Abstract
Discovery of therapeutics for neurological diseases is hampered by the lack of predictive in vitro and in vivo models. Traditionally, in vitro assays rely on engineered cell lines grown two-dimensionally (2D) outside a physiological tissue context, which makes them very amenable for large scale drug screening but reduces their relevance to in vivo neurophysiology. In recent years, three-dimensional (3D) neural cell culture models derived from human induced pluripotent stem cells (iPSCs) have been developed as an in vitro assay platform to investigate brain development, neurological diseases, and for drug screening. iPSC-derived neural spheroids or organoids can be developed to include complex neuronal and glial cell populations and display spontaneous, synchronous activity, which is a hallmark of in vivo neural communication. In this report we present a proof-of-concept study evaluating 3D iPSC-derived cortical neural spheroids as a physiologically- and pharmacologically-relevant high-throughput screening (HTS) platform and investigate their potential for use for therapeutic development. To this end, a library of 687 neuroactive compounds were tested in a phenotypic screening paradigm which measured calcium activity as a functional biomarker for neural modulation through fluctuations in calcium fluorescence. Pharmacological responses of cortical neural spheroids were analyzed using a multi-parametric approach, whereby seven peak characteristics from the calcium activity in each well were quantified and incorporated into principal component analysis and Sammon mapping to measure compound response. Here, we describe the implementation of the 687-compound library screen and data analysis demonstrating that iPSC-derived cortical spheroids are a robust and information-rich assay platform for HTS.
Keywords: iPSC, neural spheroids, high-throughput screen
Introduction
Neurological disorders are the leading cause of disability and second leading cause of death worldwide 1,2. In recent years, diagnoses for both psychiatric and neurodegenerative diseases have increased due to factors such as increased aging populations and ineffective treatment options 2,3. Although there is increased investment in research and development for neurological diseases, still only around 10% of treatments in clinical trials reach approval by the United States Food and Drug Administration 4-6. While this high failure rate is in part because of the cost and complexity of clinical trials, inadequate disease modeling and drug-screening platforms play a significant role as well 5,7. One way to address this treatment deficit is through the development of drug discovery platforms that have improved predictability of efficacy and safety.
Three-dimensional (3D) cellular organotypic models, including cortical neural spheroids and organoids derived from human induced pluripotent stem cells (iPSCs), have emerged as an alternative drug discovery platform to traditional two-dimensional (2D) in vitro and animal model approaches 8,9. Traditional 2D cellular models rely on engineered cell lines which, although powerful as tools to develop technically robust assays for large scale drug screening, do not capture the molecular biology of cells in their native physiological tissue context, thus reducing in vivo neurophysiological relevance. While animal models have provided valuable insight into the function of neural circuits, species differences exist that often limit the scope of these studies 10,11. These differences are particularly pronounced in brain areas such as cerebral cortex which, as the most recently evolved brain structure, is significantly more complex in both structure and function in humans compared to other animals 12,13.
Three-dimensional iPSC-derived cortical spheroids and organoids, in contrast to 2D in vitro models, acquire a more brain-like neural cell complexity to model in vivo neural connections and may therefore represent more physiologically-relevant models 14-16. Both cerebral and patterned brain organoids are generated by iPSC growth and differentiation to form large complex cellular arrangements with some brain-like spatial architecture. While complexity varies between the preparations, both have demonstrated utility over 2D models 8-9,17. While organoids may have higher physiological relevance and biomimicry than spheroids, their production requires long incubation times, they are heterogenous in cell composition and size, and their large size can generate a necrotic core and makes the development of functional assays more challenging for high-throughput screening (HTS) 8. Spheroids are generated by cellular self-assembly and include different neural cell-types. Though spheroids are generated with less structural organization compared to organoids, their assembly requires shorter incubation times and produces smaller, highly homogenous, functional cellular aggregates, which make them more amenable for use in HTS applications 17,18.
Additionally, 3D cortical spheroids show both longer neurite growth and increased population neural activity compared to 2D models 15,17. Dimensionality differences provide advantages to 3D models, enabling the structural formation of synapses that facilitate neuronal communication through the release of neurotransmitters and ions that produce action potentials within single cells. Local field potentials (LFPs) represent the summation of these spontaneously generated action potentials among groups of neurons, and this activity occurs as synchronous oscillations within a system that are generated spontaneously to drive neuronal communication between brain regions in vivo 19,20. Previous reports have shown that 3D cortical spheroids display spontaneous, synchronized activity that is more reproducible than what is observed in 2D cultures, suggesting that these systems are more translatable than 2D models 15,17. Network activity is a critical aspect of in vivo neuronal communication and disease modeling, given that synchronous cortical neuronal activity regulates cognitive behaviors such as attention and working memory, which are impaired in diseases like schizophrenia and addiction 21-24. This spontaneous, synchronized neuronal activity can be measured through changes in intracellular calcium concentrations, which correlate with the electrophysiological properties of neurons, and measured with a calcium dye on a fluorescent imaging plate reader (FLIPR) 17,25. Moreover, global cell population activity is impaired in disease states but can also be modulated and controlled through pharmacological manipulation 17,18. As such, calcium imaging is an advantageous method for HTS since changes in population neural cell activity after drug treatment can be assessed in hundreds of wells simultaneously.
Here we present a proof-of-concept study investigating the applicability of the iPSC-derived cortical spheroids as a neural assay platform for high-throughput drug screening. To do this, we used the StemoniX microBrain platform, which has been previously characterized by reports showing these spheroids contain neurons and astrocytes that produce functional responses, as measured by both calcium and activity and LFPs, that can be modulated pharmacologically 17,26-28. Here, a fluorescent imaging plate reader (FLIPR) was used to conduct a calcium fluorescence assay to measure changes in spheroid neural activity. We created a focused library of 687 neuroactive compounds in order to establish the pharmacological profile of the cortical spheroids as well as evaluate their robustness in the context of drug screening. In line with the objectives of the Helping End Addiction Long-term (HEAL) initiative at the National Center for Advancing Translational Sciences, a focus was placed on choosing compounds that could be used for future disease modeling and screening for opioid use disorder (OUD). Therefore, an emphasis was placed on compounds targeting opioid receptors given that chronic opioid use disrupts the homeostasis of reward signaling through increased activation of these receptors 29. Additionally, psychoactive compounds that play a role in depression, anxiety, and analgesia were included. We also developed a data analysis pipeline to classify pharmacological activity profiles from multiparametric calcium wave measurements. We found that our cortical spheroid assay platform performed robustly in a 384-well plate format and enabled the classification of compounds based on multiparametric calcium activity profiles.
Materials and Methods
Human iPSC-derived Cortical Spheroids
Pre-formed human iPSC-derived cortical spheroids were obtained from StemoniX, Inc. in a 384-well format, with four 384-well plates used for compound screening and one plate used to test seven concentrations of select compounds (3D microBrain platform, StemoniX, Inc., Cat. No. BSARX-AA-0384; Fig. S2). Plates containing spheroids that had been matured for 9.5 weeks were shipped from StemoniX, Inc. after and maintained following manufacturer’s instructions upon arrival. Specifically, plates were centrifuged at 400 x g for 2 minutes after receiving plates but prior to removal of shipping lid. The presence of spheroids was confirmed in each well after removal of the shipping lid by manual visual inspection, and 25 μL existing cell culture media was exchanged with 25 μL fresh cell culture media using an automated liquid handler (Biotek MultiFlo™ FX). The removal and addition process was repeated a total of 3 times, leaving a total of 50 μL of media per well. Cell culture media for the initial media change as well as for maintenance of spheroids consisted of BrainPhys Neuronal Medium and SM1 kit (StemCell Tech Cat #05792), supplemented with 20 ng/mL brain-derived neurotrophic factor (BDNF, StemCell Technologies, Cat.# 78005), 20 ng/mL glial-derived neurotrophic factor (GDNF, StemCell Technologies, Cat.# 78058), and 1X penicillin/streptomycin (Gibco). Plates containing spheroids were incubated in a standard tissue culture incubator, and 25 μL of cell culture media was exchanged every 2-3 days for one week prior to screening. Therefore, spheroids had been matured for 10.5-weeks at the time of the compound screen was conducted.
