostly comprised clinical or hospital environment isolates and accumulated a wide range of antimicrobial resistance genes, including various extended-spectrum β-lactamase and carbapenemase genes, and fluoroquinolone target site mutations, leading to a high proportion of MDR strains. This finding suggests that clades 1 and 2 represent hospital-adapted lineages in the S. marcescens complex although their potential virulence is currently unknown. These data provide an important genomic basis for reconsidering the classification of this group of bacteria and reveal novel insights into their evolution, biology and differential importance in clinical settings.
Keywords: Serratia marcescens complex, genomics, population structure, hospital-adapted lineage, antimicrobial resistance
Impact Statement
Serratia marcescens causes various opportunistic infections. Although S. marcescens and its close relatives are widely distributed in natural environments, their genomic differences have yet to be elucidated. Here, we present the results of a large-scale and comprehensive genome analysis of S. marcescens and its closely related species (referred to as the ‘ S. marcescens complex’ in this study). Our analysis revealed the global population structure of the complex, comprising 14 distinct clades, which will be an important basis for its reclassification in the future, and we identified two hospital-adapted clades. These two clades are clearly distinguished from S. marcescens sensu stricto and should be paid special attention because of their marked accumulation of a wide range of antimicrobial resistance genes, including various extended-spectrum β-lactamase and carbapenemase genes and mutations conferring fluoroquinolone resistance.
Data Summary
The raw sequences obtained in this study have been deposited in GenBank/EMBL/DDBJ under the BioProject accession number PRJDB10568 and Sequence Read Archive (SRA) accession numbers DRR253307 to DRR253531. All supporting data, codes and protocols have been provided within the article or through supplementary data files. Twelve supplementary tables and nine supplementary figures are available with the online version of this article.
Introduction
Serratia marcescens (Sma) is a Gram-negative bacterium that is now recognized as an important opportunistic pathogen. It causes a range of nosocomial infections, such as pneumonia, urinary tract infections, bacteremia, meningitis and endocarditis, particularly in immunocompromised patients [1]. Many hospital outbreaks of this opportunistic pathogen, which sometimes occur recurrently, have also been reported [2–8]. While Sma is known to be naturally resistant to penicillins and first- and second-generation cephalosporins, strains showing higher levels of antimicrobial resistance (AMR) have also emerged and spread widely [9, 10]. In particular, strains producing extended-spectrum β-lactamases (ESBLs) and carbapenemases are highly problematic, as they are resistant to a wide range of β-lactams, thus posing serious problems in treating infections caused by this bacterium [11, 12].
Sma is ubiquitous in nature, being found in both water and soil, and it is associated with plant surfaces, insects and animals [13, 14]. Starting from the first whole-genome sequence (WGS) determination and detailed genomic comparison of two Sma strains (Db11, an isolate from an insect source; and SM39, a multidrug-resistant clinical isolate) [15], both of which were used as representative strains of Sma in this study, several comparative genome analyses have been reported. They included a large-scale genomic analysis of multidrug-resistant (MDR) clones in the UK and Ireland [9] and more recently published studies that analysed many publicly available WGSs [16–18]. The results of three recent studies indicated the presence of clinical isolate-enriched lineages, but WGSs of environmental isolates were limited in the database. Thus, it appears that genomic differences between clinical isolates and those inhabiting natural environments have not yet been fully understood.
Taxonomically, the genus Serratia has recently been moved from the family Enterobacteriaceae to the newly proposed family Yersiniaceae [19], but there are also various debates regarding the species/subspecies classification in the genus, in which more than 21 species/subspecies have been proposed. For example, although S. marcescens subspecies sakuensis (Sma_sak) has been proposed, recent studies indicated that there is no merit in its separation to subspecies status [20–22]. Although average nucleotide identity (ANI) analysis using WGSs is now widely used in bacterial and archaeal taxonomy [23, 24], the relationship of Sma and its close relatives, such as S. nematodiphila (Sne) and S. ureilytica (Sur), has not yet been well elucidated even in the above-mentioned comparative genome analyses [16–18, 22].
Herein, we performed a large-scale genome analysis of Sma and closely related species, which we referred to as the ‘ S. marcescens complex (Sma complex)’, including 108 soil and 117 clinical isolates collected and sequenced in this study. Our analysis revealed their phylogenetic relationships and global population structure, which consisted of 14 clades defined based on the results of whole-genome ANI analysis. The 14 clades showed different genomic properties and distinct trends related to isolation sources. Based on the results, we propose the presence of two hospital-adapted clades in the Sma complex. The marked accumulation of AMR genes and mutations conferring fluoroquinolone resistance (FQR) is also demonstrated in these clades.
Methods
Analyses of the type strains of genus Serratia
We downloaded the genome sequences of the type strains of 16 species and one subspecies belonging to genus Serratia from the NCBI Assembly database (last access: 21 January 2021). These sequences and those of two completely sequenced Sma strains, SM39 and Db11 [15], were used for core-gene-based phylogenetic analysis, with the type strain of Y. enterocolitica (NCTC 12982T) as an outgroup (see Table S1, available in the online version of this article, for the list of type strains used). The core genes, which were defined as those present in all 20 strains (n=801), were identified using Roary version 3.7.0 [25] with a 60 % blastp identity cutoff, and a core-gene alignment was generated using mafft [26]. Using the 296 627 SNPs identified in the alignment, a maximum-likelihood (ML) phylogenetic tree was constructed using RAxML [27] with the GTR-GAMMA model of nucleotide substitution and 500 bootstrap replicates. The tree was displayed and annotated using ggtree version 2.2.4 [28] in the R package. ANI was calculated using PYANI version 0.2.7 [29] in ANIb mode. The 16S rRNA sequences of each strain were identified by blastn searches using the consensus 16S rRNA reference sequence of Sma SM39, which was obtained by aligning the sequences of seven copies using Clustal Omega [30] and created with the cons command in the emboss package [31]. The 16S rRNA consensus sequences of other strains were obtained via the same procedure. As the finished genome sequence of the Sma-type strain was not available, we used the sequence of strain SM39 as a reference in this analysis.
Isolation and genome sequencing of Sma strains from soil samples
We collected soil samples (approximately 10 g for each) in several regions in Japan and in Toulouse, France. Each sample was suspended in 30 ml saline in a 50 ml tube, vortexed for 3 min, and centrifuged at 100 g for 5 min at 4 °C. The supernatant was filtered through several sheets of sterilized gauze, centrifuged at 4000 g for 15 min at 4 °C, and inoculated onto a deoxyribonuclease-toluidine blue-cephalothin (DTC) agar plate [32], followed by a 24 h incubation at 37 °C under aerobic conditions. When colonies with a purple halo formed, one of the colonies was picked and inoculated onto a fresh DTC agar plate. After a 24 h incubation at 37 °C, a single colony was isolated from the plate and used for subsequent analyses. For species identification by 16S rRNA sequence determination, colonies were suspended in 50 µl of 10 mM Tris-HCl (pH 8.0) buffer in 1.5 ml tubes, boiled at 100 °C for 5 min and rapidly cooled on ice. Samples were centrifuged at 18 700 g for 10 min at room temperature to obtain supernatants to be used as PCR templates. PCR was performed using the KAPA Taq EXtra PCR kit (Nippon Genetics, Tokyo, Japan) and the 27F and 1492R universal primers. Each reaction mixture for PCR (15 µl) contained 1 µl of template DNA, 0.75 µl of each primer (10 µM), 0.45 µl of dNTPs, 1.5 µl of MgCl2, 3 µl of 5×EXtra buffer, and 0.075 µl of KAPA Taq EXtra DNA polymerase. The PCR conditions employed were as follows: 96 °C for 2 min; 20 cycles of 1 min at 96 °C, 1 min at 55 °C, and 2 min at 72 °C; and 72 °C for 5 min. The PCR products were analysed by 1.5 % agarose gel electrophoresis to confirm the generation of ca. 1.5 Kb amplicons. The amplicons were sequenced with capillary sequencers (Applied Biosystems 3130xL or 3500xL Genetic Analyzers, Thermo Fisher Scientific Japan, Tokyo, Japan) and compared to the 16S rRNA sequence of SM39 by using blastn. Isolates showing >99 % sequence identity were used as Sma strains in this study. According to this procedure, we collected 108 soil strains in Japan and France. In addition, we obtained 117 clinical strains from various hospitals in Japan and France.
For genome sequencing, strains were cultured in Lysogeny broth overnight at 37 °C, and genomic DNA was purified from the cultures using a DNeasy Blood and Tissue kit (Qiagen, CA, USA). Sequencing libraries were prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, USA) and sequenced using the Illumina MiSeq platform to generate 300 bp paired-end reads.
Collection of publicly available genome sequences
We searched the NCBI database (https://www.ncbi.nlm.nih.gov) for the genomes registered as Sma, Sma_sak, Sne and Sur and found 844 Sma, 4 Sma_sak, 8 Sne and 2 Sur genomes (last access: 23 January 2019). From this dataset, we selected 644 genomes with records on country information and isolation sources. In particular, we included the genomes of type strains even if their strain information was not available.
Genome assembly, quality filtering and annotation
The genomes of 171 Japanese and 54 French strains sequenced in this study were assembled using Platanus_B assembler version 1.1.0 [33]. Of the 644 genome sequences obtained from the NCBI database, only assembled sequences were available for 176 genomes. As raw-read sequences were available for the remaining 468 genomes, these sequences were also assembled using the Platanus_B assembler. To remove low-quality sequences from those assembled from short reads, we first removed the sequences if the total number of scaffolds was greater than 300 or the read depth was less than 20 (n=13). Then, 79 genomes judged to be of low quality by CheckM [34] with a cutoff of <99 % completeness or >3 % contamination were removed (see Fig. S1 for the entire process of genome assembly and quality filtering). Two additional genomes were removed because they shared identical core-gene sequences with other strains in the strain set. Thus, we analysed a total of 775 strains (Table S2). For annotation, we used the DNA Data Bank of Japan (DDBJ) Fast Annotation and Submission Tool (DFAST) [35]. The sequencing statistics of the strains sequenced in this study are shown in Table S3.
