Abstract
Background:
Inhaled corticosteroid (ICS) response among patients with asthma is influenced by genetics, but biologically actionable insights based on associations have not been found. Various glucocorticoid response omics datasets are available to interrogate their biological effects.
Objective:
We sought to identify functionally relevant ICS response genetic associations by integrating complementary multiomics datasets.
Methods:
Variants with p-values<10−4 from a previous ICS response genome-wide association study were re-ranked based on integrative scores determined from: i) glucocorticoid receptor (GR)- and ii) RNA polymerase II (RNAP II)-binding regions inferred from ChIP-Seq data for three airway cell types, iii) glucocorticoid response element (GRE) motifs, iv) differentially expressed genes in response to glucocorticoid exposure according to 20 transcriptomic datasets, and v) expression quantitative trait loci (eQTLs) from GTEx. Candidate variants were tested for association with ICS response and asthma in six independent studies.
Results:
Four variants had significant (q-value<0.05) multiomics integrative scores. These variants were in a locus consisting of 52 variants in high LD (r2≥0.8) near GR-binding sites by the gene BIRC3. Variants were also BIRC3 eQTLs in lung, and two were within/near putative GRE motifs. BIRC3 had increased RNAP II occupancy and gene expression with glucocorticoid exposure in two ChIP-Seq and 13 transcriptomic datasets. Some BIRC3 variants in the 52-variant locus were associated (p-value<0.05) with ICS response in three independent studies and others with asthma in one study.
Conclusion:
BIRC3 should be prioritized for further functional studies of ICS response.
Clinical Implication:
Genetic variation near BIRC3 may influence ICS response in people with asthma.
Keywords: Asthma, ChIP-Seq, genome-wide association study, glucocorticoid response, glucocorticoid receptor, inhaled corticosteroids, integrative analysis, multiomics, transcriptomics
Capsule summary
Multiomics data integration found that genetic variants near the BIRC3 gene may influence glucocorticoid response in people with asthma by modulating BIRC3 gene expression via altered GR-binding.
Introduction
Inhaled corticosteroids (ICS) are glucocorticoids that are commonly used for the treatment of asthma1. Although use of ICS according to clinical guidelines successfully controls symptoms in most patients with persistent asthma, up to 30% of patients have severe asthma that requires additional medications2. The inter-individual variability in clinical response to ICS is not fully understood, but previous pharmacogenetics studies suggest that it is influenced by genetics3–11. Genome-wide association studies (GWAS) have yielded reproducible loci associated with many traits, including asthma12, but their application to the study of ICS response has not yielded consistent findings despite the identification of promising loci such as GLCCI13. The lack of large study populations to study ICS response genetics, and the variability in study designs among available studies, including use of different outcomes (e.g., improved lung function, reduced number of asthma exacerbations), are among the issues that have hampered ICS response pharmacogenetics studies. The largest ICS response GWAS to date was based on 2,672 asthma patients treated with fluticasone furoate and fluticasone propionate from clinical studies conducted by GlaxoSmithKline (GSK)11. This study, in which response to ICS was measured as the change in forced expiratory volume in one second (FEV1) from baseline to 8–12 weeks after initiation of ICS treatment, found little evidence that common genetic variants underlie ICS response in asthma patients, as no associations reached the genome-wide significance threshold of p-value<5×10−8. Given the relatively small sample size of this and other ICS response GWAS, some of the nominally significant loci could represent biologically relevant findings.
At a cellular level, glucocorticoids bind to the glucocorticoid receptor (GR), a transcription factor that forms GR-GR dimers in cell nuclei that directly bind to glucocorticoid response elements (GREs) where they induce the transcription of genes encoding anti-inflammatory proteins13. GRs also interact with other transcription factors (e.g., nuclear factor kappa B (NF-κB)) to reduce the transcription of genes encoding pro-inflammatory cytokines and cytokine-induced proteins13. The binding of GRs in tissues such as airway smooth muscle (ASM), BEAS-2B and A549 cells has been characterized via ChIP-Seq studies to occur at thousands of genomic regions and to change with exposure to glucocorticoids14–16. Similarly, the differential expression of thousands of genes in response to glucocorticoid exposure has been measured in various asthma-relevant tissues using gene expression microarrays and RNA-Seq17,18. These omics studies have begun to identify cell- and tissue-type specific responses to glucocorticoids that occur at the level of transcription and GR-binding17,19–21.
The integration of multiomics data has provided insights into pulmonary diseases beyond those available in single-modality studies22,23. The combination of two types of omics data has shown promise to 1) prioritize ICS response-related GWAS variants according to transcriptomic data24, and 2) distinguish primary from secondary targets of the GR via the combination of GR ChIP-Seq and transcriptomic data15,20. Although several relevant omics datasets related to glucocorticoid mechanisms of action in asthma-relevant tissues are currently available, including GR ChIP-Seq, glucocorticoid response-related transcriptomics, expression quantitative trait locus (eQTL) and genomic location of GREs, studies have not yet leveraged them to gain insights via their integration. Here, we assigned ICS response GWAS variants with integrative scores based on functional evidence from glucocorticoid response-related multiomics layers to identify ICS response loci whose nominally significant associations were supported by functional evidence.
Methods
Detailed methods are provided in the Online Supplement.
ICS Response GWAS Results
Summary statistics of a previously published GSK ICS response GWAS11, whose outcome consisted of change in FEV1 after ICS treatment in 2,672 asthma patients, were obtained. Subjects were primarily of European ancestry (1,787; 67%), and the available summary data included effect sizes and p-values for 10,215,080 variants derived from the hg19 reference genome obtained with an additive model that included relevant covariates. Variants having inconsistent information with 1000 Genomes Project phase 3 (1000G) data were excluded. Among filtered variants, common nominally associated ones with GWAS p-value<10−4 were selected. The set of nominally significant variants was expanded to include bi-allelic variants that were in high linkage disequilibrium (LD) with them (r2≥0.8 based on 1000G European (EUR) genotype data). The hg19 coordinates were converted to hg38 coordinates using the UCSC LiftOver tool. Because several GSK subjects were not of European ancestry, an additional panel of variants was selected based on LD according to the 1000G cosmopolitan reference panel11.
Transcriptomic Data Analysis
Twenty transcriptomic datasets related to glucocorticoid response involving 11 cell types were retrieved from Gene Expression Omnibus (GEO) and analyzed with the RAVED pipeline (https://github.com/HimesGroup/raved)19. Differential expression analysis was performed for 21 comparisons of glucocorticoid exposure versus vehicle control in vitro and two comparisons of glucocorticoid treatment versus placebo in human studies. Genes/probes tested were considered significant if they had Benjamini-Hochberg adjusted p-values (i.e., q-values) <0.05.
ChIP-Seq Data Analysis
ChIP-Seq datasets involving the GR with dexamethasone exposure for ASM (GSE95632)14, BEAS-2B (GSE79803)16 and A549 cell lines (SRP000762)15 were retrieved from GEO or Sequence Read Archive (SRA) and analyzed with the brocade pipeline (https://github.com/HimesGroup/brocade)20, which included differential binding analysis comparing GR- or RNA Polymerase II (RNAP II)-occupancy changes in cells exposed to dexamethasone versus vehicle control. Protein-binding sites with Benjamini-Hochberg adjusted p-values (i.e., q-values)<0.05 were considered significant. Target genes were restricted to those having GR-binding sites within 20kb of the gene’s transcription start site (TSS), a distance that has been shown to identify well-known glucocorticoid-responsive genes20, while also having either (1) RNAP II-binding sites within ±5kb to the genes’ TSS (i.e. promoter region) in a matched ChIP-Seq dataset, or (2) significant differential expression results in at least one of the transcriptomic datasets. The FIMO tool25 was used to identify putative GRE motifs within GR-binding sites as determined by sequences having p-values<10−4. Protein-coding genes were annotated to protein-DNA binding sites based on GENCODE Human annotation data (v32). Overlap between GWAS variants, GR-binding sites and GRE motifs was performed using the bedtools intersect function26.
eQTL Data
Single-tissue cis-eQTL results for whole blood, lung, and skeletal muscle tissues were obtained from the GTEx V8 release (https://gtexportal.org/home/datasets)27.
Multiomics Integrative Score
A multiomics integrative score was assigned to each variant based on the sum of scores of evidence from five layers: 1) GR-binding, 2) GRE occurrence, 3) RNAP II-binding, 4) differential expression in responses to glucocorticoids and 5) eQTL. Significance of integrative multiomics scores was determined via permutation and subsequent adjustment using the Benjamini-Hochberg approach. Variants with adjusted p-values (i.e., q-values) <0.05 were considered significant. See Online Supplementary Methods for more detailed description.