A compound library targeting relevant neural signaling pathways
The compound library consisted of 687 compounds targeting neural targets and signaling pathways. A full annotated list of the compound library can be found in excel file in supplemental material (Table S1). The library was designed with an emphasis on opioids that spanned across two 384-well plates, with two central columns (32 wells) on each plate dedicated to control compounds, and 704 wells dedicated to test compounds, which were run in duplicates. Control treatments consisted of 0.1% dimethyl sulfoxide (DMSO) as vehicle control, 10 μM muscimol as an inhibitory functional control (i.e. inhibiting functional activity), and 10 μM 4-aminopyridine (4-AP) as a stimulatory functional control (i.e. increasing functional activity). Compounds were screened in duplicate over a total of four plates such that technical repeats were spread across two different plates. Most compound concentrations were 10 μM with some exceptions (Table S1).
To investigate the effects of compound concentration on activity, control compounds along with a select group of compounds that were included in the library, were tested at seven concentrations using a seven point serial dilution (0.1, 0.4, 1.2, 3.7, 11.1, 33.3, and 100 μM; Fig. S2). For this experiment, compounds were tested in triplicate for each concentration at n=2 biological replicates such that six total replicates were tested for each concentration of each compound. On each batch of spheroids tested, 21 control wells were treated with 0.1% DMSO. Compounds tested included the stimulatory control, 4-AP, inhibitory control, muscimol, along with Cutamesine (sigma receptor agonist), dihydrexidine (dopamine 1/5 receptor agonist), DNQX (AMPAR antagonist), MK-801 (NMDAR antagonist), and kainic acid (excitatory amino acid).
Fluorescent Imaging Plate Reader (FLIPR) Assay
To examine neural cell activity changes in response to compound treatment, calcium fluorescence was assessed using a Fluorescent Imaging Plate Reader (FLIPR) Tetra (Molecular Devices). On the same day of imaging but prior to the addition of Calcium 6 (Cal6) dye, a fluorophore that binds to intracellular calcium ions to act as a calcium indicator, media was exchanged for phenol-free BrainPhys Neuronal Medium (StemCell Technologies, Cat. No. 05791). First, 30 μL of media was aspirated per well. Next, 20 μL of phenol-free media was added per well, and then 20 μL was aspirated. This process was repeated a total of 3 times, again checking to ensure spheroids were settled prior to aspiration. Two hours before baseline imaging, the FLIPR Calcium 6 assay kit (Cal6, Molecular Devices, Cat. No. R8190) was used according to manufacturer’s instructions, where Cal6 dye was dissolved and vortexed to mix with 10 mL of cell culture media. Then, 20 μL of Cal6 reagent was added to each well using a 16-channel manual pipette, leaving a total volume of 40 μL per well, required to minimize media spillage in the subsequent acoustic dispensing step. Spheroid plates were incubated at 37°C in standard incubation conditions for 2-hours to allow for reagent penetration. A fluorescent imaging plate reader (FLIPR) Tetra system was used for kinetic imaging of calcium dynamics in the cortical spheroids. The FLIPR incubator stage was pre-warmed to 37°C approximately 30 minutes prior to imaging. Standard filter sets were used for Cal6 imaging (excitation 470-495 and emission 515-575 nm). FLIPR gain was set to 2000, exposure time to 0.5330 seconds, excitation intensity to 30%, and gate opening time to 6%. Fluorescent image reads were taken every 0.6 seconds for a total of 800 reads over a total run time of 8 minutes. A baseline read was taken for each spheroid plate prior to drug treatment. Baseline reading and subsequent drug treatments were staggered such that all drug treatment incubations were 90 minutes.
Drug Treatment Protocol
Compound solutions from the library compound plates were acoustically dispensed into the cortical spheroid-containing plates using Echo 655 Liquid Handling System (Beckman Coulter), 90-min before calcium activity was assessed. Briefly, compounds were prepared in DMSO at a concentration 1000X the final treatment concentration. The acoustic dispenser was programmed to transfer a compound volume of 40 nL into spheroid-containing wells with 40 μL media. Inside the acoustic dispenser, the source plate was oriented in a standard orientation, and the spheroid plate was flipped upside down to receive the transferred droplet from the source plate. Because of this spheroid plate movement after acoustic dispensing, spheroid plates were centrifuged at 1485 x g for 2 minutes after dispensing to re-center spheroids for subsequent FLIPR imaging. Following centrifugation, spheroids were incubated at 37°C in standard incubation conditions for 90 minutes to allow for compound penetration. Spheroid plates were then read on the FLIPR using the previously described imaging settings.
Immunostaining
Immunostaining of 3D neurospheroids was performed as previously described 30. Briefly, spheroids were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) overnight at 4°C, washed several times with PBS, and permeabilized in 0.2% Triton-X in PBS for 30 minutes at room temperature. All wash steps and solution changes were accomplished via a two-thirds solution exchange protocol (i.e., aspirate to 25μL/well, add 50μL/well fresh solution). Blocking solution (2% BSA + 0.2% Triton-X in PBS) was then added and spheroids were incubated at 37°C for >1 hour under gentle agitation. Primary antibodies (MAP2, Synaptic Systems 188 004, 1:1000; GFAP, Novus Biologicals, NBP1-05198, 1:1000) were added in blocking solution and incubated overnight at 37°C with gentle agitation followed by 6 washes using PBS + 0.2% Triton-X. Spheroids were blocked again for >1 hour and incubated with secondary antibodies (Invitrogen, A-21450 and A-11039) diluted 1:500 in blocking solution with DAPI (Invitrogen, 1:5000) overnight at 37°C with gentle agitation, protected from light. Spheroids were then washed 6 times using PBS, transferred to flat bottom plates and either stored at 4°C or imaged using an Operetta CLS spinning disk confocal microscope (Perkin Elmer) at 20x magnification (Fig. S1).
FLIPR Data Analysis
The FLIPR software ScreenWorks PeakPro 1.0 was used for the initial analysis of calcium activity from the compound screen. Using the kinetic reduction configuration function, peaks were identified using the following settings: smooth width of 5 data points, fit width of 10 data points, slope threshold of 0.001/s, and a dynamic amplitude threshold of 100 relative light units. For the experiment testing seven concentrations of compounds, ScreenWorks PeakPro 2.0 was used. Here, the vector length was set to 13 for all wells, and both trigger level and dynamic threshold were set to 10% above baseline for peak detection. With these peak detection settings, average peak statistics over the 8-min recording for each well were exported from PeakPro to Microsoft Excel. Exported statistics included: peak count, peak width, peak width standard deviation (SD), peak amplitude, peak amplitude SD, peak spacing, peak spacing SD, peak rise time, peak rise time SD, peak decay time, peak decay time SD, and peak width at 10% amplitude (Fig. 1b).
