Abstract
A notable number of acute lymphoblastic leukemia (ALL) patients develop CD19-positive relapse within 1 year after receiving chimeric antigen receptor (CAR) T cell therapy. It remains unclear if the long-term response is associated with the characteristics of CAR T cells in infusion products, hindering the identification of biomarkers to predict therapeutic outcomes. Here, we present 101,326 single-cell transcriptomes and surface protein landscape from the infusion products of 12 ALL patients. We observed substantial heterogeneity in the antigen-specific activation states, among which a deficiency of T helper 2 function was associated with CD19-positive relapse compared with durable responders (remission, >54 months). Proteomic data revealed that the frequency of early memory T cells, rather than activation or coinhibitory signatures, could distinguish the relapse. These findings were corroborated by independent functional profiling of 49 patients, and an integrative model was developed to predict the response. Our data unveil the molecular mechanisms that may inform strategies to boost specific T cell function to maintain long-term remission.
TH2 functionality is indispensable in maintaining 5-year relapse-free durable remission in CAR T therapy.
INTRODUCTION
Children with chemotherapy-resistant and/or refractory (r/r) acute lymphoblastic leukemia (ALL) have dismal prognosis (1, 2). Emerging as a potential immunotherapeutic approach, autologous chimeric antigen receptor (CAR) T cellular therapy targeting the B cell surface protein CD19 (CART19) has produced remarkable outcomes in the treatment of B cell malignancies (3–7). Multicenter clinical trials have demonstrated that 70 to 90% of patients with B-ALL achieve complete remission after receiving CART19 products (6–9), providing a potentially curative option for these patients. However, the remissions in a notable fraction of subjects are short-lived, and 30 to 60% of treated patients relapse within 1 year (8–11), let alone ~20% of ALL patients and over half of patients with large B cell lymphoma (LBCL) or chronic lymphocytic leukemia (CLL) fail to enter initial remission after CART19 infusion (12–15).
These disparities in therapeutic efficacy have evoked the efforts to interrogate the molecular determinants of long-term remission. It has been well accepted that robust in vivo CAR T cell engraftment and expansion is a prerequisite for inducing antitumor efficacy (6, 16), which is affected by several CAR T product characteristics such as CAR construct design (16, 17) and early memory T cell abundance (18–20). By characterizing the phenotype of premanufacture starting T cells, an elevated frequency of CD27+CD45RO−CD8+ T cells (21), a higher percentage of naïve and early memory T cell composition (22), and a decreased frequency of LAG-3+/TNF-αlo CD8+ T cells were found to be associated with sustained remission (23). In addition, impaired death receptor signaling in tumor cells (24), high pretreatment disease burden (8), and the detection of minimal residual disease (MRD) in the bone marrow by next-generation sequencing (25) also contributed to observed therapeutic failure and reduced remission duration.
However, the molecular mechanisms that underlie the acquired resistance to the CAR T cell treatment are still unclear. Although antigen loss could explain a majority of CD19-negative disease relapse after receiving CART19 therapy (26–28), the mechanisms of CD19-positive relapse remain elusive. We hypothesized that the functional capacity of CAR T cells in the infusion product could be an essential factor determining long-term therapeutic response. By combining single-cell RNA sequencing (scRNA-seq) and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) (29), we have investigated the single-cell activation landscape of CAR T infusion products from 12 pediatric r/r ALL patients upon CAR-specific stimulation or T cell receptor (TCR)–mediated activation, in comparison with unstimulated controls. We find that CD19-positive relapsed patients (n = 5) have a deficit of CAR T cells capable of inducing a T helper type 2 (TH2) functional response and of maintaining stem cell–like memory and central memory states compared with very durable responders (n = 5), whereas the expression levels of activation- or coinhibitory-related proteomic markers are comparable between two groups. We have performed an independent functional validation of these findings in two cohorts comprising 49 patients by means of flow cytometry and a multiplexed secretomic assay (30). In addition, we have incorporated the identified biomarkers into a predictive index using a binomial logistic regression. These results provide new insights into the molecular characteristics that underlie CD19-positive relapse via single-cell multiomics profiling of CAR T activation states and computational dissection of functional modules and biological pathways associated with clinical outcomes.
RESULTS
Single-cell multiomics profiling of CD19 CAR T infusion products from ALL patients
The infusion products of 12 pediatric and young adult patients (median age, 11.15 years; range, 4.9 to 21.5 years) from a phase 1/2A CART19 clinical trial of r/r B-ALL at the Children’s Hospital of Philadelphia were analyzed (table S1) (6). Patients were divided into two groups based on their initial response to the CAR T therapy: 10 patients (responders) had a complete remission defined by morphologic assessment of the bone marrow as M1 (<5% leukemic blasts), whereas the remaining 2 patients did not show an objective response to the therapy [nonresponders (NR)]. The responders were then subdivided into those who had a very durable complete remission (CR; >54 months, n = 5) and those who had a CD19-positive relapse during the course of the trial (RL; n = 5; median relapse-free remission duration = 9.6 months).
We sought to assess whether the intrinsic molecular characteristics of the infusion products could be predictive of clinical outcomes, reasoning that differences in T cell states and subtype proportions may determine both initial and long-term responses. Since the study of CAR T cells at baseline might not suffice to gain insights into the dynamics of early responses upon CAR engagement, we developed a stimulation method to emulate CAR-specific activation, for example, by B-ALL leukemic cells. To that end, CAR T cells in infusion products were analyzed upon CAR-specific stimulation with human CD19-expressing antigen-presenting cells (APCs) and compared them to three additional conditions: TCR-mediated stimulation, nontarget APC stimulation, and unstimulated controls (Fig. 1A). We engineered an NIH3T3 mouse cell line to stably express the human proteins CD19, CD86, and 4-1BB ligand (CD19-3T3 cells) (fig. S1A), and a control cell line expressing human mesothelin instead of CD19 (MSLN-3T3 cells) (fig. S1B). The infusion product of each patient was cocultured with the engineered cell lines for 6 hours at a 1:1 ratio. In addition to these experiments, we stimulated the same infusion batches using anti-CD3/CD28–coated beads for the same duration to examine the TCR-mediated activation states.
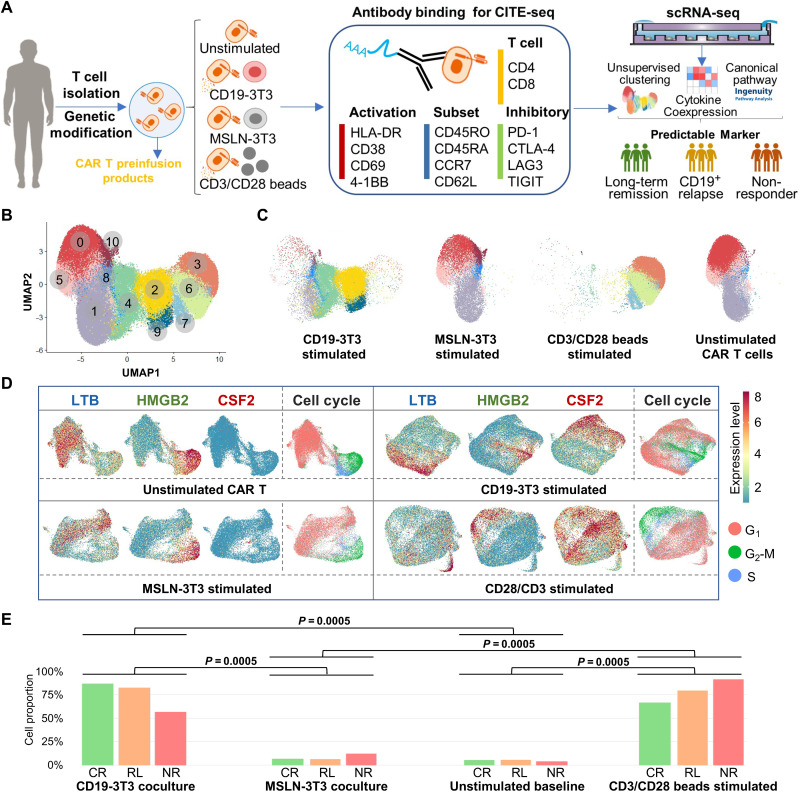
Fig. 1. A functional landscape and activation states of CD19 CAR T preinfusion products from patients with ALL.
(A) Schematic of experimental design. MSLN, mesothelin. (B) Uniform Manifold Approximation and Projection (UMAP) plot of 97,981 single CAR T cells collected from 12 patients. Eleven clusters are identified through unsupervised clustering. (C) UMAP distribution of all profiled cells separated by stimulation conditions. (D) Clustering and differential expression analysis of single CAR T cells revealed three major populations, characterized respectively by high expression levels of HMGB2, LTB, and CSF2. The distribution of the three marker genes and the cell cycle expression pattern of four experimental conditions were shown. (E) Quantification of fully active CSF2+ cell proportion in three response groups at the four conditions. Upon stimulation with CD19-3T3 cells or anti-CD3/CD28 beads, a large population of cells express high level of CSF2 and other cytokines, including IFNG, IL2, IL13, CCL3/CCL4, and XCL1/XCL2. The P values were calculated with Mann-Whitney test.
We performed scRNA-seq of the CAR T infusion products of the 12 patients under these four experimental conditions using a microwell-based approach (31). CITE-seq was adapted to this platform to concurrently profile 14 surface proteins through sequencing of antibody-derived DNA tags (ADTs), including T cell markers (CD4 and CD8), differentiation and subtype markers (CD45RO, CD45RA, CCR7, and CD62L), activation markers (HLA-DR, CD69, CD38, and 4-1BB), and coinhibitory markers (PD-1, CTLA-4, LAG-3, and TIGIT). In our coculture experiments, CD19-3T3 cells formed strong immune synapses with CAR T cells to induce CAR-specific activation. To separate the transcriptome of CAR T cells and APCs, we mapped the sequencing reads to the concatenation of the CAR construct, human, and mouse reference genomes and classified each read based on the genome it best aligned to.
A global transcriptomic landscape of single CAR T cell activation states
In total, we profiled the transcriptome of 101,326 cells from CAR T infusion products, of which 97,981 cells were analyzed after quality control and filtering. Integrating and clustering the scRNA-seq data across all patients and conditions identified 11 subpopulations with distinct transcriptomic signatures, which were visualized using Uniform Manifold Approximation and Projection (UMAP) (Fig. 1B). As expected, stimulated cells separated from their unstimulated or control counterparts in this representation and MSLN-3T3 cocultured cells overlapped with unstimulated cells (Fig. 1C), indicating experimental consistency and a minimal impact of antigen-negative costimulatory ligand-expressing mouse APCs. To exclude the possibility of xenogeneic stimulation by the NIH3T3 cells, we confirmed that the expression of genes associated with major immune functions in each individual patient was significantly higher in CD19-3T3 cocultures as compared with control MSLN-3T3 cocultures (fig. S2). Ingenuity Pathway Analysis (IPA) (32) was performed to explore the biological themes associated with the differentially expressed genes (DEGs) of each subpopulation, and we found that CAR T cells were highly heterogeneous in terms of their basic cell programs, cell cycle regulations, metabolic activities, T cell activation spectra, and effector functions that resulted upon CAR-specific activation (fig. S3).
