Abstract
Multicellular organisms rely on cell–cell communication to exchange information necessary for developmental processes and metabolic homeostasis. Cell–cell communication pathways can be inferred from transcriptomic datasets based on ligand–receptor expression. Recently, data generated from single-cell RNA sequencing have enabled ligand–receptor interaction predictions at an unprecedented resolution. While computational methods are available to infer cell–cell communication in vertebrates such a tool does not yet exist for Drosophila. Here, we generated a high-confidence list of ligand–receptor pairs for the major fly signaling pathways and developed FlyPhoneDB, a quantification algorithm that calculates interaction scores to predict ligand–receptor interactions between cells. At the FlyPhoneDB user interface, results are presented in a variety of tabular and graphical formats to facilitate biological interpretation. To illustrate that FlyPhoneDB can effectively identify active ligands and receptors to uncover cell–cell communication events, we applied FlyPhoneDB to Drosophila single-cell RNA sequencing data sets from adult midgut, abdomen, and blood, and demonstrate that FlyPhoneDB can readily identify previously characterized cell–cell communication pathways. Altogether, FlyPhoneDB is an easy-to-use framework that can be used to predict cell–cell communication between cell types from single-cell RNA sequencing data in Drosophila.
Keywords: single-cell genomics, cell–cell communication, Drosophila, signaling pathways, scRNA-seq, bioinformatics resources
Introduction
Single-cell RNA sequencing (scRNA-seq) is increasingly being used for high-throughput and high-precision studies to characterize cell states and cell types and, in particular, is quickly becoming the method of choice to study developmental and physiological processes. scRNA-seq avoids the limitation of only detecting average expression levels of RNAs as in traditional bulk RNA-seq approaches, and allows studies of sample heterogeneity at the single-cell level. scRNA-seq methods have also improved in terms of throughput and scalability, such that it is now possible to analyze tens of thousands of cells in 1 experiment (Fan et al. 2015; Klein et al. 2015; Macosko et al. 2015). Further, improvements in throughput and accuracy have expanded potential applications of scRNA-seq technology. For example, scRNA-seq is gradually being used to gain a deep understanding of entire development processes, including, through cluster analysis of all cells, definition of new subgroups, comparisons of subgroup heterogeneity, reconstruction of pseudo-time trajectories, generation of regulatory networks during development processes, and prediction of cell–cell interactions (Ghosh et al. 2020; Hung et al. 2020; Tattikota et al. 2020).
Several methods have been developed to predict cell–cell communication from scRNA-seq data, including CellPhoneDB, NicheNet, and CellChat (Browaeys et al. 2020; Efremova et al. 2020; Jin et al. 2021). All of these methods start by generating a single-cell gene expression matrix of ligand–receptor (L–R) pairs that is then used to predict the strength of interactions between cells. Each method has its own strengths. CellPhoneDB in its predictions of cell–cell communication not only considers L–R interactions but also considers coexpression of components of multisubunit L–R complexes (Efremova et al. 2020). NicheNet adds another layer of complexity as it predicts ligand–target interactions by integrating L–R information with downstream signal transduction and gene regulatory network information (Browaeys et al. 2020). CellChat considers signal cofactors, including inhibitory and stimulatory membrane-bound coreceptors, soluble agonists, and antagonists of cell–cell communication (Jin et al. 2021). All of these tools rely on the annotation of ligands and receptors while some also rely on the annotation of signaling pathways. Among all the existing resources/tools, only CellPhoneDB and CellChat are possible to be configured to analyze Drosophila data if user provides the L–R annotation, however, it is not easy to use for this purpose. For example, preparing files in the format needed for analysis requires extra work and does not provide ways for users to view activities of individual core pathway components.
Here, we developed FlyPhoneDB, a quantification algorithm that calculates interaction scores to predict L–R interactions between cells in Drosophila (Fig. 1a). We first established a high-confidence list of L–R pairs specific for the major Drosophila signaling pathways and stored the annotation in FlyPhoneDB. Next, we developed a pipeline that calculates interaction scores in scRNAseq data, which is then used to make predictions of L–R interactions between cells. We demonstrate the utility of the tool by analyzing 3 published scRNAseq datasets from the adult Drosophila midgut, abdomen, and immune system (Ghosh et al. 2020; Hung et al. 2020; Tattikota et al. 2020). Finally, we developed a web-based FlyPhoneDB user interface (http://www.flyrnai.org/tools/fly_phone), where users can browse data, upload additional L–R pairs, and analyze their own data sets.
Fig. 1.
