Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has infected almost 200 million people worldwide by July 2021 and the pandemic has been characterized by infection waves of viral lineages showing distinct fitness profiles. The simultaneous infection of a single individual by two distinct SARS-CoV-2 lineages may impact COVID-19 disease progression and provides a window of opportunity for viral recombination and the emergence of new lineages with differential phenotype. Several hundred SARS-CoV-2 lineages are currently well phylogenetically defined, but two main factors have precluded major coinfection/codetection and recombination analysis thus far: (i) the low diversity of SARS-CoV-2 lineages during the first year of the pandemic, which limited the identification of lineage defining mutations necessary to distinguish coinfecting/recombining viral lineages; and the (ii) limited availability of raw sequencing data where abundance and distribution of intrasample/intrahost variability can be accessed. Here, we assembled a large sequencing dataset from Brazilian samples covering a period of 18 May 2020 to 30 April 2021 and probed it for unexpected patterns of high intrasample/intrahost variability. This approach enabled us to detect nine cases of SARS-CoV-2 coinfection with well characterized lineage-defining mutations, representing 0.61 % of all samples investigated. In addition, we matched these SARS-CoV-2 coinfections with spatio-temporal epidemiological data confirming its plausibility with the cocirculating lineages at the timeframe investigated. Our data suggests that coinfection with distinct SARS-CoV-2 lineages is a rare phenomenon, although it is certainly a lower bound estimate considering the difficulty to detect coinfections with very similar SARS-CoV-2 lineages and the low number of samples sequenced from the total number of infections.
Keywords: codetection, coinfection, COVID-19, genomics
Impact Statement
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is the etiological agent of the global pandemic that in approximately 2 years has caused a large public health emergency leading to the death of more than 5 million people worldwide. Despite the vast literature about the SARS-CoV-2 genomics, there is still a knowledge gap regarding the intrahost nucleotide diversity during SARS-CoV-2 infection and detection of coinfection and recombination of different viral lineages. Our results, based on the largest dataset of raw sequenced reads assembled so far from Brazil, shows nine coinfection events from patients of different Brazilian regions. Knowledge of these events allows a more detailed understanding of how we can identify them, its impact on disease progression, the likelihood of new recombining lineage emergence and early detection of circulating lineages before official reports.
Data Summary
The raw fastq data of codetection cases are deposited on gisaid.org and are associated to the following GISAID codes: EPI_ISL_1068258, EPI_ISL_2491769, EPI_ISL_2491781, EPI_ISL_2645599, EPI_ISL_2661789, EPI_ISL_2661931, EPI_ISL_2677092, EPI_ISL_2777552, EPI_ISL_3869215. Supplementary Material are available on Figshare at https://doi.org/10.6084/m9.figshare.19361270.v1 [1]. The workflow code used in this study is publicly available on: https://github.com/dezordi/ViralFlow.
Introduction
SARS-CoV-2, the etiological agent of the COVID-19 pandemic, has a relatively low mutation rate compared to other RNA viruses [2], and most viral lineages are normally defined by only a few synapomorphic SNPs (n<10) [3]. However, the pervasiveness of SARS-CoV-2 infections during the COVID-19 pandemic provided substantial opportunities for the virus to explore the fitness landscape through single nucleotide substitutions and/or indels, giving rise to a range of more transmissible variants of concern (VOCs). These lineages are characterized by an unusual pattern of lineage-defining SNPs along the genome (n>15) [4–6].
Coinfection is defined as a single host infection by more than one pathogen or virus lineage simultaneously. Despite being a rare phenomenon, it may provide opportunity for genetic recombination, an event known to occur in viruses of the Coronaviridae family [7, 8] including SARS-CoV-2-like viruses [9]. Recombinant viruses may, in turn, trigger the emergence of new lineages with enhanced biological properties, including the capacity to infect new hosts (expansion of viral host range) [10–13]. The frequency of coinfection and its role to promote recombination-driven SARS-CoV-2 evolution and the emergence of SARS-CoV-2 lineages is still poorly understood. The low variability found in SARS-CoV-2 lineages and the few well-defined lineage-specific SNPs until the second half of 2020 probably hindered the identification of coinfection and recombination events of SARS-CoV-2 lineages so far. In contrast, the emergence of VOC lineages carrying a substantial number of additional SNPs may provide enough markers to currently detect these events. A number of coinfection cases were reported for SARS-CoV-2, including lineages B.1.1.28/B.1.1.33 and B.1.1.91/B.1.1.28 [14] in Brazil, several variants of interest (VOIs) and VOCs [15], and different lineages in the UK [16, 17]. Moreover, putative coinfections were indirectly inferred from North America and Europe patients by detecting recombinant genomes [18–20].