Differential expression analysis between each subpopulation and the three clusters enriched for unstimulated cells (clusters 0, 1, and 5) revealed that subpopulations associated with stimulated cells (clusters 2, 3, 4, 6, 7, and 9) had significant (q < 0.05) up-regulation of CSF2 [colony-stimulating factor 2; granulocyte-macrophage colony-stimulating factor (GM-CSF)], IFNG [interferon-γ (IFN-γ)], and GZMB (granzyme B) and down-regulation of CCL5 (C-C motif chemokine 5), CD52, LTB (lymphotoxin β), and STMN1, suggesting that CAR T cells in these subpopulations are active functional effectors (fig. S4A). Two C-C motif chemokines, CCL3 [macrophage inflammatory protein-1α (MIP-1α)] and CCL4 (MIP-1β), and two C motif chemokines, XCL1 and XCL2, were also up-regulated in five of these clusters. In addition, cells cocultured with CD19-3T3 APCs separated from anti-CD3/CD28–stimulated cells in the UMAP representation (Fig. 1C), suggesting differences in the gene expression programs induced by TCR- and CAR-mediated T cell activation. Consistent with this observation, we found a deficit of IL13 and IL2 expression in subpopulations enriched for TCR-stimulated cells (clusters 3, 6, and 7), among other differences (fig. S4B).
The CAR T cell activation states and their association with initial treatment responses
The presence of lentiviral vector elements in the CAR construct allowed us to differentiate the expression of the CAR from that of homologous endogenous TCR and costimulatory elements (fig. S5A). On the basis of the fraction of cells with detected CAR construct expression among active cells in the stimulated samples, it was estimated that the sensitivity to detect the expression of the CAR construct in the scRNA-seq data was 47%. Using this estimate, we inferred that 30% [90% confidence interval (CI): 21 to 41%] of the T cells in the infusion products were CAR+ (fig. S5B), of which 57 to 68% and 34 to 53% (90% CI) were CD4+ and CD8+, respectively (fig. S5C), in agreement with previous reports of CAR transduction efficiency (6, 33).
On the basis of these results, our analysis was restricted to only CAR+ cells in the infusion products and each experimental condition was examined separately. Clustering and differential expression analysis of cells at baseline revealed two major populations of CAR T cells, characterized respectively by high expression levels of HMGB2 (high mobility box group 2) and LTB (Fig. 1D and table S2). The population of HMGB2-expressing CAR T cells was marked by high expression of cell cycle genes (table S2), consistent with these cells being actively proliferating. In contrast, the population of LTB-expressing cells consisted mostly of cells in the G1 phase of the cell cycle (Fig. 1D) and had expression of CCL5 (table S2). The same analysis in the single-cell data from CAR- or TCR-stimulated conditions revealed the presence of the same HMGB2+ and LTB+ cell populations across all experimental conditions, although their size was smaller upon CAR or TCR stimulation. In addition, in the experiments involving CAR or TCR stimulation, our analysis revealed a large population of cells with high expression of CSF2 (Fig. 1D) and other cytokines, including IFNG, IL2, IL13, CCL3 and CCL4, and XCL1 and XCL2 (table S2). Thus, upon stimulation with CD19-3T3 cells or anti-CD3/CD28 beads, the proportion of CSF2+ CAR T cells significantly increased for all patients [mean fold change (MFC) = 22.2 and 20.7 for CAR and TCR stimulation, respectively; Fig. 1E]. This increase was not observed upon coculture with MSLN-3T3 control cells, consistent with CSF2 expression being induced by the CAR or TCR activation. On the basis of these results, CSF2+ CAR T cells were identified as CAR T cells in a functionally active state.
We hypothesized that the number of functionally active CAR T cells upon stimulation might be related to disparities in the clinical responses. We compared the proportion of CSF2+ cells between the clinical response groups defined above. However, there was no significant difference in the abundance of CSF2+ cells between the three groups of patients (Fig. 1E), although we did observe a significant anticorrelation between the duration of the response to the therapy and the proportion of HMGB2+ CAR T cells upon stimulation with CD19-3T3 cells (Spearman’s r = − 0.65, P = 0.02).
Failure to sustain a long-term response to the therapy is associated with a deficit of TH2 function in antigen-specific CAR T activation
We next investigated the functional heterogeneity of the CAR T cell activation states upon stimulation with CD19-3T3 cells. We performed an unsupervised analysis to identify modules of coexpressed cytokines. This analysis identified multiple functional cytokine coexpression modules, including two type 1–like cytokine modules comprising an array of type 1 response genes like IFNG and IL2, a type 2 cytokine module consisting of IL4, IL5, IL9, IL13, and IL31, and a module consisting of beta chemokines like CCL3, CCL4, and related factors (Fig. 2A). These modules were highly consistent with the global landscape of single-cell transcriptional states of healthy donor CAR T cells that we identified in a previous study (34). Our analysis also identified a module consisting of LTB, CCL5, IL16, IL32, and other cytokine genes related to the LTB-expressing cell subpopulation described above. In addition, we considered a literature-derived expression module for terminally differentiated CD8 T cells (35), which was characterized by the expression of the transcriptional repressor ZEB2. After removing cells that did not belong to these modules, we generated UMAP representations to delineate the distribution of each functional module, in which cells were located according to their inferred cytokine module activities (Fig. 2B). Expectedly, MSLN cocultured control cells separated in the UMAP representation and showed high expression of the LTB module (Fig. 2C).
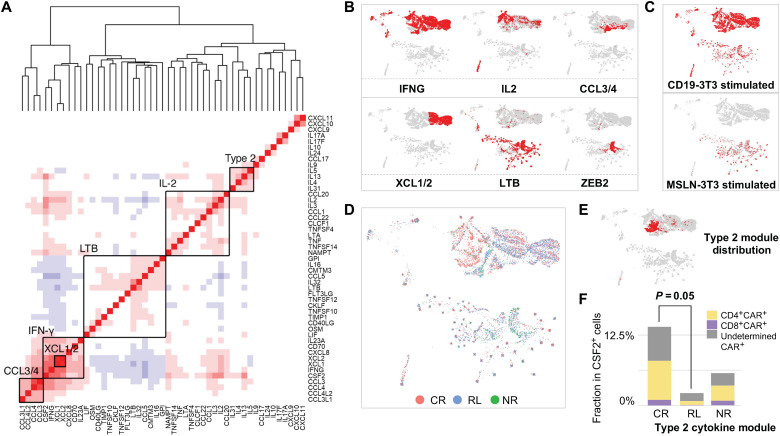
Fig. 2. Coexpressed cytokine module analysis of CD19-specific stimulated CAR T cells identified functional heterogeneities and a deficit of TH2 function in CD19-positive relapsed patients.
(A) Coexpressed cytokine modules identified in CD19-specific stimulated CAR+ cells through unsupervised analysis. Genes are ordered by hierarchical clustering. Multiple functional cytokine coexpression modules, including two type 1–like, a type 2, and two chemokine modules, were identified. (B) Expression distribution of the identified modules on the UMAP. (C) Integrated cytokine module representation UMAP split by experimental conditions. (D) Localization of cells from each response group on the integrated UMAP. (E) Distribution of type 2 (TH2) module. (F) Comparison of type 2 (TH2) module–expressed CSF2+CAR+ active cell proportion between very durable remission patients (CR) and CD19+ relapsed patients (RL). The P values were calculated with Mann-Whitney test.
In the landscape of major responsive states, CAR+ cells from CR, RL, and NR patients were localized in the representation (Fig. 2D), suggesting a distinct cytokine module expression profile for these cells. Notably, the type 2 (TH2) module was found to be enriched in a region containing mostly cells from CR patients (Fig. 2E), implying that TH2 function might be indispensable for maintaining a long-term remission in CAR T therapy. A quantitative comparison between the landscape of CD19-3T3–stimulated CAR T cells identified a significant depletion of CAR+ cells expressing the type 2 cytokine module in RL patients compared to CR patients (MFC = 4.14; Mann-Whitney test, P = 0.05) (Fig. 2F), whereas other functional modules remained comparable between the two response groups (fig. S6). These data prompted us to perform more detailed investigations of the differences between the activation states of CAR T cells from CR and RL patients.
Functional immune programs in activated CAR+ cells do not distinguish CD19-positive relapse from CR subjects, except for TH2-related pathways, genes, and upstream regulators
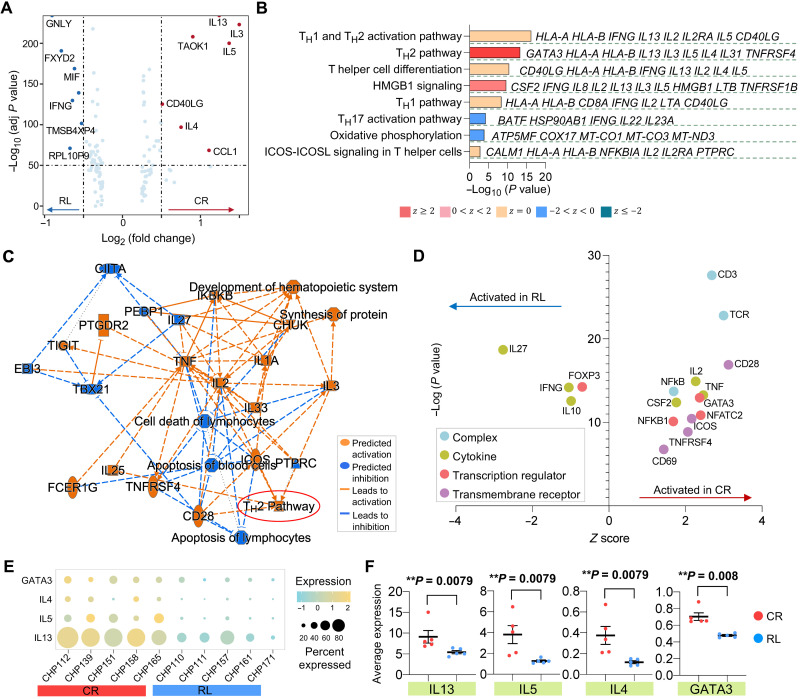
To identify the characteristics of functional CAR T cell states associated with clinical responses, we performed differential expression analyses between the CAR+ cells from CR and RL patients at the unstimulated basal level or upon CAR stimulation. The analysis of baseline CAR T cells only revealed the up-regulation of eukaryotic initiation factor 2 (EIF2) signaling in the CR group (fig. S7), which might represent active protein synthesis and translation initiation in these cells (36). Comparing CAR T cells between CR and RL patients upon antigen-specific stimulation showed the up-regulation of genes associated with TH2-related cytokine production, such as IL4, IL5, and IL13, in CR patients (Fig. 3A), consistently with our previous module analysis. Other immune pathways like TH1 cytokine production, TH differentiation, and ICOS-ICOSL signaling were comparable between the two response groups (Fig. 3B). In line with these findings, a network analysis of canonical pathways, upstream regulators, and biological functions regulated by the DEGs showed that CD28, ICOS, interleukin-3 (IL-3), and IL-25 were collectively operative in promoting the activation of the TH2 pathway in CAR T cells from CR patients (Fig. 3C). To explore the cascade of upstream regulators and assist the interpretation of the observed gene expression differences, we also performed IPA upstream analysis. The transcription regulator FOXP3 and the cytokines IL27, IFNG, and IL10 were particularly active in cells from RL patients. By contrast, GATA3, a master transcription factor mediating the activation of TH2 signature genes, and several genes involved in T cell activation, including NFKB1, NFATC2, TNF, CD28, and CD69, were up-regulated in CR patients (Fig. 3D). To ensure that the above observations were not biased by any single subject, we assessed the expression of IL13, IL5, IL4, and GATA3 in each of the CR and RL patients. The results of this analysis showed ubiquity of expression enrichment of the four genes in CR patients compared with relapsed subjects (Fig. 3, E and F).
