Abstract
Study on sialidases as antiviral agents has been widely performed, but many types of sialidase have not been tested for their antiviral activity. Pasteurella multocida NanB sialidase is one such sialidase that has never been isolated for further research. In this study, the activity of NanB sialidase was investigated in silico by docking the NanB sialidase of Pasteurella multocida to the Neu5Acα(2–6)Gal and Neu5Acα(2–3)Gal ligands. Additionally, some local isolates of Pasteurella multocida, which had the NanB gene were screened, and the proteins were isolated for further testing regarding their activity in hydrolyzing Neu5Acα(2–6)Gal and Neu5Acα(2–3)Gal. Silico studies showed that the NanB sialidase possesses an exceptional affinity towards forming a protein–ligand complex with Neu5Acα(2–6)Gal and Neu5Acα(2–3)Gal. NanB sialidase of Pasteurella multocida B018 at 0.129 U/mL and 0.258 U/mL doses can hydrolyze Neu5Acα(2–6)Gal and Neu5Acα(2–3)Gal better than other doses. In addition, those doses can inhibit effectively H9N2 viral binding to red blood cells. This study suggested that the NanB sialidase of Pasteurella multocida B018 has a potent antiviral activity because can hydrolyze sialic acid on red blood cells surface and inhibit the H9N2 viral binding to the cells.
Subject terms: Drug discovery, Microbiology, Molecular biology
Introduction
Avian influenza virus infection prevention and treatment have been frequently studied. Various types of vaccines and antiviral agents have been developed to reduce the severity of viral infection. Vaccination is performed to stimulate the development of a body's immune system that can neutralize the virus before it replicates1. Avian influenza virus is an RNA virus that allows the virus to mutate through an antigenic shift or antigenic drift quickly2. Mutation cause vaccination to be less effective because the antibodies formed cannot recognize the mutant viruses3.
On the other hand, the antivirals to treat avian influenza infection are also considered ineffective4. Various reports suggest that severe infections occur due to the inability of antivirals to inhibit the multiplication of viruses in cells adequately5. Resistance towards multiple classes of antivirals makes the virus difficult to contain6. A previous study showed that most avian influenza viruses isolated from human cases were resistant to oseltamivir and amantadine groups7.
Recently, various studies have been carried out to address the avian influenza virus infection problem, one of which is through sialidases8. The use of sialidases in inhibiting viral infection was first investigated by Malakhov et al.9 later known as DAS18110. Sialidase DAS181 is derived from the bacterium Actinomyces viscosus, part of the normal floras in human teeth. Further studies showed that sialidase DAS181 could prevent the death of 100% of the mouse population infected with the pathogenic H5N1 avian influenza virus and prevent the spread of infection in 70% of the total mice in a single population10. The success of DAS181 in suppressing the spread of avian influenza infection results in a prevailing interest in sialidases as antiviral agents. In 2010, sialidase from Clostridium perfringens bacteria from poultry was successfully isolated and was later known as sialivac. The administration of sialivac in avian influenza infection livestock outbreaks can prevent the spread of the virus infection11.
In overcoming avian influenza infection, each drug has a different mechanism of action. Generally, antivirals work after the virus enters the host cell, in contrast to sialidases which functions to degrade sialic acid (Neu5Ac) found on the surface of host cells. Sialidases prevent hemagglutinin (HA) protein from binding to sialic acid to perform endocytosis12. The unavailability of sialic acid is thought to disrupt the budding viral process after replication in the host cell, resulting in the inability of the virus to infect other cells13. Studies related to sialidases also showed a decrease in the severity of symptoms in chickens infected with avian influenza.
In addition to Actinomyces viscosus and Clostridium perfringens, the presence of sialidase was also detected in several other types of bacteria such as Salmonella typhimurium, Vibrio cholerae, Corynebacterium diphtheria, Streptococcus pneumonia to Pasteurella multocida14–18. Molecularly, these bacteria have genes encoding sialidase with different targets for sialic acid hydrolysis activity. For example, Vibrio cholera tends to hydrolyze Neu5Acα(2–3)Gal compared to Neu5Acα(2–6)Gal, whereas Corynebacterium diphtheria tends to hydrolyze Neu5Acα(2–6)Gal compared to Neu5Acα(2–3)Gal16.
Pasteurella multocida uses sialidase to obtain nutrients in hydrolyzed sialic acid19. Of the many sialidase-producing bacteria, Pasteurella multocida is one with two genes encoding sialidase, namely NanH and NanB14. These genes encode sialidase protein which is a bacterial virulence factor in infecting the host. Several studies have screened the presence of NanH and NanB in Pasteurella multocida isolates and showed varied results, although the reason for these disparities is yet to be established20.
Although NanB and NanH are both sialidases derived from Pasteurella multocida bacteria, they have been shown to tend to hydrolyze different types of sialic acid. NanH tends to hydrolyze Neu5Acα(2–3)Gal, generally found in avian cells, while NanB sialidase is a broad-spectrum sialidase19. This is evidenced by the ability of NanB of Pasteurella multocida to hydrolyze Neu5Acα(2–3)Gal despite its resemblance to NanH of Actinomyces viscosus, which tends to hydrolyze Neu5Acα(2–6)Gal21.
Previous studies19 have shown the sialidase activity of Pasteurella multocida by expressing recombinant NanB sialidase in Escherichia coli bacteria, but the pure NanB sialidase protein has yet to be isolated. Therefore, in this study, in addition to predicting in silico the binding of NanB sialidase to Neu5Acα(2–6)Gal and Neu5Acα(2–3)Gal, authors screened the presence of the gene encoding sialidase in archival local isolates from Indonesia and characterize the NanB sialidase protein. Pure NanB sialidase is expected to hydrolyze sialic acid well so that it can be used in further research to study sialidase activity in inhibiting avian influenza infection.
Materials and methods
Bacteria, virus, NanB sialidase gene and ligand
Neu5Acα(2–6)Gal and Neu5Acα(2–6)Gal ligands were downloaded from the Protein Data Bank database web server (https://pubchem.ncbi.nlm.nih.gov) in 3D and optimized with Open Babel. The 3D structure of NanB sialidase from Pasteurella multocida isolate 86–1913 (Accession number AF274868) was obtained by entering the amino acid to Raptor X program. Then the molecular weight prediction of the NanB sialidase protein was carried out by entering the amino acid sequence at https://www.bioinformatics.org/sms/protmw.html. Meanwhile, to isolate pure NanB sialidase, screening was carried out on Pasteurella multocida, a local isolate archive of PT. Medika Satwa Laboratoris which only has one type of sialidase, namely NanB sialidase. Pure avian influenza subtype H9N2 strain A/Layer/Indonesia/WestJava04/2017 (MG957203) was used to show the relationship between the efficient hydrolysis of α2-6 and α2-3 bonds by NanB sialidase and anti-influenza activity.
Analysis of the bonding of NanB sialidase Pasteurella multocida with Neu5Ac2-6Gal by in silico test
NanB and ligands were prepared using AutoDockTools 1.5.6 by removing water molecules and adding nonpolar hydrogens, charges, and atoms. The grid was arranged by making a gridbox that covers the target protein's surface followed with treatment by the auto grid program linked to the application. The molecular docking process of the ligand with NanB sialidase from Pasteurella multocida was carried out using the auto grid program. The docking output was the docked ligand's structure on the enzyme active site and its respective affinity score. Analysis of the docking results was carried out on residues interacting with the ligands, Gibbs free binding energy (∆G), structural conformation, affinity, and hydrogen bonding between NanB sialidase and the ligands22.
Visualization of molecular docking results between ligands and proteins was carried out using Edu PyMOL software. Visualization using Edu PyMOL software aimed to clarify the binding site of the ligand to the protein22.
Culture, identification, and confirmation of Pasteurella multocida
A total of nine archival isolates were re-cultured using Brain Heart Infusion (BHI) Broth media (Himedia) and subsequently with Blood agar (BA) media (Himedia). Incubation was carried out overnight at 37 °C. The growing colonies were observed macroscopically and microscopically, followed by the catalase test23, oxidase test24, indole test25, and molecular confirmation through polymerase chain reaction (PCR) assay.