In this study, we assessed amplicon sequencing reads of 2263 SARS-CoV-2 samples from Brazilian patients generated by the Fiocruz Genomic Surveillance Network. We identified nine coinfection cases through the identification of an unusual pattern of intrahost single nucleotide variant (iSNV) sites and phylogenetic reconstruction of alternative SARS-CoV-2 genomes generated from well supported major and minor allele frequency nucleotide variants. Moreover, epidemiological trends of circulating lineages in each Brazilian state supported that the SARS-CoV-2 VOIs and VOC lineages found in these coinfected samples were also cocirculating at the time of sampling, thus providing further plausibility for our findings.
Methods
SARS-CoV-2 sequences and ethical aspects
The sequencing data was obtained from the genomic survey of SARS-CoV-2 positives samples sequenced by FIOCRUZ’s Genomic Surveillance Network between 18 May 2020 and 30 April 2021. SARS-CoV-2 genomes were amplified and sequenced using previously described Illumina protocols [21–23] (Table S1, available in the online version of this article). The frequency of lineages obtained from Brazilian states was evaluated using data recovered from GISAID (gisaird.org) on 23 July 2021.
Genome assembly and intrahost variant analysis
The genomic analysis were performed with ViralFlow v.0.0.5 [24], through the following steps: removal of duplicated reads, adapters and read extremities with less than 20 of phred score quality with the fastp tool [25]; reference genome assembly using BWA [26] to map reads against the SARS-CoV-2 Wuhan reference genome (NC_045512.2); the consensus genomes generation with samtools mpileup [27] and iVar [28], using a threshold quality score of 30 and calling SNPs and indels present as major allele frequencies. After the consensus generation, the bam-readcount tool [29] retrieve the proportion of each base (A, C, T, G) present in each position of the bam file and an in house python script (intrahost.py) identifies iSNV sites following the specific rules: the minor variant (MinV) should represent at least 5 % of total position depth and should appear in both sense and antisense reads (at least 5 % in each sense) with a depth of at least 100 reads. Two consensus genomes were generated as output based on the major and minor allele frequency of each iSNV site: the major variant (MajV) with the nucleotide present in major allele frequency in each genomic position, and the MinV with the nucleotide present in the lower allele frequency at that same positions. The consensus genomes were automatically analysed with PangoLineage v1.1.23 with pangoLEARN updated at 28 May 2021 [30] and to Nextclade [31] tools. Only genomes with more than 95 % coverage breadth and 100 reads of average coverage depth (Table S2) were considered.
If the MajV and MinV genomes were assigned to the same viral lineage, we may assume that the variability observed likely resulted from: (I) de novo generation of intrahost variants that emerged during viral replication; or (II) coinfection with two viruses of the same lineage. Conversely, if MajV and MinV genomes were assigned to different lineages, the intrahost variability observed is more likely derived from a codetection event. All samples in which alternative genomes where assigned to different pango lineages were manually curated with Interactive Genomic Viewer [32], indels related to intrahost variants into specific genomes that change the coding frame were discarded. Additional evidence of codetection was searched on the raw sequence reads: (I) if the proportion of reads supporting lineage-specific defining SNPs are similar it suggests codetection while if the proportion is drastically different the variability is likely derived from de novo intrahost variability; (II) if SNPs and/or intrahost variants are restricted to some specific SARS-CoV-2 genomic region it likely indicates a recombination event. Otherwise, if iSNV sites are distributed along the entire SARS-CoV-2 genome it is likely to be derived from the codetection of different SARS-CoV-2 genomes in the same sample.
Recombination analysis
To identify putative recombination events, we compared the set of mutations present in each genome with the expected set of mutations of the lineage assigned by PANGO lineage. In this step, the common mutations (present in at least 75 % of genomes per lineage deposited on GISAID) in the 33 lineages identified in our samples are established based in the Lineage|Mutation Tracker available on outbreak.info, updated on 08 November 2021. The excess or lack of mutations are then compared with the mutations annotated with NextClade using an in house R script (https://github.com/dezordi/SARS-CoV-2_tools/ blob/master/compare_mutation.R). Samples with qc.overallStatus equal to ‘good’ and with ten amino acid mutations missing or in excess, were separated to a manual curation of lineage-specific mutations with the same information from outbreak.info. Samples with signals of mutations from two different lineages (parental lineages) were then analysed for recombination following the methodology of previous published studies [18, 19].
