Abstract
Peptides play a key role in controlling many physiological and neurobiological pathways. Many bioactive peptides require a C-terminal α-amide for full activity. The bifunctional enzyme catalyzing α-amidation, peptidylglycine α-amidating monooxygenase (PAM), is the sole enzyme responsible for amidated peptide biosynthesis, from Chlamydomonas reinhardtii to Homo sapiens. Many neuronal and endocrine functions are dependent upon amidated peptides; additional amidated peptides are growth promoters in tumors. The amidation reaction occurs in two steps, glycine α-hydroxylation followed by dealkylation to generate the α-amide product. Currently, most potentially useful inhibitors target the first reaction, which is rate-limiting. PAM is a membrane-bound enzyme that visits the cell surface during peptide secretion. PAM is then used again in the biosynthetic pathway, meaning that cell-impermeable inhibitors or inactivators could have therapeutic value for the treatment of cancer or psychiatric abnormalities. To date, inhibitor design has not fully exploited the structures and mechanistic details of PAM.
Keywords: peptidylglycine α-amidating monooxygenase, peptide, copper, oxygen, ascorbate
1. Introduction
1.1. Bioactive peptides play a key role in controlling many complex pathways including blood glucose (insulin and glucagon, for example), salt and water balance (vasopressin, atrial natriuretic peptide), appetite (cholecystokinin, α-melanocyte stimulating hormone, leptin), reproduction (gonadotropin hormone releasing hormone, follicle stimulating hormone) and gastrointestinal function (gastrin, motilin). Each of these peptides is synthesized from a newly synthesized precursor protein as it moves from its site of synthesis in the endoplasmic reticulum, through the secretory pathway lumen and into the secretory granules from which it is released in response to the appropriate combination of stimuli (Chrétien & Mbikay, 2016; Clark & Lowry, 2016; Kumar, Mains, & Eipper, 2016). A limited set of subtilisin-like endoproteases, carboxypeptidase B-like exoproteases and other post-translational processing enzymes convert these precursors, which are often inactive, into active products as they move through the secretory pathway (Fig.1A). The activity of many bioactive peptides requires α-amidation of their C-terminal residue; lacking an amidated C-terminus, their ability to bind to their receptors (generally a G Protein Coupled Receptor) is greatly reduced. Consistent with this, Peptidylglycine α-Amidating Monooxygenase (PAM), the only enzyme known to catalyze the formation of α-amidated peptides, is an essential enzyme. Mice lacking PAM survive only until mid-gestation (Czyzyk et al., 2005) and neither flies nor zebrafish lacking PAM are viable (Kolhekar, Roberts, et al., 1997; Kumar et al., 2018). Mice having a single functional Pam gene exhibit a wide variety of deficits, ranging from altered inhibitory synaptic neurotransmission to increased anxiety-like behavior and an inability to maintain body temperature in a cold environment (Bousquet-Moore et al., 2010; Gaier, Eipper, & Mains, 2014; Gaier et al., 2013). Genetic studies have identified PAM as a risk factor for type 2 diabetes (Steinhorsdottir et al., 2014; Thomsen et al., 2018).
Figure 1. Peptide processing basics, the amidation reaction and amidation enzymes.
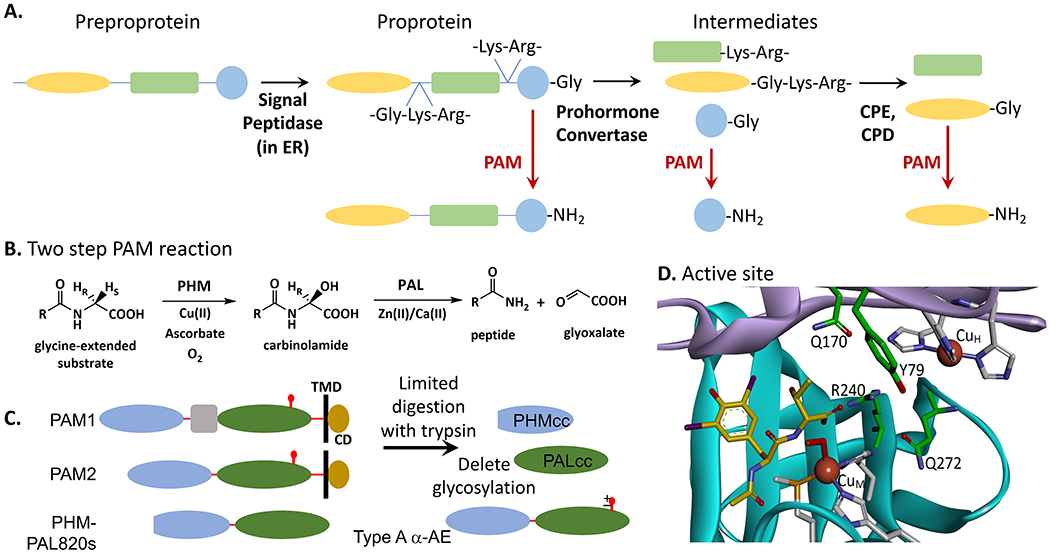
A. The preprotein peptide precursor is processed by endoproteases (prohormone convertases such as PCSK1, PCSK2, PCSK3) and an exoprotease (such as CPE or CPD) to present to PAM the immediate precursor (peptide-Gly), which yields the final amidated peptide product. B. The two-step PAM reaction. Step 1 is performed by PHM and Step 2 is performed by PAL. The two copper residues bound to PHM are reduced by ascorbate, peptide and dioxygen bind to the reduced enzyme, a proton is abstracted and the peptide is hydroxylated by PHM. The N-C bond is then cleaved by PAL, liberating the α-amidated peptide plus glyoxalate. C. Proteins used for enzymology and structural determination. In mammals, two major forms of PAM are the bifunctional membrane enzymes PAM1 and PAM2, each with a transmembrane domain (TMD) and short cytoplasmic domain (CD). The proteins used thus far for structural analyses are the recombinant catalytic cores, PHMcc and PALcc. The reactions have been studied with purified natural proteins from various sources, recombinant pure rat PHMcc and PALcc, and with two pure recombinant bifunctional proteins, rat PHM-PAL820s and Type A α-AE. References given in the text. Residues (using NP_037132.2): PHMcc (42-356), PALcc (498-820, lacking N-glycosylation [S767A]), PHM-PAL820s (42-820 lacking Exon 16 [residues 393-498], lacking N-glycosylation [S767A]), Type A PAM (27-820 lacking Exon 16). D. Structure of reduced PHM with N-acetyl-diiodo-tyrosyl-D-threonine bound ((Prigge et al., 2004); PDB: 1SDW). Residues 46 to 198 illustrating the Cu(I)H subdomain are shown in lilac and residues 199 to 356 illustrating the Cu(I)M subdomain are in cyan. The residues that coordinate the bound Cu(I) atoms are shown in gray. The peptide substrate analog is in yellow and several residues important in substrate binding are shown in green. R240 forms a salt bridge with the glycine carboxylate of the substrate, Q170 and Q172 function in organizing hydrogen bond networks for substrate binding, and Y79 has a role in substrate binding.
1.2. PAM is a bifunctional enzyme: its monooxygenase domain (peptidylglycine α-hydroxylating monooxygenase, PHM: EC 1.14.17.3) catalyzes the copper- and ascorbate-dependent α-hydroxylation of its peptidylglycine substrates; its lyase domain (peptidyl-α-hydroxyglycine α-amidating lyase, PAL: EC 4.3.2.5; also called peptidylamidoglycolate lyase) then generates the α-amidated product peptide plus glyoxylate (Fig.1B). C-terminal amidation can alter peptide structure, confer resistance to proteolytic degradation and reduce the effects of pH on receptor binding (Kahns & Bundgaard, 1991; Luxmi, Mains, King, & Eipper, 2021). Extensive mechanistic studies and structural studies facilitated the development of PAM inhibitors. In this brief review we will summarize several successful approaches taken to develop inhibitors of PHM and discuss observations indicating that PAM may serve as a useful therapeutic target or biomarker.
2. Peptide α-Amidation in Two Steps
2A.1. The PHM Reaction:
Progress in studies on the formation of a-amidated peptides depended crucially on three key findings. First, studies conducted from the 1950’s to the early 1980’s made it clear that the proprotein precursors to peptide-amides always had a Gly residue located to the C-terminal side of the residue that was to be α-amidated (reviewed in (Kumar, Mains, et al., 2016)) (Fig.1A and B). Second, the development of the first useable enzymatic assay of peptide amidation by Bradbury, Finnie and Smyth (Bradbury, Finnie, & Smyth, 1982) rapidly led to papers detailing the tissue distribution of PHM activity, the copper dependence of the PHM reaction, and the purification of the PHM protein (Eipper et al., 1991; Murthy, Keutmann, & Eipper, 1987; Murthy, Mains, & Eipper, 1986; Tamburini, Young, Jones, Palmesino, & Consalvo, 1990). Third, the realization that pituitary cells maintained in serum-free culture medium efficiently stored and secreted Proopiomelanocortin (POMC) products, but were unable to produce a-amidated peptides, led to the identification of ascorbate as a required cofactor in the PHM reaction (Eipper, Glembotski, & Mains, 1983). These observations led to the cloning and expression of PAM cDNAs from several species (Bertelsen et al., 1990; Eipper et al., 1987; Glauder, Ragg, Rauch, & Engels, 1990; Mizuno et al., 1987; Ouafik et al., 1992; Stoffers, Green, & Eipper, 1989; Stoffers, Ouafik, & Eipper, 1991). Although the lyase reaction catalyzed by PAL can occur at elevated pH in the absence of enzyme, phylogenetic studies support the presence of a bifunctional, integral membrane PAM protein (Fig.1C) in the last eukaryotic common ancestor (Bäck, Mains, & Eipper, 2021; Kumar, Mains, et al., 2016).
2A.2. Most of the enzymological and all of the crystallographic studies of PHM and PAL have been carried out using the soluble, protease-resistant catalytic cores of PHM (PHMcc) and PAL (PALcc) (Fig.1C). Endoproteolytic digestion of lysates prepared from mammalian cells expressing exogenous PAM and of tissues that express high levels of endogenous PAM revealed that both catalytic activities were remarkably resistant to proteolytic degradation (Husten & Eipper, 1994; Husten, Tausk, Keutmann, & Eipper, 1993). For rat PAM1 (Uniprot P14925), PHMcc consists of residues 42–356 (Eipper, Quon, Mains, Boswell, & Blackburn, 1995; Kolhekar, Keutmann, Mains, Quon, & Eipper, 1997) and PALcc consists of residues 498–820 (Kolhekar et al., 2002; Kolhekar, Quon, Berard, Mains, & Eipper, 1998). The crystal structures of recombinant PHMcc and PALcc have been extensively studied (Chufan, De, Eipper, Mains, & Amzel, 2009; Prigge, Eipper, Mains, & Amzel, 2004; Prigge, Kolhekar, Eipper, Mains, & Amzel, 1997; Siebert et al., 2005). PAM, PHM, and PAL purified from natural sources and soluble, recombinant PHM, PAL and bifunctional PAM have provided additional insight into the reaction mechanism (Fig.1C) (Bertelsen et al., 1990; Miller et al., 1992). To date, it has not been possible to obtain high resolution structural data for any bifunctional PAM proteins.
2A.3. Studies of the PHM and PAL components of bifunctional PAM are accomplished by employing the appropriate assays: O2 consumption from a glycine-extended peptide is specific for PHM and glyoxylate production from a carbinolamide is specific for PAL. Although few studies have utilized human PAM, PHM or PAL, the highly conserved nature of the PHM and PAL active sites (Bäck et al., 2021; Kumar, Mains, et al., 2016) suggests that the catalytic properties of all of the various PAM, PHM and PAL proteins mentioned above are quite similar. Readers are encouraged to review the cited references if the exact source of the proteins is important to their research goals. Despite many attempts to express fully active PAM in bacterial, yeast, and insect systems, mammalian cell lines remain the system of choice. PHMcc has been expressed in E. coli with an N-terminal fusion to thioredoxin, but with a yield of only ~100 μg/liter of E. coli (Handa, Spradling, Dempsey, & Merkler, 2012). Production of recombinant rat PHMcc, PALcc and rat medullary thyroid carcinoma (MTC) PAM in Chinese hamster ovary (CHO) cells has yielded >100 mg of pure enzyme (Bauman, Ralle, & Blackburn, 2007; Miller et al., 1992), sufficient enzyme to support spectroscopic studies (Blackburn, Rhames, Ralle, & Jaron, 2000; Eipper et al., 1995; Evans, Blackburn, & Klinman, 2006; Jaron & Blackburn, 1999), structural biology (Chufan et al., 2009; Prigge et al., 1997; Prigge, Kolhekar, Eipper, Mains, & Amzel, 1999), and amidated peptide production at the multi-gram level (Ray et al., 1993).
2A.4. PHM function requires two bound copper ions, CuH (with three His ligands) and CuM (with two His and one Met ligand) (Maheshwari et al., 2018; Prigge et al., 2004; Prigge et al., 1999; Siebert et al., 2005) (Fig.1D). Copper is easily lost from PHM, meaning that assaying PHM activity in crude lysates or in serum samples (Gaier et al., 2012) requires the addition of exogenous copper and analysis of purified PHM and PAM often requires replacement of copper lost during purification (Blackburn et al., 2000; Evans et al., 2006; Jaron & Blackburn, 1999; Murthy et al., 1986). The first step in the amidation reaction is the reduction of Cu(II)H and Cu(II)M to Cu(I) by ascorbate (Fig.2A, Step 1). Studies show that the kinetic mechanism is equilibrium ordered, with the glycine-extended substrate binding first to reduced enzyme (Fig. 2A, Step 2), followed by the binding of O2 to Cu(I)M (Fig.2A, Step 3). Formation of the (S)-α-hydroxyglycine derivative (Cowley, Tian, & Solomon, 2016; Ping, Mounier, & May, 1995), which is the PHM product and PAL substrate, proceeds from the PHM-Cu(I)H-Cu(I)M-substrate-O2 complex (Fig. 2A, Step 4) and regenerates oxidized PHM-Cu(II)H-Cu(II)M, preparing PHM for the next catalytic cycle (Fig.2A, Step 5).
