Abstract
The nor-1 gene is involved in aflatoxin biosynthesis in Aspergillus parasiticus and was predicted to encode a norsolorinic acid ketoreductase. Recombinant Nor-1 expressed in Escherichia coli converted the 1′ keto group of norsolorinic acid to the 1′ hydroxyl group of averantin in crude E. coli cell extracts in the presence of NADPH. The results confirm that Nor-1 functions as a ketoreductase in vitro.
Background.
The anthraquinones norsolorinic acid (NA) and averantin (AVN) accumulate in some mutant strains of Aspergillus parasiticus and are intermediates in the aflatoxin (AF) pathway (4, 8, 9, 10). Disruption of the nor-1 gene in A. parasiticus resulted in NA accumulation (5), confirming the function of Nor-1 in AF synthesis and suggesting that NA is a substrate for this protein. The nor-1 mutant also accumulates small amounts of AF, suggesting that another enzyme can bypass the step catalyzed by Nor-1. Based on nucleotide sequence data, nor-1 was proposed to encode a short-chain alcohol dehydrogenase that could convert the 1′ keto group of NA to the 1′ hydroxyl group of AVN (12).
Our objective in this study was to identify the enzymatic function of the Nor-1 protein. We observed that recombinant Nor-1 expressed in Escherichia coli converted NA to AVN in the presence of NADPH in vitro. These data support the hypothesis that the reaction catalyzed by Nor-1 is an important pathway for the conversion of NA to AVN in A. parasiticus.
Construction of pMN1, a recombinant Nor-1 expression vector.
The plasmid pNOR contained a nor-1 cDNA cloned into the EcoRI/XhoI sites of pBluescript SK(−) (Stratagene Cloning Systems, La Jolla, Calif.). Nucleotide sequence analysis of the nor-1 cDNA was performed with a commercial product (Sequenase II; U.S. Biochemical Corp., Cleveland, Ohio). The cDNA sequence was identical to the genomic DNA sequence reported previously under accession no. L278801 except for 6 nucleotides at the 5′ end of the predicted coding region. These were either not present in the mRNA or deleted during cDNA production or cloning.
The nor-1 cDNA was removed from pNOR with BamHI and KpnI and cloned into the BamHI/KpnI sites of pQE31 (QIAexpress; Qiagen, Inc., Chatsworth, Calif.). The nor-1 cDNA was excised from pQE31 with EcoRI and SalI and cloned into the EcoRI/SalI sites of the expression vector pMAL-c2 (New England Biolabs, Inc., Beverly, Mass.). The resulting vector was cut with EcoRI, the protruding ends were filled with deoxynucleoside triphosphates with Klenow fragment, and the plasmid was recircularized to generate the expression vector pMN1. In pMN1, the nor-1 cDNA was fused in frame with the 3′ end of malE, which encodes a maltose-binding protein (MBP). Nucleotide sequencing confirmed this construction. Plasmid constructs were amplified in E. coli DH5α F′ e[F′ endA1 hsdR17(rK− mK+) upE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 (m80 lacZΔM15)] by standard methods (1).
Preparation of recombinant Nor-1 proteins MBP–Nor-1c and Nor-1c.
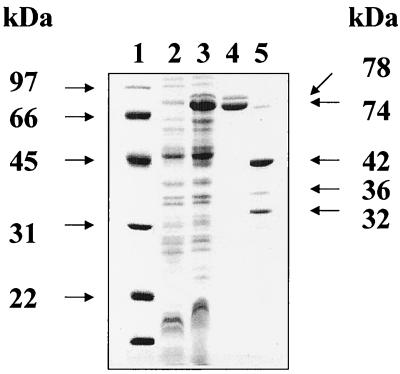
pMN1 was transformed into E. coli DH5α and induced to express fusion proteins by standard methods (1). Recombinant fusion proteins (known as MBP–Nor-1c proteins) produced by cultures of selected transformants were purified by standard procedures described in the Protein Fusion and Purification system (New England Biolabs, Inc.). Two MBP–Nor-1c proteins with apparent 74- (major) and 78-kDa (minor) masses were detected in E. coli cell extracts by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Fig. 1). These proteins were not observed in extracts from noninduced cells, in control cells containing pMAL-c2, or in cells without vectors, suggesting that they are recombinant Nor-1 proteins expressed from nor-1 cDNA in pMN1. According to cDNA and genomic sequence data, MBP–Nor-1c proteins lacked the first two amino acid residues (M and T) of the Nor-1 coding sequence (12) which were replaced by eight amino acids (I, S, E, L, I, R, H, and E) from pMAL-c2 during vector construction.