Fig. 3. TH2 function–related pathways, genes, and upstream regulators are collectively down-regulated in activated CAR T cells from patients with CD19-positive relapse.
(A) Volcano plot of DEGs between CAR+ cells from CR and RL patients. (B) Corresponding canonical pathways regulated by the highly differential genes identified in (A). Pathway terms are ranked by –log10 (P value). The side listed gene names represent symbolic molecular marker related to the pathway. A statistical quantity, called z score, is computed and used to characterize the activation level.Z score reflects the predicted activation level (z < 0, inhibited; z > 0, activated; z ≥ 2 or z ≤ − 2 can be considered significant). (C) Graphical network of canonical pathways, upstream regulators, and biological functions regulated by DEGs identified in (A). (D) Predicted activation of upstream regulators, including complex, cytokine, transcription regulator, and transmembrane receptor, in CR or RL patients. (E) Dot plot of TH2-related gene expression of each patient in CR and RL groups. The size of circle represents the proportion of single cells expressing the gene, and the color shade indicates the normalized expression level. (F) Average expression level of genes IL13, IL5, IL4, and GATA3 across all single cells in each patient and their comparison between CR and RL groups. Each scatter point represents the average expression value of all single cells of specific patient. The P values were calculated with Mann-Whitney test. Scatter plots show means ± SEM.
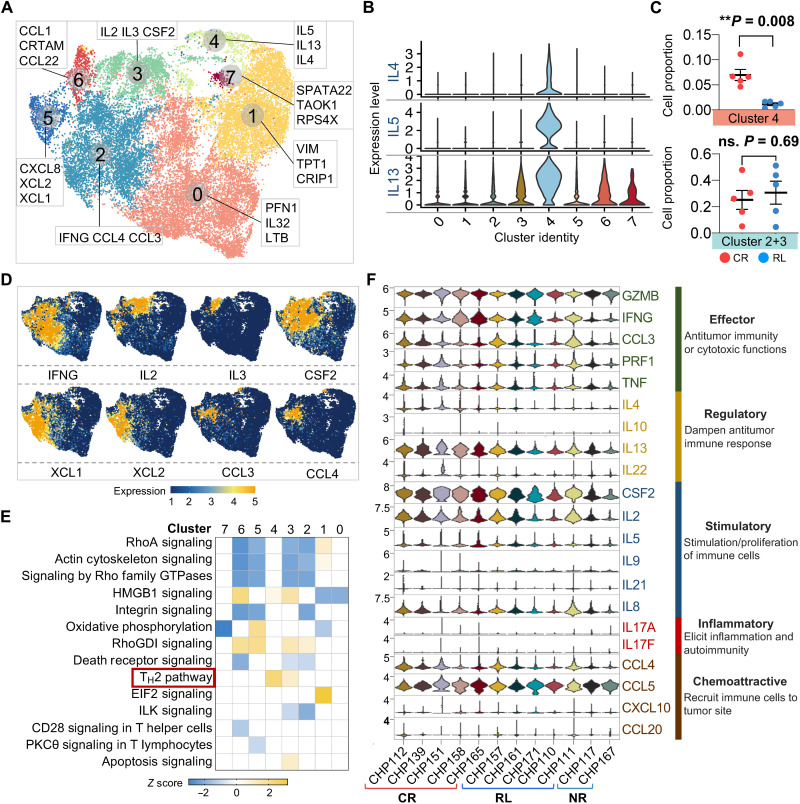
We next asked whether the proportion of cells with specific functional signatures affect the sustainability of CAR T therapy. Clustering activated CAR+ cells from CR and RL samples identified eight transcriptionally distinct subpopulations (Fig. 4A). The top three genes differentiating each cluster indicated that CAR T cells in cluster 4 mainly functioned as TH2 helpers (Fig. 4B and fig. S8), and cells in clusters 2 and 3 had vigorous expression of cytokine and chemokine genes, including IL2/3, IFNG, CSF2, XCL1/2, and CCL3/4 (Fig. 4D). The cell proportion of cluster 4 was significantly elevated in CR patients, and no fraction difference was observed for the combination of clusters 2 and 3 (Fig. 4C), suggesting that the lack of TH2 function, rather than TH1 response, could induce CD19-positive relapse. Consistently, the comparison of canonical pathway activities between clusters did not detect differences in other major immune programs (Fig. 4E). To further verify these CAR+ cell functional profiles at the transcriptional level, we looked at the single-cell gene expression levels of key immunologically relevant molecules across major categories, including effector, regulatory, stimulatory, inflammatory, and chemoattractive functions (Fig. 4F). No apparent differences were found when comparing CR individuals with RL, with the exception of IL4 and IL13. Together, these results show that upon antigen-specific stimulation of CAR T infusion products, the TH2-related genes, upstream regulators, and pathways are collectively down-regulated in RL patients as compared with their long-term CR counterparts, whereas other major immune programs remain comparable.
Fig. 4. Single-cell transcriptomic clustering of CAR T cells identifies comparable functional immune profiles between CR and RL patients, except for TH2 function.
(A) UMAP reclustering of CAR+ cells from CR and RL patients and the top three dominant genes defining each cluster. (B) Expression of IL4, IL5, and IL13 in each identified cluster. (C) Comparison of cell proportion in particular cluster between CR and RL patients. The P values were calculated with Mann-Whitney test. ns., not significant. Scatter plots show means ± SEM. (D) Distribution feature plot of functional genes on the UMAP. (E) Canonical signaling pathway comparison between all the identified clusters. DEGs of each cluster are used to identify the biological pathways. A statistical quantity, called z score, is computed and used to characterize the activation level.Z score reflects the predicted activation level (z < 0, inhibited; z > 0, activated; z ≥ 2 or z ≤ − 2 can be considered significant). (F) Single-cell expression level violin plot of all key immunologically relevant cytokine genes from CAR+ cells in each patient.
Independent functionality assessment in an expanded cohort confirms the deficiency of TH2 function in CD19-positive relapse patients
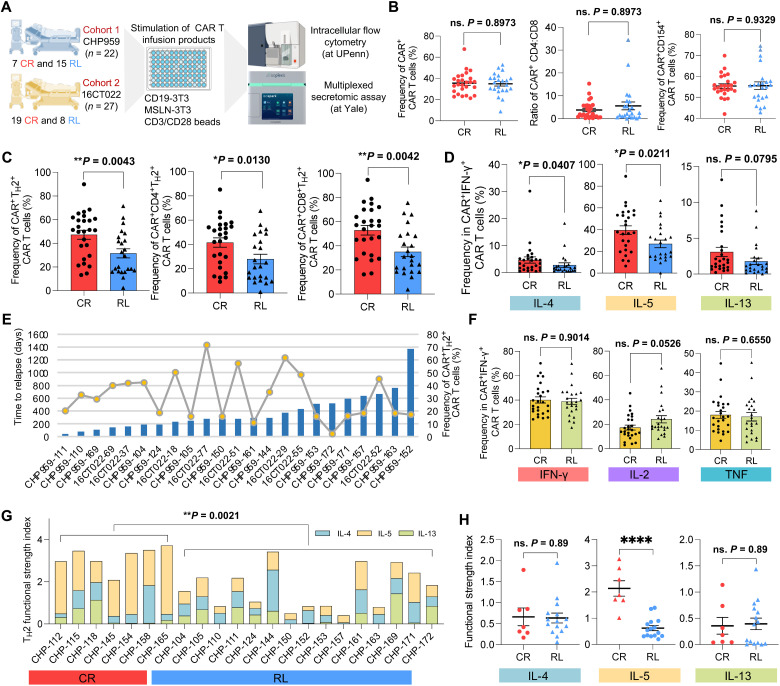
To further validate and refine our findings, we expanded our study to 49 B-ALL patients enrolled in two anti-CD19 CAR T cell therapy trials (Fig. 5A). Twenty-six patients had robust CR response with median relapse-free remission duration of 33.7 months, and the other 23 patients developed CD19-positive relapse at a median time point of 9.3 months (table S3). We performed an independent evaluation on the preinfusion CAR T cells of these patients using intracellular flow cytometry and a multiplexed secretomic assay at separate research centers. In the flow cytometry dataset, fully active CAR+ IFN-γ+ T cells were gated for downstream analysis (fig. S9). Comparing the frequencies of IFN-γ+, IL-2+, CD154+, and TNF+ cells between MSLN-3T3 and CD19-3T3 or anti-CD3/CD28 bead coculture conditions provided further confirmation of the absence of xenogeneic stimulation from the NIH3T3 mouse cells (figs. S10 and S11).
Fig. 5. Independent functional evaluation in expanded cohorts validates the deficiency of TH2 function in RL patients.
(A) Schematic of validation experimental design. (B) Comparison of CAR+ CAR T cell frequency, CD4:CD8 ratio, and CD154+ CAR T cell frequency between CR and RL patients. (C) Comparison of CAR+TH2+ CAR T cell frequency between CR and RL patients. Left, combined CD4+ and CD8+; middle, CD4+; right, CD8+. TH2 measures the combined frequency of IL-4+, IL-5+, and IL-13+ cells. (D) Frequency comparison of each TH2-related cytokine+ CAR T cells between CR and RL patients. (E) Correlation between TH2+ CAR T cell frequency and the relapse-free duration to the therapy. Bar plots show the days to relapse of each RL patient. Line and dot plots show the frequency of CAR+TH2+ cell of each RL patient. (F) Frequency comparison of major TH1-related cytokine+ CAR T cells between CR and RL patients. (G) Comparison of the TH2 functional strength index (FSI) between CR and RL groups, which was measured by multiplexed secretomic assay using Isoplexis devices. FSI denotes to the frequency of cells secreting specific cytokine multiplied by the average signal intensity of this cytokine. (H) PSI comparison of IL-4, IL-5, and IL-13 between CR and RL groups. All the P values were calculated with Mann-Whitney test. Scatter plots show means ± SEM. ****P < 0.0001.