Overview of FlyPhoneDB. a) Framework of FlyPhoneDB with input, statistical analysis, and user interface. b) FlyPhoneDB requires users to provide gene-cell matrix and metadata that contains barcode and cluster information. c) FlyPhoneDB provides various visualization tools to show ligand–receptor interactions between cell types. d) FlyPhoneDB website contains 4 different sections: (1) Browse the annotation database; (2) Submit your own annotations; (3) Browse an example; and (4) Analyze new datasets.
Materials and methods
Manual curation of L–R interactions for inclusion in FlyPhoneDB
To establish a high-confidence list of Drosophila L–R pairs, we manually curated information in the GLAD database (Hu et al. 2015; Upadhyay et al. 2017; Gontijo and Garelli 2018), FlyBase (Larkin et al. 2021), and QuickGO (Huntley et al. 2015) and imported these data into FlyPhoneDB. Other core components of major signaling pathways, such as transcription factors, were directly imported from GLAD.
Processing of Drosophila scRNA-seq data
The Drosophila midgut scRNA-seq dataset reported by Hung et al. (2020) was chosen for this study because a number of signaling pathways that maintain gut homeostasis have been well characterized, thus allowing us to evaluate the accuracy of FlyPhoneDB. The gene expression matrix and metadata were retrieved from GEO (accession code: GSE120537). The gene expression matrix was stored in the file “GSE120537_counts.csv.gz” in which rows are genes, columns are barcodes, and values are raw counts. The metadata was stored in the file “GSE120537_metadata.csv.gz” in which barcodes and cell type columns were extracted from this file (Fig. 1b). This information can also be extracted from Seurat Object with the function GetAssayData and seuratObj@meta.data (Stuart et al. 2019). The abdomen and blood datasets were obtained from GEO (GSE147601 and GSE146596, respectively) and processed similarly. The wild-type condition was analyzed from abdomen dataset and the wounded condition was analyzed from the blood dataset.
Calculation of L–R interaction scores and specificity
Expression values for each gene were normalized by dividing them by the total expression in each cell and then multiplying by the scale factor 10,000. Ligand and receptor expression levels were extracted from this normalized matrix based on the L–R pair database. Next, the average ligand and receptor expression values for each cell type were calculated by combining cell type information from the input metadata with the L–R expression matrix. The interaction score was calculated as the product of log transformed average ligand expression plus a pseudocount of 1 in the “sender cell” with the log transformed average receptor expression plus a pseudocount of 1 in the “receiver cell.” Specificity was calculated using a permutation test by random shuffling of the original cell type assignments from the metadata (1,000 times by default) and then recalculating the interaction scores. P-values were computed based on the interaction score distribution of randomly shuffled cell types. P-values <0.05 were considered significant.
Construction of the database and website
The L–R pairs and core components information was stored in a MySQL database. The back end of the website was written in PHP and the front end was written in HTML. The JQuery JavaScript library and the DataTables plugin were used in user interface and displaying tables. Both the database and website are hosted on the O2 high-performance computing cluster at Harvard Medical School that is supported by Research Computing group.
Results
Overview of FlyPhoneDB
FlyPhoneDB uses single-cell gene expression matrix and metadata that contains cell annotation as the input to calculate interaction scores based on gene expression and L–R pairs. To implement FlyPhoneDB, we first manually curated signaling pathway information from GLAD, FlyBase, and QuickGO to generate a list of high-confidence L–R pairs. This resulted in a set of 196 L–R pairs with the majority representing the EGFR, PVR, FGFR, Hedgehog, Hippo, Insulin, Notch, JAK/STAT, TGF-β, TNFα, Wnt, Toll, and Torso signaling pathways. Next, to identify the specificity of L–R interactions, FlyPhoneDB permutes the cell annotation and recalculates the interaction score as background for P-value calculation. The lower P-values (<0.05), the more specific the interaction between 2 cell clusters is predicted to be.