FIG. 1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis detection of MBP–Nor-1c and Nor-1c. Expression vector pMN1, containing the nor-1 cDNA fused with malE, which encodes a maltose-binding protein (MBP), was transformed into E. coli DH5α. Under isopropyl-β-d-thiogalactopyranoside (IPTG) induction, major and minor MBP–Nor-1c fusion proteins were produced. Lanes: 1, molecular mass standard (kDa); 2, total crude extract (10,000 × g supernatant) from E. coli DH5α; 3, total crude extract from E. coli DH5α transformed with expression vector pMN1; 4, MBP–Nor-1c 74-kDa (major) and 78-kDa (minor) fusion proteins, purified by affinity chromatography; 5, 32-kDa (major) and 36-kDa (minor) Nor-1c proteins and MBP (42 kDa) obtained by factor Xa cutting of the two Nor-1c–MBP fusion proteins.
The 74- and 78-kDa MBP–Nor-1c proteins were purified by affinity chromatography with amylose resin. These proteins could not be purified from E. coli control cultures (i.e., in cells with no vector, cells with pMAL-c2, or noninduced cells with pMN1), providing additional evidence that these MBP–Nor-1c fusion proteins are expressed from the cloned nor-1 cDNA in pMN1. The resulting proteins in a column buffer (10 mM phosphate, 0.5 M NaCl, 1 mM azide, 10 mM 2-mercaptoethanol, and 1 mM EGTA [pH 7.2]) were concentrated to 1 mg/ml (Centriprep concentrator; Amicon, Inc., Beverly, Mass.), dialyzed against Factor Xa buffer (20 mM Tris-HCl, 100 mM NaCl, 2 mM CaCl2, 1 mM azide [pH 8.0]) and cut with Factor Xa. This protease, supplied with the pMAL-c2 expression kit, removes MBP to generate MBP (42 kDa) and two recombinant Nor-1 proteins (Nor-1c) with apparent molecular masses of 32 (major) and 36 (minor) kDa (Fig. 2). Since Factor Xa cuts upstream from the amino acid sequence ISELIRHE, these eight residues were predicted to be present in purified Nor-1c.
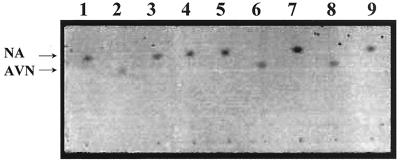
FIG. 2.
Enzyme activity assay of Nor-1c–MBP and Nor-1c proteins. All reactions were conducted at 37°C in the dark. The reaction products were resolved by TLC with benzene-ethyl acetate (7:3) as the developing system. The photograph was taken under white light. The reaction components for each lane are as follows. Lane 1, NA (0.9 mM); lane 2, AVN (0.9 mM); lane 3, NA (0.9 mM) plus NADPH (2.3 mM); lane 4, NA (0.9 mM) plus NADPH (2.3 mM) plus crude cell extract (35 μg) from E. coli DH5α; lane 5, NA (0.9 mM) plus NADPH (2.3 mM) plus crude cell extract (70 μg) from E. coli DH5α containing pMAL-c2; lane 6, NA (0.9 mM) plus NADPH (2.3 mM) plus crude cell extract (70 μg) from E. coli DH5α containing pMN1; lane 7, NA (0.9 mM) plus crude cell extract from (70 μg) E. coli DH5α containing pMN1; lane 8, NA (0.9 mM) plus NADPH (2.3 mM) plus crude cell extract (35 μg) from E. coli DH5α plus purified Nor-1c (35 μg); lane 9, NA (0.9 mM) plus crude cell extract (35 μg) from E. coli DH5α plus purified Nor-1c (35 μg). All crude cell extracts are the 10,000 × g supernatant fraction.