ANI-based clustering of Sma complex strains and clade definition
ANI analysis was performed using pyani version 0.2.7 as described above. Based on the all-to-all ANI matrix, strains were clustered via the complete linkage clustering method, and a dendrogram was generated with the hclust command in R. Clades were defined based on minimum within-node pairwise ANI scores with a 97 % threshold.
Phylogenetic analysis and pangenome clustering of the Sma complex strains
Core genes (n=2,488), defined as those present in 99 % of the strains analysed, were identified using Roary version 3.7.0 with an 80 % blastp identity cutoff, and a core-gene sequence alignment was generated using mafft. Based on the 182 520 SNPs identified in this alignment, an ML phylogenetic tree was constructed using RAxML, and displayed and annotated using ggtree version 2.2.4, as described above. Clustering analysis based on the presence/absence matrix of the pangenome (34 342 genes) determined by Roary version 3.7.0 was performed using Ward’s clustering method [36] and Euclidean distance with the hclust command in R, and the pangenome dendrogram was displayed using ggtree version 2.2.4. The presence/absence matrix of the pangenome is shown in Table S4.
Analysis of the phylogenetic relationships of the strains used in this study with the type strains in the Sma complex
To analyse the phylogenetic relationships of the strains used in this study with the type strains of the Sma complex and its close relatives, we selected all strains in clades 13 (n=9) and 14 (n=1), the strains that were registered as Sma_sak, Sne, or Sur (strain IDs; DL654, DL655 and DL659, respectively), one representative strain from each clade other than clades 13 and 14, and the type strains of Sma, Sma_sak, Sne, Sur and S. surfactantfaciens (Ssu). The type strain of S. ficalia was also included as an outgroup. In this strain set, core genes (n=2790) defined as those present in all 30 strains were identified using Roary version 3.7.0 with a 60 % blastp identity cutoff, and a core-gene sequence alignment was generated using mafft. An ML phylogenetic tree was constructed based on the SNPs (n=562 767) identified in the alignment using RAxML as described above. Pairwise ANI scores were also calculated using pyani as described above.
Analysis of the distribution of the prodigiosin biosynthesis gene cluster
The prodigiosin biosynthetic cluster [37] was identified with diamond version v0.9.21 [38] in blastx mode, with the amino acid sequences of the pigA - pigN gene products of the Sma strain ATCC 274 (GCA_009936295.1) [39] as a reference (Table S5). blastx hits with ≥50 % coverage and ≥80 % sequence identity were considered to indicate presence. The nucleotide sequence identities of the pig gene clusters (from the start codon of pigA to the stop codon of pigN) were further analysed in all the pig-positive strains except for two (DL253 and DL256; both belonging to clade 10) whose clusters were separated into two scaffolds. The all-to-all nucleotide sequence identities of the pig gene clusters of 85 strains were determined based on the alignment generated by Clustal Omega with the sequences of strains ATCC 274 and ATCC 39006 (GCA_000463345.3) as references.
Strain clustering based on SNP distances (determination of SNP10 clusters)
The strain set used in this study included many subsets comprising very closely related strains, which could affect statistical analyses. To reduce this potential effect of strain bias, we performed strain clustering based on the SNP distances between each strain. The SNP distances in the core-gene alignment used for phylogenetic analysis were calculated using snp-dists version 0.4 (https://github.com/tseemann/snp-dists). Based on the SNP distances, the strains were clustered by Ward’s method, and a dendrogram was generated with the hclust command in R. The maximum SNP distance in each cluster was identified, and clusters with a maximum within-cluster SNP distance of ≤10, ≤20 or ≤30 were defined as SNP10, SNP20 and SNP30 clusters, respectively. We randomly selected one strain from each multimember SNP10 cluster as a representative strain for analyses. The details of the SNP10, SNP20 and SNP30 clusters and the representative strains used are shown in Table S2.
Analyses of plasmid replicons, integrase genes, potentially virulence-related genes and the accessary genes specifically conserved in clades 1 and 2
Plasmid replicons were identified by PlasmidFinder [40]. Integrase genes were identified in the pangenome data generated by Roary. To identify the potentially virulence-related genes described in a previous study that compared the SM39 and Db11 genomes [15], the annotation data of SM39 and Db11 genomes (locus IDs) were converted to those in the DFAST annotation data, and corresponding genes/gene clusters were identified in the pangenome data. To identify the accessary genes specifically conserved in clades 1 and 2, accessary genes that were significantly more frequently present in the two clades than the other clades were identified by Scoary version 1.6.16 (Bonferroni P<0.01) [41], and among these genes, those present in >90 % of the strains in both clades 1 and 2 were identified.
Statistical analyses
Statistical analyses other than the above-mentioned analysis performed by Scoary were performed in R version 4.1.0 [42]. One-way analysis of variance (ANOVA) followed by the Tukey–Kramer multiple comparison test was performed for the comparison of genome sizes between clades and that of GC contents between clades. Simple linear regression analysis was performed to analyse the relationship between the genome sizes and GC contents of each strain. Fisher’s exact test [43] with Benjamini–Hochberg correction [44] was performed to determine the difference in the ratio of clinical/hospital environment strains between the entire dataset and each clade.
Identification of AMR genes, FQR mutations and MDR strains
AMR genes were identified by SRST2 [45] using ARG-ANNOT_v3 (https://www.mediterranee-infection.com/wp-content/uploads/2019/03/arg-annot-nt-v3-march2017.txt) as a database. To analyse the genome sequences for which short-read sequences were not available, we used the Illumina MiSeq v3 100 bp paired-end simulated short-read data of each genome (insert size; 300 bp with a standard deviation of ±10 bp, read depth; 100×) generated by art version 2.5.8 [46]. When a gene was predicted as ‘-?-’ by SRST2, the presence of the gene was further analysed by a blastn search [47] against the assembled genome sequence, and the presence/absence decision was made with the thresholds of sequence identity ≥90 % and coverage ≥80 %. To search for FQR mutations, the amino acid sequences of the gyrA, gyrB, parC and parE gene products of each strain were aligned with those of Escherichia coli strain K-12 (NC_000913.3) using Clustal Omega [30], and FQR-associated point mutations in the quinolone-resistance-determining regions (QRDRs) of these gene products were identified [48, 49]. MDR strains were defined according to the definition proposed by Magiorakos et al. [50] as strains that were predicted to potentially be resistant to three or more classes/categories of antimicrobials based on the presence of AMR genes and FQR mutations. In this analysis, because Sma is known to be naturally resistant to penicillins, penicillins+β-lactamase inhibitors, nonextended spectrum cephalosporins (first- and second-generation cephalosporins) and polymyxins [51], these classes were excluded from the analysis. In addition, because the presence of the aac(6)-Ic gene is not associated with aminoglycoside resistance due to its low expression and Sma strains are usually sensitive to aminoglycosides [51], aac(6)-Ic was excluded. Therefore, among the 77 AMR genes identified in the entire strain set, those conferring resistance to the following seven classes of antimicrobials were used to define MDR: aminoglycosides, extended-spectrum cephalosporins (third- and fourth-generation cephalosporins), carbapenems, phenicols, tetracyclines, sulfonamides/trimethoprim and fluoroquinolones. We considered a strain as being resistant to an antimicrobial class if the strain possessed at least one resistance gene related to the class (in the case of FQR, at least one FQR mutation in gyrA).
Programs, parameter settings and codes used in this study
Programmes, parameter settings and codes to run each analysis pipeline used in this study are listed in Table S6. The in-house programmes and their codes used are available in GitHub (https://github.com/itTan-git/SMAscript).
Results
Phylogenetic relationships of Sma with closely related Serratia species
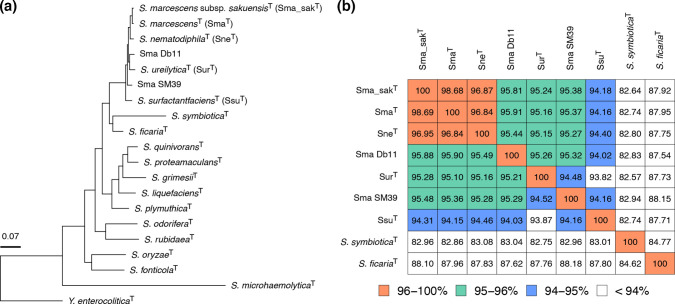
To investigate the phylogenetic relationships between Sma and its close relatives in the genus Serratia , we performed a core-gene-based phylogenetic analysis of the type strains of 17 Serratia species/subspecies and two completely sequenced Sma strains (SM39 and Db11) using the type strain of Y. enterocolitica as an outgroup (Table S1). In the constructed ML tree, the type strains of Sma, Sma_sak, Sne, Sur and Ssu formed a cluster distinct from those of other Serratia species. Strains SM39 and Db11 were included in this cluster, but they did not form a subcluster with the Sma-type strains (Fig. 1a). Although the type strain of Ssu showed ANI scores lower than 95 % to all other strains, the pairwise ANI scores between the other strains were over 95%, except for that between SM39 and the type strain of Sur (94.5 %) (Fig. 1b). Based on these results, we tentatively defined the three species, Sma, Sne and Sur, as the ‘Sma complex’, and we collected and analysed the genome sequences of strains belonging to this group.
Fig. 1.
Phylogenetic relationships between Serratia species. (a) An ML phylogenetic tree based on the core gene SNPs (296 627 sites in 801 genes) of the type strains of 17 Serratia species/subspecies and two completely sequenced S. marcescens strains (SM39 and Db11), with the Y. enterocolitica type strain as an outgroup. The core genes that are present in all strains were identified using Roary with a 60 % blastp identity cutoff. (b) ANI matrix among the nine strains of S. marcescens (Sma) and its close relatives. All-to-all ANI values were calculated using PYANI in ANIb mode, and cells are coloured according to ANI values. Strains other than Db11 and SM39 are type strains of each species/subspecies.