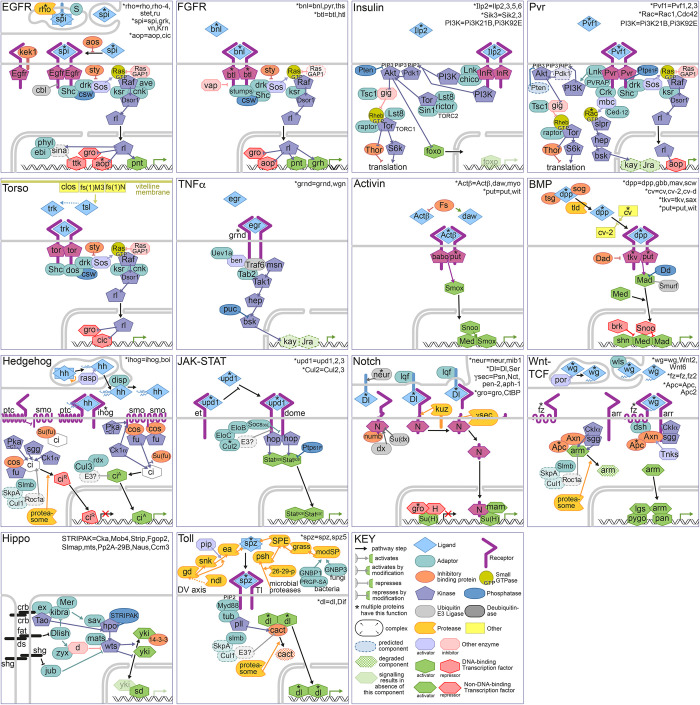
The FlyPhoneDB online analysis pipeline as well as the standalone program supports a variety of visualizations, including circle plots, dot plots, and heatmaps (Fig. 1c). For example, circle plots are provided for each signaling pathway, which depict all of the potential cell–cell communication events, with each node representing a unique cell type and each edge representing a communication event. The thickness of an edge reflects the interaction strength of the communication event, such that users can quickly identify cell types of interest for each pathway. In addition, we calculate the score of each L–R pair from one cell type to another and vice versa. If the score from one cell type to another is higher than the score from the reverse direction, we infer the direction accordingly, which is illustrated by the arrowhead on the edge. Dot plots are designed to provide additional detail. For example, in cases where there are multiple ligands and/or receptors involved in a pathway we use 1 dot plot to illustrate the statistical outputs for all of the L–R pairs between any 2 cell types. After users identify pathways and related cell types from a circle plot, users can zoom into more detailed information on the relevant dot plots. On the other hand, heatmaps, based on the average expression level of all major components, i.e. other than ligands and receptors, in all cell types and for each pathway are also provided. Lists of additional major components for each pathway were obtained from GLAD database (Hu et al. 2015). These heatmaps provide a quick way for users to compare expression levels among different cell types. We also provide thumbnails of the main pathways in EGFR, FGFR, Insulin, Pvr, Torso, TNFα, TGF-β/BMP, TGF-β/Activin, Notch, Wnt-TCF, Hedgehog, JAK–STAT, Hippo, and Toll, which are available at FlyBase (http://flybase.org/; release FB2021_04) (Fig. 2).
Fig. 2.
Thumbnails of the main signaling pathways. A set of thumbnails representing EGFR, FGFR, Insulin, Pvr, Torso, TNFα, TGF-β/BMP, TGF-β/Activin, Notch, Wnt-TCF, Hedgehog, JAK–STAT, Hippo, and Toll signaling pathways.
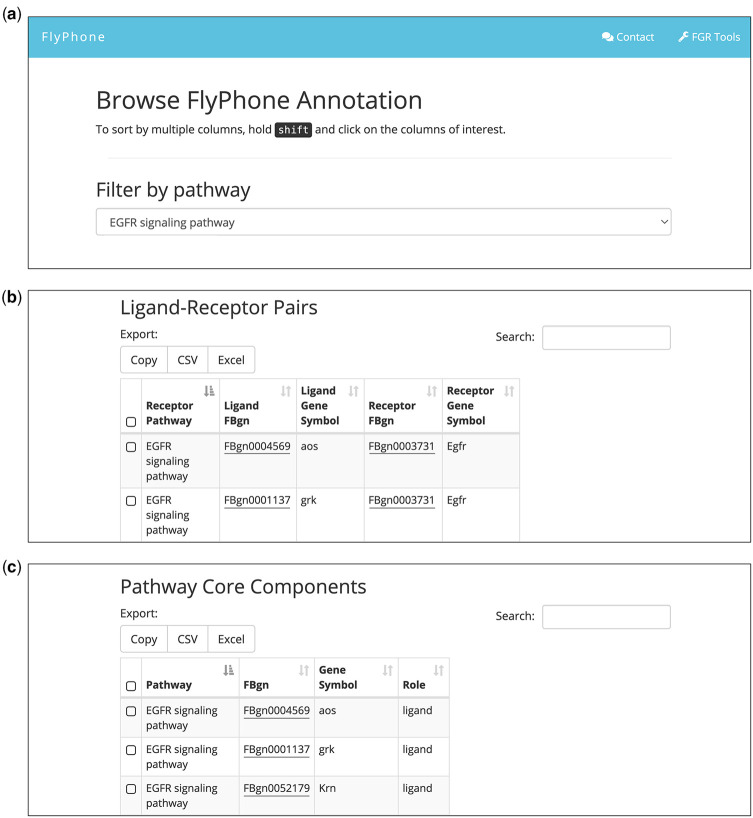
FlyPhoneDB website
The FlyPhoneDB site contains 4 different sections that allow users to do the following (Fig. 1d): (1) Browse the annotation of L–R pairs as well as the core component for each pathway with an option to download the information (Fig. 3); (2) Provide their own annotations by uploading a file with new information. Users have the option to upload 1 pair at a time or upload in batch mode using a template file. An upload triggers an email message to FlyPhoneDB. The information will then be reviewed by staff and updated on the backend database; (3) Browse the analysis result of scRNA-seq dataset from the Drosophila midgut as user case. Users can browse L–R interactions illustrated by dot plots and view expression patterns of all core components on a heatmap; and (4) Analyze new datasets. Users can upload a file of expression matrix obtained from a scRNA-seq dataset and the corresponding metadata file, and then analyze the dataset using the FlyPhoneDB pipeline. An email message is sent automatically when the analysis is completed and result files, including a table of statistics and visualizations, is made available for download.