Phylogenetic analysis
A reference alignment was created using mafft [33] with the 6167 genomes, which represents the genomes used in Nextstrain [34] global phylogeny with N content less than 5 % accessed on 24 May 2021 and Brazilian genomes obtained through a cd-hit-est [35] clusterization of genomes present on GISAID at 16 March 2021 with high-quality and with more than 99.8 % sequence identity and from the same Brazilian state. The reference alignment was edited to mask UTR regions and to maintain the indel regions. The 32 MajV and MinV consensus genomes were aligned to reference alignment with mafft add, and we performed a maximum-likelihood phylogenetic analysis with IQtree2 [36] employing the aLRT branch support evaluation method and the GTR+F+R5 nucleotide substitution model. The PANGO lineages were evaluated with pangolin and used to annotate the tree with iTOL [37].
Results and discussion
Our initial analysis revealed that 1462 out of 2263 genomes had enough sequencing breadth and depth to be able to consistently detect and characterize the viral genomic variability at the sequencing reads level. In total, 1150 out of 1462 SARS-CoV-2 positive samples investigated showed at least one iSNV site, that is, at least one genomic position with more than 100 reads supporting a minimum of two alternative nucleotides. Those samples showed an average coverage depth of 1817.46 (sd=908.59) and an average coverage breath supported by at least 100 reads of 99.66 (sd=1.10) (Table S2). In addition, we estimated a mean of 2.57 iSNVs per genome (Table S3). MajV and MinV consensus sequences representing the viral genome variability found in each sample were generated for all samples bearing well supported alternative nucleotides and then assessed for lineage assignment using the PANGOlineage tool.
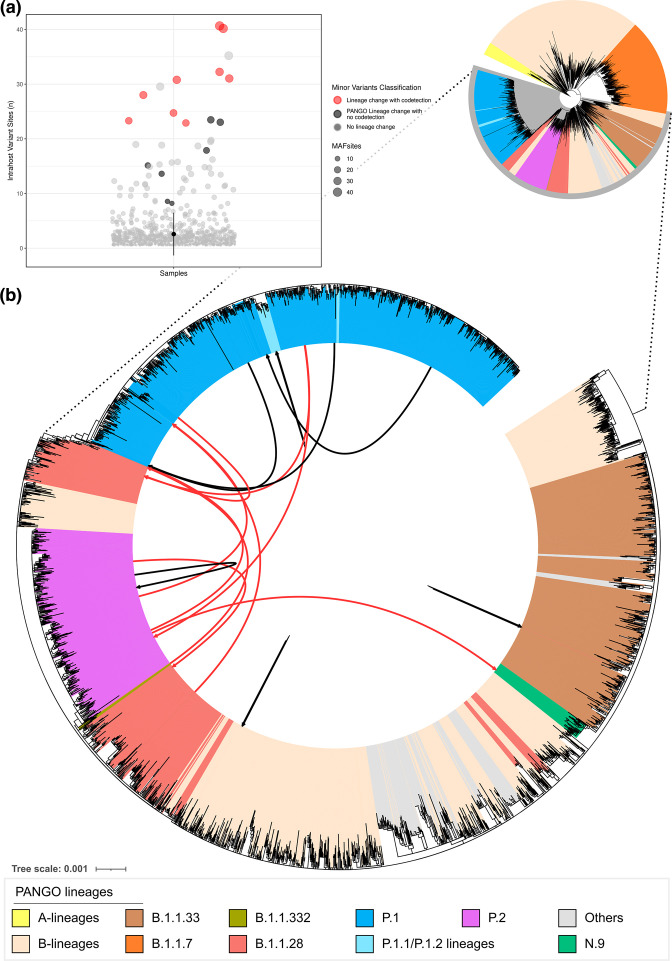
We detected 16 instances in which MajV and MinV were assigned to distinct lineages (iSNV sites: mean=24, sd=9.75), including former VOIs and now known as variants for further monitoring (VFM) N.9 and P.2 as well as the VOC P.1 (Table S4). To further confirm the lineage assigned by the PANGOlineage tool, we performed a phylogenetic analysis of representative lineages including both MajV and MinV genomes. Nine alternative genomes were confidently repositioned into distinct lineages (Fig. 1, red arrows, mean iSNVs sites 30.44, sd=6.63), while the remaining alternative genomes branched within the same lineage (Fig. 1, black arrows, mean iSNVs 23.06, sd=10.54). Seven out of nine putative coinfection events involve the VOC Gamma (P.1 lineage) (Table 1). In four cases, Gamma (lineage P.1) represented the MajV genome, while in the remaining three cases, it corresponded to the MinV genome. The large proportion of codetection events with P.1/Gamma is likely a result of the higher number of lineage-defining SNPs characteristic of this VOC that facilitate the distinction between coinfecting SARS-CoV-2 lineages. As more distinct lineages, bearing many lineage-defining SNPs, coinfect the same host, it becomes increasingly more likely to objectively distinguish coinfections through the reconstruction of alternative intrasample viral genomes.