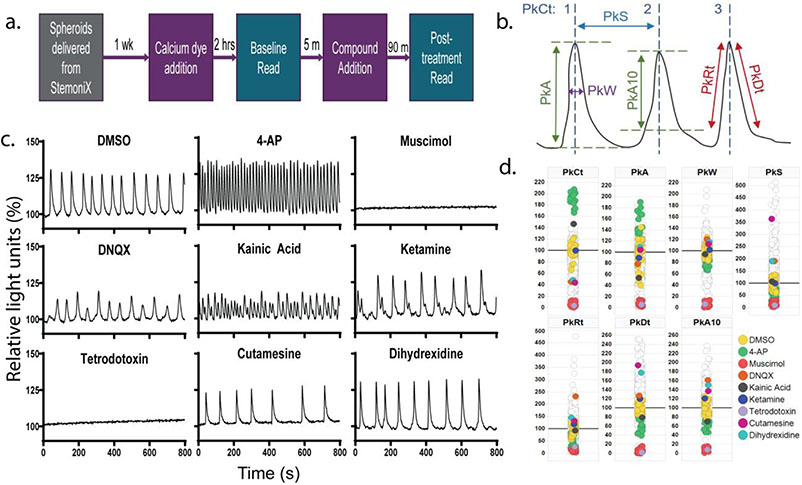
Figure 1.
A multiparametric calcium fluorescence assay for assessing neural cell activity in cortical spheroids. (a.) Schematic showing the timeline of the experiment. StemoniX cortical spheroids are maintained for 1-week prior to imaging with the FLIPR. Calcium 6 (Cal6) dye is added and 2-hrs later, baseline activity is recorded; 5-min after the baseline recording, compounds are added to spheroids via acoustic dispensing, and calcium activity is recorded again 90-min after compound treatment. FLIPR recordings are 8-min. (b.) Schematic defining peak parameters obtained from Peak Pro 1.0 analysis of calcium imaging data. PkCt = Peak Count; PkS = Peak Spacing; PkW = Peak Width; PkA = Peak Amplitude for whole peak; PkA10 = Peak Amplitude beginning at 10% peak height; PkRt = Peak Rise Time; PkDt = Peak Decay Time. (c.) Representative time series plots showing calcium activity after treatment with control compounds that should reliably show activity changes based on cell types represented. Plots are displayed as percent change in calcium fluorescence over baseline, which is considered the first 2 seconds of the recording. (d.) Scatter plots showing the percent change from the DMSO average for all compounds tested across each peak parameter used in the HCP analysis. Compounds from 1c are highlighted, and data is shown in comparison to DMSO-treated wells in iPSC-derived cortical spheroids are highlighted, along with stimulatory (4-AP) and inhibitory (muscimol) controls.
To determine which peak parameters were included in the analysis, coefficient of variation (CV) values were calculated for each plate across each peak parameter by dividing the standard deviation (SD) by the mean for each plate’s DMSO control wells (Table S2). An upper cutoff of 30% for each plate was applied to determine which peak parameter to use in further analysis and, as such, peak count, amplitude, width, spacing, rise time, decay time, and amplitude at 10% were used, whereas SD values for these parameters were excluded from future analysis due to CV values >30%. To normalize the data, averages of DMSO-containing wells were obtained for each parameter, and the percent change from DMSO average was calculated for all wells on that plate. Then, technical repeats were averaged for further analysis such that each compound, though run as n=2, was represented as n=1 in future analysis.
Multiplexed data analysis and activity classifications
Normalized FLIPR data obtained from Peak Pro 1.0 was analyzed using both R Studio and TIBCO Spotfire’s High Content Profiler (HCP) platform. One-way between subjects ANOVA was performed in R Studio to investigate compound response from a select group of compounds. Here, the compound group, based on primary mechanism of action, was the between-subjects factor, and significant main effects of group (F-values) were followed up with Tukey’s post hoc testing for multiple comparisons. Multiple comparisons results were reported as t-values, and degrees of freedom are listed as subscripts for both main effect and post hoc data. The aov function was used for ANOVA and the package lsmeans was used for post hoc tests. To analyze calcium activity changes across all wells, TIBCO Spotfire’s High Content Profiler (HCP) was run using standard settings, including principle component analysis (PCA) data exploration, self-organizing map (SOM) class discovery, and z-prime robust for feature selection. Given that data had previously been normalized to DMSO-treated wells in Microsoft Excel, data was not further normalized for the HCP analysis. Only Peak Pro statistics values that were below the CV cutoff of 30% for each plate were used for the HCP analysis and, therefore, SD values for each peak parameter were excluded from analysis due to high variability. Graphs were created using either GraphPad Prism or TIBCO Spotfire.
Results
A multiparametric calcium fluorescence assay for neural cell activity in cortical spheroids
Cortical spheroids (3D microBrain, StemoniX) were characterized by showing that they consist of neurons and astrocytes, and that their activity can be differentially modulated depending on concentration of compound used (Fig. S1,S2). Prior to the proof-of-concept screen, spheroids were cultured for one week before assaying for synchronized neural activity, as measured using a calcium fluorescence assay paired with the FLIPR HTS reader. The current work assessed acute compound effect on synchronized neural activity. Fig. 1a shows an assay schema: on the day of the screen, the Cal6 dye was added to phenol red-free BrainPhys media, after two hours baseline calcium activity reads were taken, and then compounds added for 90 minutes, before reading the signal again for 8 min.
Calcium fluorescence peak analysis was done using ScreenWorks PeakPro to extract summary information on peak features across the 8-min FLIPR recordings for each well (Fig. 1b). Here, information on peak count (PkCt), amplitude (PkA), width (PkW), spacing (PkS), rise time (PkRt), and decay time (PkDt) was obtained along with the standard deviation (SD) for each of these parameters (Fig. 1b). Percent coefficients of variance (CV) values were calculated from each plates’ DMSO-treated wells, and parameters with values under 30% were used in future analysis (Table S2).
Given that in vivo cortical neurons are primarily glutamatergic and GABAergic neurons, control compounds targeting relevant receptor subtypes were chosen for initial observation of activity changes across peak features examined (Fig 1c). Kainic acid, a glutamate receptor (GluR) agonist, reliably increased PkCt while DNQX, an AMPAR antagonist, reduced PkCt, suggesting an enhancement and reduction, respectively, in neural activity (Fig. 1c,d). However, ketamine, an NMDAR antagonist, did not show changes in calcium fluorescence across any of the analyzed features. Muscimol, a GABAAR agonist, led to a total inhibition of calcium activity, indicated across all peak features (Fig. 1c,d). Additionally, the presence of voltage-gated ion channels was confirmed through calcium activity changes after exposure to 4-AP, a voltagegated potassium (Kv)-inhibitor, and tetrodotoxin, a voltage-gated sodium (Nav)-blocker. 4-AP, which induces hyperactivity in cortical neurons through Kv channel blockade in vivo 31, increased activity features such as PkCt and PkA while reducing PkW and PkS (Fig. 1c,d). In contrast, tetrodotoxin, which can induce neuronal inactivity in vivo 32, led to an overall inhibition across all peak features (Fig. 1c,d). Cutamesine, a sigma receptor agonist, and dihydrexidine, a dopamine 1/5 receptor (D1R) agonist, both showed similar increases in features including PkS, PkDt, and PkA10 (Fig. 1c,d). Together, this data shows that the cortical spheroids reliably respond to control compounds targeting relevant receptors.