Fig. 3.
FlyPhoneDB annotation panel. a) Browser of FlyPhoneDB annotation. b) Annotation of L–R pairs. c) Core components information for each signaling pathway. Users can filter, sort, and download the various tables.
Using FlyPhoneDB to identify signaling pathways in adult Drosophila organs
To assess FlyPhoneDB, we tested whether the tool could make accurate predictions, from existing scRNA-seq data sets, of cell–cell communication pathways that have been reported in the literature. We first focused our analysis on the fly gut, as a number of specific signaling pathways have been shown to regulate the maintenance of the adult midgut in an autocrine or paracrine manner.
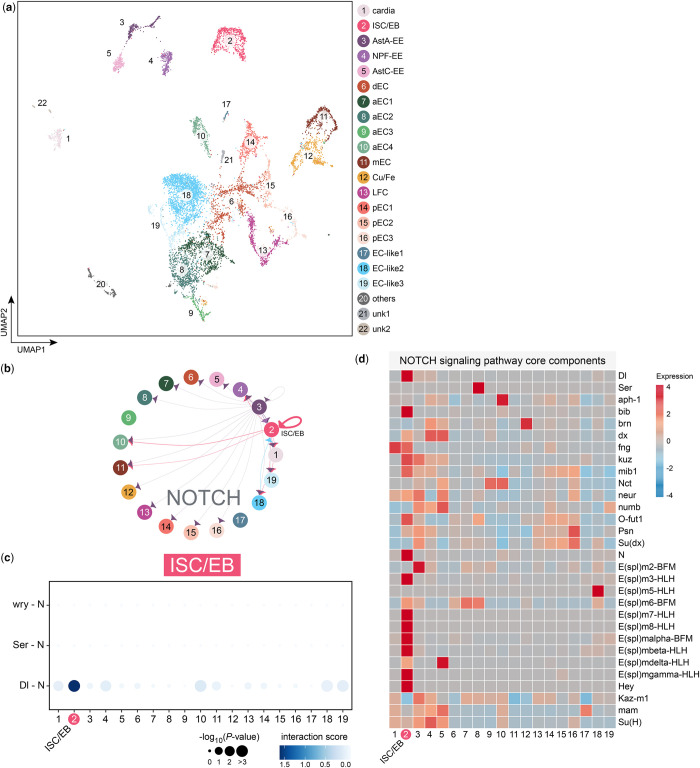
The adult midgut largely consists of absorptive enterocytes (ECs) and secretory enteroendocrine cells (EEs) that are replenished by proliferative intestinal stem cells (ISCs). These 3 major cell types can be further categorized into subtypes according to their spatial or transient states (Buchon et al. 2013; Dutta et al. 2015; Guo et al. 2019; Hung et al. 2020, 2021). For example, ISCs can differentiate into enteroblasts (EBs), which is a transient state before becoming ECs. Previously, we identified 22 clusters in scRNA-seq data acquired from whole Drosophila midguts (Hung et al. 2020). These clusters included 1 cluster annotated as ISC/EBs, 14 clusters annotated as ECs, 3 clusters annotated as EEs, 1 cluster annotated as cardia, and 3 clusters of unidentified cell types. Among the EC clusters, 4 clusters (aEC1–4) correspond to the anterior midgut, 1 (mEC) maps to the middle midgut, 3 (pEC1–3) to the posterior, one to copper and iron cells, one to large flat cells (LFC), and one to differentiating EC (dEC), and 3 were annotated as “EC-like” clusters. We applied FlyPhoneDB to this dataset and constructed heatmaps to visualize the expression levels for each of the 13 major signaling pathways (Supplementary Fig.1, a–c). We also generated visual representations of the interaction score and statistical significance of all possible combinations of L–R cell–cell interaction pairs.
