Abstract
Microbes can convert inexpensive renewable substrates to valuable metabolites by their natural metabolic pathways. To maximize the productivity, the pathways yet require further optimization, which remains challenging for our limited knowledge of complex biology. Genetically encoded biosensors are able to detect metabolite concentrations or environmental changes and transfer these inputs to measurable or actionable outputs, thus providing enabling regulation and monitoring tools for complicated pathway optimization. Here, we review recent advances in biosensor-mediated dynamic regulation and strain screening for the highest microbial production of diverse desirable products.
Keywords: biosensor, pathway optimization, dynamic regulation, strain screening
1. Introduction
Metabolic engineering harnesses microbes to produce value-added chemicals and maximize the productivity to fulfill the requirement of industrial production. However, microbial cellular machinery is naturally designed to seek for optimal cell fitness rather than maximal metabolite yield [1, 2]. While advancements have been made with the emerging of revolutionary strategies and technologies, metabolic engineering of microorganisms remains laborious and difficult due to the complexity of metabolic systems [3].
Biosensors, including transcriptional factor-based, nucleic acid-based and protein level biosensors, throw light on simplifying the engineering process [4–6]. These protein, DNA or RNA molecules can sense various metabolites or environmental signal changes and generate measurable or actionable responses. In this review, we focus on the most commonly used transcriptional factor-based biosensors (TF biosensors) and nucleic acid-based biosensors. Specifically, TF biosensors experience conformational changes once the ligand-binding domains are stimulated by inducers or environmental signals, relieving or triggering the interaction with their corresponding promoters, and thus activating or repressing the expression of downstream genes (Figure 1a, 1b) [4]. Similarly, nucleic acid-based biosensors such as riboswitches, ribozymes and aptamers also change their structures as responses to certain ligands, further regulating their interacting mRNAs or downstream genes at transcriptional or translational level (Figure 1c, 1d and 1e) [6]. Although biosensors usually cannot be directly used for their limited sensitivity and dynamic range, biosensor engineering can optimize the properties and fulfill the requirements for application [7, 8]. These naturally developed regulation mechanisms can subsequently be utilized to manipulate gene expression by manually controlling the signal inputs [9, 10], or endow microbes the intelligence to implement self-control by installing biosensors recognizing the signals generated by their own cell metabolisms [11, 12]. Such dynamic regulation strategy has been proven to be effective in balancing carbon fluxes to address the conflicts and stresses brought by the intention shifting from growth to production [13, 14]. Meanwhile, when controlling the expression of easily detectable reporter genes, biosensors can easily be repurposed as monitors to rapidly sense and estimate chemical production for high-throughput library screening. In addition, genetic circuits using biosensors to connect productivity with growth fitness allow efficient evolution to yield overproduction strains [15].
Figure 1.
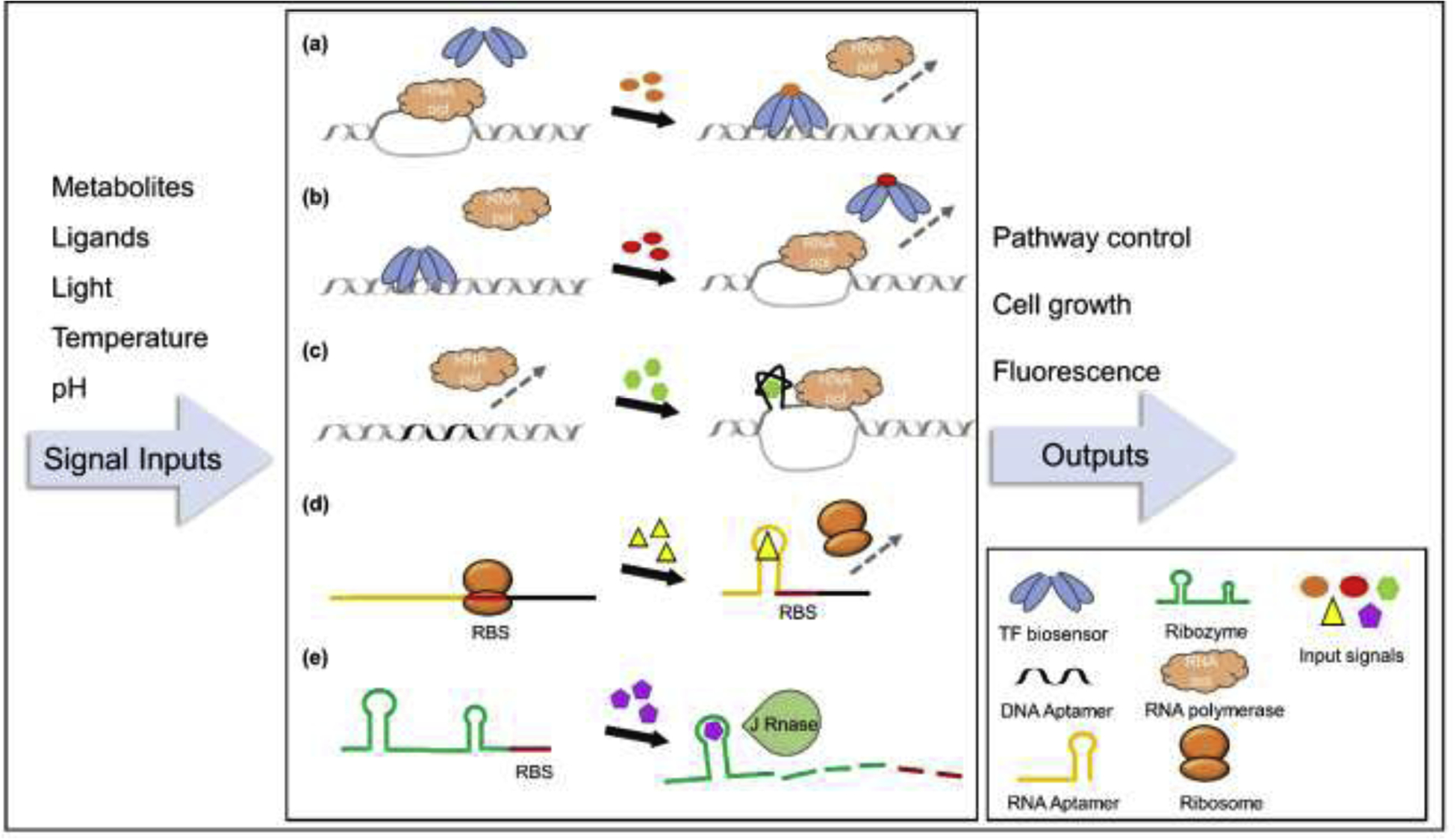
Application of TF and nucleic acid biosensors in pathway optimization and the regulation mechanisms. (a) TF biosensor-enabled transcription switch-on. The signal input activates the TF biosensor to interact with its corresponding promoter sequence, blocking the accession of RNA polymerase. (b) TF biosensor-enabled transcription switch-off. The signal causes some conformational changes of the TF biosensor, releasing it from the promoter and thus allowing the transcription by RNA polymerase. (c) DNA aptamer-promoted transcription switch-on. Signal input triggers the conformational changes of the DNA aptamer, promoting the unwinding of promoter sequence for RNA polymerase binding. (d) RNA aptamer-enabled translation switch-off. The conformational changes of the RNA aptamer inhibit the targeting of ribosome to RBS site. (e) Ribozyme-mediated translation switch-off. The ribozyme experiences self-cleavage with the signal input, leading to the digestion of the downstair mRNA. Input signals include diverse metabolites and environmental changes, enabling the generation of versatile regulation and screening strategies for metabolic engineering.