Preparation of reaction substrate NA.
ΔNor-1, a nor-1-disrupted strain of A. parasiticus (13), was used to prepare NA as a potential substrate for Nor-1 activity. Conidia (∼108) of ΔNor-1 were cultured in 2 liters of yeast extract-sucrose liquid medium for 2 days at 29°C with constant shaking (150 rpm) in the dark; they were then incubated without shaking at 30°C in the dark for an additional 13 days. Red-colored mycelia (250 g [wet weight]) were collected by filtration and extracted three times with 300 ml of acetone. The acetone extracts were combined and dried with a Rotovapor R110 (Brinkmann Instruments, Inc., Westbury, N.Y.) at 65°C. The resulting solid was resuspended in 100 ml of chloroform which was concentrated (Rotovapor R110; 72°C) to 6 ml and extracted again with 100 ml of acetone. Purple-red NA was purified by preparative thin-layer chromatography (TLC) with PKF silica gel 60 Å chromatography plates (20 by 20 cm; Whatman, Inc., Clifton, N.J.) with chloroform-acetone (9:1) as the developing system. NA (as a brown-colored spot) was scraped from the TLC plate with a razor blade, and the NA was extracted with a small volume of chloroform.
This compound was confirmed to be NA by TLC (with a benzene-ethyl acetate [7:3] developing system) and by UV/visible light (VIS) spectroscopy, mass spectroscopy, and nuclear magnetic resonance (NMR) spectroscopy. UV/VIS spectroscopy was conducted with a UV/VIS spectrophotometer (CARY 3E; Varian-Australia Parity, Ltd.). Mass spectrometry was carried out on a double-focusing mass spectrometer (JEOL AX-505H; Varian-Australia Parity, Ltd.) under the following conditions: electron energy, 70 eV, and scan range, m/z 45 to 600. 1H proton NMR spectroscopy was performed with an NMR spectrometer (VX12-500; Varian-Australia Parity, Ltd.) under the following conditions: proton probe, 80 mHz; range, 0 to 12 ppm; solvent, deuterated dimethyl sulfoxide. The UV/VIS spectrum (200 to 600 nm), the mass spectrum, and the NMR spectrum of the purified compound were consistent with published data for NA (7).
Enzyme activity of recombinant Nor-1.
The enzyme assay was conducted according to a published method (13) with modifications. The reaction mixture included protein (70 to 105 μg), NA (90 μmol), NADPH (0.23 μmol), KH2PO4 (90 mM [pH 7.5]), and 10% (vol/vol) glycerol in a total volume of 100 μl. Reactions were conducted at 37°C in the dark for 30 to 90 min, and stopped by addition of 900 μl of ethyl acetate. The ethyl acetate was air dried, the residue was extracted by adding 400 μl of chloroform, and the reaction products in chloroform extract were resolved by TLC (HPK silica gel 60 Å [10 by 10 cm]; Whatman, Inc.) with benzene-ethyl acetate (7:3) as the developing system (Fig. 2).
In the presence of NADPH, an E. coli crude cell extract containing MBP–Nor-1c (Fig. 2, lane 6), affinity-purified Nor-1c plus a control cell extract (10,000 × g supernatant of E. coli DH5α without pMN1) (lane 8), and affinity-purified MBP–Nor-1c plus control cell extract (results not shown) converted NA to a compound which comigrated with AVN in TLC analysis. The control cell extract (lane 4) and a crude cell extract of E. coli containing pMAL-c2 (lane 5) were unable to convert NA to AVN in the presence of NADPH. NADPH alone failed to convert NA (lane 3). These data suggested that recombinant Nor-1 used NA as a substrate and converted it to AVN. The data also suggested that E. coli provided a helper activity required for Nor-1c activity in vitro. Cell fractionation demonstrated that the helper activity was found in the soluble fraction (105,000 × g supernatant) and not in the membrane fraction (105,000 × g pellet) (data not shown).
