Abstract
In the gut ecosystem, microorganisms regulate group behaviour and interplay with the host via a molecular system called quorum sensing (QS). The QS molecule 3-oxo-C12:2-HSL, first identified in human gut microbiota, exerts anti-inflammatory effects and could play a role in inflammatory bowel diseases where dysbiosis has been described. Our aim was to identify which signalling pathways are involved in this effect. We observed that 3-oxo-C12:2-HSL decreases expression of pro-inflammatory cytokines such as Interleukine-1β (− 35%) and Tumor Necrosis Factor-α (TNFα) (− 40%) by stimulated immune RAW264.7 cells and decreased TNF secretion by stimulated PBMC in a dose-dependent manner, between 25 to 100 µM. Transcriptomic analysis of RAW264.7 cells exposed to 3-oxo-C12:2-HSL, in a pro-inflammatory context, highlighted JAK-STAT, NF-κB and TFN signalling pathways and we confirmed that 3-oxo-C12:2-HSL inhibited JAK1 and STAT1 phosphorylation. We also showed through a screening assay that 3-oxo-C12:2-HSL interacted with several human bitter taste receptors. Its anti-inflammatory effect involved TAS2R38 as shown by pharmacologic inhibition and led to an increase in intracellular calcium levels. We thus unravelled the involvement of several cellular pathways in the anti-inflammatory effects exerted by the QS molecule 3-oxo-C12:2-HSL.
Subject terms: Cell biology, Cell signalling, Inflammatory bowel disease, Inflammation, Microbial communities
Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are chronic relapsing inflammatory conditions of the gastrointestinal tract leading to bowel damages and increased risk of intestinal cancer1. The incidence and prevalence of CD and UC are increasing and an estimated 0.3% of the European population suffers from IBD, representing 2.5–3 million people. IBD, which affects young adults and is long-lasting, accounts for substantial costs to the health care system and society. Disease management has changed significantly over the last decade with the use of parenterally administrated biologic agents and new small oral molecules such as pan-Janus kinase (JAK) inhibitor2. Immunosuppressants are used more widely and earlier in the progression of the disease. However, this strategy is not always effective, is quite expensive and exposes patients to adverse effects, mainly serious infections and malignancies3. Thus, there is both a need and a place for more “gut restricted” and physiological therapeutic approaches mostly based on a better knowledge of the gut microbiota.
As a matter of fact, gut microbiota has been strongly implicated in the pathogenesis of IBD4,5 where microbial unbalance (dysbiosis) has been described6. Thus microbiota-based treatments, such as faecal microbiota transplantation, are currently being studied7,8. In this setting, investigating gut microbiota-derived molecules remains an attractive strategy to control gut inflammation and prevent IBD flares. In fact, gut microbiota reciprocally interacts with co-evolved host epithelial and immune cells. This results in a beneficial mutualistic relationship in which microbiota synthesizes metabolites that can modulate the host’s immune responses9–12.
Diffusible signal compounds represented by bacterial quorum sensing (QS) molecules, called autoinducers have attracted increasing interest due to their effects on host cells demonstrated in various models13–15. Acyl-Homoserine Lactones (AHL) are a class of autoinducers that can impact the human host, as part of the inter-kingdom signalling16. Our team identified for the first time several AHLs in the gut ecosystem, and among them, the most prominent was the 3-oxo-C12:2-HSL, a molecule that had never been described17. 3-oxo-C12:2-HSL was decreased in IBD patients compared to healthy subjects and its presence was correlated with normobiosis17. 3-oxo-C12:2-HSL is structurally close to 3-oxo-C12-HSL synthesized by Pseudomonas aeruginosa, a molecule whose effects on host cells are well reported in the literature15,16. We previously described the anti-inflammatory and dose-dependent effects of 3-oxo-C12:2-HSL on intestinal epithelial cells Caco-2/TC7 stimulated by interleukin-β17. In addition, we have demonstrated that 3-oxo-C12:2-HSL protects against tight junctions disruption induced by pro-inflammatory cytokines and thus has a beneficial role on intestinal barrier function18. In the present study, we aim to characterise the effects of 3-oxo-C12:2-HSL on immune cells and to identify cellular targets of the autoinducer. Using the macrophage cell line RAW264.7 as well as peripheral blood mononuclear cells (PBMC) stimulated by lipopolysaccharides (LPS) and interferon-γ (IFNγ), we assessed how 3-oxo-C12:2-HSL modulates host-inflammatory response and the mechanisms involved. Our results show that this AHL exerts anti-inflammatory effects by down-regulating pro-inflammatory cytokines secretion. Using transcriptomic analysis, we have identified several pathways involved, one of them being the JAK-STAT pathway. 3-oxo-C12:2-HSL is able to prevent RAW264.7 macrophages from JAK-STAT signalling activation induced by LPS and IFNγ. Additionally, based on the interaction of 3-oxo-C12-HSL from P. aeruginosa with bitter taste receptor TAS2R3819, we demonstrated that several bitter taste receptors are potential membrane receptors for 3-oxo-C12:2-HSL.
Results
3-oxo-C12:2-HSL reduces expression and secretion of pro-inflammatory cytokines by activated immune cells
We first observed the effect of 3-oxo-C12:2-HSL on cytokine secretion by immune cells. We exposed RAW264.7 murine macrophages cells to lipopolysaccharides (LPS) and Interferon-γ (IFNγ) to trigger classical activation of M1 macrophages20. Cells were treated with or without 50 µM of 3-oxo-C12:2-HSL followed by analysis of cytokines levels in the supernatant (Fig. 1a,b). Lower secretion of pro-inflammatory Tumor Necrosis Factor-α (TNFα) (− 40%, Fig. 1a) and Interleukine-1β (IL-1β) (− 35%, Fig. 1b) was observed in presence of 3-oxo-C12:2-HSL compared with the control. We then investigated the effects on inflammation at the transcript level. mRNA levels of the two cytokines mentioned above were reduced in activated macrophages exposed to 3-oxo-C12:2-HSL, to the same extent as the protein (Fig. 1c,d). Anti-inflammatory cytokine IL-10 expression was strongly increased when cells were exposed to the AHL (Fig. 1e).
Figure 1.
3-oxo-C12:2-HSL decreases pro-inflammatory cytokine secretion by immune cells. Secreted TNFα (a) and IL-1β (b) levels were measured by ELISA from supernatant of RAW264.7 macrophages stimulated with LPS and IFNγ in absence (control) or presence of 50 µM 3-oxo-C12:2-HSL for 6 h. Unpaired t test, *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. control. mRNA levels for Tnf (c), Il1b (d), Il10 (e), genes measured by RT-qPCR in RAW264.7 macrophages stimulated with LPS and IFNγ in absence (control) or presence of 50 µM 3-oxo-C12:2-HSL for 2 h. Results are expressed in arbitrary units as a ratio of the target gene to cyclophilin (Ppib) used as housekeeping gene. Unpaired t test, *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. control. (f) Secreted TNFα measured by ELISA from supernatant of PBMC stimulated with LPS (10 ng/mL) and exposed to a concentration range of 3-oxo-C12:2-HSL (0–100 µM). Results are expressed as mean ± SEM of triplicates from 3 independent experiments. One Way ANOVA Dunnett’s post-test. 3oC12:2 stands for 3-oxo-C12:2-HSL.
