Abstract
Pseudomonas aeruginosa (P. aeruginosa) is an important opportunistic pathogen that is responsible for many clinical infections in both animals and humans. This study aimed to detect the prevalence of P. aeruginosa in dairy farm's that possess a great importance to dairy industry where it shares in milk spoilage. Evaluation of the efficacy of commonly used disinfectants to control the pathogen in dairy environment and finding a way to overcome high resistance to the used agents. Samples (n = 250) were collected from different environmental components, milk, and milkers' hands. Pathogens were isolated, biofilm was detected and their sensitivity against two commonly used disinfectants and against silver nanoparticles and Virokill AgNPs at different concentrations and contact times were tested. The pathogen significantly prevailed in milk samples (70.0%, P < 0.001). 50 out 74 isolates were biofilm-forming that was significantly obtained from environment (71.8%, P < 0.001). P. aeruginosa showed variable degree of resistance to tested disinfectants but it was significantly sensitive to Virokill AgNPs (200/1000) mg/l at exposure time 24 h (P < 0.001). It was concluded that using Virokill AgNPs in regular sanitation and disinfection of dairy farms, this helps the control of P. aeruginosa subsequently increasing milk quality and improving dairy industry and protecting human health.
Subject terms: Biochemistry, Microbiology, Molecular biology, Environmental sciences
Introduction
Pseudomonas spp. is a ubiquitous pathogen that is commonly found in dairy farms' environment1,2. P. aeruginosa is a Gram-negative motile bacterium that is associated with various diseases in both humans and animals. In humans it is responsible for many infections such as pneumonia, septicemia, and necrosis in immunocompromised individuals3, meanwhile in animals it causes dermatitis, otitis, and urinary tract infection4. It also has been associated with many cases of clinical and subclinical mastitis in dairy ruminants5,6. Due to its low nutritional requirement and its ability to form a biofilm, Pseudomonas sp. were able to survive in different environments and they were allowed to grow and survive on equipment and utensils used in dairy production such as bulk milk tank, milking machines, pipelines, soiled bedding, humid soil, in air and water; subsequently increasing the risk of spreading infections to other animals and humans7–9 and very much for the same reasons the World Health Organization has listed it as a critical priority pathogen10.
P. aeruginosa possess several and variable virulence factors and antimicrobial determinants that are harbored in its genome, these factors provide the pathogen with metabolic flexibility and the ability to adapt adverse conditions11. For instance polymerized flagellin (fliC) gene is not only responsible for microbial motility but also responsible for binding to the membrane glycolipid asialo-GM1 on the apical surface of the lung epithelial cells12. Besides Flagellar attachment provided by fliC it helps in initial biofilm establishment, where motility allows cell dispersal13.
Additionally, the exotoxin A encoded tox A gene play a potential role in of P. aeruginosa pathogenicity where it is responsible for its cytotoxic capacity. It is a protein involved in pro-inflammatory cytokine synthesis stimulation. These virulence factors are mainly cooperated during P. aeruginosa colonization of the host cell and biofilm formation, allowing for further infection, and increasing bacterial pathogenicity14.
Biofilm formation is a microbial feature that increases' the bacterial resistance to antimicrobial agents (both antibiotics and disinfectants), biofilm is difficult to remove also it consider as a source of contamination to dairy products and subsequently human consumers with such biofilm producing pathogens15,16. Pseudomonas spp. is one of biofilm producing microorganisms. Pseudomonas's antimicrobial resistance is adapted by the formation of biofilms on the contaminated surfaces that act as a diffusion barrier that limit the access of these agents to the bacterial cells and hinder their action17.
Providing hygienic measures during milking process and using disinfectants to keep the environment surrounding dairy animals clean is an important measure to reduce the contamination with such pathogen18. Despite the availability of many commercial disinfectants used in the cleaning and sanitation process in dairy farms, they show weak efficacy to limit contamination of both raw milk and/or dairy products with such pathogen due to biofilm formation7,15. Moreover, it represents a significant public health issue in hospitals as well as medical care due to the intensified resistance to antimicrobial agents19,20.
As a result of upsurge and worldwide antimicrobial resistance, nanomaterials have been used as disinfectants that have proven their efficacy and among these various nanomaterials, copper, silver, and gold21. Silver ions and silver-based nano compounds are well known materials that have been used in medicine science 1000 BC22. Nano-based silver disinfectants have been proven to have more effective antimicrobial properties due to high surface exposure of the disinfectant to the microbe23.