During the past several years, advancements in discovering or engineering biosensors for novel signal recognition, higher sensitivity, or expanded dynamic range further contributed to the development of metabolic engineering [16, 17]. Here, we review recent studies regarding the application of TF and nucleic acid biosensors in dynamic regulation and overproducer screening and discuss the future perspectives on the biosensor-boosted metabolic engineering.
2. Biosensor enabled dynamic regulation
2.1. Metabolite-responsive biosensor
Biosensors response to wide variety of metabolites existing among various types of living microbes to balance cellular metabolism [14]. Over the past few decades, increasing number of metabolites have been identified as signal molecules for application of biosensor in dynamic circuits as multiple technologies being explored for biosensor mining and optimizing [7, 8]. Metabolites-responsive biosensors transfer input molecular signals to the expression levels of downstream operons. During this process, production is enhanced as the result that carbon flux is rewired to balance cell fitness and production dynamically, also to prevent the accumulation of toxic intermediates [14, 18]. GlcN6P responsive sensor glmS ribozyme switch was developed as a tool to dynamically regulate the production of N-acetylglucosamine (GlcNAc) (Figure 1e). With the control of glmS ribozyme, pfkA and glmM or pgi were inhibited according to GlcN6P accumulation to balance the GlcN6P level and enhanced GlcNAc titer up to 18.45 g/L [11]. HucR is a natural biosensor responsive to uric acid. A library of HucR variants was constructed by saturation mutagenesis, and effector specificity changed variants that response to ferulic acid and vanillin were identified. By using the modified HucR system, enhanced vanillin production was achieved through balancing the growth phase and production phase.
In recent years, bifunctional dynamic regulation circuits have been designed by applying antisense RNA and CRISPRi. With the strategy of bifunctional dynamic regulation, muconic acid (MA)-responsive biosensor CatR was used to activate genes within the MA synthesis pathway and simultaneously guide a RNAi system to inhibit the central metabolism (Figure 2a). The MA titer reached 1.8 g/L under such dynamic regulation [19]. Another bifunctional circuit was designed using TF biosensor GamR (Figure 2b) to control the expression of GlcN6P N-acetyltransferase transferring GlcN6P to GlcNAc while GlcN6P accumulating.
Figure 2.
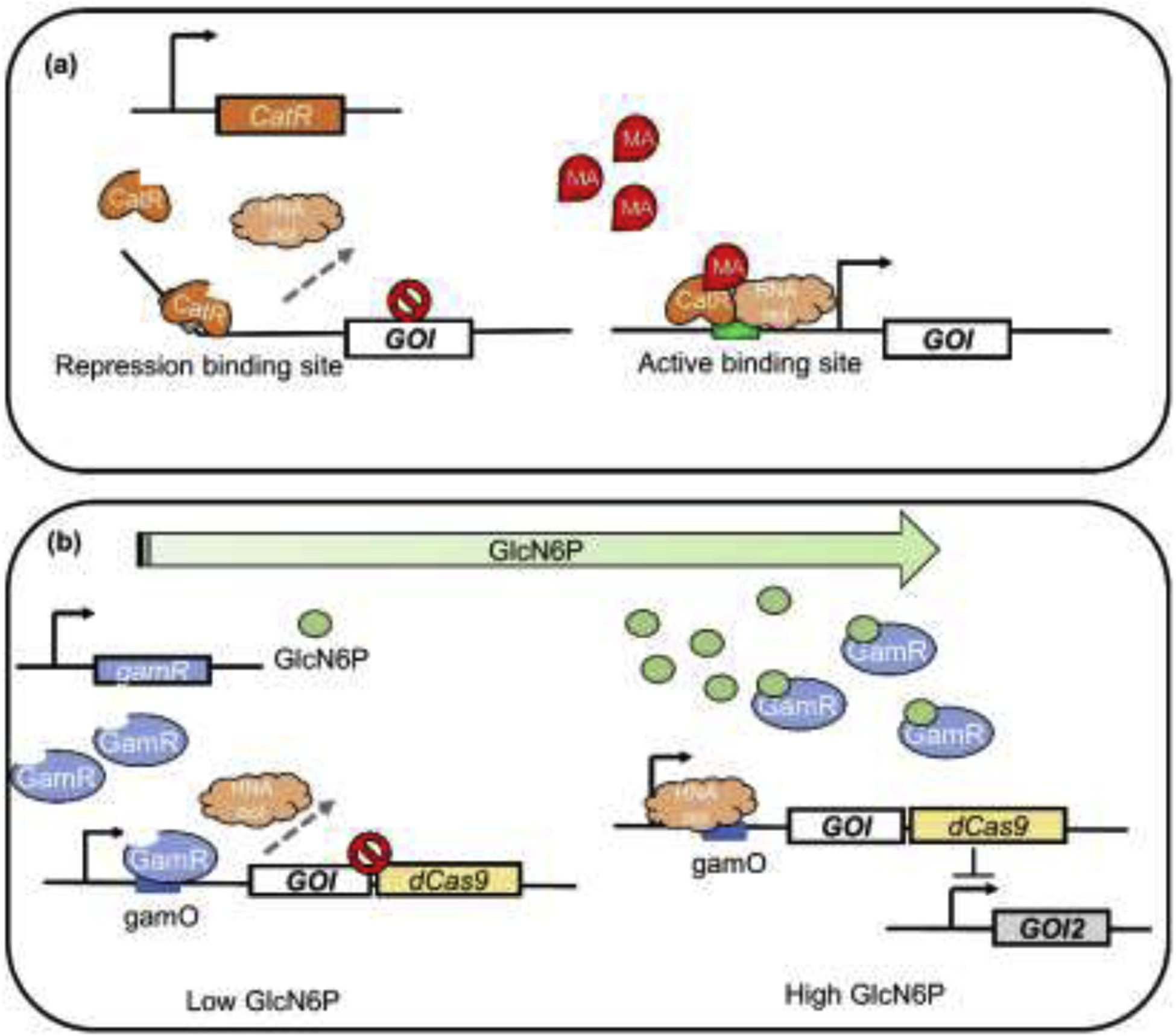
Application of metabolite-responsive biosensors for dynamic pathway regulation. (a) The mechanism of MA-responsive biosensor CatR. The CatR tetramer binds to a specific repression site, triggering the bending of DNA chain and blocking its interaction with RNA polymerase when the signal molecule MA is absent. Once MA combines with CatR and causes its conformational changes, CatR binds to another active site to release the bending of DNA and retrieve transcription. MA, muconic acid. (b) GlcN6P-responsive biosensor enabled bifunctional dynamic regulation. When the intracellular concentration of GlcN6P is low, TF biosensor GamR binds to gamO sites to block the binding of RNA polymerase, inhibiting the transcription of GOI (gene of interest) and CRISPR/dCas9. When GlcN6P concentration reaches high level, GamR combines with GlcN6P, and the allosteric effect of GlcN6P decreases the affinity of GamR to DNA and activates the expression of GOI and CRISPR/dCas9. The dCas9 coupled with sgRNA represses GOI2, thus realizing simultaneous activation and repression.