Quantification of the CAR transduction efficiency, CD4+/CD8+ ratio, and activation level in the CAR T cells did not reveal any significant differences between CR and RL patients (Fig. 5B). By contrast, the combined proportions of IL-4+, IL-5+, and IL-13+ CAR T cells (TH2+) were significantly higher in the CR group compared with RL patients, with consistent discrimination observed in CD4+ or CD8+ subpopulation (Fig. 5C). The analysis of each individual TH2-related cytokine also showed clear differences between CR and RL groups, except for IL-13+ cells, which were not significantly enriched in any of the two groups (Fig. 5D). We did not observe a significant correlation between the duration of the effective responses in RL patients and their proportion of CAR+TH2+ cells (Fig. 5E). For example, patient CHP959-152 had the longest relapse-free duration in our RL cohort, whereas the TH2+CAR+ frequency of this patient ranked behind most of the other RL patients. We also compared the frequencies of CAR+ cells expressing TH1-related cytokines, chemokines, or cytotoxic enzymes (Fig. 5F and fig. S12). In this case, we did not find any significant difference between CR and RL patients, in agreement with our previous analysis of coexpressed cytokine gene modules.
To robustly characterize the role of TH2 functionality in maintaining long-term CR, we conducted a multiplexed secretomic assay on the infusion products of 22 patients (CHP959 trial) upon CD19-3T3 stimulation. We used the functional strength index (FSI) to describe the specific functionality profile of CAR T cells, which was defined as the frequency of cells secreting a cytokine multiplied by the average signal intensity of the cytokine. We found a significantly higher TH2 FSI in CR patients as compared with RL patients (Fig. 5G). The average intensity of TH2 cytokines alone was also able to discriminate CR from RL patients, whereas no significant difference was found in the secretion frequency (fig. S13). In addition, the predictive power of the IL-5 FSI was greater than that of the combined TH2 FSI (Fig. 5H). Together, these data demonstrate a general and robust association between TH2 function deficiency and CD19-positive relapse.
Phenotypic proteomic profiling reveals that early memory states, rather than activation spectra or coinhibitory signatures, could predict CD19-positive relapse
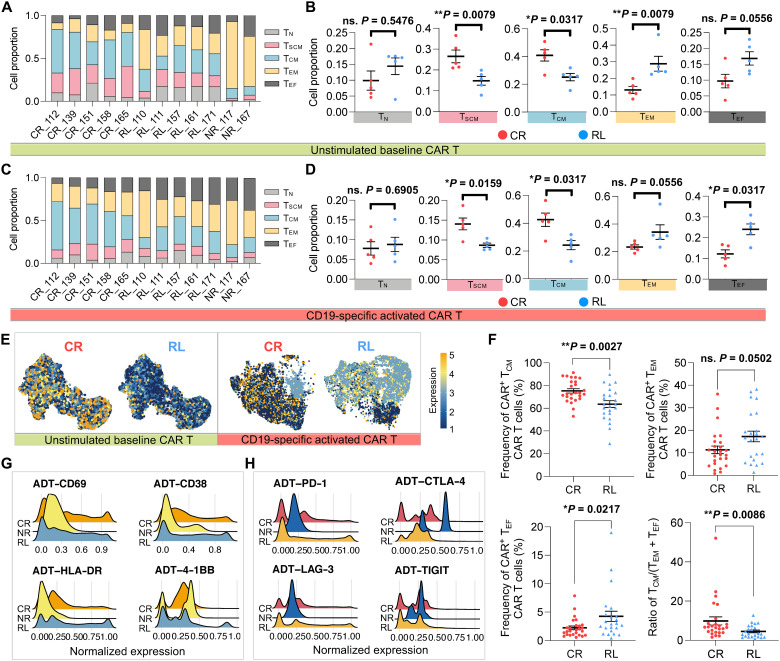
Given the role of cellular stemness and memory formation of CAR T cells in mediating the efficacy and persistence of the therapy, we investigated the composition of naïve (TN), stem cell–like memory (TSCM), central memory (TCM), effector memory (TEM), and effector (TEF) subsets in the infusion products from the three clinical response groups based on CD62L, CCR7, CD45RA, and CD45RO cellular protein expression from our CITE-seq data (fig. S14) (37). For unstimulated baseline CAR T cells, we found significantly higher proportions of TSCM and TCM and a lower proportion of TEM cells in the five CR patients (Fig. 6, A and B). We also observed that most of the CAR T cells derived from NR patients developed late memory signatures (Fig. 6A). Upon CAR stimulation, the lower abundance of TSCM and TCM cells was still significantly associated with the relapsed subjects (Fig. 6, C and D). There was no difference in the frequency of TEM CAR T cells, yet a higher proportion of cells in the RL group differentiated into a TEF state compared with the CR group. The expression of the memory-related surface marker CCR7 explained the disparities of CAR T differentiation state composition between long-term responders and relapsed patients, both at baseline and upon CAR stimulation (Fig. 6E). Our flow cytometry analysis in the expanded cohort confirmed a significant increase of the TCM and a decrease of the TEF cell proportion in CR patients as compared with RL patients, thereby providing further validation of our findings (Fig. 6F).
Fig. 6. Phenotypic proteomic profile reveals the dampened capacity of maintaining early memory T cell states in CAR T cells from relapsed patients.
(A) Differential state composition of unstimulated baseline CAR T cells in each patient based on the expression of CD62L, CCR7, CD45RA, and CD45RO from CITE-seq data. TN, naïve; TSCM, stem cell–like memory; TCM, central memory; TEM, effector memory; TEF, effector T cells. The x axis represents responsive state and patient ID. (B) Proportion comparison of unstimulated CAR T cell phenotypic subsets between CR and RL groups. (C) Differential state composition of CD19-specific activated CAR+ cells in each patient. (D) Proportion comparison of activated CAR+ cell phenotypic subsets between CR and RL groups. (E) Feature plot of cellular protein CCR7 expression in unstimulated or activated CAR T cells, split by responsive groups. (F) Frequency of CAR+ cells with different differential states in the expanded validation cohort. (G) Single-cell expression distribution of the T cell activation–related surface protein markers of each group. (H) Single-cell expression distribution of coinhibitory surface protein markers of each group. All P values were calculated with Mann-Whitney test. Scatter plots show means ± SEM.
We also evaluated the surface protein markers of T cell activation and coinhibition in activated CAR+ cells across patient groups. Of the four major activation markers in the panel, a higher proportion of cells from NR patients had a relatively low expression of CD69, CD38, and HLA-DR, whereas the expression levels were not significantly different between the CR and RL groups (Fig. 6G and fig. S15, A and B). Likewise, the genes coding for these activation markers were expressed at similar levels (fig. S15, C and D). A similar analysis of T cell coinhibitory signatures associated with dampened proliferation and effector functions (PD-1, CTLA-4, LAG-3, TIGIT, and their corresponding genes) did not reveal any significant differences between CR and RL patients (Fig. 6H and fig. S16). In summary, our phenotypic analysis indicates that CD19-positive relapse is associated with deficiencies in the capacity of CAR T cells to maintain early memory states.
An integrative model establishes the predictive power of the combined TH2 functionality strength and early memory potential
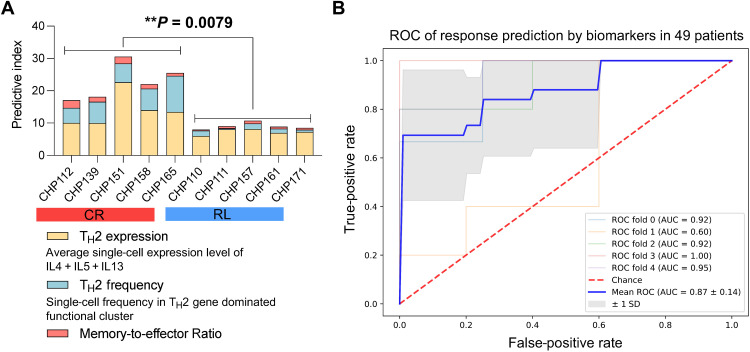
Given the various biomarkers that we have identified in the preinfusion CAR T products, we asked whether we could integrate them into a single predictive model of clinical response to the therapy. To evaluate if the combination of some of these biomarkers into a single predictive index could be a viable strategy, we aggregated three of the major biomarkers, consisting of the TH2 expression level, the TH2 expression frequency, and the memory-to-effector ratio [defined as the ratio of (TSCM + TCM)/(TEM + TEF)], into a single index. The values of the index were strongly associated with the clinical response in the initial cohort (Fig. 7A), suggesting that these markers could be efficiently combined to predict durable responses. However, the small size of the initial cohort does not allow us to use regression and cross-validation to rigorously optimize and test this approach. Thus, we devised a similar index that was exclusively based on flow cytometry inputs so that we could train and test it in the larger validation cohort of patients. Three biomarkers, including CAR+TH2+ frequency, TCM frequency, and the (TEM + TEF) frequency, were integrated into a logistic regression model using the clinical response (CR or RL) as the response variable. We assessed the discriminative power of this model to predict the clinical responses by stratified fivefold cross-validation in the extended cohort of patients profiled with flow cytometry. Overall, the trained model successfully discriminated CR and RL patients based on the integrated index [receiver operator characteristic area under the curve (ROC AUC) = 0.87 ± 0.14], achieving a sensitivity of ~70% to predict long-term remission with a false-positive rate of <5% (Fig. 7B). These results demonstrate the viability of this approach to predict the long-term remission or CD19-positive relapse of patients treated with CAR T therapy based on our key findings.
Fig. 7. Integrated model analysis demonstrates that the combination of TH2 strength and early memory potential is predictive of patient response durability.
(A) Response predictive index of each patient in the initial discovery cohort (n = 10) and the comparison between CR and RL groups. The index combines TH2 gene expression level, TH2 gene expression frequency, and early memory cell proportion. (B) ROC curve for response prediction based on an integrative biomarker consisting of CAR+TH2+ frequency, TCM frequency, and the (TEM + TEF) frequency in the validation cohort (n = 49). A binomial logistic regression was used to fit the model with CR or RL as the response variable, and a stratified fivefold cross-validation was implemented to compute the ROC and AUC. P value was calculated with Mann-Whitney test.