Fig. 1.
Number of intrahost variant sites from 1150 SARS-CoV-2 samples and phylogenetic analysis of alternative MajV and MinV consensus genomes recovered from the same sample. (a) Dot plot with number of iSNVs per sample; (b) maximum-likelihood phylogenetic tree. Others: R, S, U, L, D, C lineages. Red arrows represent samples with alternative genomes showing lineage change while black arrows indicate alternative samples with no lineage change.
Table 1.
Summary of coinfection events
|
Sample |
State |
City |
iSNVs |
Breadth* |
Depth† |
Lineages‡ |
Collection date |
First MajV§ lineage available on GISAID |
First MinV§ lineage available on GISAID |
|---|---|---|---|---|---|---|---|---|---|
|
CE-FIOCRUZ-00657 |
CE |
Fortaleza |
23 |
98.95 |
762.04 |
P.2/P.1 |
2021-01-20 |
2020-11-20/2020-04-15 |
2021-01-07/2021-01-07 |
|
AM-FIOCRUZ-21142481RG |
AM |
Manaus |
31 |
98.9 |
1105.46 |
P.1/B.1.1.28 |
2021-01-13 |
2020-12-03/2020-12-03 |
2020-04-13/2020-04-13 |
|
RS-FIOCRUZ-2060 |
RS |
Canoas |
25 |
99.69 |
3509.92 |
P.2/B.1.1.28 |
2021-01-07 |
2021-01-26/2020-08-31 |
2020-05-21/2020-03-16 |
|
BA-FIOCRUZ-4739 |
BA |
Salvador |
31 |
99.73 |
5354.25 |
P.2/N.9 |
2021-01-08 |
2020-10-26/2020-06-26 |
2020-12-10/2020-11-12 |
|
ES-FIOCRUZ-6993 |
ES |
Aracruz |
32 |
99.67 |
2222.7 |
B.1.1.28/P.1 |
2021-01-09 |
2020-10-13/2020-10-13 |
2021-04-09/2021-01-22 |
|
CE-FIOCRUZ-6559 |
CE |
Fortaleza |
41 |
99.85 |
4090.76 |
P.1/P.2 |
2021-01-24 |
2021-01-07/2021-01-07 |
2020-11-20/2020-04-15 |
|
SC-FIOCRUZ-10891 |
SC |
Porto Belo |
28 |
98.72 |
2153.25 |
B.1.1.332/B.1.1.28 |
2021-02-22 |
2021-02-22/2021-02-22 |
2020-03-18/2020-03-18 |
|
BA-FIOCRUZ-10781 |
BA |
Salvador |
40 |
99.73 |
1996.63 |
P.2/P.1 |
2021-02-10 |
2020-10-26/2020-06-26 |
2020-12-27/2020-12-27 |
|
AM-FIOCRUZ-21890619RGS |
AM |
Manaus |
23 |
96.59 |
1018.89 |
P.1/B.1.1.28 |
2021-01-13 |
2020-12-03/2020-12-03 |
2020-04-13//2020-04-13 |
*Coverage breadth supported by 100 reads.
†Average coverage depth. AM: Amazonas; BA: Bahia; ES: Espírito Santo; CE: Ceará; RS: Rio Grande do Sul; SC: Santa Catarina.
‡MajV/MinV Pango lineages supported by the the phylogenetic analysis.
§Date of the first genome deposited on GISAID of each variant into the specific municipality/state, updated on 26 July 2021.
