Abstract
The SARS-CoV-2 instigated “cytokine storm” elicited upon infection is known to majorly cause lung injury and even mortality in severe cases. Early clinical prognosis to alleviate the exaggerated release of inflammatory cytokines is thus looked upon. Considering the recent attention and advantages of saliva as a clinical specimen, i.e. ease and painlessness of collection, which does not require trained staff and could allow self-sampling, the present study attempts to explore saliva for detection of IL-6, TNF-α and IL-10 which constitute major inflammatory genes that are elevated in COVID-19 using RT-PCR. Blood specimens of the same patients were also parallelly assessed to compare and validate the inflammatory marker expression. A total of 64 COVID-19 subjects who met the inclusion criteria were enrolled in this pilot study. Paired samples of blood and saliva from each patient were collected as per standard sampling protocols. RNA from all specimens were extracted using Qiagen RNA Blood Mini Kit and subjected to RT-PCR. IL-6, TNF-α and IL-10 expression were assessed in Ct (cycle threshold) values. It was observed that all 64 (100%) patients expressed IL-6 gene and TNF-α gene, whereas only 7 (5.19%) patients expressed IL-10 in both blood and saliva samples. The mean Ct values of IL-6 gene expressed in blood and saliva were 26.68 ± 2.26 and 28.53 ± 3.11 respectively. Similarly, the mean Ct values of TNF-α gene expressed in blood and saliva were 27.98 ± 2.45 and 28.92 ± 3.70 respectively. The observed mean Ct values of IL-10 gene expressed in blood and saliva were 31.26 ± 3.96 and 30.11 ± 4.12 respectively. Accordingly, the results indicate that inflammatory genes IL-6, TNF-α and IL-10 were detectable in both patient saliva as well as in blood. Moreover, mean Ct values of IL-6, TNF-α and IL-10 in both samples were found to be comparable. This finding thus suggests the possible use of saliva as an alternative specimen to blood for monitoring inflammation in COVID-19 patients.
KEY WORDS: human saliva, COVID-19, inflammation, IL-6, TNF-Α, IL-10.
INTRODUCTION
A growing body of clinical data acknowledge that an exaggerated immune response known as “cytokine storm” is associated with COVID-19 severity and is also considered to be an undeniable cause of death [1, 2]. In the absence of an effective cure for COVID-19, there is a demanding need to understand the cytokine storm that governs disease severity [1]. Cytokines have long been thought to play a crucial role in viral infection immunopathology. The initial line of defence against viral infection is a quick and well-coordinated innate immune response. However, excessive and dysregulated immune responses lead to damage to the body [3]. In vitro cell experiments demonstrated release of cytokines and chemokines in respiratory epithelial cells, macrophages and dendritic cells at the early stage of SARS-CoV-2 infection followed by low levels of antiviral factors and interferons (IFNs) and high levels of proinflammatory cytokines (interleukin (IL)-1β, IL-6 and tumour necrosis factor (TNF)) and chemokines (C–C motif chemokine ligand (CCL-2, CCL-3 and CCL-5)) secreted by the cells [3]. Several investigations also report the utility of inflammatory markers in COVID-19 severity and progression. Severe cases of COVID-19 exhibited increased plasma levels of IL-2, IL-6, IL-7, IL-10, GSCF, IP-10, MCP-1, MIP-1A and TNF-α compared to mild cases, indicating that measurement of inflammatory cytokine release is critical in understanding COVID-19 progression and severity [4]. Among these cytokines, IL-6 and IL-10 are noteworthy predictors of COVID-19 severity and are reported to be elevated in severe infections compared to mild or moderate cases [5, 6].
An early research on COVID-19 also reported that proinflammatory cytokines such as IL-1, IL-6 and TNF-α and even IL-10 (which is otherwise thought be an anti-inflammatory molecule in major diseases) [6] were released by activated mast cells in respiratory tract submucosa which led to worsening of inflammatory state and pathogenesis [7–9].
The reported detection of these inflammatory markers in the mentioned studies was mainly from human specimens such as serum [10], plasma [11–13] and BALF [4] of COVID-19 patients.