GamR also regulated a CRISPRi system inhibiting genes related to cell growth and byproduct generation. Such dual-control circuit constructed in Bacillus subtilis improved GlcNAc production to 131.6 g/L [20]. Central metabolism can also be the target of dynamic regulation. A pyruvate-responsive biosensor PdhR was developed to regulate glucaric acid synthesis, ino1 gene expression was activated to enhance yield, and antisense RNAs targeting pgi and zwf were employed to inhibit glycolysis and pentose pathway to construct bifunctional circuit based on PdhR [21].
As more biosensors responsive to various molecules being identified, dynamic regulation circuit design tends to be more complex and multi-layered. Naringenin responsive sensor FdeR is an activator targeting on FdeO site when binding with naringenin. Leucine-auxotrophic strain with LEU2 under the control of PfdeO was constructed to achieve a naringenin addicted growth manner. Additionally, fatty acid responsive promoter controlled CRISPRi negatively regulates the competing lipogenic pathway in this study [22]. p-Coumaric acid can inactivate regulator PadR. By coupling FdeR and PadR system, a multiple-layered circuit was constructed to enhance naringenin production [23]. In another layered dynamic regulation circuit, both quorum sensing (QS) system and metabolite-responsive biosensor were applied to increase the production of glucaric acid. With the QS system dynamically downregulates glycolysis pathway and the TF biosensor IpsA responsive to myo-inositol (MI) inducing the pathway gene Miox as MI accumulating, the yield of glucaric acid reached almost 2 g/L [24].
2.2. Environmental signal-responsive biosensor
2.2.1. Quorum sensing
Quorum sensing (QS) functions to coordinate gene expression according to population density through TF biosensors responsive to extracellular signaling molecules in cell communication [25]. A typical QS circuit is composed of a synthase responsible for the synthesis of signal molecules as cells grow, and a signal-responsive transcription regulator controlling specific genes [26]. As a pathway-independent dynamic control, QS systems have been applied for the induction of gene expression in metabolic engineering. The reported QS systems mostly utilized in metabolic engineering include EsaI/EsaR system from Pantoea stewartia [27] and LuxI/LuxR system from Vibrio fscheri [28]. In the EsaI/EsaR system, PesaS promoter can be activated by the binding of transcriptional regulator EsaR. The accumulation of 3-oxohexanoylhomoserine lactone (AHL), which is synthesized by EsaI, can disrupt the EsaR binding and deactivate the transcription of PesaS (Figure 3a). As an example, EsaI/EsaR system was applied in Escherichia coli to redirect glycolytic flux from competing pathway into heterologous pathways by switching off Pfk-1 expression at desired times and cell densities. The final titer of myo-inositol increased 5.5-fold and titer of glucaric acid increased from unmeasurable to over 0.8 g/L [12]. Similar strategy has also been demonstrated for further increasing glucaric acid titer, resulting in a 5-fold increase in glucaric acid titer [24]. To release the toxicity of the heterologous pathway, EsaI/EsaR system was also applied to activate the 4-Hydroxyphenylacetic acid (4HPAA) synthesis pathway with the accumulation of AHL in E. coli, and the final titer exhibited a 46.4% improvement compared to the statically controlled pathway [29]. EsaI/EsaR system was optimized continually and applied to the production of 5-aminolevulinic acid (ALA) and poly-β-hydroxybutyrate (PHB). The optimized QS switch can achieve inhibition and activation of specific genes simultaneously, resulting in a 6- and 12-fold increase in the titer of PHB and ALA [30]. As another well-studied system, LuxR can bind with AHL, which is synthesized by LuxI, to activate the transcription of PluxI promoter (Figure 3b). Examples of utilizing the LuxI/LuxR circuit for dynamic regulation include autonomous control of metabolic state to increase the titer of bisabolene (1.1 g/L) [31] and redirection of carbon flux from central metabolic pathway into isopropanol production [32]. For the synthesis of bisabolene, related genes were controlled by PLuxI to activate their expression at specific time with specific concentration of AHL. In another case, LuxI/LuxR circuit was combined with a positive feedback loop to rewire the carbon flux from TCA cycle into isopropanol synthesis pathway. More challenging pathway may require layered dynamic regulation to up- and down-regulate related genes simultaneously. EsaI/EsaR and LuxI/LuxR systems were also combined to up-regulate the genes for naringenin synthesis and down-regulate assumption of malonyl-CoA for competing pathway. The final titer of naringenin was 463 ± 1 μM naringenin, exhibiting a 140% increasement compared to the original strain [33]. In addition to the aforementioned systems, there are many other QS circuits used in other model organisms. A bifunctional Phr60-Rap60-Spo0A QS system was applied in B. subtilis to fine-tune the synthesis of menaquinone-7 dynamically, which caused a 40-fold improvement in the final titer [34]. Similarly, a bifunctional comQXPA-PsrfA QS system was developed for dynamic control of gene expression in Corynebacterium glutamicum [35].
Figure 3.