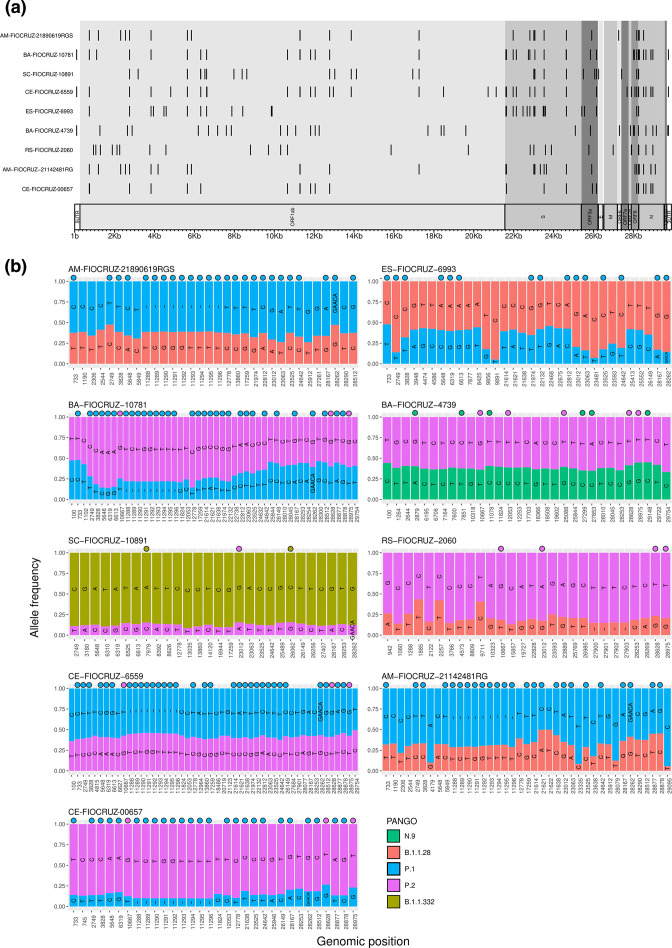
Intrahost single nucleotide variant sites identified showed several lineage defining SNPs spread across the whole SARS-CoV-2 genome, and the sequencing read depth was roughly similar throughout the genome (Fig. 2a, Table S5). Considering the lineage defining SNPs present in lineages assigned into MajV and MinV genomes and the absence of lineage defining SNPs of different lineages interlaced in the same genome (Fig. 2b, Table S3), our results indicate that the coinfection cases detected here did not generate hybrid recombinant genomes. Moreover, the analysis of missing and extra mutations of all 1150 consensus genomes did not reveal any putative recombined genome (Table S6). In order to assess if codetection could be a result of sample contamination we reassessed sample AM-FIOCRUZ-21142481RG from RNA extraction, library preparation and sequencing. We confirmed the intrahost variability for 25 out of 31 sites present in the first sequencing run (Table S7). Lineage assignment, phylogenetic reconstruction and the detection of SNP defining mutations confirmed the codetection status of that sample (Table S4) suggesting that the intrahost variability found in this sample did not result from laboratory contamination. However, this reassessment did not rule out contamination during sample collection and cannot be extrapolated to the other eight coinfection samples.
Fig. 2.
MajV and MinV of samples with codetection of different SARS-CoV-2 lineages. (a) Karyoploter with iSNV sites across the SARS-CoV-2 genome. (b) iSNV sites with read-depth frequency supporting MajV and MinV. Defining SNPs based on data of outbreak.info update on 24 July 2021, are indicated with a circle. Karyoplots depicting iSNV-site sequencing depth can be accessed in File S1, and raw depth values can be accessed in Table S5.