This study aimed to point out the prevalence of biofilm producing P. aeruginosa in dairy farm environments passing through different points in milking process and then to evaluate the efficacy of some commonly used disinfectant in the routine disinfection and cleaning regimes in the farm under the study to the efficacy of silver nano-based disinfectants.
Material and methods
Study area and period
The current study was performed in a private dairy cattle farm at Beni-Suef locality (coordinates 29° 04ʹ N–31° 05ʹ E), Egypt from December 2019 till September 2020. Dairy cows (n = 70) were milked in abreast parlor with two milking machines that were located at the northern part of the farm and near to the yards where the cows were housed on earthy floor that was partially sheltered provided with common water trough and food manager for each group of animals (n = 14). The hygienic condition that prevailed in the farm and the dairy was fair.
Study design
The work in this study was done in two successive steps the first was to investigate the prevalence of biofilm-forming P. aeruginosa from different sources by collecting samples such as milk samples, environmental samples besides, human hand swabs that were cultured for the recovery of P. aeruginosa and their identification was done by molecular identification using 16S rDNA that is specific for P. aeruginosa and determination of toxA and fliC genes, as the most known virulence genes in the identified isolates, then identification of biofilm formation using Congo Red Assay (CRA). The second step was to control this pathogen using dependable disinfectants that are commonly used in the field and loaded disinfectant on sliver nanoparticles, using the broth micro-dilution method. Animal sampling design and protocol was carried out based upon the International Animal Care and Use Committee (IACUC), Ref. No: IORG 0001080), of Beni-Suef University, Egypt, while human samples did not require any approval of Institutional Review Board (IRB) since hand swabs were only collected after oral consent of farm workers. Additionally, the authors confirmed that all methods illustrated in the manuscript were carried out in accordance with relevant guidelines and regulations. All data and results were recorded and statistically analyzed.
Sample collection
Milk samples (10 ml) (n = 50), environmental samples including milk machine, bulk milk tank, water, water trough, feeding manager, feed stuff and flies' samples were screened (n = 25 each), and hand swabs (n = 25) were obtained from the farm workers under investigation after taking their oral consent, all the workers were apparent healthy at the time of sampling. All samples were collected aseptically according to Munoz et al.24 then preserved on ice box to be transported to the lab of Animal Hygiene and Zoonoses on Faculty of Veterinary Medicine, Beni-Suef University, where the bacteriological examination was done.
Bacteriological and molecular techniques
Milk samples (one ml each) were pre-enriched into tubes containing 9 ml of the tryptic soy broth while all sample swabs were directly inoculated into tubes containing 9 ml of tryptic soy broth (Oxoid, Basingstoke, UK) and incubated at 37 °C for 24–72 h then a loopful from each tube was inoculated on the surface of freshly prepared cetrimide agar (consisted of peptone, 20 g; Mg chloride, 1.4 g; Potassium sulphate 10 gm; cetrimide 0.3 g; glycerol, 10 ml; agar 13 g per one liter of distilled water). Plates showing large blue–green or green–yellow colonies with characteristic sweet-grape like odor were picked up and biochemically identified25. The biochemical tests used to confirm the recovery of P. aeruginosa were oxidase, catalase, gelatin hydrolysis and hydrolysis of polysaccharides tests, fermentation of various sugars, pigment production on tryptic soya agar and growth either in 4 °C or 42 °C26. The biochemically confirmed isolates were sent to the biotechnology center in the animal health research institute, Egypt for molecular characterization. Where the amplification of P. aeruginosa specific 16S rDNA as well as detection of virulence (toxA and fliC) genes were performed27–29 respectively. Data of primer sequences and target genes were illustrated in Table 1.
Table 1.
Oligonucleotide and primer sequences specific for P. aeruginosa investigated during the study.
Molecular identification
Firstly, DNA was individually extracted from each biochemically confirmed colonies using QIAamp DNA Mini kit (Qiagen, Germany, GmbH). Briefly, 200 µl of the sample suspension was incubated with both 10 µl of proteinase K and 200 µl of lysis buffer at 56 °C for 10 min. After incubation, 200 µl of the absolute ethanol was added up to the lysate. The samples were washed and centrifuged following the manufacturer’s instructions. Lastly, the nucleic acid of each sample was eluted with 100 µl of elution buffer supplied with the kit. For conventional-type PCR amplification, the primers were utilized in a 25 µl volume reaction including 12.5 µl of EmeraldAmp Max PCR Master Mix (Takara, Japan), 1 µl of each primer of 20 pmol concentration, 4.5 µl of water as well as 6 µl of DNA template. The reaction was performed in an Applied biosystem 2720 thermal cycler.