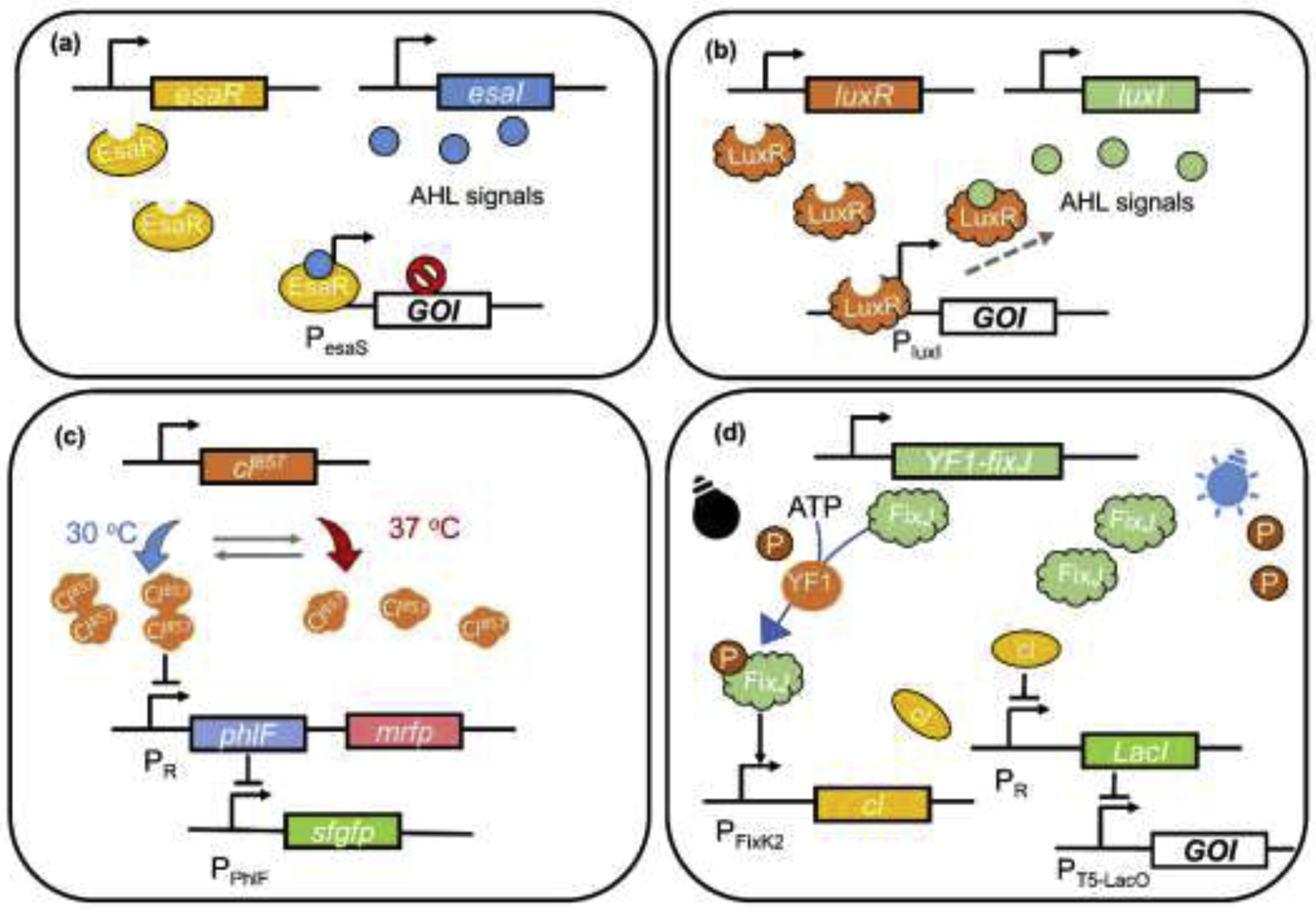
Application of biosensors responsive to environmental changes for dynamic regulation. (a) The diagram of EsaI/EsaR system. The transcription regulator EsaR activates the downstream expression from PesaS. The binding of AHL generated by EsaI releases EsaR to deactivate the transcription from PesaS. (b) The diagram of LuxI/LuxR system. The transcription regulator LuxR activates the transcription from Pluxl when combining with the AHL produced by LuxI. (c) Mechanism of the T-Switch system. Dimers of Cl857 are formed under low temperature (30 °C), which inhibit the transcription of PhlF from PR and maintain the transcription of sfgfp from PPhlF at normal level. When the temperature rises (37 °C), Cl857 dimers decompose to monomers and thus relieve the repression on PhlF and mrfp. Transcription of sfgfp is then blocked since the inhibition of PhlF on PPhlF. (d) Mechanism of the OptoLAC system. Under dark, FixJ phosphorylated by YF1 (a photosensory histidine kinase) activates the expression of Cl, which inhibits the expression of LacI and allowing the expression of GOI form the LacO containing promoter. Bule light induces the dephosphorylation of FixJ, stopping the CI expression and removing the inhibition on LacI. Expressed LacI thus can repress the expression of GOI.
2.2.2. Other environmental changes
Some biosensors are sensitive to environmental stimulus including certain chemical or bio-molecules, pH, lights and temperature, and thus can serve as inducible components to design dynamic regulation circuits. For example, Deng et al. [36] reported using human thrombin-responsive DNA/RNA aptamers to design bifunctional dynamic regulation circuits at not only transcriptional but also translational level (Figure 1c, 1d). They further used the circuits for the biosynthesis of 2′-fucosyllactose (2′-FL) in B. subtilis, increasing the titer from 24.7 to 674 mg/L. Bañares et al. [37] adopted an engineered transmembrane transcriptional factor CadCΔ responsive to pH in constructing a dynamic regulation circuit controlling the accumulation of D-xylonic acid to overcome the culture acidization problem in utilizing xylose oxidative pathway, resulting in 170% higher glycerol production.