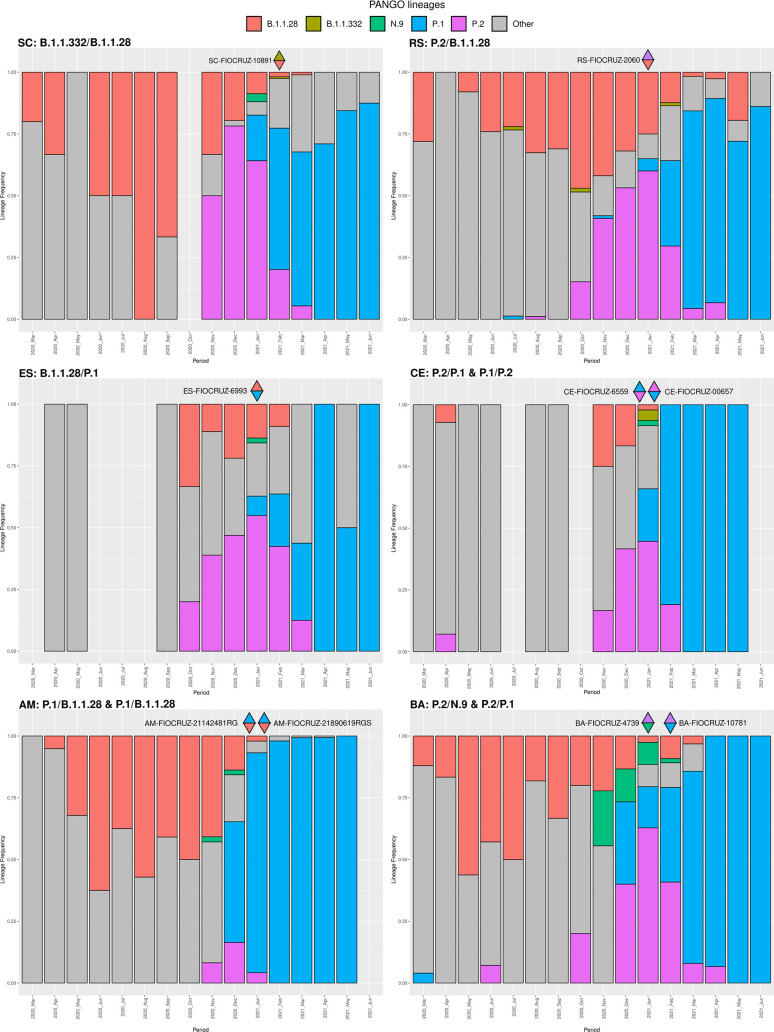
The plausibility of the codetection events was further validated by the fact of all MajV and MinV alternative genome lineages identified by our study were cocirculating in their sampling location, overlapping in time and space, as well as matching with SARS-CoV-2 lineage information form their respective geographical states (Fig. 3). The MajV genome corresponding to the predominant lineages circulating in Rio Grande do Sul, Bahia and Amazonas states were recovered in our analysis. On the other hand, in Santa Catarina, both lineages involved in codetection were present at lower frequency than the dominant lineages at the same location and period of sampling. Only one event of VOC circulation without previous notification was detected, the VOC Gamma was detected as a MinV genome in sample ES-FIOCRUZ-6393 from Aracruz city, Espirito Santo state, collected on 9 January 2021. The earliest record on GISAID of the VOC Gamma in Aracruz municipality was on 9 April 2021 and in Espirito Santo was on 22 January 2021 (Table 1). Of note, the genomic sequence of this specimen available on GISAID under EPI_ISL_2645599 code confirmed the assignment of the MajV genome to lineage B.1.1.28. These findings highlight that the analysis of MinV genome revealed the early spread of cryptically circulating lineages not detected by the analysis of MajV consensus genomes alone.
Fig. 3.
SARS-CoV-2 lineage proportion through time in different Brazilian states with codetection cases. Data were recovered from GISAID on 23 July 2021, raw data can be accessed in Table S8. Upper triangles coloured with the lineage of major consensus genomes and lower triangles with minor consensus genomes lineages.
Finally, the antiviral mechanism mediated by APOBEC-like host proteins against SARS-CoV-2 is known to induce a high frequency of ‘C→U transition’ in SARS-CoV-2 genomes [38] and may affect the identification of coinfection events based on the analysis of SARS-CoV-2 intrahost diversity. Our results showed a twofold difference of ‘C→U’ with respect to U→C or G→U mutations, a fourfold difference with respect to G→A or A→G and a 16-fold difference when compared with other transitions and transversions in single infection samples (File S2a), which is in line with other findings [16, 38]. By contrast, the frequency of intrahost changes in the nine coinfection cases showed a similar proportion between C→U and U→C (File S2b) mutations, suggesting that several lineage defining SNPs correspond to non-C→U mutations. Therefore, although the C→U bias has likely blurred the distinction of some lineage defining mutations, the remaining non-C→U lineage defining mutations are still sufficient to detect such events [38].