The amplification of P. aeruginosa specific 16S rDNA as well as detection of virulence (toxA and fliC) genes were performed. The parameter of thermocycling began an initial denaturation cycle at 94 °C for 5 min followed by 30 cycles of the subsequent program, 94 °C for 30 s, the annealing temperatures were 52, 55 and 56.2 °C for 45 s for each primer27–29, respectively. The final extension step was at 72 °C for 7 min. The products of PCR were disconnected by electrophoresis on 1.0% agarose gel (Applichem, Germany, GmbH) in 1 × Tris/Borate/EDTA (TBE) buffer at room temperature using gradients of 5 V/cm. For gel evaluation, 40 µl of the PCR products was inserted in each gel slot and the gel was portrayed by a gel documentation system (Alpha Innotech, Biometra) and the data was evaluated using the computer software of DigiDoc-It Imaging System.
Screening biofilm formation capacity
It was qualitatively investigated using the Congo Red Assay (CRA), by plating on the surface of Congo Red agar (prepared by mixing of brain heart infusion broth (37 g), sucrose (5 g), Congo red dye (0.8 g) and agar (15 g) per a liter of distilled water). Congo red agar plates were inoculated with isolates of P. aeruginosa, and incubated aerobically for 24–48 h 37 °C. Following the incubation period, the colonies' color was noticed and interpreted30.
Synthesis and characterization of silver nanoparticle and Virokill silver nanoparticle / composite
The silver colloid was prepared according to Sileikaite et al.31 using chemical reduction method through dissolving 0.085 g of silver nitrate in 500 ml distilled water, and then this mixture was heated till boiling. Then 1 g of trisodium citrate was dissolved in 100 ml distilled water and 5 ml of trisodium citrate was added drop by drop to the prepared solution of silver nitrate then it was vigorously mixed. The mixture was then left to heat at 90 °C for 2 h on a hot plate, and then it was left to cool at room temperature. Where the solution color turn into reddish green (end point). The crystal structure and crystallinity of materials were detected using X-ray diffraction (XRD) (Fig. 2), while the vibration and the chemical bonds of the materials were screened by Fourier-transform infrared spectrum (FT-IR) (Fig. 3). The average size and morphological shape of Virokill/AgNPs was characterized by Transmission Electron Microscopy (TEM) (Fig. 4a,b) at the National Research Center (NRC), Egypt.
Figure 2.
X-ray Diffraction (XRD) of silver nanoparticles before and after Virokill loading, showing characteristic peaks for silver and its oxides AgO and Ag2O, indicating successfully incorporated of Virokill disinfectant within the silver nanoparticles.
Figure 3.
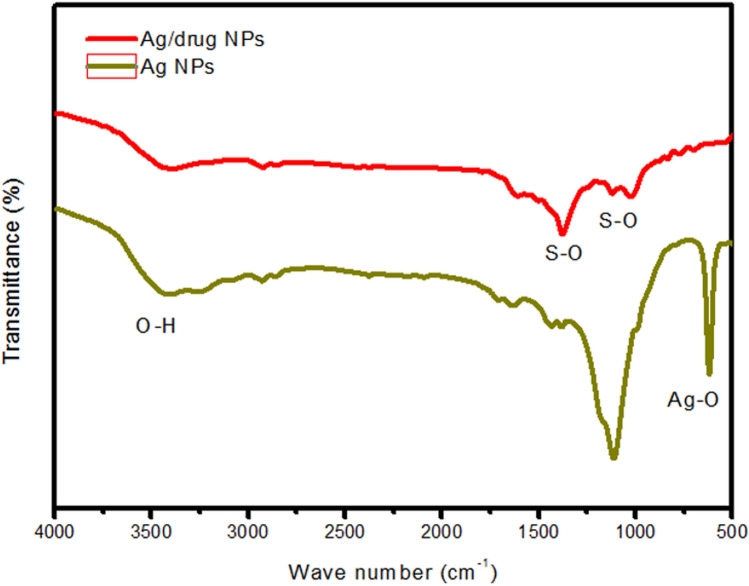
Fourier-transform infrared spectra (FTIR) of silver nanoparticles before and after Virokill disinfectant loading.
Figure 4.