Multiomics Analysis of Previously Reported ICS Response-Associated Variants
A secondary analysis was performed to include variants reported by other GWAS3–10 and candidate gene studies24,28–31, in which these variants were combined with the GSK GWAS variants. The same criteria as outlined above was applied to expand variant selection based on LD, except that LD structure for variants included was computed using 1000G genotype data that matched the population described in the paper originally describing the association. The multiomics score was computed on this expanded set of variants.
Genetic Association Analysis of ICS Response in Replication Studies
The association analyses between ICS response and 52 BIRC3 locus variants selected for replication were conducted in the following studies:
GERA cohort.
The Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort32,33 consisted of 5,710 individuals of European ancestry. The outcome of ICS response was defined as lack of asthma exacerbation while using ICS (i.e. the use of oral corticosteroid (OCS) bursts within 30 days after receiving an ICS prescription).
GALA II and SAGE studies.
The Genes-environments & Admixture in Latino Americans Study (GALA II) consisted of 854 Hispanic/Latino individuals and the Study of African Americans, Asthma, Genes and Environments (SAGE)10,34,35 consisted of 493 African American individuals. The outcome of ICS response was defined as asthma exacerbation in the prior year while using ICS.
SLOVENIA cohort.
The SLOVENIA cohort8 consisted of 166 patients of European ancestry. ICS response was defined as a binary outcome of ICS non-responders versus responders based on whether change in FEV1 in patients after 6 weeks of ICS therapy was less than 8%.
Any variants with p-values<0.05 in a study population were considered significant, as these variants were considered representative of the single BIRC3 genetic locus identified.
Genetic Association Analysis of Asthma in Replication Studies
We sought to determine whether the ICS-associated variants were also associated with asthma given that differences in ICS response might contribute to asthma development and symptoms and thus, influence the ascertainment of asthma cases in large biobank studies. The association analyses between asthma susceptibility and 52 BIRC3 locus variants selected for replication were conducted in the following studies:
UK Biobank.
The UK Biobank study consisted of 19,216 individuals with asthma and 162,637 healthy controls of European ancestry from the UK Biobank36.
EVE Consortium study.
The EVE asthma consortium study37 included 3,246 cases, 3,385 controls, 1,702 case-parent trios, and 355 family-based cases and 468 family-based controls. Meta-analysis results of asthma GWAS in 3,459 European Americans, 992 African Americans/African Caribbeans, and 1,180 Latino Americans were previously reported37.
Meta-analysis
We performed meta-analysis using Liptak’s method separately for ICS response and asthma studies, where p-value-derived z statistics were weighted based on each study’s sample size and a combined p-value for each variant was derived from the resultant z-statistics’ distribution38.
Results
Single Modality Glucocorticoid Response Omics Results
Characteristics of subjects from the previously published GSK ICS response GWAS are in Table I. Of 8,198,689 variants that passed quality control criteria, 6,700,204 (81.7%) common variants (EUR MAF≥0.01) were selected for analyses. No variants reached a genome-wide significance threshold of 5×10−8. The top-ranked associations, with p-values<1×10−6, were in loci near genes PXDC1, C11orf72, and RORA (Fig. E1, Table E1). Based on the 1000G EUR reference panel, a set of 4,468 GWAS variants was selected: 820 variants were nominally associated with ICS response (p-value<10−4) and 3,648 variants were in high LD (r2≥0.8) with them. Based on the 1000G cosmopolitan reference panel, a set of 2,822 GWAS variants was selected: 883 variants were nominally associated with ICS response and 1,939 variants were in high LD (r2≥0.8) with them. We next report results based on the 4,468 GWAS variants obtained with the EUR reference panel. Integrative analysis results using the 2,822 variant set corresponding to the 1000G cosmopolitan reference panel can be found in the Online Repository.
Table I.
Characteristics of participants in ICS response studies.
| A. GSK cohort | |||
|---|---|---|---|
| Characteristics | Phase IIB studies | Phase IIIA studies | HZA 106837 |
| Total patients | 1,125 | 679 | 868 |
| Age (y) | 42.1±16.8 | 44.2±15.6 | 42.7±14.7 |
| Sex: %female | 61.2 | 58.6 | 68.6 |
| BMI (kg/m2) | 27.9±6.7 | 27.9±5.2 | 27.6±5.9 |
| Height (cm) | 167.9±13.4 | 165.9±7.8 | 163.9±8.8 |
| Asthma duration (y) | 17.7±13.4 | 14.9±13.0 | 15.7±11.8 |
| FEV1 at baseline (% predicted) | 69.9±10.1 | 69.2±10.4 | 71.7±11.8 |
| FEV1 change (% predicted) | 7.2±13.4 | 6.9±13.0 | 6.1±11.8 |
| B. GERA cohort | ||
|---|---|---|
| Characteristics | Asthma patients with exacerbation | Asthma patients without exacerbation |
| Total patients | 3,097 | 2,613 |
| Age (y) | 63.8±14.0 | 63.4±15.2 |
| Sex: %female | 2168 (70.0%) | 1765 (67.5%) |
| BMI (kg/m2) | 29.2±6.2) | 28.6±6.3 |
| Smoking status: %ever smoker | 1378 (45.9%) | 1027 (40.9%) |
| Allergic rhinitis status | 1404 (45.3%) | 1113 (42.6%) |
| Gastroesophageal reflux disease | 983 (31.7%) | 651 (24.9%) |
| C. GALA II study | ||
| Characteristics | Asthma patients with exacerbation | Asthma patients without exacerbation |
| Total patients | 567 | 287 |
| Age (y) | 11.9±3.1 | 12.5±3.3 |
| Sex: %female | 246 (43.4%) | 119 (41.5%) |
| D. SAGE study | ||
| Characteristics | Asthma patients with exacerbation | Asthma patients without exacerbation |
| Total patients | 256 | 237 |
| Age (y) | 13.2±3.4 | 13.8±3.4 |
| Sex: %female | 123 (48.0%) | 103 (43.5%) |
| E. SLOVENIA cohort | ||
| Characteristics | ICS non-responders | ICS responders |
| Total patients | 94 | 72 |
| Age (y) | 10.7±3.2 | 11.2±3.5 |
| Sex: %female | 35 (37.2%) | 33 (45.8%) |
| F. UK Biobank participants | ||
| Characteristics | Asthma patients | Healthy controls |
| Total patients | 19,216 | 162,637 |
| Age (y) | 55.8±8.4 | 57.1±8.3 |
| Sex: %female | 62.2 | 55.4 |
| BMI (kg/m2) | 27.9±5.0 | 27.1±4.7 |
| Smoking status: %ever smoker | 57.4 | 58.1 |
Considering the ChIP-Seq data alone, 46,867 GR-binding sites had significantly increased GR occupancy and 7,238 RNAP II-binding sites had significantly different RNAP II occupancy with glucocorticoid exposure in at least one airway cell type (q-value<0.05). Among the many glucocorticoid-responsive GR-binding sites, 2,003 were within 20kb of a gene TSS while also having an associated RNAP II-binding event (RNAP II-binding site within 5kb of the gene’s TSS). These 2,003 GR-binding sites with evidence of active transcription corresponded to 851 genes. Of the 46,867 GR-binding sites, 13,165 (<30%) had putative GRE motifs.
Analysis of 20 transcriptomic datasets (Table E2) identified 13,580 differentially expressed genes (q-value<0.05) in response to glucocorticoids in at least one of the 11 cell types considered. 4,672 differentially expressed genes in airway cells (ASM, A549, BEAS-2B, bronchial epithelial cells) and 2,324 genes in remaining cell types also had nearby GR-binding sites, suggesting that they were primary GR targets. Detailed results of individual studies are available in the web application REALGAR (http://realgar.org/)39.
1,262,229 cis-eQTLs in lung, 1,277,338 in whole blood and 1,406,351 in skeletal muscle were reported in GTEx.