We first looked at the Notch signaling pathway (Fig. 4). ISCs produce the ligand Delta (Dl), which activates the Notch (N) receptor, triggering ISCs to develop into EBs (Micchelli and Perrimon 2006; Ohlstein and Spradling 2006). Subsequently, EBs differentiate toward the EC lineage. Accordingly, our expression profile shows enrichment in the ISC/EB cluster of Dl, N, and other key components of the Notch signaling pathway, including kuzbanian (kuz), an ADAM family metalloprotease that plays a role in the cleavage of N (Lieber et al. 2002), and many of the enhancer of split [E(spl)] genes. In addition, ISC/EB > ISC/EB signaling had the strongest Dl–N interaction score of all pairwise interactions.
Fig. 4.
Analysis of signaling pathways in the adult Drosophila midgut. a) UMAP of the Drosophila midgut (data from Hung et al. 2020). b) Circle plot showing the significant interaction between the Delta (Dl) ligand and Notch (N) receptor in ISC/EB. c) Dot plot showing the significant ligand–receptor pairs of Dl–N in ISC/EB. d) Heatmap showing that Notch signaling in the entire gut is used only in ISC/EBs. It also gives a view of the components of the Notch network that is being used.
We next looked at the FlyPhoneDB output related to the JAK/STAT pathway. The L–R pairs for this pathway consist of binding interactions between any of the 3 unpaired ligands (Upd1–3) and the Domeless (Dome) receptor. In the gut, all Upds are upregulated upon injury (Jiang et al. 2009). Specifically, Upds produced from ECs activate JAK/STAT signaling in Dome-expressing ISCs to promote cell division. Upds are also required for proliferation of ISCs under homeostatic conditions (Osman et al. 2012). Accordingly, we observed upd2-3 expression in a few EC clusters, with the strongest expression in the pEC3 cluster, and we observed dome expression in the ISC/EB cluster (Supplementary Fig. 1a). Notably, for the L–R pair Upd3-Dome, the cell interaction score for aEC1 and pEC3 cluster signaling to the ISC/EB cluster was greater than that of ISC/EB signaling to these EC clusters. This indicates that the directionality of signaling can be inferred from the cell interaction scores. We also found that ISC/EB signaling to itself (ISC/EB > ISC/EB) had significant interaction scores for both the Upd2-Dome and Upd3-Dome L–R pairs, suggesting an autocrine role for Upd2 and Upd3.
We then looked at the EGFR signaling pathway. Upon damage or infection of the midgut, EGFR signaling in ISCs is enhanced by the upregulation and secretion of the EGFR ligands Vein (Vn), Spitz (Spi), and Kernen (Krn) from surrounding cells to promote proliferation (Jiang et al. 2011). Moreover, suppression of EGFR signaling during homeostasis also leads to a loss of EBs and ISCs (Jiang et al. 2011). This suggests that the ECs > ISCs L–R interactions of the EGFR pathway are active during homeostasis, albeit at a lower level. Consistent with this, we observed expression of Krn in 3 EC clusters (aEC4, mEC, and pEC2-3) and spi in the ISC/EB cluster. In addition, we also observed Vn expression in pEC2 and pEC3. Similar to the JAK/STAT pathway, cell interaction scores for EC to ISC/EB signaling for Vn-, Krn-, or Spi-Egfr L–R pairs were greater than those of ISC/EB to EC signaling (Supplementary Fig. 1b). This suggests that FlyPhoneDB can accurately predict the directionality of L–R pairs between cell types. Furthermore, EGFR signaling in the midgut is likely to be active at a basal level during homeostasis and could be primed for ISC proliferation in the event of damage.
Interestingly, among the many interactions observed in the FlyPhoneDB output for the midgut dataset, we found very strong interaction scores originating from various EE clusters that were directed toward other cell types. For example, all EE to ISC/EB interaction scores were strong with regards to the Spi-Egfr and Krn-Egfr L–R interactions. This suggests that EEs may send signals to ISCs to promote proliferation. Among other L–R pairs, Tk-TkR99D and Dh31–Dh31R have strong interaction scores between AstA-EE or NPF-EE and many EC or EC-like clusters. The Tk-TkR99D observation is consistent with expectation, as the secretion of Tk by EEs is thought to regulate lipid metabolism in ECs (Song et al. 2014). Less is known about the interaction of Dh31–Dh31R between EE and ECs, suggesting this is a topic that could be explored further. Finally, we also observed high interaction scores when comparing EEs between itself and other EEs cluster interaction scores for the L–R pair, Sema1a-PlexA. In the nervous system, Sema1a binds to its receptor PlexA and triggers a cascade of downstream events that lead to a repulsive growth cone response (Winberg et al. 1998; Winberg et al. 2001). This raises the interesting possibility that a similar mechanism is used by EEs to ensure that they are evenly distributed along the midgut. Altogether, these findings show that FlyPhoneDB can generate new predictions that can be further tested experimentally.