Strain dataset for the phylogenetic analysis of the Sma complex
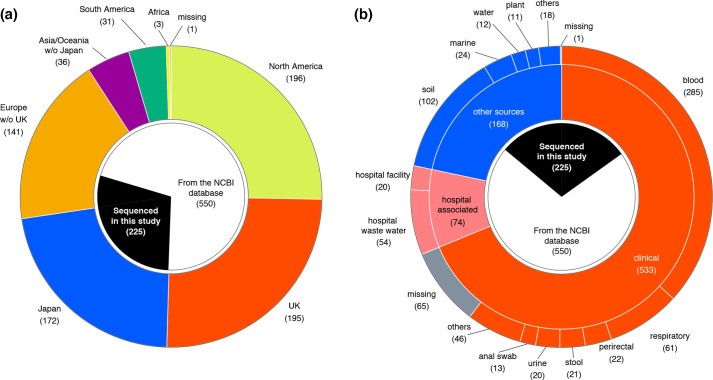
We collected the genome sequences of strains belonging to the Sma complex from various geographic regions and isolation sources (see Methods and Fig. S1). We isolated 108 Sma strains from soil samples collected in Japan and France and 117 Sma clinical strains from multiple hospitals in Japan and France and obtained their draft genome sequences. In addition, we collected the genome sequences of 644 strains from the NCBI database that were registered as species/subspecies belonging to the Sma complex along with their metadata (countries and sources). The type strains of the Sma complex species and completely sequenced strains were included in the dataset even if sufficient metadata were not recorded. After quality filtering and removing strain redundancy, the final dataset included 775 strains (225 sequenced in this study and 550 from the NCBI database; Table S2). These strains were isolated in 29 countries (divided into seven geographic regions in Fig. 2a). Approximately 43 % of the strains were isolated in Europe (n=336), of which more than half of them were from the UK (n=195), many of which were analysed in a population study of Sma isolated from cases of bacteremia [9]. Regarding the isolation sources, 68.8 % were clinical isolates (n=533), and 9.5 % were isolated from hospital environments (n=74). The remaining 21.7 % (n=168) were isolated from other sources, mostly from natural environments (Fig. 2b). The strains sequenced in this study represented 64.7 % of the last group, thus markedly expanding the genome resources of environmental strains in the Sma complex.
Fig. 2.
Regions and sources of the isolation of the Sma complex strains. (a) Distribution of the regions where strains were isolated. Regions were classified into seven geographic regions: the UK, Japan, North America, South America, Europe other than the UK (Europe w/o UK), Asia/Oceania other than Japan (Asia/Oceania w/o Japan), and Africa. (b) Distribution of isolation sources. Subcategories that contained <10 strains were grouped as ‘others’. The proportion of strains that were sequenced in this study is indicated in the most inner circles in both panels.
ANI-based clustering of the Sma complex strains
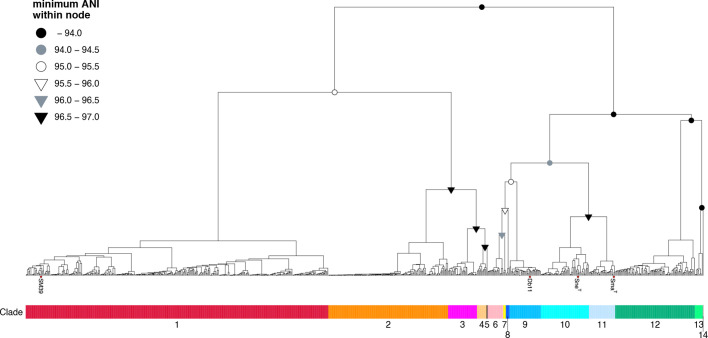
We first classified the 775 strains based on their pairwise ANI scores. By using the minimum within-node ANI of 97 % as a threshold, which was employed because the ANI score between the type strains of Sma and Sne was 96.84 %, the strains were divided into 14 clades (Fig. 3). In this study, we isolated and performed genome sequencing of the strains whose 16S rRNA sequences showed >99 % identity to that of strain SM39, and we collected the genome sequences of strains registered in the NCBI database as the species/subspecies belonging to the initially defined Sma complex using a >95 % ANI threshold. However, the minimum ANI scores between clades were below 95 % in several cases (indicated by black and grey filled circles in Fig. 3). This result indicated that our strain set included more diverse strains than we initially expected and highlighted the difficulty of the species identification of strains belonging to the Sma complex and its close relatives.
Fig. 3.
ANI-based clustering of the Sma complex strains. Complete-linkage unsupervised hierarchical clustering of the Sma complex strains was performed based on their all-to-all ANI analysis. The type strains of S. marcescens (SmaT), S. nematodiphila (SneT), and two Sma strains (Db11 and SM39) are indicated by their names. Nodes whose minimum pairwise ANI scores were <97 % are indicated by coloured symbols according to minimum within-node ANI scores. Fourteen clades defined according to the minimum within-node ANI threshold of 97 % are indicated under the tree.
Core-gene-based phylogenetic analysis of the Sma complex and the relationships between clades and known species
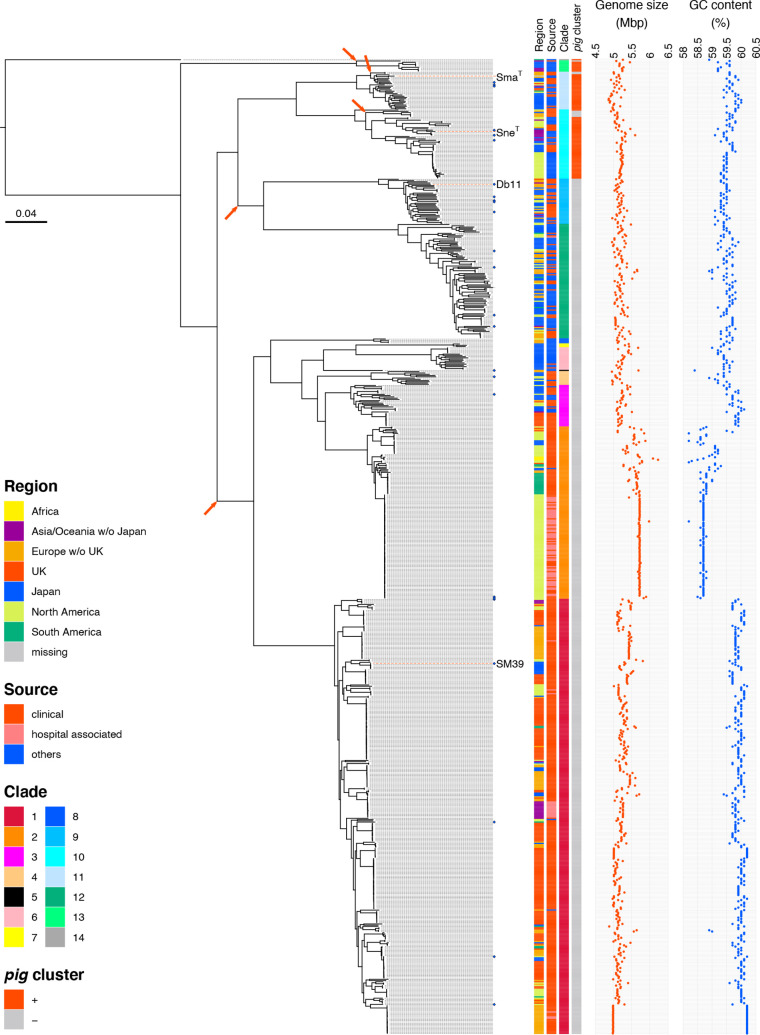
Next, we performed a core-gene-based phylogenetic analysis of our strain dataset (Fig. 4). By mapping the clade information to the phylogenetic tree, we confirmed that the strains belonging to the same clade clustered together and formed monophyletic clusters.
Fig. 4.
Phylogenetic relationships among the Sma complex strains. The phylogenetic relationship between the Sma complex strains analysed in this study (n=775) is shown. An ML tree was constructed based on the 182 520 SNP sites identified in 2488 core genes. The root was defined by a similar phylogenetic analysis using the type strain of S. ficalia as an outgroup. Blue diamonds indicate the strains whose complete genome sequences are available. The type strains of Sma and Sne and two Sma strains (Db11 and SM39) are indicated by their names and red dotted lines. For each strain, the isolation region, isolation source, clade, presence/absence of the pig gene cluster, total genome size and GC content are shown. Branches where the deletion of the pig gene cluster occurred are indicated by arrows.
The type strains of Sma (ATCC 13880T) and Sne (CGMCC 1.6853T) belonged to clades 11 and 10, respectively. Although the type strains of Sma_sak (DSM 17174T) and Sur (CCUG 50595T) were not included in the strain set because their sequences were not available when we designed this study, the phylogenetic analysis of these type strains with representative strains of each clade revealed that the Sma_sak type strain belonged to clade 11 together with the Sma-type strain and indicated that the Sur-type strain belonged to clade 12 (Fig. S2). The former finding is consistent with a recent report [22]. Notably, strain CRK0003, which was registered as Sne in the database, belonged to clade 3 (not to clade 10) and showed 94.95 % ANI to the Sne-type strain belonging to clade 10 (Fig. S2). On the other hand, strain K27, which was registered as Sma_sak, belonged to clade 10 and showed 98.44 % ANI to the Sne-type strain but 96.72 and 96.69 % to the type strains of Sma and Sma_sak, respectively.