TCR-mediated bead stimulation of infusion products fails to capture clinical responses
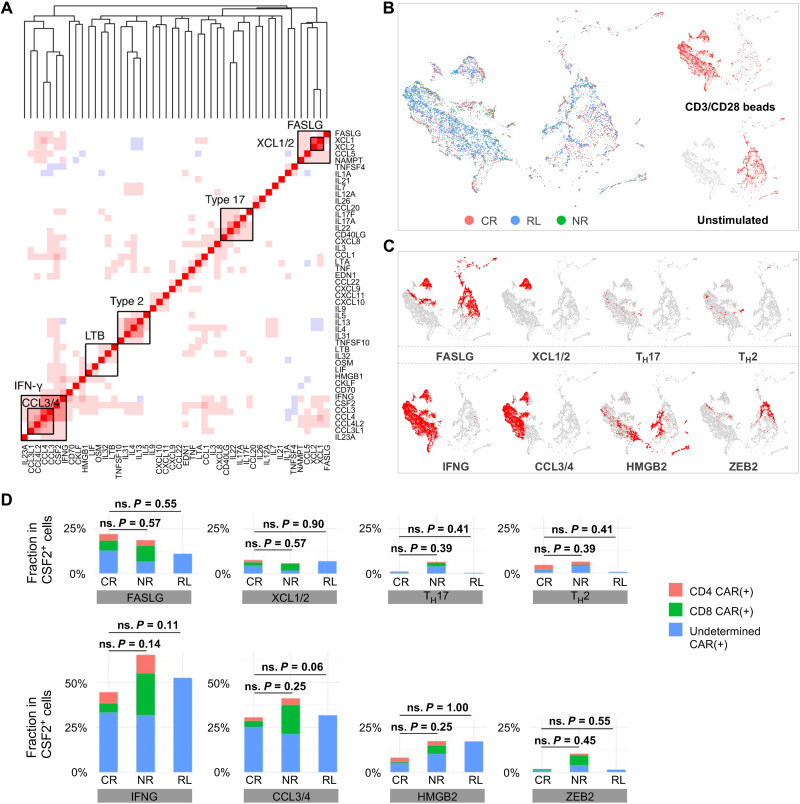
The findings described in previous sections show that the landscape of CART19 activation states upon CAR-mediated ex vivo stimulation is associated with the clinical response to the therapy. To assess whether stimulation via anti-CD3/CD28 beads was sufficient to uncover the determinants of response, we used the same computational approach to search for differences in the gene expression programs associated with the CAR T cells of CR, RL, and NR patients upon TCR-mediated stimulation (Fig. 8, A and B). This analysis revealed that, overall, the cytokine coexpression modules were consistent with those identified in CD19-stimulated CAR T cells, except for a TH17-like module consisting of IL17F, IL17A, IL22, IL26, and CCL20 that was uniquely observed upon TCR-mediated simulation (Fig. 8A). Nevertheless, the number of cells expressing this module was extremely low (Fig. 8C). We also observed a type 1 module in correlation with the expression of the apoptosis antigen ligand FASLG, possibly reflecting substantial activation-induced cell death upon bead stimulation. However, contrary to what we observed in the case of CAR-mediated stimulation, we did not find any significant difference among CR, RL, and NR patients in the proportion of cells expressing each cytokine coexpression module upon TCR-mediated stimulation (Fig. 8D), suggesting that this stimulation mechanism was incapable of uncovering the biology of CAR T cells that underlies differences in clinical responses. Consistently, we did not observe any difference on the gene expression levels of major cytokines across the three response groups (fig. S17). When examining the cytometry data in our larger validation cohort, we still observed a significant higher frequency of CAR+TH2+ population in bead-stimulated CAR T cells in the CR group, although the frequency of individual TH2-related cytokine did not show any statistically significant difference (fig. S18). Last, to check whether CD19-3T3 and anti-CD3/CD28 beads activate distinct pathways, we performed IPA analysis and found an overall similarity between the two conditions, although the activation or inhibition level and cell number of specific pathways differed substantially (fig. S19).
Fig. 8. TCR-mediated anti-CD3/CD28 beads stimulation of CAR T cells fails to predict clinical responses.
(A) Coexpressed cytokine modules identified in TCR-mediated stimulation. Genes are ordered by hierarchical clustering. (B) An integrated cytokine module representation UMAP. Cells are colored by responsive states, and UMAPs are split by experimental conditions. (C) The distribution of the identified modules on the UMAP. (D) Quantification of identified modules among CR, NR, and RL patients. The P values were calculated with Mann-Whitney test.
DISCUSSION
CART19 cell therapy is highly effective in pediatric and adult patients with B-ALL, with 90% patients responding initially to the therapy (6). Despite this high response rate, relapse occurs in nearly 50% of patients, among which a large fraction is CD19 positive (8–10, 25). Owing to the costly and lengthy process to manufacture and administer CAR T therapy in a patient-specific fashion, predictive biomarkers are urgently needed to better weigh the cost and value of treatment and specific strategies are expected to ameliorate CAR T products to effectively extend the remission durability. Significant efforts have been made toward identifying patient characteristics, including tumor burden and immune status, to correlate with efficacy and/or toxicities of CAR T therapy (7, 38). Early expansion of peripheral blood CAR T cells in vivo has been associated with favorable therapeutic outcome (7, 11), but it is not able to predict the response or relapse before infusion. Furthermore, to date, the mechanisms inducing CD19-positive relapse remain an open question. Since the dose of CAR T cells approved as a standard of care for B cell malignancies can vary up to 10-fold and the sustained remission is largely independent of dosage in long-term follow-up studies (6), the composition of the infusion product appears to be more critical than the quantity of CAR-transduced T cells. Thus, it is crucial to understand if the characteristics of infused CAR T cells contribute to the CD19-positive relapse and therapeutic failure.
We conducted single-cell transcriptional profiling of CD19-targeting 4-1BB/CD3 signaling CAR T cells in infusion products of B-ALL patients upon stimulation with human CD19-expressing APCs and observed significant intrinsic functional heterogeneity. Through unbiased analyses of distinct coexpressed cytokine modules on a fully activated CAR T subset marked by high CSF2 expression, we identified two type 1–like cytokine modules comprising an array of TH1 response genes including IFNG and IL2; a type 2 cytokine module consisting of IL4, IL5, IL9, and IL13; and a module consisting of beta chemokines like CCL3 and CCL4. A partially activated subset marked by the expression of LTB and other cytokines like CCL5, IL16, and IL32 was also found, which constituted the dominant feature for MSLN cocultured CAR T cells. Single-cell multiplexed cytokine profiling was previously used to characterize healthy donor 4-1BB/CD3 signaling CART19 cells stimulated with CD19-coated beads and showed highly heterogeneous functional states (39). The most polyfunctional subset produces GM-CSF, the protein encoded by CSF2, type 1 cytokine IFN-γ, type 2 cytokine IL-13, and beta chemokines like MIP-1α and MIP-1β, encoded by CCL3 and CCL4 genes, respectively, in both CD4 and CD8 CAR T cells, which is in general agreement with what we observed in this study.
Comparing the coexpressed cytokine expression profile of CD19-positive relapsed patients with that of durable responders, we found that CAR T cells from RL patients had a significant deficit of TH2 module expression. Differential gene expression analyses identified that all major genes encoding TH2 functions, including IL13, IL5, IL4, and the upstream regulator GATA3, were up-regulated in long-term CR patients compared with RL patients. In contrast, these analyses did not identify differences in the expression of TH1 or beta chemokine response–related genes, suggesting the need for both type 1 and type 2 functions to achieve a sustained long-term remission. To systematically validate these predictive biomarkers, we assessed a larger retrospective cohort of patients (n = 49) with flow cytometry, and our results confirmed a significant increase of CAR+TH2+ frequency in CR patients as compared with RL patients. In addition, we evaluated the TH2 functionality strength in 22 patients using a multiplexed secretomic assay and found consistent results, further enhancing the significance of our observations. These findings suggest a potential strategy to improve the therapeutic outcome by reengineering the infusion product with augmented type 2 signaling or by boosting TH2 functions after infusion.
Recently, Deng et al. (20) performed scRNA-seq of autologous axi-cel anti-CD19 CAR T cell infusion products from 24 patients with LBCL. Their data revealed threefold higher frequencies of CD8 T cells expressing memory signatures in patients who achieved a complete response than patients with partial response or progressive disease. Moreover, CD8 T cell exhaustion was associated with a poor molecular response, suggesting that both T cell memory status and immune function capacity in infusion products can contribute to therapeutic outcome. By using our CITE-seq surface proteomic data, we identified T cell differentiation subsets and found that CAR T cells from relapsed patients had significantly lower proportion of TSCM and TCM early memory states under both unstimulated baseline and CD19-specific activated conditions. This finding was further confirmed in our expanded validation cohort. Combining the memory states with the TH2 level, an integrative regression model demonstrated a sensitivity of ~70% in predicting long-term responses and a false-positive rate of <5%. We expect that further prospective investigations in different stage/multicenter cohorts will provide larger training datasets to refine this predictive model.
We observed that neither unstimulated nor TCR-stimulated CAR T cell characteristics were significantly associated with therapeutic outcomes, but only CAR-specific signaling and cytokine responses elicited by antigen-specific stimulation could predict CR, RL, and NR, indicating the necessity to characterize infusion products via CAR activation to develop more informative and accurate predictive biomarkers. In this regard, one of the limitations of our study is the use of an artificial system based on CD19-expressing murine APCs. While this experimental setting reduces the consumption of precious patient-derived autologous B cells and facilitates the bioinformatic separation of human CAR T cells and murine APC cells, our model only captures the reactivity to the human CD19 and does not recapitulate other aspects of the immune synapse.
An outstanding question is whether the observed functional heterogeneity and the activation modules of CAR T cells in the infusion product can be solely attributed to the characteristics of the patient immune cells to start with or can be improved through optimization of the manufacturing process by analyzing apheresis products in comparison with the corresponding infusion products. In-depth analysis of proteomic (surface epitopes and cytokines), transcriptional (mRNAs), genomic (i.e., integration sites), and epigenomic (chromatin state) landscapes of single CAR T cells throughout the manufacturing process may further unveil the mechanisms of their functional changes with time and inform new strategies to improve manufacturing or control the quality of autologously transferred CAR T cells for individual patients. Recently, Wang et al. (40) reported on a method for joint profiling of chromatin accessibility and integration sites of CAR T cells at the population and single-cell level, which may address the question whether CAR T integration at certain genomic regions can rewire the regulatory landscape of T cells, but it is yet to be applied to patient-derived CAR T cells in correlation with clinical outcomes. Last, the clonal expansion in vivo after infusion constantly alters the heterogeneity of circulating CAR T cells in patients (41), and a finite number of clones are the major effector cells inducing therapeutic remission and long-term surveillance to completely eliminate tumor cells. However, it remains to be revealed how the immune function landscape correlates with clonal evolution and the persistence in patients throughout the treatment and the long-term remission.
