Abstract
Glioblastoma is believed to originate from nervous system cells; however, a putative origin from vessel-associated progenitor cells has not been considered. We deeply single-cell RNA–sequenced glioblastoma progenitor cells of 18 patients and integrated 710 bulk tumors and 73,495 glioma single cells of 100 patients to determine the relation of glioblastoma cells to normal brain cell types. A novel neural network–based projection of the developmental trajectory of normal brain cells uncovered two principal cell-lineage features of glioblastoma, neural crest perivascular and radial glia, carrying defining methylation patterns and survival differences. Consistently, introducing tumorigenic alterations in naïve human brain perivascular cells resulted in brain tumors. Thus, our results suggest that glioblastoma can arise from the brains’ vasculature, and patients with such glioblastoma have a significantly poorer outcome.
Neural network defines GBM features to be of neural crest perivascular or radial glia lineage.
INTRODUCTION
Glioblastoma is the most common brain tumor (1), and it has an invariably poor prognosis despite aggressive therapy. A combination of high-throughput genomic and epigenetic data with bioinformatic analyses has provided a comprehensive view of genetic mechanisms underlying glioblastoma oncogenesis and progression (2, 3). Analyzing transcriptional intertumor heterogeneity within The Cancer Genome Atlas (TCGA) project identified three main subtypes, which are tightly associated with genomic alterations: TCGA-classical, TCGA-proneural, and TCGA-mesenchymal (TCGA-mes) (4). However, there is also notable intratumoral heterogeneity where different cells from the same tumor can be classified into different TCGA subtypes (5).
Gliomas are believed to arise from one of the two major types of neural cells of the brain: neuronal or glial by a reactivation of stem-like developmental gene programs. This cancer stem cell (CSC) hypothesis implicates a hierarchical continuum of differentiating cells within the tumor, with the CSC at the apex, having tumor-initiating and -propagating properties with resistance to therapy (6). Single-cell RNA sequencing (scRNA-seq) studies support this conjecture, and transcriptional profiles of various types of gliomas are consistent with neural progenitor–like, oligodendrocyte precursor (OPC)–like, or astrocytic-like cells (5, 7–10). Introducing identical glioblastoma driver mutations into human glial or neuronal progenitor cells results in molecular distinct subtypes, highlighting the importance of the originating cell lineage for tumor phenotype and stratification (11, 12). However, less is known of the cellular origin of the highly malignant glioblastoma with mesenchymal features (5, 13).
Thus, previous computational cell-of-origin classifications mapped most glioblastoma to neuronal and glial cell types (5, 9, 10) and additional studies have identified possible mechanisms for these to transition into mesenchymal-like glioblastoma. However, the relation of mesenchymal glioblastoma to alternative nonneural progenitor cells residing in the brain has not been explored. Perivascular mural cells of the brains’ blood vessels are of neural crest origin (14, 15). As blood vessels descend into the brain parenchyma during development, vessel-attached neural crest–derived cells differentiate into the different perivascular cell types, with those remaining behind differentiating into leptomeningeal cells (14, 16). Recently, a previously unknown perivascular fibroblast (vFB)–like cell type was identified (17), which appears to function as a restricted stem-like cell type that generates pericytes and mesenchymal smooth muscle cells (SMCs) in both the developing and adult brain (18, 19).
Here, we deeply sequenced 4073 glioblastoma progenitor cells from 18 patients and integrated data from an additional 8443 tumor cells from 16 patients with low-grade glioma and 60,979 tumor cells from 66 patients with glioblastoma in the analysis. A novel neural network–based projection was used to learn the transcriptional features from normal brain cell types and thereafter used to assign individual tumor cells as well as deconvoluted bulk tumors at the level of both the cellular steady state and the developmental trajectory dynamics. Our analysis revealed two principal cell lineage patterns in glioblastoma—neural and perivascular. The most undifferentiated adult naïve cell type correlate in the neural cell lineage pattern was radial glia (Rgl), and in the vascular, it is the vFB cell type. Patients with perivascular glioblastoma exhibited significantly poorer survival. Animals with xenografts of naïve human perivascular cells harboring targeted genetic changes observed in glioblastoma present with tumors, indicating that the brain perivascular cells are competent to initiate brain tumors.
RESULTS
Neural network classifier maps glioblastoma tumor progenitor cells to two principally different endogenous cell lineages of the brain
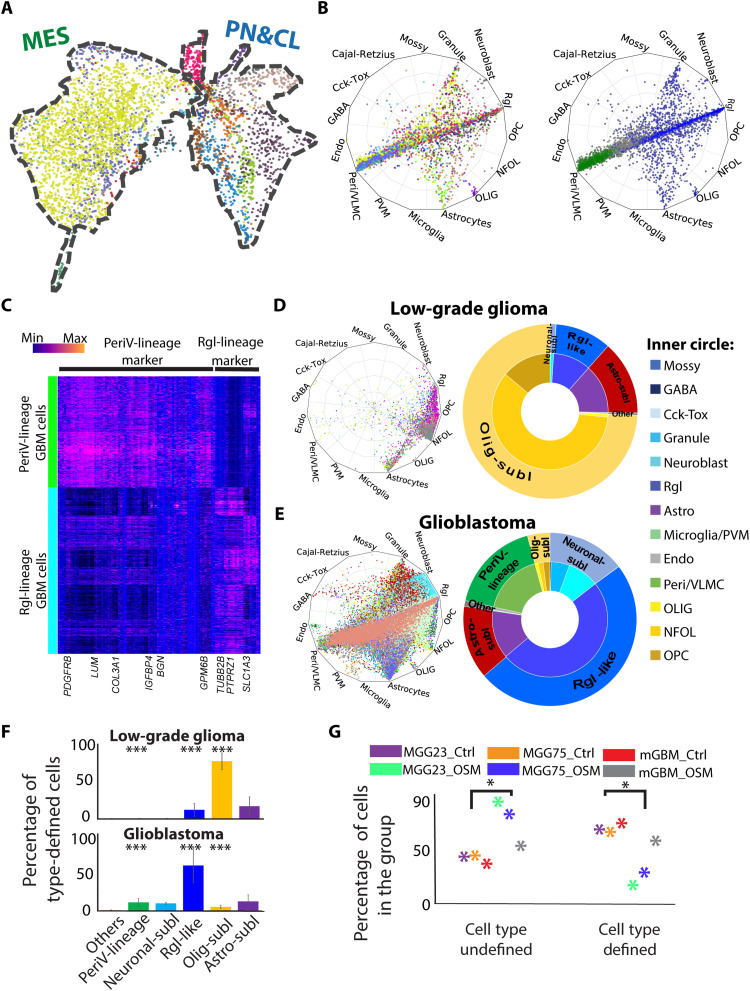
We enriched tumor progenitor cells from 18 patients of high-grade glioblastoma for scRNA-seq (data file S1) and validated the tumorigenicity of these cells by intracranial orthotopic xenografts with follow-up histological analyses (fig. S1A). Fourteen of the 18 patient samples reduced overall survival in the xenograft experiment (fig. S1B). A total of 4073 high-quality single cells (median 2.87 million total reads per cell; fig. S1C) were included in a copy number variation (CNV) analysis, confirming alterations associated with brain tumors (data file S1) and subsequently clustered. Excluding a cluster of CD45+ immune cells, the remaining 19 clusters were assigned into TCGA subtypes by a neural network classifier trained by the original TCGA data and subsequently named after TCGA subtype names (MS1-8, CL1-8, PN1-2, and NL1) (fig. S1D). Most clusters dominantly differed among individual patients, except for two cell clusters of TCGA-mes subtypes (MS3 and MS5) that spanned across different patients (fig. S1E, left). The cell clusters were organized into two clouds of coclustered cells when using Uniform Manifold Approximation and Projection (UMAP). Cells of the TCGA-mes subtype were in one cloud, while cells of all other TCGA subtypes were located in another cloud (Fig. 1A and fig. S1E, right).
Fig. 1. Cell-type assignment of high- and low-grade glioma revealed that perivascular lineage tumor cells are present only in high-grade glioma.
(A) UMAP visualization of patient-derived glioblastoma cells. Color coding based on cell clusters. The contours of two main clouds of cells outlined with a dashed line and labeled with TCGA subtypes on the top. CL, classical; MES, mesenchymal; PN, proneural. (B) Radar plot visualization of the cell-type scores of glioblastoma cells in relation to the trained reference brain cell types. Color coding based on cell clusters (left) or cell-type lineages (right, blue: Rgl-lineage; green, PeriV-lineage). The position of each dot indicates the cell-type score between that cell and the trained reference cell types, which are indicated outside each wheel bend. Abbreviations are as in fig. S1F. (C) Heatmap of differential gene expression between PeriV-lineage and Rgl-lineage glioblastoma cells. Selected gene symbols are at the bottom. Color bar indicates the expression intensity at the top left. (D and E) Left: Radar plots show the cell-type scores of low-grade glioma and glioblastoma cells in relation to the trained reference brain cell types. Right: Donut charts show the quantitative distribution of cell type–defined glioblastoma cells. The inner donut layer represents the reference cell types that tumor cells are assigned to, and the outer layer represents the normal cell-type lineages. (F) The distribution of low-grade glioma and glioblastoma cells to defined reference cell-type lineages. ***P < 0.001. (G) Scatter chart represents the significant cell-type score of control (Ctrl) and oncostatin M (OSM)–treated glioblastoma multiforme (GBM) cells against each defined reference brain cell type. “Cell type defined” represents glioblastoma cells with high cell-type scores above the cutoff, and “cell type undefined” represents cells with low scores. Dot colors are indicated at the top. *P < 0.05.
To identify the endogenous brain cell-type correlates of the patients’ glioma cells, we applied the machine learning classifiers with learned transcriptional features from normal brain reference cell types derived from the neurogenic niche of the developing mouse brain (20). After comparing four classifiers driven by logistic regression, support vector machine, vanilla neural network, and node-level graph neural networks, we decided to use a vanilla neural network classifier for further studies according to the prediction accuracy, time consumption, and overfitting control, as described in Materials and Methods. The classifier accuracy was further validated by an independent integrated dataset of normal cells from human embryonic midbrain (21) and cortex (fig. S1F) (22), and a randomized expression matrix (fig. S1G). Throughout the study, we refer to previously annotated cell types as “reference” cell types, and such closely related reference cell types were annotated in this study into cell lineages on the basis of the known differentiation trajectories. Using this neural network classifier, most tumor cells of the TCGA-mes subtype were assigned to the reference pericytes and vascular leptomeningeal cells (VLMCs), both of the perivascular lineage, while tumor cells of other TCGA subtypes were similar to reference neuronal or glial cells (i.e., reference Rgl, neuroblasts, astrocytes, oligodendrocyte cells, and immature granule neurons) (Fig. 1B and fig. S1H). Cells that failed to assign into one single cell type were located in the center of the radar plot, indicating cells of unknown cell type or a transcriptional plasticity of multiple cell types.
The neural crest–derived perivascular cells (reference pericytes and VLMCs) of the brain and the reference radial glia–derived neural cells (all neuronal and glial cell types of the brain) represent entirely different developmental cell lineages. When stratifying patients into either an Rgl-lineage type or a perivascular (PeriV)–lineage type based on the dominant cell percentage of one type and nonsignificant cell percentage of the other type in each patient, we did not observe significant differences of overall survival in the xenograft experiment (fig. S1, A and B). To further increase the resolution of reference brain cell types, we applied the machine learning classifier with learned transcriptional features from human developing brain cell types (23) and validated the observation of the existence of both Rgl-lineage–type and PeriV-lineage–type glioblastoma cells (fig. S1I). Thus, these results suggest that glioblastoma cells share molecular features with either the Rgl-lineage [including Rgl-like tumor progenitor cells; a neuronal sublineage including neuroblasts and neurons; an oligodendrocyte-sublineage (Olig-sublineage) including oligodendrocytes and its precursors, the OPCs and newly formed oligodendrocytes (NFOL); and an astrocyte-sublineage including differentiating and adult astrocytes] or the PeriV-lineage including perivascular cells and VLMCs.
Analysis of the differentially expressed genes between Rgl-lineage– and PeriV-lineage–type glioblastoma cells that were also expressed in their respective naïve cell types (i.e., normal reference brain Rgl and PeriV cells) revealed the existence of mutually exclusive expression between lineages but highly shared features with their corresponding endogenous reference cell types of each lineage in glioblastoma cells (Fig. 1C) and in the naïve cell types of the developing mouse brain (fig. S1J).
Perivascular lineage–type tumors exclusive to high-grade glioma
The previously analyzed cells were from high-grade glioma. We therefore made use of scRNA-sequenced cells obtained from resected and dissociated high- and low-grade gliomas (5, 7, 9, 10, 24–26) to validate our results and to compare the cell-type composition of PeriV- and Rgl-lineage tumor cells between high- and low-grade gliomas. A total of 8443 cells from low-grade glioma and 65,052 cells from glioblastoma originally defined as tumor cells were applied for the neural network classifier described in Fig. 1B. We found that low-grade glioma contains tumor cells with higher cell-type similarity to native reference cell types (high cell-type probability) than high-grade glioblastoma (Fig. 1, D and E, left, and fig. S1K, left). To exclude the fact that this result is caused by variability of sequencing quality between platforms of scRNA-seq, and to exclude a bias due to required threshold in the similarity scoring, we also validated this observation using only data generated from the same technical platform and applied different threshold requirements (fig. S1K, right). Low-grade glioma cells were most similar to reference Rgl, OPCs/NFOLs, and astrocytes, which together accounted for 99.48% of all tumor cells (Fig. 1D, right). In contrast, almost all glioblastomas were composed of multiple cell types, including high similarity to reference pericytes/VLMCs, to Rgl (i.e., Rgl-like tumor cells), as well as substantial numbers to the more differentiated progenies (astrocytes of the Astro-sublineage; OPC, NFOLs, and oligodendrocytes of the Olig-sublineage; neuroblasts and immature granule cells “Granule” of the Neuronal-sublineage) (Fig. 1E, right). Among the glioblastoma cells, 11.1% were assigned to the reference PeriV-lineage, while none of the low-grade glioma cells were assigned to these (Fig. 1F and fig. S1L). Thus, the existence of glioma assigning to the PeriV-lineage reference cells is specific to high-grade glioma among all 100 patients.