Clades 13 (n=9) and 14 (n=1) were distantly related to the other clades; their ANI scores relative to all other clades were lower than 94 %. The phylogenetic analysis of these strains with representative strains of other clades and the type strains of species closely related to the Sma complex (Ssu and S. ficalia) revealed that clade 13 corresponded to Ssu (Fig. S2), while the 16S rRNA sequences of the clade 13 strains and the Ssu-type strain showed >99.44 % identity to the sequences of the Sma complex (Fig. S3). This analysis also revealed that clade 14 (single-member clade) represented a lineage that was more distantly related to the initially defined Sma complex (clades 1–12) than Ssu (clade 13) and was most closely related to S. ficalia among the 14 clades, but the 16S rRNA sequence also showed >99.03 % identity to those in the finished genomes in the Sma complex and 98.77 % to that of the S. ficaria -type strain. The clade 14 strain was isolated from Rhyncholacis pedicillata in the Guayana region of Venezuela [52] and was shown to have anti-oomycete, antifungal and antibacterial activities [53].
Based on these findings, we redefined the Sma complex to include Ssu (clade 13) and clade 14.
Pangenome clustering of Sma complex strains
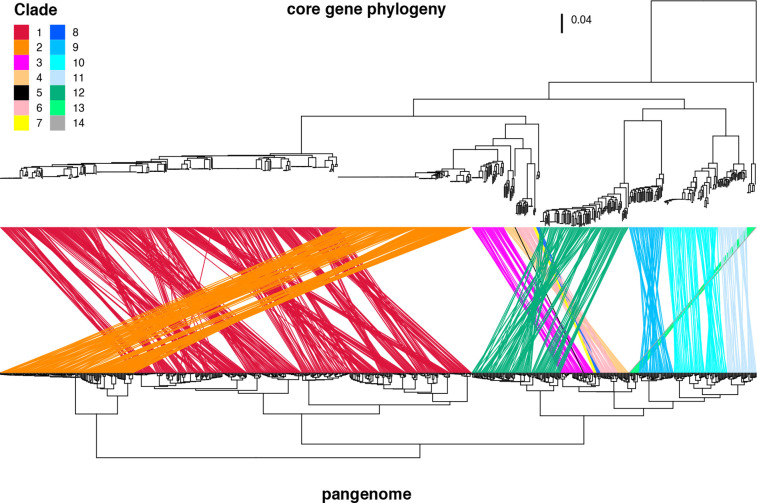
To examine the differences and similarities of the accessary gene contents within each clade and between clades, we performed a pangenome-based clustering analysis of the Sma complex strains (Fig. 5). The clustering of the strains by this method was consistent with that obtained via the ANI-based approach; strains belonging to the same clade clustered together. This result indicates the presence of accessary genes unique to each clade or shared by clades in an evolutionary context and corroborated the presence of the 14 clades defined based on the results of ANI analysis.
Fig. 5.
Relationship between the core-gene-based phylogeny and the pangenome-based clustering of the Sma complex strains. The upper tree is the core-gene-based ML tree shown in Fig. 4, and the lower dendrogram was constructed based on the cluster analysis of the pangenome (n=34 342) identified by Roary. Ward’s clustering method with Euclidean distance was used for the cluster analysis. The same strains present in the two trees are connected by coloured lines according to their clades.
Distribution of the pig gene cluster
The production of a red pigment known as prodigiosin is a well-known characteristic of Sma [1], but it was reported that the prodigiosin biosynthesis gene cluster (pigA - pigN) is not present in all Sma strains [54]. The analysis of the distribution of the pig gene cluster in our strain dataset revealed that the pig gene cluster was distributed only in clades 10 (corresponding to Sne), 11 (Sma and Sma_sak), 13 (Ssu) and 14 (Fig. 4). All pig-positive strains contained a full set of pig gene cluster comprising 14 genes in the chromosome region upstream of the copA gene encoding a cupper-translocating P-type ATPase. The nucleotide sequences of these gene clusters were highly conserved, showing >98.2 % identity to the cluster of Sma strain ATCC 274 [39], except for the genes of clade 14 (92.4 %) and one clade 10 strain (ADJS-2C_Purple, SAMN08373108; 80.4%), but notably lower similarities (<60 %) to the pig gene cluster found in the Serratia sp. strain ATCC 39006 [55]. The low sequence identity of the ADJS-2C_Purple genes appeared to be due to low sequence quality of this chromosome region. These findings indicate that the pig gene cluster was acquired by the common ancestor of the Sma complex and has been deleted from most clades in the Sma complex. The distribution of pig-negative strains in the phylogenetic tree shown in Fig. 4 indicated that the deletion of the pig gene cluster occurred at least five times in the strain set analysed in this study (indicated by arrows in Fig. 4, including the deletions in three sublineages/strain in clades 10, 11 and 13).
Inter- and within-clade variations in genome size and GC content related to horizontal gene transfer
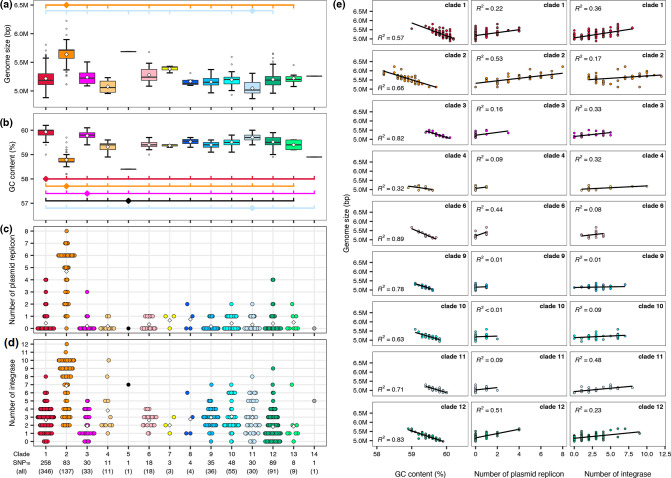
Notable interclade differences in genome sizes and GC contents were observed (Fig. 4). However, clades 1 and 2 contained many subclades comprising very closely related strains. To reduce such strain bias, we grouped strains with less than a 10-SNP distance into one cluster (named the SNP10 cluster) and then randomly selected one strain from each multimember cluster for clade comparison. Among the 619 SNP10 clusters identified, 515 were single-member clusters and 93 contained two or three members, while 11 clusters contained four or more members (up to seven). Notably, limited numbers of SNP10 clusters showed marked within-cluster variations in genome size and GC content (Fig. S4). In addition, there were no clusters that contained strains isolated in different countries while several clusters comprised strains that were isolated 8–10 years apart in different countries when the SNP20 distance metric was used (see Table S2 for all strain-clustering information).
Statistical analysis using representative strains from the 619 SNP10 clusters revealed that the genome size of clade 2 was significantly larger than those of other clades (excluding clades 5, 7 and 14, which were single- or three-member clades), and clade 11 contained significantly smaller genomes than the seven other clades (Fig. 6a). Conversely, the GC content of clade 2 was significantly lower than those of all other clades except for two single-member clades, and the GC contents of clades 1 and 3 were higher than those of most other clades (Fig. 6b). The GC contents of the clade 11 genomes were lower than those of clades 1 and 3 but higher than those of seven clades. Further analysis of nine clades containing >10 members revealed a negative correlation between genome size and GC content in all nine clades (R2 =0.32–0.89) (Fig. 6e).
Fig. 6.
Interclade differences in genome size and GC content. Genome size ranges (a) and GC content ranges (b) of SNP10 clusters belonging to each clade. For multimember SNP10 clusters, one strain was randomly selected from each cluster. Open diamonds in the boxplot indicate the mean values of each clade. Clades that show significant differences (P<0.01) compared to the clades marked by diamonds are indicated by short vertical lines. Lines are coloured according to the clade indicated by diamonds. Only the combinations that show significant interclade differences are shown. The numbers of total strains and SNP10 clusters in each clade are shown at the bottom of (d). (c, d) The numbers of plasmid replicons and integrase genes found in the SNP10 clusters belonging to each clade. Diamonds indicate the averages in each clade. (e) Correlations between genome size and GC content (left) and between genome size and the numbers of plasmid replicons (middle) or integrase genes (right) in nine clades that contained >10 members. Linear regression lines and R-squared results are shown. In all panels, colours for each clade are the same as those in Figs. 3–5.
Because the Sma complex members showed relatively high GC contents (ranging from 58.2–60.2 % in this study), it is most likely that the lower GC contents of large genomes are due to the acquisition of more genes from other species via horizontal gene transfer [56, 57]. To confirm this possibility, we analysed plasmid replicons and integrase genes (genetic markers of prophages and other integrative elements) in each SNP10 cluster. As shown in Fig. 6(c, d) (see Figs. S5 and S6, Table S7 for the distribution of the identified 35 plasmid replicons and 130 integrase genes in the Sma complex), clade 2 contained apparently more plasmids and prophages (and other integrative elements) than the other clades, indicating that the larger genome size of clade 2 is attributed considerably to the acquisition of these mobile genetic elements. In addition, notable within-clade variations in the numbers of plasmid replicons and integrase genes, particularly that of integrase genes, were also observed. Further analysis of the nine clades revealed a positive correlation between genome size and the numbers of plasmid replicons (clades 2, 6 and 12) or those of integrase genes (clades 1, 3, 4 and 11) in seven of the nine clades (R2 >0.32) (Fig. 6e). This result indicates that the acquisition of different types of mobile genetic elements contributed to the generation of observed within-clade variations in genome size (and GC content) in each clade.