Intersection of ICS GWAS and Single Modality Omics Results
Of the 4,468 GWAS variants considered, 29 were located within 39 of the 46,867 GR-binding sites (Fig. 1, Table E3). Nine genes (B4GALT1, BIRC3, CSF1R, EMILIN2, GAB1, OXSR1, PKP1, SLC22A14 and TIMP4) had GWAS variants within GR-binding sites that were within 20kb of their TSS. Twenty-two of the 39 GR-binding sites had putative GRE motifs, and SNP rs11225201 was the only variant located within a GRE motif in a GR-binding site. 501, 577 and 430 of the 4,468 GWAS variants were cis-eQTLs in lung, whole blood and skeletal muscle, respectively. Among the 29 GWAS variants located within GR-binding sites, eight were eQTLs: four for BIRC3 in lung, whole blood and/or skeletal muscle tissues; three for SIL1 in lung; and one for B4GALT1 in skeletal muscle tissue.
Figure 1.
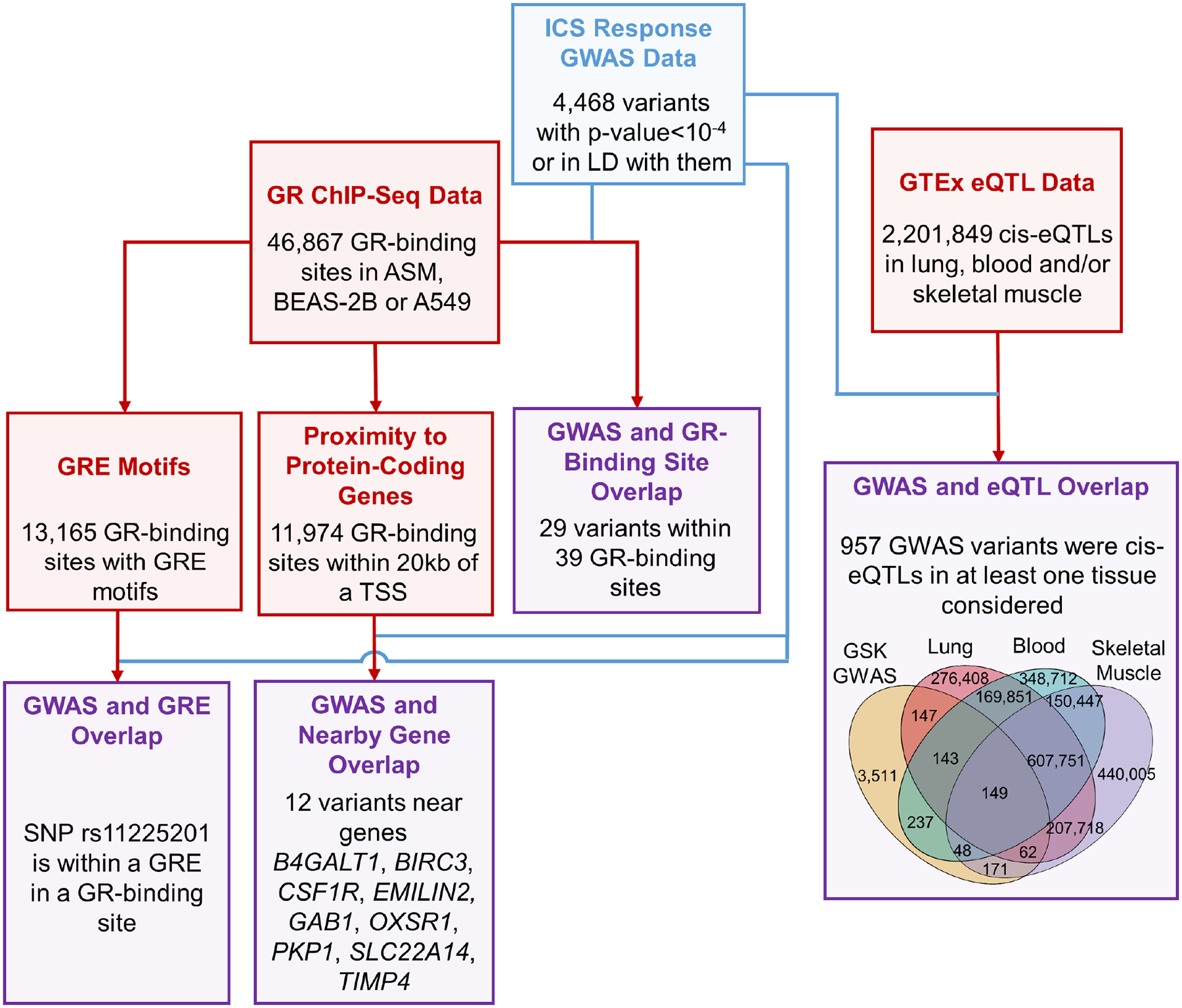
Intersections between ICS response GWAS and single modality omics results. GWAS results, results from other layers and intersection results are shown in blue, red, and purple boxes, respectively. GR: glucocorticoid receptor; GRE: glucocorticoid response element; TSS: transcription start site.
Integrative Analysis of Multiomics Glucocorticoid Response Data Supports a Locus Near BIRC3 as Involved in ICS Response
Multiomics integrative scores for the 4,468 GWAS variants selected found that four single nuclear polymorphisms (SNPs) near the gene BIRC3 were significant (score range 8.5 to 12; q-value<0.05) (Fig. 2, Fig. E2, Table E4). These four SNPs were in high LD (r2≥0.8 based on 1000G EUR reference panel) with each other, and SNP rs2846858 had the smallest (4.35×10−5) GWAS p-value among them (Table II). The four SNPs overlapped with six glucocorticoid-responsive GR-binding sites. BIRC3 had differential RNAP II-binding sites within 5kb of its TSS with exposure to glucocorticoids in BEAS-2B and A549 cells (q-value<0.05), suggesting that GR-binding events led to changes in gene transcription activity (Fig. 3A). The four BIRC3 SNPs were also eQTLs in lung and blood, and two were additionally BIRC3 eQTLs in skeletal muscle. SNP rs11225201 was located within a putative GRE motif, and SNP rs17878708 was 231bp from another putative GRE motif. According to transcriptomic data, BIRC3 was differentially expressed in response to glucocorticoid exposure (q-value<0.05), with increased expression observed in ASM, BEAS-2B, A549, bronchial epithelium, childhood acute lymphoblastic leukemia, lymphoblastoid cells, and U2OS bone osteosarcoma epithelial cells and decreased expression observed in macrophages (Fig. 4). To select variants for replication, we expanded the BIRC3 region to include all variants in high LD (r2≥0.8 based on 1000G EUR reference panel) with rs2846858 that were within 30kb upstream to 5kb downstream of the BIRC3 TSS, resulting in a set of 52 variants (GWAS p-values ranged from 2.12×10−5 to 5.56×10−4; Table E5). The minor alleles of these variants were associated with decreased change in FEV1 after ICS treatment, and hence, decreased ICS response; minor alleles were associated with increased gene expression of BIRC3 in GTEx lung tissues.
Figure 2.