In summary, our single-cell multiomics profiling of preinfusion CAR T products provides valuable insights for understanding the mechanistic determinants of long-term durable CAR T cell therapy. We developed a systematic in vitro model to characterize CAR T functionalities upon CD19-specific activation and correlated them with clinical response. We identified that TH2 deficiency and overdifferentiation to effector phenotype were significantly associated with CD19-positive relapse, which was confirmed through independent evaluation in a larger cohort using different assays in a separate research center. Our data thus illuminate potential strategies to improve the response duration to CAR T therapy by precisely strengthening specific CAR T functionality and restoring early memory phenotype.
MATERIALS AND METHODS
Patient samples
Samples were acquired from patients with relapsed/refractory B-ALL who enrolled in pilot clinical trials designed to assess the safety and feasibility of CD19-dericted CAR (CTL019) T cell therapy, which was conducted at the Children’s Hospital of Philadelphia and the University of Pennsylvania (ClinicalTrials.gov, number NCT01626495). Besides, other patients in the expanded validation cohort were those who enrolled in a pilot study of the tocilizumab optimization timing for CART19-associated cytokine release syndrome (ClinicalTrials.gov, number NCT02906371). Written informed consent for participation was obtained from patients or their guardians according to the Declaration of Helsinki. All laboratory operations were under principles of International Conference on Harmonization Guidelines for Good Clinical Practice with established standard operating procedures and protocols for sample receipt, processing, freezing, and analysis. All ethical regulations were strictly followed.
T cell isolation, vector production, and generation of CTL019 cells
Autologous peripheral blood mononuclear cells were collected by standard leukapheresis. T cells were enriched by mononuclear cell elutriation and then washed and activated with anti-CD3/CD28–coated paramagnetic beads. A lentiviral vector containing a previously described CD19-specific CAR with 4-1BB/CD3ξ transgene was constructed and produced (33), which was then used to transduce the cells during activation and was washed out 3 days after the culture initiation (42). A rocking platform device (WAVE Bioreactor System) was used to expand cells for 8 to 12 days, and the beads were then magnetically removed. CTL019 cells were harvested and cryopreserved in infusible medium. The final product release criteria are listed as follows (6): cell viability ≥ 70%, CD3+ cells ≥ 80%, residual paramagnetic anti-CD3/CD28–coated paramagnetic beads ≤ 100 per 3 × 106 cells, endotoxin ≤ 3.5 EU/ml, mycoplasma negative, bacterial and fungal cultures negative, residual bovine serum albumin ≤ 1 μg/ml, vesicular stomatitis virus-G (VSV-G) DNA as a surrogate marker for replication-competent lentivirus ≤ 50 copies per microgram of DNA, transduction efficiency by flow cytometry ≥ 2%, and transduction efficiency by vector DNA sequence = 0.02 to 4 copies per cell.
Generation of human CD19 and mesothelin NIH3T3 cells
NIH3T3 mouse fibroblasts were originally obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (Gibco) with glutamate and supplemented with 10% fetal bovine serum (FBS; Gibco) in a humidified incubator (37°C in an atmosphere of 5% CO2). The cells were transduced with a lentiviral vector encoding human CD19 (CD19-3T3), and a negative control was NIH3T3 cells expressing mesothelin (MSLN-3T3). Cells were sorted on FACSAria (BD) to reach a >99% purity after transgene introduction. Mycoplasma and authentication were routinely performed before and after molecular engineering.
In vitro coculture assay
CTL019 cells were thawed and cultured in OpTmizer T-Cell Expansion Basal Medium supplemented with OpTmizer T-Cell Expansion Supplement and GlutaMAX Supplement (Gibco) in the humidified incubator on day 1 for overnight rest. On day 2, dead cells were removed using the EasySep Dead Cell Removal Kit (STEMCELL Technologies) as per the manufacturer’s instructions, and a certain number of cells were counted using a hemocytometer before coculture assays. For the stimulation with CD19-3T3 cells or MSLN-3T3 cells (target cells), 1 × 106 CTL019 cells were mixed with the same number of target cells in 2 ml of medium; for the stimulation with anti-CD3/CD28 beads, 1 × 106 CTL019 cells were mixed with 3 × 106 beads in 1 ml of medium; for the evaluation of unstimulated condition, 1 × 106 cells were prepared. All the suspensions were cultured in RPMI 1460 medium (Gibco) containing 10% FBS in a tissue culture–treated 48-well plate (Fisher Scientific) for 6 hours in the incubator. After coculture, the anti-CD3/CD28 beads were removed by passage over a magnetic field.
Intracellular cytokine detection assay
The cocultured cells were washed twice in phosphate-buffered saline (PBS) (Life Technologies) and stained for 20 min at room temperature (RT) with LIVE/DEAD Blue detection reagent (Thermo Fisher Scientific) diluted 1:800 in PBS. Cells were washed two times in fluorescence-activated cell sorting (FACS) staining buffer and then stained for surface molecules for 20 min at RT. Cells were fixed using the Cytofix/Cytoperm Kit (BD Biosciences) for 20 min at RT protected from light. Cells were washed twice with 1× Perm/Wash buffer and then stained for CAR19 (ACROBiosystems) and intracellular cytokines with antibodies in Perm/Wash buffer. Staining was done for 20 min at RT in the dark. Cells were washed twice more with Perm/Wash buffer and resuspended in FACS staining buffer. Samples were run on Cytek Aurora, and analysis was performed using FlowJo.
Multiplexed secretomic assay
After stimulation with CD19-3T3 cells, the activated CTL019 cells were collected, washed, and stained with CAR19 (ACROBiosystems) antibody in FACS buffer, followed by magnetic labeling and separation using magnetic-activated cell sorting (MACS) columns (Miltenyi Biotec). Approximately 30,000 CAR+ cells were then stained with membrane stain (included in the IsoPlexis Kit) before being loaded onto the IsoCode chip (IsoPlexis) consisting of 12,000 chambers prepatterned with an array of 32 cytokine capture antibodies. The chip was further incubated in the IsoLight machine for 16 hours at 37°C with 5% CO2 supplement. A cocktail of detection antibodies was used to detect the secreted cytokines followed by the fluorescent labeling. The fluorescent signals were analyzed by the IsoSpeak software to calculate the numbers of specific cytokine-secreting cells and the intensity level of each cytokine. The raw data of TH2-related cytokines, including IL-4, IL-5, and IL-13, were extracted for the downstream analyses.
Cell staining with DNA-barcoded antibodies for CITE-seq
After 6-hour coculture, for each condition, cells were resuspended in 100 μl of cell staining buffer (BioLegend) added with 5 μl of Human TruStain FcX Fc Blocking reagent (BioLegend). Cell suspensions were then incubated at 4°C for 10 min, during which the antibody pool was prepared using 1 μg of each TotalSeq antibody (BioLegend). After 30-min incubation at 4°C, cells were washed two times with 1 ml of PBS (Life Technologies) and finally resuspended in PBS at 5 × 105 cells/ml for the downstream loading. TotalSeq antibodies for human T cell characterization panels are the following: anti-CD4 (clone SK3), anti-CD8 (clone SK1), anti-CD45RO (clone UCHL1), anti-CD45RA (clone HI100), anti-CCR7 (clone G043H7), anti-CD62L (clone DREG-56), anti–HLA-DR (clone L243), anti-CD38 (clone HB-7), anti-CD69 (clone FN50), anti–4-1BB (clone 4B4–1), anti–PD-1 (clone EH12.2H7), anti–CTLA-4 (clone BNI3), anti-LAG3 (clone 11C3C65), and anti-TIGIT (clone A15153G).
Device preparation and microfluidic operation of scRNA-seq
The detailed materials and methods for preparing and operating the scRNA-seq device have been described in our previous report (31, 43). Briefly, the device consisted of a microwell array layer and a microfluidic channel layer, both of which were made by casting polydimethylsiloxane (PDMS) over the SU8 master wafers followed by degassing and curing at 80°C for 6 to 8 hours. After curing, PDMS was peeled off, and the two layers were cut to proper sizes and then plasma-bonded to assemble onto a glass slide. Before cell loading, the device was pressurized to remove air bubbles inside the microwells using a manually operated syringe with outlet closed, and then primed for 1 hour at RT with 1% bovine serum albumin (Sigma-Aldrich) in PBS. A total of 100 μl of cell suspension with a density of 5 × 105 cells/ml was pipetted on the inlet reservoir and withdrawn into the device. Next, barcoded beads, lysis buffer, and fluorinated oil were loaded sequentially to seal microwells. After cell and bead loading, the device was placed in a petri dish and exposed to three cycles of freeze and thaw, each of which included freezing at −80°C freezer or dry ice for 5 min followed by thawing at RT for 5 min. To capture mRNA onto beads, the device was then incubated for 1 hour inside an aluminum foil–covered wet chamber. After incubation, the beads were retrieved by 6× saline sodium citrate (SSC) buffer flushing. Last, collected beads were washed twice with 6× SSC buffer and then proceeded to the reverse transcription step.
scRNA-seq library preparation and sequencing
Library preparations were performed as described in the DropSeq protocol (version 3.1) (44). Briefly, the captured mRNA was reverse-transcribed using Maxima H Minus reverse transcriptase (Thermo Fisher Scientific) with a custom template switching oligo. Then, the beads coated with complementary DNA (cDNA) were treated using exonuclease I (New England Biolabs) for 1 hour at 37°C with rotation to chew away any unbound mRNA capture probes. The cDNA was then amplified using a 13-cycle polymerase chain reaction (PCR) whole-transcriptome amplification. At this step, 0.4 μl of “additive” primers was added to cDNA PCR to increase the yield of ADT (cDNA derived from the TotalSeq antibodies) products. After the amplification, ADT-derived cDNAs (180 base pairs) and mRNA-derived cDNAs were separated using SPRIselect (Beckman Coulter) beads at 0.6 ratio. After a standard Nextera tagmentation, PCRs (Nextera XT, Illumina), and another round of 0.6× purification using AMPure XP beads (Beckman Coulter), the mRNA library was ready to be sequenced. For ADT-derived cDNAs, 2× SPRI purifications and then an eight-cycle PCR were performed to amplify ADT sequencing library, followed by another round of 1.6× SPRI purification as per the manufacturer’s protocol. The quality of both the mRNA and ADT libraries was assessed by high-sensitivity bioanalyzer test (Agilent Inc.). To obtain sufficient read coverage for both libraries, we pooled 10% of ADT and 90% of cDNA library into one lane. The libraries were sequenced on HiSeq4000 (Illumina) at medium depth (average of 20,000 to 40,000 reads per cell) with three samples pooled into one sequencing lane.