To confirm these results in a more physiological model, we tested the impact of the AHL on Peripheral Blood Mononuclear Cells (PBMC) from healthy donors. Inflammation was triggered with LPS (10 ng/mL), and we exposed PBMC to an increasing dose of 3-oxo-C12:2-HSL. We showed that TNFα secretion was strongly decreased in a dose-dependent manner, between 25 to 100 µM (Fig. 1f).
Of note, cytotoxicity was monitored through lactate dehydrogenase release for all experiments and there was no significant difference compared to the control for the doses of AHL used (Supplementary Fig. 1).
3-oxo-C12:2-HSL modulates gene expression in RAW264.7 cells
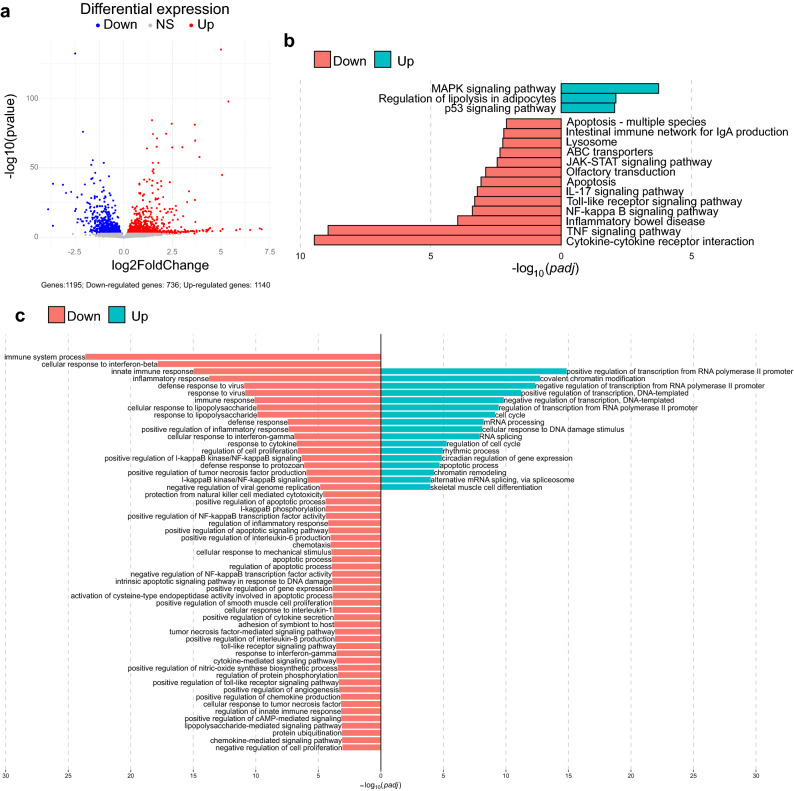
In an attempt to understand the mechanisms underpinning the immunomodulatory effects of 3-oxo-C12:2-HSL on immune cells, we performed a transcriptomic analysis of the activated RAW264.7 cells exposed to 3-oxo-C12:2-HSL. We observed that 3-oxo-C12:2-HSL down-regulated 736 genes and up-regulated 1140 genes compared with control cells treated with DMSO (Fig. 2a).
Figure 2.
3-oxo-C12:2-HSL modulates gene expression in RAW264.7 macrophage cells. Cells were stimulated with LPS and IFNγ in absence (control) or in presence of 50 µM 3-oxo-C12:2-HSL for 2 h. (a) Volcano plot of differentially expressed genes between the activated cells cultured in absence and in presence of 3-oxo-C12:2-HSL. Blue, grey and red dots represent down regulated, not significantly regulated and up regulated genes respectively. (b) Significant KEGG pathways involved in the inflammation process and differentially modulated by 3-oxo-C12:2-HSL were identified by EGSA method (p < 0.01). Red and green bars represent down-regulated and up-regulated pathways respectively. All significant modulated pathways are displayed in Supplementary Fig. 2. (c) Significant Gene Ontology (GO) biology processes enriched by differentiated expressed genes modulated by 3-oxo-C12:2-HSL. Terms were annotated using the Database for Annotation, Visualization and Integrated Discovery (DAVID). Red and green bars represent down-regulated and up-regulated pathways respectively. GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes.
Differentially expressed genes were annotated by Gene Ontology (GO) analysis (Fig. 2c). GO analysis revealed that down-regulated genes were enriched in “immune system process”, “innate immune response” and “inflammatory response” categories, in accordance with the anti-inflammatory effects of 3-oxo-C12:2-HSL observed in Fig. 1. In terms of molecular function, we identified that the downregulated genes were enriched in the NF-κB and Toll Like Receptor (TLR) and TNF signalling pathways. The latter correlated with the decrease of TNFα protein secretion observed in Fig. 1a,f.
A KEGG pathway analysis was carried out to classify and group differentially expressed genes in cellular processes. A large number of pathways were modulated by the AHL (Supplementary Fig. 2), among which, the most down-regulated pathways were “Cytokine–cytokine receptor interaction” and “TNF signalling pathway”. A selection of pathways related to inflammation and IBD modulated by 3-oxo-C12:2-HSL is displayed on Fig. 2b. This includes crucial pathways directly involved in cell signalling during inflammation such as the “JAK-STAT signalling pathway”, the “NF-κB signalling pathway” and the “TNF signalling pathway”. This also includes pathways related to IBD such as “apoptosis” and “ABC transporters”. Interestingly, 3-oxo-C12-HSL produced by P. aeruginosa was reported to up-regulate both apoptosis and expression of ABC receptors21, suggesting that these two AHL do not exert similar effects.
From both GO and KEGG pathway analyses, we were able to pinpoint specific cellular pathways involved in the signalling of 3-oxo-C12:2-HSL. As JAK-STAT pathway is shared by several cytokine receptors and known to mediate key deleterious effects of pro-inflammatory cytokines in IBD22,23, we further investigated it in presence of 3-oxo-C12:2-HSL.
3-oxo-C12:2 prevents the activation of the JAK-STAT pathway
To explore the effects of 3-oxo-C12:2-HSL on the JAK-STAT pathway, we analysed the phosphorylation level of proteins involved in this pathway. As the latter is composed of a family of JAK and STAT proteins, we studied several of them in RAW264.7 macrophages activated by LPS and IFNγ and exposed or not to 3-oxo-C12:2-HSL (Fig. 3). When cells were treated with the pro-inflammatory cocktail, we observed an increase of the phosphorylation of JAK1, JAK2, STAT1, STAT3 proteins (Fig. 3a–e). When activated macrophages were exposed to 3-oxo-C12:2-HSL, a reduced phosphorylation level of JAK1 (− 50%) and STAT1 (− 40%) protein was observed, in comparison with activated cells without AHL treatment (Fig. 3a,b). However, JAK2 and STAT3 phosphorylation levels were not significantly altered upon exposure to 3-oxo-C12:2-HSL.
Figure 3.
3-oxo-C12:2-HSL prevents activation of the JAK-STAT pathway. Cells were stimulated with LPS and IFNγ in absence (control) or in presence of 50 µM 3-oxo-C12:2-HSL for 2 h. The levels of phosphorylated proteins P-JAK1 (a), STAT1 (b), JAK2 (c), STAT3 (d) were normalized to their respective unphosphorylated forms. Results are expressed as mean ± SEM from 4 independent experiments. One-Way ANOVA, Dunnett’s post-test **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. control. (e) Reconstructed images from Simple Western analysis of protein levels and actin used as housekeeping protein are displayed. They are based on the area under the chemiluminescence signal curve obtained for one experiment, representative of 4 independent experiments performed in duplicates. Molecular markers are indicated on the left (kDa). 3oC12:2 stands for 3-oxo-C12:2-HSL.