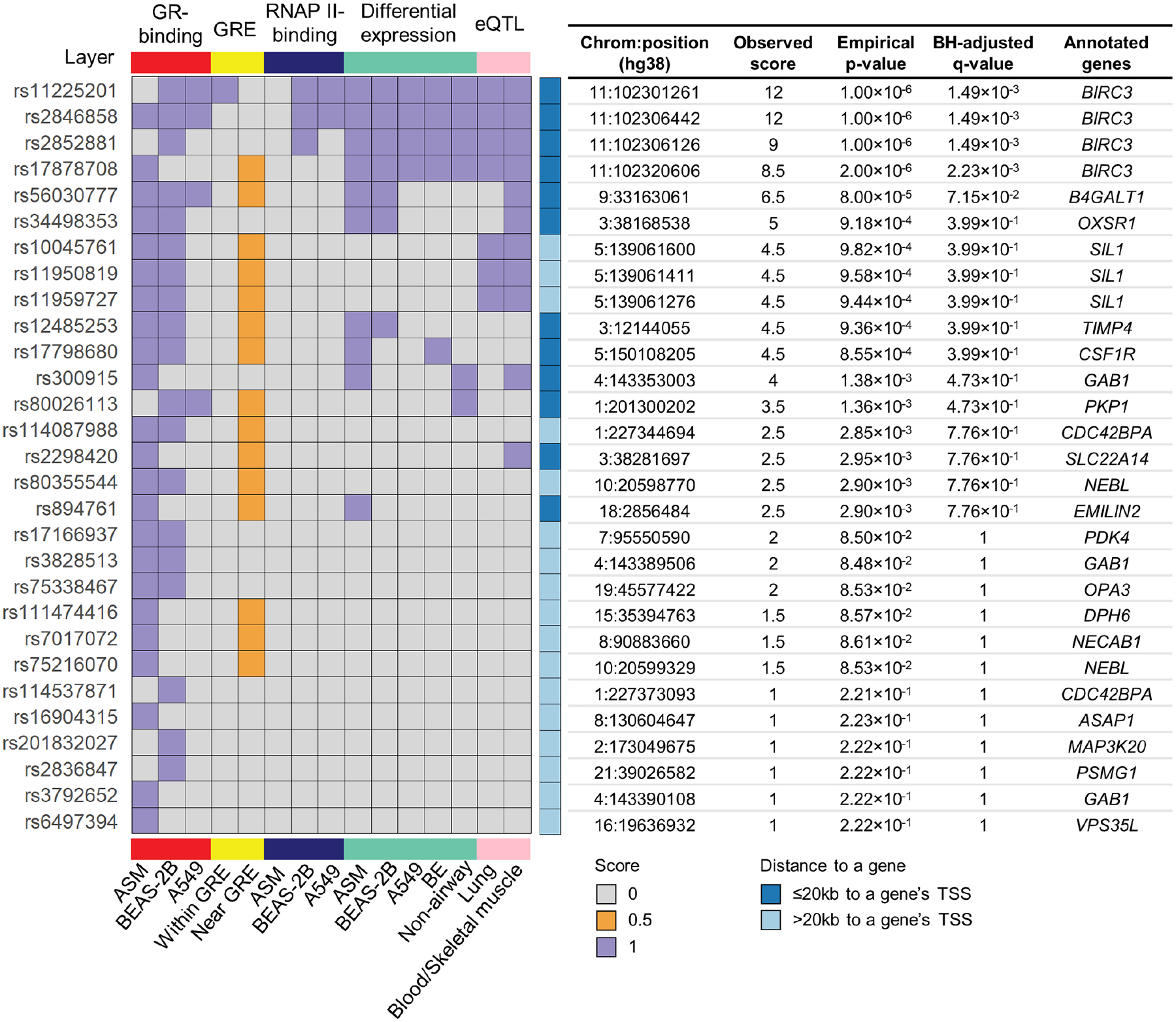
Multiomics integrative scores for 29 GR-binding site-overlapping SNPs. ASM: airway smooth muscle; BE: bronchial epithelium; BH: Benjamini-Hochberg; GR: glucocorticoid receptor; GRE: glucocorticoid response element; RNAP II: RNA polymerase II; TSS: transcription start site.
Table II.
Characteristics of four variants with significant multiomics scores that were in a locus near BIRC3.
| rsID | Position | Ref | Alt | Alternative allele frequency (EUR/cosmopolitan) | eQTL Tissue | GSK ICS Response Associations | GR-Binding Site Characteristics | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect size | P-value | Peak start to end | Log2 Fold change | Q-value | Tissue | Distance to TSS | GRE motif start to end | ||||||
| rs11225201 | 102301261 | T | G | 0.330/0.341 | L,M,B | −0.043 | 1.16×10−4 | 102301030–102301522 | 5.56 | 4.70×10−8 | BEAS-2B | −15,928 | 102301253–102301271 |
| 102301132–102301325 | 3.68 | 2.85×10−3 | A549 | −16,125 | |||||||||
| rs2852881 | 102306126 | C | T | 0.314/0.283 | L,B | −0.045 | 6.82×10−5 | 102305878–102307125 | 4.49 | 1.64×10−67 | BEAS-2B | −10,325 | - |
| rs2846858 | 102306442 | C | T | 0.315/0.327 | L,M,B | −0.046 | 4.35×10−5 | 102306146–102306808 | 4.78 | 1.05×10−7 | ASM | −10,642 | - |
| 102305878–102307125 | 4.49 | 1.64×10−67 | BEAS-2B | −103,25 | |||||||||
| 102306154–102306657 | 6.32 | 7.05×10−31 | A549 | −107,93 | |||||||||
| rs17878708 | 102320606 | G | A | 0.330/0.291 | L,B | −0.044 | 1.36×10−4 | 102320563–102321139 | 7.11 | 3.83×10−10 | ASM | 3,113 | 102320838–102320855 |
Genomic coordinates are based on hg38 on chromosome 11.
Effect size refers to the alternative allele.
eQTLs in lung (L), whole blood (B), and skeletal muscle (M) tissues are selected.
GR-binding sites containing putative GRE motifs are reflected by presence of information in the column “GRE motif start to end”.
SNP rs11225201 is located within a putative GRE.
Figure 3.
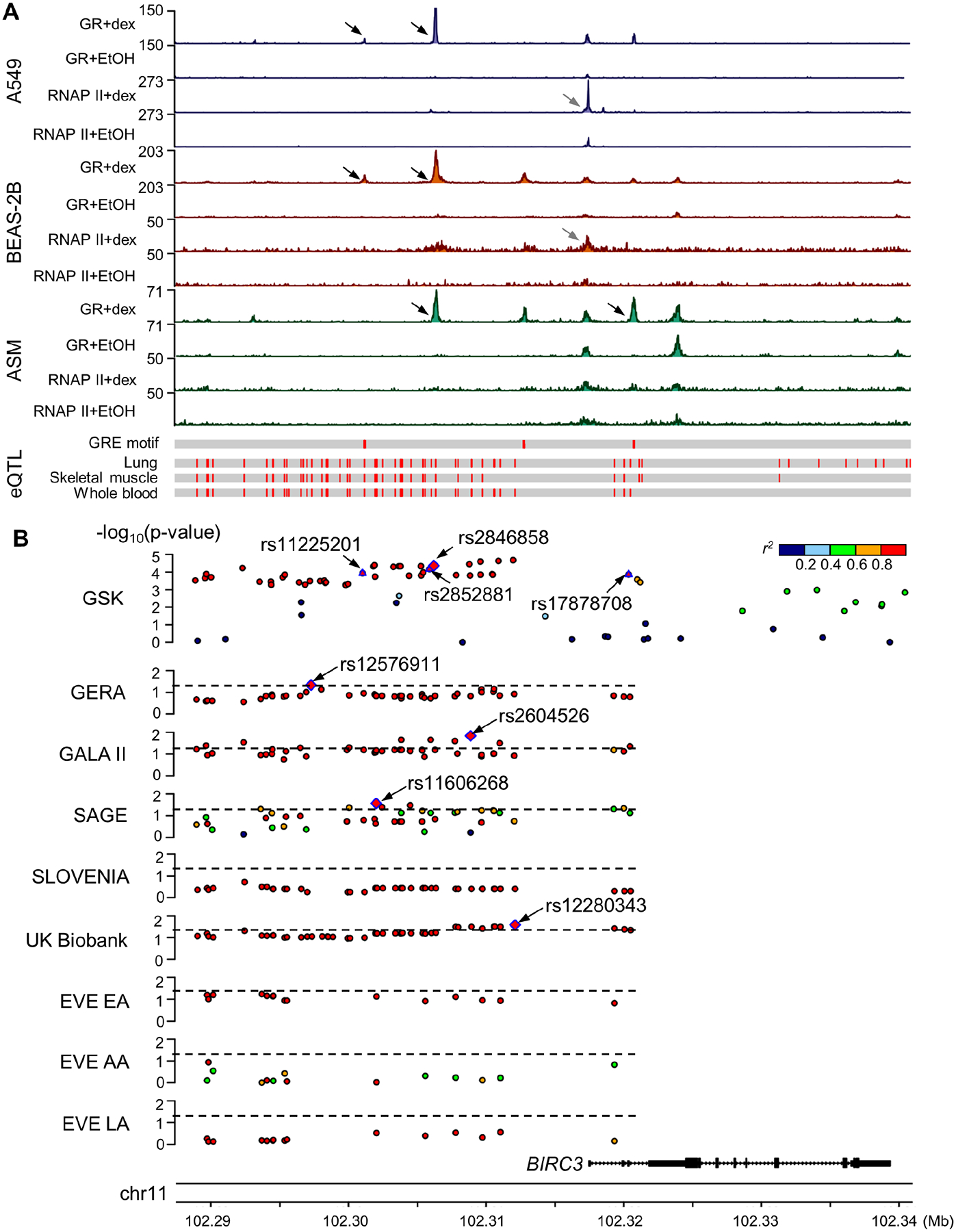
Multiomics evidence supporting BIRC3 as a glucocorticoid response-associated gene. A) ChIP-Seq (top), eQTL (bottom) and B) genetic association results in studies of ICS response and asthma susceptibility near the BIRC3 gene. Black arrows in ChIP-Seq genomic tracks indicate differential binding sites for GR where BIRC3 variants were located and grey arrows indicate differential binding sites for RNAP II. Pairwise r2 was computed between rs2846858 and corresponding variants based on 1000G genotype data that matched the subjects’ race/ethnicity in each study. Four GR-binding site-overlapping SNPs in the GSK study and the top SNPs with p-value<0.05 in replication studies are annotated. Dashed lines indicate p-value of 0.05. AA: African Americans/African Caribbeans; ASM: airway smooth muscle; Dex: dexamethasone; EA: European Americans; EtOH: ethanol; GR: glucocorticoid receptor; GRE: glucocorticoid response element; LA: Latino Americans; RNAP II: RNA polymerase II.