Pure colonies were extracted to obtain pure DNA using PrestoTM Mini gDNA Bacteria Kit (Geneaid). The primer used in the confirmation PCR test for Pasteurella multocida was an KMT1-specific primer pair with a target amplicon of 460 bp (Table 1). The PCR process was carried out using the KAPA2G Fast Hotstart Readymix PCR Kit (Merck) according to the relevant procedures with a final reaction volume of 10 μl and annealing temperature optimized for 55 °C for 40 cycles. The amplified sample was then visualized by electrophoresis using 1.5% agarose gel (1st base) and stained using 0.5 g/ml ethidium bromide (Vivantis). The marker used was 100 bp (VC 100 bp Plus DNA Ladder Vivantis) as a standard measure. Isolates identified as Pasteurella multocida through PCR testing were then tested on the hyaD-hyaC and bcbD genes (Table 1) to differentiate the identified serotypes of Pasteurella multocida27.
Table 1.
List of Primers for Identification of Pasteurella multocida.
| Genes | Sequences | Amplicon (bp) |
|---|---|---|
| KMT127 | T7(F) 5′-GCGTTTCATTCAAAGCATCTC-3′ | 460 |
| SP6(R) 5′-ATGACCGCGTAACGACTTTC-3′ | ||
| hyaD-hyaC28 | CapA(F) 5′-TGCCAAAATCGCAGTGAG-3′ | 1044 |
| CapA(R) 5′-TTGCCATCATTGTCAGTG-3′ | ||
| bcbD28 | CapB(F) 5′-CATTTATCCAAGCTCCACC-3′ | 758 |
| CapB(R) 5′-GCCCGAGAGTTTCAATCC-3′ | ||
| NanB29 | (F) 5′-GTCCTATAAAGTGACGCCGA-3′ | 554 |
| (R) 5′-ACAGCAAAGGAAGACTGTCC-3′ | ||
| NanH29 | (F) 5′-GAATATTTGGGCGGCAACA-3′ | 360 |
| (R) 5′-TTCTCGCCCTGTCATCACT-3′ |
Identification and molecular characterization of sialidase-coding genes Pasteurella multocida
Identification of the presence of the gene encoding Pasteurella multocida sialidase was carried out on isolates that had been identified as Pasteurella multocida type A. Screening for the presence of the sialidase gene was carried out using the NanB and NanH primer (Table 1). The KAPA2G Fast Hotstart Readymix PCR Kit (Merck) was used in the PCR process with a total PCR reaction of 50 μl and an annealing temperature of 56 ℃30. The sequencing of amplicons that only showed positive NanB were performed by 1st base sequencing service agency, Malaysia. Sequencing data were analyzed using MEGA X and Bioedit software to compare the genetics of the study isolate NanB with the NanB gene belonging to Pasteurella multocida 86-1913.
Native production of NanB sialidase Pasteurella multocida
Pasteurella multocida bacteria containing only the gene encoding NanB sialidase was propagated in the main batch of Brain Heart Infusion (BHI) broth (Himedia) 1000 ml and then centrifuged at 4 °C at a speed of 4000×g for 45 min. The bacterial pellet was used for the subsequent production of NanB sialidase31.
Some of the methods used are the chloroform method with a slight modification32, the glycine method33, freeze-thaw34, and osmotic shock35. In this study, several modifications of osmotic shock were used to divide them into the original method, the addition of Ca2+, the addition of lysozyme, and a combination of Ca2+ and lysozyme. Based on the results of sialidase production, the method that gave the highest specific activity value of sialidase was used as the sialidase production method, followed by anion exchange chromatography and affinity chromatography.
Anion exchange chromatography was performed using Q-sepharose with the resulting fractions labeled as F0, F1, F2, F3, F4, and F5, respectively. The fraction showing the highest specific activity was purified by column (5 ml) affinity chromatography using N-(p-Aminophenyl)oxamic acid–Agarose (Sigma-Aldrich) according to the relevant procedures. The estimated molecular weight of protein sialidase was carried out using SDS PAGE at 12% separating gel and 4% stacking gel36. At each stage of purification, protein count was performed using the Bradford method and tested for sialidase activity using the Neuraminidase assay kit (Sigma-Aldrich) according to the relevant procedures.
Optimum pH, temperature and sialidase incubation period tests
To find the optimum temperature, 80 μl of the Neuraminidase assay kit (Sigma-Aldrich) reaction mixture and 20 μl of NanB sialidase protein fraction were mixed and then incubated at different temperatures, namely 20, 25, 30, 37, 40, 45 and 50 °C. The sialidase activity was subsequently calculated37.
The determination of the optimum pH was tested by incubating 20 µl of sialidase in pH 3 to pH 10 (1:1) in 80 µl of the Neuraminidase assay kit (Sigma-Aldrich) reaction mixture at 37 °C. The sialidase activity was subsequently calculated as Units/ml in the same manner as the previous protocol. Meanwhile, the determination of sialidase activity in a certain incubation period was carried out by incubating sialidase at 37 °C and then calculating its activity at 24, 48, and 72 h of incubation37.
Sialidase toxicity test on red blood cells
NanB sialidase toxicity test was carried out on chicken and rabbit red blood cells. A total of 500 μl of sialidase in graded doses (0 U/mL, 0.032 U/mL, 0.064 U/mL, 0.129 U/mL and 0.258 U/mL) was added to 500 μl of chicken and rabbit red blood cells with a concentration of 1% each and then incubated at 37 °C for 2 h. Centrifugation was carried out at a speed of 6000×g for 3 min, then transferred the supernatant to a microplate and read using a microplate reader with a wavelength of 562 nm. As a lysis control, lysis buffer (Geneaid) was used. Toxicity was calculated using the following formula: 100 × (OD of sample—OD of negative control/ OD of lysis control—OD of negative control)38.
Sialidase specificity test for sialic acid in chicken and rabbit red blood cells
Red blood cells of chickens and rabbits were treated with the addition of graded sialidase concentrations (0 U/mL, 0.032 U/mL, 0.064 U/mL, 0.129 U/mL and 0.258 U/mL) for 2 h at 37 °C. Cells were washed three times with PBS and fixed with 0.05% glutaraldehyde in PBS. Cell-based enzyme-linked lectin assay measured the amounts of (2,6)-linked sialic acid and (2,3)-linked sialic acid. The fixed cells were blocked with 3% bovine serum albumin (BSA) in PBS and streptavidin–biotin blocking reagent (Vector Laboratories, Burlingame, CA) to block endogenous streptavidin- and biotin-binding sites. Cells were rinsed once with PBS-0.1% Tween 20 (PBST) and incubated with 2 µg biotinylated SNA lectin (Vector Laboratory)/ml and 20 µg biotinylated MAA lectin (Vector Laboratory)/ml at 37 °C. SNA (Sambucus nigra) is specific for Neu5Acα(2–6)Gal while MAA (Maackia amurensis) is specific for Neu5Acα(2–3)Gal. The cells were washed four times with PBST. Secondary detection of bound lectins was carried out by incubating 5 g streptavidin-HRP/ml for 1 h at 37 °C. Cells were washed five times in PBST, added in tetramethylbenzidine (TMB; Sigma), and suspended in 1 M H2SO4. The absorbance was measured at 450 nm, and the percentage of sialic acid remaining was calculated using the following calculation: 100% × [(absorbance of treated cells − background)/(absorbance of treated cells − background)]. Cells treated with streptavidin-HRP alone without lectins were used as a background control9.