Rgl-lineage glioblastoma cells acquire higher cellular plasticity after mesenchymal transition but rarely transition into PeriV-lineage cell types
The acquisition of a mesenchymal transcriptional profile in glioblastoma cells can be forced by the microenvironment or by an intrinsic transition under certain selective pressure (13). To examine whether the PeriV-lineage tumor cells can transition from Rgl-lineage glioblastoma cells, we applied the neural network classifier on a recent published scRNA-seq dataset containing spontaneous mouse glioblastoma that was initiated from glial fibrillary acidic protein (GFAP)–expressing cells (27). In this model, a mesenchymal cell transition from the GFAP+ Rgl-lineage could be induced by oncostatin M (OSM) (27). Thus, if the identified PeriV-lineage glioblastoma represents a transition from the Rgl-lineage through this mechanism, we expected to identify PeriV-lineage glioblastoma cells in this dataset. Nearly all GFAP-derived glioblastoma cells were assigned to reference Rgl-lineage cells (Rgl, neuroblasts, and granule cells), but none to pericytes/VLMCs (fig. S1M). Because OSM induced a mesenchymal transition of these glioblastoma cells (27, 28), we compared three glioblastoma cell lines with or without OSM treatment in our classifier of TCGA subtypes and observed that OSM significantly increased mesenchymal features and inhibited proneural features (fig. S1N), in line with previous findings. Nevertheless, our classifier of endogenous reference brain cells did not recognize the OSM-transformed mesenchymal cells as PeriV cells, and instead assigned these mesenchymal cells into an undefined state (Fig. 1G and fig. S1O). These results corroborate that OSM initiates plasticity of glioblastoma cells including initiation of mesenchymal features and that this mechanism could account for some glioblastoma classified as mesenchymal. However, our results suggest that glioblastoma with perivascular features as defined using our classifier cannot be explained by an OSM-driven cell state transition.
Clinical relevance and CpG methylation of PeriV-lineage and Rgl-lineage glioblastoma
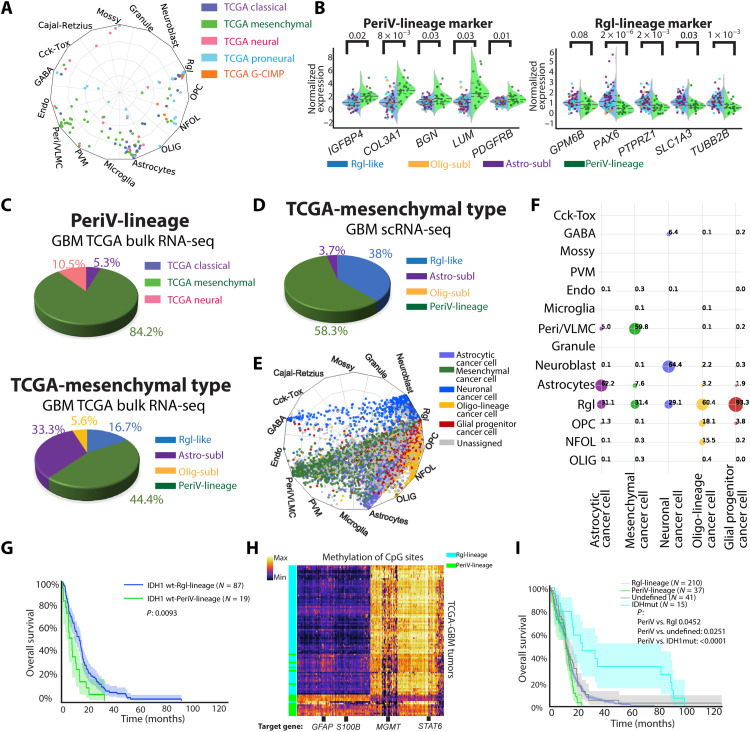
To explore the clinical relevance of tumors with PeriV-lineage and Rgl-lineage signatures, we scored the data of 161 bulk RNA-sequenced glioblastoma from the TCGA using the classifier. However, the bulk data reflect transcriptional features of multiple cell types (fig. S2A) that are highly heterogeneous, consistent with previous results (5). To identify the dominant cell types, the bulk data were transformed (29) and deconvoluted into single-cell resolution (fig. S2B) (30), and the deconvoluted data were then scored and visualized in a radar plot (Fig. 2A). The majority of the TCGA classified glioblastoma subtypes (TCGA-mes, TCGA-proneural, TCGA-classical, and TCGA-neural) were robustly assigned into four endogenous reference brain cell types: 19 tumors were assigned to reference cells of the PeriV-lineage (perivascular cells and VLMCs) and the remaining tumors were assigned to Rgl-lineage reference cells, including 53 to astrocytes, 32 to Rgl, and 9 to OPCs/NFOLs, accounting for 70.19% of all tumors. The lack of assignment of tumors to reference granule and neuroblast cells in bulk sequenced data likely reflects that these differentiated cells are rare in the tumors and might therefore become dwarfed when bulk-sequenced. In line with previous results obtained from scRNA-seq data, 9 of 10 top scRNA-seq enriched marker genes of PeriV-lineage–type and Rgl-lineage–type reference cells (data file S2) were found to be differentially expressed between PeriV-lineage–type and Rgl-lineage–type glioblastoma tumors sequenced in the TCGA framework (Fig. 2B). We next examined the relation between PeriV- and Rgl-lineage tumor types to TCGA subtypes by cross annotation. PeriV-lineage glioblastoma was overwhelmingly composed from the TCGA-mes subtype (Fig. 2C, top). In contrast, only 44.4% of TCGA-mes subtypes were of the PeriV-lineage, while the rest were most similar to the reference Rgl-lineage (including Rgl-like cells and cells in sublineages of Rgl) (Fig. 2C, bottom), indicating that the TCGA-mes subtype might consist of two different transcriptional states, one but not the other showing high similarity to the reference PeriV cells. The TCGA-classified proneural and glioma cytosine-phosphate-guanine (CpG) island methylator phenotype (G-CIMP) subtype mostly shared features with reference Rgl, while TCGA-classical and TCGA-neural subtypes mostly shared features with reference astrocyte cells (fig. S2C). To exclude that this finding was a result of a distortion due to analysis of bulk RNA-sequenced data, we classified the merged set of all scRNA-seq high-grade glioblastoma cells into TCGA subtypes and thereafter cross-annotated the cells of the TCGA-mes subtype to native reference brain cell types (Fig. 2D and fig. S2D). This analysis confirmed that glioblastoma cells of the TCGA-mes subtype are mainly assigned to PeriV cells, with most of the remaining cells showing the greatest similarity to reference Rgl and astrocytes of the brain. Furthermore, we re-examined glioblastoma cells from a public dataset (7) in our classifier of endogenous brain cells. In this study, tumor cells were assigned as “glial progenitor cancer cell,” “oligo-lineage cancer cell,” “astrocytic cancer cell,” “mesenchymal cancer cell,” and “neuronal cancer cell” on the basis of the similarity to developing brain cell types (7). Our classifier confirmed these previous results (Fig. 2E) and, in addition, corroborated that their annotated mesenchymal cancer cells are assigned to either PeriV-lineage or Rgl-lineage reference cells (Fig. 2F).
Fig. 2. Tumor subtype assignment, methylation status, and survival of deconvoluted bulk tumor data from TCGA/DFKN.
(A) Radar plot visualizes the cell-type scores for deconvoluted bulk glioblastoma in relation to trained reference brain cell types. Colors represent the TCGA-defined subtype of each tumor. (B) Violin swarm plot of the original gene expression of selected marker genes in the PeriV-lineage and Rgl-lineage of TCGA glioblastoma; blue background represents Rgl-lineage tumors and green background represents PeriV-lineage tumors. Dot colors represent the defined reference brain cell types of each tumor in (A). The dashed line in each violin plot represents the distribution quartiles. P value of Student’s t test on top. Abbreviations are as in fig. S2C. (C and D) Pie plots representing the composition of TCGA-classified subtypes in the PeriV-lineage (C, top), cell-type sublineages identified in the TCGA-mes subtype (C, bottom) of bulk glioblastoma, or cell-type sublineages identified in the TCGA-mes subtype of scRNA-seq glioblastoma cells (D). (E) Radar plot visualizes cell-type scores of state-defined glioblastoma cells in relation to trained reference brain cell types. (F) Dot plot represents the percentage of the defined cell states of glioblastoma cells in each originally defined cell-type state. Dot sizes from small to big represent the percentage from low to high. (G) Patient survival of isocitrate dehydrogenase 1 (IDH1) wild-type glioblastoma from the TCGA assigned as belonging to the Rgl-lineage and PeriV-lineage. (H) Heatmap representing the differential methylated site–based hierarchical clustering of the TCGA glioblastomas assigned to the PeriV-lineage and Rgl-lineage type. Selected target genes of the methylated sites are listed at the bottom. Color bar indicates the expression intensity at the top left. STAT6, signal transducer and activator of transcription 6. (I) Patient survival of glioblastoma from TCGA assigned to Rgl-lineage, PeriV-lineage, IDH1-mutant types, and nonclassified based on methylation.
In the bulk RNA-sequenced glioblastoma of the TCGA, 106 of 113 cell type–defined IDH1 wild-type (wt) glioblastoma patients with survival information were used for survival analysis. Glioblastoma with a dominant PeriV-lineage–type phenotype predicted markedly shorter survival than the Rgl-lineage type, and 18 of 19 patients’ life spans were <24 months (Fig. 2G). This observation was further validated when stratifying the Rgl-lineage into sublineages on the basis of assignment to the dominating reference cell types (Rgl-like, Astro-sublineage, and Olig-sublineage) (fig. S2E).
We next explored the mutational burden among the glioblastoma defined by PeriV-lineage– and Rgl-lineage–type signatures. Thirty-two genes with high frequency of mutation were significantly enriched (fig. S2F and data file S3). PeriV-lineage–type and Rgl-lineage–type glioblastoma carried a shared enrichment in mutations of TTN, PKHD1, TP53, PTEN, and FLG genes, and a differential mutational burden with NF1 gene strongly associated to the PeriV-lineage type and EGFR gene to the Rgl-lineage type, especially the astrocyte subtype.
In addition to the transcriptional level, we tested if the methylation status can be used to predict the lineage-based classification of glioblastoma. We first enriched the differential methylation sites with PeriV-lineage–type and Rgl-lineage–type signatures. Hierarchical clustering using these signature methylation sites confirmed a classification congruent to transcription for nearly all patients (Fig. 2H, fig. S2G, and data file S4). Examining the signature methylation sites revealed that tumors of the PeriV-lineage type displayed, for example, increased methylation of GFAP gene and S100B gene, while MGMT gene and STAT6 (signal transducer and activator of transcription 6) gene were more unmethylated, indicating a suppression of glial genes and an enhanced malignant expression pattern. In agreement, STAT6 has been shown as a unique marker and driver of meningeal hemangiopericytoma, a type of brain tumor that originates from pericytes (31). Thus, the methylation signatures reflected the innate cell-type features of PeriV-lineage– and Rgl-lineage–type glioblastoma.
We examined if the methylation status can predict tumor type using machine learning. A neural network classifier was generated by training transcriptionally defined PeriV-lineage– or Rgl-lineage–type glioblastoma with the methylation signatures. Similar to the hierarchical clustering (Fig. 2H), the methylation-based classifier assigned the majority of tumors to the corresponding transcriptionally defined PeriV-lineage–type and Rgl-lineage–type glioblastoma with high accuracy (fig. S2H). Next, we used this trained classifier for scoring 559 glioblastomas from a merged TCGA/DKFZ dataset (data file S4) and evaluated patient survival. Consistent with previous studies, isocitrate dehydrogenase 1 (IDH1)–mutant glioblastoma predicted a better outcome. In the remaining 288 IDH1 wt patients that include life span information, the PeriV-lineage type predicted the poorest patient survival with 0% 2-year survival (Fig. 2I). We also applied the same classifier for an independent dataset of 151 patients from the CGGA (Chinese Glioma Genome Atlas) (32) and further evaluated the IDH1 wt patient survival. A comparable survival to that of the TCGA/DKFZ studies was observed. Although the difference was not significant, none of the glioblastoma patients with PeriV-lineage–type signatures were alive after 2 years (fig. S2I).
Perivascular lineage–type glioblastoma consists of cells similar to vFBs, pericytes, and vascular SMCs
To examine whether cells of PeriV-lineage glioblastoma cells can be assigned to a specific perivascular cell type, we used a high-quality dataset of reference brain vascular cells, generated by Smart-seq2 scRNA-seq (17). Thus, we trained a neural network classifier with learned features from this dataset (fig. S3, A and B), and then assigned the merged dataset of low- and high-grade glioma cells to the reference vascular cell types. Consistent with our previous finding (Fig. 1), glioblastoma cells that were previously assigned to pericytes/VLMCs (fig. S3C, left) were robustly assigned to one of the three perivascular cell types: the immature stem-like vFBs, SMCs, and pericytes. Bulk sequenced data from TCGA were robustly assigned to vFBs (fig. S3C, middle). In contrast, low-grade glioma cells were rarely assigned to any vascular cell types (fig. S3C, right).