Difference in isolation sources between clades and identification of hospital-adapted clades
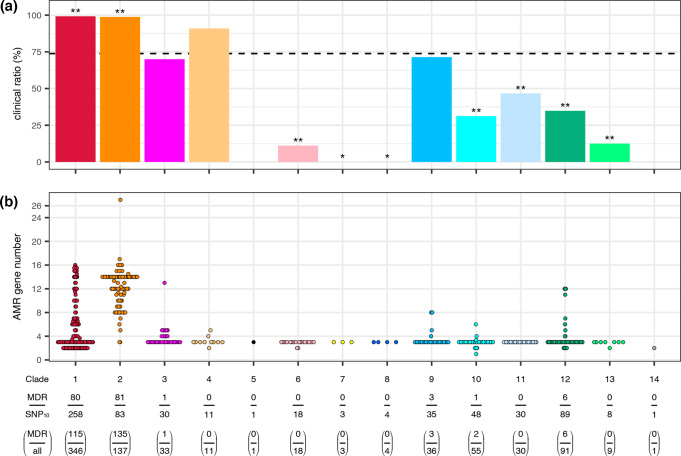
We next analysed the differences in isolation sources among clades using the 619 strains representing each SNP10 cluster (Fig. 7a). Importantly, when the SNP10 distance was used as the threshold for strain clustering, 33.8 % of the hospital environment isolates (25/74) formed SNP10 clusters with clinical isolates while no isolates from the other sources did not form SNP10 clusters with any clinical isolates (note that, when the SNP20 distance was used as the threshold, 79.9 % of the hospital environment isolates clustered with clinical isolates while only two strains from the other sources clustered with clinical isolates; see Table S2 for the details). Therefore, hospital environment isolates in our strain set were unable to be distinguished from clinical isolates. More importantly, nearly all SNP10 clusters in clades 1 and 2 were clinical or hospital environment isolates (99.2 and 98.8 %, respectively), and these ratios were significantly higher than the average ratio for the entire strain set (73.8 %) (P<0.01 by Fisher’s exact test after false discovery rate correction by the Benjamini–Hochberg method). In contrast, the ratios of such SNP10 clusters in seven clades (clades 6–8 and clades 10–13) were significantly lower (P<0.01 for clades 6, 10, 11, 12 and 13; P<0.05 for clades 7 and 8). As mentioned before, clades 10, 11, 12 and 13 correspond to Sne, Sma (and Sma_sak), Sur and Ssu, respectively. This result suggests that clades 1 and 2 represent hospital-associated clades in the Sma complex.
Fig. 7.
Proportions of the SNP10 clusters isolated from clinical/hospital environmental sources in each clade and numbers of AMR genes in each SNP10 cluster belonging to each clade. (a) The proportions of the SNP10 clusters isolated from clinical/hospital environment samples in each clade. The dotted line indicates the proportion of clinical/hospital environment clusters in the entire strain set (73.8%). Note that the isolation sources of the strains belonging to the same SNP10 clusters were the same for all multimember clusters. *; P<0.05, **; P<0.01. (b) The numbers of AMR genes in each SNP10 cluster belonging to each clade. The numbers of AMR genes in multimember SNP10 clusters were calculated as the average numbers in each SNP10 cluster. The numbers of MDR strain-containing SNP10 clusters, total SNP10 clusters, MDR strains and total strains belonging to each clade are indicated at the bottom. In both panels, colours for each clade are the same as those in Figs. 3–6.
Accumulation of AMR genes and fluoroquinolone target-site mutations in hospital-adapted clades
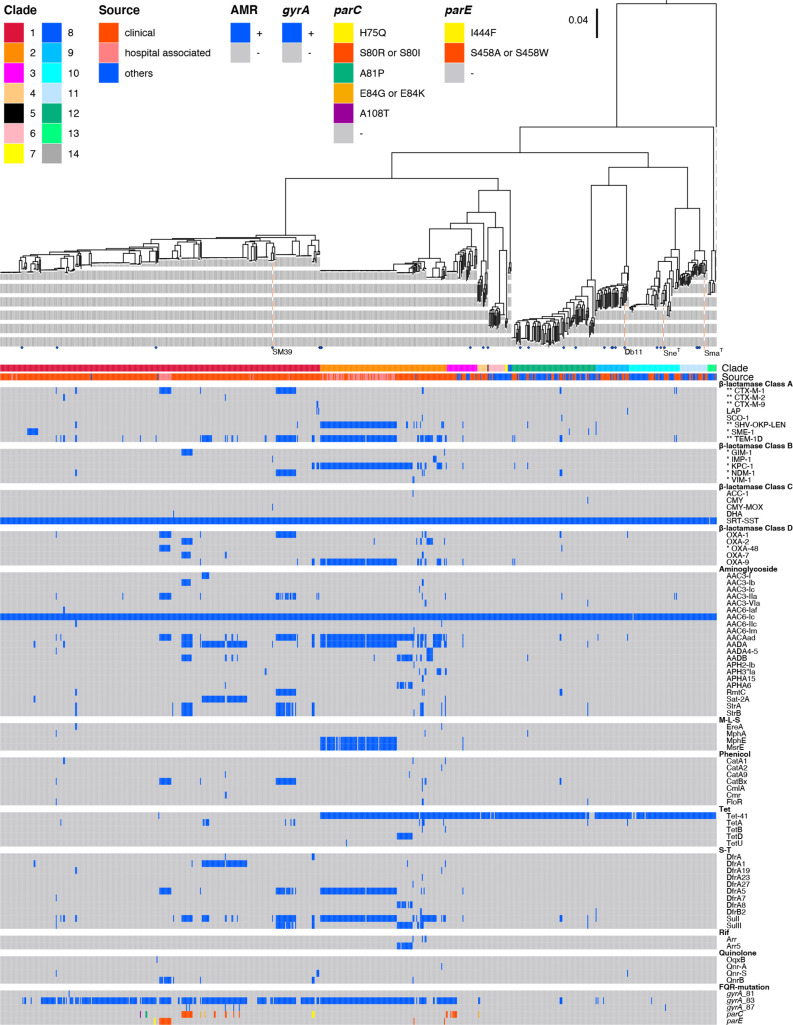
As two hospital-associated clades were identified, we analysed the distribution of AMR genes and FQR-associated mutations (FQR mutations) in the gyrA, gyrB, parC and parE genes [48, 49] in our strain set (Fig. 7b, Fig. 8, Tables 1 and 2 and S8). In this section, although 23 out of the 104 multimember SNP10 clusters showed some variation in the repertoire of AMR genes and FQR mutations (Table S9), a SNP10 cluster was counted as positive for the gene/mutation of interest if one of the cluster members contained it.
Fig. 8.
Distributions of AMR genes and FQR-conferring mutations in the Sma complex strains. The same ML tree and strain information on the clade and isolation source presented in Fig. 4 are shown here. Different mutations in the parC and parE genes are indicated by different colours. Carbapenemase and ESBL genes are marked by * and **, respectively.
Table 1.
Distribution of FQR mutations
|
Entire strain set |
Clades |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
1 |
2 |
3 |
4 |
9 |
10 |
12 |
|||
|
Number of SNP10 clusters (number of strains) |
619 (775) |
258 (346) |
83 (137) |
30 (33) |
11 (11) |
35 (36) |
48 (55) |
89 (91) |
|
|
gyrA |
total |
264 (393) |
174 (248) |
71 (124) |
9 (11) |
2 (2) |
3 (3) |
2 (2) |
3 (3) |
|
Gly81Asp |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ser83Ile |
186 (268) |
106 (134) |
69 (121) |
9 (11) |
0 |
2 (2) |
0 |
0 |
|
|
Ser83Arg |
78 (123) |
68 (113) |
3 (3) |
0 |
2 (2) |
1 (1) |
1 (1) |
3 (3) |
|
|
Asp87Tyr |
3 (3) |
3 (3) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Asp87Asn |
2 (2) |
1 (1) |
0 |
0 |
0 |
0 |
1 (1) |
0 |
|
|
Asp87Gly |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
double mutations in gyrA* |
total |
5 (5) |
5 (5) |
0 |
0 |
0 |
0 |
0 |
0 |
|
Ser83Ile + Asp87Tyr |
3 (3) |
3 (3) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ser83Ile + Asp87Asn |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ser83Ile + Asp87Gly |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
parC |
total |
30 (35) |
23 (26) |
1 (1) |
5 (7) |
1 (1) |
0 |
0 |
0 |
|
His75Gln |
2 (3) |
2 (3) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ser80Ile |
22 (25) |
16 (17) |
1 (1) |
5 (7) |
0 |
0 |
0 |
0 |
|
|
Ser80Arg |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ala81Pro |
1 (2) |
1 (2) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Glu84Lys |
2 (2) |
1 (1) |
0 |
0 |
1 (1) |
0 |
0 |
0 |
|
|
Glu84Gly |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ala108Thr |
1 (1) |
1 (1) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
parE |
total |
13 (17) |
11 (15) |
2 (2) |
0 |
0 |
0 |
0 |
0 |
|
Ile444Phe |
2 (2) |
2 (2) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ser458Trp |
9 (13) |
9 (13) |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
Ser458Ala |
2 (2) |
0 |
2 (2) |
0 |
0 |
0 |
0 |
0 |
|
|
gyrA + parC |
total |
30 (35) |
23 (26) |
1 (1) |
5 (7) |
1 (1) |
0 |
0 |
0 |
|
gyrA+ parE |
total |
13 (17) |
11 (15) |
2 (2) |
0 |
0 |
0 |
0 |
0 |
|
OqxB, QnrA, S, or B |
total |
19 (25) |
16 (22) |
2 (2) |
0 |
0 |
1 (1) |
0 |
0 |
|
without FQR mutations |
4 (4) |
3 (3) |
0 |
0 |
0 |
1 (1) |
0 |
0 |
|
All of the five strains additionally contained Ser80Ile or Ser80Arg mutations in parC.
*The clusters/strains containing double gyrA mutations were also included in the count of each mutation.
Table 2.