H9N2 virus binding inhibition test on post- red blood cells administration of sialidase
The H9N2 virus binding inhibition test on red blood cells was carried out in six repetitions with graded sialidase concentrations (0 U/mL, 0.032 U/mL, 0.064 U/mL, 0.129 U/mL and 0.258 U/mL). A total of 500 μl of graded sialidase concentration was added to 500 μl of 1% concentration of chicken and rabbit red blood cells and then incubated at 37 °C for 2 h. After incubation, 500 μl 9 log 2 HAU of avian influenza H9N2 virus was poured into a mixture of sialidase and red blood cells and then incubated for 30 min at room temperature. Then centrifuged at 5000 rpm for 5 min. The supernatant was discarded and the pellet in the form of red blood cells was extracted by RNA using the Total RNA Mini Kit (Geneaid) and then followed by qRT-PCR to count the number of RNA copies of the virus that were still bound to the red blood cells. The qRTPCR process was carried out using the Sensifast SYBR Lo-ROX One-Step Kit (Bioline) kit according to the protocol for use.
Statistical analysis
Data were analysed in Graphpad Prism 9.1.2. The mean and standard error of the mean (SEM) or standard deviation (SD) were used to display normally distributed data. The data comparison between the groups were analysed using analysis of variance (ANOVA).
Ethics declarations
No experimental animals were used in this study, however this study has carefully reviewed and approved by The Ethics Committee of the Faculty of Medicine, University of Indonesia (No. KET-1215/UN2.F1/ETIK/PPM.00.02/2020).
Results
In silico bonding of NanB sialidase Pasteurella multocida with Neu5Acα(2–6)Gal ligand
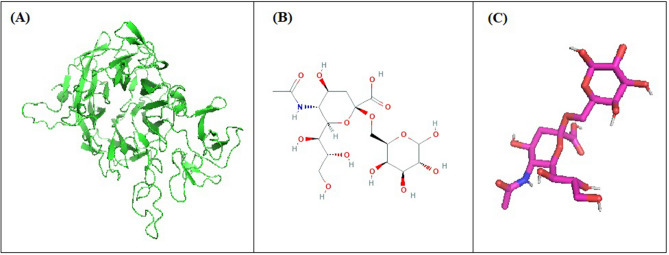
The 3D structure of NanB sialidase from Pasteurella multocida was generated from the Raptor X program (Fig. 1A), while the Neu5Acα(2–6)Gal ligand was successfully prepared from the 2D form (Fig. 1B) to 3D form (Fig. 1C) with the Open Babel program. Based on the results of the formation of 3D NanB sialidase, there were differences in the number of amino acid sequences from 1070 amino acids to 503 amino acids. These results also affect the prediction of the molecular weight of the NanB sialidase protein which https://www.bioinformatics.org/sms/prot_mw.html analyzed. The results of the analysis of the amino acid composition of Raptor X obtained that the molecular weight of NanB sialidase was 56.44 kDa, very different from the initial amino acid prediction of 119.81 kDa.
Figure 1.
Some of the structures required in the docking process. (A) NanB sialidase protein generated from the Raptor X program; (B) 2D structure of Neu5Acα(2–6)Gal; (C) Neu5Acα(2–6)Gal 3D structure.
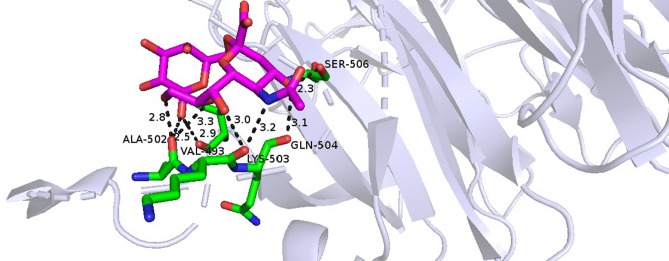
Based on the docking results between NanB sialidase and the Neu5Acα(2–6)Gal ligand, the Gibbs free energy or ∆G of − 5.56 kcal/mol was obtained. While Ki value produced by the recent docking process in this study was 84.26 μM. This was reinforced by the conformation between the ligand binding and the receptor, indicating that the Neu5Acα(2–6)Gal and NanB sialidase ligands can bind to each other. In this study, the hydrogen bonding interaction of NanB sialidase with Neu5Acα(2–6)Gal was shown at Val 493, Ala 502, Lys 503, Gln 504, Ser 506 with an overall distance below 3.9 Å (Fig. 2).
Figure 2.
Visualization of interactions between Neu5Acα(2–6)Gal and NanB sialidase using Pymol showing hydrogen bonds, residues and their interaction distance with Neu5Acα(2–6)Gal in 3D.
NanB sialidase and the Neu5Acα(2–3)Gal ligand showed − 7.15 kcal/mol for the Gibbs free energy value or ∆Gand the Ki value was 5.70 μM. There were some hydrogen bonding interactions of NanB sialidase with Neu5Acα(2–3), including Arg69, Gln169 and Gly321.
Culture, identification, and confirmation of Pasteurella multocida
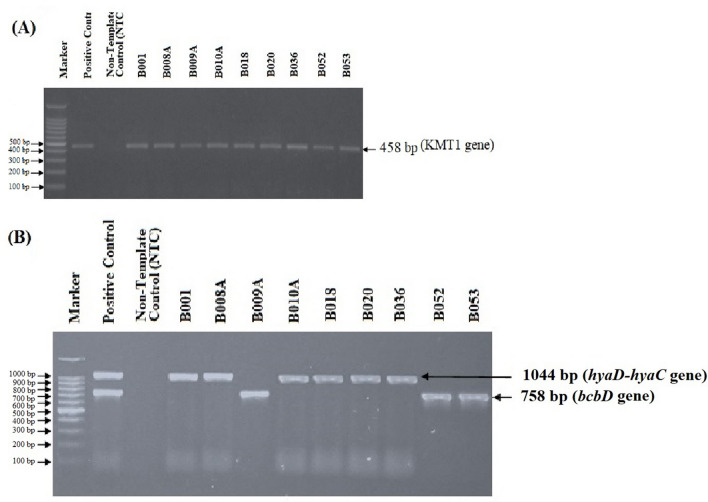
Identification of Pasteurella multocida was carried out using macroscopic, microscopic, and biochemical tests, further continued by the PCR test (Table 2). The results showed that all isolates were Pasteurella multocida bacteria (Fig. 3A), while the results of type classification through PCR showed that only six of the nine isolates were Pasteurella multocida type A (Fig. 3B).
Table 2.
Results of culture, identification and confirmation of Pasteurella multocida with various test methods.
| No | Isolate | Macroscopic | Microscopic | Catalase test | Oxidase test | Indole test | PCR test (ompH) | Multiplex PCR test | |
|---|---|---|---|---|---|---|---|---|---|
| CapA | CapB | ||||||||
| 1 | B001 | + | + | + | + | + | + | + | − |
| 2 | B008A | + | + | + | + | + | + | + | − |
| 3 | B009A | + | + | + | + | + | + | − | + |
| 4 | B010A | + | + | + | + | + | + | + | − |
| 5 | B018 | + | + | + | + | + | + | + | − |
| 6 | B020 | + | + | + | + | + | + | + | − |
| 7 | B036 | + | + | + | + | + | + | + | − |
| 8 | B052 | + | + | + | + | + | + | − | + |
| 9 | B053 | + | + | + | + | + | + | − | + |
Figure 3.
PCR test results confirm Pasteurella multocida and its serotypes. (A) amplification of the ompH gene from all isolates showed a position of 460 bp; (B) PCR test results to distinguish serotypes from Pasteurella multocida. The positive control is the result of DNA extraction from locale isolates of Pasteurella multocida types A and B mixed that showed two bands of the gel electrophoresis.