These results demonstrate that 3-oxo-C12:2-HSL prevents the activation of the JAK-STAT pathway, by specifically reducing the phosphorylation of JAK1 and STAT1 proteins.
3-oxo-C12:2-HSL interacts with bitter taste receptors
It was previously reported that the AHL 3-oxo-C12-HSL synthesized by P. aeruginosa might exert some of its effects through its interaction via the human bitter taste receptor TAS2R3819,24. Bitter taste receptors are G protein-coupled receptor and their role beyond oral taste has been extensively studied throughout the last decade25. Taste receptors are all expressed along the digestive tract26–29, but also by immune cells19,30,31. Based on the similarities in the chemical structure of 3-oxo-C12-HSL and 3-oxo-C12:2-HSL, we adopted a targeted approach to identify potential eukaryotic receptors for intestinal 3-oxo-C12:2-HSL and hypothesized that 3-oxo-C12:2-HSL could also interact with the bitter taste receptor TAS2R38.
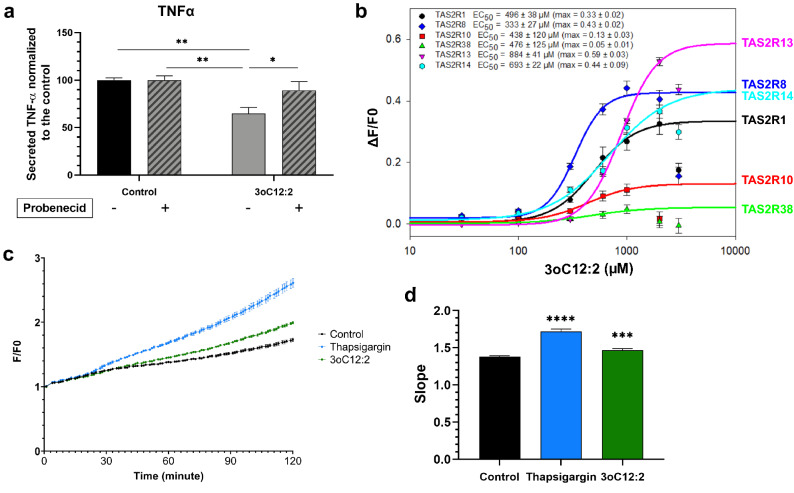
We pre-exposed RAW264.7 cells to probenecid, a known allosteric inhibitor of TAS2R3832,33 and its mouse orthologue TAS2R138 then added LPS and IFNγ and 3-oxo-C12:2-HSL (Fig. 4a). First, we noted that probenecid treatment had no effect on the TNFα secretion in activated cells cultured in absence of 3-oxo-C12:2-HSL (control). When exposed to probenecid and 3-oxo-C12:2-HSL in a pro-inflammatory context, more TNFα was secreted compared to cells only exposed to 3-oxo-C12:2-HSL without inhibitor treatment. This result shows that when TAS2R138 is inhibited, the 3-oxo-C12:2-HSL anti-inflammatory effect is abolished.
Figure 4.
3-oxo-C12:2-HSL interacts with bitter taste receptors. (a) RAW264.7 macrophages were pre-treated or not with 1 mM Probenecid, an allosteric inhibitor of the bitter taste receptor TAS2R138 one hour before their stimulation with LPS and IFNγ in absence (control) or in presence of 3-oxo-C12:2-HSL for six hours. Mean ± SEM from 3 independent experiments performed in triplicate, Two Way ANOVA Tukey’s post-test. (b) Calcium response in HEK293T cells expressing reporter human bitter taste receptor exposed to several concentrations of 3-oxo-C12:2-HSL (0–1000 µM). Fluorescence (F) was measured after exposure to the AHL and normalized to basal fluorescence (F0). Mean ± SEM from 3 independent experiments performed in triplicate. (c) Intracellular calcium flux in RAW264.7 cells exposed to 1000 µM 3-oxo-C12:2-HSL or 10 µM thapsigargin as a positive control or DMSO (0.5%) as a negative control. Fluorescence (F) monitoring the increase in intracellular concentration of calcium was measured for two hours and normalized to basal fluorescence (F0). Each curve is representative of 3 independent experiments of 8 replicates. (d) Quantification of the slope of the curves displayed in (c). Values are mean ± SEM of 3 independent experiments (8 replicates each). For (a) and (d): Two Way ANOVA Tukey’s post-test. *p ≤ 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. control. 3oC12:2 stands for 3-oxo-C12:2-HSL.
To elucidate if 3-oxo-C12:2-HSL was able to activate human TAS2R38 and other bitter taste receptors, we screened 25 known human bitter taste receptors in HEK293T reporter-cells (Fig. 4b)34. Among these bitter taste receptors, the AHL was able to activate TAS2R1, TAS2R8, TAS2R10, TAS2R13, TAS2R14, and TAS2R38. TAS2R13 displayed the highest maximum amplitude (max ampl = 0.59 ± 0.03), while having the highest half maximal effective concentration (EC50 = 884 ± 41 µM). TAS2R1, TAS2R8 and TAS2R14 showed similar maximum amplitude values (around 0.45), but TAS2R8 was the most sensitive to 3-oxo-C12:2-HSL (EC50 = 333 ± 27 µM), making it a good candidate for further analysis. TAS2R10 and TAS2R38 exhibited similar EC50 values (EC50 = 438 ± 120 µM and 476 ± 125 µM respectively) but had much lower amplitude responses (max ampl = 0.13 ± 0.03 and 0.05 ± 0.01 respectively).
Transduction of taste signalling results in the opening of IP3R-dependant calcium channels and the intracellular Ca2+ release25. Therefore, we investigated the effect of 3-oxo-C12:2-HSL with a Fluo-4 probe in the basal state, using the endoplasmic reticulum Ca2+ ATPase inhibitor thapsigargin as a positive control. We observed an increase in intracellular calcium level when RAW264.7 cells were exposed to 3-oxo-C12:2-HSL compared with the control (Fig. 4c,d). The quantification of calcium response shows that in presence of 3-oxo-C12:2-HSL the slope of the curve was significantly higher than that observed in the control condition (Fig. 4d).
Discussion
As the human gut is one of the most densely populated microbial ecosystems, studying QS molecules, related to bacterial density, seems legitimate in this environment. AHLs are the most studied QS auto-inducers and, using mass spectrometry, our team previously identified one that had never been documented before in the gut ecosystem, 3-oxo-C12:2-HSL17. Our initial work showed that 3-oxo-C12:2-HSL correlated with normobiosis and was less prevalent in IBD associated dysbiosis.
Our group observed that 3-oxo-C12:2-HSL decreased IL-8 production in the intestinal epithelial cell line Caco-2/TC7 stimulated by IL-1β17. We therefore sought to investigate how 3-oxo-C12:2-HSL can modulate inflammatory response of immune cells. Here, we demonstrate that 3-oxo-C12:2-HSL exerts anti-inflammatory effects on immune cells by preventing JAK-STAT pathway activation and by activating bitter taste signalling (Fig. 5).
Figure 5.