Processing of sequencing data
Reads from the mRNA libraries were aligned to the concatenated human (hg38) and mouse (mm10) reference genomes and the sequence of the transduced CAR construct (33) using STAR 2.6.1a. A unique molecule identifier (UMI) count matrix was built using the DropSeq core computational protocol (version 2.0.0) (44). As reads were aligned to the concatenated human and mouse reference genomes, we were able to classify each uniquely aligned UMI as originating from a human or mouse cell. CITE-seq count was used to count UMIs in the ADT libraries. We filtered out cells with less than a specified total number of mRNA UMIs. This number was determined in a sample-specific manner by examination of log-scale barcode rank plots, where a steep drop-off was taken to indicate the boundary between empty wells and wells containing a cell. This fraction was between 1 × 102.8 and 1 × 103.2 for all samples. For coculture experiments, we filtered out mouse cells by removing cells with more than a specified number of mouse-associated UMIs. Again, this number was determined in a sample-specific manner by examination of plots of mouse versus human UMI counts. After removing wells that contained a mouse cell from the analysis, the residual background expression of mouse genes was removed by setting counts to zero. The mRNA expression data were log-normalized using Seurat (45), and the Seurat v3 anchor-based integration workflow was used to integrate data from different experiments. We used 20 canonical correlation analysis (CCA) dimensions and the default number of anchor neighbors and reduced the integrated mRNA data to two dimensions using UMAP for visualization (46). The ADT libraries were normalized using Seurat v3.
Estimation of CAR+ cell fraction
To estimate the sensitivity to detect the expression of the CAR, we counted the fraction of CAR mRNA-expressing cells. We focused on cells from the CD19 APC coculture experiments expressing CSF2, on the assumption that all CSF2-expressing cells had been successfully activated via CAR binding to CD19. To estimate the CAR+ fraction in the infusion samples, we used the sensitivity defined above and assumed a false-positive detection rate of 0. We then estimated 90% CIs using a normal sampling distribution.
Cytokine coexpression modules
Cytokine coexpression modules were identified separately for CD19-3T3–stimulated samples and anti-CD3/CD28 bead–stimulated samples. We computed Pearson correlation on the concatenated normalized count matrices, removing any cytokines expressed in fewer than 10 cells and removing any associations with a Benjamini-Hochberg adjusted q value greater than 0.05. The resulting correlation matrix was then clustered using hierarchical clustering. For each of the identified cytokine coexpression modules, AUCell was used to identify cells expressing that module (47). AUCell uses the AUC to calculate whether a module of genes is enriched within the expressed genes for each cell. First, for each module, a continuous AUC score is computed on the basis of the rank expression of the genes in the module. From these continuous scores, a Boolean assignment to each of these modules was determined for every cell by examining the distribution of AUC scores across all the cells and setting a threshold above which the module is considered “on.” To build integrated visual representations of cells across experiments based on the activity of these cytokine coexpression modules, the continuous AUC vectors representing the relative levels of module activity for each cell were reduced to two dimensions using UMAP. For each of the identified cytokine coexpression modules, Wilcoxon rank sum tests were performed between CR, RL, and NR patients to determine statistically significant differences in the proportions of cells expressing that module. We only considered CAR+ cells in these tests.
Differential expression analysis
All differential expression analyses were performed using Seurat v3 (48). The function “FindMarkers” was used for pairwise comparison between groups of cells (samples or clusters). A log fold change threshold of 0.25 was applied in later steps to select genes as differentially expressed. IPA (QIAGEN) (32) was used to pathway enrichment analysis based on the DEGs associated with each cluster. Ingenuity knowledge base (genes only) was used as reference set to perform core expression analysis. T cell–related signaling pathways were selected from the top identified pathways to represent major functional profiles of each cluster. The z score was used to determine the activation or inhibition level of specific pathways.
Ingenuity Pathway Analysis
IPA (QIAGEN) was used to understand underlying signaling pathways (32). Here, DEGs distinguishing each identified cluster and the corresponding fold change value of each gene were loaded into the dataset. Ingenuity knowledge base (genes only) was used as reference set to perform core expression analysis. T cell–related signaling was selected from identified top canonical pathways to represent major functional profiles of each cluster. The z score was used to determine the activation or inhibition level of specific pathways.
Integrative model analysis
Logistic regression was used to classify patients based on their frequencies of specific biomarkers using their response (CR or RL) status after receiving CAR T cell therapy as the response variable. We used LogisticRegression() from scikit-learn v0.23.2 in python to train the classifier using a stratified fivefold cross-validation as implemented in StratefiedKFold() and to compute the ROC AUC.
Statistics
Analyses were performed with Prism 9 (GraphPad). Mann-Whitney test was used to compare the specific observations between two groups. A P value of <0.05 was considered statistically significant.
Acknowledgments
The microfluidic devices were fabricated in the Yale School of Engineering and Applied Science cleanroom and Yale West Campus cleanroom. The sequencing service was conducted at Yale Stem Cell Center Genomics Core facility, which was supported by the Connecticut Regenerative Medicine Research Fund and the Li Ka Shing Foundation or Yale Center for Genomics Analysis (YCGA). Computational data analysis was conducted with the Yale High Performance Computing clusters (HPC).
Funding: The research was supported by Stand-Up-to-Cancer (SU2C) Convergence 2.0 Grant (to R.F.) and the Packard Fellowship for Science and Engineering (grant number 2012-38215 to R.F.). This material is based on work supported under a collaboration by Stand Up To Cancer, a program of the Entertainment Industry Foundation and the Society for Immunotherapy of Cancer.
Author contributions: Conceptualization: J.J.M., P.G.C., and R.F. Investigation: Z.B., Z.Z., R.A., D.K., S.L., A.B., and H.S. Methodology: Z.B., S.W., and Z.Z. Data analysis: S.W., Z.B., K.G., P.G.C., Y.D., and Y.X. Resources: D.M.B., R.M.M., S.A.G., and C.H.J. Writing—original draft: Z.B., P.G.C., R.F., and J.J.M. Writing—review and editing: All the authors.
Competing interests: R.F. is scientific founder and adviser for IsoPlexis, Singleron Biotechnologies, and AtlasXomics. The interests of R.F. were reviewed and managed by Yale University Provost’s Office in accordance with the University’s conflict of interest policies. J.J.M. and C.H.J. hold patents related to CAR T cell manufacturing and biomarker discovery. S.A.G. reports grants, personal fees, and other support from Novartis; grants from Kite and Servier; grants and other support from Vertex; personal fees from Roche, GSK, Humanigen, CBMG, and Janssen/JnJ; and other support from Jazz, Adaptimmune, TCR2, Cellectis, Juno, Allogene, and Cabaletta; in addition, S.A.G. has a patent for Toxicity management for antitumor activity of CARs, WO 2014011984 A1 issued. D.M.B. reports that his employment in Tmunity Therapeutics Inc. has no relation to this work, and no financial conflicts are present. The remaining authors declare no other competing interests.
Data and materials availability: All scRNA-seq data used in this study can be accessed in the NCBI Gene Expression Omnibus (GEO) database under the accession number GSE197215. The other relevant data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 and S3
Figs. S1 to S19
Other Supplementary Material for this manuscript includes the following:
Table S2
REFERENCES AND NOTES
- 1.Bhojwani D., Pui C.-H., Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 14, e205–e217 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Raetz E. A., Bhatla T., Where do we stand in the treatment of relapsed acute lymphoblastic leukemia? Hematology Am. Soc. Hematol. Educ. Program 2012, 129–136 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Kalos M., Levine B. L., Porter D. L., Katz S., Grupp S. A., Bagg A., June C. H., T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 3, 95ra73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter D. L., Levine B. L., Kalos M., Bagg A., June C. H., Chimeric antigen receptor–modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochenderfer J. N., Wilson W. H., Janik J. E., Dudley M. E., Stetler-Stevenson M., Feldman S. A., Maric I., Raffeld M., Nathan D.-A. N., Lanier B. J., Morgan R. A., Rosenberg S. A., Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 116, 4099–4102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maude S. L., Frey N., Shaw P. A., Aplenc R., Barrett D. M., Bunin N. J., Chew A., Gonzalez V. E., Zheng Z., Lacey S. F., Mahnke Y. D., Melenhorst J. J., Rheingold S. R., Shen A., Teachey D. T., Levine B. L., June C. H., Porter D. L., Grupp S. A., Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 371, 1507–1517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee D. W., Kochenderfer J. N., Stetler-Stevenson M., Cui Y. K., Delbrook C., Feldman S. A., Fry T. J., Orentas R., Sabatino M., Shah N. N., Steinberg S. M., Stroncek D., Tschernia N., Yuan C., Zhang H., Zhang L., Rosenberg S. A., Wayne A. S., Mackall C. L., T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: A phase 1 dose-escalation trial. Lancet 385, 517–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J. H., Rivière I., Gonen M., Wang X., Sénéchal B., Curran K. J., Sauter C., Wang Y., Santomasso B., Mead E., Roshal M., Maslak P., Davila M., Brentjens R. J., Sadelain M., Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 378, 449–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner R. A., Finney O., Annesley C., Brakke H., Summers C., Leger K., Bleakley M., Brown C., Mgebroff S., Kelly-Spratt K. S., Hoglund V., Lindgren C., Oron A. P., Li D., Riddell S. R., Park J. R., Jensen M. C., Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood 129, 3322–3331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maude S. L., Laetsch T. W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M. R., Stefanski H. E., Myers G. D., Qayed M., de Moerloose B., Hiramatsu H., Schlis K., Davis K. L., Martin P. L., Nemecek E. R., Yanik G. A., Peters C., Baruchel A., Boissel N., Mechinaud F., Balduzzi A., Krueger J., June C. H., Levine B. L., Wood P., Taran T., Leung M., Mueller K. T., Zhang Y., Sen K., Lebwohl D., Pulsipher M. A., Grupp S. A., Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turtle C. J., Hanafi L.