Figure 2. The Reactions Catalyzed by PHM and PAL.

A. Bifunctional PAM is compromised of the two separate catalytic units, PHM and PAL (Reaction A). Steps 4 and 5 in reaction A represent a collection of steps in the PHM mechanism (Cowley et al., 2016; Prigge et al., 2000; Wu et al., 2019). B. and C. Additional reactions catalyzed by PHM; any involvement of PAL in these reactions has not been specifically addressed. The S-dealkylation reaction, shown in 2B, is consistent with the finding of glyoxylate as a minor product during the sulfoxidation reaction. A sulfoxide/glyoxylate ratio of 8 was reported, but no mercaptan was found (Katopodis & May, 1990a). D. The formation of glyoxylate from the imino-oxy acetate (bottom line) is PAL-independent.
2A.5. The chemical mechanism for PHM catalysis has been intensively debated (Chen, Bell, Eipper, & Solomon, 2004; Cowley et al., 2016; Francisco, Merkler, Blackburn, & Klinman, 1998; Jaron & Blackburn, 1999; Klinman, 2006; Kulathila, Merkler, & Merkler, 1999; Ping et al., 1995); it is agreed that a substrate-based radical on the α-carbon of the glycine is an intermediate and that a copper-superoxo species, CuM(II)-O2•, is responsible for hydrogen atom abstraction from the substrate glycine. Recent work from Wu et al. (Wu et al., 2019) suggests that an unusual (μ-O•)(μ-OH)Cu(II)MCu(II)H is the species responsible for hydrogen atom abstraction from substrate. In addition to the uncertainties about the reduced oxygen species responsible for hydrogen atom abstraction, there remain questions about the structure of the bifunctional PAM, binding site(s) for ascorbate, the electron transfer pathways between the two copper atoms and from the copper atoms to the substrate during catalysis, potential domain motion in PHM to reduce the 11Å distance between the Cu(II), and the possibility of carbinolamide channeling between the PHM and PAL active sites in PAM. Note that development of the current cohort of PHM inhibitors (discussed below) has progressed without definitive answers to these questions. Thus, the PHM and PAL inhibitors developed to date are substrate analogs. The rational design of second and third generation PHM inhibitors would benefit from a resolution of the mechanistic uncertainties, fostering the design of tight-binding transition-state analog inhibitors.
2B.1. The PAL Reaction:
Like PHM, PAL is a metalloenzyme; PAL contains a bound Zn(II) and a bound Ca(II) (Bell et al., 1997; Chufan et al., 2009; De, Bell, Blackburn, Mains, & Eipper, 2006). PAL catalyzes the dealkylation of the (S)-α-hydroxyglycine derivative to the corresponding amide and glyoxylate (Fig.2A, Step 5) (Katopodis, Ping, & May, 1990; Perkins, Husten, & Eipper, 1990). To date, evidence about channeling of the peptidyl-α-hydroxyglycine product from PHM to PAL is conflicting (Francisco et al., 1998; Moore & May, 1999). One major site for PHM and PAL catalysis is the acidic secretory granules with an internal pH~5 (Mains & May, 1988). The α-hydroxyglycine derivative (a carbinolamide) is stable at this pH (Bundgaard & Kahns, 1991); thus, PAL is required for effective amide formation. The bound Ca(II) has a structural role in PAL, while the bound Zn(II) is likely involved in catalysis (Chufan et al., 2009; Takahashi et al., 2009). The precise role played by Zn(II) is not yet clear; suggestions include Zn(II)-bound water or hydroxide serving as a general base for proton abstraction from the hydroxyl group (Bell et al., 1997; Takahashi et al., 2009) and Zn(II)-coordination to the hydroxyl group to lower its pKa, facilitating proton transfer to a tyrosyl phenolate in the active site (Chufan et al., 2009). Model studies by Tenn et al. (Tenn et al., 2007) suggest that carbinolamide dealkylation is not base-catalyzed and is independent of the pKa of the hydroxyl group of the carbinolamide. Instead, these authors argue that carbinolamide dealkylation is dependent upon the nucleofugality (the tendency of a leaving group to retain an electron pair upon departure) of the departing amide. If so, the PAL-bound Zn(II) could assist in the departure of the amidated product. Additional studies of PAL catalysis are required to address these mechanistic questions.
2B.2. In the few cases examined, tissue lysates contained substantially more α-amidated peptide than peptidylglycine precursor and levels of peptidyl-α-hydroxyglycine intermediate were well below peptidylglycine levels (Yin et al., 2011). These observations are consistent with the higher Vmax values reported for PAL vs PHM. As discussed below, the PAL domain plays an essential role in the ability of cells to retrieve membrane PAM from the cell surface. This suggests that inhibitors targeted to the active site of PAL might provide an effective means of inhibiting peptide amidation by reducing the amount of PAM available to amidate newly synthesized peptidylglycine substrates entering immature secretory granules. It is intriguing that the PAM loci most frequently associated with human diseases are in the PAL domain, not the PHM domain (Steinhorsdottir et al., 2014; Thomsen et al., 2018).
3. Effective PHM and PAL Inhibitors Were Developed.
3.1. For a period of about 25 years, from 1990 to 2015, there was considerable interest in developing PHM inhibitors and inactivators (Ali et al., 2015; Bolkenius, Ganzhorn, Chanal, & Danzin, 1997; Cao et al., 2011; Katopodis & May, 1990a; Langella et al., 2010). The hope was that a PHM-targeted compound would be therapeutically useful in the treatment of human disease and/or would contribute to our understanding of PHM. Compounds that would intercept the radical intermediate were developed (Zabriskie, Cheng, & Vederas, 1992; Zabriskie, Klinge, Szymanski, Cheng, & Vederas, 1994) and PAM labeling with mechanism-based inactivators (suicide substrates) was used to try to identify the active site amino acids critical to catalysis. There was less interest in developing PAL-targeted compounds, the thought being that the non-enzymatic dealkylation of the accumulated carbinolamide intermediate would supply sufficient levels of the amidated product to alleviate PAL inhibition; as discussed above, PAL-targeted compounds that alter the trafficking of PAM could still prove useful in manipulating amidation in vivo.
3.2. Many drugs exhibit cell toxicity.
Such therapeutics have greater toxicity towards diseased cells relative to healthy cells and must be used properly in treating a disease. In addition, ongoing research will lead to “molecular zip codes”, a protein or a small molecule that binds tightly and specifically to uniquely targeted diseased cells (Dai et al., 2018; Enbäck & Laakkonen, 2007; Kumar, Zhang, & Liang, 2013; Li & Zhang, 2019). The attachment of a drug to a molecular zip code would enable its delivery specifically to the diseased cells – a strategy enabling the safe use of highly toxic drugs to treat human disease. A toxic PHM-specific inhibitor could be valuable therapeutically if appended to the appropriate molecular zip code. The production of amidated autocrine growth factors by tumor cells suggests that high-affinity cell-impermeant PHM inhibitors might prove useful in controlling the growth of these cells. Most neurons and endocrine cells cleave PHM from PAL within secretory granules and release PHM along with peptide. These cells would presumably ingest less of the toxic PHM-specific inhibitor than would cancerous cells, since tumor cells usually leave much of the PAM they produce intact while making the amidated autocrine growth factors which enhance tumorigenicity.
3.3. Another use of biomedical significance for a PHM-targeted compound would be as a PAM-specific imaging agent. PHM is a biomarker for specific cancers (Jiménez et al., 2003; Rocchi et al., 2004; Scopsi et al., 1998; Thouënnon et al., 2007), post-polio syndrome (Gonzalez et al., 2009), and neural dysfunction (Bousquet-Moore et al., 2010). In sum, the toxicity concern about a PHM inhibitor/inactivator is short-sighted, is limiting work to develop PHM-dependent imaging agents and provides few options when a PHM-specific molecular zip code becomes available (Bäck et al., 2017; McKay et al., 2006; Scopsi et al., 1998; Thouënnon et al., 2007).
3A.1. PHM Inhibitors:
The inhibitory effects of divalent metal ion chelators on peptide amidation became apparent as soon as different buffers were tested for use in enzyme assays of tissue lysates (Eipper, Mains, & Glembotski, 1983). Inhibition by diethyldithiocarbamate was reversed only by the addition of Cu(II), providing the first evidence that PAM/PHM catalysis was copper-dependent; no other divalent metal, including Zn(II), restored activity (Eipper, Mains, et al., 1983; Eipper, Park, Keutmann, & Mains, 1986). Other metal chelators like EDTA (Merkler, Kulathila, Young, Freeman, & Villafranca, 1993) and disulfiram inhibit PAM/PHM reversibly (Mains, Park, & Eipper, 1986) (Fig.3A). Disulfiram (Antabuse) is used for the treatment of alcohol abuse (Kranzler & Soyka, 2018) and has anti-cancer activity (Corsello et al., 2020). The possible link between the anti-cancer activity and PHM inhibition has not been directly investigated. Disulfiram has been used to inhibit α-amidated peptide production in cultured mammalian cells and in rats (Mains et al., 1986; Mueller & Altarac, 1995; Rondeel et al., 1995).
Figure 3. Ligands that Bind to PHM and PAL.
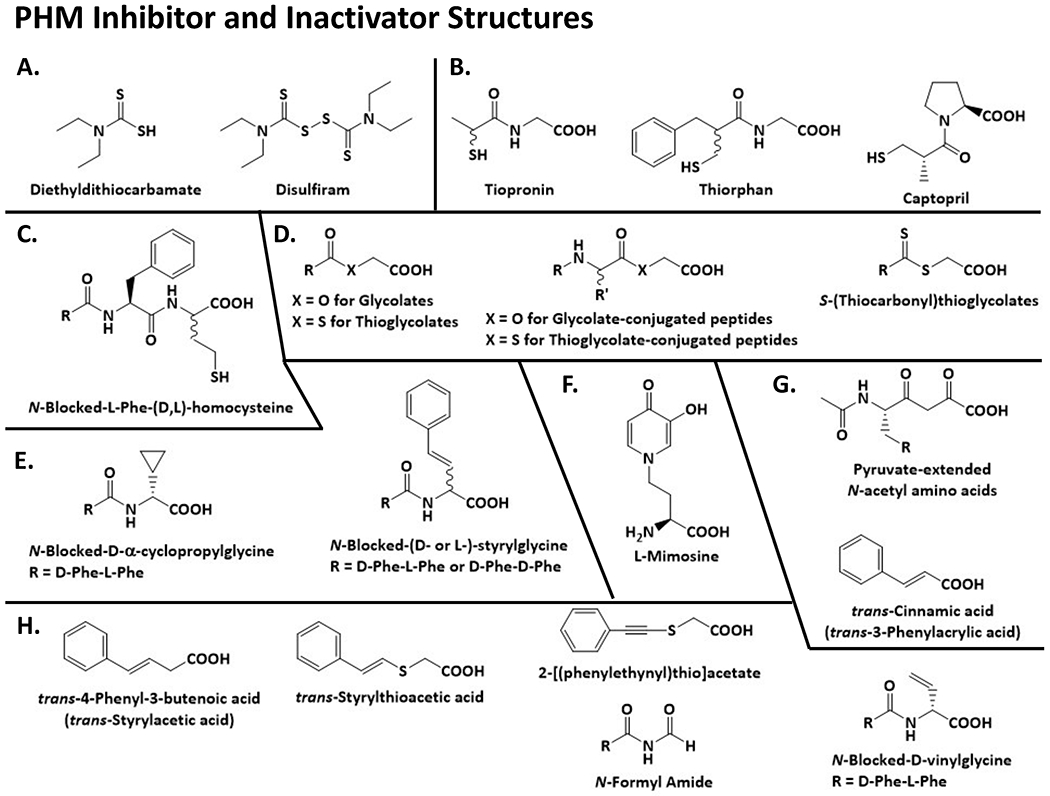
Structures for the inhibitors and activators discussed in this review.
3A.2. Most of the PHM inhibitors developed to date likely bind to the peptide substrate site within the PHM active site (Prigge et al., 1997). A review of the substrates oxidized by PHM is required to provide context for the PHM inhibitors. Only peptides with a C-terminal glycine are PHM substrates in vivo (Johnson, Paisley-Flango, Tangarone, Porter, & Rouse, 2007). Synthetic peptides with a C-terminal D-Ala are substrates leading to the corresponding α-amidated peptide and pyruvate (Landymore-Lim, Bradbury, & Smyth, 1983). However, the D-Ala-extended peptides are relatively poor substrates with low Vmax/Km ratios. The Vmax/Km ratio provides an estimate of the first-order rate constant for the conversion of substrate to product and is obtained using a substrate concentration far below its Km value; when comparing different substrates, it provides a measure of enzyme specificity (Johnson, 2019). For example, the Vmax/Km ratio for N-dansyl-Tyr-Val-D-Ala is 0.06 times the ratio for N-dansyl-Tyr-Val-Gly; the Vmax/Km ratio for N-benzoyl-D-Ala is 0.0005 times the ratio for N-benzoylglycine (Merkler et al., 2008). Systematic studies to determine the effect of the penultimate amino acid on the kinetics of amidation showed that glycine-extended peptides with a penultimate aromatic, hydrophobic, or sulfur-containing amino acid had the highest Vmax/Km ratios while those with a penultimate Lys or Arg had the lowest Vmax/Km ratios (Merkler et al., 1993; Tamburini et al., 1990). α-Amidated peptides with a C-terminal amino acid amide for all 20 of the common amino acids are known (Eipper, Stoffers, & Mains, 1992). Studies aimed at producing amidated peptides in cell lines using endogenous or exogenous PAM indicate that the general rules for effective amidation developed by Tamburini et al. are not directly applicable to the conditions encountered as PAM and its substrates move through the secretory pathway in cells (Chew et al., 2005). A free carboxylate is required on the C-terminal glycine for amidation; methyl or ethyl esters are not substrates, but are inhibitors of relatively low affinity (Tamburini et al., 1988). Similarly, the amidated products are low affinity inhibitors (Glembotski, Eipper, & Mains, 1984).