To further evaluate the strength of FlyPhoneDB predictions, we next applied the FlyPhoneDB analysis approach to a second dataset. The dataset was generated using single nuclei RNA-seq (snRNAseq) on whole abdomens and predominantly identified 3 clusters consisting of adipocytes, muscle cells, and oenocytes (Ghosh et al. 2020, 2021). In that study, we reported that the muscle produces the VEGF ligand Pvf1, which acts on its receptor, Pvr, in the oenocytes to inhibit lipid synthesis (Ghosh et al. 2020). FlyPhoneDB expression profile analysis accurately identified this cell–cell communication pathway, as Pvf1 and Pvr are enriched in the muscles and oenocytes, respectively. Accordingly, the Pvf1–Pvr interaction score for muscle to oenocyte signaling is greater than found for oenocyte to muscle signaling (Supplementary Fig. 2).
Finally, we applied FlyPhoneDB to a third study, which surveyed hemocytes in unwounded, wounded, and parasitic wasp-infested larvae (Tattikota et al. 2020). The study identified 17 clusters including 12 plasmatocyte (PM) clusters, 2 clusters of lamellocytes (LM), 2 clusters of crystal cells (CC), and 1 nonhemocyte cluster. Pathway enrichment analysis revealed that the FGF receptor breathless (btl) and its only ligand, branchless (bnl), was enriched in the LM2 and CC2 clusters, respectively. Functional analyses further showed that Bnl–Btl L–R communication between LM2–CC2 is crucial for the melanization of parasitoid wasp eggs. Consistent with this, the FlyPhoneDB expression profile indicates that Bnl is enriched in LM2 and Btl in CC2. Furthermore, the Bnl–Btl interaction score for LM2 to CC2 signaling is greater than that of CC2 to LM2 signaling (Supplementary Fig. 3).
In summary, by testing FlyPhoneDB with existing scRNA-seq datasets for well-studied tissues, we found that we were able to identify known L–R interactions, validating the effectiveness of the approach. In all the cases that we examined, the directionality of L–R between cell types, when known, was accurately predicted by selecting the higher of the 2 directional cell-to-cell interaction scores for a pair of cell types. Altogether, our reanalysis using FlyPhoneDB of established directional and autonomous L–R signaling events suggest that the approach can be used to uncover novel cell communication events.
Comparison of FlyPhoneDB with CellPhoneDB and CellChat
CellPhoneDB and CellChat each provide a standalone program at github without any web-based portal. CellChat provides ligand and receptor annotations for mouse whereas CellPhoneDB provides these annotations for human. Users have to install the program on a local computer to analyze datasets either using the mouse/human L–R annotations provided with the tool or by uploading L–R annotations for other species. In comparison, FlyPhoneDB provides: (1). a knowledgebase of manually curated high-quality L–R relationships for Drosophila, which is currently not provided by any of the existing resources; (2) a standalone program; and (3) a web portal for scientists to upload and analyze scRNA-seq data directly without a need to set up and run a program locally. The third option is quite important for bench scientists, who often lack the expertise to install and configure a complicated software package.