HTS of a focused library of small molecules regulators of neural signaling pathways
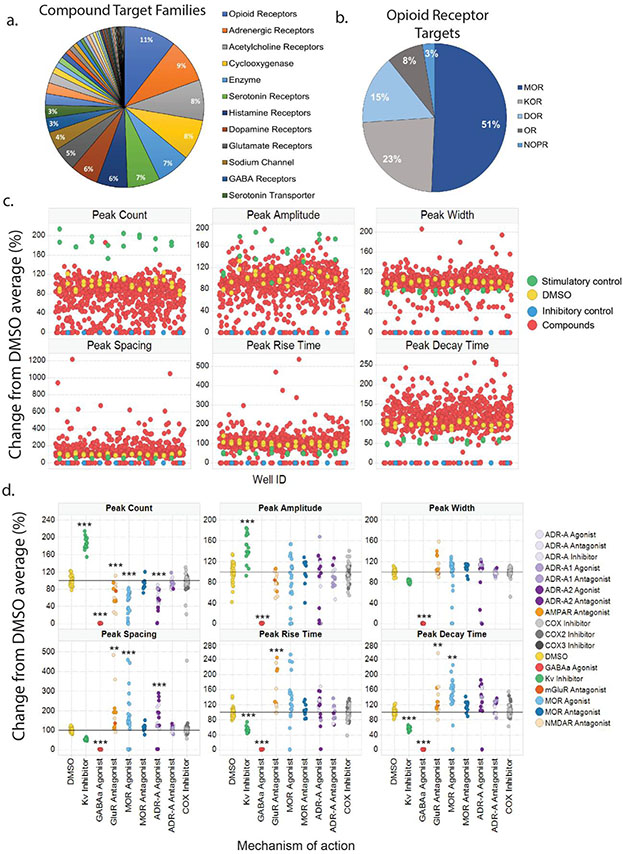
To investigate whether iPSC-derived cortical spheroids are suitable as a robust and pharmacologically-relevant drug discovery assay platform for neurological diseases, a library consisting of 687 neuroactive compounds was assembled and screened (Table S1 excel file). Compounds with various mechanisms of action relevant to neural signaling were included with the breakdown of compound target families shown in Fig. 2a. The largest group of compounds were those targeting opioid receptors, adrenergic receptors, serotonin receptors and transporters, along with dopamine receptors. Given the significance of the opioid epidemic in the United States, 11% of the library consisted of compounds targeting opioid receptor subtypes. Fig. 2b shows the breakdown of the percentage of compounds targeting opioid receptors, most of them targeting MORs, which is responsible for the rewarding and addictive aspects of OUD (23). Furthermore, other families of control compounds with activity in the brain, but perhaps not directly targeting neural cell activity, such as cyclooxygenase (COX) inhibitors, accounted for around 10% of the library. The distribution of the changes in calcium activity of all compounds compared to the DMSO vehicle is shown in Fig. 2c for the different calcium peak parameters used in the HCP analysis including peak count (PkCt), peak amplitude (PkA), peak width (PkW), peak spacing (PkS), peak rise time (PkRt), and peak decay time (PkDt).
Figure 2.
A proof-of-concept screen of a focused compound collection using calcium fluorescence multiparametric assay on cortical spheroids. (a.) Pie chart indicating major compound target families within the library that spheroids were screened against. (b.) Pie chart showing the breakdown of compounds targeting opioid receptor subtypes. (c.) Scatter plot showing the distribution of spheroid responses to compound, measured by calcium activity, for parameters including peak count, amplitude, width, spacing, rise time, and decay time. (d.) For each peak parameter used in the HCP analysis, specific mechanisms of action (MOAs) were analyzed against the average of DMSO-treated wells. Control wells, 4-AP and muscimol, were significantly different from DMSO across all parameters except peak width. Glutamate receptor (GluR) antagonists were different from DMSO across all measures aside from peak width. Mu opioid receptor (MOR) and alpha-adrenergic receptor (ADR-A) agonists were different from DMSO across all parameters aside from peak width and amplitude at 10%. MOR and ADR-A antagonists, along with cyclooxygenase (COX) inhibitors were no different from DMSO-treated wells for any peak parameter. DMSO (n=32); 4-AP (n=17); muscimol (n=17); GluR antagonists: AMPAR (n=5), NMDAR (n=7), mGluR (n=1); MOR agonists (n=24); MOR antagonists (n=11); ADR-A agonists (n=6); ADR-A1 agonists (n=3); ADR-A2 agonists (n=10); ADR-A antagonists (n=5); ADR-A1 antagonists (n=6); ADR-A2 antagonists (n=2); COX inhibitors (n=42); COX2 inhibitors (n=14); COX3 inhibitor (n=1). * p < 0.05, ** p < 0.01, *** p < 0.001
One way between-subjects ANOVA was conducted within each peak parameter obtained from the PeakPro analysis to investigate differences in control compounds and compounds with mechanisms of action (MOAs) of interest (Fig. 2d, S3). Here, the stimulatory control compound 4-AP and the inhibitory control compound muscimol are indicated by their mechanism of actions, Kv-inhibitor and GABAAR agonist, respectively. Compounds targeting MORs and alpha-adrenergic receptors (ADRs) were analyzed given their role in both addiction and pain disorders. GluR antagonists were analyzed given that cortical spheroids should contain glutamatergic neurons, and COX inhibitors were used as a negative control. Analysis was performed on compounds sharing the same primary mechanism of action, and specific names of the compounds within each group can be found in Table S1.
Here, compound group was the between-subjects factor, and follow-up post hoc testing compared each compound group to DMSO-treated controls. There was a significant main effect of compound group for peak count (PkCt), and Tukey’s post hoc revealed that the Kv-inhibitor used as a stimulatory control, significantly increased PkCt while the GABAA agonist used as an inhibitory control, significantly decreased PkCt compared to DMSO-treated controls (F(8, 193) = 138.7, p < 0.0001; Kv-inhibitor: t(193) = 15.6, p < 0.0001; GABAA agonist: t(193) = 17.9, p <0.0001, Fig. 2d). Additionally, mu opioid receptor (MOR) and ADR-A agonists significantly reduced PkCt compared to DMSO-treated wells, while MOR and ADR-A antagonists had no impact (MOR agonists: t(193) = 11.2, p < 0.0001; ADR-A agonists: t(193) = 9.2, p < 0.0001, Fig. 2d). GluR antagonists also significantly reduced PkCt compared to DMSO controls but COX inhibitors had no effect (GluR antagonists: t(193) = 5.2, p < 0.0001, Fig. 2d). A significant main effect of compound group was observed for peak amplitude, and post hoc tests revealed that both 4-AP and muscimol significantly changed PkA compared to DMSO controls, though no other group showed significant differences (F(8,193) = 31.8, p < 0.0001; Tukey’s post hoc, Kv inhibitor: t(193) = 5.6, p < 0.0001; GABAA agonist: t(193) = 11.8, p < 0.0001, Fig. 2d). For peak width (PkW), a significant main effect of compound group was followed up with Tukey’s post hoc, which showed that only the GABAA agonist, muscimol, was significantly different than DMSO controls (F(8,193) = 50.2, p < 0.0001; muscimol: t(193) = 16.8, p < 0.0001). However, Kv-inhibitor, 4-AP, showed a statistical trend toward a reduction in PkW compared to DMSO, though this was not significant (t(193) = 3.02, p = 0.069, Fig. 2d). One way ANOVA for peak spacing (PkS) showed a significant main effect of compound group, and post hoc testing showed a significant reduction in width by GABAA agonist, muscimol (F(8,193) = 14.1, p < 0.0001; Tukey’s post hoc, GABAA agonist: t(193) = 5.3, p < 0.0001, Fig. 2d). Similar to PkCt, MOR and ADR-A agonists along with GluR antagonists displayed significantly different PkS compared to DMSO controls (Tukey’s post hoc: ADR-A agonist: t(193) = 7.2, p < 0.0001; MOR agonist: t(193) = 4.1, p = 0.002; GluR antagonist: t(193) = 3.97, p = 0.003, Fig. 2d). A main effect of compound group was observed for peak rise time (PkRt, F(8,193) = 29.6, p < 0.0001), and both control compounds as well as GluR antagonists significantly altered PkRt compared to DMSO controls (Tukey’s post hoc, Kv-inhibitor: t(193) = 4.75, p = 0.0001; GABAA agonist: t(193) = 10.01, p < 0.0001; GluR antagonist: t(193) = 5.1, p < 0.0001, Fig. 2d). One way ANOVA run on peak decay time (PkDt) also showed a significant main effect of group, and both control compounds showed significant differences compared to DMSO-treated wells (F(8,193) = 35.87, p < 0.0001; Kv-inhibitor: t(193) = 4.7, p = 0.0002; GABAA agonist: t(193) = 10.9, p < 0.0001). Furthermore, GluR antagonists and MOR agonists significantly increased PkDt and while ADR-A agonists showed a statistical trend toward a significant increase, this was not significant (Tukey’s post hoc: MOR agonist: t(193) = 4.1, p = 0.002; GluR antagonist: t(193) = 0.009; ADR-A agonists: t(193) = 3.1, p = 0.06, Fig. 2d). Taken together, both control compounds displayed significant differences compared to DMSO controls across measures of PkCt, PkA, PkRt, and PkDt. Furthermore, MOR and ADR-A agonists displayed significant changes in functional activity on Ca2+ waveform features such as PkCt and PkS whereas MOR and ADR-A antagonists remained similar to DMSO controls.