3A.3. α-Amidated peptide hormones range considerably in length, from 3 amino acids for TRH to 52 amino acids for adrenomedullin. Much larger proteins that terminate with a C-terminal amide and generate α-amidated chemomodulatory peptides have been identified in the green algae, Chlamydomonas reinhardtii (Luxmi, Kumar, Mains, King, & Eipper, 2019). When expressed exogenously, for therapeutic purposes, monoclonal antibodies whose heavy chains terminate with a glycine are often α-amidated (Skulj, Pezdirec, Gaser, Kreft, & Zorec, 2014); elimination of the low levels of PAM expressed in the mammalian cells used to produce these monoclonal antibodies eliminates their α-amidation (Skulj et al., 2014). Dipeptides, X-Gly, are poor PHM substrates, exhibiting low Vmax/Km ratios (Wilcox et al., 1999) and, in general, longer peptide substrates exhibit higher Vmax/Km ratios (Chew et al., 2005). Modeling of N-α-acetyl-3,5-diiodotyrosylglycine into the structure of PHMcc indicates that the carboxylate of the glycyl residue forms a salt-bridge with Arg240 (Prigge, Mains, Eipper, & Amzel, 2000). This arginine is highly conserved across PHM sequences and likely provides a rationale for the importance of the free carboxylate for substrate binding. PHMcc is comprised of two domains of approximately the same size that are connected by a single strand; the single copper atom bound to each domain is poised at the edge facing the solvent-filled cleft separating the domains (Prigge et al., 1997). The interiors of both domains are hydrophobic and the copper atoms are separated by 11Å (Prigge et al., 1997). Upon reduction, there is virtually no change in PHMcc structure (Prigge et al., 2004). The large, hydrophobic interdomain region of PHMcc is necessary for the amidation of relatively large peptide substrates and is consistent with a preference for penultimate hydrophobic amino acids in the substrate. How electron transfer and radical stability are accomplished in the large, solvent accessible interdomain region of PHM are amongst the unanswered questions about PHM catalysis. A PHM structure with a large peptide substrate bound might provide insight about these issues.
3A.4. Strikingly, PHM and PAL will accept a wide variety of non-peptide substrates possessing a moiety that is equivalent to the C-terminal glycine, including the N-acylglycines, hippurate (N-benzoylglycine) and substituted hippurates (Merkler et al., 2008), and the bile acid glycine conjugates (King et al., 2000). A carbinolamide intermediate is known to form as these substrates are converted into the amide and glyoxylate. Consistent with the preference for a hydrophobic penultimate amino acid for peptide substrates, the Vmax/Km ratio for N-acylglycine amidation increases as acyl-chain length increases. Early studies by Katopodis and May demonstrated that PHM could also catalyze sulfoxidation, amine N-dealkylation, and O-dealkylation (Katopodis & May, 1990a) (Fig. 2B, C, D). The PHM catalyzed O-dealkylation of the imino-oxy acetates yields the corresponding oximes and glyoxylate in a PAL-independent manner (McIntyre, Lowe, Battistini, Leahy, & Merkler, 2016; McIntyre, Lowe, & Merkler, 2009; Schade, Kotthaus, Hungeling, Kotthaus, & Clement, 2009). A complete understanding of PHM catalysis must account for the alternative reactions catalyzed by this enzyme.
3A.5. A series of N-blocked Phe-(D,L)-homocysteine peptide analogs were synthesized and evaluated as PAM inhibitors (Erion, Tan, Wong, & Jeng, 1994; Jeng, Fujimoto, Chou, Tan, & Erion, 1997) (Fig. 3). Members of this series with IC50 values of 8-15 nM possess an N-hydrocinnamoyl, N-2-naphthoyl, or N-2-indolyl moiety conjugated to the Phe residue. Synthesis of the benzyl ester of N-2-naphthoyl-L-phenylalanyl-(D,L)-homocysteine as a prodrug increased the IC50 from 10 to 8,000 nM, but 10 μM free carboxylate or benzyl ester showed approximately 50% inhibition of PHM in cultured rat dorsal root ganglion neurons, (Jeng et al., 1997). The potency of this series of compounds mirrors the substrate specificity information from the glycine-extended peptides. The partial PHM inhibition observed in the dorsal root ganglion neurons identifies cell penetration as a challenge for the future clinical use of these compounds. The disappointing results observed using dorsal root ganglion neurons for the benzyl ester suggest that the benzyl ester is not the optimal prodrug, at least, for this series of PHM inhibitors. The binding of tiopronin (KM = 33 μM), thiorphan (KM = 82 μM), and captopril (Ki ~100 μM), all compounds with a free sulfhydryl group (Fig.3B), was attributed to coordination of the sulfur atom to one of PHM-bound Cu(II) atoms (McIntyre, Lowe, Chew, Owen, & Merkler, 2006; Mueller, Driscoll, & Eipper, 1999). The tight-binding of the homocysteine peptide analogs likely results from coordination of the sulfur atom to CuM (Fig. 3C).
3A.6. One class of compounds investigated as PHM inhibitors are the glycolates, R-CO-O-CH2-COOH; the R-groups described include an acyl group (short- and long-chain), a benzoyl group, a phenylacetyl-group, an N-acetyl-amino acid, and N-acetyl-peptides (Barratt et al., 2004; Cao et al., 2011; Katopodis & May, 1990a; Morris et al., 2012; Ping et al., 1995) (Fig. 3D). The glycolates are analogs of the glycine-extended substrates and were clear substrate analogs for consideration as inhibitors. The glycolates are PHM-specific and have no effect on PAL (Moore & May, 1999). We have summarized the data for the glycolate and structurally similar inhibitors in Table 1A and, from these data, we note the following trends. A number of the glycolate inhibitors exhibit Ki or IC50 values <5 μM, meaning that this class of compounds represent a good starting point for the future development of higher affinity PHM inhibitors. While the C-terminal glycolate analogs of two α-amidated peptide precursors (oxytocin and calcitonin) exhibit IC50 values of 2-12 μM, the structurally simpler O-acylglycolates, like O-lauroylglycolate (CH3-(CH2)10-CO-O-CH2-COOH), exhibit sub-μM affinities, against PHM proteins from cultured mammalian cells. Thus, a future tight-binding glycolate inhibitor might be possible without the complications associated with peptide synthesis. Another encouraging observation is the differences in affinity between PHM proteins for some of the glycolate inhibitors. For example, O-lauroylglycolate exhibits an IC50 of 60 nM for human lung cancer PHM and an IC50 of 35 μM for frog PHM, a ratio of IC50 values of ~600. This suggests that tissue-, organism-, or disease-state-specific PHM inhibitors may be possible. The stereochemistry of the penultimate group has a dramatic effect on the affinity of the glycolate inhibitors. The ratio of the Ki values for the D-amino acid glycolate/L-amino acid glycolate is 40-50 for the N-acetyl-D-Phe-O-CH2-COOH/N-acetyl-L-Phe-O-CH2-COOH pair and for the N-acetyl-D-Leu-O-CH2-COOH/N-acetyl-L-Leu-O-CH2-COOH pair. An even stronger preference for an L-amino acid at the penultimate position was identified for the glycine-extended peptide substrates; N-Acetyl-L-Phe-Gly binds to PHM ≥700-fold more tightly than N-acetyl-D-Phe-Gly (Ping et al., 1995). Future tight-binding glycolate inhibitors could take advantage of the stereochemical preference for the penultimate position.
Table 1.
Glycolate (A), Thioglycolate (B), and S-(Thiocarbonyl)thioglycolate (C) Inhibitors of PHM
| A. Glycolates R-CO-O-CH2-COOH | Ki or IC50 (μM) | Source of Enzymea | Ref. |
|---|---|---|---|
| R = CH3-CO | 1100 | DMS53 cells – (E)b,c | Morris et al., 2012 |
| 1100 | DMS53 cells – (M)b,c | ||
| R = C6H5 | 250 | Frog | Barratt et al., 2004 |
| R = 4-CH3O-C6H4 | 480 | Bovine | Katopodis & May, 1990 |
| R = C6H5-CH2 | 510 | Frog | Cao et al., 2011 |
| 2 | H889 cells – (E)b,d | ||
| R = C6H5-NH | 50 | Rat MTCe | Merkler et al., 2008 |
|
O-Benzamidoglycolate C6H5-CO-NH-O-CH2-COOH |
1000 | Rat MTCe | Merkler et al., 2008 |
| R = CH3-(CH2)4 | 780 | Frog | Cao et al., 2011 |
| R = CH3-(CH2)6 | 250 | Frog | Cao et al., 2011 |
| R = CH3-(CH2)8 | 40 | Frog | Cao et al., 2011 |
| 0.05 | H889 cells – (E)b,d | ||
| 0.04 | DMS53 cells – (E)b,c | ||
| 30 | DMS53 cells – (M)b,c | ||
| 120 | PC3 cells – (E)b,f | ||
| 30 | SW1783 cells - (M)b,g | ||
| R = CH3-(CH2)10 | 40 | Frog | Cao et al., 2011 |
| 0.06 | H889 cells – (E)b,d | ||
| R = CH3-(CH2)12 | 30 | Frog | Cao et al., 2011 |
| R = N-acetyl-D-Leu | 2110 | Bovine | Ping et al., 1995 |
| R = N-acetyl-L-Leu | 60 | Bovine | Ping et al., 1995 |
| R = N-acetyl-D-Phe | 2250 | Bovine | Ping et al., 1995 |
| R = N-acetyl-L-Phe | 45 | Bovine | Ping et al., 1995 |
| 6 | H889 cells – (E)b,d | Cao et al., 2011 | |
| >2000 | Frog | ||
| 5 | DMS53 cells – (E)b,c | Morris et al., 2012 | |
| 11 | DMS53 cells – (M)b,c | ||
| R = N-benzoyl-L-Val | 500 | Frog | Barratt et al., 2004 |
| R = N-acetyl-Gly (R = CH3CO-NH-CH2) |
1250 | Bovine | Ping et al., 1995 |
| 210 | DMS53 cells – (E)b,c | Morris et al., 2012 | |
| 1800 | DMS53 cells – (M)b,c | ||
| R = N-acetyl-L-Phe-L-Phe | 50 | Frog | Barratt et al., 2004 |
| R = N-acetyl-L-Leu-Gly | 120 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 200 | DMS53 cells – (M)b,c | ||
| R = N-acetyl-L-Leu-Phe | 3 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 9 | DMS53 cells – (M)b,c | ||
| R = N-acetyl-L-Pro-L-Leu-Glyh | 76 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 180 | DMS53 cells – (M)b,c | ||
| R = N-acetyl-L-Pro-L-Leu-Pheh | 6 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 38 | DMS53 cells – (M)b,c | ||
| R = CYIQNCPLGh All amino acids are L- and a -S-S-(disulfide bond) between the two Cys residues | 12 | DMS53 cells – (E)b,c | Morris et al., 2012 |
| 71 | DMS53 cells – (M)b,c | ||
| R = CYIQNCPLFh All amino acids are L- and a -S-S-(disulfide bond) between the two Cys residues |
2 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 23 | DMS53 cells – (M)b,c | ||
| R = L-Pro | 600 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 500 | DMS53 cells – (M)b,c | ||
| R = L-Phe | 1100 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 1600 | DMS53 cells – (M)b,c | ||
| R = L-Ala-L-Pro | 370 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 290 | DMS53 cells – (M)b,c | ||
| R = L-Ala-L-Phe | 350 | DMS53 cells – (E)b,c |
Morris et al., 2012
|
| 370 | DMS53 cells – (M)b,c | ||
| R = Gly-L-Ala-L-Proi | 980 | DMS53 cells – (E)b,c | Morris et al., 2012 |
| 1100 | DMS53 cells – (M)b,c | ||
| R = Gly-L-Ala-L-Phei | 560 | DMS53 cells – (E)b,c | Morris et al., 2012 |
| 160 | DMS53 cells – (M)b,c | ||
| B. Thioglycolates R-CO-S-CH2-COOH | Ki or IC50 (μM) | Source | Ref. |
| R = C6H5 | 150 | Frog | Cao et al., 2011 |
| R = C6H5-CH2 | 20 | Frog | Cao et al., 2011 |
| 45 | H889 cells – (E)b,d | ||
| R = CH3-(CH2)8 | 9 | Frog | Cao et al., 2011 |
| 7 | H889 cells – (E)b,d | ||
| R = N-acetyl-L-Met | 50 | Frog | Cao et al., 2011 |
| R = N-acetyl-L-Leu | 260 | Frog | Cao et al., 2011 |
| R = N-acetyl-L-Phe | 25 | Frog | Cao et al., 2011 |
| 45 | H889 cells – (E)b,d | ||
| C. S-(Thiocarbonyl)thioglycolates R-CS-S-CH2-COOH | Ki or IC50 (μM) | Source | Ref. |
| R = C6H5 | 40 | Rat MTC | Merkler et al., 2008 |
| 92 | German cockroachj | ||
| R = 4-CH3-C6H4 | 4 | Rat MTC | Merkler et al., 2008 |
| 14 | German cockroachj | ||
| R = C6H5-CH2 | 8 | Rat MTC | Merkler et al., 2008 |
| 4 | German cockroachj | ||
| R = C6H5-NH | 9 | Rat MTC | Merkler et al., 2008 |
| 5 | German cockroachj | ||
| R = C6H5-CH2-CH2 | 9 | Rat MTC | Merkler et al., 2008 |
| 0.9 | German cockroachj | ||
| R = CH3-(CH2)10 | 0.5 | Rat MTC | Merkler et al., 2008 |
See the indicated references to determine if purified PHM, partially purified PHM, or lysates were used in the specific study.
PHM is obtained from either a cell extract (E) or cell-conditioned medium (M). Much work has shown that PHM, PAL and bifunctional PAM are quite stable in cell-conditioned medium, the source of all the purified recombinant PAM-related proteins (Bauman et al., 2007; Chufan et al., 2009; Miller et al., 1992; Prigge et al., 1997). In contrast, cell extracts often contain a complex mixture of partially processed PAM proteins (Kumar et al., 2016).