Temperature sensitive- and optical biosensors grant researchers the opportunity to use temperature and lights to manually induce dynamic regulation, which are more inexpensive and reversible compared to chemical inducers. The thermal sensitive repressor CI and its modified version CI857 have been extensively applied in temperature controlled dynamic regulation circuits. They inhibit transcription at the corresponding PR-PL promoter by forming dimer complexes at low temperature, and the inhibition can be eliminated by raising temperature. Fang et al. [9] designed a thermal switch system with the CI repressor to overexpress the L-threonine synthesis pathway or switch off the L-alanine synthesis pathway according to temperature. The highest yield of L-threonine reached 124.03% molar yield. Wang et al.[38] developed a thermal-controlled bifunctional dynamic regulation system, namely T-switch, by combining CI857 and a TetR-family repressor PhlF. This system demonstrated the reversibility of temperature-control by swapping between 30 °C and 37 °C (Figure 3c) and was successfully applied to regulate the fraction of 3-hydroxybutyrate and 4-hydroxybutyrate in synthesizing di-block polyhydroxyalkanoates (PHA). Compared to temperature, light-controlled biosensors can generate more tunable regulation circuits. An optogenetic circuit with transcriptional factor EL222 controlling GAL system was firstly applied to improve the production of valuable chemicals in Saccharomyces cerevisiae [10]. This system was further optimized in recent studies to obtain amplified induction fold (OptoAMP [39]) and rapid response time (OptoINVRT7 [40]). Both systems were applied for the biosynthesis of lactic acid and isobutanol, with the latter improved the production of lactic acid and isobutanol by more than 50% and 15%. Optogenetic switch was also developed for metabolic engineering in E. coli. Tandar et al. [41] adopted chromatic acclimation sensor/regulator (CcaSR), which inhibits gene expression when exposed to green light and relives the inhibition with red light, to rewire the carbon fluxes between the Embden-Meyerhof-Parnas and oxidative pentose phosphate pathways with different ratios. Wu et al.[42] combined EL222 with dCpf1-mediated CRISPRi system to construct a light controlled CRISPRi platform. With the opto-CRISPRi dynamically repressing central metabolism and competing pathway, muconic acid production was increased by 130%. Very recently, Lalwani et al. [43] designed a series of OptoLAC systems using light as the replacement of IPTG to induce the lac operon in E. coli (Figure 3d). Through this, light generated stronger and more tunable induction than IPTG, and applying the OptoLAC systems resulted in mevalonate and isobutanol productions respectively 24% and 27% higher than IPTG induction.
3. Biosensor-aided strain screening
3.1. High-throughput screening
In addition to rationally rewiring carbon fluxes, biosensor involved high-throughput screening is a promising approach to dig out the potential productivity of the chassis host [44]. Such strategy shows the superiority of time-saving, high-efficiency, and easy-monitoring compared to the traditional screening methods by transferring productivity to easily readable signals [45]. The well-studied TF biosensors efficiently assisted to screen strains or enzymes for the high production of diverse valuable compounds. Yang et al. [46] repurposed type III polyketide synthase as a malonyl-CoA biosensor, which converts malonyl-CoA to a direct colorimetric indicator, flaviolin. The repurposed biosensor enabled the screening of an 1,858 synthetic regulatory RNA library to identify knockdown sites that would improve the malonyl-CoA level. The authors then rapidly obtained a strain with the highest production of 6-methylsalicylic acid (440.3 mg/L). Qiu et al. [47] adopted an erythritol-responsive TF biosensor EryD to control the expression of eGFP (Enhanced green fluorescent protein). Such design converts erythritol production to fluorescence level and efficiently screened a mutated strain library of 1,152 mutants, receiving an erythritol overproducer reaching over 148 g/L.
Biosensor coupled fluorescence activated cell sorting (FACS) system can sort high and low-productive variants automatically and accurately. Yeom et al. [48] reported a caprolactam-detecting genetic enzyme screening system (CL-GESS) with an optimized lactam-detecting biosensor NitR-L117F and FACS (Figure 4a). By applying this rapid screening toolbox to screen an enormous marine metagenome, they successfully found and characterized a cyclase, CF3HBD, that had the capacity to converted 6-ACA (6-aminocaproic acid) to ε-caprolactam. Wang et al. [49] built a screening system based on a BenM biosensor system coupled with FACS system in S. cerevisiae to obtain high cis,cis-muconic acid (CCM) production variants. As a result, a variant Mut131 possessing 49.7% higher CCM titer was screened which could be used for related pathway optimization. By similar strategy, Bentley et al. [50] screened a muconate-productive variant by harnessing another muconate biosensor CatM with FACS system in Pseudomonas putida. Aptamer-riboswitch-based biosensors were also used to rapidly screen strains with increased titer of target products. Liu et al. [51] constructed a high-throughput screening platform combining a riboswitch-based tryptophan biosensor and fluorescence-activated droplet sorting (FADS). After screening the ARTP (atmospheric and room-temperature plasma)-based whole-genome random mutation library, 192 colonies with higher fluorescence readout were chosen as overproducer candidates. The best strain K3mu1 produced 2.19 g/L tryptophan, which is 155.1% higher than the parent strain.