High Content Profiler Analysis
Multiparametric analysis was run using TIBCO Spotfire’s High Content Profiler (HCP) on the normalized data to group compounds by activity profiles. Only FLIPR peak parameters with CV values below 30% were used in the HCP analysis (Table S2). As such, the features PkCt, PkA, PkW, PkS, PkRt, PkDt, and PkA10 were included in the HCP analysis. Negative controls were DMSO-treated wells, and the Kv-inhibitor, 4-AP, was used as a stimulatory control while the GABAA agonist, muscimol, was used as an inhibitory control. Spotfire HCP was run using standard settings, including principal component analysis (PCA) data exploration, self-organizing map (SOM) class discovery, and z-prime robust for feature selection.
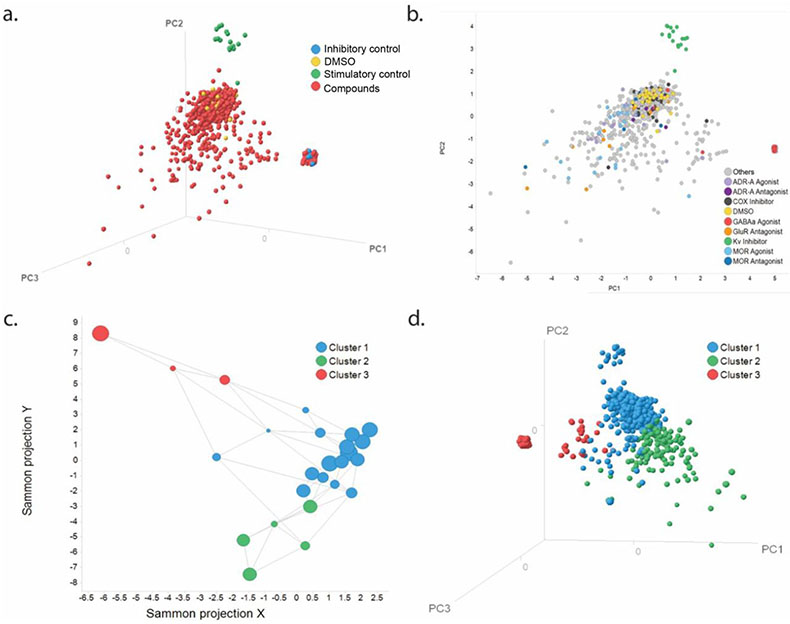
PCA was created and initially colored by sample type. The PCA analysis showed that wells treated with 4-AP and muscimol occurred in clusters that were separated from each other as well as from most of the screened samples and DMSO control wells (Fig. 3a). Furthermore, most of the samples tested clustered around the DMSO control, but there were samples with clear different activities. The PCA showed that ADR-A and MOR agonists as well as GluR antagonists were further apart from DMSO-treated wells compared to ADR-A and MOR antagonists along with COX inhibitors (Fig. 3b).
Figure 3.
High content profiler analysis of library screen on iPSC-derived cortical spheroids. (a.) Principal component analysis (PCA), represented as a 3D scatter plot, showing the spatial distribution of sample types including DMSO, stimulatory control (4-AP), inhibitory control (muscimol), and all other compounds. (b.) PCA, represented as a 2D scatter plot, showing the distribution of compounds with mechanisms of action (MOA) of interest in relation to DMSO, 4-AP, and muscimol control compounds. All other compounds are indicated by gray symbols. (c.) Sammon mapping plot displaying groupings of compounds, based on calcium activity after compound exposure, identified by the HCP analysis. All smaller clusters are grouped into three major clusters. (d.) PCA, represented as a 3D scatter plot, showing the spatial distribution of Sammon groupings by color.
Sammon connections network was used to group compounds together with similar activity profiles. This resulted in 24 small clusters that shared similar activity profiles, which were then grouped into three large clusters to simplify further analysis. Large clusters were formed using the automated k-means classification feature, whereby smaller clusters with similar characteristics were grouped together (Fig. 3c). The grouping of the three Sammon clusters was examined on the PCA, and three distinct groups were revealed, suggesting that the Sammon connections network was able to group compounds used in the screen (Fig. 3d).