DMS53 cells are human small cell lung cancer cells
H889 cells are human small cell lung cancer cells
MTC = medullary thyroid cancer
PC3 cells are human prostate cancer cells
SW1783 cells are human brain cancer cells
Pro-oxytocin related peptide sequences
Pro-calcitonin related peptide sequences
Blattella germanica
3A.7. Cao et al. (Cao et al., 2011) synthesized a series of thioglycolates, R-CO-S-CH2-COOH, to determine any effect of the sulfur atom on the affinity of the PHM inhibitor (Fig. 3A and Table 1B). In general, the thioglycolates inhibit with higher affinity, with a decrease in the IC50 value by a factor of 2-25 relative to the corresponding glycolate. The greatest difference was observed for the N-acetyl-Phe-O-CH2-COOH/N-acetyl-Phe-S-CH2-COOH pair, with a ratio of IC50 values > 80 for frog PHM. Again, these results suggest that sufficient differences exist between PHM proteins to enable the design of an inhibitor targeted against PHM from a specific species.
3A.8. PHM inhibition by the glycolates and the thioglycolates coupled with the discovery of N-acylglycines as PHM substrates lead to a structurally related set of PHM inhibitors, the S-(thiocarbonyl)thioglycolates (R–CS–S–CH2–COOH) (Fig. 3D and Table 1C). S-(Thiolauroyl)thioglycolate, CH3-(CH2)10-CS-S-CH2-COOH, is the tightest binding inhibitor within this set of compounds, with a Ki = 540 nM, a relatively high affinity for a ground-state, substrate analog. A direct comparison between structurally related glycolates, thioglycolates, and the S-(thiocarbonyl)thioglycolates to define whether PHM binds most tightly to either the-CS-S-group, the-CO-S-group, or the-CO-O-group is not possible because the PHM protein used in each study was different. For example, the differences in Ki or IC50 values for C6H5-CH2-CS-S-CH2-COOH (8 μM for rat MTC PHM), C6H5-CH2-CO-S-CH2-COOH (20 μM for frog PHM), and C6H5-CH2-CO-O-CH2-COOH (2 μM for human H889 cell PHM) might result from the different sources of PHM. The S-(thiocarbonyl)thioglycolate study does provide a direct comparison of Ki values between rat MTC PHM and German cockroach PHM for six of these inhibitors; differences were found, with C6H5-CH2-CS-S-CH2-COOH binding to cockroach PHM having ~10-fold higher affinity (Table 1). Again, these data point towards the future development of inhibitors targeting PHM in a species-specific manner.
3A.9. In their focus on the synthesis of PHM inactivators designed to capture the substrate-based radical that likely forms during catalysis, the Vederas group identified new PHM inhibitors. Included in this group are D-Phe-L-Phe-α-cyclopropylglycine (Andrews, O’Callaghan, & Vederas, 1997) and tripeptides with a C-terminal D- or L-styrylglycine (Zabriskie et al., 1992) (Fig. 3E). PHM is inactivated by trans-4-phenyl-3-buteonate (trans-styrylacetate) (Bradbury, Mistry, Roos, & Smyth, 1990). The cyclopropylglycine-containing tripeptide does not inactivate PHM and binds with low affinity, Ki > 5 mM, additional evidence that PHM does not readily accommodate any substitution on the glycyl α-carbon. None of the styrylglycine-containing peptides inactivate PHM, but all inhibit with IC50 values of 100 to 450 μM. This indicates that the styryl moiety must be optimally positioned in the PHM active site for inactivation to occur. Even a small repositioning of the styryl group eliminates its ability to inactivate PHM; neither N-acetyl-D- nor N-acetyl-L-styrylglycine inactivates PHM. Little difference in the IC50 values of the L- and D-styrylglycine containing inhibitors was found (Zabriskie et al., 1992). Thus, the importance of the stereochemistry at the penultimate position for PHM affinity found for the glycine-extended substrates and the glycolate inhibitors (Ping et al., 1995) was not observed for the styrylglycine-containing inhibitors. The significance of these differences is currently unknown and could be resolved by in silico modeling or crystallographic analysis of appropriate PHM-ligand complexes. The information gained from such studies could prove useful for the future design of tight-binding PHM inhibitors.
3A.10. One clear outcome from the work carried out on PHM inhibitors is preference for hydrophobicity in groups attached to the glycine or glycine analog. Consistent with this observation are reports of hydrophobic organic acids that inhibit PAM with relatively low affinity Examples from this group of PHM inhibitors include 1-(carboxymethyl)-3,5-diphenyl-2-methylbenzene (IC50 = 550 μM) (Cutler et al., 1998), urocanic acid (KI = 10 mM) (Merkler et al., 2008), and 4-pentenoic acid (IC50 = 60 mM) (Rhodes & Honsinger, 1993).
3A.11. Mimosine, a toxic, heterocyclic, non-protein amino acid produced by plants (Fig. 3F) (Nguyen & Tawata, 2016), inhibits both PHM and dopamine β-monooxygenase (DBM), a structurally and mechanistically-related enzyme (Klinman, 2006; Southan & Kruse, 1989; Hashiguchi & Takahashi, 1977). Docking of ascorbate to the open form of DBM suggests that specific binding requires the presence of dopamine, with ascorbate positioned between the two copper atoms (Vendelboe et al., 2016). In the PHM reaction, mimosine (KI 4.0 μM) acts as a competitive inhibitor of ascorbate (Miller et al., 1992), which plays an essential role in both PHM and DBM. Recent structural and modeling studies suggest that defining the manner in which mimosine binds to PHM and to DBM may facilitate the design of novel, mimosine-based inhibitors (Maheshwari et al., 2018; Wu et al., 2019; Vendelboe et al., 2016). The earliest structural studies of PHM revealed a single open state, with the CuH and CuM sites separated by an 11 Å cleft (Prigge et al., 1997; Prigge et al., 2004; Prigge et al., 2000). In contrast, both open and closed states were observed for DBM, with 12.7 Å separating its two copper ions in the open state, but only 5.1 Å separating them in its closed state (Vendelboe et al., 2016) (PDB ID: 4ZEL). Transition between these two states requires rotation of the CuH subdomain of DBM (Vendelboe et al., 2016). Structural studies focused on mutations targeted to the CuH site of PHM revealed that crystallization of PHM H108A in the presence of citrate produced a similar rotation and structural reconfiguration, yielding a closed state, with His107, through citrate, coordinating to CuM (Maheshwari et al., 2018) (PDB ID: 6AO6). Modelling studies suggest that the binuclear copper site found in the closed conformation of DBM is essential for catalysis, with DBM readily transforming between its open and closed states (energy barrier ~1.3 kcal/mol); this energy barrier is higher for PHM (~2.1 kcal/mol), with its copper atoms separated by 6.7 Å (Wu et al., 2019).
3B.1. PAL inhibitors:
The metals bound to PAL remain bound during its purification. Like PHM, metal chelators can inhibit PAL by removing the bound Zn(II). Activity is restored to inactive apo-PAL by the addition of Zn(II) and other divalent metal ions (Bell et al., 1997). Unlike PHM, PAL does not exhibit stereochemical preference at the penultimate position; the kinetic constants for N-acetyl-L-Phe-α-hydroxyglycine and N-acetyl-D-Phe-α-hydroxyglycine are very similar (Ping et al., 1995). C-Terminal pyruvate-extended amino acids, R-CO-CH2-CO-COOH, are inhibitors of PAL with Ki values >15 μM (Fig.3G). N-Acetyl-L-Phe-pyruvate is the best PAL inhibitor reported, with a Ki of 0.24 μM. The pyruvate-extended amino acids weakly inhibit PHM, being competitive with ascorbate and a ~100-fold lower potency (Mounier et al., 1997). The related compound, C6H5-CH=CH-SO-CH2-COOH (trans isomer), shows no inhibition or inactivation of PHM at 3 mM, but was not evaluated as a PAL inhibitor (Casara, Banzhorn, Philippo, Chanal, & Danzin, 1996). We are not aware of any other work to develop a PAL-specific inhibitor.
4.1. PHM Inactivators
There are important differences between an enzyme inhibitor (I) and an enzyme inactivator (IN). An inhibitor forms a reversible enzyme-inhibitor complex: E + I ⇆ E-I. An inhibitor shows no time-dependence in the percent inhibition at any inhibitor concentration. An inactivator forms an irreversible enzyme-inactivator complex (E-IN)* from the reversible E-IN complex: E + IN ⇆ E-IN → (E-IN)*. Typically, the (E-IN)* complex results from the formation of a covalent bond between the inactivator and the enzyme. At any concentration of inactivator, the degree of inhibition increases over time as higher concentrations of (E-IN)* accumulate (Morrison & Stone, 1985).
4.2. The first report of a PHM inactivator was trans-4-phenyl-3-butenoate (PBA) (Bradbury, Mistry, Roos, et al., 1990) (Fig.3H). In addition to PBA and ring-substituted PBAs (Langella et al., 2010), other PHM inactivators include trans-styrylthioacetate (Casara et al., 1996), 2-[(phenylethynyl)thio]acetate (Casara et al., 1996), acrylates (Foster, Oldham, & May, 2011; Katopodis & May, 1990b; Rhodes & Honsinger, 1993), monoethyl fumarate (Katopodis & May, 1990b), 2-, 3-, and 2,4-alkenoates (Rhodes & Honsinger, 1993), cinnamate and ring-substituted cinnamates (Bradbury, Mistry, & Smyth, 1990; McIntyre et al., 2016), N-formyl amides (Klinge, Cheng, Zabriske, & Vederas, 1994), and peptides with a C-terminal vinylglycine (Zabriskie et al., 1994). Treatment of cultured mammalian cells and rats with PBA inhibits PHM activity and the biosynthesis of α-amidated peptides (Abou-Mohamed et al., 2000; Ogonowski et al., 1997). The methyl ester prodrug of PBA was ~10-fold more effective than PBA in inhibiting the growth of tumorigenic rat liver epithelial cells (WB-Ras cells) (Sunman, Foster, Folse, May, & Matesic, 2004).
4.3. Most of the PHM inactivators were designed as mechanism-based inactivators to trap the radical intermediate that is likely to form during catalysis (Cao & Easton, 2013; Cowley et al., 2016). These are mechanism-based inactivators because O2 and ascorbate are required for inactivation and peptide substrates protect against inactivation. However, the chemistry of inactivation is unclear (Cao & Easton, 2013; Driscoll et al., 2000; McIntyre et al., 2016; Zabriskie et al., 1992). As expected PAL is unaffected by the PHM inactivators (McIntyre et al., 2016; Moore & May, 1999). Attempts to label PHM using either a 14C-labeled or fluorescently-tagged inactivator have been unsuccessful, with the exception of one early study of PBA-mediated inactivation, in which the labeled protein was not subjected to peptide mapping. Peptide-mapping of cinnamate-inactivated PHM showed no differences compared to the untreated control (McIntyre et al., 2016). We are unaware of any PAL-specific inactivators.
5.1. What have we learned towards the design of a PAM targeted therapeutic
PAM is gaining acceptance as a reliable biomarker for certain slow-growing tumors. A number of markers have been reported in neuroendocrine neoplasms, including several peptide hormones (e.g. adrenocorticotropic hormone [ACTH], growth hormone, adrenomedullin) and secretory granule proteins (e.g. chromogranins and synaptophysin) (Horton et al., 2020; Kaufmann, Bergmann, & Melander, 2021; Scopsi et al., 1998). Recent interest has focused on PAM as a reliable marker for this set of neoplasms whose prevalence exceeds the combined numbers of various gastrointestinal cancers (Horton et al., 2020; Kaufmann et al., 2021; Scopsi et al., 1998). A fascinating finding is that PAM appears to be quite protective in some studies, with lower PAM levels correlating with increased risk of fatality (Horton et al., 2020; Kaufmann et al., 2021). Earlier studies found that the highest PAM enzyme activity and protein levels were found in tumors producing amidated peptides, which were often the slowest growing tumors (Horton et al., 2020; Kaufmann et al., 2021; Scopsi et al., 1998; Vos, Scott, Iwai, & Treston, 1996). Elevated serum PAM activity has also been reported to correlate strongly with high blood pressure and cardiovascular mortality (Horton et al., 2020; Kaufmann et al., 2021).
5.2. When viewed collectively, patterns emerge from the studies of the PHM inhibitors and inactivators. Clearly, PHM prefers a free carboxylate conjugated to a hydrophobic moiety positioned as close as possible to the penultimate amino acid for the glycine-extended peptide substrates. Incorporation of a sulfur-containing group that can coordinate with one of the PHM-bound copper atoms would likely increase binding affinity. Other possibilities to increase binding affinity in future PHM inhibitors/inactivators would be to link mimosine (or a mimosine analog) to a compound that binds at the peptide site to create a bifunctional inhibitor. The identification of a putative mimosine-binding site in conjunction with the published PHM and DBM structures (Francisco, Blackburn, & Klinman, 2003; Prigge et al., 2004; Vendelboe et al., 2016) and the wealth of structure-activity data for PHM substrates and inhibitors provide an excellent starting point for in silico modeling of high affinity PHM inhibitors or inactivators. Another possible bifunctional inhibitor could include a copper-chelator linked to a compound that binds at the peptide site in PHM. Inhibitors/inactivators with high affinity unique to PAM could possess a PHM binder linked to the PAL-specific pyruvate-extended amino acids. Without a structure for bifunctional PAM, a series of compounds with different length spacers between the PHM inhibitor and the PAL inhibitor would be required to define the compound with the highest affinity for PAM. The appropriate incorporation of mimosine into a PAM inhibitor could yield a tight-binding trifunctional inhibitor with a group that binds into the mimosine site of PHM, the peptide site of PHM, and the substrate site of PAL.