Proposed mechanisms of 3-oxo-C12:2-HSL effects on inflammation. In immune cells, 3-oxo-C12:2-HSL is able to activate bitter taste receptors, which are G-protein coupled receptors, triggering a signalling cascade resulting in the release of calcium from the endoplasmic reticulum. In addition, in murine activated macrophages the presence of 3-oxo-C12:2-HSL attenuates the activation of the JAK-STAT signalling pathways, by specifically preventing JAK1 and STAT1 phosphorylation. This leads to a decrease in pro-inflammatory cytokine secretions and an overall reduced inflammatory response. Part of the effects of 3-oxo-C12:2-HSL on inflammatory response is dependent on bitter taste receptors. Created with BioRender.com.
AHLs have been described to modulate host’s immunity, in particular 3-oxo-C12-HSL from Pseudomonas aeruginosa, structurally close to 3-oxo-C12:2-HSL. A number of studies have reported that 3-oxo-C12-HSL downregulated cytokine secretion by macrophages35–37, dendritic cells38, T lymphocytes35,39,40 as well as epithelial cells17,41. In the present study, we show that the intestinal unsaturated 3-oxo-C12:2-HSL displays anti-inflammatory effects by downregulating secretion of pro-inflammatory cytokine TNFα and IL-1β, as well as their respective gene expression by RAW264.7 murine macrophages. Interestingly, this AHL was also able to increase anti-inflammatory cytokine IL-10 expression, suggesting a potent anti-inflammatory effect in immune cells. These findings were confirmed using a more physiological model, PBMC stimulated by LPS, in which TNFα was strongly decreased by 3-oxo-C12:2-HSL exposure, in a dose-dependent manner. We activated RAW264.7 cells with LPS and IFNγ, leading the macrophages to a M1 profile20, similar to what is observed during IBD physiopathology. Indeed, in the mucosa of patients, there is an influx of pro-inflammatory macrophages42. Under such conditions, activation of the JAK-STAT pathway is enhanced, with a particular involvement of the STAT1 protein43–45. One study previously reported that 3-oxo-C12-HSL is able to modulate the JAK-STAT pathway in breast carcinoma cells46, but our group is the first to report AHL modulation of this pathway in immune cells. 3-oxo-C12:2-HSL was able to prevent the phosphorylation of JAK1 and STAT1, which is particularly relevant in the field of IBD. Indeed, 3-oxo-C12:2-HSL, which is decreased in IBD patients17, appears as a microbial product able to modulate IBD pathways involved in chronic inflammation. Novel IBD treatments are targeting the JAK proteins23. Current IBD drugs target one or more cytokines (such as anti-TNF)23 while new oral small molecules act by inhibiting either pan-JAK or specific JAK pathways. However, clinical trials with JAK inhibitors revealed serious adverse events23. Alternative approaches using molecules targeting the gut are appealing, with drugs exerting a strong anti-inflammatory effect while being less prone to side effects. The use of AHL as modulators of gut inflammation is thus an interesting and promising path. Moreover, 3-oxo-C12:2-HSL was shown to stabilize tight junctions proteins involved in gut barrier18, thereby counterbalancing the disruption of the gut barrier in IBD47. Altogether, these results pave the way for future therapeutic development of this type of molecule. In this setting, it remains crucial to study the mechanisms of action by which a given AHL signals its effect on epithelial and immune cells. Our future studies will focus on the JAK-STAT pathway and explore how 3-oxo-C12:2-HSL is able to prevent phosphorylation of JAK1 and STAT1 proteins.
Interkingdom effects of AHL raise the question of eukaryotic AHL receptors. Some proteins were identified as target of 3-oxo-C12-HSL from P. aeruginosa: the scaffolding protein IQGAP1f., the aryl hydrocarbon receptor (AhR)48, PPARγ49,50, the bitter taste receptor TAS2R3819,24,51,52. The latter has aroused our curiosity due to the growing interest in bitter taste receptors, particularly in their extra-oral expression25. In this study, we demonstrate that TAS2R38 signalling is needed for 3-oxo-C12:2-HSL to exert anti-inflammatory effects on immune cells. Taste receptor signalling, through the second messenger IP3, activates calcium release from the endoplasmic reticulum25. We demonstrated that 3-oxo-C12:2-HSL is able to induce calcium flux in RAW264.7 macrophages, thus reinforcing the hypothesis that 3-oxo-C12:2-HSL signals through taste receptors. Furthermore, through a screening of all human bitter taste receptors, we identified six taste receptors that were activated in the presence of 3-oxo-C12:2-HSL. The six receptors displayed different response amplitudes and maximal effective concentration, with TAS2R8 being the most sensitive (lowest EC50) to the AHL. Some of the tested BTR have numerous known ligands, such as TAS2R14 which can bind to more than 150 compounds, while others only have two known synthetic agonists like TAS2R13, according to the BitterDB database53. Among the receptors we identified as activated by 3-oxo-C12:2-HSL, TAS2R8, TAS2R10 TAS2R13 and TAS2R14 are closely related, while TAS2R1 and TAS2R38 are more distant (Supplementary Fig. 3). BTR seem to share high homology in their intra-membrane and intra-cytoplasm domains but their extra-cellular domain is more diverse, possibly to allow the recognition of several agonists34. A study described precisely the interactions between AHLs and BTR at the amino-acid level, and showed that the AHLs tested bind to the same extra-cellular orthosteric site on different BTR (TAS2R4, TAS2R14, TAS2R20)24. Future structure–function studies on those identified domains could allow determining their involvement in the interaction with 3-oxo-C12:2-HSL. Advances in knowledge of BTR involvement in inflammatory pathways are needed to link our findings with AHLs signalling in immune cells.
In the present study, we restricted our investigation to the effect of 3-oxo-C12:2-HSL on host immune cells. One could imagine that 3-oxo-C12:2-HSL also exerts an effect on gut microbiota. Since we have previously shown that the absence of 3-oxo-C12:2-HSL is associated with the differential representation of several bacterial taxa in faecal microbiota from IBD patients17, we could hypothesise that 3-oxo-C12:2-HSL participates in gut normobiosis.
Gut dysbiosis is now a well-recognized feature in IBD. This condition can promote or inhibit specific bacteria species of interest, which have been demonstrated to play a role in the pathophysiology54. Therefore, elucidating the origin of 3-oxo-C12:2-HSL in the intestine would allow a major advance in understanding its possible involvement in microbiota eubiosis and the physiology of the host. Several teams have attempted to use a bioinformatics approach to determine a system of bacterial synthesis of AHLs in the intestine17,55–58. Among the genes of the microbiome that have been sequenced and listed in the databases, no solid lead has been highlighted and such investigations have to be pursued. Concerning specifically 3-oxo-C12:2-HSL, it could be the product of a trophic chain, where host enzymes would be involved, in particular to generate the double unsaturations.
It is worth noting that in the complex gut ecosystem, several QS auto-inducers coexist and may have opposite effects. Most effects exerted by QS molecules are detrimental to the host; for instance 3-oxo-C12-HSL from P. aeruginosa disrupts barrier function15,18, AI-3 synthesized by enterohemorrhagic E. coli exerts pro-inflammatory effects on host-cells59 and the universal QS molecule AI-2 produced by Fusobacterium nucleatum promotes pro-inflammatory macrophages60. On the opposite, a study showed that AI-2, delivered by modified E. coli, increased Firmicutes proportions of the microbiota following an in vivo antibiotic challenge and was able to recapitulate gut microbiota61. In the same way, the role of 3-oxo-C12:2-HSL in shaping gut microbiota needs to be examined. Its putative effects induced in the gut microbiota, which must be investigated, could be additive to its effects on host cells, leading to an overall beneficial effect in gut homeostasis.