Transmission electron microscopy (TEM) images of silver nanoparticles before loading, showing a spherical like shape with diameters ranging amongst 36–60 nm (a) while in after loading (b), appears that the Virokill is completely encapsulates the silver nanoparticles.
Evaluation of germicidal efficacy of tested disinfectants
The germicidal power of two commercially disinfectants proven their efficacy and were approved by food industry32; Virokill (potassium per-oxy-mono-sulfate 50.0%, NaCl 3.0%, UBM, Egypt), Peracetic acid (6th October 3rd Industrial Area, Egypt), were tested using different concentration of them against 30 strains of P. aeruginosa isolated from milk, human and environmental samples using broth macro-dilution33 with different concentrations: Virokill (0.25, 0.5 and 1.0%), peracetic acid (0.25 and 0.5%), after contact times (30 min, 1 h and 24 h).
Evaluation of germicidal silver nanoparticle and Virokill silver nanoparticle / composite
Different concentrations of silver oxide Ag2O NPs (100,150 and 200 mg/L), in addition conc. of 150 and 200 mg/L of Ag NPs were added to 1.0% of Virokill, were tested against 30 traits of P. aeruginosa obtained from milk, human and environmental samples using broth macro-dilution method34. Ag NPs were added immediately before use and was vigorously shaken using magnetic stirrer to increase the dispersion and prevent settling of NPs over the incubation periods (30 min, 1 h and 24 h).
Statistical analysis
Data obtained were recorded and the frequency of P. aeruginosa in the collected samples as well the germicidal efficacy of tested disinfectants and silver nanoparticles composite were calculated using non-parametric tests (Chi-Square Test) using SPSS (Inc. version 22.0, Chicago, IL, USA).
Results and discussion
Pseudomonas spp. are incontrovertible psy-chrotolerant bacteria that are commonly found in natural environment, such as water, inner side of bulk tanks, cows' teat and surfaces that may associate with the contamination of raw milk8,35. Therefore, the searching for probable sources of contamination and adoption of hygienic measures during milking process had become a necessity in the growing dairy industry18,35.
The obtained results shown in Table 2 revealed that the total number of positive samples for P. aeruginosa isolation were 74 (29.6%) that were recovered from different sources; and this pattern of isolation confirms the ubiquitous nature of the pathogen and variety of media in which it can survive in various conditions36.The pathogen was mainly recovered from milk samples (70.0%) followed by swabs of milk machine, hand swabs and milk tank (32.0, 28.0 and 24.0%, respectively) at X2 = 156.584 at P < 0.001. Much lower results were recorded by Banerjee et al.36 who found that the percentage of Pseudomonas sp. isolation was only 6.5%, and to some extent Banda et al.37 could isolate Gram negative rods (Pseudomonas sp.) by the percentage of 10.2%. Conversely to our finding Banda et al.37 did not isolate Pseudomonas spp. from milk samples but it was isolated (13.3 and 13.0%) from water samples and environmental swabs, respectively. Also, in a study made by Ma et al.38 revealed that P. aeruginosa was detected in the following samples from workers, milk, feed stuff, floor surface and water samples by (1.0, 10.0, 4.0, 4.0 and 6.0%, respectively). Similarly, Vidal et al.39 mentioned that they were able to isolate Pseudomonas spp. from teat cups of milking machine and bulk tank (95.0%), milk samples (90.0%), from milkers' hands (80.0%), teat surface (50.0%), and water sample (70.0%).
Table 2.
Prevalence of P. aeruginosa recovered from animals, their environment, and humans during the study period.