Reconstruction of glioblastoma cells along the developmental trajectory of the radial glia and neural crest cell lineages
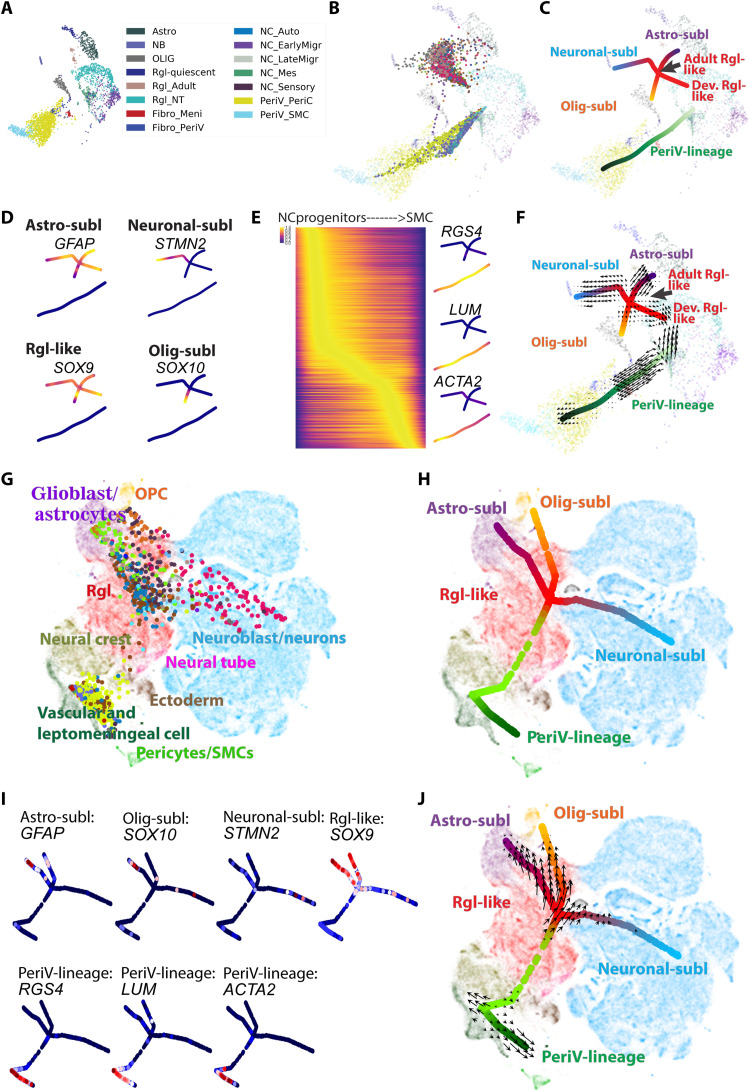
Meningeal cells as well as the brain perivascular cell types arise from mesenchymal neural crest cells (15, 19) attaching to blood vessels descending into the brain parenchyma during development (19). We therefore next examined the similarity of glioblastoma cells to cranial neural crest and neural tube cells captured from the developing mouse embryo at the time when neural crest cells delaminate from the neural tube (33) to meningeal cells (34) and to perivascular cells (17), as well as cells of the Rgl-lineage including adult Rgl, neuroblasts (35), oligodendrocytes, and astrocytes. All these data were generated using the Smart-Seq2 platform. On the basis of our previous analyses, these cell types together represent the endogenous cell types that glioblastoma displays similarities to. To track the developmental location of each glioblastoma cell along the lineage trajectory of brain cells, we developed a neural network–based projection model, SWAPLINE (Single-cell Weighted Assignment and Projection on developmental LINEages) (fig. S3D). We first visualized the normal reference brain cell types in a UMAP (Fig. 3A). Each cell-type cluster’s position in the UMAP reflects its transcriptional status in the relatively flattened topology in partition-based graph abstraction (PAGA) and the predicted cells must be assigned according to the limited PAGA nodes supervised by machine learning (fig. S3E). Nevertheless, the result is consistent with previous experimental lineage tracing studies, confirming the validity of the model. Consistently, all assigned tumor cells via SWAPLINE exhibited marker expression consistent with their position and naïve reference cell types (see below). This UMAP was later used as reference map for the projection of glioblastoma cells onto the brain’s normal differentiation trajectories.
Fig. 3. Relation of glioblastoma cells to the developing central nervous system and neural crest.
(A) Plot of reference cells. UMAP visualization of cell clusters from the developing central nervous system and neural crest lineages (17, 33–35). Abbreviations are as in fig. S3E. (B) Projection of all glioblastoma cells to the reference plot. Reference cells are indicated by “×” and glioblastoma cells are indicated by “dot,” which represent the projected developmental position of the individual glioblastoma cells to native reference cell types. (C) Principal tree plot summarizing the developmental status trajectory of the glioblastoma cells. Lineages are indicated by colors and text. Abbreviations are as in fig. S3M. (D) Visualization of normalized expression in tumor cells of pseudo-time marker genes for branches in the Rgl-lineage. (E) Left: Heatmap shows the normalized expression of pseudo-time genes according to the voltage peak along the neural crest trajectory. Right: Projection of the normalized expression in tumor cells of selected marker genes on the branching tree plot. Dark purple to yellow represents the minimal to maximal expression. (F) Quiver visualization of RNA velocity of glioblastoma cells on the branching tree plot. The arrow of each glioblastoma cell points to the direction of future status, extrapolated from RNA velocity estimates. (G and H) SWAPLINE projection and branching tree visualization of glioblastoma cells onto developmental mouse brain and neural crest reference plot from the mouse developmental brain atlas (16). Abbreviations are as in fig. S4J. (I) Marker gene expression in glioblastoma cells and visualized in the branching tree projected on the reference developmental mouse brain plot. Dark blue to red represents the minimal to maximal gene expression. Abbreviations are as in fig. S4J. (J) Quiver visualization of RNA velocity of glioblastoma cells onto developmental mouse brain and neural crest reference plot.
The accuracy of the SWAPLINE model was tested and confirmed using the independent sets of human brain cells (fig. S3, F and G) (21, 22). SWAPLINE assigned cells correctly in the lineage trajectories, while unrelated control cells (endothelial cells and microglia) were filtered out automatically in the model because of low scores. Next, we applied the model to project each glioblastoma cell into the differentiation trajectories of brain cell types (fig. S3H). The relative tumor cell position in relation to the background map plot of reference developmental/endogenous cell types was visualized (Fig. 3B). To disentangle the transcriptional roadmap of glioblastoma cells, we generated a statistical ensemble of principal branching tree trajectories (36) from the high-dimensional transcriptional space (Fig. 3C). The main tree structure summarized glioblastoma cell distribution and comprehensively showed the progression of glioblastoma cells along each developmental lineage trajectory. Two main glioblastoma lineage structures were observed with differentiated cells at termini, after which each branch was named. One lineage was organized around a shared center of Rgl reference cells with branches of cancer cells toward reference astrocytes (Astro-sublineage glioblastoma cells), neuroblasts (Neuronal-sublineage glioblastoma cells), and oligodendrocyte cells (Olig-sublineage of glioblastoma cells). Here, reference Rgl from two developmental stages was included (adult Rgl and developmental Rgl). The other lineage structure was the PeriV-lineage represented as a single line structure, with PeriV-lineage glioblastoma cells positioned from the most undifferentiated early reference migratory neural crest cells to differentiated reference perivascular mural cells.
Cross-annotation of patients and lineage branches revealed that all patients dominantly contained glioblastoma cells assigned either to the reference Rgl-lineage (Astro-sublineage, Neuronal-sublineage, or Olig-sublineage) or to the reference PeriV-lineage cells (fig. S3I). For patients with an Rgl-lineage–type glioblastoma, all subbranches coexisted in all patients, although at different proportions, revealing the intratumor lineage heterogeneity among patients with an Rgl-lineage signature.
To further explore the most similar cell type of PeriV-lineage glioblastoma cells along the differentiation trajectory from undifferentiated reference migratory neural crest cells to differentiated reference perivascular mural cells, we constructed a new cranial neural crest cell reference dataset via integrating the migrating cranial neural crest cells, neural crest mesenchymal progenitor cells (33), meningeal cells (34), and brain perivascular cells (17), which should represent all known neural crest derivatives in the brain region. After training with this reference dataset in the neural network model, we found that the PeriV-lineage tumor cells are most similar to vFBs and migrating neural crest cells (fig. S3J).
The existence of two lineages in glioblastoma cells was further confirmed by SWAPLINE lineage reconstruction for two independently published glioblastoma datasets, including (5) (fig. S3, K to N) and (7) (fig. S3, O to R). Moreover, we applied SWAPLINE assignment for glioblastoma cells with or without OSM treatment and found that almost all cells were assigned to Rgl-lineage cells (fig. S3, S and T), indicating that the cell-type state of glioblastoma cells remains conserved even after the OSM-induced transition to a more mesenchymal-like state. However, OSM-treated cells exhibited an increased feature of delaminating neural crest cell (fig. S3U) and reduced feature of radial glia, suggesting that the mesenchymal signature induced by OSM reflects features of the epithelial-mesenchymal transition of premigratory neural crest cells (37).
Next, we enriched pseudo-time marker genes that associated with each branch trajectory (data file S5), and the normalized expression of the selected marker genes along the Rgl-lineage branches was visualized in the branching tree (Fig. 3D). For example, STMN2 and SOX10 were specifically expressed in glioblastoma cells at the distal part of the neuronal- and Olig-sublineages, respectively, suggesting the existence of stable transcriptional status along these two branches. In contrast, Rgl-like tumor cells and glioblastoma cells at the distal part of the Astro-sublineage and Rgl-enriched SOX9 and GFAP were, albeit at lower levels, also expressed across all branches, indicating lack of unique markers for these glioblastoma cells. Consistently, RGS4, which is transiently expressed during neural crest differentiation (38), was also expressed in PeriV-lineage glioblastoma, specifically enriched in the progenitor-like cells of such tumors (Fig. 3E), while expression of lumican (LUM) and actin alpha 2, smooth muscle (ACTA2) was consistently enriched in glioblastoma cells corresponding to the more differentiated brain vFBs and SMCs, respectively.
Cell cycle and differentiation potential along differentiation branches of glioblastoma cells
Tumor initiation and propagation requires cell division. In our dataset and two independent glioblastoma datasets (5, 7), cycling tumor cells were mainly observed at the region of reference Rgl and between the reference migrating neural crest and vFB cells, while tumor cells in all branch termini were relatively quiescent (fig. S4, A to C). These observations suggest that the mitotic hyperactivity of progenitor-like tumor cells is a general rule for tumors with an Rgl-lineage–type and PeriV-lineage–type transcriptional signature. Mitotic events developmentally couple with cell differentiation and fate decision (39). RNA velocity analysis (40) revealed that the main trend of differentiational status change along each sublineage branch was from the progenitor region to differentiated termini (Fig. 3F and fig. S4D). Both the neuroblast and the oligodendrocyte branch of glioblastoma cells showed reduced differentiation at the developmental terminus, consistent with pseudo-gene results in Fig. 3D. Tumor cells at the terminus of the astrocyte branch exhibited lineage reversal, indicating bidirectional glioblastoma cell differentiation along the reference Rgl to astrocyte differentiation trajectory. In the PeriV-lineage, the main differentiation trend of glioblastoma cells was from reference migrating neural crest cells to perivascular cells. We also found that some of the most undifferentiated glioblastoma cells assigned to the PeriV-lineage displayed differentiation vectors toward reference spinal cord Rgl cells.
The most undifferentiated glioblastoma cells are expected to be enriched at the regions of the reference Rgl and neural crest cells (fig. S4E). To enhance the resolution of the reference map for a subsequent annotation of the most undifferentiated stem-like glioblastoma cells, we extracted these cells according to the density estimation and performed a zoom-in projection on the recently released mouse developmental brain atlas (16) again using the SWAPLINE projection (fig. 3G and fig. S4, F to I). The summarized tree structures and RNA velocity estimation further disentangled the progression of glioblastoma progenitor-like cells along each embryonic developmental brain lineage (Fig. 3, H to J, and fig. S4J). Confirming the above results, some tumor cells clustered with reference Rgl cells as well as along branches of reference cell differentiation into astrocytes, neuroblasts, and oligodendrocytes. Other glioblastoma cells were mainly located at the reference embryonic neural crest/VLMC region of the map with a branch toward reference perivascular cells. Reference cell lineage markers further confirmed that the tumor cells assigned to a developmental position also expressed the expected markers of naïve cells in that differentiation branch of the embryonic brain (Fig. 3I and data file S5). Furthermore, the relation of glioblastoma cells to these reference embryonic developmental lineages was further validated by SWAPLINE lineage reconstruction for two independent published glioblastoma datasets from (5) (fig. S4, K to M) and (7) (fig. S4, N to P), with similar results. To enhance the resolution of the reference brain cell types, we applied the machine learning classifiers with learned transcriptional features from early human developing brain cell types (fig. S4Q) (41), further validating our observation (fig. S4R). Combined, these results indicate that heterogeneity in glioblastoma can be explained by two main cell-type lineages of the brain, the radial glia and the PeriV-lineage, with tumor cell transcriptional programs at large recapitulating normal transcriptional routes of differentiation.