Though human saliva is reported to be used for detection of numerous inflammatory markers such as IL-1, IL-6, TNF, arachidonate 5-lipoxygenase (5-LOX), chemokines, vascular endothelial growth factor (VEGF), prostaglandin-endoperoxide synthase 2 (COX-2), matrix metalloproteinases (MMPs), twist-related protein (TWIST) and cell surface adhesion molecules in patients affected with chronic diseases [14, 15], we could not find any study reporting the detection of salivary inflammatory markers in COVID-19 patients.
Considering the recent attention towards saliva as a diagnostic specimen with regard to its ease and painlessness of collection, which does not require trained staff, this study aimed to explore the detection of three major inflammatory markers, i.e. IL-6, TNF-α and IL-10 in the saliva of COVID-19 patients for monitoring inflammation, using RT-PCR. Additionally, gene expression of inflammatory cytokines, IL-6, TNF-α and IL-10, in patient saliva was also compared to whole blood, individually, to further validate the finding.
The demonstrated approach of using saliva as a specimen to detect and monitor inflammation could serve early detection and systematic monitoring of inflammatory markers which in turn provides scope for an easy and time effective prognosis which could possibly aid early medical intervention for effective patient management and care.
METHODOLOGY
Study Population
Subjects over 18 to 75 years of age of either sex, who were diagnosed with COVID-19 via RT-PCR, were recruited between May 2021 and July 2021. Exclusion criteria included patients of age less than 18 years and more than 75 years; patients with a COVID-19-positive test done more than 48 h prior to enrolment in study; pregnancy and lactation; severe or complicated course of COVID-19 disease; presence of acute hypoxic respiratory failure/need for intensive care unit (ICU) stay/patients who need mechanical ventilation; any uncontrolled systemic disease/infection; and those with serious cardiovascular, cerebrovascular, respiratory, liver or renal disease or any other disorder/conditions, which in the opinion of the investigators made the patient unsuitable for enrolment or could interfere in adherence of the study protocol such as difficulty in providing blood or saliva samples.
The protocol was approved by the Orchid Specialty Hospital Ethics committee, and the trial was registered in Clinical Trials Registry India (CTRI—CTRI/2021/04/033143). Informed consent was obtained from all participants.
Sample Collection
From 135 subjects, both saliva and blood samples were collected using Umbrella Life Science Saliva Collection Device (ULSD019) and BD Vacutainer® EDTA Tubes in the same sitting. A standard venipuncture was performed to collect blood from patients. Saliva samples were collected in Umbrella Life Science saliva collection tubes by following the manufacturer’s instructions. Patients were provided with a pair of gloves, a paper towel and saliva collection tube which had a funnel connected to a tube filled with buffer. The patient was instructed to expel 2 mL of saliva through the funnel, which collected in the attached buffer tube. The funnel was removed and discarded by them and the tube was tightly closed. Patients were asked to shake the tube for 5 s to mix the buffer and sample well. A nurse received the tubes wearing gloves and deposited it into a cooler containing ice packs for transport to the laboratory. Quality control of saliva specimen and other preparations before saliva collection was ensured by following the recommended guidelines [14]. All samples were processed within the same day.
RT-PCR Testing
Both saliva and blood samples underwent RNA extraction (200 μL of the sample) using Qiagen RNeasy Protect Saliva mini kit and Qiagen RNA Blood Mini kit as per the manufacturer’s instruction. Extracted RNA extracted was then subjected to RT-PCR analysis on a Qiagen Rotor-Gene Q PCR machine (Qiagen Inc., Valencia, CA, USA) using commercially available One-step multiplex RT-PCR kit (PROGNOSEEZ ™, India) for detection of IL-6, TNF-α and IL-10, as per the manufacturer’s instruction. Briefly, the reaction set up for each sample included 12.5µL of Master mix, 2.0 µL of RT mix, 4.0 µL of PP mix and 6.5 µL RNA template (extracted mRNA). β-actin served as the reference gene. Each run was performed in triplicates with positive and non-template controls to ensure test quality. For each gene, assessment of quality and specificity was performed by examining PCR melt curves. All results inferred according to the manufacturer’s specifications and are represented in Ct values.