Our work is part of an innovative approach highlighting the role of bacterial QS molecules in the gut apart from the interaction with a pathogen. Our results provide a first set of data on the interactions of the gastrointestinal tract with AHL and we propose a model laying the foundation for the effect of 3-oxo-C12:2-HSL on immune cells (Fig. 5). 3-oxo-C12:2-HSL is able to activate bitter taste receptors, resulting in calcium release from the endoplasmic reticulum. Besides, 3-oxo-C12:2-HSL prevents from the activation of the JAK-STAT pathways in activated macrophages by specifically preventing the phosphorylation of JAK1 and STAT1. Finally, the effect of 3-oxo-C12:2-HSL on immune cells highlights the role of this type of molecule in the pathophysiology of puzzling diseases such as IBD where dysbiosis is involved in gut inflammation. Investigating QS molecules and their interkingdom effect therefore remains an unavoidable pursuit when searching for links between inflammation and gut microbiota.
Methods
Cell culture
RAW264.7 murine macrophage cells (ATCC, Manassas, Virginia, USA) were cultured between passage 12 and 25 in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum and 1% l-glutamine (ThermoFisher Scientific). The macrophages were seeded at 35,000 cells per well in 12-well plates (Falcon) for ELISA experiments or 110,000 cells per well in 6-well plates (Falcon) for cell lysates, upon reaching 80–90% confluence after 3-day culture. Cells were maintained at 37 °C with 5% CO2/air atmosphere and the media were changed every other day.
Human peripheral blood mononuclear cell isolation
Peripheral Blood Mononuclear Cells (PBMC) were extracted from whole blood of anonymous donors and collected by Etablissement Français du Sang of Paris, France.
PBMC were cultured in Roswell Park Memorial Institute medium (ThermoFisher Scientific) with 10% heat-inactivated fetal calf serum, 1% l-glutamine, 1% non-essential amino acids, 1% sodium pyruvate, penicillin (100 IU/mL), and streptomycin (100 µg/mL). Peripheral blood was harvested in EDTA tubes, blood was diluted with PBS 1:1, layered over Histopaque-1077® (Sigma) and centrifuged for 20 min at 400g at room temperature. Extracted PBMC were washed twice with sterile PBS then centrifuged at 300g for 5 min and resuspended in complete medium. For experiments, cells were seeded at 300,000 cells per well in 200 µL of medium in 96-well plates (ThermoFisher Scientific).
Cell treatments
Cells were exposed to 3-oxo-C12:2-HSL (synthesized as previously described17) or DMSO 0.1% as a control for different durations depending on the experiment, in the presence of the lactonase inhibitor 2-hydroxyquinoline (100 µM, Sigma-Aldrich), in order to limit the hydrolysis of the AHL lactone ring, as described previously17,18.
In RAW264.7 cells, inflammation was triggered with LPS (10 ng/mL, Sigma) and IFNγ (10 U/mL, R&Dsystems) for 2 h or 6 h, depending on the experiments. PBMC were exposed to LPS (10 ng/mL) and AHL for 24 h. For TAS2R38 inhibition experiments, cells were exposed to TAS2R38 allosteric inhibitor probenecid (1 mM, Sigma) for one hour before adding AHLs and the inflammatory cocktail, without changing the media.
Lactate dehydrogenase release
Release of lactate dehydrogenase into the medium was measured to assess cellular toxicity. LDH concentration was measured using the Cytotoxicity Detection Kit (Roche, Boulogne-Billancourt, France) according to the manufacturer’s instructions. The media were centrifuged 5 min at 250g to remove any cells. 50 µL of reaction mixture was added to 50 µL of media supernatant in 96-well plates. After 10 min at room temperature, 25 µL of stop solution was added. Absorbance at 490 nm was determined with a microplate spectrometer (FLUOstar Omega; BMG Labtech). Results are expressed as absorbance arbitrary units.
Enzyme-linked immunosorbent assay
At the end of cell treatment, the medium was centrifuged at 300g for 5 min at 4 °C. Supernatant was frozen at − 80 °C until ELISA assays. TNFα and IL-1β ELISA kits were purchased from R&D systems and performed according to the manufacturer’s instructions. Briefly, plates were coated with a capture antibody and incubated overnight. Wells were blocked using 1% bovine serum albumin-PBS solution (Sigma), then exposed to cell supernatants at room temperature for two hours. Wells were washed and incubated with the detection antibody for an additional two hours. After thorough washing, Streptavidin-HRP solution was added for 20 min. Wells were washed, then incubated with TMB (Biolegend) solution for 20 min and the reaction was stopped using 1 M H2SO4. Absorbance was measured by a spectrometer (FLUOstar Omega; BMG Labtech) at 450 nm and the absorbance at 540 nm was subtracted.
RNA extraction for RNA sequencing
Cells were treated for 2 h and then washed once with PBS before RNA extraction using RNeasy minikit (Qiagen), according to the manufacturer’s instructions. Briefly, cells were lysed and 1 volume of ethanol was added. The sample was transferred into a spin column and centrifuged at 8000g for 15 s. The column was washed several times and then eluted with water. RNA quality was assessed using NanoChip (Agilent) on a chip reader (Bioanalyzer 2100, Agilent).
mRNA sequencing
After extraction, total RNA were qualified with the AGILENT tapeStation 2200. Preparation of mRNA libraries was performed according to the manufacturer’s recommendations (KAPA mRNA HyperPrep Kit from ROCHE). Final samples pooled into a library were sequenced on ILLUMINA Novaseq 6000, corresponding to 2 × 28 Millions of 100 bases reads per sample after demultiplexing.
RNA-Seq data analysis workflow
FastQC v 0.11.962 was used for the quality control of raw paired-end fastq data set. Illumina’s adapters were removed from reads using cutadapt v2.1063. Trim_galore v0.6.464 was used to trim bad quality bases (Phred < 20). Salmon v 1.4.065 was used to quantify paired-end reads against a mapping-based index built from the Ensembl GRCm38 transcript set for all samples. Downstream analysis was handled in R platform v4.0.266 and its add-on packages. Quantified transcripts were imported into R using the package tximport v1.16.167. Gene-level DESeqDataSet object was built from previously imported transcript level abundances using the package DESeq268 to perform the differential expression analysis. A pre-filtering was applied to remove genes with less than 5 counts in one sample. For visualisation, raw counts were transformed using the “variance stabilizing transformation” method implemented in the package DESeq2. For an overview of the variability among samples, Principal Component Analysis was performed on variance stabilizing transformation and normalised count dataset using the package ade4 v 1.7-1669. Differential expression analysis was carried out by the DESeq function of the DESeq2 package. Briefly, the size factor was estimated for each sample, the dispersion was estimated for each gene, counts were fitted to a Negative Binomial Generalized Linear Model and Wald’s test was conducted to test the difference in expression between biological conditions. The Benjamini–Hochberg method70 was used for controlling the False-discovery rate (FDR). Significance level was fixed at type I error alpha = 0.01. Volcano plots were used to display the results of differential analysis using the ggplot2 v3.3.371 package. All significant gene lists were annotated for enriched biological functions and pathways using the DAVID platform through the RDAVIDWebService v1.28.072 package for gene ontology73 and Kyoto Encyclopedia of Genes and Genomes terms74. Significant terms had adjusted p-value, according to the Benjamini–Hochberg method, below 0.05. In addition to the traditional gene by gene analysis, count dataset was analysed at KEGG pathway level using the package Ensemble of Gene Set Enrichment Analysis (EGSEA) v1.16.075 that combines results from twelve algorithms to improve the biological relevance of pathways. Thus, significant pathways had adjusted p-value below 0.01. Gene expression heatmaps were produced by the pheatmap v1.0.1276 package. We used Venn diagrams to globally visualize the overlap between all significant genes as well as all significant pathways in the biological condition comparisons.