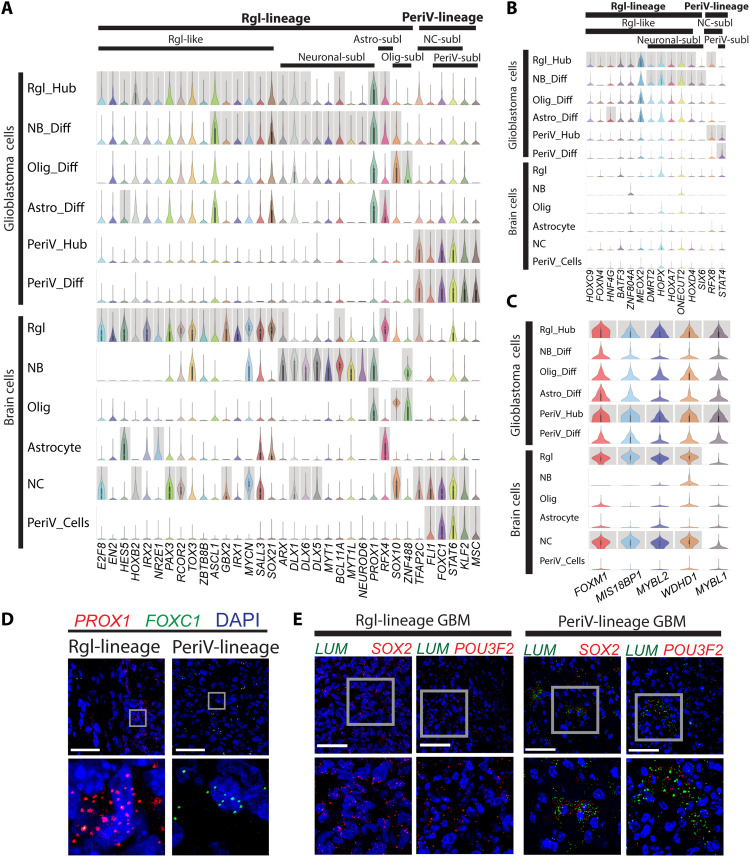
The direct lineage relationship of glioblastoma cells to developmental and adult brain cells indicates that transcription factors (TFs) that define cell types and thereby drive differentiation in the developing brain also contribute to the diversity of glioblastoma cells along the lineage trajectories. Thus, we divided our tumor cells into six lineage clusters according to their lineage branches and progenitor feature relationship to reference cells. Subsequently, we enriched the differentially expressed TFs from each glioblastoma lineage cluster as described in fig. S4E. Next, we applied the same enrichment for the published glioblastoma dataset (5), as well as for the annotated reference dataset of normal brain cell types (20). By comparing these three datasets, we identified unique TFs defining Rgl-lineage (30 TFs) and PeriV-lineage tumor cells (6 TF genes: FLI1, FOXC1, STAT6, KLF2, TFAP2C, and MSC) shared with normal development and a few glioblastoma-specific factors within each of the lineages (Fig. 4, A and B, and data file S6). Next, we applied SCENIC for identifying gene networks regulated by master TFs (regulon activity) in both Rgl-lineage and PeriV-lineage cells. After comparing the enriched TFs, 20 master TFs were identified with significant regulon activity (fig. S4S). The Rgl-lineage consisted of 14 TF regulons, including some known Rgl-specific TF genes, such as HES5, RFX4, and SOX10. We identified six TF regulons specific for PeriV-lineage, including STAT4, STAT6, TFAP2C, FOXC1, FLI1, and MSC. Furthermore, analysis showed that shared features between the two lineages (PeriV and Rgl) all relate to the cell cycle, including five cell cycle–regulating TFs (FOXM1, MIS18BP1, MYBL1, MYBL2, and WDHD1) (Fig. 4C and data file S6). Two lineage-specific TFs, PROX1 for Rgl-lineage and FOXC1 for PeriV-lineage, were validated in the tumor tissue of patient-derived xenografts (Fig. 4D). SOX2 and POU3F2 are driver genes in glioblastoma-propagating cells (42) that are induced during oncogenesis since they are not expressed in normal perivascular cells but present in migrating neural crest (43). Therefore, we also validated these two genes as lineage-shared TFs (Fig. 4E).
Fig. 4. Conserved TF signatures between naïve brain and neural crest cells with Rgl- and PeriV-lineage glioblastoma.
(A to C) Violin plot of TF expression shared between tumor cells and normal reference cell types (A), of TFs unique to glioblastoma cells (B), and of TFs shared between Rgl- and PeriV-lineage glioblastoma cells (C). y axis, the relative expression level; x axis, TF gene names. Cell types and lineages are indicated at the top of the chart. Gray columns represent the significantly differential expression. “Diff” indicates tumor cells at the distal differentiation of the sublineage trajectories and “Hub” indicates stem-like cells of the Rgl and perivascular lineages corresponding to native radial glia and neural crest cells, respectively. (D and E) Validation of PROX1 and FOXC1 mRNA expression in Rgl-lineage– and PeriV-lineage–type patient-derived glioblastoma xenografts, respectively (D). Validation of SOX2 and POU3F2 mRNA expression in both PeriV-lineage–type and Rgl-lineage–type patient-derived glioblastoma xenografts, LUM was used as a marker of PeriV-lineage tumor (E). Tumor lineage type and gene names are at the top. Each bottom figure is a higher magnification from the gray frame of the top figure. Scale bars, 50 μm.
Initiation of PeriV brain tumors from perivascular cells
Mouse models have indicated that glioblastoma can efficiently be initiated from the glial and stem cell compartments of the brain (11). The notable similarity of PeriV-lineage–type tumor cells to endogenous reference perivascular cells suggests that perivascular cells can also be susceptible for malignant transformation. To test whether perivascular cells might initiate brain tumors when carrying genetic alterations mimicking glioblastoma, we first investigated the expression profiles of the spontaneous glioblastoma tumors from both Nes-CreERt2 Pten/Trp53/Nf1 KO mice and NG2-CreERt2 Pten/Trp53/Nf1 KO mice (11, 12). Nestin is predominantly expressed in neural stem cells (i.e., radial glia cells), but NG2 is typically expressed in oligodendrocytes as well as in perivascular cells in the mouse brain (44). Thus, we hypothesized that tumors from NG2-CreERt2 Pten/Trp53/Nf1 KO mice can arise from either naïve oligodendrocytes or perivascular cells of the brain, while tumors from Nes-CreERt2 Pten/Trp53/Nf1 KO mice should arise only from radial glia cells. Hierarchical clustering revealed that two of the seven sequenced tumors derived from NG2+ cells were PeriV-lineage and the other five were Rgl-lineage. Furthermore, none of the seven glioblastomas induced from Nes+ cells carried any perivascular signature pattern (fig. S5A).
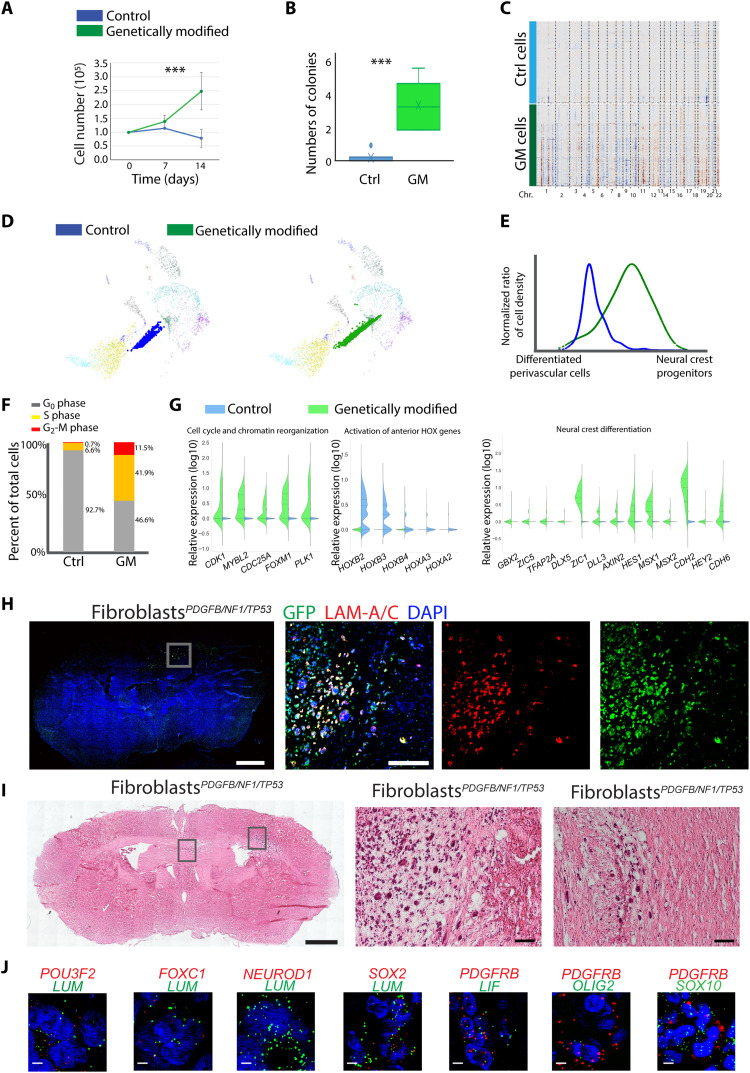
Platelet-derived growth factor (PDGF) acting through PDGF receptors induces proliferation and migration of perivascular cells (45). We therefore estimated the tumorigenesis potential of human brain perivascular cells by introducing PDGFB and depleting CDKN2A (p16INK4A and p14ARF) in primary human brain pericytes (PeriPDGFB/CDKN2A) and introducing PDGFB and co-depleting NF1/TP53 in human primary brain vFBs (fibroblastPDGFB/NF1/TP53) with green fluorescent protein (GFP) introduced into both cell types (fig. S5, B to D). These alterations led to marked increases in in vitro growth compared to naïve cells and significantly promoted the colony formation in vFBs (Fig. 5, A and B, and fig. S5E). To explore the consequences of these genetic alterations on cell identity, we scRNA-sequenced vFBs with and without the alterations. We observed comprehensive CNV changes in genetically modified vFBs (Fig. 5C and fig. S5F), with the significant deletion of Chr.4q, 1q, 9q, and 18q, and amplification of Chr.12q and 5q, indicating that a few founding mutations can lead to large genetic alterations. In particular, the alterations of Chr.18q and 5q have been identified in mesenchymal glioblastoma (5) and meningioma (46)—another type of brain tumor derived from the neural crest lineage. SWAPLINE projection of the control and genetically modified vFBs in the developmental adult reference plot revealed a marked dedifferentiation of the modified vFBs toward reference neural crest progenitors (Fig. 5, D and E). Consistently, more G2-M cycling cells were observed in modified vFBs (Fig. 5F and fig. S5G). By comparing the transcriptional profile between control and modified vFBs, we identified 773 up-regulated and 638 down-regulated genes (data file S7). Pathway enrichment revealed that “cell cycle and chromatin reorganization” and “neural crest differentiation” were significantly increased, while “HOX gene–related tissue patterning” was suppressed, indicating a dedifferentiation toward a neural crest stem cell state and a loss of anterior-posterior positioning information (Fig. 5G and data file S7). The cells were introduced into the brain in the orthotopic mouse model to test for tumor initiation. Both the modified pericytes and vFBs generated tumors, and the mice exhibited poorer tumor-associated survival than the control group receiving naïve cells (fig. S5H). Consistently, none of the control groups transplanted with the corresponding naïve cell types had a confirmed brain tumor by histological analysis, while all genetically altered perivascular cells did. Fluorescence staining confirmed that the brain tumors were of human cell origin (anti-human lamin A/C and GFP; Fig. 5, H and I, and fig. S5, I to K). Both PeriPDGFB/CDKN2A mice and fibroblastPDGFB/NF1/P53 mice exhibited extensive neoplastic growth and most animals displayed a diffuse and infiltrative phenotype. The xenograft tumor tissue exhibited cellular mitotic activity (Ki67), altered microvascular patterns (CD31), and abnormal remodeling of extracellular matrix proteins (fibronectin and collagen VI) (fig. S5L). Furthermore, the expression of PeriV-lineage tumor marker genes (POU3F2, FOXC1, SOX2, and LIF) in the tumor tissue of the grafted mice was observed, while Rgl-lineage genes NEUROD1 and OLIG2 were rarely observed (Fig. 5J). We observed some tumor cells coexpressing the neural crest progenitor marker SOX10, in line with our in silico observation of a cellular dedifferentiation in transformed tumor cells (Fig. 5, D and E).
Fig. 5. In vivo initiation of tumors from perivascular cells.
(A and B) In vitro proliferation (A) and colony formation (B) of brain vFB with/without carrying genetic alterations of patient-derived glioblastoma [genetically modified (GM), green]. Means ± SD, three independent measurements. Student’s t test, ***P < 0.001. (C) CNV analysis of control (blue) and GM fibroblasts (green). (D) Projection of control and genetic modified fibroblasts to the reference plot of normal reference cell types from Fig. 3A. (E) Quantification of the differentiation status of control (blue) and GM fibroblasts (green) along the developmental trajectory of in vivo differentiation of reference perivascular cells. The y axis represents the normalized cell density of projected fibroblasts in (D). The x axis represents the linearized developmental position between differentiated brain perivascular cells and neural crest progenitors. (F) Quantification of cycling phases of control (Ctrl) and GM fibroblasts. (G) Gene expression of top significant pathways enriched by up- and down-regulated genes in GM fibroblasts as compared to the naïve fibroblasts. (H and I) Representative fluorescence (H) or hematoxylin and eosin (I) staining of the coronal section from mouse xenograft of GM fibroblasts. Magnified tumor regions boxed. Green, GFP; red, anti-human lamin (LAM) A/C; blue, 4′,6-diamidino-2-phenylindole (DAPI). Scale bars (H and I): 1000 μm, whole section; 100 μm, magnified figures. (J) In vivo mRNA expression of indicated marker genes in xenograft tumor tissues of genetic modified fibroblasts. Human LUM and PDGFRB were used to label tumor cells. Gene names and color are indicated in each panel. Scale bars, 10 μm.