Statistical Analysis
The statistical analysis was performed by IBM SPSS 26.0 software. Categorical variables are expressed using frequency and percentage. Continuous variables are presented by mean and standard deviation.
RESULTS
Gene expression of IL-6, TNF-α and IL-10 from blood and saliva samples were assessed in cycle threshold (Ct) values (Fig. 1). In total, 64 (100%) patients expressed IL-6 and TNF-α gene, whereas only 7 (5.19%) patients expressed IL-10 in both blood and saliva specimens. The mean Ct value of IL-6 gene expressed in blood and saliva was 26.68 ± 2.26 and 28.53 ± 3.11 respectively. Similarly, the mean Ct value of TNF-α gene expressed in blood and saliva was 27.98 ± 2.45 and 28.92 ± 3.70 respectively. The observed mean Ct value of IL-10 gene expressed in blood and saliva was 31.26 ± 3.96 and 30.11 ± 4.12 respectively (Table 1). Additionally, the Ct values of SARS-CoV-2 detected in these patients at day 0 of COVID-19 diagnosis is also given in Table 2 for better understanding of the levels of the inflammatory markers detected in these patients at the time. Thus, the results indicate that the mean Ct values of inflammatory genes IL-6, TNF-α and IL-10 detected in patient saliva and blood specimens are comparable. This finding supports the use of saliva as an alternative specimen to blood.
Fig. 1.
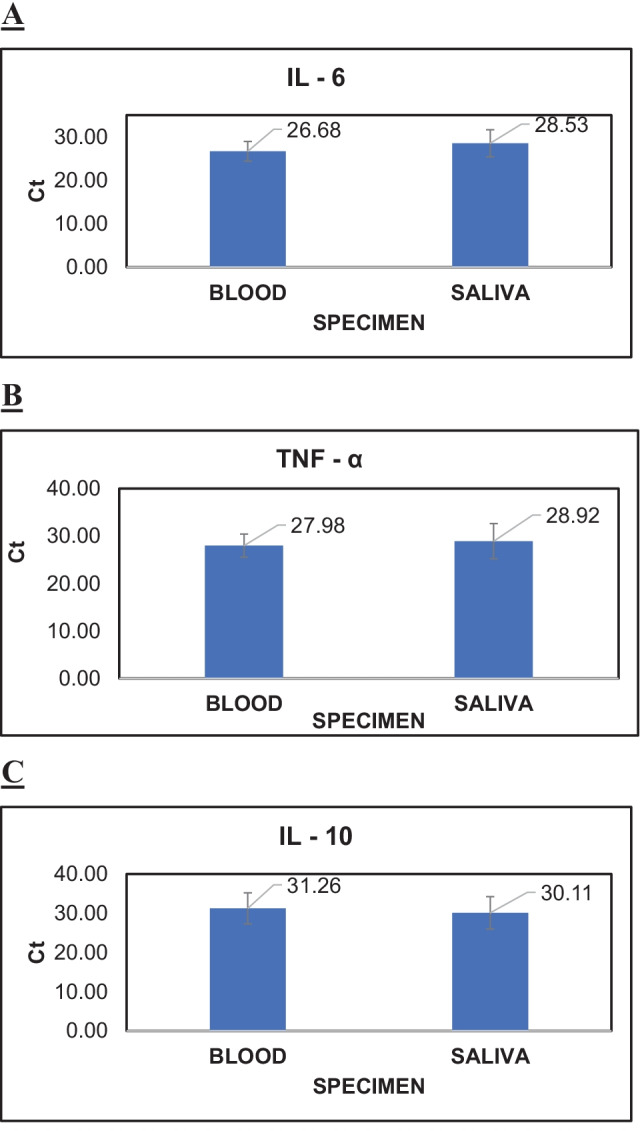
A–C Graphical representation indicating the expression of IL-6, TNF-α and IL-10 gene in both blood and saliva samples of the same patient.
Table 1.