-A., Berger C., Gooley T. A., Cherian S., Hudecek M., Sommermeyer D., Melville K., Pender B., Budiarto T. M., Robinson E., Steevens N. N., Chaney C., Soma L., Chen X., Yeung C., Wood B., Li D., Cao J., Heimfeld S., Jensen M. C., Riddell S. R., Maloney D. G., CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Invest. 126, 2123–2138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter D. L., Hwang W.-T., Frey N. V., Lacey S. F., Shaw P. A., Loren A. W., Bagg A., Marcucci K. T., Shen A., Gonzalez V., Ambrose D., Grupp S. A., Chew A., Zheng Z., Milone M. C., Levine B. L., Melenhorst J. J., June C. H., Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med. 7, 303ra139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neelapu S. S., Locke F. L., Bartlett N. L., Lekakis L. J., Miklos D. B., Jacobson C. A., Braunschweig I., Oluwole O. O., Siddiqi T., Lin Y., Timmerman J. M., Stiff P. J., Friedberg J. W., Flinn I. W., Goy A., Hill B. T., Smith M. R., Deol A., Farooq U., McSweeney P., Munoz J., Avivi I., Castro J. E., Westin J. R., Chavez J. C., Ghobadi A., Komanduri K. V., Levy R., Jacobsen E. D., Witzig T. E., Reagan P., Bot A., Rossi J., Navale L., Jiang Y., Aycock J., Elias M., Chang D., Wiezorek J., Go W. Y., Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster S. J., Bishop M. R., Tam C. S., Waller E. K., Borchmann P., McGuirk J. P., Jäger U., Jaglowski S., Andreadis C., Westin J. R., Fleury I., Bachanova V., Foley S. R., Ho P. J., Mielke S., Magenau J. M., Holte H., Pantano S., Pacaud L. B., Awasthi R., Chu J., Anak Ö., Salles G., Maziarz R. T., Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 380, 45–56 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Schuster S., Bishop M. R., Tam C. S., Waller E. K., Borchmann P., McGuirk J. P., Jaeger U., Jaglowski S., Andreadis C., Westin J. R., Fleury I., Bachanova V., Foley S. R., Ho P. J., Mielke S., Magenau J. M., Holte H., Van Besien K., Kersten M. J., Teshima T., Tobinai K., Corradini P., Anak O., Pacaud L. B., del Corral C., Pharma D., Awasthi R., Tai F., Salles G., Maziarz R. T., Primary analysis of Juliet: A global, pivotal, phase 2 trial of CTL019 in adult patients with relapsed or refractory diffuse large B-cell lymphoma. Blood 130, 577 (2017). [Google Scholar]
- 16.Long A. H., Haso W. M., Shern J. F., Wanhainen K. M., Murgai M., Ingaramo M., Smith J. P., Walker A. J., Kohler M. E., Venkateshwara V. R., Kaplan R. N., Patterson G. H., Fry T. J., Orentas R. J., Mackall C. L., 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawalekar O. U., O’Connor R. S., Fraietta J. A., Guo L., McGettigan S. E., Posey A. D. Jr., Patel P. R., Guedan S., Scholler J., Keith B., Snyder N. W., Blair I. A., Milone M. C., June C. H., Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Singh N., Perazzelli J., Grupp S. A., Barrett D. M., Early memory phenotypes drive T cell proliferation in patients with pediatric malignancies. Sci. Transl. Med. 8, 320ra3 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Blaeschke F., Stenger D., Kaeuferle T., Willier S., Lotfi R., Kaiser A. D., Assenmacher M., Döring M., Feucht J., Feuchtinger T., Induction of a central memory and stem cell memory phenotype in functionally active CD4+ and CD8+ CAR T cells produced in an automated good manufacturing practice system for the treatment of CD19+ acute lymphoblastic leukemia. Cancer Immunol. Immunother. 67, 1053–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng Q., Han G., Puebla-Osorio N., Ma M. C. J., Strati P., Chasen B., Dai E., Dang M., Jain N., Yang H., Wang Y., Zhang S., Wang R., Chen R., Showell J., Ghosh S., Patchva S., Zhang Q., Sun R., Hagemeister F., Fayad L., Samaniego F., Lee H. C., Nastoupil L. J., Fowler N., Eric Davis R., Westin J., Neelapu S. S., Wang L., Green M. R., Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 26, 1878–1887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraietta J. A., Lacey S. F., Orlando E. J., Pruteanu-Malinici I., Gohil M., Lundh S., Boesteanu A. C., Wang Y., O’Connor R. S., Hwang W.-T., Pequignot E., Ambrose D. E., Zhang C., Wilcox N., Bedoya F., Dorfmeier C., Chen F., Tian L., Parakandi H., Gupta M., Young R. M., Johnson F. B., Kulikovskaya I., Liu L., Xu J., Kassim S. H., Davis M. M., Levine B. L., Frey N. V., Siegel D. L., Huang A. C., Wherry E. J., Bitter H., Brogdon J. L., Porter D. L., June C. H., Melenhorst J. J., Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G. M., Chen C., Das R. K., Gao P., Chen C.-H., Bandyopadhyay S., Ding Y.-Y., Uzun Y., Yu W., Zhu Q., Myers R. M., Grupp S. A., Barrett D. M., Tan K., Integrative bulk and single-cell profiling of pre-manufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 11, 2186–2199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finney O. C., Brakke H., Rawlings-Rhea S., Hicks R., Doolittle D., Lopez M., Futrell B., Orentas R. J., Li D., Gardner R., Jensen M. C., CD19 CAR T cell product and disease attributes predict leukemia remission durability. J. Clin. Invest. 129, 2123–2132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh N., Lee Y. G., Shestova O., Ravikumar P., Hayer K. E., Hong S. J., Lu X. M., Pajarillo R., Agarwal S., Kuramitsu S., Orlando E. J., Mueller K. T., Good C. R., Berger S. L., Shalem O., Weitzman M. D., Frey N. V., Maude S. L., Grupp S. A., June C. H., Gill S., Ruella M., Impaired death receptor signaling in leukemia causes antigen-independent resistance by inducing CAR T-cell dysfunction. Cancer Discov. 10, 552–567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulsipher M. A., Han X., Maude S. L., Laetsch T. W., Qayed M., Rives S., Boyer M. W., Hiramatsu H., Yanik G. A., Driscoll T., Myers G. D., Bader P., Baruchel A., Buechner J., Stefanski H. E., Kalfoglou C., Nguyen K., Waldron E. R., Thudium Mueller K., Maier H. J., Kari G., Grupp S. A., Next-generation sequencing of minimal residual disease for predicting relapse after tisagenlecleucel in children and young adults with acute lymphoblastic leukemia. Blood Cancer Discov. 3, 66–81 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlando E. J., Han X., Tribouley C., Wood P. A., Leary R. J., Riester M., Levine J. E., Qayed M., Grupp S. A., Boyer M., de Moerloose B., Nemecek E. R., Bittencourt H., Hiramatsu H., Buechner J., Davies S. M., Verneris M. R., Nguyen K., Brogdon J. L., Bitter H., Morrissey M., Pierog P., Pantano S., Engelman J. A., Winckler W., Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat. Med. 24, 1504–1506 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Gardner R., Wu D., Cherian S., Fang M., Hanafi L.-A., Finney O., Smithers H., Jensen M. C., Riddell S. R., Maloney D. G., Turtle C. J., Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 127, 2406–2410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh N., Orlando E., Xu J., Xu J., Binder Z., Collins M. A., O’Rourke D. M., Melenhorst J. J., Mechanisms of resistance to CAR T cell therapies. Semin. Cancer Biol. 65, 91–98 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P. K., Swerdlow H., Satija R., Smibert P., Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Z., Su G., Fan R., Single-cell analysis technologies for immuno-oncology research: From mechanistic delineation to biomarker discovery. Genom. Proteom. Bioinform. 19, 191–207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dura B., Choi J.-Y., Zhang K., Damsky W., Thakral D., Bosenberg M., Craft J., Fan R., scFTD-seq: Freeze-thaw lysis based, portable approach toward highly distributed single-cell 3′ mRNA profiling. Nucleic Acids Res. 47, e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krämer A., Green J., Pollard J. Jr., Tugendreich S., Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milone M. C., Fish J. D., Carpenito C., Carroll R. G., Binder G. K., Teachey D., Samanta M., Lakhal M., Gloss B., Danet-Desnoyers G., Campana D., Riley J. L., Grupp S. A., June C. H., Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 17, 1453–1464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai Z., Lundh S., Kim D., Woodhouse S., Barrett D. M., Myers R. M., Grupp S. A., Maus M. V., June C. H., Camara P. G., Melenhorst J. J., Fan R., Single-cell multiomics dissection of basal and antigen-specific activation states of CD19-targeted CAR T cells. J. Immunother. Cancer 9, e002328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez C. X., Amezquita R. A., Guan T., Marshall H. D., Joshi N. S., Kleinstein S. H., Kaech S. M., The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adomavicius T., Guaita M., Zhou Y., Jennings M. D., Latif Z., Roseman A. M., Pavitt G. D., The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10, 2136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Golubovskaya V., Wu L., Different subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers (Basel) 8, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kochenderfer J. N., Somerville R. P. T., Lu T., Shi V., Bot A., Rossi J., Xue A., Goff S. L., Yang J. C., Sherry R. M., Klebanoff C. A., Kammula U. S., Sherman M., Perez A., Yuan C. M., Feldman T., Friedberg J. W., Roschewski M. J., Feldman S. A., McIntyre L., Toomey M. A., Rosenberg S. A., Lymphoma remissions caused by anti-CD19 chimeric antigen receptor T cells are associated with high serum interleukin-15 levels. J. Clin. Oncol. 35, 1803–1813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Q., Bettini E., Paczkowski P., Ng C., Kaiser A., McConnell T., Kodrasi O., Quigley M. F., Heath J., Fan R., Mackay S., Dudley M. E., Kassim S. H., Zhou J., Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J. Immunother. Cancer 5, 85 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W., Fasolino M., Cattau B., Goldman N., Kong W., Frederick M. A., McCright S. J., Kiani K., Fraietta J. A., Vahedi G., Joint profiling of chromatin accessibility and CAR-T integration site analysis at population and single-cell levels. Proc. Natl. Acad. Sci. U.S.A. 117, 5442–5452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheih A., Voillet V., Hanafi L.-A., DeBerg H. A., Yajima M., Hawkins R., Gersuk V., Riddell S. R., Maloney D. G., Wohlfahrt M. E., Pande D., Enstrom M. R., Kiem H.-P., Adair J. E., Gottardo R., Linsley P. S., Turtle C. J., Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 11, 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine B. L., Humeau L. M., Boyer J., MacGregor R.-R., Rebello T., Lu X., Binder G. K., Slepushkin V., Lemiale F., Mascola J. R., Bushman F. D., Dropulic B., June C. H., Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. U.S.A. 103, 17372–17377 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai Z., Deng Y., Kim D., Chen Z., Xiao Y., Fan R., An integrated dielectrophoresis-trapping and nanowell transfer approach to enable double-sub-poisson single-cell RNA sequencing. ACS Nano 14, 7412–7424 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., McCarroll S. A., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. McInnes, J. Healy, J. Melville, Umap: Uniform manifold approximation and projection for dimension reduction. arXiv:1802.03426 [stat.ML] (9 February 2018).
- 47.Aibar S., González-Blas C. B., Moerman T., Huynh-Thu V. A., Imrichova H., Hulselmans G., Rambow F., Marine J.-C., Geurts P., Aerts J., van den Oord J., Atak Z. K., Wouters J., Aerts S., SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S3
Figs. S1 to S19
Table S2