DISCUSSION
scRNA-seq has provided unparalleled insights into the molecular nature of glioblastoma cells and has offered new means to explain the cell of origin, tumor phenotype, cell heterogeneity, and patient outcome (47). In this study, we combined the application of a neural network classifier and the trajectory analysis of native brain cells to identify the relation of glioblastoma cells to normal brain cells. Our results identified that some glioblastomas display high similarities to radial glia and its progenies (Rgl-lineage), consistent with previous studies assigning tumor cells to neural cell types using a list of defined marker genes, hierarchical clustering, or reference cells in principal components analysis (PCA) (5, 7, 8, 10, 26). Unexpectedly, we identified the remaining glioblastoma to be similar to perivascular cells (PeriV-lineage), and consistently, tumor cells were robustly allocated along one of the two cell lineages. Furthermore, we validated the tumor-propagating ability of naïve brain perivascular cells. According to our neural network classification of scRNA-seq data as well as deconvolution of bulk data, glioblastoma of a PeriV-lineage type represents a proportion of the TCGA-mes subtype. Furthermore, consistent results were obtained on patient survival using gene expression– or methylation-based patient stratification into Rgl-lineage or PeriV-lineage. Patients with a PeriV-lineage–type signature show significantly poorer survival than those with an Rgl-lineage type. Combined, our results suggest the existence of a subgroup of glioblastoma with similarities to perivascular cells of the brain, which is distinct from the Rgl-lineage.
Although transcription can be affected by both mutations driving transformation as well as the microenvironment (5), the originating cell lineage can represent an important determinant of glioblastoma molecular characteristics (12). Among the conserved markers expressed in most cell types of each of the lineage (Fig. 1C), there is a high expression in Rgl-lineage cells of PTPRZ1 and SLC1A3, which previously have been shown to contribute to glioblastoma initiation and progression (10). Furthermore, the expressions of PeriV-lineage markers, LUM and platelet-derived growth factor receptor beta (PDGFRB), have also been previously evidenced in glioblastoma (48, 49). Because glioblastoma tumors exhibit cells with features consistent with precursor populations, shared developmental determinants of the progenitor cell fates could contribute to oncogenesis. Cell cycle analysis along the lineage trajectories revealed both Rgl-lineage and PeriV-lineage tumor cells to be rapidly dividing with markedly reduced proliferation of the more differentiated cells within each lineage. When we identified shared features between the two progenitor cell populations, nearly all shared genes were cell cycle–regulating transcriptional activators. This suggests that a major shared feature in the progenitor cells of the two lineages (PeriV- and Rgl-lineage) involves cell cycle control. Thus, transcriptional determinants contributing to oncogenesis in the two different lineages unrelated to cell cycle control are for the most part unique to each lineage and coincides with those in normal brain lineage trajectories.
RNA-velocity analyses show that the main flow in glioblastoma is from progenitor cells to differentiated cell types, and hence, glioblastoma develops along conserved neurodevelopmental gene programs, in agreement with a recent similar analysis (7). However, unlike that study, we find lineage reversal of tumor cells in the astrocyte branch of differentiation as well as of PeriV-lineage tumor cells carrying similarity to reference vFB cells. This difference may be a consequence of the fact that we performed a comprehensive RNA velocity with all assigned glioblastoma cells on the lineage branching tree plot, instead of on selected individual patients or selected reference brain cell types, thus overall increasing resolution. Furthermore, the standard dimensional reduction (such as PCA and t-distributed stochastic neighbor embedding) in a previous analysis could be too strict for estimating RNA velocity across tumor patients, due to the individual variance (5, 10). Instead, a score-based branch plot may better reflect the roadmap of developmental programs for cancer studies (50). The finding of lineage reversal of some more differentiated cells is consistent with a high degree of plasticity observed in glioblastoma cells (5, 8, 10) and suggests that, within glioblastoma, tumor cells with astrocyte and vFB features along with the glioblastoma resident progenitor populations can be originators of the cancer cell hierarchy and, thus, driving cancer growth. This is also consistent for the PeriV-lineage–type glioblastoma in experimental data, since recapitulating in perivascular cells genetic changes of glioblastoma is sufficient to initiate tumors with perivascular cell expression features in orthotopic grafted mice, including a derepression of the stemness maintenance factor SOX2 (51).The profound impact of a limited set of TFs on the fate of perivascular cells is illustrated by the direct reprogramming of pericytes to neurons through a neural stem cell intermediate by forced expression of SOX2 and the proneural ASCL1 TF (52), suggesting that re-expression of SOX2 alone is sufficient for a dedifferentiation of pericytes to a stem-like cell state from which ASCL1 induces neurogenesis. Thus, our results are consistent with the notion that some glioblastoma can originate from neural crest–derived leptomeningeal and perivascular cells. It appears that, within these, a few acquired mutations can start a process involving genetic instability and re-expression of developmental TFs shifting differentiated perivascular cells into more progenitor-like cells within the differentiation trajectory of the neural crest.
MATERIALS AND METHODS
The reagents, software, and public datasets are listed in data file S8. The machine learning models, training datasets, testing datasets, main lineages, sublineages, and assigned cell types are listed in data file S9.
Human GC cultures
Surgical tissue samples and clinical information for glioma patients were obtained from Karolinska Hospital in accordance with the protocol approved by the regional ethical review board. An informed written consent was obtained from all patients. We have used 18 human glioblastoma cell lines between passages 1 and 5. Tumors were classified by a neuropathologist on the basis of the World Health Organization classification. Human glioblastoma tissues were cultured as previously described (53) with some modification. The tissue was minced with a scalpel, digested in Accutase/TrypLE (1:1) at 37°C for 15 min, and triturated through 18G and 21G needles. The dissociated cells were resuspended in NeuroCult NS-a basal medium (STEMCELL Technologies) with the addition of 1% B27 (Invitrogen), 0.5% N2 (Invitrogen), and 10 ng/ml each of EGF and fibroblast growth factor 2 (PeproTech), plated on laminin-coated Primaria dishes (Corning), and cultured as adherent cells.
Lentiviral-based genetic modifications of human pericytes and fibroblasts
Human brain vascular pericytes (HBVPs) and human brain vascular adventitial fibroblasts (HBVAFs) were purchased from ScienCell and cultured following the instructions provided by the company. The lentiviral construct, shCDKN2A pGFP-c-shLenti vector, was purchased from OriGene Technologies, and shNF1/P53 dual shRNA (CS-LvRU6GP) expressing GFP and pEZ-Lv151 vector expressing PDGFB were purchased from GeneCopoeia. The viral particles were produced in 293T cells through cotransfection of pMD2.G and psPAX2 at a ratio of 4:2:3. Supernatants were harvested 48 and 72 hours after transfection and concentrated using Lenti-X Concentrator solution (ClonTech). Viral pellets were resuspended in phosphate-buffered saline (PBS) and stored at −70°C until further use. HBVPs or HBVAFs were infected for 48 hours and then selected.
Colony formation assay
A total of 1 × 104 cells were mixed in 1.5 ml of 0.4% agarose as the top layer with a bottom base of 1.5 ml of 0.6% agarose, cultured in a six-well plate. The 0.4% and 0.6% agarose are the mixtures of low–melting point agarose and NeuroCult NS-a basal medium above. Every culture well is photographed for at least two views randomly; then, the pictures were counted for colony numbers after 20 days. The average counts were taken as counts of one sample. Triplicate wells were included in each analysis and at least three independent experiments were conducted.
Intracranial transplantation
Animal experiments were performed in accordance with the rules and regulations of Karolinska Institute and approved by the local animal ethics committee. Intracranial transplantation of human germinal center (GC) cultures was performed in neonatal nonobese diabetic–severe combined immunodeficient (NOD-SCID) mice as previously described (54). Human GCs were dissociated in TrypLE, and the number of cells was determined using a Coulter Counter (Coulter Electronics). Stereotaxic injections of 2 × 105 genetic-modified HBVP or HBVAF cells in 4 μl of Dulbecco’s PBS were performed on 8- to 10-week-old female NOD-SCID mice. The coordinates were 0.5 mm anterior of bregma, 1.1 mm lateral, and 2.5 mm ventral. Injected mice were monitored every second day and euthanized upon symptoms of disease. After euthanizing the mice, their brain was collected and fixed with 4% paraformaldehyde in PBS for overnight. The tissue was then washed with PBS and incubated with 15% sucrose for 24 hours, and 30% sucrose for another 24 hours. After that, the tissue was embedded into optimal cutting temperature compound (Sakura Biotech) in a Cryomold (Sakura Biotech) and frozen using liquid nitrogen. The frozen tissue blocks were stored in −80°C. Ten- to 12-μm-thin cryo-sections of xenograft tumor tissue were prepared on Superfrost Plus slides and slides were either stored in −80°C or processed immediately for immunofluorescence, fluorescence in situ hybridization, or hematoxylin and eosin staining.
Immunofluorescence analysis of mouse brains
Frozen sections were blocked in PBS containing 0.2% Triton X-100 (PBS-T), 3% bovine serum albumin, and 5% normal goat serum and incubated with primary antibodies for 1 hour at room temperature or at +4° for 4 hours in a humidified chamber. The sections were then washed with PBS-T three times and incubated with secondary antibodies (1:500) at +4° for 4 hours. After finally washing three times in PBS-T, sections were mounted in Immu-Mount (Thermo Fisher Scientific) containing 4′,6-diamidino-2-phenylindole. The pictures were taken using an LSM 700 confocal microscope (Carl Zeiss).
Fluorescence in situ hybridization (RNAscope)
Transcripts were detected using the RNAscope assay for fresh-frozen tissue (Advanced Cell Diagnostics). The probes were designed and provided commercially by Advanced Cell Diagnostics Inc. For the complete list of probes and genes, see Resource and Reagent List. The staining was performed using the RNAscope Fluorescent Multiplex Reagent Kit (catalog no. 320850), reagents, and probes according to the manufacturer’s instructions. Imaging was performed using LSM 700 confocal microscopes (Carl Zeiss).
Single-cell isolation and cDNA synthesis
A Fluidigm C1 Autoprep System microfluidic chip was used to capture the cells. Immediately after the image acquisition, cell lysis, reverse transcription, and polymerase chain reaction (PCR) amplification were performed as previously described (55). The amplified cDNA was harvested with 13 μl of Harvest Reagent and cDNA library quality was measured on an Agilent Bioanalyzer.
Preparation of sequencing library and Illumina sequencing
For patient-derived glioblastoma cells, we used 5′ single-cell–tagged reverse transcription sequencing (STRT-seq). Cell barcoding and fragmentation were performed in a single step using Tn5 DNA transposase (“tagmentation”) as described previously. One microliter of Dynabeads MyOne Streptavidin C1 beads (Invitrogen) was resuspended in binding and blocking buffer (10 mM tris, 250 mM NaCl, 5 mM EDTA, and 0.5% SDS) at the ratio of 1:20 and then added to each well. After incubation at room temperature for 15 min, all wells were pooled, and the beads were washed once with 100 μl of washing buffer (10 mM tris–150 mM NaCl and 0.02% Tween 20), once with 100 μl of QIAGEN Qiaquick PB, and then twice with 100 μl of washing buffer. Restriction was performed to cleave 3′ fragments: The beads were incubated in 100 μl of restriction mix [1× NEB CutSmart and PvuI-HF enzyme (0.4 U/μl)] for 1 hour at 37°C. Last, the beads were washed three times with the washing buffer, and then resuspended in 30 μl of ddH2O and incubated for 10 min at 70°C to elute the DNA. AMPure beads XP (Beckman Coulter) were used at 1.8× volume and eluted in 30 μl to remove short fragments. The molar concentrations of the libraries were determined with KAPA Library Quant qPCR (Kapa Biosystems) and the size distribution was evaluated after PCR (12 cycles) using an Agilent Bioanalyzer. Sequencing was performed on an Illumina HiSeq 2000 with C1-P1-PCR2 as read 1 primer and C1-TN5-U as index read primer. Reads of 50 base pairs (bp) as well as 8-bp index reads corresponding to the cell-specific barcodes were generated. For genetic-modified perivascular cells, the scRNA-seq was performed by using Chromium Single Cell 3′ Reagent Kits (10x Genomic, version 3) according to the manufacturer’s instruction.
Bioinformatics preprocessing, copy number analysis, and clustering
For STRT-seq, the reads were aligned by STAR using GRCh38.p12 genome assembly and processed as described previously (55). The cells harboring less than 1000 detected transcripts or less than 450 detected genes were filtered out. After these quality control procedures, 4073 cells were left with the median detected protein coding genes of 3531 counts. For 10x scRNA-seq, data preprocessing was performed via Cell Ranger. The copy number analysis was performed with CONICS following the instruction (56). Briefly, genes expressed in <5 cells were excluded. After centering the gene expression in each cell around the mean, the z-score of the centered gene expression was calculated across all cells. Next, the bimodal distribution of gene expression in any regions across cells was determined by a Gaussian mixture model mode, and the regions containing more than 100 expressed genes were identified for the next step. Then, the reported mixture models were chosen following the criteria of the Bayesian information criterion >5 and the P value of likelihood ratio test <0.05. To detect the existence of CNVs, the threshold of posterior probabilities was set as 0.55, and the gain or loss was determined by comparing the average expression in the normal cells. The heatmap visualizations of chromosomal alterations were generated in every single cell across the genome for all calculated patients.
Before clustering, we removed the cell cycle–related genes and then computed the coefficient variation (CV) (SD divided by the mean) versus the predicted CV (estimated by a nonlinear noise model) and applied the fit of noise distribution to select the most variable features that are greater than the expected CV. Support vector regression (SVR) from scikit-learn package was used for this analysis. The most variable features were used for calculating the top 20 PCs, and the top 10 nearest neighbors, 0.5 minimum distance, and Euclidean distance were used for UMAP.
The most variable genes were then used for cell clustering via different algorithms including the DBSCAN algorithm (Seurat V1.2) and the Louvain method for community detection with a resolution value of 1 (Seurat V3.0+) (55, 57). Furthermore, we applied several rounds of clustering, zoom-in clustering, and cluster recombining to make sure that all clusters are biologically meaningful and exhibited significant markers. Eventually, cells were grouped into 20 clusters, and the marker genes of every cluster were determined via enrichment score as described in (44). The enrichment score Ei,j for gene i and cluster j was defined as
Here, αi, j represents the score of nonzero expression for the cells in this cluster, and represents the score of nonzero expression for the cells that are not in this cluster. βi, j represents the mean expression for the cells in this cluster, and represents the mean expression for cells that are not in the cluster. A small value of the constants ε1 and ε2 is added to prevent the divisor from having a value of zero.