Distribution of ESBL and carbapenemase genes
|
Entire strain set |
Clades |
|||||||
|---|---|---|---|---|---|---|---|---|
|
1 |
2 |
3 |
9 |
10 |
12 |
|||
|
Number of SNP10 clusters (number of strains) |
619 (775) |
258 (346) |
83 (137) |
30 (33) |
35 (36) |
48 (55) |
89 (91) |
|
|
ESBLs |
total |
122 (191) |
47 (69) |
66 (112) |
1 (1) |
1 (1) |
1 (2) |
6 (6) |
|
TEM-1D |
104 (167) |
37 (54) |
60 (105) |
1 (1) |
0 |
1 (2) |
5 (5) |
|
|
SHV-OKP-LEN |
52 (97) |
2 (3) |
47 (91) |
1 (1) |
1 (1) |
0 |
1 (1) |
|
|
CTX-M1 |
25 (45) |
20 (39) |
2 (2) |
0 |
0 |
1 (2) |
2 (2) |
|
|
CTX-M2 |
3 (3) |
3 (3) |
0 |
0 |
0 |
0 |
0 |
|
|
CTX-M9 |
1 (2) |
1 (2) |
0 |
0 |
0 |
0 |
0 |
|
|
multiple ESBLs* |
total |
62 (121) |
15 (30) |
43 (86) |
1 (1) |
0 |
1 (2) |
2 (2) |
|
TEM-1D + SHV-OKP-LEN |
43 (86) |
1 (1) |
41 (84) |
1 (1) |
0 |
0 |
0 |
|
|
TEM-1D + CTX-M1 |
15 (30) |
10 (24) |
2 (2) |
0 |
0 |
1 (2) |
2 (2) |
|
|
TEM-1D + CTX-M2 |
3 (3) |
3 (3) |
0 |
0 |
0 |
0 |
0 |
|
|
TEM-1D + SHV-OKP-LEN + CTX-M1 |
1 (2) |
1 (2) |
0 |
0 |
0 |
0 |
0 |
|
|
carbapenemases |
total |
122 (198) |
45 (67) |
68 (122) |
1 (1) |
1 (1) |
0 |
7 (7) |
|
KPC-1 |
68 (122) |
4 (6) |
61 (113) |
1 (1) |
0 |
0 |
2 (2) |
|
|
NDM-1 |
14 (29) |
9 (24) |
2 (2) |
0 |
0 |
0 |
3 (3) |
|
|
SME-1 |
14 (15) |
11 (12) |
0 |
0 |
1 (1) |
0 |
2 (2) |
|
|
GIM-1 |
12 (13) |
11 (12) |
1 (1) |
0 |
0 |
0 |
0 |
|
|
OXA-48 |
10 (13) |
9 (12) |
0 |
0 |
0 |
0 |
1 (1) |
|
|
IMP-1 |
3 (5) |
1 (1) |
2 (4) |
0 |
0 |
0 |
0 |
|
|
VIM-1 |
2 (2) |
0 |
2 (2) |
0 |
0 |
0 |
0 |
|
|
multiple carbapenemases* |
total |
1 (1) |
0 |
0 |
0 |
0 |
0 |
1 (1) |
|
NDM-1 + OXA-48 |
1 (1) |
0 |
0 |
0 |
0 |
0 |
1 (1) |
|
|
ESBLs + carbapenemases* |
total |
82 (148) |
21 (41) |
54 (100) |
1 (1) |
1 (1) |
0 |
5 (5) |
|
TEM-1D & KPC-1 |
13 (15) |
2 (3) |
10 (11) |
0 |
0 |
0 |
1 (1) |
|
|
CTX-M1 & OXA-48 |
9 (12) |
9 (12) |
0 |
0 |
0 |
0 |
0 |
|
|
CTX-M9 & KPC-1 |
1 (2) |
1 (2) |
0 |
0 |
0 |
0 |
0 |
|
|
SHV-OKP-LEN & KPC-1 |
4 (4) |
0 |
3 (3) |
0 |
0 |
0 |
1 (1) |
|
|
SHV-OKP-LEN & VIM-1 |
1 (1) |
0 |
1 (1) |
0 |
0 |
0 |
0 |
|
|
SHV-OKP-LEN & SME-1 |
1 (1) |
0 |
0 |
0 |
1 (1) |
0 |
0 |
|
|
TEM-1D + CTX-M1 & NDM1 |
11 (25) |
8 (22) |
1 (1) |
0 |
0 |
0 |
2 (2) |
|
|
TEM-1D + SHV-OKP-LEN & KPC-1 |
42 (85) |
0 |
41 (84) |
1 (1) |
0 |
0 |
0 |
|
|
TEM-1D & NDM-1 + OXA-48 |
1 (1) |
0 |
0 |
0 |
0 |
0 |
1 (1) |
|
|
TEM-1D & NDM-1 + OXA-49 |
1 (2) |
1 (2) |
0 |
0 |
0 |
0 |
0 |
|
*The clusters/strains containing these combinations of genes were also included in the count of each gene.
AMR genes:
We detected 77 AMR genes in the entire strain set. Among these, bla SRT-SST (ampC) and aac(6)−1 c, which are known to be chromosomally encoded in Sma [1], were highly conserved in the set, including clades 13 and 14. The tet41 gene, encoding a tetracycline-specific efflux pump [58], was also widely distributed but was absent in all but one strain of clade 1, indicating that tet41 was lost from clade 1. Many of the SNP10 clusters in clade 1 (190/258) contained additional AMR genes that were probably acquired horizontally (up to 13). More strikingly, nearly all clade 2 clusters (81/83) contained additional AMR genes (up to 23). Thus, AMR genes have undergone marked accumulation in these two clades. In contrast, in the other clades, a small number of clusters contained additional AMR genes (28 clusters in five clades). All 28 clusters were clinical isolates except for a bovine isolate in clade 12 (Fig. S7).
FQR mutations:
As summarized in Table 1, FQR mutations in gyrA were detected in 264 SNP10 clusters. Mutations in parC (30 clusters) and parE (13 clusters) were also detected, but all of these clusters contained gyrA mutations. Similar accumulation of FQR mutations is often found in Gram-negative bacteria and is known to enhance FQR [59]. Among the identified gyrA mutations, the majority were located at Ser83, Ser83Ile (186 clusters) or Ser83Arg (78 clusters). In five clusters, an additional gyrA mutation was present at Asp87. Such double mutations are also associated with high levels of FQR [59].
Notable accumulation of FQR mutations also occurred in clades 1 and 2. In clade 1, 67.4 % of the 258 clusters contained gyrA mutations, with 23 and 11 clusters additionally carrying parC or parE mutations. The five aforementioned clusters that contained double gyrA mutations all belonged to clade 1. In clade 2, 85.5 % of the 83 clusters contained gyrA mutations, but only one or two clusters additionally contained mutations in parC and parE, respectively. In the other clades, while gyrA mutations were detected in nine clade 3 clusters (9/30), including five clusters that additionally contained mutations in parC, only a few or no clusters contained FQR mutations in the remaining clades.
In addition to FQR mutations, four FQR-associated AMR genes (oqxB, qnrA, qnrB or qnrS) were found in 19 clusters, but most of them (15/19) contained gyrA mutations.
MDR strains:
The accumulation of horizontally acquired AMR genes and FQR mutations in clades 1 and 2 resulted in a high proportion of MDR strains in these clades; 31.0 % of the SNP10 clusters in clade 1 and 97.6 % of the clade 2 SNP10 clusters were genotypically defined as MDR (resistant to three or more categories of antimicrobials; see Methods for the definition of MDR). The higher proportion of MDR clusters in clades 1 and 2 was not a simple reflection of a higher proportion of clinical and hospital environment isolates in these clades because the proportions of MDR strains among the clinical isolates of the other clades were much lower than those of clades 1 and 2 (11 out of the 202 clinical clusters in all other clades).
ESBL and carbapenemase genes:
We analysed these genes in more detail because they are the clinically most important genes worldwide (Table 2). We detected five ESBL genes in 122 clusters and seven carbapenemase genes in 122 clusters, among which 82 clusters carried both ESBL and carbapenemase genes. These clusters, particularly ESBL/carbapenemase double-positive clusters (77/82), frequently contained gyrA mutations (Fig. 8).
Among the detected ESBL genes, bla TEM-1D was the most prevalent (104 clusters), followed by bla SHV-OKP-LEN (52 clusters) and bla CTX-M1 (25 clusters), with a few clusters containing bla CTX-M2 and bla CTX-M9. A notable number of clusters contained multiple ESBL genes (62 clusters, one of which contained three ESBL genes). Among the carbapenemase genes detected, bla KPC-1 was the most frequently found (68 clusters), followed by bla NDM-1 (14 clusters), bla SME-1 (14 clusters), bla GIM-1 (12 clusters) and bla OXA-48 (10 clusters), with a few clusters containing bla IMP-1 or bla VIM-1. Notably, one cluster in clade 12 contained two carbapenemase genes (bla NDM-1 and bla OXA-48).
Strains carrying ESBL and/or carbapenemase genes were highly enriched in both clades 1 and 2, but a clear difference in the repertoire of these genes was observed between the two clades (Table 2). In clade 1, among the 47 ESBL-positive clusters, bla TEM-1D (37 clusters) and bla CTX-M1 (20 clusters) were predominant, and 11 clusters contained both of these genes (one of them additionally contained bla SHV-OKP-LEN). Among the 45 carbapenemase-positive clade 1 clusters, bla SME-1, bla GIM-1, bla NDM-1 and bla OXA-48 were predominant (found in 11 or nine clusters). In addition, 21 clusters contained both ESBL and carbapenemase genes. In clade 2, much higher proportions of clusters contained ESBL and carbapenemase genes than in clade 1. ESBL genes were found in 79.5 % of the clade 2 SNP10 clusters. bla TEM-1D (60 clusters) and bla SHV-OKP-LEN (47 clusters) were the most prevalent of these genes and were frequently present together (41 clusters). Carbapenemase genes were found in 68 clusters (81.9%), and most of them (61/68) contained bla KPC-1. The proportion of ESBL/carbapenemase double-positive clusters was also much higher than in clade 1 (65.1 vs. 8.1 %).
The distribution of potentially virulence-related genes in the Sma complex and the identification of accessary genes specifically conserved in the hospital-adapted clades
The virulence factors of Sma have not been well defined except for several factors, such as the ShyA haemolysin [60]. However, a set of potentially virulence-related genes/operons were listed up in a previous study where a detailed genomic comparison of strains SM39 and Db11 [15]. We therefore analysed the distribution of these genes/operons in the strain set to search for potentially virulence-related genes/operons specifically conserved in clades 1 and 2 (Fig. S8 and Table S10). Although this analysis identified the genes/operons that are relatively well conserved across the Sma complex and those exhibited variable distributions, only an operon encoding a haemolysin or contact-dependent inhibition (CDI)-related protein and a protein for its secretion (cdiA2 and shlB_3 in Fig. S8) and two fimbriae operons (represented by ecpC and yehB in Fig. S8) showed a distribution somewhat biassed to clades 1 and 2.