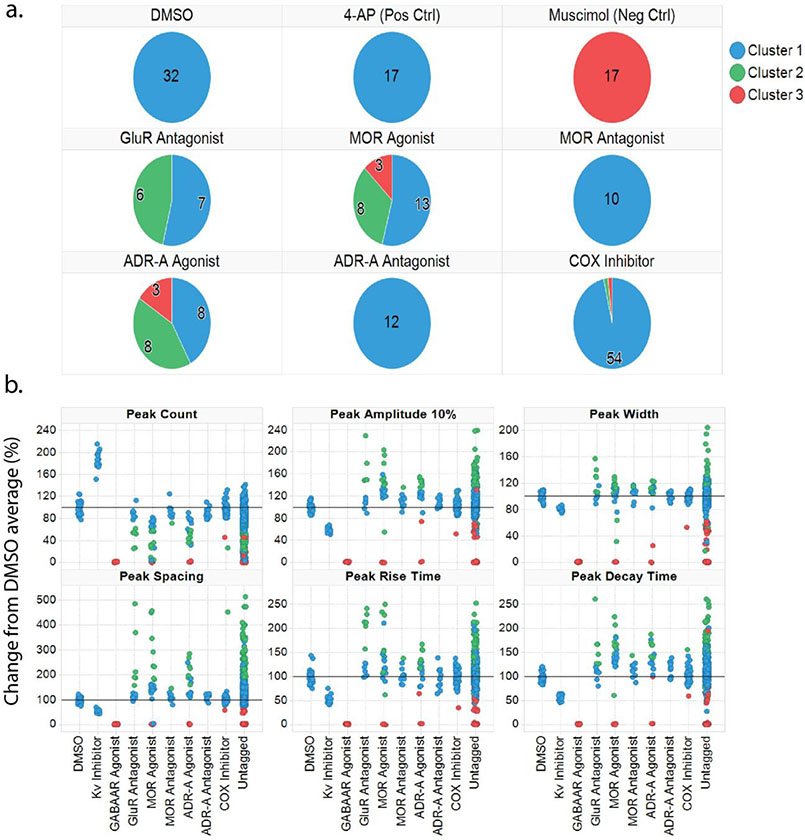
Finally, pie charts and scatter plots were generated to show where control compounds as well as some compounds with target mechanisms of interest fell within the three Sammon clusters (Fig. 4). Here, it was revealed that all wells treated with DMSO, 4-AP, MOR antagonists, ADR-antagonists, and the COX inhibitors were both grouped into Sammon cluster 1, suggesting that compounds with these activity profiles should fall into cluster 1 (Fig. 4a). Additionally, all wells treated with muscimol fell into cluster 3, suggesting compounds with inhibitory activity profiles should fall into cluster 3. Compounds that were GluR antagonists, MOR agonists, and ADR-A agonists resulted in activity profiles that were evenly distributed across all three Sammon clusters, suggesting a wide range of activity profiles produced by these compounds (Fig. 4a). To investigate which compounds within each target of interest were clustering, scatter plots were produced to show the activity features for compounds within each cluster (Fig. 4b). This plot revealed that, despite the increased distribution of clusters among compounds with significant changes in activity profiles compared to DMSO (i.e., MOR agonists, GluR antagonists, ADR-A agonists), the change could be distinguished by cluster. Given that compounds within these target classes produced significant, yet variable responses compared to DMSO, it is likely that this variability contributed to the increased number of clusters that these compound groups were distributed across. Together, this shows that the HCP analysis was able to group different compounds by activity features, using both PCA and Sammon connections network, and showed that compounds with responses further from DMSO control wells are distributed across different clusters.
Figure 4.
High Content Profiler analysis reliably clusters compounds by response-based activity profiles. (a.) Pie charts trellised by compounds with mechanisms of interest, colored by Sammon grouping cluster, show that DMSO and stimulatory control (4-AP) cluster together in Sammon cluster 1. Inhibitory control, muscimol, clusters separately in cluster 3. MOR and ADR-A antagonists, along with COX inhibitors, fall in cluster 1 along with vehicle and positive controls. GluR antagonists along with MOR and ADR-A agonists are distributed across more clusters, with compounds in these groups clustering evenly across 2-3 Sammon clusters. (b.) Scatter plots showing changes in activity features among compounds with specific mechanisms of interest, color-coded based on the Sammon cluster they fall into.
Discussion
In the present study, a library consisting of 687 neuroactive compounds was created to conduct a proof-of-concept screen using iPSC-derived cortical neural spheroids. Activity changes in response to compound exposure were measured through fluctuations in calcium fluorescence using a fluorescent imaging plate reader (FLIPR). Activity features of calcium dynamics were extracted through analysis in PeakPro, and peak features were chosen based on low variability in DMSO-treated control wells. These activity features were then incorporated into the high content profiler (HCP) analysis that was used to analyze compound responses across the entire screen. From this, compounds were clustered into three major groups based on activity profile. The findings from this study demonstrate that human iPSC-derived cortical spheroids could potentially serve as a high-throughput screening (HTS) platform for human-based drug discovery. Additionally, we present analysis using a multiparametric approach to analyze neural activity and use Sammon mapping to cluster compounds as a novel way to identify compounds eliciting responses.
While readouts such as viability assays and reporter gene expression, which have been traditionally used in HTS studies, can identify functional changes of a compound in a screening library, these readouts lack information about changes in neural signaling 33. Multielectrode arrays (MEAs) are useful for measuring electrophysiological properties through local field potentials (LFPs) but are not yet ideal for measuring these LFPs in high-throughput (HT) formats with spheroids since these plates are not designed beyond a 96-well plate format and the size and shape of spheroids can present challenges for attachment to electrodes in a traditional MEA plate with 8 electrodes in a 96-well plate (Axion Cytoview, Lumos MEA). While high-density electrode arrays address this problem, they are not yet compatible with multi-well plates larger than six wells per plate and are, therefore, not ideal for HTS studies (Maxwell Biosystems). Despite MEAs being a more direct readout of neuronal activity, studies in vivo have shown that intracellular calcium oscillations correlate with these electrophysiological properties and serve as a reliable activity readout 25. As such, we utilized the FLIPR to assess basal activity and compound-induced phenotypic functional changes via a calcium fluorophore in each well of 384-well plates simultaneously and identified functional endpoints with suitable basal reproducibility between wells (i.e. low CV), suggesting that calcium-based functional biomarker activity can be used as a reliable HTS readout. Calcium imaging as a HT readout also has better temporal resolution than traditional HTS readouts since time-dependent changes in activity following compound exposure can be observed. Furthermore, a previous report showed the activity response of these cortical spheroids changes based on the concentration of compound used, suggesting this calcium assay is sensitive to the degree of pharmacological action (Fig. S2) 17. Therefore, the findings from the current study also highlight the fact that, in addition to using human iPSC-derived cortical spheroids as a relevant biological platform, calcium imaging is an advantageous HT readout for neural cell activity compared to traditional methods.
Given the implication of cortical neurons in addiction, anxiety, and pain disorders, compounds with mechanisms of action related to these pathologies were examined prior to HCP analysis. MOR agonists and antagonists were chosen for their dual role in both pain and addiction, and compounds targeting alpha adrenergic receptors (ADR-A) were chosen for their role in depression and anxiety disorders 29, 34-35. Here we showed that treatment with MOR agonists and ADR-A agonists led to functional activity responses that were significantly different than DMSO-controls, while MOR and ADR-A antagonists had no effect. Specifically, pharmacologically activating MORs and ADR-As produced inhibitory activity, measured through reductions in peak count and increases in peak spacing. This is in line with the in vivo responses to MOR agonism, since studies have shown inhibition of prefrontal cortical neuronal activity after heroin infusions along with synaptic loss after morphine administration in animal studies 36,37. Furthermore, ADR-A agonists cause analgesia and sedation through reductions in norepinephrine release, which is associated with reduced neuronal excitability 34-35, 38. These data suggest that the cortical spheroids respond reliably and predictably to compounds targeting receptor subtypes relevant to neurological disorders impacting cortical circuitry.