Scoring analysis of cell-type identity
For this analysis, our goal was to score the probabilistic cell identity of each cell relative to the defined cell types at the transcriptional level (21). We built an L2-regularized logistic regression model, a C-support vector classification model, and a vanilla neural network model (PyTorch framework with Skorch package) for classification tasks and trained the model to learn the general prototypes of defined cell types. To train the model, we removed the cell cycle–related genes, and then computed the CV (SD divided by the mean) versus the predicted CV (estimated by a nonlinear noise model), and applied the fit of noise distribution to select the most variable features that are greater than the expected CV. SVR from the scikit-learn package was for this analysis. The overdispersed genes were further ranked by two heuristics for the cell-type specificity of both fold change and enrichment score change (44). For TCGA subtype classification, the originally defined TCGA subtypes were used as reference cell types, and the originally identified marker genes of the four subtypes were manually added as feature genes for training the neural network classifier. For the lineage classification based on the differential methylation sites, the defined lineages at the transcriptional level were used as the reference cell types, and the identified differential methylation sites were used as the features for training the neural network classifier. The cross-species alignment was performed as described in (21). To compare the data from UMI-based platforms and the Smart-seq2 platform, data were scaled by SD owing to the potentially larger gene variation in Smart-seq2 (58). Subsequently, the ranked marker genes of the defined cell types were log-transformed and scaled by Minmax normalization, and then used for the different learning models:
1) The L2-regularized logistic regression model was as described in (59).
2) To test the adequate strength of the regularization in the C-support vector classification model, the C regularization parameter and three kernel types, “linear,” “sigmoid,” and “rbf,” were inspected via GridSearchCV. The classifier accuracy was estimated by a k-fold cross-validation, of which the dataset was randomly split (25% test_size). The value of the C regularization parameter and the kernel type were chosen corresponding to the maximum point of the learning curve reaching the accuracy plateaus.
3) The neural network model contains an input layer with the number of neuron nodes being the same as the number of marker genes, a hidden layer with the number of neuron nodes being the same as 20% of marker gene numbers, and an output layer with the number of neuron nodes being the same as the number of defined cell types. Linear regression was performed between each layer, and 30% of dropouts were set to reduce the overfitting. Rectified linear unit (ReLu) was used as the activation function of the hidden layer, and Softmax was used for the output layer to evaluate the probabilities. Nesterov momentum was used as a stochastic gradient descent (SGD) optimizer. To choose the adequate regularization strength, the classifier accuracy and the loss value were inspected against epoch numbers. The classifier accuracy was estimated by a k-fold cross-validation, of which the dataset was randomly split (k = 3). The learning rate, epoch number, and momentum were chosen corresponding to the maximum point of the learning curve reaching the accuracy plateaus.
4) The node-level graph neural network (GNN) model contains an input layer with the number of node features being the same as the number of marker genes, two hidden layers with the number of neuron nodes being the same as 25% of marker gene numbers, and an output layer with the number of neuron nodes being the same as the number of defined cell types. The edge indexes were selected as the top 10 nodes upon K-nearest neighboring (KNN) calculation of the top 30 principal components.
GCNConv (message passing) was performed between each layer, and 20% of dropouts were set. ReLu was used as the activation function of the hidden layer, and Softmax was used for the output layer to evaluate the probabilities. Momentum γ was set to 0.9 in the SGD optimizer. To choose the adequate regularization strength, the classifier accuracy and the loss value (CrossEntropyLoss) were inspected against epoch numbers. The learning rate, epoch number, and momentum were chosen corresponding to the maximum point of the learning curve reaching the accuracy plateaus.
We set the same learning steps for all four models and found that the learning accuracy and running period were 97.62% and 1390.83 s for the L2-regularized logistic regression model; 97.59% and 3084.81 s for the C-support vector classification model; 99.6% and 131.14 s for the vanilla neural network model; and 99.13% and 349.81 s for the node-level GNN. Thus, the ready vanilla neural network model was further used to predict the probabilities of each cell belonging to each trained reference cell type. The permutation test of dataset was applied to qualify the significance of the prediction, and the P value was calculated by false discovery rate. The prototype threshold of a defined cell type was determined as the larger value of significant probability (P < 0.05) and dominant probability (>60). If the probability of a predicted cell to one cell type is over this cell type’s prototype threshold, this predicted cell was considered as “cell type defined” and was assigned to this cell type. Data were visualized in the radar plot. The radar plot consists of a sequence of equiangular polygon spokes with the distal vertex representing each trained reference cell type. The distance between the polygon center and each vertex of the polygon represents the relative probabilities of each trained reference cell assigned to the defined reference cell types. Thus, the position of each predicting cell was calculated as a linear combination of the probabilities against all reference cell types and then visualized as the relative position to all vertices of the polygon.
Deconvolution of bulk tumor RNA sequencing
A bulk tumor tissue contains both the malignant cells and various microenvironment cells that disturb the transcriptional profile of the endogenous tumor cells. In addition, the intratumor heterogeneity of glioblastoma tissue further blurs the expression matrix. To enrich/denoise the gene expression of the dominant tumor cells from glioblastoma bulk tissue, we applied the deconvolution method via the power-law transformations and the autoencoder of convolutional neural network (CNN) (60). The RNA-seq data of TCGA were obtained from the UCSC Cancer Browser, and our scRNA-seq data were used as the reference dataset for deconvolution. Genes in the reference dataset were prefiltered by the count frequency as described in BACKSPIN (55), and then used for the deconvolution of bulk tissue. Each gene was scaled by Minmax normalization and visualized by a curved line plot; the x axis represents the cell/sample that was sorted by the expression value of the gene. Thus, we obtained the distribution of gene expression of these datasets and visualized them in a curve line plot. The mean values of all curves were calculated for the least squares polynomial approximation via Numpy, and the square root was used as weights to find the γ value of the curve. By comparing the γ values of both bulk tissue data and reference glioblastoma single-cell data, the expression matrix of bulk sequencing was fit to the same distribution of single-cell sequencing via power-law transformations (fig. S2B, step 1).
Next, the CNN autoencoder was applied for denoising the transformed datasets. The autoencoder contains two layers of convolution and four layers of transposed convolution in the PyTorch framework. The hyperbolic tangent activation function (Tanh) was used as the activation function between each layer, and sigmoid was used for the output layer. The mean squared error between each element in the input (MSELoss) was evaluated against the epoch. The learning rate and epoch number were chosen corresponding to the minimum point of loss_value curve after reaching the loss_value plateaus (fig. S2B, step 2). After the training of the reference glioblastoma scRNA-seq data, the model was performed for the deconvolution of the transformed dataset of glioblastoma bulk tissue. The deconvoluted dataset was scaled and visualized in a curve line plot as described above for evaluation and subsequently used for further analysis.
Single-cell Weighted Assignment and Projection on developmental LINEages
The aim of SWAPLINE is to place each test cell into a trajectory position of normal developmental lineage(s), via combining both KNN and the scoring of probabilistic cell identity. The workflow is described in fig. S3D.
To construct the reference lineage trajectory, the endogenous mouse brain cell types were from developmental brain atlas (16) or collected from different datasets generated via the Smart-seq2 scRNA-seq platform, including adult Rgl/neural stem cells, neuroblasts (35), meningeal cells derived from neural crest (34), neural crest and neural tube cells captured from the developing embryo (33), and oligodendrocytes, astrocytes, and perivascular mural cells (17). These cell types should together represent possible endogenous brain cell types to which glioblastoma cells display similarities. Meningeal cells, embryonic neural crest cells, and perivascular mural cells theoretically belong to neural crest lineage in brain, while other cell types follow the CNS neural development. UMAP was used to build the reference plot that reflects the transcriptional relations among all reference cell types. PAGA analysis (61) further confirmed the lineage relations among the reference cell types. Subsequently, two steps of quantification were applied in parallel: First, we used all these reference cell types to perform the cell scoring of probabilistic similarity. Next, we divided the prototype probabilities into two groups according to the developmental lineages of the reference cell types: a neural crest lineage and a CNS neural lineage as described above. For each predicted cell type, the mean value of the prototype probabilities of the two lineage groups was used to estimate the lineage similarity of this predicted cell type, the higher lineage probability assigned, and the predicted cell type into this lineage for further lineage-specific SWAPLINE analysis. Since there are two major lineages in the reference cells during neural/neural crest development, we assigned each predicted cell type into its normal developmental lineage by referring to the top N (N = 3 or 4 here) closest reference cell type in PAGA. For each lineage, the top connected reference cell types and predicted cells were used for probabilistic scoring. The permutation test was applied as the negative control and background noise. Second, we used KNN to evaluate the putative position of each predicted cell corresponding to every reference cell types in the UMAP. Briefly, we first calculated the top principal components of all cells following the Elbow method, and then used these principal components to access the pairwise distances of Euclidean metric among all cells. For each predicted cell, we selected the top 25 nearest cells in each reference cell type and calculated the median UMAP coordinates of these top nearest cells. Thus, we obtained the KNN putative positions of the predicted cells in each top N connected reference cell type. Furthermore, the prototype probabilistic score of each cell was normalized to the median value of randomized probabilities that were generated from the permutation test and further rescaled by Minmax. The cells with global prototype similarity (putatively low-quality cells or extremely high-plasticity cells) were excluded if one predicted cell’s SD of probability among prototypes was lower than the permutation test. Subsequently, a linear combination of both KNN putative positions and cell probabilities of top N related and connected reference cell types represents the developmental trajectory position of each predicted cell: Let N be the total number of prototypes, let pm be the probability of a cell belonging to prototype m, let cmj be the coordinate of nearest neighboring cell j of the predicted cell from prototype m, and let k be the top closest constant; the predicted coordinates of test cell upon the origin of coordinates then was defined
Disentangling trajectory analysis of the branching tree
The principal branching tree was constructed to elucidate the fundamental lineages of glioblastoma cells via a simplified elastic principal graphs. Elastic principal graphs are a generalization of the elastic map algorithm for approximating principal manifolds from the data with a given topology (36). A principal manifold is an undirected graph (B) composed of nodes (N) and edges (E). The nodes are embedded into the data space by minimizing both the approximation error (mean squared distance) to the data points and the elastic energy [Uϕ(B)], defined as
k-star in graph G defines a subgraph that contains k + 1 nodes, n0,1,...,k ∈N, and k edges {(n0, ni)|i = 1, ..., k}. D represents the structured data points, and Num is the number of data points. ϕ (Nj) is the map Ф: N → Rm, which represents an embedding of each j node in the data space. The data point partitioning Pn was defined as Pn(i) = arg minj = 1…|V| (Di − ϕ(Vj))2, and it provides an index of a node that is the closest to the ith data point in the graph. Each iteration provides the initial guess of ϕ, the partitioning Pn(i) is computed, and U ϕ (D, B) is minimized via exploring new node positions in the data space. Tr represents the trimming radius, a distance dropout parameter in the limit, of which the data points were used for graph optimization. For the comprehensive evaluation, we set Tr as infinite here. The edges among the nodes define the elastic energy, which serves as a penalty for the graph embedding. The elastic energy is manifested by two main factors: the stretching and non-equal distance of node-to-node positions [, weighted by the λ] and the deviation from harmonic embedding [, weighted by μ], defined as
An elastic principal tree contains selected families of k-stars Sk. Each graph edge E(i) has two nodes E(i)(0) and E(i) (1). Sk(j)(0) to Sk(j)(k) denote the nodes of a star Sk(j) in the graph, and Sk(j)(0) represents the center node that links to all other nodes. According to the equation, the elastic energy is regulated by two weighted factors: λ, regularizing the overall length of the edges, and μ, the deviation of the star nodes from harmonic embedding. Thus, we evaluated the construction of a principal tree upon different combinations of λ and μ. Besides these two, the parameter α independently regulates the appearance of branches via perturbing the edges of higher-order star nodes. To avoid excessive branching, we use a small value (0.01) here according to the formal description. As the SWAPLINE coordinates of each glioblastoma cell represent its status within the developmental trajectory of normal brain cells, we use the SWAPLINE coordinates to perform the low-dimensional construction of the principal branching tree. To test the robustness of the principal graph, we inspected different combinations of the elastic stretching (λ; range, 0.001 to 0.02) and the deviation from harmonicity penalty (μ; range, 0.05 to 0.5). A total of 2565 rounds of the principal graph were tested and visualized. To obtain the minimum branching and the maximum elastic stretching, we chose the principal tree produced with λ = 0.01 and μ =0.2 for subsequent analyses; alternatively, the principal tree can be obtained from the PCA of the parameter tests described above. Each edge of the principal tree was smoothened by one-dimensional interpolation via the interp1d package from SciPy. In addition, the small branch with only one single link between two nodes was merged into the neighboring larger branch. Next, we used the Shapely package to project all cell dots onto the principal edge by evaluating the shortest distance at the two dimensions and adjusted the cell positions to keep the same intercellular distance along each branch. To identify the branching related genes, we separated the principal tree to five branches according to the branch point and the branch lineage. For each branch, the smoothed expression for each gene along the branch was determined by using a Gaussian filter or a generalized linear model (SciPy package). Significant branching genes were determined by three heuristics: (i) significant distribution based on the cumulative distribution function comparing the branching position and the smoothed expression, (ii) significant correlation (Spearman’s) between the branching position and the smoothed expression before and after peak value, and (iii) the gene expression should fit the criteria that at least 5% of the cells express two molecular counts and at least 20% of the cells express one molecular count. All smoothed expression was normalized to the central branching point for further comparison.