Figure 4.
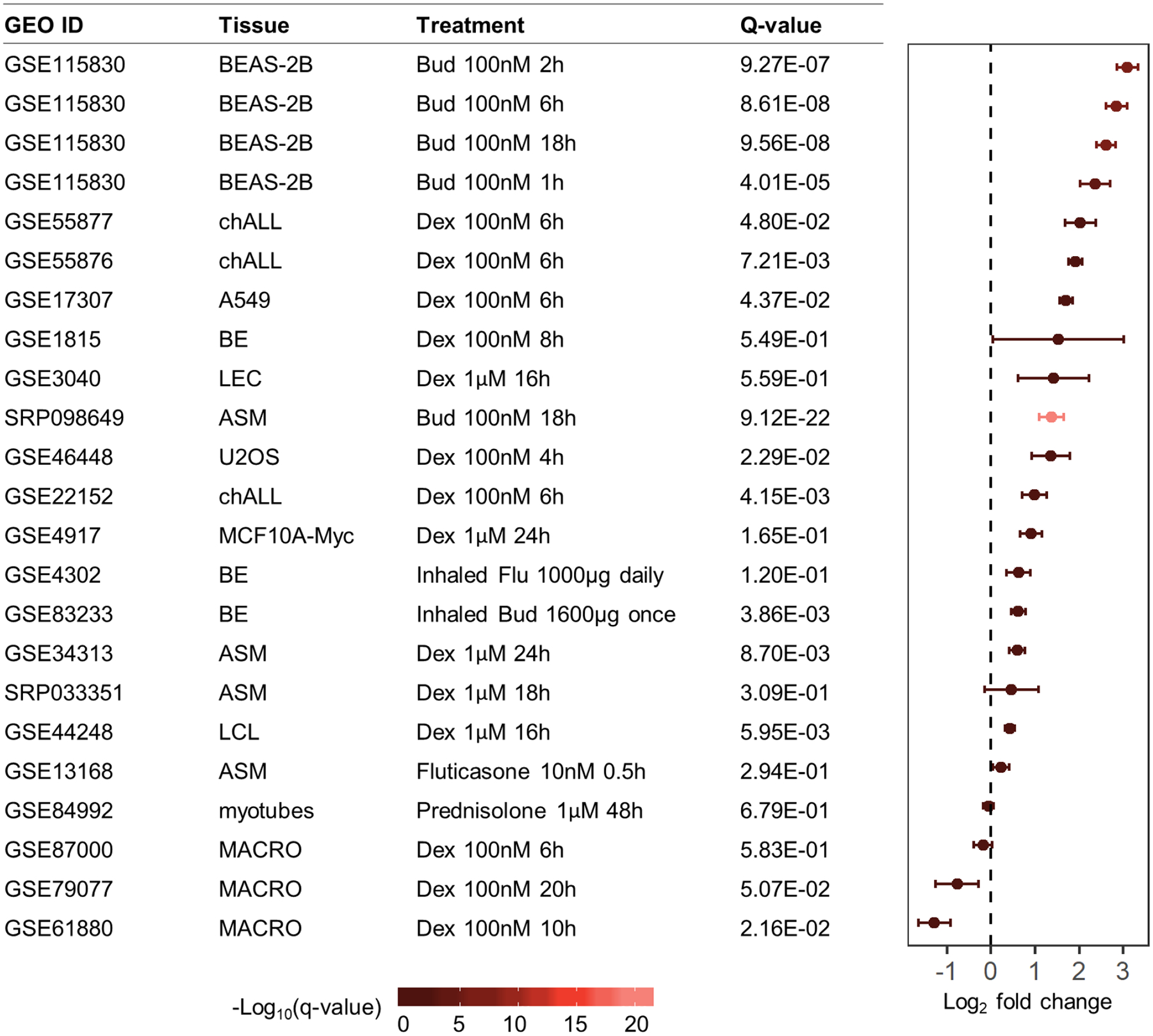
Differential expression results for BIRC3 in various tissues exposed to glucocorticoids versus control. ASM: airway smooth muscle; BE: bronchial epithelium; Bud: budesonide; chALL: childhood acute lymphoblastic leukemia; Dex: dexamethasone; LEC: lense epithelial cells; LEL: lymphoblastoid cell line; MACRO: macrophages; MCF10A-Myc: immortalized human mammary epithelial cell line MCF10A overexpressing c-Myc; U2OS: human bone osteosarcoma epithelial cell line.
Previously Reported ICS Response-Associated Variants Lack Evidence of Altering GR-binding
The set of GSK variants was expanded to 4,271 candidate variants by including 118 variants reported in other ICS response genetics studies3–10,24,28–31 and 4,153 variants in high LD with them (Table E6). None of the newly included variants overlapped with GSK GWAS variants. Two SNPs from the new set of variants overlapped with GR-binding sites (Table E3): 1) rs3794492, which was within 20kb of the TSS of LDHAL6B, was in high LD (r2≥0.8 based on 1000G EUR reference panel) with a GWAS SNP (p-value<5×10−8) reported as associated with dose-dependent response to ICS6, and 2) rs76390075, which was located 339kb of the TSS of LTBP1, was nominally associated (p-value<10−4) with asthma exacerbation despite ICS use in a prior study of Hispanic/Latino and African-American children24. Neither of these GR-binding sites had evidence of active transcription activity in the corresponding genes, nor were the eQTLs of the genes. Results with this expanded set of variants were generally consistent with those based on the GSK variants alone, and the same four variants near BIRC3 identified with GSK data remained the only significant ones according to multiomics integrative scores for the 8,739 variants (q-values<0.05) (Fig. E2, Table E6). Of note, multiomics integrative scores of variants in candidate genes GLCCI13, CRHR128, MAPT30, STIP131, and TBX2129 were not greater than those expected by chance (range 0 to 2, q-value≥0.05).
Attempted Replication of BIRC3 Variant Associations with ICS Response
Characteristics of subjects of four replication studies are in Table I. Forty-three of 52 BIRC3 variants had results available in these cohorts, and within cohorts, the effect size directions were consistent (Table E5). In the GERA cohort, minor alleles of SNP rs12576911 were associated with increased ICS response (p-value=0.046) (Fig. 3B, Table III): having copies of its minor alleles increased time to a subsequent asthma exacerbation (hazard ratio=0.942 for alternative (i.e., minor) allele compared to the reference (i.e., major) allele). Nine variants in the GALA II study and seven variants in the SAGE study were associated with having an asthma exacerbation in the prior year despite ICS use (p-value<0.05), with SNPs rs2604526 and rs11606268, respectively, having the smallest p-values. Having copies of minor alleles of BIRC3 locus variants corresponded to decreased asthma exacerbations in GALA II (odds ratio of rs2604526=0.752), and increased asthma exacerbations in SAGE (odds ratio of rs11606268=1.39). In the SLOVENIA cohort, none of the BIRC3 variants showed significant associations (p-value>0.05). Meta-analysis results found that, of 35 BIRC3 variants that had results in the five ICS response studies, 33, including the four GR-binding site-overlapping BIRC3 variants, had p-value<0.05 (Table E5).
Table III.