Mean Ct Value of IL-6, TNF-α and IL-10 Gene in Blood and Saliva
| Variable | Samples | Mean ± SD |
|---|---|---|
| IL-6 | Blood | 26.68 ± 2.26 |
| Saliva | 28.53 ± 3.11 | |
| TNF-α | Blood | 27.98 ± 2.45 |
| Saliva | 28.92 ± 3.70 | |
| IL-10 | Blood | 31.26 ± 3.96 |
| Saliva | 30.11 ± 4.12 |
Table 2.
Ct Values of SARS-CoV-2 Detected in the Study Population at Day 0 of COVID-19 Diagnosis
| Patient ID No | SARS-CoV-2 RT-PCR Ct values on day 0 |
Patient ID No | SARS-CoV-2 RT-PCR Ct values on day 0 |
|---|---|---|---|
| 1 | 22.4 | 33 | 23 |
| 2 | 16.3 | 34 | 27 |
| 3 | 18 | 35 | 23 |
| 4 | 22 | 36 | 25 |
| 5 | 18 | 37 | 28 |
| 6 | 20 | 38 | 25 |
| 7 | 24 | 39 | 26 |
| 8 | 19 | 40 | 20 |
| 9 | 19 | 41 | 22 |
| 10 | 19 | 42 | 13 |
| 11 | 23 | 43 | 29 |
| 12 | 13 | 44 | 26 |
| 13 | 15 | 45 | 20 |
| 14 | 16 | 46 | 19 |
| 15 | 22 | 47 | 23 |
| 16 | 16 | 48 | 24 |
| 17 | 21 | 49 | 26 |
| 18 | 18 | 50 | 25 |
| 19 | 19 | 51 | 19 |
| 20 | 27 | 52 | 23 |
| 21 | 21 | 53 | 28 |
| 22 | 28 | 54 | 15 |
| 23 | 19 | 55 | 18 |
| 24 | 21 | 56 | 29 |
| 25 | 22 | 57 | 23 |
| 26 | 27 | 58 | 30 |
| 27 | 17 | 59 | 30 |
| 28 | 26 | 60 | 18 |
| 29 | 27 | 61 | 30 |
| 30 | 16 | 62 | 27 |
| 31 | 26 | 63 | 16 |
| 32 | 17 | 64 | 30 |
DISCUSSION
COVID-19 has been an undeniable cause of death recently urging all health facilities to take immediate and effective medical action to curb disease spread. The alarming rate of mortality among infected persons is majorly due to lethal pneumonia and acute respiratory distress syndrome (ARDS) [16]. Even though the mechanisms of COVID-19-induced lung damage are still being discovered, an exaggerated response of innate immunity system known as “cytokine storm” has become one profound term used both in scientific literature as well as media. Broadly speaking, the so-called storm denotes a hyperactive immune response that involves the release of interleukins, interferons, tumour-necrosis factors, chemokines and several other mediators. This storm poses to be a threat to the host [16]. Various studies which analysed cytokine profiles from COVID-19 patients suggested that the exaggerated cytokine storm indicated lung injury, multi-organ failure and unpropitious prognosis of severe COVID-19. Huang et al. and several other investigators reported that compared to non-ICU patients, ICU patients had higher plasma levels of IL-2, IL-7, IL-10, GSCF, IP10, MCP1, MIP1A and TNF-α [12, 17, 18]. All cytokine profile studies performed to date were based on protein level analysis from serum/plasma of subjects, none of the studies emphasized on gene level analysis and the use of easy to procure and non-invasive salivary diagnostics in this context. Saliva has been regarded as a potential specimen in both diagnosis and prognosis of numerous diseases by investigators around the world [14]. Also, saliva sample collection is relatively much easier, non-invasive, requires less stress and promotes low-cost storage than any other body fluid. Oral fluid sampling promotes low-cost storage [19]. Moreover, salivary analysis makes frequent monitoring easy, especially in infants, children and elderly patients, as well as in conditions where urine or blood sampling is not practical. However, despite its numerous advantages, saliva is not yet considered to be an established analytical specimen due to the insufficient information about its biochemical composition and correlation with plasma levels [14, 20]. In our study, we have tried to fill this gap area by analysing the inflammatory genes, IL-6, TNF-α and IL-10 from blood and saliva samples of 64 subjects who were tested positive for SARS-CoV-2 not more than 2 days before enrolling to study,out of the total study population, all 64 (100% patients expressed IL-6 gene and TNF-α, whereas 7 (5.19% patients expressed IL-10 in both blood and saliva samples. Therefore, the acquired Ct values of blood and saliva samples seem to be comparable when the expression of IL-6, TNF-α and IL-10 genes is analysed, supporting our recommendation to use saliva over blood to monitor and detect inflammatory genes in COVID-19 patients.