To analyze functional responses induced by compounds in the entire library, HCP Sammon networking analysis clustered all compounds tested into three major clusters based on functional response profiles. Relevant compound groups (i.e., those in line with the Helping End Addiction Long-term (HEAL) objectives) were evaluated further, though all compounds received a cluster assignment, which can be found in Table S1. DMSO, 4-AP (the stimulatory control), MOR antagonists, ADR-A antagonists, and COX inhibitors were contained entirely within Cluster 1; muscimol (the inhibitory control) fell entirely within Cluster 3. GluR antagonists fell within Clusters 1 and 2 while MOR agonists and ADR-A agonists fell across Clusters 1, 2, and 3. These results indicate that Cluster 1 represents a different functional output than Clusters 2 and 3 and that peak spacing, which was unchanged for compounds in Cluster 1 and altered for compounds in Clusters 2 and 3, may be one of the key differentiators. Furthermore, MOR and ADR-A agonists showed activity profiles that were grouped across all three clusters. These compounds had individual parameter profiles that were clearly different from both DMSO controls, compounds without effect, as well as each other, thus justifying their presence across multiple Clusters (Fig. 4b). It is also interesting to note that the MOR impact was grouped across all three Clusters. This highlights the potential for polypharmacology among these compounds, which could be attributed to factors including only one concentration tested per compound, the fact that compounds were tested in duplicate rather than triplicate, and the potential for toxicity of some of the compounds to produce an activity profile that would cluster separately from compounds with similar mechanisms of action. It is possible that the 10 μM concentration used for most compounds was too high, causing indirect mechanisms to occur which may have affected the activity profile and caused noise that may have impacted the clustering during analysis. Furthermore, its possible that testing the compounds in triplicate would have reduced variability between compound responses, which could have improved the clustering. Indeed, variability as measured by the standard deviation (SD) values was significantly greater compared to DMSO-treated controls for certain compound groups such as glutamate receptor antagonists (Fig. S3). Last, running a counter cytotoxicity screen may have identified compounds producing toxic effects. Secondary screens incorporating multiple doses, each run in triplicate, with a counter-toxicity screen will be necessary to identify hit compounds and potentially improve the clustering method.
Interestingly, 4-AP significantly impacted cortical spheroid function and was still grouped in Cluster 1 with DMSO control and compounds without effect while other compounds with significant impacts (i.e. MOR agonists, ADR-A agonists, and GluR antagonists) were distributed across different clusters. There are a few reasons this may have occurred. First, grouping 4-AP with DMSO controls may have occurred due to the regularity of the 4-AP oscillation that was qualitatively similar to control, i.e. no irregularities in peak width or spacing which may have provided a strong, non-weighted foundation for a similar classification. Second, grouping the other compounds across Clusters may have arisen due to polypharmacology and multiple mechanisms of action (MOA; Table S1). As the calcium activity profile is a phenotypic readout, it represents the sum of all activity within the cortical spheroid, thus the presence of active secondary and tertiary MOA combine to create a heterogenous holistic response with components of all three Clusters. A third possibility is that additional endpoints such as sub-peaks and inter-peak activity, which may have been present in the waveforms but not captured in the six primary endpoints examined in this study, could provide additional information that would aid in dissecting and isolating more granular class-specific responses. Taken together, the current study confirms the utility of cortical spheroids as a platform for future HTS studies.
Recent reports have shown the utility of iPSC-derived cortical spheroids as a high-throughput (HT) drug and neurotoxicity screening model 17, 39-41. Specifically, one study developed a disease model for Pelizaeus-Merzbacher disease, a genetic disease associated with the loss of myelinating oligodendrocytes, through generating oligocortical spheroids and subjecting them to a HT chemical screen that was able to identify compounds capable of rescuing pathology-related deficits 42. Other reports have demonstrated that human iPSC-derived cortical spheroids exposed to neurotoxicants showed susceptibility profiles dependent on differentiation stage, suggesting these could be used to evaluate developmental neurotoxicity 39, 43. Further, a study exposed human iPSC-derived cortical spheroids to the LOPAC®1280 library and showed changes in spheroid activity in response to compounds, demonstrating the suitability of cortical spheroids as a screening platform 17. The present study replicates these findings, demonstrating the potential for human iPSC-derived cortical spheroids to be used for phenotypic and target-based HTS studies utilizing calcium activity after exposure to compounds as a functional biomarker. In the present study, Sammon clustering analysis identified 210 compounds with calcium activity profiles that clustered separately from DMSO-treated wells, suggesting that around 30% of this library produced activity changes in these cortical spheroids distinct from control compounds. While the Woodruff et al. study mentioned above identified only 8.7% of compounds that produced a response, this discrepancy is likely because the library used in the current study comprised all neuroactive compounds 17. It is expected that only ~30% of compounds would produce a response given that some compounds may require receptors present on other neuronal or glial cell types that may not have been present in these cortical spheroids, which should contain primarily glutamatergic and GABAergic neurons. Furthermore, 10% of the library consisted of COX inhibitors which were used as a negative control. Overall, this result is very encouraging because it suggests that cortical neurons can have selective responses to a library enriched for neural cell activity. Therefore, the responses obtained indeed reflect potential interventions into relevant physiology.
Recent studies have begun developing deep learning based neural networks and live image analysis to improve both speed and accuracy of measurements such as cell viability and morphology, suggesting that learning could be implemented response-based clustering approaches 44,45. Future improvements in analysis platforms for HTS datasets will help with clustering approaches implemented in the current study. Overall, this report shows that activity of iPSC-derived cortical spheroids can be measured through fluctuations in calcium activity, that this activity can be modulated through pharmacological manipulation in an HTS format, and the resulting compound-induced changes can be separated from each other at a fundamental level. As such, the current study shows that these iPSC-derived spheroids may serve as a suitable platform for HT drug screening for neurological diseases and presents an analysis method that could potentially be used for response-based screening.
Supplementary Material
Acknowledgements
This research was supported by the Helping End Addiction Longterm (HEAL) NIH program at the National Center for Advancing Translational Sciences. BVH, AL, RG, OG, and CC are employees of Stemonix Inc. The study design, data collection, and data analysis was done by scientists at NCATS, with neural spheroids purchased from Stemonix Inc, and with technical support and advice during data acquisition and multiparametric data analysis from Stemonix Inc scientists. Special thanks to Kelli Wilson Henderson Tozer, Kevin Diaz, Oved Nativi, John Braisted for help with the TIBCO Spotfire set up and analysis, and to Blake Anson for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Feigin VL; Vos T; Nichols E; et al. The Global Burden of Neurological Disorders: Translating Evidence into Policy. Lancet Neurol. 2020, 19, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2015 Neurological Disorders Collaborator Group; Global, Regional, and National Burden for Neurological Disorders during 1990-2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet Neurol. 2017, 16, 877–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehm J; Shield KD; Global Burden of Disease and the Impact of Mental and Addictive Disorders. Curr Psychiatry Rep. 2019, 21, 10. [DOI] [PubMed] [Google Scholar]