Identification of the reaction end product.
Standard AVN was prepared and purified by the method of Bennett et al. (2) with modifications. A. parasiticus ATCC 56774 accumulates AVN and was used to produce this AF pathway intermediate (2). The culture conditions were the same as described above for NA purification. Orange-colored mycelia (210 g [wet weight]) were collected by filtration and extracted with acetone until colorless. Water was added (30%, vol/vol) to the combined acetone extracts, the solution was washed with hexane (1:1 ratio), and the orange pigments were extracted from the acetone phase with chloroform (1:1 ratio). The chloroform extract was concentrated to 5 ml at room temperature and the AVN was purified by preparative TLC (HPK silica gel 60 Å [20 by 20 cm]; Whatman, Inc.) with chloroform-acetone (9:1) as the developing system. AVN (as a yellow-brown-colored spot) was scraped from the TLC with a razor blade and extracted with a small volume of chloroform.
To identify the end product of the reaction catalyzed by Nor-1c, the reaction was scaled up 10-fold. The end product was tentatively identified as AVN by TLC (with benzene-ethyl acetate [7:3] as the developing system) because it had the same Rf value and color as the pure AVN standard. The end product was scraped from the TLC plate and analyzed by UV/VIS spectroscopy, mass spectroscopy, and NMR spectroscopy to confirm its identity. The UV/VIS spectrum (200 to 600 nm) and the NMR spectrum were consistent with data for AVN published by Bennett et al. (2). The NMR spectrum was less consistent with AVN data reported by Birkinshaw et al. (4), although differences may be attributable to differences in solvent system and instrumentation. The mass spectrum was also consistent with published data for AVN (2) except for the molecular ion at m/z 371 which was reported at m/z 372, the predicted mass/charge ratio for AVN. This discrepancy is not unusual. Often, (M - 1)+ ions are observed in mass spectroscopy due to a loss of hydrogen atom(s) from the molecular ion M+; indeed, the (M - 1)+ ion may be more abundant than the expected molecular ion M+ (11). Of critical importance, the mass spectrum of the end product and standard AVN were identical. The preponderance of physical data is consistent with the ability of Nor-1c to convert NA (370 Da) to AVN (372 Da) by keto reduction.
Discussion.
Recombinant Nor-1c catalyzed the conversion of the 1′ keto group of NA to the 1′ hydroxyl group of AVN in the presence of NADPH, confirming that the protein is an NADPH-dependent NA ketoreductase (alcohol dehydrogenase) in vitro. In previous studies, disruption of nor-1 resulted in the accumulation of NA; complementation of a nor-1 mutant restored AF synthesis (13). These data support the hypothesis that native Nor-1 catalyzes the conversion of NA to AVN in A. parasiticus. Although it is clear that another enzyme can bypass the Nor-1-catalyzed step, the gene encoding this activity has not been identified. Cloning of this gene or purification of the enzyme activity should allow us to define more clearly the potential pathways for the conversion of NA to aflatoxin B1 in the fungus.
We observed that a soluble compound in E. coli DH5α was required for Nor-1c activity. The simplest explanation is that modification of the amino terminus of Nor-1c resulted in dependence on the soluble factor. Surprisingly, purification of a 31-kDa NA reductase has not been reported in previous attempts to purify a NA reductase (3, 6). One alternative explanation is that a cofactor(s) is required for activity of the native Nor-1 protein and is lost during a step in the purification scheme in previous studies, resulting in inability to purify this enzyme. Attempts to identify this cofactor in E. coli and the proposed analogous factor in Aspergillus may provide clues about the cellular environment in which this enzyme functions.