To further explore the genetic features potentially underlying the hospital adaptation of clades 1 and 2, we searched for accessary genes that are highly conserved in both clades 1 and 2 (present in >90 % of SNP10 clusters) but significantly less frequently present in the other clades (Bonferroni P<0.01) and identified 287 such genes (Table S11; the distribution of top 100 genes in the Sma complex is shown in Fig. S9). This gene set included not only the genes in the above-mentioned three operons but also many genes/operons for various metabolic functions, but it is currently unknown whether or how these genes are involved in the adaptation of clades 1 and 2 to hospital-related environments.
Discussion
In this study, we performed a large-scale genomic analysis of Sma and its close relatives (the Sma complex), including newly sequenced 108 soil and 117 clinical isolates, and obtained several taxonomically, biologically or medically important findings.
First, our analysis clarified the global population structure and complex phylogenetic relationships of Sma and its close relatives. According to the results of ANI-based clustering, we defined 14 clades in which the minimum within-clade ANI values were >97 % (Fig. 3). This clade structure was supported by core-gene-based phylogenetic analysis and pangenome-based clustering (Figs. 4 and 5). Among the 14 clades, clades 10, 11, 12 and 13 included the type strains of Sne, Sma, Sur and Ssu, respectively (Fig. S2); thus, the four clades represent these species. The type strain of Sma_sak belonged to clade 11, together with the Sma type strain, confirming their very close relationship pointed out recently [61]. The other clades each represent distinct lineages in this complex. These results indicate not only the marked diversity but also the insufficient classification status of the Sma complex, thus providing an important basis for reconsidering the classification of this complex in the future. It should also be emphasized the difficulty to accurately identify the species in the Sma complex via 16S rRNA sequence analyses that has a much lower resolution power than whole-genome-ANI analysis; sequence identities of 16S rRNA genes within the Sma complex were over 99 %.
Second, the comparison of the 14 clades revealed notable interesting inter- and within-clade variations in genome size and GC content, and we found that the acquisition of mobile genetic elements such as plasmids and prophages is related to the generation of these variations (Figs. 4 and 6). In particular, clade 2 strains contained larger genomes with lower GC contents than the other clades, which was attributable to the acquisition of more plasmids and prophages (and other integrative elements) than the other clades. The within-clade variation in genome size and GC content and a negative correlation of these two genomic parameters were observed in not only clade 2 but also other clades, and interestingly, it appears that different types of mobile elements contributed to the generation of the variations observed in different clades, which may be related to the difference in main environments where each clade inhabits or in other biological or genetic features of each clade. However, some cautions are required to interpret the current results because our preliminary results of plasmid analysis suggest that considerable numbers of plasmid replicons are unbale to be detected by PlasmidFinder used in this study (one third of plasmids was not detected in some strains) and because the genetic features of each mobile element are largely unknown at present.
The medically most important finding was the identification of two clades (clades 1 and 2) that comprised almost exclusively clinical or hospital environment isolates (Figs. 4 and 7). As the higher ratios of such strains in these clades were statistically significant, they can be regarded as hospital-adapted lineages in the Sma complex. In this context, the marked accumulation of a wide range of AMR genes and FQR mutations observed in the two clades is interesting (Figs. 7 and 8, Tables 1 and 2). Multiple genes encoding ESBLs and carbapenemases were found in many strains in these clades, and strains carrying both ESBL and carbapenemase genes were also frequently identified, particularly in clade 2. The accumulation of AMR genes was also observed in several strains of the other clades, but all such strains were clinical isolates with a single exception. Thus, it is most likely that the accumulation of AMR genes in clades 1 and 2 is the result of repeated exposure to multiple antibiotics due to their long persistence in clinical settings. This notion is supported by the wide distribution of the Ser83Phe (or Leu) substitution in gyrA and the presence of additional mutations in parC and parE in notable numbers of strains of these clades. The accumulation of AMR genes in clades 1 and 2 also suggests that these clades can be important reservoirs of AMR genes in hospitals. Regarding the mechanisms underlying the adaptation of clades 1 and 2 to hospital-related environments, we have identified a set of accessary genes that were specifically conserved in both clades (Table S11 and Fig. S9) although it is currently unknown whether or how these genes are involved in this adaptation. It is also unknown whether clades 1 and 2 have a higher potential virulence than other clades. However, our data will be a basis for investigating these important issues in future studies. Considering that many potentially virulence-related functions are encoded by core genome (Table S10 and Fig. S8) as previously suggested [15], it will be also important to search for core genes that are under positive selection in clades 1 and 2 [62].
Finally, we should mention the relationships of the clades defined in this study and the (sub)clades/clusters/lineages defined in the recently published three genome-based analyses [16–18]. In the study by Abreo and Altier [16], 45 strains analysed were classified into two clades (clades 1 and 2) each containing several subclades based on the data of whole-genome multilocus strain typing (wgMLST) analysis, and the two clades were mostly comprised of environmental (clade 1) or clinical (clade 2) isolates. In particular, subclades 2c and 2d included only clinical strains. Of the 45 strains, 34 were included in our strain set and it appears that subclades 2c and 2d correspond to clades 2 and 1 of this study, respectively (see Table S12 for more details of the comparison). On the other hand, Saralegui et al. divided 452 strains into nine clusters by ANI-based clustering [18]. Of the 452 strains, 338 were included in our strain set and it appears that cluster 1 corresponds to clades 10/11 of this study; clusters 3–5 to clades 2, 9 and 12, respectively; clusters 6/7/8 to clade 1; and cluster 9 to clade 10, but cluster 2 included strains from multiple clades of this study (see Table S12 for more details). More recently, Matteoli et al. analysed 642 genomes and divided them into 12 lineages, three of which (lineages Sm5, Sm6 and Sm7) were comprised of only human-associated strains [17]. In addition, the authors reported several interesting findings, such as the presence of more AMR genes in lineages Sm5 and Sm7 as well as lineage Sm9, which is also a human-associated strain-enriched lineage, the wide distribution of bla KPC-2 in lineage Sm7, and the presence of more plasmids in lineage Sm9. Although 340 out of the 601 strains analysed by Matteoli et al. were included in our strain set, we could not analyse the relationships between their lineages and the clades of this study because the strain compositions of the 12 lineages were not provided in their paper.
In conclusion, through a large-scale genome analysis, we revealed the global population structure of the Sma complex, comprising 14 distinct clades, which will be an important basis for its reclassification in the future, and we further identified two hospital-adapted clades. Although the potential virulence of these clades are currently unknown, special attention needs to be paid to these clades because of the marked accumulation of a wide range of AMR genes, including ESBL and carbapenemase genes, and mutations that confer FQR.
Supplementary Data
Funding information
This work was supported by KAKENHI from the Japan Society for the Promotion of Science (18K16175) granted to TH. DSN was supported by a Japanese Government Scholarship from MEXT (Ministry of Education, Culture, Sports, Science and Technology).
Acknowledgements
We thank Mai Horiguchi, Yuriko Satoh and Kie Ozaki for providing technical assistance and the members of the T. Itoh and E. Oswald laboratories for collecting soil samples.
Author contributions
Conceptualization: N.G., T.H. Methodology: T.O., I.T., D.Y., T.I., T.H. Validation: T.O., I.T., K.N., Y.G., Y.O., T.H. Formal analysis: T.O., I.T., M.P.S., T.H. Investigation: T.O., I.T., D.S.N., M.P.S., A.I., K.M., A.S., T.H. Resources: T.O., R.N., K.M., D.Y., T.I., A.S., D.D., E.O., N.G., T.H. Data curation: T.O., I.T., D.S.N., T.H. Writing – original draft: T.O., I.T., T.H. Writing – review and editing: T.O., I.T., K.N., Y.O., E.O., N.G., T.H. Visualization: T.O., I.T., T.H. Supervision: K.N., Y.G., Y.O., N.G., T.H. Project administration: N.G., T.H. Funding acquisition: T.H.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; ANI, average nucleotide identity; ANOVA, analysis of variance; CDI, contact-dependent inhibition; DFAST, DNA Data Bank of Japan Fast Annotation and Submission Tool; DTC, deoxyribonuclease-toluidine blue-cephalothin; ESBL, extended-spectrum β-lactamase; FQR, fluoroquinolone resistance; MDR, multidrug-resistant; ML, maximum-likelihood; QRDR, quinolone-resistance-determining region; Sma, Serratia marcescens; Sma_sak, Serratia marcescens subspecies sakuensis; Sne, Serratia nematodiphila; Ssu, Serratia surfactantfaciens; Sur, Serratia ureilytica; wgMLST, whole-genome multilocus strain typing; WGS, whole-genome sequence.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Nine supplementary figures and twelve supplementary tables are available with the online version of this article.