- 4.Hay M; Thomas DW; Craighead JL; et al. Clinical Development Success Rates for Investigation Drugs. Nat Biotechnol. 2014, 32, 40–51. [DOI] [PubMed] [Google Scholar]
- 5.DiMasi JA; Grabowski HG; Hansen RW; Innovation in the Pharmaceutical Industry: New Estimates of R&D Costs. J Health Econ, 2016, 47, 20–33. [DOI] [PubMed] [Google Scholar]
- 6.Wong CH; Siah KW; Lo AW; Estimation of Clinical Trial Success Rates and Related Parameters. Biostatistics. 2019, 20, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffat JG; Vincent F; Lee JA; et al. Opportunities and Challenges in Phenotypic Drug Discovery: An Industry Perspective. Nat Rev Drug Discov. 2017, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- 8.Kelava I; Lancaster MA; Stem Cell Models of Human Brain Development. Cell Stem Cell. 2017, 18, 736–748. [DOI] [PubMed] [Google Scholar]
- 9.Di Lullo E; Kriegstein AR; The Use of Brain Organoids to Investigate Neural Development and Disease. Nat Rev Neurosci. 2017, 18, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Worp HB; Howells DW; Sena ES; et al. Can Animal Models of Disease Reliably Inform Human Studies? PloSMed. 2010, 7, e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drude NI; Gamboa LM; Danziger M; et al. Improving Preclinical Studies Through Replications. Elife. 2021, 10, e62101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodge RD; Bakken TE; Miller JA; et al. Conserved Cell Types with Divergent Features in Human versus Mouse Cortex. Nature. 2019, 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Essen DC; Donahue CJ; Glasser MF; Development and Evolution of Cerebral and Cerebellar Cortex. Brain Behav Evol. 2018, 91, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasca AM; Sloan SA; Clarke LE; et al. Functional Cortical Neurons and Astrocytes from Human Pluripotent Stem Cells in 3D Culture. Nat Methods. 2015, 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrasekaran A; Avci HX; Ochalek A; et al. Comparison of 2D and 3D Neural Induction Methods for the Generation of Neural Progenitor Cells from Human Induced Pluripotent Stem Cells. Stem Cell Res. 2017, 25, 139–151. [DOI] [PubMed] [Google Scholar]
- 16.James OG; Selvaraj BT; Magnani D; et al. iPSC-derived Myelinoids to Study Myelin Biology of Humans. Dev Cell. 2021, 56, 1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodruff G; Phillips N; Carromeu C; et al. Screening for Modulators of Neural Network Activity in 3D Human iPSC-derived Cortical Spheroids. PLoS One. 2020, 15, e0240991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nzou G; Wicks RT; Wicks EE; et al. Human Cortex Spheroid with a Functional Blood Brain Barrier for High-Throughput Neurotoxicity and Disease Modeling. Sci Rep. 2018, 8, 7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitureira N; Goda Y; Cell Biology in Neuroscience: The Interplay Between Hebbian and Homeostatic Synaptic Plasticity. J Cell Biol. 2013, 203: 175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoepfer KJ; Xu Y; Wilber AA; Wu W; Kabbaj M; “Sex Differences and Effects of the Estrous Stage on Hippocampal-Prefrontal Theta Communications. Physiol Rep. 2020, 8: e14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim T; Thankachan S; McKenna JT; et al. Cortically Projecting Basal Forebrain Parvalbumin Neurons Regulate Cortical Gamma Band Oscillations. Proc Natl Acad Sci USA. 2015, 112, 3535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaeser AS; Connors BW; Nurmikko AV; Spontaneous Dynamics of Neural Networks in Deep Layers of Prefrontal Cortex. J Neurophysiol. 2017, 117, 1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu MY; Wong AHC; GABAergic Inhibitory Neurons as Therapeutic Targets for Cognitive Impairment in Schizophrenia. Acta Pharmacol Sin. 2018, 39, 733–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruijnen CJHW; Dijkstra BAG; Walvoort SJW; et al. Prevalence of Cognitive Impairment in Patients with Substance Use Disorder. Drug Alcohol Rev. 2019, 38, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ah F; Kwan AC; Interpreting in vivo Calcium Signals from Neuronal Cell Bodies, Axons, and Dendrites: A Review. Neurophotonics. 2020, 7, 011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirenko O; Parham F; Dea S; et al. Functional and Mechanistic Neurotoxicity Profiling Using Human iPSC-Derived Neural 3D Cultures. Toxicol Sci. 2019, 167: 58–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negraes PD; Trujilo CA; Yu NK; et al. Altered Network and Rescue of Human Neurons Derived from Individuals with Early-Onset Genetic Epilepsy. Mol Psychiatry. 2021, doi: 10.1038/s41380-021-01104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park Y; Franz CK; Ryu H; et al. Three-Dimensional, Multifunctional Neural Interfaces for Cortical Spheroids and Engineered Assembloids. Sci Adv. 2021, 7: eabf9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koob GF; Neurobiology of Opioid Addiction: Opponent Process, Hyperkatifeia, and Negative Reinforcement. Biol Psychiatry. 2020, 87, 44–53. [DOI] [PubMed] [Google Scholar]
- 30.Boutin ME; Voss TC, Titus SA; et al. A High-Throughput Imaging and Nuclear Segmentation Analysis Protocol for Cleared 3D Culture Models. Sci Rep. 2018, 8: 11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenzel M; Hamm JP; Peterka DS; et al. Reliable and Elastic Propagation of Cortical Seizures In Vivo. Cell Rep. 2017, 19, 2681–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schonfeld-Dado E; Fishbein I; Segal M; Degeneration of Cultured Cortical Neurons Following Prolonged Inactivation: Molecular Mechanisms. J Neurochem. 2009, 110, 1203–13. [DOI] [PubMed] [Google Scholar]
- 33.Little D; Ketteler R; Gissen P; et al. Using Stem Cell-derived Neurons in Drug Screening for Neurological Diseases. NeurobiolAging. 2019, 78, 130–141. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X; Norton J; Carriere I; et al. Preliminary Evidence for a Role of the Adrenergic Nervous System in Generalized Anxiety Disorder. Sci Rep. 2017, 7, 42676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schramm NL; McDonald MP; Limbird LE; The alpha(2a)-Adrenergic Receptor Plays a Protective Role in Mouse Behavioral Models of Depression and Anxiety. J Neurosci. 2001, 21, 4875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang JY; Zhang L; Janak PH; et al. Neuronal Responses in Prefrontal Cortex and Nucleus Accumbens During Heroin Self-Administration in Freely Moving Rats. Brain Res. 1997, 754, 12–20. [DOI] [PubMed] [Google Scholar]
- 37.Robinson TE; Kolb B; Morphine Alters the Structure of Neurons in the Nucleus Accumbens and Neocortex of Rats. Synapse. 1999, 33, 160–2. [DOI] [PubMed] [Google Scholar]
- 38.Giovannitti JA; Thoms SM; Crawford JJ; Alpha-2 Adrenergic Receptor Agonists: A Review of Current Clinical Applications. Anesth Prog. 2015, 62, 31–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobolak J; Teglasi A; Bellak T; et al. Human Induced Pluripotent Stem Cell-Derived 3D Neurospheres are Suitable for Neurotoxicity Screening. Cells. 2020, 9, 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vliet SM; Ho TC; Volz DC; Behavioral Screening of the LOPAC1280 Library in Zebrafish Embryos. Toxicol Appl Pharmacol. 2017, 329, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas AG; Sattler R; Tendyke K; et al. High-Throughput Assay Development for Cystine-Glutamate Antiporter (xc-) Highlights Faster Cystine Uptake than Glutamate Release in Glioma Cells. PLoS One. 2015, 10, e0127785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elitt MS; Shick HE; Madhaven M; et al. Chemical Screening Identifies Enhancers of Mutant Oligodendrocyte Survival and Unmasks a Distinct Pathological Phase in Pelizaeus-Merzbacher Disease. Stem Cell Reports. 2018, 11, 711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slavin I; Dea S; Arunkumar P; et al. Human iPSC-Derived 2D and 3D Platforms for RapidlyAssessing Developmental, Functional, and Terminal Toxicities in Neural Cells. Int. J. Mol. Sci. 2021, 22, 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benning L; Peintner A; Finkenzeller G; et al. Automated Spheroid Generation, Drug Application and Efficacy Screening Using Deep Learning Classification: A Feasibility Study. Sci Rep. 2020, 10, 11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alsehli H; Mosis F; Thompson C; et al. An Integrated Pipeline for High-Throughput Screening and Profiling of Spheroids Using Simple Live Image Analysis of Frame to Frame Variations. Methods. 2020, S1046-2023, 30300–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.