Acknowledgments
This work was supported by the Michigan State University Agricultural Experiment Station and grant CA 52003 from the National Institutes of Health.
We thank Matthew Rarick for help in preparation of the figures, B. Chamberlin (Department of Biochemistry, Michigan State University) for help with mass spectrometry, and K. Johnson (Department of Chemistry, Michigan State University) for help with 1H proton NMR spectroscopy. Aflatoxin B1 antibodies were kindly provided by J. Pestka (Department of Food Science and Human Nutrition, Michigan State University). The plasmid pNOR was kindly provided by Perng-Kuang Chang (USDA ARS SRRC).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. I. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 2.Bennett J W, Lee L S, Shoss S M, Boudreaux G H. Identification of averantin as an aflatoxin B1 precursor: placement in the biosynthetic pathway. Appl Environ Microbiol. 1980;39:835–839. doi: 10.1128/aem.39.4.835-839.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatnagar D, Cleveland T E. Purification and characterization of a reductase from Aspergillus parasiticus SRRC 2043 involved in aflatoxin biosynthesis. FASEB J. 1990;4:A2164. [Google Scholar]
- 4.Birkinshaw J H, Roberts J C, Roffey P. Studies in mycological chemistry. XIX. “Product B” (averantin) [1,3,6,8-tetrahydroxy-2-(1-hydroxyhexyl)anthraquinone], a pigment from Aspergillus versicolor (Vuillemin) Tiraboschi. J. Chem Soc Perkin Trans 1. 1966;9:855–857. doi: 10.1039/j39660000855. [DOI] [PubMed] [Google Scholar]
- 5.Chang P-K, Skory C D, Linz J E. Cloning of a gene associated with aflatoxin biosynthesis in Aspergillus parasiticus. Curr Genet. 1992;21:231–233. doi: 10.1007/BF00336846. [DOI] [PubMed] [Google Scholar]
- 6.Chuturgoon A A, Dutton M F. The affinity purification and characterization of a dehydrogenase from Aspergillus parasiticus involved in aflatoxin B1 biosynthesis. Prep Biochem. 1991;21:125–140. doi: 10.1080/10826069108018008. [DOI] [PubMed] [Google Scholar]
- 7.Cole R J, Cox R H, editors. Handbook of toxic fungal metabolites. New York, N.Y: Academic Press, Inc.; 1981. pp. 111–112. [Google Scholar]
- 8.Donkersloot J A, Mateles R I, Yang S S. Isolation of averufin from a mutant of Aspergillus parasiticus impaired in aflatoxin biosynthesis. Biochem Biophys Res Commun. 1972;47:1051–1055. doi: 10.1016/0006-291x(72)90939-4. [DOI] [PubMed] [Google Scholar]
- 9.Dutton M F. Enzymes and aflatoxin biosynthesis. Microbiol Rev. 1988;52:274–295. doi: 10.1128/mr.52.2.274-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee L S, Bennett J W, Goldblatt L A, Lundin R E. Norsolorinic acid from a mutant strain of Aspergillus parasiticus. J Am Oil Chem Soc. 1971;48:93–94. doi: 10.1007/BF02635696. [DOI] [PubMed] [Google Scholar]
- 11.Rose M E, Johnstone R A W. Mass spectrometry for chemists and biochemists. Cambridge, England: Cambridge University Press; 1982. Structure elucidation; pp. 185–246. [Google Scholar]
- 12.Trail F, Chang P-K, Cary J, Linz J E. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl Environ Microbiol. 1994;60:4078–4085. doi: 10.1128/aem.60.11.4078-4085.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabe K, Nakamura Y, Nakajima H, Ando Y, Hamasaki T. Enzymatic conversion of norsolorinic acid to averufin in aflatoxin biosynthesis. Appl Environ Microbiol. 1991;57:1340–1345. doi: 10.1128/aem.57.5.1340-1345.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]