5.3. The development of a high affinity PHM (or PAM) inhibitor will encounter significant hurdles that could hinder clinical use. One concern is delivery. PHM is found within the lumen of the secretory pathway (Kumar, Mains, et al., 2016), a challenging site for drug delivery. As discussed above, the clinical use of a PHM inhibitor is likely dependent upon a molecular zip code for the secretory system or the surface of specific cell types. Perhaps an engineered version of Shiga toxin might enable the delivery of a high affinity PHM/PAM inhibitor to the secretory system (Luginbuehl, Meier, Kovar, & Rohrer, 2018). Another concern is the diversity of the amidated products produced in vivo by PHM. The inhibition of PHM would produce unselective blocking of the biosynthesis of many amidated peptides and lipids. One solution to this concern is not a PHM inhibitor, but the development of inhibitor that binds selectively and with high affinity to one glycine-extended substrate, which inhibits the amidation of only one PHM substrate (Weiss, McIntyre, McLaughlin, & Merkler, 2006).
5.4. Inactivators require at least one trans-olefinic bond positioned β- to a carboxylate for the most efficient inactivation. Inactivators with the highest affinity have the inactivating moiety attached to the C-terminus of a peptide or a hydrophobic group like a phenyl group. The hydrophobic group must be appropriately spaced away from the inactivating species for the highest affinity, exactly as was observed for the glycine-extended substrates and the PHM inhibitors. Again, the available structure-activity data and PHM structures provide an excellent backdrop for the in silico design of high affinity inactivators. The decoration of such high affinity inactivators with the appropriate imaging reagent could yield a PHM-specific imaging reagent, demonstrating that the molecular targeting was in fact successful. Another application would be decoration with biotin to enable the profiling of PHM similar to the activity-based profiling strategies developed by Cravatt et al. (Cravatt, Wright, & Kozarich, 2008). Additional research is required to unravel uncertainties in the PHM inactivation chemistry to fully exploit the development of a PHM-specific imaging reagent.
5.5. As stated above, resistance to the clinical use of a PAM inhibitor results from concerns over toxicity and delivery. The potential therapeutic uses of a PAM inhibitor indicate that solutions to these concerns should be sought. A PAM inhibitor would find use as a treatment of cancer, specifically cancers of the prostate (Merkler et al., 2008; Rocchi et al., 2004) and lung (Iwai et al., 1999; Sunman et al., 2004). In a study of >140 neuroendocrine tumors, Scopsi et al. (Scopsi et al., 1998) found >20 tumors that expressed relatively high levels of PAM and were shown to produce α-amidated peptides; there were also tumors not shown to produce such peptides, suggesting that PAM inhibitors could find therapeutic use in treating a wide range of tumors. The first report of a rationally designed PAM inhibitor was targeted to block the formation of substance P (Erion et al., 1994), an 11-amino acid α-amidated peptide involved the perception of pain, rheumatoid arthritis, and inflammatory disease (Chang, Leeman, & Niall, 1971; Mashaghi et al., 2016; Zieglgänsberger, 2019). Subsequent studies have shown that decreasing the production of substance P by inhibiting PAM can be useful in the treatment of pain, inflammation, and rheumatoid arthritis (Bauer et al., 2007; Ogonowski et al., 1997).
6.1. The bigger picture – PAM from green algae to human
PAM and other granule membrane proteins are deposited on the cell surface during the process of exocytosis (Fig.4A). Immature granules contain PAM and soluble cargo proteins. During the maturation process, cargo proteins are further modified; the granules move from the Golgi region towards the cell surface and acquire the cytosolic proteins needed to make them responsive to the right stimuli. Any PAM protein in the granule membrane appears on the cell surface as the soluble cargo proteins are released into extracellular space (Fig.4A, Exocytosis). Under normal conditions, PAM is rapidly removed from the plasma membrane (Fig.4A, Endocytosis) and returned to the secretory granules via a complex series of steps through the endocytic pathway. Clathrin coated vesicles mediate the removal of PAM from the cell surface and detailed studies of PAM trafficking in corticotrope tumor cells have tracked the internalized protein as it moves from early endosomes into late endosomes (Bäck et al., 2017; Rajagopal, Stone, Francone, Mains, & Eipper, 2009). Whether the PAM protein that exits late endosomes is degraded in lysosomes or is recycled to immature granules (Fig.4A) is determined by O-linked sugars attached to the linker region between PHM and PAL, by a conserved pH-sensitive region that follows PHMcc and by the phosphorylation status of the PAM cytosolic domain (Rajagopal et al., 2009; Rao, Zavala, Deb Roy, Mains, & Eipper, 2019; Vishwanatha, Bäck, Mains, & Eipper, 2014; Vishwanatha, Bäck, Lam, Mains, & Eipper, 2016). Targeting its O-linked sugars or its pH-sensitive region could provide a means of preventing the return of PAM to the secretory pathway.
Figure 4.

A. PAM trafficking in neuroendocrine cells. As newly synthesized PAM and soluble cargo proteins exit the trans-Golgi, they accumulate in immature secretory granules. Granule maturation involves vesicular trafficking and acquisition of the cytosolic proteins needed to respond to secretagogues. Upon fusion of the secretory granule membrane with the plasma membrane, soluble content proteins are released and membrane PAM appears on the cell surface. Clathrin-mediated endocytosis means that less than 5% of the PAM protein in a cell typically resides on the plasma membrane. PAM retrieved from the cell surface can be degraded or returned to the secretory pathway for re-use. B. In C. reinhardtii, PAM is localized to the Golgi Complex, small vesicular structures and the ciliary membrane. The ciliary budding process that generates ectosomes is illustrated, with the localization of CrPAM and one of its amidated products (Cre03.g20450) illustrated (Luxmi et al., 2018; Luxmi et al., 2019). C. The movement of PAM through late endosomes and into the intraluminal vesicles (ILVs) that form in multivesicular bodies (MVBs) was determined using ectodomain antibodies (Bäck et al., 2017; Rajagopal, Mains, & Eipper, 2012). Upon fusion with the plasma membrane, PAM-containing exosomes are released. D. The species-specific roles of PAM in ciliogenesis are summarized. E. The non-catalytic effects of PAM are summarized. The ability of PAM to alter gene expression is thought to require the generation of sfCD through γ-secretase-catalyzed regulated intramembrane proteolysis (RIP) (Rajagopal, Stone, Mains, & Eipper, 2010). Neither the ability of PAM to support the formation of atrial granules or its ability to interact with actin require its catalytic activity.
6.2. In a number of species, genes encoding membrane PAM and soluble PAM, PHM and/or PAL are expressed. This is especially true in species which rely on amidated peptides to attack their prey and produce a multitude of very species-specific venoms which usually do not harm the species producing the venom (Mackieh et al., 2021; Terlau & Olivera, 2004; Turner, Kaas, & Craik, 2020; Ul-Hasan et al., 2013). Scorpions, for example, have retained separate genes encoding integral membrane PAM, soluble PHM and soluble PAL (Delgado-Prudencio, Possani, Becerril, & Ortiz, 2019), leading the authors to speculate that the PHM secreted in scorpion venom might catalyze some of these additional reactions.
6.3. Compounds that stimulate the secretion of peptide hormones and neurotransmitters stored in granules increase the amount of PAM protein delivered to the plasma membrane (Fig.4A, lightning bolt). Tumor cells often secrete autocrine growth factors, many of which are amidated peptides (Jimenez et al., 2001). The appearance of PAM on the surface of these cells provides an opportunity for cell-impermeant PHM inhibitors or PAM-targeted tags that can be tracked in vivo to bind to its ectodomain and enter the endocytic pathway along with PAM. Internalization of a PHM inhibitor would be expected to reduce the synthesis of bioactive amidated growth factors while ectodomain antibodies or active-site targeted tags might facilitate tumor cell localization (Fig.4A). An inactive prodrug might be expected to bind to PAM while it is exposed on the cell surface, with drug activation designed to occur in the lower pH environments encountered during endocytic trafficking. As discussed above, with an understanding of the structure of bifunctional PAM, the design of dual function drugs (targeting PHM and PAL) might provide a tool capable of preventing the endocytic trafficking needed to return PAM to the secretory pathway. Outside of its active sites, the sequences of insect and human PHM may be divergent enough to design species specific inhibitors useable as insecticides.
6.4. Phylogenetic studies have suggested several different situations in which controlling PAM activity or localization could be beneficial. While the active sites of C. reinhardtii PAM (CrPAM) include each of the residues identified as essential in vertebrate PHM and PAL, this unicellular green alga does not store peptides in secretory granules (Fig.4B). CrPAM is localized to the membranes of the Golgi complex, as observed in vertebrates, but the presence of two prominent motile cilia made it clear that CrPAM is also localized to the ciliary membrane (Fig.4B) (Kumar, Blaby-Haas, et al., 2016). Unlike the enzymes used by C. reinhardtii to respond to the need for various nutrients, neither PAM activity nor PAM protein is secreted through the classical secretory pathway (Kumar et al., 2017; Luxmi et al., 2019). In vertebrate systems, endoproteolytic cleavages that separate the PHM and PAL domains from the transmembrane domain result in their secretion under both basal and stimulated conditions (Kumar, Mains, et al., 2016). During vegetative growth, C. reinhardtii rely on bioactive ectosomes released from their cilia (Fig.4B, Inset) to destroy the mother cell wall (Wood, Huang, Diener, & Rosenbaum, 2013). Ciliary ectosomes also play a key role during sexual reproduction in C. reinhardtii (Cao et al., 2015). Active CrPAM is present in mating ectosomes, along with its amidated products, one of which acts as a chemomodulator (Luxmi et al., 2019) (Fig.4B). PAM was subsequently identified in the ciliary membranes of vertebrate motile and sensory cilia (Kumar et al., 2017). The formation of ciliary ectosomes (Fig.4B) is topologically similar to the formation of intraluminal vesicles in the endocytic pathway (Fig.4C). Fusion of a multivesicular body containing intraluminal vesicles results in their release as exosomes (Fig.4C). PAM has been identified in intraluminal vesicles (Bäck et al., 2017) and in exosomes isolated from human saliva and urine (Gonzalez-Begne et al., 2009; Principe et al., 2013; Wang, Hill, Luther, Hachey, & Schey, 2012). Much remains to be learned about the role played by ectosomes/exosomes, but the presence of PAM in these structures offers a means of isolating and characterizing that subset.
6.5. When PAM expression was reduced in C. reinhardtii, the cells were unable to form cilia (Fig.4D) (Kumar et al., 2017). Instead of their normal pair of motile 12 μm long cilia (Fig.4B), PAM knockdown cells had ciliary stubs containing microtubule fragments. The ciliary axoneme did not form and the appearance of the transition zone was altered. In humans, fully 5% of the genome is thought to be involved in the formation and function of cilia and ciliopathies are associated with many congenital diseases including obesity, blindness and kidney disease (Engle, Bansal, Antonellis, & Berbari, 2021; Leroy et al., 2021; Nager et al., 2017). The effects of reducing PAM levels on ciliogenesis are species specific, but occur in planaria, zebrafish and mouse (Fig.4D). The hypothalamic neurons that produce proopiomelanocortin must have cilia and the melanocortin 4 receptor must be localized to those cilia in order to regulate appetite normally (Wang et al., 2021). Much remains to be learned about the sensory and signaling roles of cilia and tools that allow the manipulation of ciliary PAM would be of great utility in these studies.
6.6. It is now known that the PAM protein has several non-catalytic functions (Fig.4E). Regulated intramembrane proteolysis of PAM releases a soluble fragment of its cytosolic domain (sfCD). Nuclear accumulation of sfCD depends on its phosphorylation status and is thought to contribute to the ability of PAM to alter gene expression in a tissue-specific manner (Rajagopal et al., 2009). In atrial myocytes, which express PAM at levels higher than those observed in other cells, the PAM protein (whether active or inactive) plays an essential role in the formation of atrial granules, which store natriuretic peptides (Bäck, Luxmi, Powers, Mains, & Eipper, 2020). The ability of PAM to function as a re-usable luminal cargo receptor for proatrial natriuretic peptides early in the secretory pathway is thought to be essential to its ability to support granulogenesis.
Acknowledgments
We thank the many lab members and colleagues over several decades whose insights and hard work contributed to these studies. Supported by: NIH R01-DK032948 and R01-DK032949 (BAE, REM); NIH R01-GM125606 (BAE, Stephen M. King); The Daniel Schwartzberg Fund (BAE, REM); NIH R21-GM140390 and R15-GM073659 (DJM); Eppley Foundation for Research (DJM); Milheim Foundation for Cancer Research (DJM); Unigene Laboratories, Inc. (DJM).
Declaration of transparency and scientific rigor
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigor of preclinical research as stated in the BJP guidelines for Design and Analysis, and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
Abbreviations:
- ACTH
adrenocorticotropic hormone
- CHO
Chinese hamster ovary
- CrPAM
Chlamydomonas reinhardtii PAM
- CuH
copper bound to the Histidine (N-terminal half) site in PHM
- CuM
copper bound to the Methionine-Histidine (C-terminal half) site in PHM
- PAL
peptidyl-α-hydroxyglycine α-amidating lyase
- PALcc
catalytic core of PAL
- PAM
peptidylglycine α-amidating monooxygenase
- PBA
trans-4-phenyl-3-butenoate
- PDB
Protein Data Bank; https://www.rcsb.org
- PHM
peptidylglycine α-hydroxylating monooxygenase
- PHMcc
catalytic core of PHM
- POMC
proopiomelanocortin
- sfCD
soluble fragment of the cytosolic domain of PAM
Footnotes
Nomenclature of Targets and Ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, and are permanently archived in the Concise Guide to PHARMACOLOGY 2021/22 (Alexander et al., 2021).
Conflict of Interest
All authors declare no conflict of interest.