Conclusions
In line with other studies, we showed that SARS-CoV-2 has a low intrahost variability overall. Our in-depth analysis revealed at least nine codetection events, which are corroborated by epidemiological data from cocirculating lineages in different Brazilian states. Codetection/coinfection events occurred at a lower rate in Brazil (0.61%) compared to Europe (1–4 %) [16, 17]. However, this is certainly a lower bound estimate due to the limitation of detecting coinfection events with the same viral lineage or with low-divergent viral lineages that dominated the first year of the pandemic. The large number of genomic sites carrying alternative nucleotides in codetection events may generate artificial hybrid consensus genomes, therefore a careful inspection of consensus sequences against sequencing read polymorphisms is warranted to generate robust consensus sequences. Considering the large case numbers of SARS-CoV-2 infections worldwide and that coinfection are more likely to happen in high-transmission settings, all efforts should be placed to limit SARS-CoV-2 transmission hence reducing the likelihood of coinfection and the probability of emergence of novel recombinant hybrid lineages with altered phenotype.
Supplementary Data
Funding information
Financial support was provided by FAPEAM (PCTIEmergeSaude/AM call 005/2020 and Rede Genômica de Vigilância em Saúde - REGESAM); Ministério da Ciência, Tecnologia, Inovações e Comunicações/Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq/Ministério da Saúde - MS/FNDCT/SCTIE/Decit (grants 402457/2020–9 and 403276/2020-9); FAPERJ (CNE E-203.074/2017 and E-26/210.196/2020) Inova Fiocruz/Fundação Oswaldo Cruz (Grants VPPCB-007-FIO-18-2-30 and VPPCB-005- FIO-20-2-87) and INCT-FCx (465259/2014-6). Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (Covid 10 MCTI 402457/2020-0 and CNPQ BRICS STI 4441080/2020-0) This work was also supported by the Pan American Health Organization, Brazil Country Office and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001. FGN, GLW, GB and MMS are supported by the CNPq through their productivity research fellowships (306146/2017-7, 303902/2019, 302317/2017-1, 313403/2018-0, respectively).
Acknowledgements
We thank the Fiocruz COVID-19 Genomic Surveillance Network for sharing this large dataset and embracing such collaborative work. We also thank all the researchers around the world that are working and generating data of SARS-CoV-2 in those difficult times. The acknowledgement info of all SARS-CoV-2 genomes from GISAID and used in this work are present in File S3.
Author contributions
Conceptualization: F.Z.D. and G.L.W. Methodology: F.Z.D. and G.L.W. Formal analysis: F.Z.D. Investigation: F.Z.D., P.C.R., F.G.N., V.A.N., V.C.S., A.C.D.P., L.A., R.S.L., A.C.F.M., A.S.B.R., T.M.M.V., E.C.P., M.H.S.P., C.D., M.F.B., L.C.M., R.S.S., T.S.G., L.G.M., F.M.P., D.B.R., S.B.F., R.R., T.O.C., J.C.S., F.M., E.D., T.G., G.B., M.M.S. Resources: P.C.R., F.G.N., F.M., E.D., T.G., G.B., M.M.S., G.L.W. Writing: F.Z.D., P.C.R., F.G.N., V.A.N., V.C.S., A.C.D.P., L.A., R.S.L., A.C.F.M., A.S.B.R., T.M.M.V., E.C.P., M.H.S.P., C.D., M.F.B., L.C.M., R.S.S., T.S.G., L.G.M., F.M.P., D.B.R., S.B.F., R.R., T.O.C., J.C.S., F.M., E.D., T.G., G.B., M.M.S., G.L.W.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All samples used in this work are approved by the Ethics Committee of the Aggeu Magalhaes Institute Ethical Committee—CAAE 32333120.4.0000.5190, the Ethics Committee of Amazonas State University (no. 25430719.6.0000.5016), the Ethics Committee of FIOCRUZ-IOC (68118417.6.0000.5248) and the Brazilian Ministry of Health SISGEN (A1767C3).
