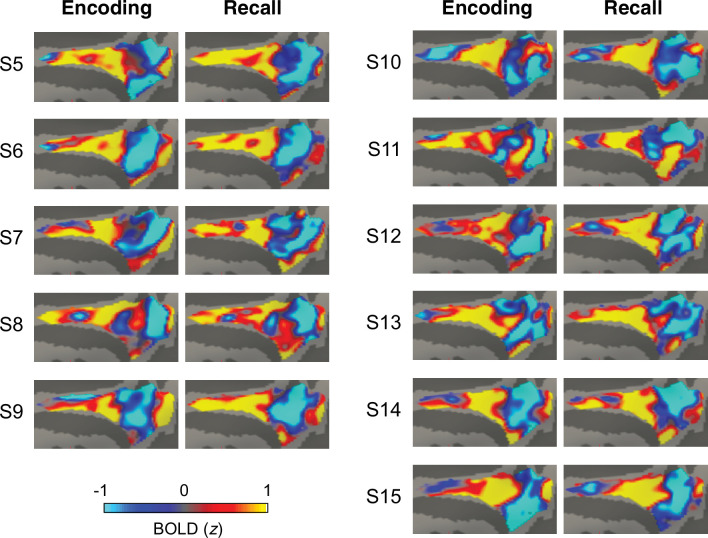
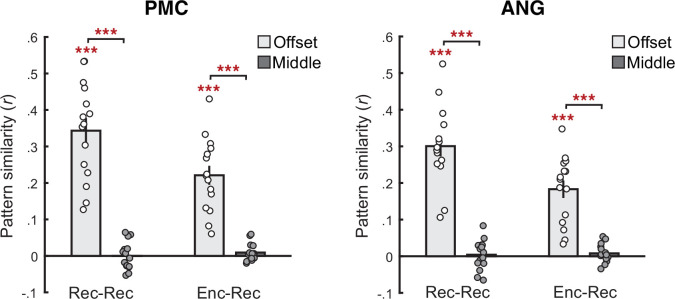
Figure 3. Boundary pattern in the posterior medial cortex (PMC).
(A) PMC activation pattern similarity (Pearson correlation) between the 10 movie stimuli (M1–10), conditions (offset = boundary, middle = non-boundary), and experimental phases (encoding, recall), averaged across all subjects. The boundary pattern of a movie was defined as the mean pattern averaged across the 15 s window following the offset of the movie. The non-boundary pattern was defined as the mean pattern averaged across the 15 s window in the middle of a movie. The time windows for both boundary and non-boundary patterns were shifted forward by 4.5 s to account for the hemodynamic response delay. PMC regions of interest (ROIs) are shown as white areas on the inflated surface of a template brain. (B) Subject-specific mean activation patterns associated with between-movie boundaries during encoding (left) and recall (right). The boundary patterns were averaged across all movies and then z-scored across vertices within the PMC ROI mask, separately for each experimental phase. PMC (demarcated by black outlines) of four example subjects (S1–4) are shown on the medial surface of the right hemisphere of the fsaverage6 template brain. (C) Within-phase (recall-recall) and between-phase (encoding-recall) pattern similarity across different movies, computed separately for the boundary (offset) and non-boundary (middle) patterns in PMC. Bar graphs show the mean across subjects. Circles represent individual subjects. Error bars show SEM across subjects. ***p<.001. (D) Time-point-by-time-point PMC pattern similarity across the encoding phase and recall phase activation patterns around between-movie boundaries, averaged across all subjects. The time series of activation patterns were locked to either the onset (left) or the offset (right) of each movie. During encoding, the onset of a movie and the offset of the preceding movie were separated by a 6 s title scene. During recall, onsets and offsets of recalled movies were separated by, on average, a 9.3 s pause (boundaries concatenated across subjects, SD = 16.8 s). Dotted lines on the left and right panels indicate the mean offset times of the preceding movies and the mean onset times of the following movies, respectively. Note that in this figure zero corresponds to the true stimulus/behavior time, with no shifting for hemodynamic response delay. Areas outlined by black lines indicate correlations significantly different from zero after multiple comparisons correction (Bonferroni corrected p<0.05). Time–time correlations within each experimental phase can be found in Figure 3—figure supplement 4.