Literature
- Abou-Mohamed GA, Huang J, Oldham CD, Taylor TA, Jin L, Caldwell RB, … Caldwell RW (2000). Vascular and Endothelial Actions of Inhibitors of Substance P Amidation. J. Cardiovascular Pharmacol, 35, 871–880. [DOI] [PubMed] [Google Scholar]
- Ali A, Burns TJ, Lucrezi JD, May SW, Green GR, & Matesic DF (2015). Amidation inhibitors 4-phenyl-3-butenoic acid and 5-(acetylamino)-4-oxo-6-phenyl-2-hexenoic acid methyl ester are novel HDAC inhibitors with anti-tumorigenic properties. Invest. New Drugs, 33, 827–834. [DOI] [PubMed] [Google Scholar]
- Andrews MD, O’Callaghan KA, & Vederas JC (1997). Synthesis of Tripeptide Inhibitors of Peptidylglycine α-Amidating Monooxygenase (PAM) Containing D- and L-Styrylglycine. Tetrahedron, 52, 8295–8306. [Google Scholar]
- Bäck N, Kanerva K, Vishwanatha KS, Yanik A, Ikonen E, Mains RE, & Eipper BA (2017). The endocytic pathways of a secretory granule membrane protein in HEK293 cells: PAM and EGF traverse a dynamic multivesicular body network together. Eur.J.Cell.Biol, 86, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäck N, Luxmi R, Powers KG, Mains RE, & Eipper BA (2020). Peptidylglycine α-Amidating Monooxygenase Is Required for Atrial Secretory Granule Formation. Proc Natl Acad Sci U S A, 117, 17820–17831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäck N, Mains RE, & Eipper BA (2021). Organism and Cell Type Specific Uses of Peptidylglycine α-Amidating Monooxygenase (PAM). The FEBS J, doi: 10.1111/febs.16049 [DOI] [Google Scholar]
- Barratt BJ, Easton CJ, Henry DJ, Li IH, Radom L, & Simpson JS (2004). Inhibition of peptidylglycine alpha-amidating monooxygenase by exploitation of factors affecting the stability and ease of formation of glycyl radicals. J Am Chem Soc, 126(41), 13306–13311. [DOI] [PubMed] [Google Scholar]
- Bauer JD, Sunman JA, Foster MS, Thompson JR, Ogonowski AA, Cutler SJ, et al. (2007). Anti-inflammatory effects of 4-phenyl-3-butenoic acid and 5-(acetylamino)-4-oxo-6-phenyl-2-hexenoic acid methyl ester, potential inhibitors of neuropeptide bioactivation. J Pharmacol Exp Ther, 320(3), 1171–1177. [DOI] [PubMed] [Google Scholar]
- Bauman AT, Ralle M, & Blackburn NJ (2007). Large scale production of the copper enzyme peptidylglycine monooxygenase using an automated bioreactor. Protein Expr Purif, 51(1), 34–38. [DOI] [PubMed] [Google Scholar]
- Bell J, Ash DE, Snyder LM, Kulathila R, Blackburn NJ, & Merkler DJ (1997). Structural and functional investigations on the role of zinc in bifunctional rat peptidylglycine a-amidating enzyme. Biochemistry, 36, 16239–16246. [DOI] [PubMed] [Google Scholar]
- Bertelsen AH, Beaudry GA, Galella EA, Jones BN, Ray ML, & Mehta NM (1990). Cloning and characterization of two alternatively spliced rat alpha-amidating enzyme cDNAs from rat medullary thyroid carcinoma. Arch Biochem Biophys, 279(1), 87–96. [DOI] [PubMed] [Google Scholar]
- Blackburn NJ, Rhames FC, Ralle M, & Jaron S (2000). Major changes in copper coordination accompany reduction of peptidylglycine monooxygenase: implications for electron transfer and the catalytic mechanism. JBIC, 5, 341–353. [DOI] [PubMed] [Google Scholar]
- Bolkenius FN, Ganzhorn AJ, Chanal M-C, & Danzin C (1997). Selective Mechanism-based Inactivation of Peptidylglycine α-Hydroxylating Monooxygenase in Serum and Heart Atrium vs. Brain. Biochem. Pharmacol, 53, 1695–1702. [DOI] [PubMed] [Google Scholar]
- Bousquet-Moore D, Prohaska J, Nillni EA, Czyzyk TA, Wetsel WC, Mains RE et al. (2010). Interactions of peptide amidation and copper: novel biomarkers and mechanisms of neural dysfunction. Neurobiology of Disease, 37, 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury AF, Finnie MDA, & Smyth DG (1982). Mechanism of C-terminal amide formation by pituitary enzymes. Nature, 298, 686–688. [DOI] [PubMed] [Google Scholar]
- Bradbury AF, Mistry J, Roos BA, & Smyth DG (1990). 4-Phenyl-3-butenoic acid, an in vivo inhibitor of peptidylglycine hydroxylase (peptide amidating enzyme). Eur J Biochem, 189(2), 363–368. [DOI] [PubMed] [Google Scholar]
- Bradbury AF, Mistry J, & Smyth DG (1990). Fluorescent Inhibitors of Peptidylglycine Hydroxylase. In Giralt E & Andreu D (Eds.), Peptides 1990, Proceedings of the Twenty-First European Peptide Symposium. , (pp. 763–765). Leiden, The Netherlands: ESCOM Science Publishers B.V. [Google Scholar]
- Bundgaard H, & Kahns AH (1991). Chemical stability and plasma-catalyzed dealkylation of peptidyl-a-hydroxyglycine derivatives-intermediates in peptide alpha-amidation. Peptides, 12(4), 745–748. [DOI] [PubMed] [Google Scholar]
- Cao F, & Easton CJ (2013). Production and regulation of levels of amidated peptide hormones. Aust. J. Chem, 66, 297–307. [Google Scholar]
- Cao F, Gamble AB, Kim HK, Onagi H, Gresser MJ, Kerr J et al. (2011). Potent and selective inhibitors of human peptidylglycine alpha-amidating monooxygenase. Med. Chem. Commun, 2, 760–763. [Google Scholar]
- Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK et al. (2015). Uni-directional ciliary membrane trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. eLife, e05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casara P, Banzhorn A, Philippo C, Chanal M-C, & Danzin C (1996). Unsaturated thioacetic acids as novel mechanism-based inhibitors of peptidylglycine alpha-hydroxylating monooxygenase. Bioorg. Medicinal Chem. Lett, 6, 393–396. [Google Scholar]
- Chang MM, Leeman SE, & Niall HD (1971). Amino-acid sequence of substance P. Nat New Biol, 232(29), 86–87. [DOI] [PubMed] [Google Scholar]
- Chen P, Bell J, Eipper BA, & Solomon EI (2004). Oxygen activation by the noncoupled binuclear copper site in peptidylglycine alpha-hydroxylating monooxygenase: spectroscopic definition of the resting sates and the putative CuIIM-OOH intermediate. Biochemistry, 43, 5735–5747. [DOI] [PubMed] [Google Scholar]
- Chew GH, Galloway LC, McIntyre NR, Schroder LA, Wright DW, & Merkler DJ (2005). Ubiquitin and ubiquitin-derived peptides as substrates for peptidylglycine alpha-amidating monooxygenase. FENS Lett, 579, 4678–4684. [DOI] [PubMed] [Google Scholar]
- Chrétien M, & Mbikay M (2016). 60 years of POMC: From the prohormone theory to pro-opiomelanocortin and to proprotein convertases (PCSK1 to PCSK9). J Mol Endocrinol, 56(4), T49–62. [DOI] [PubMed] [Google Scholar]
- Chufan EE, De M, Eipper BA, Mains RE, & Amzel LM (2009). Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure, 17(7), 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, & Lowry P (2016). 60 years of POMC: POMC: the consummate peptide hormone precursor. J Mol Endocrinol, 56(4), E1–2. [DOI] [PubMed] [Google Scholar]
- Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG et al. (2020). Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer, 1(2), 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley RE, Tian L, & Solomon EI (2016). Mechanism of O2 activation and substrate hydroxylation in noncoupled binuclear copper monooxygenases. Proc.Natl.Acad.Sci.USA, 113, 12035–12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, & Kozarich JW (2008). Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- Cutler SJ, DeWitt Blanton C Jr., Akin DT, Steinberg FB, Moore AB, Lott JA et al. (1998). Pharmacological evaluation of 1-(carboxymethyl)-3,5-diphenyl-2-methylbenzene, a novel arylacetic acid with potential anti-inflammatory properties. Inflamm Res, 47(7), 316–324. [DOI] [PubMed] [Google Scholar]
- Czyzyk TA, Ning Y, Hsu M-S, Peng B, Mains RE, Eipper BA et al. (2005). Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev. Biol, 287, 301–313. [DOI] [PubMed] [Google Scholar]
- Dai Q, Bertleff-Zieschang N, Braunger JA, Björnmalm M, Cortez-Jugo C, & Caruso F (2018). Particle Targeting in Complex Biological Media. Adv Healthc Mater, 7(1) [DOI] [PubMed] [Google Scholar]
- De M, Bell J, Blackburn NJ, Mains RE, & Eipper BA (2006). Role for an essential tyrosine in peptide amidation. jbc, 281, 20873–20882. [DOI] [PubMed] [Google Scholar]
- Delgado-Prudencio G, Possani LD, Becerril B, & Ortiz E (2019). The Dual α-Amidation System in Scorpion Venom Glands. Toxins (Basel), 11(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll WJ, Konig S, Fales HM, Pannell LK, Eipper BA, & Mueller GP (2000). Peptidylglycine-α-Hydroxylating Monooxygenase Generates Two Hydroxylated Products from Its Mechanism-Based Suicide Substrate, 4-Phenyl-3-butneoic Acid. Biochemistry, 39, 8007–8016. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Glembotski CC, & Mains RE (1983). Selective loss of alpha-melanotropin-amidating activity in primary cultures of rat intermediate pituitary cells. J. Biol. Chem, 258, 7292–7298. [PubMed] [Google Scholar]
- Eipper BA, Mains RE, & Glembotski CC (1983). Identification in pituitary tissue of a peptide alpha-amidation activity that acts on glycine-extended peptides and requires molecular oxygen, copper, and ascorbic acid. Proc. Natl. Acad. Sci. U. S. A, 80, 5144–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper BA, Park L, Keutmann HT, & Mains RE (1986). Amidation of joining peptide, a major pro-ACTH/endorphin-derived product peptide. J. Biol. Chem, 261, 8686–8694. [PubMed] [Google Scholar]
- Eipper BA, Park LP, Dickerson IM, Keutmann HT, Thiele EA, Rodriguez H et al. (1987). Structure of the precursor to an enzyme mediating COOH-terminal amidation in peptide biosynthesis. Mol. Endocrinol, 1, 777–790. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Perkins SN, Husten EJ, Johnson RC, Keutmann HT, & Mains RE (1991). Peptidyl-α-hydroxyglycine α-amidating lyase: purification, characterization, and expression. J. Biol. Chem, 266, 7827–7833. [PubMed] [Google Scholar]
- Eipper BA, Quon ASW, Mains RE, Boswell JS, & Blackburn NJ (1995). The catalytic core of peptidylglycine alpha-hydroxylating monooxygenase: investigation by site-directed mutagenesis, Cu X-ray absorption spectroscopy, and electron paramagnetic resonance. Biochemistry, 34, 2857–2865. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Stoffers DA, & Mains RE (1992). The biosynthesis of neuropeptides: peptide alpha-amidation. Annu. Rev. Neurosci, 15, 57–85. [DOI] [PubMed] [Google Scholar]
- Enbäck J, & Laakkonen P (2007). Tumour-homing peptides: tools for targeting, imaging and destruction. Biochem Soc Trans, 35(Pt 4), 780–783. [DOI] [PubMed] [Google Scholar]
- Engle SE, Bansal R, Antonellis PJ, & Berbari NF (2021). Cilia signaling and obesity. Semin Cell Dev Biol, 110, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion MD, Tan J, Wong M, & Jeng AY (1994). Inhibition of peptidylglycine alpha-amidating monooxygenase by N-substituted homocysteine analogs. J Med Chem, 37(26), 4430–4437. [DOI] [PubMed] [Google Scholar]
- Evans JP, Blackburn NJ, & Klinman JP (2006). The catalytic role of the copper ligand H172 of peptidylglycine alpha-hydroxylating monooxygenase: a kinetic study of the H172A mutant. Biochemistry, 45(51), 15419–15429. [DOI] [PubMed] [Google Scholar]
- Foster MS, Oldham CD, & May SW (2011). Looking Glass Mechanism-Based Inhibition of Peptidyglycine α-Amidating Monooxygenase. Tetrahedon Asymmetry, 22, 283–293. [Google Scholar]
- Francisco WA, Blackburn NJ, & Klinman JP (2003). Oxygen and hydrogen isotope effects in an active site tyrosine to phenylalanine mutant of peptidylglycine alpha-hydroxylating monooxygenase: mechanistic implications. Biochemistry, 42(7), 1813–1819. [DOI] [PubMed] [Google Scholar]
- Francisco WA, Merkler DJ, Blackburn NJ, & Klinman JP (1998). Kinetic Mechanism and Intrinsic Isotope Effects for the Peptidylglycine α-Amidating Enzyme Reaction. Biochemistry, 37, 8244–8252. [DOI] [PubMed] [Google Scholar]
- Gaier ED, Eipper BA, & Mains RE (2014). Pam heterozygous mice reveal essential role for Cu in amygdalar behavioral and synaptic function. Ann. N. Y. Acad. Sci, 1314, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaier ED, Kleppinger A, Ralle M, Mains RE, Kenny AM, & Eipper BA (2012). High serum Cu and Cu/Zn ratios correlate with impairments in bone density, physical performance and overall health in a population of elderly men with frailty characteristics. Exp. Gerontol, 47, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaier ED, Miller MB, Ralle M, Aryal D, Wetsel WC, Mains RE et al. (2013). Peptidylglycine alpha-amidating monooxygenase heterozygosity alters brain copper handling with region specificity. J. Neurochem, 127, 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauder J, Ragg H, Rauch J, & Engels JW (1990). Human peptidylglycine alpha-amidating monooxygenase: cDNA, cloning and functional expression of a truncated form in COS cells. Biochem. Biophys. Res. Commun, 169, 551–558. [DOI] [PubMed] [Google Scholar]
- Glembotski CC, Eipper BA, & Mains RE (1984). Characterization of a peptide alpha-amidation activity from rat anterior pituitary. J. Biol. Chem, 259, 6385–6392. [PubMed] [Google Scholar]
- Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE et al. (2009). Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J. Proteome Res, 8, 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez H, Ottervald J, Nilsson KC, Sjogren N, Miliotis T, Von Bahr H et al. (2009). Identification of novel candidate protein biomarkers for the post-polio syndrome-Implications for diagnosis, neurodegeneration and neuroinflammation. J. Proteomics, 71, 670–681. [DOI] [PubMed] [Google Scholar]
- Handa S, Spradling TJ, Dempsey DR, & Merkler DJ (2012). Production of the catalytic core of human peptidylglycine α-hydroxylating monooxygenase (hPHMcc) in Escherichia coli. Prot. Expr. Purif, 84, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi H, & Takahashi H (1977). Inhibition of two copper-containing enzymes, tyrosinase and dopamine beta-hydroxylase, by L-mimosine. Mol Pharmacol, 13(2), 362–367. [PubMed] [Google Scholar]
- Horton TM, Sundaram V, Lee CH, Hornbacker K, Van Vleck A, Benjamin KN et al. (2020). PAM staining intensity of primary neuroendocrine neoplasms is a potential prognostic biomarker. Sci Rep, 10(1), 10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husten EJ, & Eipper BA (1994). Purification and characterization of PAM-1, an integral membrane protein involved in peptide processing. Arch. Biochem. Biophys, 312, 487–492. [DOI] [PubMed] [Google Scholar]
- Husten EJ, Tausk FA, Keutmann HT, & Eipper BA (1993). Use of endoproteases to identify catalytic domains, linker regions and functional interactions in soluble PAM. J. Biol. Chem, 268, 9709–9717. [PubMed] [Google Scholar]
- Iwai N, Vos M, Miller MJ, Vos M, Mulshine JL, & Treston AM (1999). Autocrine growth loops dependent on peptidyl α-amidating enzyme as targets for novel tumor cell growth inhibitors. Lung Cancer, 23, 209–222. [DOI] [PubMed] [Google Scholar]
- Jaron S, & Blackburn NJ (1999). Does superoxide channel between the copper centers in PAM? A new mechanism based on carbon monooxide reactivity. Biochemistry, 38, 15086–15096. [DOI] [PubMed] [Google Scholar]
- Jeng AY, Fujimoto RA, Chou M, Tan J, & Erion MD (1997). Suppression of Substance P Biosynthesis in Sensory Neurons of Dorsal Root Ganglion by Prodrug Esters of Potent Peptidylglycine α-Amidating Monooxygenase Inhibitors. J. Biol. Chem, 272, 14666–14671. [DOI] [PubMed] [Google Scholar]
- Jiménez N, Abasolo I, Jongsma J, Calvo A, Garayoa M, van der Kwast TH et al. (2003). Androgen-independent expression of adrenomedullin and peptidylglycine alpha-amidating monooxygenase in human prostatic carcinoma. Mol Carcinog, 38(1), 14–24. [DOI] [PubMed] [Google Scholar]
- Jimenez N, Jongsma J, Calvo A, van der Kwast TH, Treston AM, Cuttitta F et al. (2001). Peptidylglycine α-Amidating Monooxygenase- and proadrenomedullin-derived peptide-associated neuroendocrine differentiation are induced by androgen deprivation in the neoplastic prostate. In Int. J. Cancer (Vol. 9, pp. 28–34). (Reprinted from: Not in File). [DOI] [PubMed] [Google Scholar]
- Johnson KA (2019). New Standards for Collecting and Fitting Steady State Kinetic Data. Beilstein J. Org. Chem, 15, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA, Paisley-Flango K, Tangarone BS, Porter TJ, & Rouse JC (2007). Cation exchange-HPLC and mass spectrometry reveal C-terminal amidation of an IgG1 heavy chain. Anal. Biochem, 360, 75–83. [DOI] [PubMed] [Google Scholar]
- Kahns AH, & Bundgaard H (1991). Prodrugs of peptides. 13. Stabilization of peptide amides against alpha-chymotrypsin by the prodrug approach. Pharm Res, 8(12), 1533–1538. [DOI] [PubMed] [Google Scholar]
- Katopodis AG, & May SW (1990a). Novel Substrates and Inhibitors of Peptidyglycine α-Amidating Monooxygenase. Biochemistry, 29, 4541–4548. [DOI] [PubMed] [Google Scholar]
- Katopodis AG, & May SW (1990b). Novel substrates and inhibitors of peptidylglycine alpha-amidating monooxygenase. Biochemistry, 29(19), 4541–4548. [DOI] [PubMed] [Google Scholar]
- Katopodis AG, Ping D, & May SW (1990). A novel enzyme from bovine neurointermediate pituitary catalyzes dealkylation of alpha-hydroxyglycine derivatives, thereby functioning sequentially with peptidylglycine alpha-amidating monooxygenase in peptide amidation. Biochemistry, 29, 6115–6120. [DOI] [PubMed] [Google Scholar]
- Kaufmann P, Bergmann A, & Melander O (2021). Novel insights into peptide amidation and amidating activity in the human circulation. Sci Rep, 11(1), 15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King L 3rd, Barnes S, Glufke U, Henz ME, Kirk M, Merkler KA et al. (2000). The enzymatic formation of novel bile acid primary amides. Arch Biochem Biophys, 374(2), 107–117. [DOI] [PubMed] [Google Scholar]
- Klinge M, Cheng H, Zabriske TM, & Vederas JC (1994). Irreversible Inhibition of Mammalian and Insect Peptidylglycine α-Hydroxylating Monooxygenases (PHMs), Peptide Amidating Enzymes, by N-Formyl Amides. J. Chem. Soc., Chem. Commun, 1994, 1379–1380. [Google Scholar]
- Klinman JP (2006). The Copper-Enzyme Family of Dopamine β-Monooxygenase and Peptidylglycine α-Hydroxylating Monooxygenase: Resolving the Chemical Pathway for Substrate Hydroxylation. J Biol. Chem, 281, 3013–3016. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Bell J, Shiozaki EN, Jin L, Keutmann HT, Hand TA et al. (2002). Essential features of the catalytic core of peptidyl-alpha-hydroxyglycine alpha-amidating lyase. Biochemistry, 41, 12384–12394. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Keutmann HT, Mains RE, Quon ASW, & Eipper BA (1997). Peptidyl a-hydroxylating monooxygenase: active site residues, disulfide linkages and a two-domain model of the catalytic core. Biochemistry, 36, 10901–10909. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Quon ASW, Berard CA, Mains RE, & Eipper BA (1998). Post-translational N-glycosylation of a truncated form of a peptide processing enzyme. jbc, 273, 23012–23018. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Roberts MS, Jiang N, Johnson RC, Mains RE, Eipper BA et al. (1997). Neuropeptide Amidation in Drosophila: Separate Genes Encode the Two Enzymes Catalyzing Amidation. J. Neurosci, 17, 1363–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, & Soyka M (2018). Diagnosis and Pharmacotherapy of Alcohol Use Disorder: A Review. Jama, 320(8), 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathila R, Merkler KA, & Merkler DJ (1999). Enzymatic formation of C-terminal amides. Nat. Prod. Rep, 16, 145–154. [DOI] [PubMed] [Google Scholar]
- Kumar A, Zhang X, & Liang XJ (2013). Gold nanoparticles: emerging paradigm for targeted drug delivery system. Biotechnol Adv, 31(5), 593–606. [DOI] [PubMed] [Google Scholar]
- Kumar D, Blaby-Haas C, Merchant S, King S, Mains RE, & Eipper BA (2016). Early eukaryotic origins for cilia-associated bioactive peptide-amidating activity. J.Cell Sci, 129, 943–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Mains RE, & Eipper BA (2016). 60 years of POMC: From POMC and α-MSH to PAM, molecular oxygen, copper, and vitamin C. J.Mol.Endocrinol, 56, T63–T76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Strenkert D, Patel-King RS, Leonard MT, Merchant SS, Mains RE et al. (2017). A bioactive peptide amidating enzyme is required for ciliogenesis. eLife, 6, e25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Thomason RT, Yankova M, Gitlin JD, Mains RE, Eipper BA et al. (2018). Microvillar and ciliary defects in zebrafish lacking an actin-binding bioactive peptide amidating enzyme. Sci.Rep, 4, 4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landymore-Lim AE, Bradbury AF, & Smyth DG (1983). The amidating enzyme in pituitary will accept a peptide with C-terminal D-alanine as substrate. Biochem Biophys Res Commun, 117(1), 289–293. [DOI] [PubMed] [Google Scholar]
- Langella E, Pierre S, Ghattas W, Giorgi M, Réglier M, Saviano M et al. (2010). Probing the peptidylglycine alpha-hydroxylating monooxygenase active site with novel 4-phenyl-3-butenoic acid based inhibitors. ChemMedChem, 5(9), 1568–1576. [DOI] [PubMed] [Google Scholar]
- Leroy BP, Birch DG, Duncan JL, Lam BL, Koenekoop RK, Porto FBO et al. (2021). Leber congenital amaurosis due to cep290 mutations-severe vision impairment with a high unmet medical need: A Review. Retina, 41(5), 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, & Zhang H (2019). Nanoparticle-Based Drug Delivery Systems for Enhanced Tumor-Targeting Treatment. J Biomed Nanotechnol, 15(1), 1–27. [DOI] [PubMed] [Google Scholar]
- Luginbuehl V, Meier N, Kovar K, & Rohrer J (2018). Intracellular drug delivery: Potential usefulness of engineered Shiga toxin subunit B for targeted cancer therapy. Biotechnol Adv, 36(3), 613–623. [DOI] [PubMed] [Google Scholar]
- Luxmi R, Blaby-Haas C, Kumar D, Rauniyar N, King SM, Mains RE et al. (2018). Proteases Shape the Chlamydomonas Secretome: Comparison to Classical Neuropeptide Processing Machinery. Proteomes, 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxmi R, Kumar D, Mains RE, King SM, & Eipper BA (2019). Cilia-based Peptidergic Signaling. PLoS Biology, 17, e3000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxmi R, Mains RE, King SM, & Eipper BA (2021). Peptidylglycine α-amidation monooxygenase (PAM). In Jez J (Ed.), Encyclopedia of Biological Chemistry, (pp. in press). London: Elsevier. [Google Scholar]
- Mackieh R, Abou-Nader R, Wehbe R, Mattei C, Legros C, Fajloun Z et al. (2021). Voltage-Gated Sodium Channels: A Prominent Target of Marine Toxins. Mar Drugs, 19(10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S, Shimokawa C, Rudzka K, Kline CD, Eipper BA, Mains RE et al. (2018). Effects of copper occupancy on the conformational landscape of peptidylglycine α-hydroxylating monooxygenase. Commun.Biol, 1, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains RE, & May V (1988). The role of a low pH intracellular compartment in the processing, storage, and secretion of ACTH and endorphin. J. Biol. Chem, 263, 7887–7894. [PubMed] [Google Scholar]
- Mains RE, Park LP, & Eipper BA (1986). Inhibition of peptide amidation by disulfiram and diethyldithiocarbamate. J. Biol. Chem, 261, 11938–11941. [PubMed] [Google Scholar]
- Mashaghi A, Marmalidou A, Tehrani M, Grace PM, Pothoulakis C, & Dana R (2016). Neuropeptide substance P and the immune response. Cell Mol Life Sci, 73(22), 4249–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre NR, Lowe EW, Battistini MR, Leahy JW, & Merkler DJ (2016). Inactivation of peptidylglycine α-hydroxylating monooxygenase by cinnamic acid analogs. J.Enzyme Inhib.Med.Chem, 31, 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre NR, Lowe EW Jr., Chew GH, Owen TC, & Merkler DJ (2006). Thiorphan, tiopronin, and related analogs as substrates and inhibitors of peptidylglycine alpha-amidating monooxygenase (PAM). FEBS Lett, 580(2), 521–532. [DOI] [PubMed] [Google Scholar]
- McIntyre NR, Lowe EW Jr., & Merkler DJ (2009). Imino-oxy acetic acid dealkylation as evidence for an inner-sphere alcohol intermediate in the reaction catalyzed by peptidylglycine alpha-hydroxylating monooxygenase. J Am Chem Soc, 131(29), 10308–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JS, Bigley A, Bell A, Jenkins R, Somers R, Brocklehurst S et al. (2006). A pilot evaluation of the use of tissue microarrays for quantitation of target distribution in drug discovery pathology. Exp Toxicol Pathol, 57(3), 181–193. [DOI] [PubMed] [Google Scholar]
- Merkler DJ, Asser AS, Baumgart LE, Carballo N, Carpenter SE, & Owen TC (2008). Substituted hippurates and hippurate analogs as substrates and inhibitors of peptidylglycine alpha-hydroxylating monooxygenase (PHM). Bioorg. Med. Chem, 16, 10061–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkler DJ, Kulathila R, Young SD, Freeman J, & Villafranca JJ (1993). Amidation. In Karlin KD & Tyeklar Z (Eds.), Bioinorganic Chemistry of Copper, (pp. 196–209). New York: Chapman and Hall. [Google Scholar]
- Miller DA, Sayad KU, Kulathila R, Beaudry GA, Merkler DJ, & Bertelsen AH (1992). Characterization of a bifunctional peptidylglycine alpha-amidating enzyme expressed in Chinese hamster ovary cells. Arch. Biochem. Biophys, 298, 380–388. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Ohsuye K, Wada Y, Fuchimura K, Tanaka S, & Matsuo H (1987). Cloning and sequence of cDNA encoding a peptide C-terminal alpha-amidating enzyme from Xenopus laevis. Biochem. Biophys. Res. Commun, 148, 546–552. [DOI] [PubMed] [Google Scholar]
- Moore AB, & May SW (1999). Kinetic and inhibition studies on substrate chaneling in the bifunctional enzyme catalyzing C-terminal amidation. Biochem. J, 341, 33–40. [PMC free article] [PubMed] [Google Scholar]
- Morris KM, Cao F, Onagi H, Altamore TM, Gamble AB, & Easton CJ (2012). Prohormone-substrate peptide sequence recognition by peptidylglycine α-amidating monooxygenase and its reflection in increased glycolate inhibitor potency. Bioorg. Medicinal Chem. Lett, 22, 7015–7018. [DOI] [PubMed] [Google Scholar]
- Morrison JF, & Stone SR (1985). Approaches to the Study and Analysis of the Inhibition of Enzymes by Slow and Tight-binding Inhibitors. Comments Mol. Cell. Biophys, 2, 347–368. [Google Scholar]
- Mounier CE, Shi J, Sirimanne SR, Chen B-H, Moore AB, Gill-Woznichak et al. (1997). Pyruvate-extended Amino Acid Derivatives as Highly Potent Inhibitors of Carboxyl-terminal Peptide Amidation. J. Biol. Chem, 272, 5016–5023. [DOI] [PubMed] [Google Scholar]