Identification and molecular characterization of sialidase Pasteurella multocida
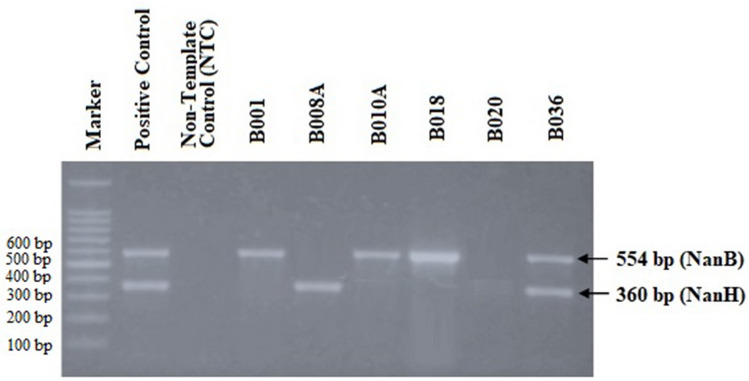
Molecular identification of the gene encoding sialidase was carried out on six isolates of Pasteurella multocida, previously confirmed as type A. The amplicon position was shown at 554 bp for isolates identified as having NanB sialidase and 360 bp for isolates having NanH sialidase. In this study, the isolates tested against the two genes gave varied results. Namely, three isolates only had NanB sidalise, one isolate only had nanH sialidase, one isolate had both types of sialidase, and another isolate did not have the gene encoding sialidase (Fig. 4).
Figure 4.
The detection results of NanB and NanH sialidase genes from six isolates confirmed as Pasteurella multocida type A. NanB is indicated by the 554 bp, while NanH in the 360 bp.
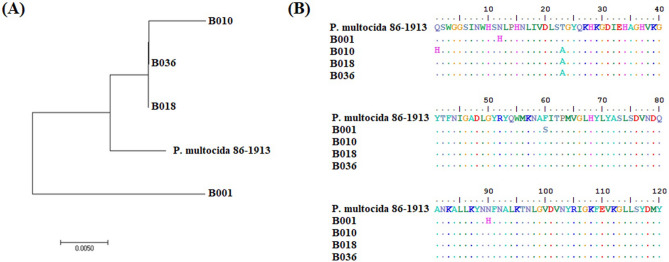
Based on the similarity of amino acid sequence and close kinship to isolate Pasteurella multocida 86–1913, some isolates in this study were selected for the isolation of NanB sialidase (Fig. 5). Isolate B018 was chosen because it has a single sialidase NanB (Fig. 4) and shows the highest amino acid sequence similarity, 99.00% with Pasteurella multocida 86–1913 (Table 3).
Figure 5.
Results of the Pasteurella multocida isolate selection test to be used in purification of protein sialidase. (A) Phylogenetic tree showing the relationship between Pasteurella multocida isolates; (B) Alignment of amino acids with the Bioedit program to determine points of difference in amino acids.
Table 3.
The results of the test of the similarity level of the NanB gene isolate with Pasteurella multocida 86–1913 as a reference isolate.
| Strain isolates | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| 1 | Pasteurella multocida 86–1913 | |||||
| 2 | B001 | 96.76 | ||||
| 3 | B010 | 98.40 | 96.33 | |||
| 4 | B018 | 99.00 | 96.96 | 99.40 | ||
| 5 | B036 | 99.00 | 96.96 | 99.40 | 100 | |
Isolation and purification of protein NanB sialidase Pasteurella multocida B018
Among the various isolation methods carried out, the results showed that the chloroform method could produce the highest specific activity of NanB sialidase, namely 0.202 U/mg (Table 4). Some methods that were also quite good in providing specific activity of NanB sialidase were Glycine and Freeze–thaw with specific activity values of 0.191 U/mg and 0.152 U/mg. In contrast to the other three methods, the osmotic shock method gave relatively lower values, both for the first supernatant containing sucrose, Tris–HCl, and EDTA, and the second supernatant in the form of reverse osmosis water (Table 4).
Table 4.
Results of testing the specific activity of NanB sialidase from various isolation methods.
| NanB sialidase isolation method | Supernatant | Supernatant volume (ml) | Protein concentration (mg/ml) | Total protein (mg) | Sialidase activity (U/ml) | Total sialidase activity (U) | Specific activity (U/mg) |
|---|---|---|---|---|---|---|---|
| Kloroform | Tris–HCl pH 8.0 | 1 | 0.823 | 0.823 | 0.166 | 0.166 | 0.202 |
| Glysin | Sterile aquades + Glysin 1% | 1 | 0.497 | 0.497 | 0.095 | 0.095 | 0.191 |
| Freeze thaw | Potasium phosphat buffer pH 6.8 | 1 | 0.611 | 0.611 | 0.093 | 0.093 | 0.152 |
| Osmotic shock | |||||||
| a. Original | sucrose, Tris–HCl, EDTA | 1 | 0.819 | 0.819 | 0.089 | 0.089 | 0.109 |
| reverse osmosis (RO) water | 1 | 0.203 | 0.203 | 0.002 | 0.002 | 0.009 | |
| b. Addition of Ca2+ | sucrose, Tris–HCl, EDTA | 1 | 0.870 | 0.870 | 0.075 | 0.075 | 0.087 |
| reverse osmosis (RO) water | 1 | 0.249 | 0.249 | 0.008 | 0.008 | 0.034 | |
| c. Addition of lysozyme | sucrose, Tris–HCl, EDTA | 1 | 0.893 | 0.893 | 0.081 | 0.081 | 0.091 |
| reverse osmosis (RO) water | 1 | 0.198 | 0.198 | 0.013 | 0.013 | 0.067 | |
| d. Combination of Ca2+ and lysozyme | sucrose, Tris–HCl, EDTA | 1 | 0.816 | 0.816 | 0.068 | 0.068 | 0.083 |
| reverse osmosis (RO) water | 1 | 0.216 | 0.216 | 0.021 | 0.021 | 0.099 | |
Crude NanB sialidase, which was isolated by chloroform method, was subsequently treated with anion exchange chromatography. An unusual result was observed at this stage: a decrease in the target protein at F0, which showed the highest sialidase-specific activity of 1.79 U/mg (Table 5). In this study, anion exchange chromatography and affinity chromatography increased purity of 6.9 and 40.13 times compared to crude NanB sialidase. The overall purification results can be seen in Table 6.
Table 5.
Results of purification of NanB sialidase by anion exchange chromatography.
| Anion exchange chromatography fraction | Supernatant volume (ml) | Total protein (mg) | Total sialidase activity (U) | Specific activity (U/mg) |
|---|---|---|---|---|
| Ccrude protein | 100 | 64.80 | 16.80 | 0.259 |
| F0 | 100 | 8.60 | 15.40 | 1.790 |
| F1 (0.2 M NaCl) | 5 | 10.66 | 0.14 | 0.013 |
| F2 (0.4 M NaCl) | 5 | 11.13 | 0.02 | 0.002 |
| F3 (0.6 M NaCl) | 5 | 1.82 | 0.01 | 0.006 |
| F4 (0.8 M NaCl) | 5 | 0.02 | 0.001 | 0.023 |
| F5 (1 M NaCl) | 5 | 0.03 | 0.001 | 0.020 |
Table 6.
Results of purification of NanB sialidase by anion exchange chromatography and affinity chromatography.
| NanB sialidase isolation method | Supernatant volume (ml) | Total protein (mg) | Total sialidase activity (U) | Specific activity (U/mg) | Sialidase activity (U/ml) | Purification fold |
|---|---|---|---|---|---|---|
| Crude protein | 100 | 64.80 | 16.80 | 0.259 | 0.168 | 1 |
| Anion exchange chromatography | 100 | 8.60 | 15.40 | 1.791 | 0.154 | 6.9 |
| Affinity chromatography | 15 | 0.372 | 3.87 | 10.40 | 0.258 | 40.13 |
The molecular weight measurements of NanB Sialidase produced in this study are novelties that have not been reported in previous studies correctly. NanB sialidase produced from Pasteurella multocida B018 showed a size of about 55 kDa (Fig. 6).
Figure 6.