Regarding the core algorithm, FlyPhoneDB is a reimplementation of CellPhoneDB. CellPhoneDB does the analysis based on the average expression level of ligand and receptor using a program written in Python, whereas FlyPhoneDB is based on the product of L–R expression levels using a package implemented in R. By contrast, the core algorithm of CellChat is quite different. CellChat quantifies the communication probability based on a mass action-based model. Given that the core algorithm of CellChat is different from that of FlyPhoneDB, and given that the performance of CellChat has only been demonstrated using human and mouse data, we analyzed a Drosophila gut dataset (Hung et al. 2020) using CellChat with the Drosophila L–R annotations used at FlyPhoneDB and compared the results. Using a P-value of <0.05 as a filter, CellChat predicted 11,879 L–R relationships of 2,520 cell–cell communication events, while FlyPhoneDB predicted 4,615 L–R relationships for 1,814 cell–cell communication events. The overlap is 729 cell–cell communication events. Since cell–cell communication is not an extensively studied area with a gold standard set of benchmarks, we focused on the Notch, JAK/STAT, and EGFR signaling pathways relevant to gut ISC/EB as 3 ground truths established by literature. In all, FlyPhoneDB and CellChat identified 49 and 46 cell–cell communication events respectively, in which 35 events are overlapping (Supplementary Table 1). For both methods, all cell–cell L–R pairs were ranked based on the corresponding measurement score. Since multiple L–R combinations exist for each individual pathway, we used the highest L–R ranking as the representation of pathway activity in any given cell–cell interaction. For the purpose of evaluating the 2 methods, we assumed that a higher ranking equates to more relevance. Comparing the best rank of the cell–cell interaction of each signaling pathway showed that both methods are quite consistent. Both methods were able to identify ISC/EB > ISC/EB as the highest best ranked score amongst all pair-wise combination of source > target within the Notch pathway. Likewise, both approaches resulted in ISC/EB > ISC/EB scores with very high ranking for the EGFR pathway (FlyphoneDB : 3, CellChat : 1). aEC/pEC > ISC/EB also had similar best ranks when comparing both methods (aEC1–3, pEC1–3, FlyphoneDB: 35, 59, 34, 18, 19, 12; CellChat: 56, 45, 22, 20, 17, 16). On the other hand, some of the overlapping cell–cell communication events have different directionality. We investigated the pathways for which CellChat was not able to infer the same directionality observed with FlyPhoneDB. The directionality inferred from FlyPhoneDB is more consistent with the literature compared to CellChat. For example, the probability score of JAK–STAT signaling pathway between most EC clusters > ISC/EB pairs were no different from the reciprocal score (ISC/EB > EC clusters). Furthermore, the score of most ISC/EB > EC cluster had a higher score for all ligand–EGFR pairs than the opposite direction. This is contrary to what is expected.
Altogether, these comparisons highlight the advantages of FlyPhoneDB, including the inclusion of manually curated high-quality L–R relationships, the convenience offered to bench scientists, who can upload their data to get analysis result without the difficulty of setting up a program on a local computer, and the usefulness of obtaining more biologically relevant results when analyzing a Drosophila dataset.
Discussion
We have developed FlyPhoneDB to explore cell–cell communication in Drosophila using scRNA-seq data. To provide a high-confidence L–R interaction set, we manually curated L–R interactions from various resources, literature, and the Drosophila community. The strength of FlyPhoneDB analyses depends on an up-to-date database of L–R interactions. To facilitate community updates, we implemented a page at which users can use a form to suggest new L–R pairs and provide the relevant publication(s) supporting the database expansion. In addition to supporting user-initiated submissions, we will continue to work with FlyBase to curate L–R interactions reported in the literature and import them into FlyPhoneDB as new knowledge emerges. Given the ability to intake community input as well as the potential annotation pipeline from FlyBase, we anticipate that FlyPhoneDB will improve and expand over time, further increasing its value to the community. To make FlyPhoneDB user-friendly, we also developed the web-based FlyPhoneDB Explorer, which allows users to easily search the L–R database, upload new L–R annotations, browse the midgut cell crosstalk example, and upload scRNA-seq data to perform their own analyses. We also note that current scRNA-seq data do not provide spatial information and note that FlyPhoneDB can help predict spatial locations of cells in specific clusters by projecting cell–cell communication pathways onto the anatomy.
Although the core algorithm of score calculation of FlyPhoneDB is similar to that of CellPhoneDB, FlyPhoneDB presents a number of new features compared to other analysis tools that predict cell–cell communication from scRNA-seq data. Whereas all existing tools were originally developed to analyze human/mouse data, FlyPhoneDB was developed specifically for Drosophila research and makes it convenient to analyze Drosophila scRNA-seq data by providing manually curated high-quality L–R annotation for Drosophila and a web portal that allows users to directly analyze a dataset online. In addition, FlyPhoneDB provides multiple ways to visualize results. This includes displaying the activity of all core components besides L–R for each pathway through a heatmap, which can help users quickly compare pathway activities between different cell types. The availability of a standalone version of FlyPhoneDB also makes it possible for users to analyze datasets obtained from other organisms. With more and more scRNA-seq datasets becoming available, we anticipate that FlyPhoneDB will play an important role in the characterization of cell–cell communication in Drosophila and beyond.
Code availability
The code is available on Github: https://github.com/liuyifang/FlyPhoneDB.
Data availability
FlyPhoneDB web server is available at https://www.flyrnai.org/tools/fly_phone/web/. The code of the standalone program is available on Github: https://github.com/liuyifang/FlyPhoneDB. Three datasets used in this study are accessible through GEO series accession number GSE120537 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120537), GSE146596 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146596), and GSE147601 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147601).
Supplemental material is available at GENETICS online.
Supplementary Material
Acknowledgments
The authors would like to thank the members of the Perrimon laboratory, Drosophila RNAi Screening Center (DRSC), Transgenic RNAi Project (TRiP) for helpful suggestions regarding the project, and Stephanie Mohr for comments on the manuscript.