Figure 4.
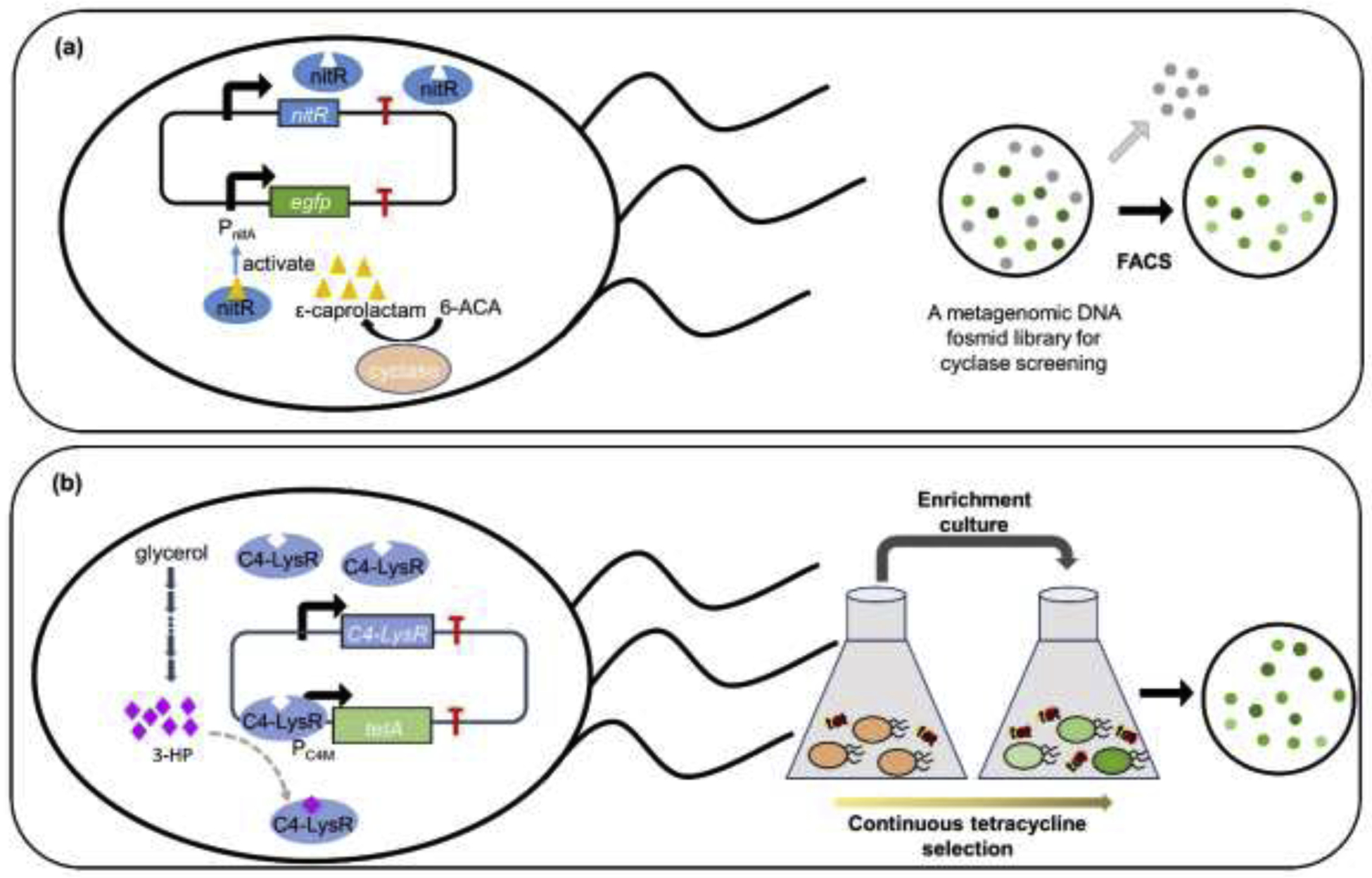
Highly efficient strain screening enabled by biosensors. (a) Schematic process of nitR enabled high-throughput enzyme screening method. ε-Caprolactam binding with nitR activates the egfp expression. Only cyclases efficiently producing ε-caprolactam from 6-ACA can generate intense fluorescence to be screened out by the FACS system. 6-ACA, 6-aminocaproic acid; FACS, fluorescence activated sorting system. (b) Schematic process of 3-HP responsive biosensor driven adaptive evolution. 3-HP induces the expression of tetracycline resistance gene, tetA. With continuous tetracycline selection, only strains with high 3-HP yield can survive. Tet, tetracycline; 3-HP, 3-hydroxypropionic acid.
3.2. Adaptive evolution
Adaptive evolution driven by biosensors is another significant screening application for productive phenotypes, in which genes affecting cell growth are controlled by biosensors to promote high producers to become dominant under selection pressure [52]. An example is the population quality control (PopQC) system using biosensors to control the tetracycline resistance gene tetA. In tetracycline-added culture, the PopQS system allows the continuous enrichment of high performers to overcome nongenetic cell-to-cell variation. The application of PopQS system employing FadR for fatty acid biosynthesis reached the titer of 21.5 g/L in fed-batch fermentation [53]. Similar continuous cell selection systems were designed with 4-hydroxybenzoate (4HB) sensor PobR [54], and with tryptophan sensor Tnac [55, 56]. Guo et al. [57] attempted to replace exogenous antibiotic with a toxin/antitoxin system hip A/hip B in cell selection, and demonstrated that it can be used in combination with a phenylalanine biosensor TyrR or a tryptophan biosensor TrpR for production improvements.
In addition to screening target overproducers, adaptive evolution also provides a unique opportunity to uncover the mechanism underlying a phenotype and provide guidance for strain engineering [58, 59]. Seok et al. adopted a synthetic biosensor C4-LysR responsive to 3-hydroxypropion (3-HP) to design similar adaptive evolution (Figure 4b). Upon further investigation, they found that mutations in global transcriptional factors rewired central metabolic flux towards 3-HP, and combining the mutations reached almost theoretical maximum 3-HP titer [60].
4. Conclusion and perspectives
In this paper, we summarize recent studies on applying biosensors in versatile dynamic regulation and strain or enzyme screening for pathway optimization. Various metabolite-responsive biosensors, in cooperation with other booming technologies, generated not only genetic circuits with diverse regulation functions, but also efficient overproducer screening methods. Meanwhile, environmental-responsive biosensors present the possibility to introduce autonomous regulation according to cell density, or the use of signals like light or temperature rather than chemical inducers that usually affect cell fitness. Overall, biosensors markedly assisted the pathway optimization process, leading to impressive production enhancements or novel biosynthesis of desired chemicals.
However, challenges remain mainly for the limited number of biosensors. The variety of recognizable signal inputs including environmental changes and more importantly cellular metabolites, are crucial for the development of biosensor-powered pathway optimization. Advances in computational methods applicable for biosensor mining and engineering can promisingly overcome these challenges. Tools such as MD (Molecular dynamics) simulations and molecular docking not only can generate guidance for experimental engineering of biosensor, but also allow in silico design of ligand-binding domains of biosensors to alter their specificity to target chemicals [61, 62]. Genome mining has been serving as an effective approach to discover various novel biosensors [62]. Further improvements might be made by coupling bioinformatic tools with recently released machine learning based structure prediction program AlphaFold [63]. In the future, we expect that a large number of biosensors with distinct specificities can be obtained more efficiently with the assistance of these advanced computational methods.