Determination of protein levels by Wes™ capillary electrophoresis
RAW264.7 cells were scraped in lysis buffer (20 mM Tris- HCl, pH 7.4, 5 mM EDTA, 0.15 M NaCl, 1% Triton X-100, 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail (Complete Mini; Roche, Boulogne-Billancourt, France) and phosphatase inhibitor cocktail (PhosSTOP, Sigma-Aldrich). Protein concentrations were determined using the BC Assay (Uptima/Interchim). Wes™ analyses (capillary electrophoresis system; ProteinSimple, San Jose, CA) were performed according to the manufacturer’s recommendations, and adequate protein concentrations and antibody dilutions were determined in preliminary assays to allow optimal quantitative conditions. The microplates were loaded with 0.8 μg/μL protein, primary antibodies (as detailed below), and reagents provided by the manufacturer: anti-JAK1 (1:25; Biotechne MAB4260), anti-Phospho-JAK1 (1:100; Invitrogen 44422G), anti-JAK2 (1:13; Cell Signalling, 3230), anti-Phospho-JAK2 (1:13; Cell Signalling 3776), anti-STAT1 (1:50; Cell Signalling 9172), anti-Phospho-STAT1 (1:40; Biotechne AF2894), anti-Actin (1:1000; Millipore MAB1501R). Data were analysed using Compass for SW3.1 software (ProteinSimple). Protein levels were determined by chemiluminescence signal (AUC) and Phospsho-Protein levels were normalized to the total level of each protein of interest, after verification that actin levels do not differ between conditions.
Bitter taste receptors screening
Bitter taste receptor cDNA were cloned into the pcDNA4 mammalian expression vector (Invitrogen) using full coding receptor sequences reported on UniProt database. The synthetic constructs were codon optimized (Genewiz) and combine the first 45 amino acids of rat somatostatin type 3 receptor at the amino terminus to improve plasma membrane targeting of the receptors in the heterologous system77. The FLAG epitope was added to the carboxy terminus without interfering with receptor functionality and can be used for immunocytochemistry. Calcium-mobilization assays were performed using human embryonic kidney cells (HEK293T) stably transfected with chimeric Gα16gust44 protein78. Cells were seeded in black, poly-d-lysine coated 96-well plates at a density of 35,000 cells/100 µL in high glucose DMEM supplemented with 10% dialysed fetal bovine serum and 1% penicillin/streptomycin. Twenty-four hours later, the cells were transiently co-transfected using Lipofectamine 2000 (Life Technologies) with one of the TAS2R synthetic optimized vectors or empty expression plasmid, as a negative control, and the pCMV-GCaMP5G construct (Addgene #31788) coding for a genetically encoded calcium indicator79,80. Twenty-four hours after transfection, cells were washed twice with C1 buffer (130 mM NaCl, 5 mM KCl, 10 mM HEPES, 2 mM CaCl2, 5 mM sodium pyruvate, pH7.4). 3-oxo-C12:2-HSL was solved in DMSO and further diluted in C1 buffer. Next, the cells were placed in a fluorometric imaging plate reader (Flexstation® 3, Molecular Devices) and stimulated with automatic injection of increasing concentrations of AHLs. Calcium responses leading to increase in fluorescence were monitored at 510 nm after excitation at 488 nm. Experiments were performed in duplicates and repeated at least four times. The recorded calcium levels of each wells receiving the same stimulus were averaged, response of mock-transfected cells were subtracted from receptor-transfected cells and net signals were normalized to background fluorescence (F/F0, F0 fluorescence light before stimulus application). The resulting dose–response curves of the averaged fluorescent signal amplitudes against the logarithm of the agonist concentration were fitted using Sigma Plot software. For data analysis, the four-parameter logistic equation [f(x) = min + (max − min)/(1 + (x/EC50)nH)] was used to calculate the half-maximal effective concentrations (EC50 values).
RNA extraction and RT-qPCR
Total RNA was extracted from RAW264.7 cells using TRIzol (Invitrogen) according to the manufacturer's instructions. Reverse Transcription (RT) was performed with 1 μg RNA using LunaScript® RT SuperMix Kit (NewEngland Biolabs). Semi-quantitative real-time PCR was performed with the Mx3000P Stratagene system using SYBR Green (Applied Biosystems) according to the manufacturer's procedures. Cyclophilin was used as the reference gene. After amplification, Ct were determined. Sequences of the oligonucleotide primers used are reported in Table 1.
Table 1.
Sequences of the primers used in this study.
| Target gene (protein) | Forward primer | Reverse primer |
|---|---|---|
| Tnf (TNFα) | CCAGACCCTCACACTGAGATC | CACTTGGTGGTTTGCTACGAC |
| Il1b (IL-1β) | AGTTGACGGACCCCAAAAG | AGCTGGATGCTCTCATCAGG |
| Il10 (IL-10) | CTGGACAACATACTGCTAACC | GGGCATCACTTCTACCAGGTA |
| Ppib (Cyclophilin B) | GCCTTAGCTACAGGAGAGAA | TTTCCTCCTGTGCCATCTC |
Intracellular calcium assay
RAW264.7 cells were seeded in black, clear-bottom 96-well plates (Costar) at 20,000 cells per well. The next day, release of intracellular Ca2+ was measured using Fluo-4 (Fluo-4 Direct Calcium Assay; Life Technologies) according to the manufacturer’s instructions. Medium was discarded and 100 µL of 1 X Fluo-4 dye was incubated at 37 °C for 1 h in 5% CO2, protected from light. Then, basal fluorescence was measured using a fluorospectrometer (FLUOstar Omega; BMG Labtech), excitation at 485 nm and emission at 520 nm. Molecules of interest were then added with an electronic pipette; thapsigargin (10 µM, Sigma) as a positive control, 3-oxo-C12:2-HSL (1000 µM) or DMSO 0.5%; with 8 replicates per conditions for every experiment. The fluorescence signal was immediately measured every 80 s for 2 h. Results are expressed as a ratio of the fluorescence x times over the basal fluorescence (F/F0).
Quantification and statistical analysis
Data are expressed as means ± standard error of the mean (SEM) unless otherwise indicated. The number of independent experiments and replicates are indicated in the figure captions. Figures and statistical analyses were performed with Graphpad® Prism 8.0 (Ritme Informatique, Paris, France). Normal distribution was tested using Shapiro–Wilk test. When normal distribution was confirmed, statistical differences between the means were assessed by ANOVA or t test otherwise by a non-parametric Kruskal–Wallis test. Post-test are indicated in the figure captions. For all results, p < 0.05 was considered statistically significant. Values are represented as follows: *p ≤ 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Ethics declarations
Peripheral Blood Mononuclear Cells (PBMC) were extracted from whole blood of anonymous donors and collected by Etablissement Français du Sang of Paris, France. Informed consent was obtained from all participants.