Replication results of variants within the BIRC3 locus with p<0.05 in at least one independent study.
| GERA ICS Response GWAS | |||||||
|---|---|---|---|---|---|---|---|
| SNP | Ref | Alt | Alt allele frequency | Hazard ratio | P-value | 1000G EUR panel based r2 | 1000G cosmopolitan panel based r2 |
| rs12576911 | A | G | 0.275 | 0.942 | 0.046 | 0.921 | 0.942 |
| GALA II ICS Response GWAS | |||||||
| SNP | Ref | Alt | Alt allele frequency | Odds ratio | P-value | 1000G EUR panel based r2 | 1000G cosmopolitan panel based r2 |
| rs2604526 | C | A | 0.297 | 0.752 | 0.014 | 0.945 | 0.687 |
| rs2852880 | G | C | 0.312 | 0.769 | 0.022 | 0.995 | 0.809 |
| rs2852881 | C | T | 0.312 | 0.769 | 0.022 | 0.995 | 0.813 |
| rs2852882 | T | A | 0.306 | 0.771 | 0.025 | 0.945 | 0.792 |
| rs7480738 | G | T | 0.299 | 0.775 | 0.029 | 0.982 | 0.685 |
| rs2852891 | G | T | 0.306 | 0.779 | 0.032 | 0.928 | 0.786 |
| rs12787745 | C | A | 0.325 | 0.790 | 0.041 | 0.912 | 0.759 |
| rs17878708 | G | A | 0.316 | 0.793 | 0.045 | 0.823 | 0.738 |
| rs150283650 | G | A | 0.340 | 0.803 | 0.050 | 0.921 | 0.889 |
| SAGE ICS Response GWAS | |||||||
| SNP | Ref | Alt | Alt allele frequency | Odds ratio | P-value | 1000G EUR panel based r2 | 1000G cosmopolitan panel based r2 |
| rs11606268 | C | A | 0.316 | 1.385 | 0.027 | 0.995 | 0.936 |
| rs2846857 | G | A | 0.314 | 1.369 | 0.034 | 1.000 | 0.943 |
| rs11600522 | A | T | 0.318 | 1.351 | 0.041 | 0.995 | 0.941 |
| rs150283650 | G | A | 0.311 | 1.350 | 0.043 | 0.921 | 0.889 |
| rs58277500 | C | G | 0.328 | 1.339 | 0.046 | 0.823 | 0.835 |
| rs12098902 | G | A | 0.322 | 1.334 | 0.049 | 0.921 | 0.888 |
| rs11602147 | C | G | 0.421 | 1.304 | 0.049 | 0.824 | 0.790 |
| UK Biobank Asthma GWAS | |||||||
| SNP | Ref | Alt | Alt allele frequency | Odds ratio | P-value | 1000G EUR panel based r2 | 1000G cosmopolitan panel based r2 |
| rs12280343 | T | C | 0.307 | 0.974 | 0.026 | 0.906 | 0.880 |
| rs2511293 | T | C | 0.299 | 0.975 | 0.032 | 0.932 | 0.882 |
| rs3132831 | C | T | 0.299 | 0.975 | 0.032 | 0.932 | 0.886 |
| rs2604526 | C | A | 0.300 | 0.975 | 0.032 | 0.945 | 0.687 |
| rs2852891 | G | T | 0.299 | 0.975 | 0.032 | 0.928 | 0.786 |
| rs2852882 | T | A | 0.300 | 0.975 | 0.033 | 0.945 | 0.792 |
| rs2846849 | G | A | 0.300 | 0.975 | 0.034 | 0.945 | 0.894 |
| rs11602147 | C | G | 0.320 | 0.976 | 0.038 | 0.824 | 0.790 |
| rs2604527 | C | T | 0.299 | 0.976 | 0.039 | 0.945 | 0.916 |
| rs2846848 | G | T | 0.299 | 0.976 | 0.039 | 0.941 | 0.891 |
| rs58277500 | C | G | 0.320 | 0.977 | 0.042 | 0.823 | 0.835 |
| rs17878708 | G | A | 0.318 | 0.977 | 0.046 | 0.823 | 0.738 |
| rs7480738 | G | T | 0.296 | 0.977 | 0.049 | 0.982 | 0.685 |
Hazard ratio and odds ratio use the alternative allele as reference. Pairwise r2 was computed between rs2846858 and the corresponding variant.
BIRC3 Variant Associations with Asthma
Characteristics of UK Biobank participants who comprised an asthma GWAS are in Table I. Forty-eight of 52 BIRC3 variants had results available, 13 of which were associated with asthma (p-value<0.05), with SNP rs12280343 having the smallest p-value of 0.026 (Fig. 3B, Table III). Having copies of minor alleles in these variants corresponded to decreased asthma risk (odds ratio of rs12280343=0.974) (Table E5). Characteristics of EVE participants can be found in Table 1 of a previous publication37. 15 of 52 variants had results available in at least one EVE racial/ethnic group sub-analysis, but none of the variants had p-value<0.05. Meta-analysis of the UK Biobank and EVE studies found that four of the 15 variants were significant overall (p-value<0.05) (Table E5).
Discussion
The discovery of reproducible glucocorticoid response genetic associations is challenged by the need to collect data on large numbers of patients with consistent and well-defined drug response phenotypes13. The largest published GWAS for ICS response that was based on 2,672 asthma patients enrolled in GSK clinical trials lacked statistically significant genetic associations, and its nominally-associated variants have not been the subject of further studies. We reasoned that some of the nominal associations may reflect true associations that did not reach genome-wide significant thresholds due to lack of statistical power stemming from its low sample size. Given that cellular-level glucocorticoid response mechanisms have been studied for many years, and various omics studies have characterized layers of relevant glucocorticoid-response data in an unbiased fashion, designing a biologically principled ad hoc integrative score to rank the nominally significant GWAS variants was possible. According to our multiomics integrative score, four GWAS SNPs that were part of a locus near BIRC3 were identified as having an influence on GR-mediated transcription changes in airway cells that was greater than that expected by chance.
To functionally support SNP-trait associations identified via GWAS, variants are often annotated with information from resources such as GTEx and ENCODE, to determine whether variants are eQTLs or overlap with regulatory elements, respectively. These resources are limited for the study of ICS response because 1) cell type-specific results for prominent asthma tissues such as ASM and airway epithelium are not available, and 2) transcriptomic and DNA-protein binding information for most tissues was measured at baseline. In the case of glucocorticoid responses, richer, trait-specific data can be obtained. We searched the repository GEO, which contains ChIP-Seq and transcriptomic data from published studies related to various cell types and treatment conditions, to identify GR ChIP-Seq datasets corresponding to airway structural cell types treated with equivalent dexamethasone dosage and exposure times, each of which had corresponding RNAP II ChIP-Seq experiments, as well as to obtain a large number of gene expression microarray and RNA-Seq studies involving glucocorticoids. We are limited by the currently available eQTL data in GTEx, which does not include that from airway smooth muscle cells. Additionally, eQTL data derived from people with asthma, and in particular, eQTL data corresponding to the same donors with GWAS and drug response data would enable more precise studies into glucocorticoid response. However, the GTEx eQTL results of asthma-relevant tissues from non-asthma donors we used have been helpful in previous asthma GWAS studies40,41, and skeletal muscle has been found to have eQTLs that were colocalized with asthma GWAS variants42, suggesting that the currently used data are of value to study glucocorticoid response in people with asthma. Although our integration results were biased according to the currently available studies in terms of cell types and choice of glucocorticoid types, dosages and time frames, by integrating all available data, our results were as robust as is currently possible to identify mechanistic hypotheses for consideration.
More broadly, there is an increasing interest in integrative multiomics approaches commensurate with the large amount of omics data being generated. Some of the existing approaches include EUGENE43, which aggregates GWAS summary statistics from independent eQTLs; PrediXcan44 and S-PrediXcan45, which use gene expression prediction models derived from eQTL data to impute gene-level expression in a GWAS dataset; and colocalization methods that identify overlapping GWAS and eQTL and/or methylation quantitative trait locus (mQTL) signals46,47. Our method is novel in that it focuses specifically on glucocorticoid responsiveness, and as such, the omics datasets selected for it uniquely address this trait. For example, inclusion of transcription factor binding information has not been used in prior multiomics efforts, but in the case of glucocorticoid response, knowing where the glucocorticoid receptor binds to DNA suggests a direct biological mechanism for DNA variants to influence response to glucocorticoid drugs. We chose to use a binary score for each omics layer based on the presence of evidence in favor of altered GR-binding and/or transcription activity partly because we selected datasets closely related to glucocorticoid response in lung, but a more generalized version of our approach could include a greater number of datasets and/or weight the individual tissues and layers differently based on prior knowledge. For example, the effect sizes of association of individual SNPs could be used as weights to preferentially select SNPs with stronger association signals. As relevant new omics data becomes available (e.g., eQTL for airway smooth muscle and/or people with asthma), the scoring metrics can be adapted to include these data to generate additional hypotheses for mechanistic studies.