References
- 1.Mahlen SD. Serratia infections: from military experiments to current practice. Clin Microbiol Rev. 2011;24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández AL, Adrio B, Martínez Cereijo JM, Martínez Monzonis MA, El-Diasty MM, et al. Clinical study of an outbreak of postoperative mediastinitis caused by Serratia marcescens in adult cardiac surgery. Interact Cardiovasc Thorac Surg. 2020;30:523–527. doi: 10.1093/icvts/ivz312. [DOI] [PubMed] [Google Scholar]
- 3.Fleisch F, Zimmermann-Baer U, Zbinden R, Bischoff G, Arlettaz R, et al. Three consecutive outbreaks of Serratia marcescens in a neonatal intensive care unit. Clin Infect Dis. 2002;34:767–773. doi: 10.1086/339046. [DOI] [PubMed] [Google Scholar]
- 4.Hejazi A, Aucken HM, Falkiner FR. Epidemiology and susceptibility of serratia marcescens in a large general hospital over an 8-year period. J Hosp Infect. 2000;45:42–46. doi: 10.1053/jhin.1999.0722. [DOI] [PubMed] [Google Scholar]
- 5.Herbert S, Halvorsen DS, Leong T, Franklin C, Harrington G, et al. Large outbreak of infection and colonization with gram-negative pathogens carrying the metallo- beta -lactamase gene blaIMP-4 at a 320-bed tertiary hospital in Australia. Infect Control Hosp Epidemiol. 2007;28:98–101. doi: 10.1086/508841. [DOI] [PubMed] [Google Scholar]
- 6.Nastro M, Monge R, Zintgraff J, Vaulet LG, Boutureira M, et al. First nosocomial outbreak of VIM-16-producing Serratia marcescens in Argentina. Clin Microbiol Infect. 2013;19:617–619. doi: 10.1111/j.1469-0691.2012.03978.x. [DOI] [PubMed] [Google Scholar]
- 7.Naumiuk L, Baraniak A, Gniadkowski M, Krawczyk B, Rybak B, et al. Molecular epidemiology of Serratia marcescens in two hospitals in Gdańsk, Poland, over a 5-year period. J Clin Microbiol. 2004;42:3108–3116. doi: 10.1128/JCM.42.7.3108-3116.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetter L, Schuepfer G, Kuster SP, Rossi M. A hospital-wide outbreak of Serratia marcescens, and Ishikawa’s “Fishbone” analysis to support outbreak control. Qual Manag Health Care. 2016;25:1–7. doi: 10.1097/QMH.0000000000000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moradigaravand D, Boinett CJ, Martin V, Peacock SJ, Parkhill J. Recent independent emergence of multiple multidrug-resistant Serratia marcescens clones within the United Kingdom and Ireland. Genome Res. 2016;26:1101–1109. doi: 10.1101/gr.205245.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stock I, Grueger T, Wiedemann B. Natural antibiotic susceptibility of strains of Serratia marcescens and the S. liquefaciens complex: S. liquefaciens sensu stricto, S. proteamaculans and S. grimesii . Int J Antimicrob Agents. 2003;22:35–47. doi: 10.1016/s0924-8579(02)00163-2. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins KL, Findlay J, Meunier D, Cummins M, Curtis S, et al. Serratia marcescens producing SME carbapenemases: an emerging resistance problem in the UK? J Antimicrob Chemother. 2017;72:1535–1537. doi: 10.1093/jac/dkw567. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova D, Markovska R, Hadjieva N, Schneider I, Mitov I, et al. Extended-spectrum β-lactamase-producing Serratia marcescens outbreak in a Bulgarian hospital. J Hosp Infect. 2008;70:60–65. doi: 10.1016/j.jhin.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Abbott S. In: Manual of Clinical Microbiology. Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Washington, DC: ASM Press; 1999. Klebsiella, Enterobacter, Citrobacter and Serratia; pp. 475–482. [Google Scholar]
- 14.Szewzyk U, Szewzyk R, Stenström TA. Growth and survival of Serratia marcescens under aerobic and anaerobic conditions in the presence of materials from blood bags. J Clin Microbiol. 1993;31:1826–1830. doi: 10.1128/jcm.31.7.1826-1830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, et al. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol. 2014;6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abreo E, Altier N. Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the links between them. Sci Rep. 2019;9:46. doi: 10.1038/s41598-018-37118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matteoli FP, Pedrosa-Silva F, Dutra-Silva L, Giachini AJ. The global population structure and beta-lactamase repertoire of the opportunistic pathogen Serratia marcescens . Genomics. 2021;113:3523–3532. doi: 10.1016/j.ygeno.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Saralegui C, Ponce-Alonso M, Pérez-Viso B, Moles Alegre L, Escribano E, et al. Genomics of Serratia marcescens isolates causing outbreaks in the same pediatric unit 47 years apart: position in an updated phylogeny of the species. Front Microbiol. 2020;11:451. doi: 10.3389/fmicb.2020.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adeolu M, Alnajar S, Naushad S, S Gupta R. Genome-based phylogeny and taxonomy of the “Enterobacteriales”: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 20.Ajithkumar B, Ajithkumar VP, Iriye R, Doi Y, Sakai T. Spore-forming Serratia marcescens subsp. sakuensis subsp. nov., isolated from a domestic wastewater treatment tank. Int J Syst Evol Microbiol. 2003;53:253–258. doi: 10.1099/ijs.0.02158-0. [DOI] [PubMed] [Google Scholar]
- 21.Cheng TH, Saidin J, Danish-Daniel M, Gan HM, Mat Isa MN, et al. Genome sequence of Serratia marcescens subsp. sakuensis strain K27, a marine bacterium isolated from sponge (Haliclona amboinensis) Genome Announc. 2018;6:e00022–18. doi: 10.1128/genomeA.00022-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doijad S, Chakraborty T. Genome-based analyses indicate that Serratia marcescens subsp. marcescens and Serratia marcescens subsp. sakuensis do not merit separation to subspecies status. Int J Syst Evol Microbiol. 2019;69:3924–3926. doi: 10.1099/ijsem.0.003706. [DOI] [PubMed] [Google Scholar]
- 23.Parks DH, Chuvochina M, Chaumeil P-A, Rinke C, Mussig AJ, et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;38:1079–1086. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- 24.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G, Smith DK, Zhu H, Guan Y, Lam T-Y. Ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 29.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 30.Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 32.Farmer JJ, Silva F, Williams DR. Isolation of Serratia marcescens on deoxyribonuclease-toluidine blue-cephalothin agar. Appl Microbiol. 1973;25:151–152. doi: 10.1128/am.25.1.151-152.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kajitani R, Yoshimura D, Ogura Y, Gotoh Y, Hayashi T, et al. Platanus_B: an accurate de novo assembler for bacterial genomes using an iterative error-removal process. DNA Res. 2020;27:27. doi: 10.1093/dnares/dsaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murtagh F, Legendre P. Ward’s hierarchical agglomerative clustering method: which algorithms implement ward’s criterion? J Classif. 2014;31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 37.Harris AKP, Williamson NR, Slater H, Cox A, Abbasi S, et al. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology (Reading) 2004;150:3547–3560. doi: 10.1099/mic.0.27222-0. [DOI] [PubMed] [Google Scholar]
- 38.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 39.Yabe S, Fukushima J. Complete genome sequence of temperature-dependent pigment-producing Serratia marcescens ATCC 274. Microbiol Resour Announc. 2020;9:e00164-20. doi: 10.1128/MRA.00164-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:238. doi: 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team Vienna, Austria: 2020. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- 43.Fisher RA. The logic of inductive inference. J R Stat Soc. 1935;98:39. doi: 10.2307/2342435. [DOI] [Google Scholar]
- 44.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 45.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang W, Li L, Myers JR, Marth GT. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Nawaz M, Sung K, Kweon O, Khan S, Nawaz S, et al. Characterisation of novel mutations involved in quinolone resistance in Escherichia coli isolated from imported shrimp. Int J Antimicrob Agents. 2015;45:471–476. doi: 10.1016/j.ijantimicag.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 51.Livermore DM, Winstanley TG, Shannon KP. Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J Antimicrob Chemother. 2001;48 Suppl 1:87–102. doi: 10.1093/jac/48.suppl_1.87. [DOI] [PubMed] [Google Scholar]
- 52.Strobel G, Li J-Y, Sugawara F, Koshino H, Harper J, et al. Oocydin A, A chlorinated macrocyclic lactone with potent anti-oomycete activity from Serratia marcescens . Microbiology (Reading) 1999;145 (Pt 12):3557–3564. doi: 10.1099/00221287-145-12-3557. [DOI] [PubMed] [Google Scholar]
- 53.Matilla MA, Udaondo Z, Krell T, Salmond GPC. Genome Sequence of Serratia marcescens MSU97, a plant-associated bacterium that makes multiple antibiotics. Genome Announc. 2017;5:e01752-16. doi: 10.1128/genomeA.01752-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakuraoka R, Suzuki T, Morohoshi T. Distribution and genetic diversity of genes involved in quorum sensing and prodigiosin biosynthesis in the complete genome sequences of Serratia marcescens . Genome Biol Evol. 2019;11:931–936. doi: 10.1093/gbe/evz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fineran PC, Iglesias Cans MC, Ramsay JP, Wilf NM, Cossyleon D, et al. Draft genome sequence of Serratia sp. strain ATCC 39006, a model bacterium for analysis of the biosynthesis and regulation of prodigiosin, a carbapenem, and gas vesicles. Genome Announc. 2013;1:e01039-13. doi: 10.1128/genomeA.01039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daubin V, Lerat E, Perrière G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 58.Thompson SA, Maani EV, Lindell AH, King CJ, McArthur JV. Novel tetracycline resistance determinant isolated from an environmental strain of Serratia marcescens . Appl Environ Microbiol. 2007;73:2199–2206. doi: 10.1128/AEM.02511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol. 2017;66:551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 60.Goluszko P, Nowacki MR. Extracellular haemolytic activity of Serratia marcescens . FEMS Microbiol Lett. 1989;52:207–211. doi: 10.1016/0378-1097(89)90198-5. [DOI] [PubMed] [Google Scholar]
- 61.Cho G-S, Stein M, Brinks E, Rathje J, Lee W, et al. Serratia nevei sp. nov. and Serratia bockelmannii sp. nov., isolated from fresh produce in Germany and reclassification of Serratia marcescens subsp. sakuensis Ajithkumar et al. 2003 as a later heterotypic synonym of Serratia marcescens subsp. marcescens . Syst Appl Microbiol. 2020;43:126055. doi: 10.1016/j.syapm.2020.126055. [DOI] [PubMed] [Google Scholar]
- 62.Kumar N, Browne HP, Viciani E, Forster SC, Clare S, et al. Adaptation of host transmission cycle during Clostridium difficile speciation. Nat Genet. 2019;51:1315–1320. doi: 10.1038/s41588-019-0478-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.