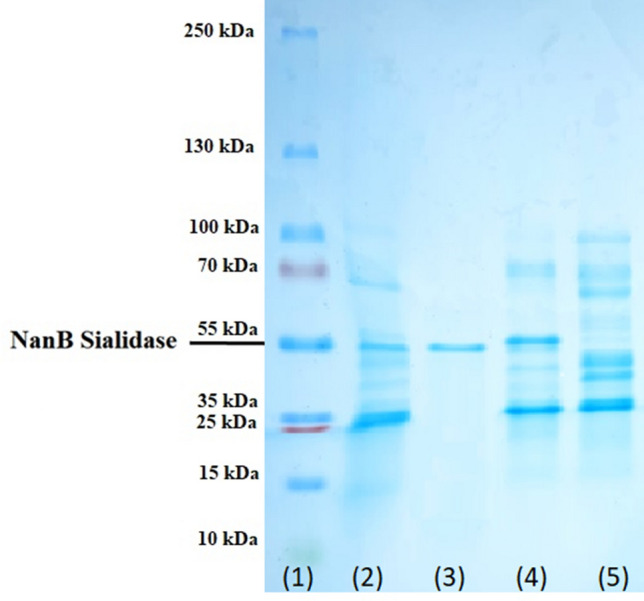
Results of SDS Page on protein at each stage of NanB sialidase Pasteurella multocida B018 purification. (1) Markers; (2) crude sel of Pasteurella multocida before chloroform method; (3) protein after Affinity chromatography; (4) protein after anion exchange chromatography; (5) initial protein supernatant crude sialidase.
Optimum temperature, pH and incubation period for NanB sialidase Pasteurella multocida B018
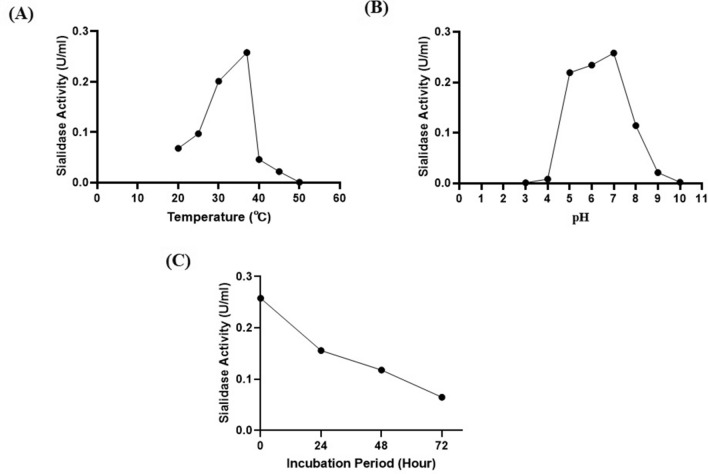
The optimum temperature in this study is defined as the temperature that shows the highest sialidase activity and stability. A temperature of 37 °C is an ideal condition for several enzymes, including NanB sialidase. Reasonably good results were also shown at the incubation temperature of 30 °C. In contrast to the two temperatures, sialidase activity was less than optimum at 20 °C and 25 °C and even showed deficient activity at 40–50 °C (Fig. 7A).
Figure 7.
Test results for optimum temperature, pH and incubation period of NanB sialidase Pasteurella multocida B018. (A) Optimum temperature; (B) optimum pH; (C) incubation period.
Tests on the optimum pH of sialidase also showed quite varied results. The optimum sialidase activity was indicated by pH 7, followed by pH 5 and 6, which were still quite good in producing sialidase activity. At pH 3 and 4, the sialidase activity decreased to zero (0). At pH 8 showed lower sialidase activity than pH 7, and pH 9–10 produced deficient sialidase activity (Fig. 7B).
After incubation for 24 h, testing of sialidase activity showed a decrease in activity from 0.258 U/ml to 0.156 U/ml. The same results were also shown at 48 and 72 h of incubation, which decreased activity to 0.118 and 0.065 U/ml. Sialidase activity decreased when incubated at 37 °C for 72 h (Fig. 7C).
NanB sialidase toxicity test results Pasteurella multocida B018
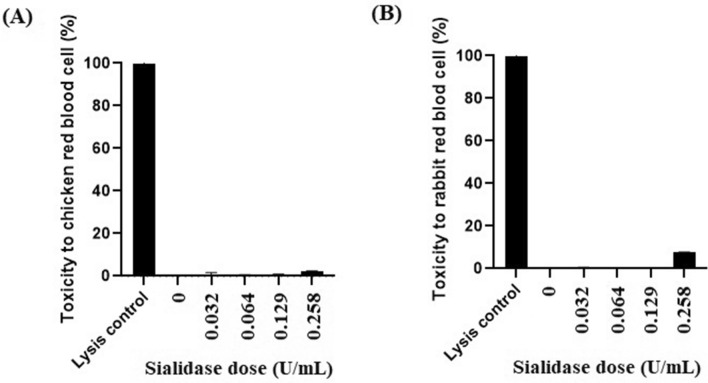
The results showed that the 0.258 U/mL sialidase dose caused the lysis of red blood cells in chickens 2.12% (Fig. 8A) as much as rabbits 7.65% (Fig. 8B). Both results represent the highest level of red blood cell damage compared to several other doses. Tests with a 0.129 U/mL dose of sialidase showed a much lower level of toxicity, namely 0.74% lysed chicken red blood cells, and 0.46% lysed rabbit red blood cells. Similarly, the two doses of sialidase, namely 0.064 U/mL and 0.032 U/mL, showed 0.42% lysis of chicken red blood cells and 0.31% and 0.08% lysis of rabbit red blood cells, respectively.
Figure 8.
Results of sialidase toxicity test on red blood cells of chicken and rabbit. (A) chicken red blood cells; (B) rabbit red blood cells.
Sialidase specificity test for sialic acid in chicken and rabbit red blood cells
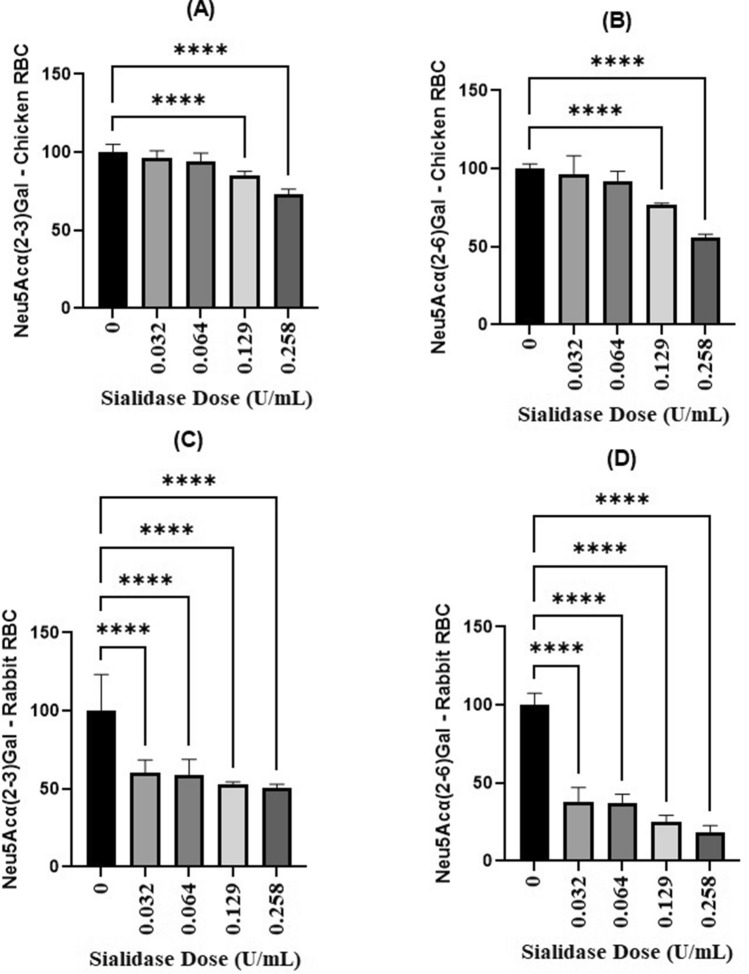
This study succeeded in demonstrating the activity of sialidase in hydrolyzing two sialic acids, namely Neu5Acα(2–6)Gal and Neu5Acα(2–3)Gal. This is evidenced by the reduced amount of these two types of sialic acid in the red blood cells of chickens and rabbits (Fig. 9).