CONCLUSION
Saliva is acknowledged as a mirror that reflects one’s physical state and is a potential specimen in diagnosis and prognostication of various diseases. The present trial was performed to study the potentiality of saliva as a specimen to detect inflammatory genes IL-6, TNF-α and IL-10 in COVID-19 patients using RT-PCR technique. The results obtained are encouraging and support the use of saliva over blood in this context. However, this study is limited by the lack of COVID-19-negative tested population which paves way for future investigations that may focus on identifying normal range levels of inflammatory genes IL-6, TNF-α and IL-10 in healthy patient’s saliva compared to those infected. This investigation also encourages more scientists/researchers to conduct studies pertaining to salivary diagnostics in various disease conditions by emphasizing on the detection of inflammatory genes using RT-PCR.
ACKNOWLEDGEMENTS
The authors thank the patients for their willingness to be part of this trial and also the treating clinicians; Dr. Ajit Mandlecha, Dr. Gous Mujawar and Dr. Aslam for their valuable contributions. The authors thank Zum Heilen Diagnostic & Therapeutics Pvt. Ltd. for the financial support in conducting this trial. The authors also thank the staff of Vishwanand Kendra and Shree Sai Hospital, Pune, Maharashtra, India, for their assistance in conducting the study
AUTHOR CONTRIBUTION
Nourin Shakeeb contributed in drafting the manuscript and interpretation of the data. Prashanth Varkey was the principal investigator of the trial with overall responsibility for conducting the trial and for medical oversight of trial implementation. Aimy Hynse organized and performed the data analysis. Amita Ajit structured the final report, conceptualized the study, contributed to the trial design, coordination of participants’ data and reviewed the interpreted data. All authors reviewed the final report.
FUNDING
This research was funded and supported by Zum Heilen Diagnostic & Therapeutics Pvt. Ltd.
AVAILABILITY OF DATA AND MATERIALS
Data supporting reported results that is not given here is available on request from the corresponding author. This data is not publicly available due to privacy requirements.
DECLARATIONS
Ethics Approval and Consent to Participate (Human Ethics, Animal Ethics or Plant Ethics)
Prior to the study, the protocol was approved by the Orchid Specialty Hospital Ethics committee, and the trial was registered in Clinical Trials Registry India (CTRI—CTRI/2021/04/033143) in compliance with the International Council on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use – Good Clinical Practice (ICH–GCP) guidelines. All participating patients were informed about the objectives and risks of participation and gave written informed consent.
Consent for Publication
All the authors mentioned in the manuscript have agreed for authorship, read and approved the manuscript, and have given consent for submission and subsequent publication of the manuscript.
Competing Interests
Dr. Prashanth Varkey is the Director of Zum Heilen Diagnostic & Therapeutics Pvt. Ltd.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nourin Shakeeb and Amita Ajit share first authorship.
Contributor Information
Nourin Shakeeb, Email: nourinshakeeb16@gmail.com.
Prashanth Varkey, Email: drpvarkey@gmail.com.
Aimy Hynse, Email: aimyhynse26@gmail.com.
Amita Ajit, Email: dr.amitaajit@gmail.com.