The ICS response outcome used in the GSK GWAS, change in FEV1 after 8–12 weeks of ICS use measured during multicenter clinical trials, is not available in other studies. We nonetheless sought support for genetic associations with ICS response and asthma using independent studies with subjects from diverse populations. In the GSK study, minor alleles of rs2846858 were associated with decreased change in FEV1 after ICS treatment, and thus, decreased response to ICS. Although rs2846858 was not significantly associated with ICS response in the GERA study, minor alleles of rs12576911, another SNP in the BIRC3 locus, were associated with increased time to a future asthma exacerbation, a direction of effect that appears opposite to that observed in the GSK study. However, it is possible that the minor alleles of variants in this locus influence change in FEV1 differently from asthma exacerbations. The SLOVENIA study, which had an outcome that resembled that of the GSK study, did not find any associations in the locus, but we note that its sample size was particularly small (n=166). The outcomes assessed in the GALA II and SAGE studies, namely exacerbations in the prior year while on ICS, resembled those of GERA, but while the direction of effect in GALA II was consistent with GERA, the direction in SAGE was opposite. These contrasting directions of effect may be due to the different racial/ethnic groups represented by these studies—GERA consisted of people of European ancestry, GALA II consisted of Hispanic/Latino subjects, while SAGE consisted of African American subjects. Specifically, the BIRC3 locus was a distinct region consisting of variants in high LD according to EUR, cosmopolitan and admixed American (AMR) reference panels, but the LD patterns of the variants differed substantially in 1000G data of African populations (AFR) (Fig. 3B). In terms of asthma, minor alleles of 13 variants within the BIRC3 locus among UK Biobank participants were associated with decreased asthma risk, indicating a protective role, though this association was not observed among EVE participants.
Our study was limited in its ability to account for differences in genetic ancestry and identify results that may be unique to some racial/ethnic groups. Participants in the original GSK study were of heterogeneous ancestral backgrounds, with 67% of participants identified as having European ancestry and remaining participants as being from other racial/ethnical groups (i.e., 15% were Hispanic, 10% were Asian, 5% were African, and less than 1% were Native American or of mixed ancestry)11. We selected GWAS variants based on having high LD (r2≥0.8) with 1000G EUR reference panel and, separately, the 1000G cosmopolitan reference panel, to determine whether our results would remain consistent based on choice of reference panel to identify independent regions of association. We found that our multiomics results did not substantially change: 1) the same four BIRC3 variants identified with the EUR panel as reference remained the only significant ones when using the 2,822 variants derived from 1000G cosmopolitan reference panel; 2) 40 variants were in high LD (r2≥0.8 based on 1000G cosmopolitan reference panel) with rs2846858—the top-ranked SNP using both reference panels—all of which were among the 52 variants in high LD with rs2846858 using the 1000G EUR reference panel (Table E5); 3) because all 40 variants from the cosmopolitan panel were among the 52 obtained with the EUR panel, variants that were nominally associated with ICS response or asthma among the 40 variants were also replicated when using the EUR reference panel. Future studies are needed to clarify differences in association results within this BIRC3 locus according to genetic ancestry. Additionally, future primary glucocorticoid response GWAS conducted in a large number of non-European subjects may identify novel ICS response loci that are specific to non-European racial/ethnic groups.
The BIRC3 gene encodes the protein baculoviral IAP repeat containing 3 (aka. cIAP2), a member of the family of inhibitor of apoptosis (IAP) proteins, which are critical suppressors of cell death and key mediators in inflammatory signaling and immunity via the activation of NF-κB pathways48. Prior publications have also reported associations with BIRC3 variants: 1) a candidate gene study found that a region not in LD with the one presently reported was associated with asthma, eosinophil abundance, and neutrophil abundance (p-value<0.05)49; and 2) a GWAS that considered protein expression levels as outcomes50, found that an intronic BIRC3 SNP was significantly associated with levels of matrix metalloproteinase-1, a protein linked to airway hyperresponsiveness and asthma severity51. Prior studies also support the involvement of BIRC3 in the modulation of asthma-relevant inflammatory pathways. Primary nasal epithelial cells exposed to IFNγ or TNFα had increased BIRC3 protein expression, while BIRC3-depleted cells had increased expression of the apoptosis protein caspase-3 with TNFα exposure, suggesting that BIRC3 protected airway epithelial cells from apoptosis when exposed to pro-inflammatory cytokines52. Birc3-deficient mice had increased susceptibility to influenza-virus infection and impaired airway epithelium integrity mediated via death-receptor-induced programmed necrosis, suggesting that BIRC3 protected airway cells via maintenance of lung tissue homeostasis and host fitness53. Transcriptomic studies reported that BIRC3 had increased gene expression levels in lymphoblastoid cell lines54 and airway cells15,55 when exposed to glucocorticoids, as well as increased expression levels in asthma patients with higher sputum neutrophils56. We found that minor alleles of rs2846858 corresponded to increased BIRC3 expression in lung according to GTEx results, and that BIRC3 gene expression increased in response to glucocorticoid exposure in airway cells (Fig. 4), suggesting that minor allele carriers may have increased expression of BIRC3 in response to glucocorticoid exposure compared to non-carriers. Future studies that capture individual BIRC3 gene expression across participants can be used to conduct mediation analysis, thereby clarifying the relationship between genetic associations, gene expression and glucocorticoid response outcomes.
In summary, our multiomics integration study identified a novel ICS response-associated locus near BIRC3 that is worth pursuing in mechanistic studies. Our demonstration that nominally-associated GWAS loci can be effectively prioritized via integration of multiomics data that is tailored to an outcome of interest, is an approach that can be applied to study other complex traits.
Supplementary Material
Acknowledgements
We thank Dr. Soumitra Ghosh, Ph.D., M.D. from GSK, and Lynn Condreay, Ph.D. and Dana J. Fraser of Parexel International, for generously providing the ICS response GWAS results, and we are grateful to Sandra Salazar for her support as GALA II and SAGE study coordinator. We acknowledge all of the patients, families, recruiters, researchers, health care providers, and community clinics for their participation in the various genetic studies. This research has been conducted using the UK Biobank Resource under Application Number 40375.
Sources of funding:
Funding was provided by National Institutes of Health (NIH) awards R01 HL133433, R01 HL141992, P30ES013508, K23HL151819, R01 HL117004, X01HL134589, R01HL155024, R01MD010443, and R01ES015794, the Spanish Ministry of Science and Innovation grant RYC-2015-17205 and fellowship FPU19/02175, the European Academy of Allergy and Clinical Immunology (EAACI) Medium-Term Research Fellowship 2021, the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished in Pharmaceutical Sciences II.
Abbreviations
- 1000G
1000 Genomes Project
- ASM
airway smooth muscle
- ICS
inhaled corticosteroids
- eQTL
expression quantitative trait locus
- EUR
1000 Genomes European ancestry population
- FEV1
forced expiratory volume in one second
- GALA II
Genes-environments & Admixture in Latino Americans Study
- GEO
Gene Expression Omnibus
- GERA
Genetic Epidemiology Research on Adult Health and Aging
- GSK
GlaxoSmithKline
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OCS
oral corticosteroid
- RNAP II
RNA polymerase II
- SAGE
Study of African Americans, Asthma, Genes, and Environments
- SNP
single nucleotide polymorphism
- SRA
Sequence Read Archive
- TSS
transcription start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no conflicts of interest.