Analysis of cell cycle
A list of genes has been assigned to two major phases (S and G2-M) of the cell cycle (9). The significant phase activation was evaluated by comparing the expression of phase-related genes and the expression of random genes as described in Seurat, with small modification. Briefly, the overdispersed genes of a dataset were evaluated by estimating the mean and coefficients of variation. The overdispersed cell cycle genes were selected for phase scoring, and the rest of the genes were ranked by the expression and separated into 25 intervals according to the rank. In each interval, we selected the first 50 genes for randomization and thus generate the random gene matrix. The phase scores were generated by estimating the differential mean expression of the phase genes and the randomized genes. Phase G0-G1 was decided if the expression of phase genes was lower than randomized values. The activation of other phases was decided by the larger value of the phase score. Thus, each cell was assigned to different phases of the cell cycle and subsequently projected to the plot.
Comprehensive RNA velocity of all glioblastoma cells on STRT-seq/STRT-seq-2i
Spliced and unspliced counts of glioblastoma cells were quantified as described by La Manno et al. (40) using the RNA velocity package, with modification for 5′ STRT-seq. We extracted the barcode and UMI with the fault tolerance of 1 base mismatch from the FASTQ file. Meanwhile, we added the first 4 bases of the transcript sequence to the original 6 bases of UMI to generate 10 new bases of UMI for each read. The barcode tag and UMI tag were defined via SAMtools (pysam). The reads were aligned by STAR using GRCh38.p12 genome assembly and processed as described previously (55). We calculated spliced and unspliced counts using the built-in package of Velocyto (session of “any technique-advanced use”) with masking expressed repetitive elements. A total of 2451 cells were selected with the criteria of 200 unspliced molecules and 200 spliced molecules, and most variable genes were filtered with the criteria of four minimum unspliced molecules detected in a minimum of three cells. PCAs were selected according to 0.55% ratio of variance explained by each of the selected components. Data were smoothened via balanced KNN imputation with K = 500, b_sight = 4*K, b_maxl = 3*K. The variance normalizing transform was performed in log value. The time step for extrapolation is 5, and kernel scaling was set as 0.05 in calculating the transition probability to project the velocity direction on the embedding. The embedding scatter plot was forked from the branching tree plot as described above, and the branching tree plot widened along each axis for better visualization.
Extraction of core/hub glioblastoma cells via density estimation
To estimate the density of glioblastoma cells in the lineage plot, the coordinate of each cell in both scatter plot and branching tree plot was stacked vertically and applied for kernel-density estimation using Gaussian kernels. Bandwidth vector was generated via the rule of thumb of Scott. Relative density was calculated by comparing the overall density in the plot. Cells with the top 50% density were defined as hub/core cells, and the rest of the cells were defined as branch cells.
SCENIC analysis
To infer the TFs and their target gene networks, SCENIC analysis was performed according to the authors’ vignette. Briefly, the TF-targeted gene sets were identified via the following criteria: first, coexpression with TFs and, second, enriched in the direct motif of the TF. Then, the regulon activities were scored and binarized to determine whether the gene sets of each regulon were significantly enriched in cells.
Quantification and statistical analysis
Statistical analysis between groups was performed using two-tailed Student’s t test. Kaplan-Meier survival was calculated via log-rank test. Experiments were representative of at least three independent and biological replicates. Error bars in figures represent means ± SEM. P values were indicated in figures or marked as *P < 0.05 and **P < 0.01.
Acknowledgments
We thank S. Linnarsson (Karolinska Institute, Sweden) and G. La Manno (École Polytechnique Fédérale de Lausanne, Switzerland) for the technical support and the discussion; P. Lönnerberg for technical assistance; and D. Usoskin for help on animal work. We thank Science for Life Laboratory, the National Genomics Infrastructure funded by the Swedish Research Council, and Uppsala Multidisciplinary Center for Advanced Computational Science for providing assistance in massively parallel sequencing and access to the UPPMAX computational infrastructure.
Funding: This research was funded by the Swedish Medical Research Council, the Knut and Alice Wallenberg Foundation (Wallenberg Scholar), the Swedish Cancer Society 18 0635 to P.E., and Swedish Society for Medical Research (SSMF) fellowship to Y.H.
Author contributions: P.E. supervised the study. P.E., Y.H., and Y.J. designed the overall study. O.P. and M.S. provided samples and clinical annotation and reviewed the clinical data. I.A. coordinated the data acquisition. Y.H., M.M.R., S.O., and A.D.S. performed and interpreted the computational analyses. Y.J., Y.H., C.K., M.H., and N.S. performed and analyzed the in vitro experiments. Y.J., Y.H., J.B., M.-D.Z., and D.L. performed the in vivo experiments. P.E., Y.H., and Y.J. interpreted the data and wrote the manuscript. All authors reviewed and approved the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The accession number for the sequencing data reported in this paper is GEO (https://www.ncbi.nlm.nih.gov/geo): GSE159416 and GSE171287. Codes and Jupyter notebooks showing key steps of the analysis are publicly available in GitHub (https://github.com/ernforslab/Hu-et-al._GBMlineage2022) and Zenodo (https://doi.org/10.5281/zenodo.6321370). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S9
REFERENCES AND NOTES
- 1.Dunn G. P., Rinne M. L., Wykosky J., Genovese G., Quayle S. N., Dunn I. F., Agarwalla P. K., Chheda M. G., Campos B., Wang A., Brennan C., Ligon K. L., Furnari F., Cavenee W. K., Depinho R. A., Chin L., Hahn W. C., Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 26, 756–784 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan C. W., Verhaak R. G. W., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O’Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G., Chin L., Benz C., Barnholtz-Sloan J., Barrett W., Ostrom Q., Wolinsky Y., Black K. L., Bose B., Boulos P. T., Boulos M., Brown J., Czerinski C., Eppley M., Iacocca M., Kempista T., Kitko T., Koyfman Y., Rabeno B., Rastogi P., Sugarman M., Swanson P., Yalamanchii K., Otey I. P., Liu Y. S., Xiao Y., Auman J. T., Chen P. C., Hadjipanayis A., Lee E., Lee S., Park P. J., Seidman J., Yang L., Kucherlapati R., Kalkanis S., Mikkelsen T., Poisson L. M., Raghunathan A., Scarpace L., Bernard B., Bressler R., Eakin A., Iype L., Kreisberg R. B., Leinonen K., Reynolds S., Rovira H., Thorsson V., Shmulevich I., Annala M. J., Penny R., Paulauskis J., Curley E., Hatfield M., Mallery D., Morris S., Shelton T., Shelton C., Sherman M., Yena P., Cuppini L., DiMeco F., Eoli M., Finocchiaro G., Maderna E., Pollo B., Saini M., Balu S., Hoadley K. A., Li L., Miller C. R., Shi Y., Topal M. D., Wu J., Dunn G., Giannini C., O’Neill B. P., Aksoy B. A., Antipin Y., Borsu L., Berman S. H., Brennan C. W., Cerami E., Chakravarty D., Ciriello G., Gao J., Gross B., Jacobsen A., Ladanyi M., Lash A., Liang Y., Reva B., Sander C., Schultz N., Shen R., Socci N. D., Viale A., Ferguson M. L., Chen Q. R., Demchok J. A., Dillon L. A. L., Shaw K. R. M., Sheth M., Tarnuzzer R., Wang Z., Yang L., Davidsen T., Guyer M. S., Ozenberger B. A., Sofia H. J., Bergsten J., Eckman J., Harr J., Myers J., Smith C., Tucker K., Winemiller C., Zach L. A., Ljubimova J. Y., Eley G., Ayala B., Jensen M. A., Kahn A., Pihl T. D., Pot D. A., Wan Y., Eschbacher J., Foltz G., Hansen N., Hothi P., Lin B., Shah N., Yoon J. G., Lau C., Berens M., Ardlie K., Beroukhim R., Carter S. L., Cherniack A. D., Noble M., Cho J., Cibulskis K., DiCara D., Frazer S., Gabriel S. B., Gehlenborg N., Gentry J., Heiman D., Kim J., Jing R., Lander E. S., Lawrence M., Lin P., Mallard W., Meyerson M., Onofrio R. C., Saksena G., Schumacher S., Sougnez C., Stojanov P., Tabak B., Voet D., Zhang H., Zou L., Getz G., Dees N. N., Ding L., Fulton L. L., Fulton R. S., Kanchi K. L., Mardis E. R., Wilson R. K., Baylin S. B., Andrews D. W., Harshyne L., Cohen M. L., Devine K., Sloan A. E., VandenBerg S. R., Berger M. S., Prados M., Carlin D., Craft B., Ellrott K., Goldman M., Goldstein T., Grifford M., Haussler D., Ma S., Ng S., Salama S. R., Sanborn J. Z., Stuart J., Swatloski T., Waltman P., Zhu J., Foss R., Frentzen B., Friedman W., McTiernan R., Yachnis A., Hayes D. N., Perou C. M., Zheng S., Vegesna R., Mao Y., Akbani R., Aldape K., Bogler O., Fuller G. N., Liu W., Liu Y., Lu Y., Mills G., Protopopov A., Ren X., Sun Y., Wu C. J., Yung W. K. A., Zhang W., Zhang J., Chen K., Weinstein J. N., Chin L., Verhaak R. G. W., Noushmehr H., Weisenberger D. J., Bootwalla M. S., Lai P. H., Triche T. J. Jr., van den Berg D. J., Laird P. W., Gutmann D. H., Lehman N. L., VanMeir E. G., Brat D., Olson J. J., Mastrogianakis G. M., Devi N. S., Zhang Z., Bigner D., Lipp E., McLendon R., The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturm D., Witt H., Hovestadt V., Khuong-Quang D. A., Jones D. T. W., Konermann C., Pfaff E., Tönjes M., Sill M., Bender S., Kool M., Zapatka M., Becker N., Zucknick M., Hielscher T., Liu X. Y., Fontebasso A. M., Ryzhova M., Albrecht S., Jacob K., Wolter M., Ebinger M., Schuhmann M. U., van Meter T., Frühwald M. C., Hauch H., Pekrun A., Radlwimmer B., Niehues T., von Komorowski G., Dürken M., Kulozik A. E., Madden J., Donson A., Foreman N. K., Drissi R., Fouladi M., Scheurlen W., von Deimling A., Monoranu C., Roggendorf W., Herold-Mende C., Unterberg A., Kramm C. M., Felsberg J., Hartmann C., Wiestler B., Wick W., Milde T., Witt O., Lindroth A. M., Schwartzentruber J., Faury D., Fleming A., Zakrzewska M., Liberski P. P., Zakrzewski K., Hauser P., Garami M., Klekner A., Bognar L., Morrissy S., Cavalli F., Taylor M. D., van Sluis P., Koster J., Versteeg R., Volckmann R., Mikkelsen T., Aldape K., Reifenberger G., Collins V. P., Majewski J., Korshunov A., Lichter P., Plass C., Jabado N., Pfister S. M., Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Wang Q., Hu B., Hu X., Kim H., Squatrito M., Scarpace L., deCarvalho A. C., Lyu S., Li P., Li Y., Barthel F., Cho H. J., Lin Y.-H., Satani N., Martinez-Ledesma E., Zheng S., Chang E., Sauvé C.-E. G., Olar A., Lan Z. D., Finocchiaro G., Phillips J. J., Berger M. S., Gabrusiewicz K. R., Wang G., Eskilsson E., Hu J., Mikkelsen T., De Pinho R. A., Muller F., Heimberger A. B., Sulman E. P., Nam D.-H., Verhaak R. G. W., Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neftel C., Laffy J., Filbin M. G., Hara T., Shore M. E., Rahme G. J., Richman A. R., Silverbush D., Shaw M. L., Hebert C. M., Dewitt J., Gritsch S., Perez E. M., Castro L. N. G., Lan X., Druck N., Rodman C., Dionne D., Kaplan A., Bertalan M. S., Small J., Pelton K., Becker S., Bonal D., Nguyen Q.-D., Servis R. L., Fung J. M., Mylvaganam R., Mayr L., Gojo J., Haberler C., Geyeregger R., Czech T., Slavc I., Nahed B. V., Curry W. T., Carter B. S., Wakimoto H., Brastianos P. K., Batchelor T. T., Stemmer-Rachamimov A., Martinez-Lage M., Frosch M. P., Stamenkovic I., Riggi N., Rheinbay E., Monje M., Rozenblatt-Rosen O., Cahill D. P., Patel A. P., Hunter T., Verma I. M., Ligon K. L., Louis D. N., Regev A., Bernstein B. E., Tirosh I., Suvà M. L., An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849.e821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parada L. F., Dirks P. B., Wechsler-Reya R. J., Brain tumor stem cells remain in play. J. Clin. Oncol. 35, 2428–2431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couturier C. P., Ayyadhury S., le P. U., Nadaf J., Monlong J., Riva G., Allache R., Baig S., Yan X., Bourgey M., Lee C., Wang Y. C. D., Wee Yong V., Guiot M. C., Najafabadi H., Misic B., Antel J., Bourque G., Ragoussis J., Petrecca K., Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat. Commun. 11, 3406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng Q., Wang J., Wang J., He D., Cheng Z., Zhang F., Verma R., Xu L., Dong X., Liao Y., He X., Potter A., Zhang L., Zhao C., Xin M., Zhou Q., Aronow B. J., Blackshear P. J., Rich J. N., He Q., Zhou W., Suvà M. L., Waclaw R. R., Potter S. S., Yu G., Lu Q. R., Single-cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell 24, 707–723.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirosh I., Venteicher A. S., Hebert C., Escalante L. E., Patel A. P., Yizhak K., Fisher J. M., Rodman C., Mount C., Filbin M. G., Neftel C., Desai N., Nyman J., Izar B., Luo C. C., Francis J. M., Patel A. A., Onozato M. L., Riggi N., Livak K. J., Gennert D., Satija R., Nahed B. V., Curry W. T., Martuza R. L., Mylvaganam R., Iafrate A. J., Frosch M. P., Golub T. R., Rivera M. N., Getz G., Rozenblatt-Rosen O., Cahill D. P., Monje M., Bernstein B. E., Louis D. N., Regev A., Suvà M. L., Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhaduri A., Lullo E. D., Jung D., Müller S., Crouch E. E., Espinosa C. S., Ozawa T., Alvarado B., Spatazza J., Cadwell C. R., Wilkins G., Velmeshev D., Liu S. J., Malatesta M., Andrews M. G., Mostajo-Radji M. A., Huang E. J., Nowakowski T. J., Lim D. A., Diaz A., Raleigh D. R., Kriegstein A. R., Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 26, 48–63.