Supplementary Information
Acknowledgements
This research was funded by Fondation pour la Recherche Médicale, grant number EQU202003010171 and by the Association François Aupetit (projects research on AHLs 2019 and 2021). G. Coquant was the recipient of a fellowship from Fondation pour la Recherche Médicale FRM ECO201806006843. D. Aguanno was the recipient of a fellowship from CORDDIM, Ile-de-France region. The authors acknowledge the Association François Aupetit for its unwavering support. The authors thank Yannick Marie and his team at the Plateforme de Genotypage/Séquençage of the Institut du Cerveau et de la Moelle épinière for the RNA sequencing.
Author contributions
G.C., S.T., P.S. conceived the experiments; G.C., D.A., L. Brot, C.B., J.D., M.M. and N.R. conducted the experiments and analysis; L.D.S., J.-.P.G., V.C., S.T., P.S., L. Briand supervised the work; G.C. and P.S. wrote the main manuscript text; G.C. prepared all the figures. Funding Acquisition: S.T., P.S. All authors reviewed the manuscript.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author upon reasonable request.
Competing interests
P Seksik declares speaker and consulting fees from Takeda, Merck MSD, Biocodex, Ferring, Janssen, Amgen, Pfizer, Pileje, Fresenius Kabi and Abbvie but has no conflict of interest linked to this work. The other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-13451-3.
References
- 1.Maaser C, et al. ECCO-ESGAR guideline for diagnostic assessment in IBD part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J. Crohn’s Colitis. 2019;13:144–164K. doi: 10.1093/ecco-jcc/jjy113. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Rocha C, Vande Casteele N. Corrigendum to ‘JAK inhibitors: Current position in treatment strategies for use in inflammatory bowel disease’. Curr. Opin. Pharmacol. 2021;58:68. doi: 10.1016/j.coph.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Hashash J. Biologic monotherapy versus combination therapy with immunomodulators in the induction and maintenance of remission of Crohn’s disease and ulcerative colitis. Ann. Gastroenterol. 2021;34:612. doi: 10.20524/aog.2021.0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.IBDMDB Investigators Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: Time to connect with the host. Curr. Opin. Gastroenterol. 2010;26:327–331. doi: 10.1097/MOG.0b013e328339536b. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y-W, Fischer M. Fecal microbiota transplantation for ulcerative colitis. Are we ready for primetime? Gastroenterol. Clin. North Am. 2020;49:739–752. doi: 10.1016/j.gtc.2020.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Saint-Antoine IBD Network Fecal microbiota transplantation to maintain remission in Crohn’s disease: A pilot randomized controlled study. Microbiome. 2020;8:12. doi: 10.1186/s40168-020-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020 doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 10.Duboc H, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 11.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quevrain E, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Spiegeleer B, et al. The quorum sensing peptides PhrG, CSP and EDF promote angiogenesis and invasion of breast cancer cells in vitro. PLoS One. 2015;10:e0119471. doi: 10.1371/journal.pone.0119471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Spiegeleer A, et al. Quorum sensing molecules as a novel microbial factor impacting muscle cells. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020;1866:165646. doi: 10.1016/j.bbadis.2019.165646. [DOI] [PubMed] [Google Scholar]
- 15.Turkina MV, Vikström E. Bacteria-host crosstalk: Sensing of the quorum in the context of Pseudomonas aeruginosa infections. J. Innate Immun. 2018;11:263–279. doi: 10.1159/000494069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coquant G, Grill J-P, Seksik P. Impact of N-acyl-homoserine lactones, quorum sensing molecules, on gut immunity. Front. Immunol. 2020;11:1827. doi: 10.3389/fimmu.2020.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landman C, et al. Inter-kingdom effect on epithelial cells of the N-Acyl homoserine lactone 3-oxo-C12:2, a major quorum-sensing molecule from gut microbiota. PLoS One. 2018;13:e0202587. doi: 10.1371/journal.pone.0202587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguanno D, et al. The intestinal quorum sensing 3-oxo-C12:2 Acyl homoserine lactone limits cytokine-induced tight junction disruption. Tissue Barriers. 2020;8:1832877. doi: 10.1080/21688370.2020.1832877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurer S, et al. Tasting Pseudomonas aeruginosa biofilms: Human neutrophils express the bitter receptor T2R38 as sensor for the quorum sensing molecule N-(3-oxododecanoyl)-l-homoserine lactone. Front. Immunol. 2015;6:369. doi: 10.3389/fimmu.2015.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreira Lopes TC, Mosser DM, Gonçalves R. Macrophage polarization in intestinal inflammation and gut homeostasis. Inflamm. Res. 2020;69:1163–1172. doi: 10.1007/s00011-020-01398-y. [DOI] [PubMed] [Google Scholar]
- 21.Bryan A, et al. Human transcriptome analysis reveals a potential role for active transport in the metabolism of Pseudomonas aeruginosa autoinducers. Microbes Infect. 2010;12:1042–1050. doi: 10.1016/j.micinf.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeshi K, et al. Revisiting inflammatory bowel disease: Pathology, treatments, challenges and emerging therapeutics including drug leads from natural products. JCM. 2020;9:1273. doi: 10.3390/jcm9051273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salas A, et al. JAK–STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:323–337. doi: 10.1038/s41575-020-0273-0. [DOI] [PubMed] [Google Scholar]
- 24.Jaggupilli A, Singh N, Jesus VCD, Duan K, Chelikani P. Characterization of the binding sites for bacterial acyl homoserine lactones (AHLs) on human bitter taste receptors (T2Rs) ACS Infect. Dis. 2018;4:1146–1156. doi: 10.1021/acsinfecdis.8b00094. [DOI] [PubMed] [Google Scholar]
- 25.Lu P, Zhang C-H, Lifshitz LM, ZhuGe R. Extraoral bitter taste receptors in health and disease. J. Gen. Physiol. 2017;149:181–197. doi: 10.1085/jgp.201611637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon T-I, Seo Y-K, Osborne TF. Gut bitter taste receptor signaling induces ABCB1 through a mechanism involving CCK. Biochem. J. 2011;438:33–37. doi: 10.1042/BJ20110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latorre R, et al. Expression of the bitter taste receptor, T2R38, in enteroendocrine cells of the colonic mucosa of overweight/obese vs. lean subjects. PLoS One. 2016;11:e0147468. doi: 10.1371/journal.pone.0147468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner A, et al. Interactions between taste receptors and the gastrointestinal microbiome in inflammatory bowel disease. J. Nutr. Intermed. Metab. 2019;18:100106. doi: 10.1016/j.jnim.2019.100106. [DOI] [Google Scholar]