Figure 9.
Results of NanB sialidase specificity test for two different sialic acids and two different blood types. (A) Neu5Acα(2–3)Gal, chicken red blood cells; (B) Neu5Acα(2–6)Gal, chicken red blood cells; (C) Neu5Acα(2–3)Gal, rabbit red blood cells; and (D) Neu5Acα(2–6)Gal rabbit red blood cells.
The calculation of residual sialic acid in chicken blood showed that there was a meaningful difference in the percentage of residual sialic acid between the control (0 U/mL) and the treatment group with the percentage of sialidase 0.129 U/mL and 0.258 U/mL (p < 0.05). In contrast, the residual amount of sialic acid in rabbit blood showed a meaningful difference in percentage between the control (0 U/mL) and the entire sialidase dose group (p < 0.05).
Inhibition of H9N2 virus binding on red blood cells after administration of sialidase
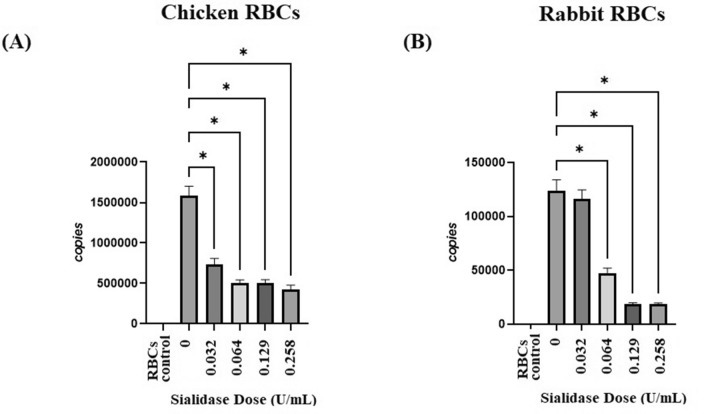
Quantitatively, the binding inhibition of the H9N2 subtype of avian influenza virus on red blood cells was assessed based on the copy number of the H9 virus gene bound to chicken and rabbit red blood cells through RT-qPCR. The results showed that there were differences in the copy number of the virus in chicken red blood cells, indicated by all treatment groups the percentage of sialidase (0.032 U/mL, 0.064 U/mL, 0.129 U/mL and 0.258 U/mL) when compared to the control (0 U/ml). With p value < 0.05. Meanwhile, in rabbit red blood cells, three doses were significantly different (p < 0.05) compared to control (0 U/ml), namely 0.064 U/ml, 0.129 U/ml and 0.258 U/ml with the best dose of 0.129 U/ml and 0.258 U/ml (Fig. 10).
Figure 10.
Viral copy number of H9 genes of the viruses that bind to red blood cells. (A) chicken red blood cells and (B) rabbit red blood cells.
Discussion
NanB sialidase is the receptor used as a docking material in this study. The enzyme was derived from Pasteurella multocida isolate 86–1913 available at GenBank with the access code AF274868 and has never been crystallized. As such, it required 3D structure prediction via Raptor X. The selection of docking results in this study was based on results that showed complete protein binding and density. X-Ray (RMSD) detected a value of 2 Å for docking of NanB sialidase and the ligands39.
Although only an approximation, − 5.56 kcal/mol for Neu5Acα(2–6)Gal and -7.15 kcal/mol for Neu5Acα(2–3)Gal of ∆G are welcome results. A negative ∆G value indicates a spontaneous formation of the protein–ligand complex due to the stability and strength of the noncovalent interactions found in the complex. Experimentally ∆G is directly related to the Ki. Thus, the value of ∆G can be used to approximate the ability of a compound to inhibit the protein. The Ki value produced by recent docking is the lowest of several docking results produced. Together with ∆G, the value of Ki determines the affinity value. The lower these two values are, the higher the affinity of the docked ligand and the more stable the bond that occurs40.
In addition to ∆G and pKi, hydrogen bonding parameters can also be used to evaluate the binding affinity of the complex formed between the receptor and the ligand41. Hydrogen bonding is the most important specific interaction in the process of interaction between ligands and receptors. Therefore, hydrogen bonding contributes to the molecule's affinity for the target protein that forms an electrostatic interaction between the hydrogen donor and acceptor42. In a hydrogen bond interaction analysis, the criteria for hydrogen bonding require a hydrogen donor and acceptor with a bond distance of 3.9 Å43. The number of hydrogen bonds produced by pairing the enzyme NanB sialidase and the Neu5Acα(2–6)Gal ligand supports the notion that the two have an excellent binding affinity. Based on the suitability of several parameters analyzed by docking, the ability of NanB sialidase to hydrolyze sialic acid was tested directly through true-experimental laboratory testing.
Identification of Sialidase in this study was carried out using two primer pairs NanB and NanH, as carried out previously by29. Based on the result of PCR, Pasteurella multocida in this study showed various NanB and NanH sialidase genes combination. Similar results were previously reported by Gharibi et al.20 63.6% of the studied isolates had NanB sialidase, and 81.8% had the gene encoding NanH sialidase. This finding was similarly reported in previous studies by Furian et al.44 and Khamesipour et al.45. Although it is known that sialidase is an enzyme that removes sialic acid conjugated with glycolipids and glycoproteins of eukaryotic cells46, the cause of genetic variation in the presence of NanB and NanH genes from each strain of Pasteurella multocida is not yet established.
In contrast to NanH sialidase, which can be quickly released from Pasteurella multocida, NanB sialidase is more challenging to isolate. This is presumably due to NanB sialidase being a transmembrane protein firmly bound to the outer cell membrane to the periplasm of the bacterium Pasteurella multocida19. Mizan et al.19 concluded that NanB sialidase is one of the proteins with an autotransporter part because it has a channel-forming C-terminal domain that is thought to be involved in protein translocation from the bacterial outer membrane. However, this study has not succeeded in isolating the sialidase released directly on the supernatant of the bacterial growth medium. Therefore, optimization of the method was carried out to determine which method could produce the highest sialidase activity.
The chloroform method was first introduced as a periplasmic protein extraction method by Ames et al.32 who stated that the chloroform method could release 16% of the total cell protein and did not damage the activity of the released protein. The same method was also applied by Sha et al.47 who isolated large amounts of catalase bound to the periplasmic bacteria Brucella abortus.
The F0 showed the highest sialidase-specific activity in anion exchange chromatography. This is presumably due to the influence of the initial isolation method using chloroform. The use of chloroform is thought to disrupt the surface charges of NanB sialidase, resulting in the inability to bind to the Q-sepharose bead. However, this unexpected finding did not meaningfully hinder this study, as evidenced by the presence of sialidase activity shown in the F0 fraction. The molecular weight of NanB sialidase was supported by the prediction results of 3D preparation in the Raptor X program which predicts that NanB sialidase Pasteurella multocida 86–1913 has a molecular weight of 56.44 kDa. Previous study reported different results, which predicted the molecular weight of NanB sialidase Pasteurella multocida of 36 kDa48, 250 kDa49, and 500 kDa50.
NanB sialidase Pasteurella multocida showed 35℃ as optimum temperature. Slightly differing results were reported in previous studies, that the optimum temperature for sialidase from Listeria monocytogenes is between 30 and 35 ℃51. In contrast, the optimum temperatures of NanH and NanJ of Clostridium perfringens type D were found to be 43 °C and 37 °C, respectively18. Based on these reports, it can be assumed that different types of sialidases from different origins also have different optimum temperature.
Previous research regarding the optimum pH of sialidase from Pasteurella multocida had an optimum pH between pH 6.2 and 6.819. However, at pH 5 and 6 in this study, sialidase activity was still found to be remarkably good. This lower pH value has previously been used as a standard by Worrall et al.11 in making sialidase-based vaccines. However, sialidase did not have good activity at pH below 5 and above 8. This was caused by damage to the enzyme structure due to the extreme pH (Fig. 7B).