Footnotes
Abbreviations: aLRT, approximate likelihood-ratio test; COVID-19, coronavirus disease 2019; iSNV, intrahost single nucleotide variant; MajV, major variant; MinV, minor variant; RNA, ribonucleic acid; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SNP, single nucleotide polymorphism; UTR, untranslated terminal region; VFM, variants for further monitoring; VOC, variant of concern; VOI, variant of interest.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
References
- 1.Dezordi FZ, Resende PC, Naveca FG, do Nascimento VA, de Souza VC, et al. Unusual SARS-CoV-2 intrahost diversity reveals lineage superinfection. Figshare. 2022. 10.6084/m9.figshare.19361270.v1 doi: 10.1099/mgen.0.000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaroszewski L, Iyer M, Alisoltani A, Sedova M, Godzik A. The interplay of SARS-CoV-2 evolution and constraints imposed by the structure and functionality of its proteins. PLoS Comput Biol. 2021;17:e1009147. doi: 10.1371/journal.pcbi.1009147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullen JL, Tsueng G, Abdel Latif A, Alkuzweny M, Cano M, et al. outbreak.info. 2021. https://outbreak.info/
- 4.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naveca FG, Nascimento V, de Souza VC, Corado A de L, Nascimento F, et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med. 2021;27:1230–1238. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- 6.Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021;124:102715. doi: 10.1016/j.jaut.2021.102715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabir JSM, Lam TT-Y, Ahmed MMM, Li L, Shen Y, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 8.Terada Y, Matsui N, Noguchi K, Kuwata R, Shimoda H, et al. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS One. 2014;9:e106534. doi: 10.1371/journal.pone.0106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein SA, Brown J, Pedersen BS, Quinlan AR, Elde NC. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. bioRxiv. 2021:2021. doi: 10.1101/2021.02.03.429646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau SKP, Li KSM, Huang Y, Shek C-T, Tse H, et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Shen L, Gu X. Evolutionary Dynamics of MERS-CoV: Potential Recombination, Positive Selection and Transmission. Sci Rep. 2016;6:25049. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin DP, Biagini P, Lefeuvre P, Golden M, Roumagnac P, et al. Recombination in eukaryotic single stranded DNA viruses. Viruses. 2011;3:1699–1738. doi: 10.3390/v3091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francisco R da S, Jr, Benites LF, Lamarca AP, de Almeida LGP, Hansen AW, et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296:198345. doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou HY, Cheng YX, Xu L, et al. Genomic Evidence for Divergent Co-infections of SARS-CoV-2 lineages. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tonkin-Hill G, Martincorena I, Amato R, Lawson AR, Gerstung M, et al. Patterns of within-host genetic diversity in SARS-CoV-2. elife. 2021;10:e66857. doi: 10.7554/eLife.66857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021;184:5179–5188. doi: 10.1016/j.cell.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haddad D, John SE, Mohammad A, Hammad MM, Hebbar P, et al. SARS-CoV-2: Possible recombination and emergence of potentially more virulent strains. PLoS One. 2021;16:e0251368. doi: 10.1371/journal.pone.0251368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brizzi A, Whittaker C, Servo LMS, Hawryluk I, Prete CA, et al. Report 46: factors driving extensive spatial and temporal fluctuations in COVID-19 fatality rates in Brazilian hospitals. medRxiv. 2021:2021.11.01.21265731. doi: 10.1101/2021.11.01.21265731. [DOI] [Google Scholar]
- 21.Nascimento VAD, Corado ALG, Nascimento FOD, Costa Á, Duarte DCG, et al. Genomic and phylogenetic characterisation of an imported case of SARS-CoV-2 in Amazonas State, Brazil. Mem Inst Oswaldo Cruz. 2020;115:e200310. doi: 10.1590/0074-02760200310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paiva MHS, Guedes DRD, Docena C, Bezerra MF, Dezordi FZ, et al. Multiple introductions followed by ongoing community spread of SARS-cov-2 at one of the largest metropolitan areas of Northeast Brazil. Viruses. 2020;12:E1414. doi: 10.3390/v12121414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Resende PC, Motta FC, Roy S, et al. SARS-CoV-2 Genomes Recovered by Long Amplicon Tiling Multiplex Approach using Nanopore Sequencing and Applicable to Other Sequencing Platforms. 2020. [DOI] [Google Scholar]
- 24.Dezordi FZ, Campos T de L, Jeronimo PMC, et al. ViralFlow: a versatile automated workflow for SARS-CoV-2 genome assembly, lineage assignment, mutations and intrahost variant detection. Viruses. 2022;2:217. doi: 10.3390/v14020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bam-Readcount The McDonnell Genome Institute. 2021. https://github.com/genome/bam-readcount
- 30.O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aksamentov I, Neher R. Nextclade. 2021. https://clades.nextstrain.org
- 32.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 36.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmonds P. Rampant C→U Hypermutation in the Genomes of SARS-CoV-2 and Other Coronaviruses: Causes and Consequences for Their Short- and Long-Term Evolutionary Trajectories. mSphere. 2020;5:e00408-20. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.