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcantara Llaguno S., Sun D., Pedraza A. M., Vera E., Wang Z., Burns D. K., Parada L. F., Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nat. Neurosci. 22, 545–555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Sun D., Chen Y.-J., Xie X., Shi Y., Tabar V., Brennan C. W., Bale T. A., Jayewickreme C. D., Laks D. R., Llaguno S. A., Parada L. F., Cell lineage-based stratification for Glioblastoma. Cancer Cell 38, 366–379.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y., Varn F. S., Park S. H., Yoon B. W., Park H. R., Lee C., Verhaak R. G. W., Paek S. H., Perspective of mesenchymal transformation in glioblastoma. Acta Neuropathol. Commun. 9, 50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etchevers H. C., Vincent C., Le Douarin N. M., Couly G. F., The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development 128, 1059–1068 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Ando K., Fukuhara S., Izumi N., Nakajima H., Fukui H., Kelsh R. N., Mochizuki N., Clarification of mural cell coverage of vascular endothelial cells by live imaging of zebrafish. Development 143, 1328–1339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Manno G., Siletti K., Furlan A., Gyllborg D., Vinsland E., Albiach A. M., Langseth C. M., Khven I., Lederer A. R., Dratva L. M., Johnsson A., Nilsson M., Lönnerberg P., Linnarsson S., Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Vanlandewijck M., He L., Mäe M. A., Andrae J., Ando K., del Gaudio F., Nahar K., Lebouvier T., Laviña B., Gouveia L., Sun Y., Raschperger E., Räsänen M., Zarb Y., Mochizuki N., Keller A., Lendahl U., Betsholtz C., A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Mravic M., Asatrian G., Soo C., Lugassy C., Barnhill R. L., Dry S. M., Peault B., James A. W., From pericytes to perivascular tumours: Correlation between pathology, stem cell biology, and tissue engineering. Int. Orthop. 38, 1819–1824 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Ando K., Wang W., Peng D., Chiba A., Lagendijk A. K., Barske L., Crump J. G., Stainier D. Y. R., Lendahl U., Koltowska K., Hogan B. M., Fukuhara S., Mochizuki N., Betsholtz C., Peri-arterial specification of vascular mural cells from naïve mesenchyme requires Notch signaling. Development 146, dev165589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochgerner H., Zeisel A., Lönnerberg P., Linnarsson S., Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci. 21, 290–299 (2018). [DOI] [PubMed] [Google Scholar]
- 21.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L. E., Stott S. R. W., Toledo E. M., Villaescusa J. C., Lönnerberg P., Ryge J., Barker R. A., Arenas E., Linnarsson S., Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodge R. D., Bakken T. E., Miller J. A., Smith K. A., Barkan E. R., Graybuck L. T., Close J. L., Long B., Johansen N., Penn O., Yao Z., Eggermont J., Höllt T., Levi B. P., Shehata S. I., Aevermann B., Beller A., Bertagnolli D., Brouner K., Casper T., Cobbs C., Dalley R., Dee N., Ding S. L., Ellenbogen R. G., Fong O., Garren E., Goldy J., Gwinn R. P., Hirschstein D., Keene C. D., Keshk M., Ko A. L., Lathia K., Mahfouz A., Maltzer Z., McGraw M., Nguyen T. N., Nyhus J., Ojemann J. G., Oldre A., Parry S., Reynolds S., Rimorin C., Shapovalova N. V., Somasundaram S., Szafer A., Thomsen E. R., Tieu M., Quon G., Scheuermann R. H., Yuste R., Sunkin S. M., Lelieveldt B., Feng D., Ng L., Bernard A., Hawrylycz M., Phillips J. W., Tasic B., Zeng H., Jones A. R., Koch C., Lein E. S., Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan X., Fu Y., Zhou X., Sun L., Yang M., Wang M., Chen R., Wu Q., Yong J., Dong J., Wen L., Qiao J., Wang X., Tang F., Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci. Adv. 6, eaaz2978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darmanis S., Sloan S. A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., Li G., Chang S. D., Connolly I. D., Li Y., Barres B. A., Gephart M. H., Quake S. R., Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venteicher A. S., Tirosh I., Hebert C., Yizhak K., Neftel C., Filbin M. G., Hovestadt V., Escalante L. E., Shaw M. K. L., Rodman C., Gillespie S. M., Dionne D., Luo C. C., Ravichandran H., Mylvaganam R., Mount C., Onozato M. L., Nahed B. V., Wakimoto H., Curry W. T., Iafrate A. J., Rivera M. N., Frosch M. P., Golub T. R., Brastianos P. K., Getz G., Patel A. P., Monje M., Cahill D. P., Rozenblatt-Rosen O., Louis D. N., Bernstein B. E., Regev A., Suvà M. L., Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, eaai8478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan J., Levitin H. M., Frattini V., Bush E. C., Boyett D. M., Samanamud J., Ceccarelli M., Dovas A., Zanazzi G., Canoll P., Bruce J. N., Lasorella A., Iavarone A., Sims P. A., Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 10, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara T., Chanoch-Myers R., Mathewson N. D., Myskiw C., Atta L., Bussema L., Eichhorn S. W., Greenwald A. C., Kinker G. S., Rodman C., Castro L. N. G., Wakimoto H., Rozenblatt-Rosen O., Zhuang X., Fan J., Hunter T., Verma I. M., Wucherpfennig K. W., Regev A., Suvà M. L., Tirosh I., Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 39, 779–792.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natesh K., Bhosale D., Desai A., Chandrika G., Pujari R., Jagtap J., Chugh A., Ranade D., Shastry P., Oncostatin-M differentially regulates mesenchymal and proneural signature genes in gliomas via STAT3 signaling. Neoplasia 17, 225–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu T., Costello C. M., Croucher P. J. P., Häsler R., Deuschl G., Schreiber S., Can Zipf’s law be adapted to normalize microarrays? BMC Bioinformatics 6, 37 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.P. Vincent, H. Larochelle, Y. Bengio, P.-A. Manzagol, Extracting and composing robust features with denoising autoencoders, in Proceedings of the 25th International Conference on Machine Learning (ACM, 2008), pp. 1096–1103. [Google Scholar]
- 31.Doyle L. A., Vivero M., Fletcher C. D., Mertens F., Hornick J. L., Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod. Pathol. 27, 390–395 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Zhao Z., Zhang K.-N., Wang Q., Li G., Zeng F., Zhang Y., Wu F., Chai R., Wang Z., Zhang C., Zhang W., Bao Z., Jiang T., Chinese Glioma Genome Atlas (CGGA): A comprehensive resource with functional genomic data for Chinese Glioma patients. Genomics Proteomic Bioinformatics 19, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soldatov R., Kaucka M., Kastriti M. E., Petersen J., Chontorotzea T., Englmaier L., Akkuratova N., Yang Y., Häring M., Dyachuk V., Bock C., Farlik M., Piacentino M. L., Boismoreau F., Hilscher M. M., Yokota C., Qian X., Nilsson M., Bronner M. E., Croci L., Hsiao W. Y., Guertin D. A., Brunet J. F., Consalez G. G., Ernfors P., Fried K., Kharchenko P. V., Adameyko I., Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, (2019). [DOI] [PubMed] [Google Scholar]
- 34.Bifari F., Decimo I., Pino A., Llorens-Bobadilla E., Zhao S., Lange C., Panuccio G., Boeckx B., Thienpont B., Vinckier S., Wyns S., Bouché A., Lambrechts D., Giugliano M., Dewerchin M., Martin-Villalba A., Carmeliet P., Neurogenic radial glia-like cells in meninges migrate and differentiate into functionally integrated neurons in the neonatal cortex. Cell Stem Cell 20, 360–373.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A., Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Albergante L., Mirkes E., Bac J., Chen H., Martin A., Faure L., Barillot E., Pinello L., Gorban A., Zinovyev A., Robust and scalable learning of complex intrinsic dataset geometry via ElPiGraph. Entropy Switz 22, 296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalluri R., Weinberg R. A., The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grillet N., Dubreuil V., Dufour H. D., Brunet J.-F., Dynamic expression of RGS4 in the developing nervous system and regulation by the neural type-specific transcription factor Phox2b. J. Neurosci. 23, 10613–10621 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jakoby M., Schnittger A., Cell cycle and differentiation. Curr. Opin. Plant Biol. 7, 661–669 (2004). [DOI] [PubMed] [Google Scholar]
- 40.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A., Fan J., Borm L. E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P. V., RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eze U. C., Bhaduri A., Haeussler M., Nowakowski T. J., Kriegstein A. R., Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 24, 584–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suva M. L., Rheinbay E., Gillespie S. M., Patel A. P., Wakimoto H., Rabkin S. D., Riggi N., Chi A. S., Cahill D. P., Nahed B. V., Curry W. T., Martuza R. L., Rivera M. N., Rossetti N., Kasif S., Beik S., Kadri S., Tirosh I., Wortman I., Shalek A. K., Rozenblatt-Rosen O., Regev A., Louis D. N., Bernstein B. E., Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schock E. N., LaBonne C., Sorting Sox: Diverse roles for sox transcription factors during neural crest and craniofacial development. Front. Physiol. 11, 606889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L. E., Manno G. L., Codeluppi S., Furlan A., Lee K., Skene N., Harris K. D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U., Linnarsson S., Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellström M., Kalen M., Lindahl P., Abramsson A., Betsholtz C., Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Weber R. G., Boström J., Wolter M., Baudis M., Collins V. P., Reifenberger G., Lichter P., Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: Toward a genetic model of meningioma progression. Proc. Natl. Acad. Sci. U.S.A. 94, 14719–14724 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suva M. L., Tirosh I., Single-cell RNA sequencing in cancer: Lessons learned and emerging challenges. Mol. Cell 75, 7–12 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Farace C., Oliver J. A., Melguizo C., Alvarez P., Bandiera P., Rama A. R., Malaguarnera G., Ortiz R., Madeddu R., Prados J., Microenvironmental modulation of decorin and lumican in temozolomide-resistant glioblastoma and neuroblastoma cancer stem-like cells. PLOS ONE 10, e0134111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balbous A., Cortes U., Guilloteau K., Villalva C., Flamant S., Gaillard A., Milin S., Wager M., Sorel N., Guilhot J., Bennaceur-Griscelli A., Turhan A., Chomel J. C., Karayan-Tapon L., A mesenchymal glioma stem cell profile is related to clinical outcome. Oncogenesis 3, e91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suva M. L., Tirosh I., The glioma stem cell model in the era of single-cell genomics. Cancer Cell 37, 630–636 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Pevny L. H., Nicolis S. K., Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 42, 421–424 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Karow M., Camp J. G., Falk S., Gerber T., Pataskar A., Gac-Santel M., Kageyama J., Brazovskaja A., Garding A., Fan W., Riedemann T., Casamassa A., Smiyakin A., Schichor C., Götz M., Tiwari V. K., Treutlein B., Berninger B., Direct pericyte-to-neuron reprogramming via unfolding of a neural stem cell-like program. Nat. Neurosci. 21, 932–940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Y., Bergström T., Jiang Y., Johansson P., Marinescu V. D., Lindberg N., Segerman A., Wicher G., Niklasson M., Baskaran S., Sreedharan S., Everlien I., Kastemar M., Hermansson A., Elfineh L., Libard S., Holland E. C., Hesselager G., Alafuzoff I., Westermark B., Nelander S., Forsberg-Nilsson K., Uhrbom L., The human glioblastoma cell culture resource: Validated cell models representing all molecular subtypes. EBioMedicine 2, 1351–1363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y., Marinescu V. D., Xie Y., Jarvius M., Maturi N. P., Haglund C., Olofsson S., Lindberg N., Olofsson T., Leijonmarck C., Hesselager G., Alafuzoff I., Fryknäs M., Larsson R., Nelander S., Uhrbom L., Glioblastoma cell malignancy and drug sensitivity are affected by the cell of origin. Cell Rep. 18, 977–990 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Zeisel A., Muñoz-Manchado A. B., Codeluppi S., Lönnerberg P., la Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., Rolny C., Castelo-Branco G., Hjerling-Leffler J., Linnarsson S., Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Muller S., Cho A., Liu S. J., Lim D. A., Diaz A., CONICS integrates scRNA-seq with DNA sequencing to map gene expression to tumor sub-clones. Bioinformatics 34, 3217–3219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W., Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 59.He B., Chen P., Zambrano S., Dabaghie D., Hu Y., Möller-Hackbarth K., Unnersjö-Jess D., Korkut G. G., Charrin E., Jeansson M., Bintanel-Morcillo M., Witasp A., Wennberg L., Wernerson A., Schermer B., Benzing T., Ernfors P., Betsholtz C., Lal M., Sandberg R., Patrakka J., Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes. Nat. Commun. 12, 2141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan Y., Bar-Joseph Z., Deep learning for inferring gene relationships from single-cell expression data. Proc. Natl. Acad. Sci. U.S.A. 116, 27151–27158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf F. A., Hamey F. K., Plass M., Solana J., Dahlin J. S., Göttgens B., Rajewsky N., Simon L., Theis F. J., PAGA: Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Data files S1 to S9