As expected, sialidase activity decreased when incubated at 37 °C for 72 h (Fig. 7C). The decreased activity may cause by accumulated damage to the protein structure due to the natural action of proteases. However, this factor can be considered in developing sialidase as a drug, namely that the treatment process will be effective because the enzyme activity still lasts for several days, even at body temperature.
In developing new drugs in the health sector, safety is a central factor that must always be considered. The toxicity test in this study aims to determine a safe dose that does not damage red blood cells, which is an indicator for further testing. Based on the toxicity testing results, NanB sialidase has very low toxicity. The highest toxicity value was shown at 0.258 U/mL dose of sialidase, which was indicated by 2.12% lysis of chicken red blood cells (Fig. 8A) and 7.65% lysis of rabbit red blood cells (Fig. 8B). However, this dose should be tested further, considering that the target of sialidase development is epithelial cells that quickly regenerate cells if there is minor damage due to high doses of sialidase. Previous research by Larson et al.52 in assessing the toxicity of sialidase concluded that sialidase did not show any toxic effect on the various cell types tested. A similar conclusion also stated that sialidase was proven to be non-toxic to human respiratory organs as a therapeutic target8.
Rabbit red blood cells are more susceptible to hydrolysis than chicken red blood cells, even with the same α2-6 and α2-3 bonds. This is presumably because there are differences in the red blood cells of the two. Chicken red blood cells possess a nucleus, which makes chicken red blood cells more stable than mammalian cells53. The study by Baier et al. concluded that the type of red blood cells of each species has a difference in thickness and adhesion that affects the strength of all cell properties. In mammalian red blood cells, adhesion increased with elevated temperatures and scaled with reported membrane sialic acid concentrations. In chicken only adhesion decreased with higher temperature. Previous study showed that NanB sialidase developed recombinantly in E. coli cells was able to hydrolyze both types of sialic acid well, with a higher tendency and ability to hydrolyze Neu5Acα(2–6)Gal19.
Conclusion
Based on the results of in silico tests and experimental studies, NanB sialidase of Pasteurella multocida B018 has been shown as an eligible candidate for further development of sialidase-based antivirals. Further studies are necessary to investigate whether red blood cells that have been treated with NanB sialidase of Pasteurella multocida B018 can still agglutinate specific viruses. Further research on the activity of NanB sialidase of Pasteurella multocida B018 on MDCK cells is required to explain the ability of NanB sialidase to inhibit the avian influenza virus.
Acknowledgements
The author appreciates the help of Dr. Sudarisman and Drh. Adin Priadi (PT Medika Satwa Laboratories, Bogor-Indonesia) for their assistance and Microbiology Laboratory facilities. This study was funded by Universitas Indonesia through PUTI Grant with contract number NKB-1291/UN2.RST/HKP.05.00/2020.
Author contributions
All authors (C.M.H.N., R.S.K., S.T., O.S.M.S., S.T.W., I.W.T.W., L.N., A.K.T., A.S.) contributed to the study conception and design. Material preparation, data collection and analysis were performed by C.M.H.N., R.S.K., and S.T. The first draft of the manuscript was written and submitted by C.M.H.N., and O.S.M.S. All authors mentioned above read, edited, and approved the final manuscript.
Funding
This article was funded by Universitas Indonesia, PUTI Grant NKB-1291/UN2.RST/HKP.05.00/2020.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Subbarao K, Joseph T. Scientific barriers to developing vaccines against avian influenza viruses. Nat. Rev. Immunol. 2007;7:267–278. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pusch EA, Suarez DL. The multifaceted zoonotic risk of H9N2 avian influenza. Vet. Sci. 2018;5:82. doi: 10.3390/vetsci5040082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Clercq, E. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov.5, 1015–1025. 10.1038/nrd2175 (2006). [DOI] [PMC free article] [PubMed]
- 5.Razonable, R. R. Antiviral drugs for viruses other than human immunodeficiency virus. Mayo Clin. Proc.86, 1009–1026. 10.4065/mcp.2011.0309 (2011). [DOI] [PMC free article] [PubMed]
- 6.Strasfeld, L. & Chou, S. Antiviral drug resistance: Mechanisms and clinical implications. Infect. Dis. Clin. North Am.24, 413–437. 10.1016/j.idc.2010.01.001 (2010). [DOI] [PMC free article] [PubMed]
- 7.Li, T. C. M., Chan, M. C. W. & Lee, N. Clinical implications of antiviral resistance in influenza. Viruses7, 4929–4944. 10.3390/v7092850 (2015). [DOI] [PMC free article] [PubMed]
- 8.Nicholls JM, Moss RB, Haslam SM. The use of sialidase therapy for respiratory viral infections. Antiviral Res. 2013;98:401–409. doi: 10.1016/j.antiviral.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malakhov MP, et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob. Agents Chemother. 2006;50:1470–1479. doi: 10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belser JA, et al. DAS181, a novel sialidase fusion protein, protects mice from lethal avian influenza H5N1 virus infection. J. Infect. Dis. 2007;196:1493–1499. doi: 10.1086/522609. [DOI] [PubMed] [Google Scholar]
- 11.Worrall, E. E., Sudarisman & Priadi, A. Sialivac: an intranasal homologous inactivated split virus vaccine containing bacterial sialidase for the control of avian influenza in poultry. Vaccine27, 4161–4168. 10.1016/j.vaccine.2009.04.058 (2009). [DOI] [PubMed]
- 12.Triana-Baltzer GB, et al. DAS181, a sialidase fusion protein, protects human airway epithelium against influenza virus infection: An in vitro pharmacodynamic analysis. J. Antimicrob. Chemother. 2010;65:275–284. doi: 10.1093/jac/dkp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenilman JM, et al. Phase 1 clinical trials of DAS181, an inhaled sialidase, in healthy adults. Antiviral Res. 2015;123:114–119. doi: 10.1016/j.antiviral.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez S, et al. Serological response to Pasteurella multocida NanH sialidase in persistently colonized rabbits. Clin. Diagn. Lab. Immunol. 2004;11:825–834. doi: 10.1128/CDLI.11.5.825-834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu G, et al. Three Streptococcus pneumoniae sialidases: Three different products. J. Am. Chem. Soc. 2011;133:1718–1721. doi: 10.1021/ja110733q. [DOI] [PubMed] [Google Scholar]
- 16.Khedri, Z. et al. Synthesis of selective inhibitors against V. cholerae sialidase and human cytosolic sialidase NEU2. Org. Biomol. Chem.10, 6112–6120. 10.1039/c2ob25335f (2012). [DOI] [PMC free article] [PubMed]
- 17.Minami A, et al. Catalytic preference of Salmonella typhimurium LT2 sialidase for N-acetylneuraminic acid residues over N-glycolylneuraminic acid residues. FEBS Open Biol. 2013;3:231–236. doi: 10.1016/j.fob.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, McClane BA. The Sialidases of Clostridium perfringens type D strain CN3718 differ in their properties and sensitivities to inhibitors. Appl. Environ. Microbiol. 2014;80:1701–1709. doi: 10.1128/AEM.03440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizan S, Henk A, Stallings A, Maier M, Lee MD. Cloning and characterization of sialidases with 2–6’ and 2–3’ sialyl lactose specificity from Pasteurella multocida. J. Bacteriol. 2000;182:6874–6883. doi: 10.1128/JB.182.24.6874-6883.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gharibi, D., Hajikolaei, M. R. H., Ghorbanpour, M. & Barzegar, S. K. Virulence gene profiles of Pasteurella multocida strains isolated from cattle and buffalo. Vet. Arh.87, 677–690. 10.24099/vet.arhiv.160727 (2017).