References
- 1.Expert Panel Working Group of the National Heart, Lung, and Blood Institute (NHLBI) administered and coordinated National Asthma Education and Prevention Program Coordinating Committee (NAEPPCC), Cloutier MM, Baptist AP, Blake KV, Brooks EG, Bryant-Stephens T, et al. 2020 Focused Updates to the Asthma Management Guidelines: A Report from the National Asthma Education and Prevention Program Coordinating Committee Expert Panel Working Group. J Allergy Clin Immunol. 2020;146:1217–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–73. [DOI] [PubMed] [Google Scholar]
- 3.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tantisira KG, Damask A, Szefler SJ, Schuemann B, Markezich A, Su J, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med. 2012;185:1286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park H-W, Dahlin A, Tse S, Duan QL, Schuemann B, Martinez FD, et al. Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. J Allergy Clin Immunol. 2014;133:664–669.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Tong C, Wang Z, Wang Z, Mauger D, Tantisira KG, et al. Pharmacodynamic genome-wide association study identifies new responsive loci for glucocorticoid intervention in asthma. Pharmacogenomics J. 2015;15:422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez-Pacheco N, Vijverberg SJ, Herrera-Luis E, Li J, Sio YY, Granell R, et al. Genome-wide association study of asthma exacerbations despite inhaled corticosteroid use. Eur Respir J. 2021;57:2003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez-Pacheco N, Gorenjak M, Li J, Repnik K, Vijverberg SJ, Berce V, et al. Identification of ROBO2 as a Potential Locus Associated with Inhaled Corticosteroid Response in Childhood Asthma. JPM. 2021;11:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park T-J, Park J-S, Cheong HS, Park B-L, Kim LH, Heo JS, et al. Genome-wide association study identifies ALLC polymorphisms correlated with FEV1 change by corticosteroid. Clin Chim Acta. 2014;436:20–6. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Pacheco N, Farzan N, Francis B, Karimi L, Repnik K, Vijverberg SJ, et al. Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clin Exp Allergy. 2019;49:789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosteller M, Hosking L, Murphy K, Shen J, Song K, Nelson M, et al. No evidence of large genetic effects on steroid response in asthma patients. Journal of Allergy and Clinical Immunology. 2017;139:797–803.e7. [DOI] [PubMed] [Google Scholar]
- 12.Kan M, Himes BE. Genetics and Pharmacogenetics of Asthma. In: Precision in Pulmonary, Critical Care, and Sleep Medicine [Internet]. 1st ed. Springer; 2020. p. 25–37. (Respiratory Medicine). Available from: https://link.springer.com/chapter/10.1007/978-3-030-31507-8_3 [Google Scholar]
- 13.Kan M, Himes BE. Insights into glucocorticoid responses derived from omics studies. Pharmacol Ther. 2020;107674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasse SK, Kadiyala V, Danhorn T, Panettieri RA, Phang TL, Gerber AN. Glucocorticoid Receptor ChIP-Seq Identifies PLCD1 as a KLF15 Target that Represses Airway Smooth Muscle Hypertrophy. Am J Respir Cell Mol Biol. 2017;57:226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadiyala V, Sasse SK, Altonsy MO, Berman R, Chu HW, Phang TL, et al. Cistrome-based Cooperation between Airway Epithelial Glucocorticoid Receptor and NF-κB Orchestrates Anti-inflammatory Effects. J Biol Chem. 2016;291:12673–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kan M, Koziol-White C, Shumyatcher M, Johnson M, Jester W, Panettieri RA, et al. Airway Smooth Muscle-Specific Transcriptomic Signatures of Glucocorticoid Exposure. Am J Respir Cell Mol Biol. 2019;61:110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, et al. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One. 2014;9:e99625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kan M, Shumyatcher M, Diwadkar A, Soliman G, Himes BE. Integration of Transcriptomic Data Identifies Global and Cell-Specific Asthma-Related Gene Expression Signatures. AMIA Annu Symp Proc. 2018;2018:1338–47. [PMC free article] [PubMed] [Google Scholar]
- 20.Diwadkar AR, Kan M, Himes BE. Facilitating Analysis of Publicly Available ChIP-Seq Data for Integrative Studies. AMIA Annu Symp Proc. 2019;2019:371–9. [PMC free article] [PubMed] [Google Scholar]
- 21.Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, et al. Distinct Properties of Cell-Type-Specific and Shared Transcription Factor Binding Sites. Molecular Cell. 2013;52:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kan M, Shumyatcher M, Himes BE. Using omics approaches to understand pulmonary diseases. Respir Res. 2017;18:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hersh CP, Adcock IM, Celedón JC, Cho MH, Christiani DC, Himes BE, et al. High-Throughput Sequencing in Respiratory, Critical Care, and Sleep Medicine Research. An Official American Thoracic Society Workshop Report. Ann Am Thorac Soc. 2019;16:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez-Pacheco N, Gorenjak M, Jurgec S, Corrales A, Jorgensen A, Karimi L, et al. Combined analysis of transcriptomic and genetic data for the identification of loci involved in glucocorticosteroid response in asthma. Allergy. 2020;76:1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13:1353–9. [DOI] [PubMed] [Google Scholar]
- 29.Tantisira KG, Hwang ES, Raby BA, Silverman ES, Lake SL, Richter BG, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004;101:18099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tantisira KG, Lazarus R, Litonjua AA, Klanderman B, Weiss ST. Chromosome 17: association of a large inversion polymorphism with corticosteroid response in asthma. Pharmacogenet Genomics. 2008;18:733–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins GA, Lazarus R, Smith RS, Tantisira KG, Meyers DA, Peters SP, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol. 2009;123:1376–1383.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlin A, Sordillo JE, Ziniti J, Iribarren C, Lu M, Weiss ST, et al. Large-scale, multiethnic genome-wide association study identifies novel loci contributing to asthma susceptibility in adults. Journal of Allergy and Clinical Immunology. 2019;143:1633–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White MJ, Risse-Adams O, Goddard P, Contreras MG, Adams J, Hu D, et al. Novel genetic risk factors for asthma in African American children: Precision Medicine and the SAGE II Study. Immunogenetics. 2016;68:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pino-Yanes M, Thakur N, Gignoux CR, Galanter JM, Roth LA, Eng C, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2015;135:228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach: Combining probabilities from many tests. Journal of Evolutionary Biology. 2005;18:1368–73. [DOI] [PubMed] [Google Scholar]
- 39.Shumyatcher M, Hong R, Levin J, Himes BE. Disease-Specific Integration of Omics Data to Guide Functional Validation of Genetic Associations. AMIA Annu Symp Proc. 2017;2017:1589–96. [PMC free article] [PubMed] [Google Scholar]
- 40.Valette K, Li Z, Bon-Baret V, Chignon A, Bérubé J-C, Eslami A, et al. Prioritization of candidate causal genes for asthma in susceptibility loci derived from UK Biobank. Commun Biol. 2021;4:700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautam Y, Afanador Y, Ghandikota S, Mersha TB. Comprehensive functional annotation of susceptibility variants associated with asthma. Hum Genet. 2020;139:1037–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z, Guo Y, Shi H, Liu C-L, Panganiban RA, Chung W, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. Journal of Allergy and Clinical Immunology. 2020;145:537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira MAR, Jansen R, Willemsen G, Penninx B, Bain LM, Vicente CT, et al. Gene-based analysis of regulatory variants identifies 4 putative novel asthma risk genes related to nucleotide synthesis and signaling. J Allergy Clin Immunol. 2017;139:1148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.GTEx Consortium, Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GTEx Consortium, Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9:1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hormozdiari F, van de Bunt M, Segrè AV, Li X, Joo JWJ, Bilow M, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet. 2016;99:1245–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silke J, Meier P. Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb Perspect Biol. 2013;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roscioli E, Hamon R, Ruffin RE, Grant J, Hodge S, Zalewski P, et al. BIRC3 single nucleotide polymorphism associate with asthma susceptibility and the abundance of eosinophils and neutrophils. Journal of Asthma. 2017;54:116–24. [DOI] [PubMed] [Google Scholar]
- 50.Folkersen L, Gustafsson S, Wang Q, Hansen DH, Hedman ÅK, Schork A, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2:1135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naveed S-U-N, Clements D, Jackson DJ, Philp C, Billington CK, Soomro I, et al. Matrix Metalloproteinase-1 Activation Contributes to Airway Smooth Muscle Growth and Asthma Severity. Am J Respir Crit Care Med. 2017;195:1000–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roscioli E, Hamon R, Ruffin RE, Lester S, Zalewski P. Cellular inhibitor of apoptosis-2 is a critical regulator of apoptosis in airway epithelial cells treated with asthma-related inflammatory cytokines. Physiol Rep. 2013;1:e00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodrigue-Gervais IG, Labbé K, Dagenais M, Dupaul-Chicoine J, Champagne C, Morizot A, et al. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 2014;15:23–35. [DOI] [PubMed] [Google Scholar]
- 54.Sharma S, Kho AT, Chhabra D, Qiu W, Gaedigk R, Vyhlidal CA, et al. Glucocorticoid genes and the developmental origins of asthma susceptibility and treatment response. Am J Respir Cell Mol Biol. 2015;52:543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leigh R, Mostafa MM, King EM, Rider CF, Shah S, Dumonceaux C, et al. An inhaled dose of budesonide induces genes involved in transcription and signaling in the human airways: enhancement of anti- and proinflammatory effector genes. Pharmacol Res Perspect. 2016;4:e00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127:153–60, 160.e1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