- 29.Yu Y, et al. Berberine induces GLP-1 secretion through activation of bitter taste receptor pathways. Biochem. Pharmacol. 2015;97:173–177. doi: 10.1016/j.bcp.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Tran HTT, Herz C, Ruf P, Stetter R, Lamy E. Human T2R38 bitter taste receptor expression in resting and activated lymphocytes. Front. Immunol. 2018;9:2949. doi: 10.3389/fimmu.2018.02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verbeurgt C, et al. The human bitter taste receptor T2R38 is broadly tuned for bacterial compounds. PLoS One. 2017;12:e0181302. doi: 10.1371/journal.pone.0181302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greene TA, et al. Probenecid inhibits the human bitter taste receptor TAS2R16 and suppresses bitter perception of salicin. PLoS One. 2011;6:e20123. doi: 10.1371/journal.pone.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wölfle U, et al. Expression and functional activity of the human bitter taste receptor TAS2R38 in human placental tissues and JEG-3 cells. Molecules. 2016;21:306. doi: 10.3390/molecules21030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 35.Telford G, et al. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 1998;66:36–42. doi: 10.1128/IAI.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glucksam-Galnoy Y, et al. The bacterial quorum-sensing signal molecule N-3-oxo-dodecanoyl-l-homoserine lactone reciprocally modulates pro- and anti-inflammatory cytokines in activated macrophages. J. Immunol. 2013;191:337–344. doi: 10.4049/jimmunol.1300368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kravchenko VV, et al. Modulation of gene expression via disruption of NF-κB signaling by a bacterial small molecule. Science. 2008;321:259. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, et al. N -3-(oxododecanoyl)-l-homoserine lactone promotes the induction of regulatory T-cells by preventing human dendritic cell maturation. Exp. Biol. Med. (Maywood) 2015;240:896–903. doi: 10.1177/1535370214564742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchie AJ, et al. The Pseudomonas aeruginosa quorum-sensing molecule N-3-(oxododecanoyl)-l-homoserine lactone inhibits T-cell differentiation and cytokine production by a mechanism involving an early step in T-cell activation. Infect. Immun. 2005;73:1648–1655. doi: 10.1128/IAI.73.3.1648-1655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hooi DSW, Bycroft BW, Chhabra SR, Williams P, Pritchard DI. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun. 2004;72:6463–6470. doi: 10.1128/IAI.72.11.6463-6470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peyrottes A, et al. Anti-inflammatory effects of analogues of N-acyl homoserine lactones on eukaryotic cells. Int. J. Mol. Sci. 2020;21:9448. doi: 10.3390/ijms21249448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isidro RA, Appleyard CB. Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;311:59–73. doi: 10.1152/ajpgi.00123.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darnell J, Kerr I, Stark G. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 44.Held TK, Weihua X, Yuan L, Kalvakolanu DV, Cross AS. Gamma interferon augments macrophage activation by lipopolysaccharide by two distinct mechanisms, at the signal transduction level and via an autocrine mechanism involving tumor necrosis factor alpha and interleukin-1. Infect. Immun. 1999;67:206–212. doi: 10.1128/IAI.67.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: Different gene signatures in M1(LPS+) vs. classically and M2(LPS–) vs. alternatively activated macrophages. Front. Immunol. 2019;10:1084. doi: 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L, Hooi D, Chhabra SR, Pritchard D, Shaw PE. Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene. 2004;23:4894–4902. doi: 10.1038/sj.onc.1207612. [DOI] [PubMed] [Google Scholar]
- 47.Odenwald MA, Turner JR. The intestinal epithelial barrier: A therapeutic target? Nat. Rev. Gastroenterol. Hepatol. 2016;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moura-Alves P, et al. Host monitoring of quorum sensing during Pseudomonas aeruginosa infection. Science (New York, N.Y.) 2019;366:eaaw1629. doi: 10.1126/science.aaw1629. [DOI] [PubMed] [Google Scholar]
- 49.Cooley MA, Whittall C, Rolph MS. Pseudomonas signal molecule 3-oxo-C12-homoserine lactone interferes with binding of rosiglitazone to human PPARγ. Microbes Infect. 2010;12:231–237. doi: 10.1016/j.micinf.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 50.Jahoor A, et al. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J. Bacteriol. 2008;190:4408–4415. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee RJ, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Investig. 2012;122:4145–4159. doi: 10.1172/JCI64240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaida MM, et al. Expression of the bitter receptor T2R38 in pancreatic cancer: Localization in lipid droplets and activation by a bacteria-derived quorum-sensing molecule. Oncotarget. 2016;7:12623. doi: 10.18632/oncotarget.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dagan-Wiener A, et al. BitterDB: Taste ligands and receptors database in 2019. Nucleic Acids Res. 2019;47:D1179–D1185. doi: 10.1093/nar/gky974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome—Searching the crime scene for clues. Gastroenterology. 2021;160:524–537. doi: 10.1053/j.gastro.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erickson DL, et al. Evidence of quorum sensing in the rumen ecosystem: Detection of N-acyl homoserine lactone autoinducers in ruminal contents. Can. J. Microbiol. 2002;48:374–378. doi: 10.1139/w02-022. [DOI] [PubMed] [Google Scholar]
- 56.Kumari A, Pasini P, Daunert S. Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem. 2008;391:1619–1627. doi: 10.1007/s00216-008-2002-3. [DOI] [PubMed] [Google Scholar]
- 57.Swearingen MC, Sabag-Daigle A, Ahmer BMM. Are there acyl-homoserine lactones within mammalian intestines? J. Bacteriol. 2013;195:173–179. doi: 10.1128/JB.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Won M-Y, Oyama LB, Courtney SJ, Creevey CJ, Huws SA. Can rumen bacteria communicate to each other? Microbiome. 2020;8:23. doi: 10.1186/s40168-020-00796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim CS, et al. Characterization of autoinducer-3 structure and biosynthesis in E. coli. ACS Cent. Sci. 2020;6:197–206. doi: 10.1021/acscentsci.9b01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, et al. Autoinducer-2 of Fusobacterium nucleatum promotes macrophage M1 polarization via TNFSF9/IL-1β signaling. Int. Immunopharmacol. 2019;74:105724. doi: 10.1016/j.intimp.2019.105724. [DOI] [PubMed] [Google Scholar]
- 61.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 62.Andrews S. fastQC. Babraham Institute; 2019. [Google Scholar]
- 63.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 64.Krueger F. Trim Galore. Babraham Institute; 2019. [Google Scholar]
- 65.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R. D. C. Team. R : A Language and Environment for Statistical Computing (2020).
- 67.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dray S, Dufour AB. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007;22:1–20. doi: 10.18637/jss.v022.i04. [DOI] [Google Scholar]
- 70.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. Methodol. 1995;57:289–300. [Google Scholar]
- 71.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; 2016. [Google Scholar]
- 72.Fresno C, Fernandez EA. RDAVIDWebService: A versatile R interface to DAVID. Bioinformatics. 2013;29:2810–2811. doi: 10.1093/bioinformatics/btt487. [DOI] [PubMed] [Google Scholar]
- 73.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alhamdoosh M, et al. Combining multiple tools outperforms individual methods in gene set enrichment analyses. Bioinformatics. 2017;33:414–424. doi: 10.1093/bioinformatics/btw623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolde, R. pheatmap: Pretty Heatmaps (2019).
- 77.Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses. 2009;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 78.Raliou M, et al. Human genetic polymorphisms in T1R1 and T1R3 taste receptor subunits affect their function. Chem. Senses. 2011;36:527–537. doi: 10.1093/chemse/bjr014. [DOI] [PubMed] [Google Scholar]
- 79.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cai B, et al. A cell-based functional assay using a green fluorescent protein-based calcium indicator dCys-GCaMP. Assay Drug Dev. Technol. 2014;12:342–351. doi: 10.1089/adt.2014.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author upon reasonable request.