- 21.Do T, Henssge U, Gilbert SC, Clark D, Beighton D. Evidence for recombination between a sialidase (nanH) of Actinomyces naeslundii and Actinomyces oris, previously named ‘Actinomyces naeslundii genospecies 1 and 2’. FEMS Microbiol. Lett. 2008;288:156–162. doi: 10.1111/j.1574-6968.2008.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panna S, et al. Isolation and molecular detection of Pasteurella multocida Type A from naturally infected chickens, and their histopathological evaluation in artifically infected chickens in Bangladesh. J. Adv. Vet. Anim. Res. 2015;2:338–345. doi: 10.5455/JAVAR.2015.B104. [DOI] [Google Scholar]
- 24.Grehn M, Müller F. The oxidase reaction of Pasteurella multocida strains cultured on Mueller-Hinton medium. J. Microbiol. Methods. 1989;9:333–336. doi: 10.1016/0167-7012(89)90075-4. [DOI] [Google Scholar]
- 25.Hunt Gerardo, S., Citron, D. M., Claros, M. C., Fernandez, H. T. & Goldstein, E. J. Pasteurella multocida subsp. multocida and P. multocida subsp. septica differentiation by PCR fingerprinting and alpha-glucosidase activity. J. Clin. Microbiol.39, 2558–2564. 10.1128/JCM.39.7.2558-2564.2001 (2001). [DOI] [PMC free article] [PubMed]
- 26.Kurnia, R. S., Indrawati, A., Mayasari, N. L. P. I. & Priadi, A. Molecular detection of genes encoding resistance to tetracycline and determination of plasmid-mediated resistance to quinolones in avian pathogenic Escherichia coli in Sukabumi, Indonesia. Vet. World11, 1581–1586. 10.14202/vetworld.2018.1581-1586 (2018). [DOI] [PMC free article] [PubMed]
- 27.Townsend KM, Frost AJ, Lee CW, Papadimitriou JM, Dawkins HJ. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J. Clin. Microbiol. 1998;36(4):1096–1100. doi: 10.1128/JCM.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap Loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 2001;39(3):924–929. doi: 10.1128/JCM.39.3.924-929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarangi LN, et al. Virulence genotyping of Pasteurella multocida isolated from multiple hosts from India. Sci. World J. 2014;14:1. doi: 10.1155/2014/814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silaen, O. S. M., Murtini, S., Pamungkas, J. & Nugroho, C. M. H. Isolation and molecular characterization of fowl aviadenovirus associated with inclusion body hepatitis from poultry in Banten and West Java, Indonesia. Vet. World13, 1940–1946. 10.14202/vetworld.2020.1940-1946 (2020). [DOI] [PMC free article] [PubMed]
- 31.Natalia, L. & Priadi, A. The use of filter paper as a transport device for serology of Pasteurella multocida infection: Analysis and comparison of protein composition of filter paper extract and serum. J. Ilmu Ternak Dan Vet.3, 182–187. 10.14334/jitv.v3i3.115 (1998).
- 32.Ames GF, Prody C, Kustu S. Simple, rapid, and quantitative release of periplasmic proteins by chloroform. J. Bacteriol. 1984;160:1181–1183. doi: 10.1128/JB.160.3.1181-1183.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaderbhai N, et al. Glycine-induced extracellular secretion of a recombinant cytochrome expressed in Escherichia coli. Biotechnol. Appl. Biochem. 1997;25:53–61. doi: 10.1111/j.1470-8744.1997.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 34.Lall SD, Eriho BE, Jay JM. Comparison for four methods for extracting periplasmic proteins. J. Microbiol. Methods. 1989;9:195–199. doi: 10.1016/0167-7012(89)90036-5. [DOI] [Google Scholar]
- 35.Neu HC, Heppel LA. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Biol. Chem. 1965;240:3685–3692. doi: 10.1016/s0021-9258(18)97200-5. [DOI] [PubMed] [Google Scholar]
- 36.Tarigan, S., Indryani, R., Darminto & Ignjatovic, J. Purification of neuraminidase from Influenza virus subtype H5N1. J. Ilmu Ternak Dan Vet.14, 75–82. 10.14334/jitv.v14i1.366 (2009).
- 37.Islam F, Roy N. Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res. Notes. 2018;11:445. doi: 10.1186/s13104-018-3558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, et al. Research articleHemolysis in human erythrocytes by Clostridium perfringens epsilon toxin requires activation of P2 receptors. Virulence. 2018;9:1601–1614. doi: 10.1080/21505594.2018.1528842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramnath, M. G., Thirugnanasampandan, R., NagaSundaram, N. & Bhuvaneswari, G. Molecular docking and dynamic simulation studies of terpenoids of I. wightii (Bentham) H. Hara against acetylcholinesterase and histone deacetylase3 receptors. Curr. Comput. Aided. Drug Des.14, 234–245. 10.2174/1573409914666180321111925 (2018). [DOI] [PubMed]
- 40.Du, X. et al. Insights into protein-ligand interactions: mechanisms, models, and methods. Int. J. Mol. Sci.17. 10.3390/ijms17020144 (2016). [DOI] [PMC free article] [PubMed]
- 41.Chen D, et al. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2:e1501240. doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itoh Y, et al. N+-C-H···O Hydrogen bonds in protein-ligand complexes. Sci. Rep. 2019;9:767. doi: 10.1038/s41598-018-36987-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J. Med. Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furian, T. Q. et al. Virulence genes and antimicrobial resistance of Pasteurella multocida isolated from poultry and swine. Braz. J. Microbiol. [publication Brazilian Soc. Microbiol.47, 210–216. 10.1016/j.bjm.2015.11.014 (2016). [DOI] [PMC free article] [PubMed]
- 45.Khamesipour F, Momtaz H, Azhdary Mamoreh M. Occurrence of virulence factors and antimicrobial resistance in Pasteurella multocida strains isolated from slaughter cattle in Iran. Front. Microbiol. 2014;5:536. doi: 10.3389/fmicb.2014.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatfaludi T, Al-Hasani K, Boyce JD, Adler B. Outer membrane proteins of Pasteurella multocida. Vet. Microbiol. 2010;144:1–17. doi: 10.1016/j.vetmic.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 47.Sha, Z. Purification, characterization and cloning of a periplasmic catalase from B abortus and the role it plays in the pathogenesis of Brucella. Retrosp. Theses Diss.11320. 10.31274/rtd-180813-12622 (1994).
- 48.Jablonski PE, Jaworski M, Hovde CJ. A minimal medium for growth of Pasteurella multocida. FEMS Microbiol. Lett. 1996;140:165–169. doi: 10.1111/j.1574-6968.1996.tb08331.x. [DOI] [PubMed] [Google Scholar]
- 49.Drzeniek R, Scharmann W, Balke E. Neuraminidase and N-acetylneuraminate pyruvate-lyase of Pasteurella multocida. J. Gen. Microbiol. 1972;72:357–368. doi: 10.1099/00221287-72-2-357. [DOI] [PubMed] [Google Scholar]
- 50.Straus DC, Jolley WL, Purdy CW. Characterization of neuraminidases produced by various serotypes of Pasteurella multocida. Infect. Immun. 1996;64:1446–1449. doi: 10.1128/iai.64.4.1446-1449.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghazaei C. Optimization and molecular detection of neuraminidase gene in Listeria monocytogenes. Int. J. Infect. 2018;5:e81884. doi: 10.5812/iji.81884. [DOI] [Google Scholar]
- 52.Larson JL, et al. A safety evaluation of DAS181, a sialidase fusion protein, in rodents. Toxicol. Sci. 2011;122:567–578. doi: 10.1093/toxsci/kfr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ajloo D, Moosavi-Movahedi AA, Sadeghi M, Gharibi H. Comparative structural and functional studies of avian and mammalian hemoglobins. Acta Biochim. Pol. 2002;49:459–470. doi: 10.18388/abp.2002_3805. [DOI] [PubMed] [Google Scholar]
- 54.Baier D, Müller T, Mohr T, Windberger U. Red blood cell stiffness and adhesion are species-specific properties strongly affected by temperature and medium changes in single cell force spectroscopy. Molecules. 2021;26:2771. doi: 10.3390/molecules26092771. [DOI] [PMC free article] [PubMed] [Google Scholar]