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM128620. We also acknowledge the support from the College of Engineering, The University of Georgia, Athens.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Lalwani MA, Zhao EM, and Avalos JL (2018). Current and future modalities of dynamic control in metabolic engineering. Curr Opin Biotechnol 52, 56–65. [DOI] [PubMed] [Google Scholar]
- 2.Liu D, Evans T, and Zhang F (2015). Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng 31, 35–43. [DOI] [PubMed] [Google Scholar]
- 3.Ko YS, Kim JW, Lee JA, Han T, Kim GB, Park JE, and Lee SY (2020). Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem Soc Rev 49, 4615–4636. [DOI] [PubMed] [Google Scholar]
- 4.Mitchler MM, Garcia JM, Montero NE, and Williams GJ (2021). Transcription factor-based biosensors: a molecular-guided approach for natural product engineering. Curr Opin Biotechnol 69, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasgow AA, Huang Y-M, Mandell DJ, Thompson M, Ritterson R, Loshbaugh AL, Pellegrino J, Krivacic C, Pache RA, Barlow KA, et al. (2019). Computational design of a modular protein sense-response system. Science 366, 1024–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michener JK, Thodey K, Liang JC, and Smolke CD (2012). Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways. Metab Eng 14, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch M, Pandi A, Borkowski O, Batista AC, and Faulon JL (2019). Custom-made transcriptional biosensors for metabolic engineering. Curr Opin Biotechnol 59, 78–84. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Liu Y, and Wang M (2017). Design, optimization and application of small molecule biosensor in metabolic engineering. Frontiers in Microbiology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y, Wang J, Ma W, Yang J, Zhang H, Zhao L, Chen S, Zhang S, Hu X, Li Y, et al. (2020). Rebalancing microbial carbon distribution for L-threonine maximization using a thermal switch system. Metab Eng 61, 33–46. [DOI] [PubMed] [Google Scholar]
- 10.Zhao EM, Zhang Y, Mehl J, Park H, Lalwani MA, Toettcher JE, and Avalos JL (2018). Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 555, 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu T, Liu Y, Li J, Koffas M, Du G, Alper HS, and Liu L (2018). Engineering a glucosamine-6-phosphate responsive glms ribozyme switch enables dynamic control of metabolic flux in Bacillus subtilis for overproduction of n-acetylglucosamine. ACS Synth Biol 7, 2423–2435. [DOI] [PubMed] [Google Scholar]
- 12.Gupta A, Reizman IM, Reisch CR, and Prather KL (2017). Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockman IM, and Prather KL (2015). Dynamic metabolic engineering: New strategies for developing responsive cell factories. Biotechnol J 10, 1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen X, Wang J, Li C, Yuan Q, and Yan Y (2019). Dynamic gene expression engineering as a tool in pathway engineering. Curr Opin Biotechnol 59, 122–129. [DOI] [PubMed] [Google Scholar]
- 15.Rogers JK, Taylor ND, and Church GM (2016). Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol 42, 84–91. [DOI] [PubMed] [Google Scholar]
- 16.Li C, Zhang R, Wang J, Wilson LM, and Yan Y (2020). Protein engineering for improving and diversifying natural product biosynthesis. Trends Biotechnol 38, 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang T, Li C, and Yan Y (2021). Optimization of a p-coumaric acid biosensor system for versatile dynamic performance. ACS Synth Biol 10, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P (2018). Production of chemicals using dynamic control of metabolic fluxes. Curr Opin Biotechnol 53, 12–19. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Lin Y, Wang J, Wu Y, Zhang R, Cheng M, Shen X, Wang J, Chen Z, Li C, et al. (2018). Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis. Nat Commun 9, 3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Chen T, Liu Y, Tian R, Lv X, Li J, Du G, Chen J, Ledesma-Amaro R, and Liu L (2019). Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis. Nucleic Acids Research 48, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Li X, Liu Y, Zhu Y, Li J, Du G, Chen J, Ledesma-Amaro R, and Liu L (2020). Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat Chem Biol 16, 1261–1268. [DOI] [PubMed] [Google Scholar]; ** A pyruvate responsive sensor PdhR was employed to construct a dynamic regulation circuit in Bacillus subtilits. As the central metabolite pyruvate accumulating, either glycolysis or pentose phosphate pathway were inhibited by antisense RNA, while the production of glucaric acid was enhanced based on the pyruvate feedback loop.
- 22.Lv Y, Gu Y, Xu J, Zhou J, and Xu P (2020). Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab Eng 61, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This reseach adopted a nariginin-responsive sensor FedR to control the leucine synthesis pathway and thus generating a flavnoid-addicted growth manner. Coupled with the lipogenetic pathway negative autoregulation layer, such design resulted in 74.8% higher nariginin production and obiously improved strain stability.
- 23.Zhou S, Yuan S-F, Nair PH, Alper HS, Deng Y, and Zhou J (2021). Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metabolic Engineering 67, 41–52. [DOI] [PubMed] [Google Scholar]
- 24.Doong SJ, Gupta A, and Prather KLJ (2018). Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proceedings of the National Academy of Sciences 115, 2964–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams TC, Averesch NJH, Winter G, Plan MR, Vickers CE, Nielsen LK, and Kromer JO (2015). Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng 29, 124–134. [DOI] [PubMed] [Google Scholar]
- 26.Tian J, Yang G, Gu Y, Sun X, Lu Y, and Jiang W (2020). Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in Streptomyces. Nucleic Acids Res 48, 8188–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minogue TD, Trebra M.W. v., Bernhard F, and Bodman S.B.v. (2002). The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Molecular Microbiology 44, 1625–1635. [DOI] [PubMed] [Google Scholar]
- 28.Whiteley M, Diggle SP, and Greenberg EP (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen YP, Fong LS, Yan ZB, and Liu JZ (2019). Combining directed evolution of pathway enzymes and dynamic pathway regulation using a quorum-sensing circuit to improve the production of 4-hydroxyphenylacetic acid in Escherichia coli. Biotechnol Biofuels 12, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu F, Jiang W, Mu Y, Huang H, Su T, Luo Y, Liang Q, and Qi Q (2020). Quorum sensing-based dual-function switch and its application in solving two key metabolic engineering problems. ACS Synthetic Biology 9, 209–217. [DOI] [PubMed] [Google Scholar]
- 31.Kim EM, Woo HM, Tian T, Yilmaz S, Javidpour P, Keasling JD, and Lee TS (2017). Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab Eng 44, 325–336. [DOI] [PubMed] [Google Scholar]
- 32.Soma Y, and Hanai T.J.M.e. (2015). Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng 30, 7–15. [DOI] [PubMed] [Google Scholar]
- 33.Dinh CV, and Prather KLJ (2019). Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proc Natl Acad Sci U S A 116, 25562–25568. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The authors demonstrated combining 2 quorum sensing systems to independently, simulateneously and dynamically activate and repress two set of genes for more effective flux rewiring.