References
- 1.Hu B, Huang S, Yin L. The cytokine storm and COVID-19. Journal of Medical Virology. 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS, Rajagopal S, Pai AR, Kutty S. Cytokine storm in COVID-19-immunopathological mechanisms, clinical considerations, and therapeutic approaches: The REPROGRAM consortium position paper. Frontiers in Immunology. 2020;11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘cytokine storm’ in COVID-19. The Journal of Infection. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, Guo D, Hu W, Yang J, Tang Z, Wu H, Lin Y, Zhang M, Zhang Q, Shi M, Liu Y, Zhou Y, Lan K, Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerging Microbes & Infections. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang F, Liu X, Sun X, Li Z. IL-10 served as an indicator in severe COVID-19 patients. Journal of Medical Virology. 2021;93(3):1233–1235. doi: 10.1002/jmv.26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhar SK, K, V., Damodar, S., Gujar, S., & Das, M. IL-6 and IL-10 as predictors of disease severity in COVID-19 patients: Results from meta-analysis and regression. Heliyon. 2021;7(2):e06155. doi: 10.1016/j.heliyon.2021.e06155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti, P., G. Ronconi, A. Caraffa, C. Gallenga, R. Ross, I. Frydas, and S. Kritas. 2020. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. Journal of Biological Regulators and Homeostatic Agents 34(2): 327–331. 10.23812/CONTI-E. [DOI] [PubMed]
- 8.Kritas, S.K., G. Ronconi, A. Caraffa, C.E. Gallenga, R. Ross, and P. Conti. 2020. Mast cells contribute to coronavirus-induced inflammation: new anti-inflammatory strategy. Journal of Biological Regulators and Homeostatic Agents 34(1): 9–14. 10.23812/20-Editorial-Kritas. [DOI] [PubMed]
- 9.Ronconi, G., G. Teté, S.K. Kritas, C.E. Gallenga, A. Caraffa, R. Ross, and P. Conti. 2020. SARS-CoV-2, which induces COVID-19, causes kawasaki-like disease in children: role of pro-inflammatory and anti-inflammatory cytokines. Journal of Biological Regulators and Homeostatic Agents 34(3): 767–773. 10.23812/EDITORIAL-RONCONI-E-59. [DOI] [PubMed]
- 10.Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, Zhang P, Liu X, Gao G, Liu F, Jiang Y, Cheng X, Zhu C, Xia Y. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerging Microbes & Infections. 2020;9(1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McElvaney OJ, McEvoy NL, McElvaney OF, Carroll TP, Murphy MP, Dunlea DM, Ní Choileáin O, Clarke J, O’Connor E, Hogan G, Ryan D, Sulaiman I, Gunaratnam C, Branagan P, O’Brien ME, Morgan RK, Costello RW, Hurley K, Walsh S, McElvaney NG. Characterization of the inflammatory response to severe COVID-19 illness. American Journal of Respiratory and Critical Care Medicine. 2020;202(6):812–821. doi: 10.1164/rccm.202005-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shakeeb N, Varkey P, Ajit A. Human saliva as a diagnostic specimen for early detection of inflammatory biomarkers by real-time RT-PCR. Inflammation. 2021;44(5):1713–1723. doi: 10.1007/s10753-021-01484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad S, Tyagi AK, Aggarwal BB. Detection of inflammatory biomarkers in saliva and urine: Potential in diagnosis, prevention, and treatment for chronic diseases. Experimental Biology and Medicine. 2016;241(8):783–799. doi: 10.1177/1535370216638770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Internal Medicine. 2020;180(9):1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 17.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan China. Intensive Care Medicine. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun, D., H. Li, X.-X. Lu, H. Xiao, J. Ren, F.-R. Zhang, and Z.-S. Liu. 2020. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World Journal of Pediatrics 1–9. 10.1007/s12519-020-00354-4. [DOI] [PMC free article] [PubMed]
- 19.Roi A, Rusu LC, Roi CI, Luca RE, Boia S, Munteanu RI. A new approach for the diagnosis of systemic and oral diseases based on salivary biomolecules. Disease Markers. 2019;2019:e8761860. doi: 10.1155/2019/8761860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corey-Bloom J, Fischer RS, Kim A, Snell C, Parkin GM, Granger DA, Granger SW, Thomas EA. Levels of interleukin-6 in saliva, but not plasma, correlate with clinical metrics in Huntington’s disease patients and healthy control subjects. International Journal of Molecular Sciences. 2020;21(17):6363. doi: 10.3390/ijms21176363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting reported results that is not given here is available on request from the corresponding author. This data is not publicly available due to privacy requirements.