- Mueller GP, & Altarac M (1995). Peptide alpha-amidation: differential regulation by disulfiram and its metabolite, diethyldithiocarbamate. Neuropept, 28, 333–340. [DOI] [PubMed] [Google Scholar]
- Mueller GP, Driscoll WJ, & Eipper BA (1999). In vivo inhibition of peptidylglycine-α-hydroxylating monooxygenase by 4-phenyl-3-butenoic acid. jpet, 290, 1331–1336. [PubMed] [Google Scholar]
- Murthy AS, Keutmann HT, & Eipper BA (1987). Further characterization of peptidylglycine alpha-amidating monooxygenase from bovine neurointermediate pituitary. Mol. Endocrinol, 1, 290–299. [DOI] [PubMed] [Google Scholar]
- Murthy AS, Mains RE, & Eipper BA (1986). Purification and characterization of peptidylglycine alpha-amidating monooxygenase from bovine neurointermediate pituitary. J. Biol. Chem, 261, 1815–1822. [PubMed] [Google Scholar]
- Nager AR, Goldstein JS, Herranz-Pérez V, Portran D, Ye F, Garcia-Verdugo JM et al. (2017). An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell, 168, 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BC, & Tawata S (2016). The Chemistry and Biological Activities of Mimosine: A Review. Phytother Res, 30(8), 1230–1242. [DOI] [PubMed] [Google Scholar]
- Ogonowski AA, May SW, Moore AB, Barrett LT, O’Bryant CL, & Pollock SH (1997). Antiinflammatory and Analgesic Activity of an Inhibitor of Neuropeptide Amidation. J. Pharm. Exp. Ther, 280, 846–853. [PubMed] [Google Scholar]
- Ouafik LH, Stoffers DA, Campbell TA, Johnson RC, Bloomquist BT, Mains RE et al. (1992). The multifunctional peptidylglycine alpha-amidating monooxygenase gene: exon/intron organization of catalytic, processing and routing domains. Mol. Endocrinol, 6, 1571–1584. [DOI] [PubMed] [Google Scholar]
- Perkins SN, Husten EJ, & Eipper BA (1990). The 108-kDA peptidylglycine alpha-amidating monooxygenase precursor contains two separable enzymatic activities involved in peptide amidation. Biochem. Biophys. Res. Commun, 171, 926–932. [DOI] [PubMed] [Google Scholar]
- Ping D, Mounier CE, & May SW (1995). Reaction Versus Subsite Stereospecificity of Peptidylglycine alpha-Monooxygenase and Peptidylamidoglycolate Lyase, and Two Enzymes Involved in Peptide Amidation. J. Biol. Chem, 270, 29250–29255. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Eipper BA, Mains RE, & Amzel LM (2004). Dioxygen binds end-on to mononuclear copper in a precatalytic enzyme complex. Science, 304, 864–867. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, & Amzel LM (1997). Amidation of bioactive peptides: the structure of peptidylglycine α-hydroxylating monooxygenase. Science, 278, 1300–1305. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, & Amzel LM (1999). Substrate-mediated electron transfer in peptidylglycine alpha-hydroxylating monooxygenase. Nature Struct. Biol, 6, 976–983. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Mains RE, Eipper BA, & Amzel LM (2000). New insights into copper monooxygenases and peptide amidation: structure, mechanism and function. Cell Mol. Life Sci, 57, 1236–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe S, Jones EE, Kim Y, Sinha A, Nyalwidhe JO, Brooks J et al. (2013). In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics, 13(10–11), 1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal C, Mains RE, & Eipper BA (2012). Signaling from the secretory granule to the nucleus. Crit Rev. Biochem. Mol. Biol, 47(4), 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal C, Stone KL, Francone VP, Mains RE, & Eipper BA (2009). Secretory Granule to the Nucleus: role of a multiply phosphorylated intrinsically unstructured domain. J.Biol.Chem, 284, 25723–25734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal C, Stone KL, Mains RE, & Eipper BA (2010). Secretion stimulates intramembrane proteolysis of a secretory granule membrane enzyme. J.Biol.Chem, 285, 34632–34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VK, Zavala G, Deb Roy A, Mains RE, & Eipper BA (2019). A pH-sensitive luminal His-cluster promotes interaction of PAM with V-ATPase along the secretory and endocytic pathways of peptidergic cells. J.Cell.Physiol, 234, 8683–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray MV, Van Duyne P, Bertelsen AH, Jackson-Matthews DE, Sturmer AM, Merkler DJ et al. (1993). Production of recombinant salmon calcitonin by in vitro amidation of an Escherichia coli produced precursor peptide. Biotechnology (N Y), 11(1), 64–70. [DOI] [PubMed] [Google Scholar]
- Rhodes CH, & Honsinger C (1993). Structure-activity relationships among inhibitors of peptidylglycine amidating monooxygenase. Ann N Y Acad Sci, 689, 663–666. [DOI] [PubMed] [Google Scholar]
- Rocchi P, Muracciole X, Fina F, Mulholland DJ, Karsenty G, Palmari J et al. (2004). Molecular analysis integrating different pathways associated with androgen-independent progression in LuCaP 23.1 xenograft. Oncogene, 23(56), 9111–9119. [DOI] [PubMed] [Google Scholar]
- Rondeel JM, Klootwijk W, Linkels E, van Haasteren GA, de Greef WJ, & Visser TJ (1995). Regulation of thyrotropin-releasing hormone in the posterior pituitary. Neuroendocrinology, 61(4), 421–429. [DOI] [PubMed] [Google Scholar]
- Schade D, Kotthaus J, Hungeling H, Kotthaus J, & Clement B (2009). The peptidylglycine alpha-amidating monooxygenase (PAM): a novel prodrug strategy for amidoximes and N-hydroxyguanidines? ChemMedChem, 4(10), 1595–1599. [DOI] [PubMed] [Google Scholar]
- Scopsi L, Lee R, Gullo M, Collini P, Husten EJ, & Eipper BA (1998). Peptidylglycine α-amidating monooxygenase in neuroendocrine tumors--its identification, characterization, quantification, and relation to the grade of morphological differentiation, amidated peptide content, and granin histochemistry. Appl. Immunohistochem, 6, 120–132. [Google Scholar]
- Siebert X, Eipper BA, Mains RE, Prigge ST, Blackburn NJ, & Amzel LM (2005). The Catalytic Copper of Peptidylglycine α-Hydroxylating Monooxygenase also Plays a Critical Structural Role. Biophysical J, 89, 3312–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulj M, Pezdirec D, Gaser D, Kreft M, & Zorec R (2014). Reduction in C-terminal amidated species of recombinant monoclonal antibodies by genetic modification of CHO cells. BMC Biotechnology, 14, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southan C, & Kruse LI (1989). Sequence similarity between dopamine beta-hydroxylase and peptide amidating enzyme: evidence for a conserved catalytic domain. FEBS Lett, 255, 116–120. [DOI] [PubMed] [Google Scholar]
- Steinhorsdottir V, Thorliefsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A et al. (2014). Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nature Genetics, 46, 294–298. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Green CB, & Eipper BA (1989). Alternative mRNA splicing generates multiple forms of peptidyl-glycine alpha-amidating monooxygenase in rat atrium. Proc. Natl. Acad. Sci. U. S. A, 86, 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers DA, Ouafik L, & Eipper BA (1991). Characterization of novel mRNAs encoding enzymes involved in peptide alpha-amidation. J. Biol. Chem, 266, 1701–1707. [PubMed] [Google Scholar]
- Sunman JA, Foster MA, Folse SL, May SW, & Matesic SF (2004). Reversal of the transformed phenotype and inhibition of peptidylglycine alpha-monooxygenase in Ras-transformed cells by 4-phenyl-3-butenoic acid. Mol. Carcinog, 41, 231–246. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Harada S, Higashimoto Y, Shimokawa C, Sato H, Sugishima M et al. (2009). Involvement of metals in enzymatic and nonenzymatic decomposition of C-terminal alpha-hydroxyglycine to amide: an implication for the catalytic role of enzyme-bound zinc in the peptidylamidoglycolate lyase reaction. Biochemistry, 48(7), 1654–1662. [DOI] [PubMed] [Google Scholar]
- Tamburini PP, Jones BN, Consalvo AP, Young SD, Lovato SJ, Gilligan JP et. (1988). Structure-Activity Relationships for Glycine-Extended Peptides and the alpha-Amidating Enzyme Derived from Medullary Thyroid CA-77 Cells. Arch. Biochem. Biophys, 267, 623–631. [DOI] [PubMed] [Google Scholar]
- Tamburini PP, Young SD, Jones BN, Palmesino RA, & Consalvo AP (1990). Peptide substrate specificity of the alpha-amidating enzyme isolated from rat medullary thyroid CA-77 cells. Int J Pept Protein Res, 35(2), 153–156. [DOI] [PubMed] [Google Scholar]
- Tenn WJ 3rd, Murphy JL, Bim-Merle JK, Brown JA, Junia AJ, Price MA et al. (2007). Amidates as leaving groups: structure/reactivity correlation of the hydroxide-dependent E1cB-like breakdown of carbinolamides in aqueous solution. J Org Chem, 72(16), 6075–6083. [DOI] [PubMed] [Google Scholar]
- Terlau H, & Olivera BM (2004). Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev, 84, 41–68. [DOI] [PubMed] [Google Scholar]
- Thomsen SK, Raimondo A, Hastoy B, Sengupta S, MacDonald PE, & Gloyn AL (2018). Type 2 diabetes risk alleles in PAM impact insulin release from human pancreatic β-cells. Nat.Genet, 50, 1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouënnon E, Elkahloun AG, Guillemot J, Gimenez-Roqueplo A-P, Bertherat J, Pierre A et al. (2007). Identification of potential gene markers and insights into the pathophysiology of pheochromocytoma malignancy. J Clin Endocrinol Metab, 92(12), 4865–4872. [DOI] [PubMed] [Google Scholar]
- Turner A, Kaas Q, & Craik DJ (2020). Hormone-like conopeptides-new tools for pharmaceutical design. RSC Med Chem, 11(11), 1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul-Hasan S, Burgess DM, Gajewiak J, Li Q, Hu H, Yandell M et al. (2013). Characterization of the peptidylglycine α-amidating monooxygenase (PAM) from the venom ducts of neogastropods, Conus bullatus and Conus geographus. Toxicon, 74, 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelboe TV, Harris P, Zhao Y, Walter TS, Harlos K, Omari KE et al. (2016). The crystal structure of human dopamine beta-hydroxylase at 2.9 Å resolution. Sci.Adv, 2, e1500980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanatha K, Bäck N, Mains RE, & Eipper BA (2014). A histidine-rich linker region in peptidylglycine alpha-amidating monooxygenase has the properties of a pH sensor. J. Biol. Chem, 289(18), 12404–12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanatha KS, Bäck N, Lam TT, Mains RE, & Eipper BA (2016). O-Glycosylation of a Secretory Granule Membrane Enzyme Is Essential for Its Endocytic Trafficking. J.Biol.Chem, 291, 9835–9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos MD, Scott FM, Iwai N, & Treston AM (1996). Expression in human lung cancer cell lines of genes of prohormone processing and the neuroendocrine phenotype. J Cell Biochem Suppl, 24, 257–268. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bernard A, Comblain F, Yue X, Paillart C, Zhang S et al. (2021). Melanocortin 4 receptor signals at the neuronal primary cilium to control food intake and body weight. J Clin Invest, 131(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hill S, Luther JM, Hachey DL, & Schey KL (2012). Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT). Proteomics, 12, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ST, McIntyre NR, McLaughlin ML, & Merkler DJ (2006). The development of molecular clamps as drugs. Drug Discov Today, 11(17–18), 819–824. [DOI] [PubMed] [Google Scholar]
- Wilcox BJ, Ritenour-Rodgers KJ, Asser AS, Baumgart LE, Baumgart MA, Boger DL et al. (1999). N-Acylglycine Amidation: Implications for the Biosynthesis of Fatty Acid Primary Amides. Biochemistry, 38, 3235–3245. [DOI] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, & Rosenbaum JL (2013). The Cilium Secretes Bioactive Ectosomes. Curr. Biol, 23, 906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Fan F, Song J, Peng W, Liu J, Li C et al. (2019). Theory Demonstrated a “Coupled” Mechanism for O(2) Activation and Substrate Hydroxylation by Binuclear Copper Monooxygenases. J Am Chem Soc, 141(50), 19776–19789. [DOI] [PubMed] [Google Scholar]
- Yin P, Bousquet-Moore D, Annangudi SP, Southey BR, Mains RE, Eipper BA et al. (2011). Probing the production of amidated peptides following genetic and dietary copper manipulations. PLoS ONE, 6, e28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabriskie TM, Cheng H, & Vederas JC (1992). Mechanism-Based Inactivation of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM) by a Substrate Analogue, D-Phenylalanyl-L-Phenylalanyl-D-Vinylglycine: Inhibition of Formation of Peptide C-Terminal Amides. J. Am. Chem. Soc, 114, 2270–2272. [Google Scholar]
- Zabriskie TM, Klinge M, Szymanski CM, Cheng H, & Vederas JC (1994). Peptide amidation in an invertebrate: purification, characterization, and inhibition of peptidylglycine alpha-hydroxylating monooxygenase from the heads of honeybees (Apis mellifera). Arch. Insect Biochem. Physiol, 26, 27–48. [DOI] [PubMed] [Google Scholar]
- Zieglgänsberger W (2019). Substance P and pain chronicity. Cell Tissue Res, 375(1), 227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]