Funding
Relevant grant support includes NIH NIGMS P41 GM132087 and BBSRC-NSF. JSSL is supported by a Croucher Fellowship for Postdoctoral Research from the Croucher Foundation. HA is supported by the British Medical Research Council grant # MR/N030117/1. NP is an investigator of Howard Hughes Medical Institute.
Conflicts of interest
None declared.
Literature cited
- Browaeys R, Saelens W, Saeys Y.. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17(2):159–162. [DOI] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FPA, Fang HY, Boquete J-P, Deplancke B, Lemaitre B.. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3(5):1725–1738. [DOI] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, Buchon N.. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 2015;12(2):346–358. [DOI] [PubMed] [Google Scholar]
- Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R.. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484–1506. [DOI] [PubMed] [Google Scholar]
- Fan HC, Fu GK, Fodor SP.. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347(6222):1258367. [DOI] [PubMed] [Google Scholar]
- Ghosh AC, Tattikota SG, Liu Y, Comjean A, Hu Y, Barrera V, Ho Sui SJ, Perrimon N.. Drosophila PDGF/VEGF signaling from muscles to hepatocyte-like cells protects against obesity. Elife. 2020;9:e56969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AC, Tattikota SG, Liu Y, Comjean A, Hu Y, Barrera V, Ho Sui SJ, Perrimon N.. Correction: drosophila PDGF/VEGF signaling from muscles to hepatocyte-like cells protects against obesity. Elife. 2021;10:e66685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontijo AM, Garelli A.. The biology and evolution of the Dilp8-Lgr3 pathway: a relaxin-like pathway coupling tissue growth and developmental timing control. Mech Dev. 2018;154:44–50. [DOI] [PubMed] [Google Scholar]
- Guo X, Yin C, Yang F, Zhang Y, Huang H, Wang J, Deng B, Cai T, Rao Y, Xi R, et al. The cellular diversity and transcription factor code of Drosophila enteroendocrine cells. Cell Rep. 2019;29(12):4172–4185.e5. [DOI] [PubMed] [Google Scholar]
- Hu Y, Comjean A, Perkins LA, Perrimon N, Mohr SE.. GLAD: an online database of gene list annotation for Drosophila. J Genomics. 2015;3:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung R-J, Hu Y, Kirchner R, Liu Y, Xu C, Comjean A, Tattikota SG, Li F, Song W, Ho Sui S, et al. A cell atlas of the adult Drosophila midgut. Proc Natl Acad Sci USA. 2020;117(3):1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Li JSS, Liu Y, Perrimon N.. Defining cell types and lineage in the Drosophila midgut using single cell transcriptomics. Curr Opin Insect Sci. 2021;47:12–17. [DOI] [PubMed] [Google Scholar]
- Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O'Donovan C.. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:D1057–D1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8(1):84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA.. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137(7):1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, Myung P, Plikus MV, Nie Q.. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW.. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, Dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, et al. ; FlyBase Consortium. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021;49(D1):D899–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16(2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N.. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439(7075):475–479. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A.. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439(7075):470–474. [DOI] [PubMed] [Google Scholar]
- Osman D, Buchon N, Chakrabarti S, Huang Y-T, Su W-C, Poidevin M, Tsai Y-C, Lemaitre B.. Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J Cell Sci. 2012;125(Pt 24):5944–5949. [DOI] [PubMed] [Google Scholar]
- Song W, Veenstra JA, Perrimon N.. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9(1):40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattikota SG, Cho B, Liu Y, Hu Y, Barrera V, Steinbaugh MJ, Yoon S-H, Comjean A, Li F, Dervis F, et al. A single-cell survey of Drosophila blood. Elife. 2020;9:e54818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A, Moss-Taylor L, Kim M-J, Ghosh AC, O’Connor MB. TGF-beta family signaling in Drosophila. Cold Spring Harb Perspect Biol. 2017;9(9):a022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg ML, Noordermeer JN, Tamagnone L, Comoglio PM, Spriggs MK, Tessier-Lavigne M, Goodman CS. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 1998;95(7):903–916. [DOI] [PubMed] [Google Scholar]
- Winberg ML, Tamagnone L, Bai J, Comoglio PM, Montell D, Goodman CS.. The transmembrane protein Off-track associates with Plexins and functions downstream of Semaphorin signaling during axon guidance. Neuron. 2001;32(1):53–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
FlyPhoneDB web server is available at https://www.flyrnai.org/tools/fly_phone/web/. The code of the standalone program is available on Github: https://github.com/liuyifang/FlyPhoneDB. Three datasets used in this study are accessible through GEO series accession number GSE120537 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120537), GSE146596 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE146596), and GSE147601 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147601).
Supplemental material is available at GENETICS online.