- 34.Cui S, Lv X, Wu Y, Li J, Du G, Ledesma-Amaro R, and Liu L (2019). Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis. ACS Synth Biol 8, 1826–1837. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Shi F, Tan S, Yu X, Lai W, and Li Y (2021). Engineering a bifunctional comqxpa-psrfa quorum-sensing circuit for dynamic control of gene expression in Corynebacterium glutamicum. ACS Synth Biol 10, 1761–1774. [DOI] [PubMed] [Google Scholar]
- 36.Deng J, Chen C, Gu Y, Lv X, Liu Y, Li J, Ledesma-Amaro R, Du G, and Liu L (2019). Creating an in vivo bifunctional gene expression circuit through an aptamer-based regulatory mechanism for dynamic metabolic engineering in Bacillus subtilis. Metab Eng 55, 179–190. [DOI] [PubMed] [Google Scholar]
- 37.Banares AB, Valdehuesa KNG, Ramos KRM, Nisola GM, Lee WK, and Chung WJ (2020). A pH-responsive genetic sensor for the dynamic regulation of D-xylonic acid accumulation in Escherichia coli. Appl Microbiol Biotechnol 104, 2097–2108. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Han JN, Zhang X, Ma YY, Lin Y, Wang H, Li DJ, Zheng TR, Wu FQ, Ye JW, et al. (2021). Reversible thermal regulation for bifunctional dynamic control of gene expression in Escherichia coli. Nat Commun 12, 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao EM, Lalwani MA, Chen JM, Orillac P, Toettcher JE, and Avalos JL (2021). Optogenetic amplification circuits for light-induced metabolic control. ACS Synth Biol 10, 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao EM, Lalwani MA, Lovelett RJ, Garcia-Echauri SA, Hoffman SM, Gonzalez CL, Toettcher JE, Kevrekidis IG, and Avalos JL (2020). Design and characterization of rapid optogenetic circuits for dynamic control in yeast metabolic engineering. ACS Synth Biol 9, 3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tandar ST, Senoo S, Toya Y, and Shimizu H (2019). Optogenetic switch for controlling the central metabolic flux of Escherichia coli. Metab Eng 55, 68–75. [DOI] [PubMed] [Google Scholar]
- 42.Wu P, Chen Y, Liu M, Xiao G, and Yuan J (2021). Engineering an optogenetic CRISPRi platform for improved chemical production. ACS Synth Biol 10, 125–131. [DOI] [PubMed] [Google Scholar]
- 43.Lalwani MA, Ip SS, Carrasco-Lopez C, Day C, Zhao EM, Kawabe H, and Avalos JL (2021). Optogenetic control of the lac operon for bacterial chemical and protein production. Nat Chem Biol 17, 71–79. [DOI] [PubMed] [Google Scholar]; ** This research used blue light as the replacement of IPTG to control the most commonly used inducible Lac operon. The developed OptoLAC circuits demonstrated better tunability, dynamic range and spatial control. Application in mevalonate and isobutanol biosynthesis improved the production by 24% and 27% respectively compared to IPTG indcution.
- 44.Zeng W, Guo L, Xu S, Chen J, and Zhou J (2020). High-throughput screening technology in industrial biotechnology. Trends Biotechnol 38, 888–906. [DOI] [PubMed] [Google Scholar]
- 45.Lim HG, Jang S, Jang S, Seo SW, and Jung GY (2018). Design and optimization of genetically encoded biosensors for high-throughput screening of chemicals. Curr Opin Biotechnol 54, 18–25. [DOI] [PubMed] [Google Scholar]
- 46.Yang D, Kim WJ, Yoo SM, Choi JH, Ha SH, Lee MH, and Lee SY (2018). Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc Natl Acad Sci U S A 115, 9835–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu X, Xu P, Zhao X, Du G, Zhang J, and Li J (2020). Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab Eng 60, 66–76. [DOI] [PubMed] [Google Scholar]; * This study demonstrated a typical workflow for enzyme screening using high-throughput screening in a large library for pathway optimization.
- 48.Yeom S-J, Kim M, Kwon KK, Fu Y, Rha E, Park S-H, Lee H, Kim H, Lee D-H, Kim D-M, et al. (2018). A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat Commun 9, 5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G, Ozmerih S, Guerreiro R, Meireles AC, Carolas A, Milne N, Jensen MK, Ferreira BS, and Borodina I (2020). Improvement of cis,cis-muconic acid production in Saccharomyces cerevisiae through biosensor-aided genome engineering. ACS Synth Biol 9, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bentley GJ, Narayanan N, Jha RK, Salvachua D, Elmore JR, Peabody GL, Black BA, Ramirez K, De Capite A, Michener WE, et al. (2020). Engineering glucose metabolism for enhanced muconic acid production in Pseudomonas putida KT2440. Metab Eng 59, 64–75. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Yuan H, Ding D, Dong H, Wang Q, and Zhang D (2021). Establishment of a biosensor-based high-throughput screening platform for tryptophan overproduction. ACS Synth Biol 10, 1373–1383. [DOI] [PubMed] [Google Scholar]
- 52.Liang C, Zhang X, Wu J, Mu S, Wu Z, Jin JM, and Tang SY (2020). Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab Eng 57, 239–246. [DOI] [PubMed] [Google Scholar]
- 53.Xiao Y, Bowen CH, Liu D, and Zhang F (2016). Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nature Chemical Biology 12, 339–344. [DOI] [PubMed] [Google Scholar]
- 54.Guo X, Li Z, Wang X, Wang J, Chala J, Lu Y, and Zhang H (2019). De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering. Biotechnol Bioeng 116, 3349–3359. [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Policarpio L, Prajapati D, Li Z, and Zhang H (2020). Developing E. coli-E. coli co-cultures to overcome barriers of heterologous tryptamine biosynthesis. Metabolic Engineering Communications 10, e00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gwon D. a., Seok JY, Jung GY, and Lee JW (2021). Biosensor-assisted adaptive laboratory evolution for violacein production. International Journal of Molecular Sciences 22, 6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Cabales A, Li Z, and Zhang H (2019). Biosensor-assisted high performing cell selection using an E. coli toxin/antitoxin system. Biochemical Engineering Journal 144, 110–118. [Google Scholar]
- 58.Li J, Kolberg K, Schlecht U, St Onge RP, Aparicio AM, Horecka J, Davis RW, Hillenmeyer ME, and Harvey CJB (2019). A biosensor-based approach reveals links between efflux pump expression and cell cycle regulation in pleiotropic drug resistance of yeast. J Biol Chem 294, 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandberg TE, Salazar MJ, Weng LL, Palsson BO, and Feist AM (2019). The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng 56, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seok JY, Han YH, Yang JS, Yang J, Lim HG, Kim SG, Seo SW, and Jung GY (2021). Synthetic biosensor accelerates evolution by rewiring carbon metabolism toward a specific metabolite. Cell Rep 36, 109589. [DOI] [PubMed] [Google Scholar]; ** This research designed biosensor-enabled adaptive evolution for 3-HP producers, and explored the underlying mechanism of production enhancement, which led to further titer improvement. It typically demonstrated the strengths of this strategy.
- 61.Libis V, Delepine B, and Faulon JL (2016). Sensing new chemicals with bacterial transcription factors. Curr Opin Microbiol 33, 105–112. [DOI] [PubMed] [Google Scholar]
- 62.Khoshbin Z, Housaindokht MR, Izadyar M, Bozorgmehr MR, and Verdian A (2021). Recent advances in computational methods for biosensor design. Biotechnol Bioeng 118, 555–578. [DOI] [PubMed] [Google Scholar]
- 63.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]