| Samples/swabs | Examined samples (No.) | Positive samples (No.) | Prevalence of P. aeruginosa (%) |
|---|---|---|---|
| Milk | 50 | 35 | 70.0 |
| Hand swab | 25 | 7 | 28.0 |
| Milk tank | 25 | 6 | 24.0 |
| Milk machine swab | 25 | 8 | 32.0 |
| water | 25 | 3 | 12.0 |
| Water trough swab | 25 | 4 | 16.0 |
| Feed stuff | 25 | 3 | 12.0 |
| Feed manager swab | 25 | 4 | 16.0 |
| flies | 25 | 4 | 16.0 |
|
Total X2 = 156.584 P. value < 0.001 |
250 | 74 | 29.6 |
Concerning the prevalence of biofilm producing capacity of isolated P. aeruginosa collected from different samples (Table 3) it showed that out of 74 recovered P. aeruginosa traits, 50 (67.5%) were biofilm producers meanwhile 24 (32.4%) were non-biofilm producers, and this prove a significant increase in the biofilm propriety of the isolated pathogen at X2 = 30.757 at P < 0.001, referring to their distribution these isolates were mainly obtained from environmental samples followed by milk and finally from human samples (23 (71.8%), 22 (62.8%), 5 (71.4%). Biofilm is a microbial property that is characterized by adhesion of the microbes to a solid surface and production of a matrix that surrounds and includes the bacterial cells and include extracellular polysaccharides, proteins, and DNA40–43 proved that biofilm protects bacteria from the most sever adverse environmental conditions including antimicrobials. Banda et al.37 recorded lower results to that found in this study where he recorded that only 14 out of 86 (16.3%) of microbial isolates were strong biofilm formers and (20.9%) did not form any biofilm. Also, Milivojevic et al.44 reported that only30 out of 202 isolates (15.0%) were biofilm former and 11 (5.0%) were non biofilm former. Meanwhile Ngo et al.45 recorded much higher results than those reported in the following study where he found that (100.0%) of tested isolates were biofilm former with different degree ranging from strong, moderate, or weak.
Table 3.
Prevalence of the biofilm producing capacity of isolated P. aeruginosa collected from samples during the study period.
| Samples/swabs | Examined samples (No.) | Biofilm-procedures | Non-biofilm procedures |
|---|---|---|---|
| Milk | 35 | 22 (62.8%) | 13 (37.1%) |
| Hand swab | 7 | 5 (71.4%) | 2 (28.5%) |
| Environment | 32 | 23 (71.8%) | 9 (28.1%) |
|
Total X2 = 30.757 P > 0.001 |
74 | 50 (67.5%) | 24 (32.4%) |
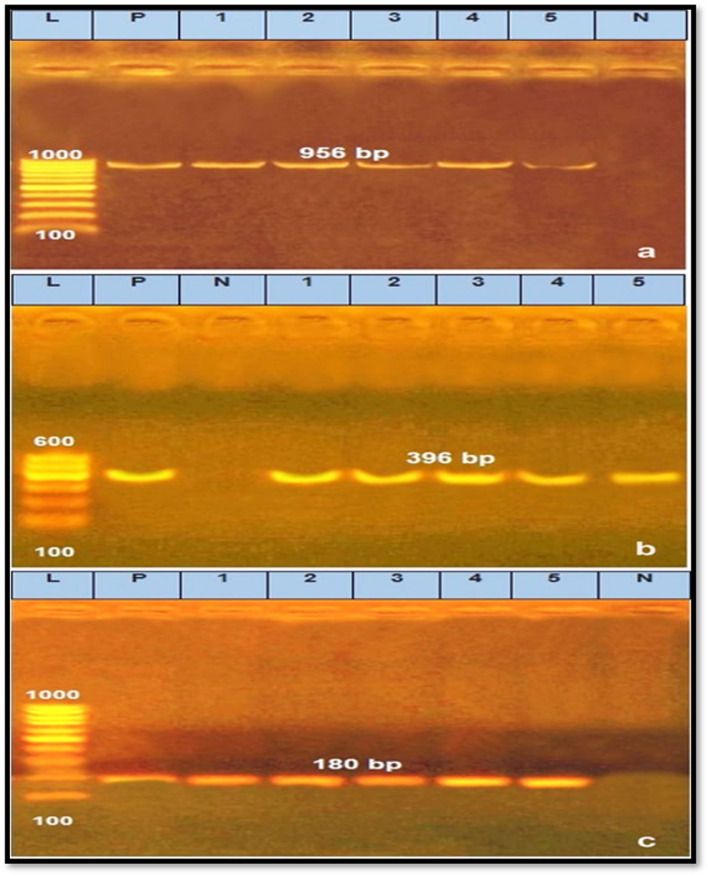
Figure 1 referring to the distribution of both tox and fliC genes; showed that all the randomly selected isolates were caring both genes. fliC (flagellin) has an important role in stimulating the immune response, with the emergence of multi-resistant strains46,47. The virulence gene exotoxin A (tox A) is secreted through Type II secretion mechanism, which secrete proteins into the extracellular environment, including lipase, phospholipase, alkaline phosphatase, and protease48,49. These results are in accordance with the results of biofilm formation of the tested isolates that also assist these pathogens to resist antimicrobial agents (both disinfectants and antibiotics).
Figure 1.
Agarose gel electrophoresis for PCR products of P. aeruginosa species identification using 16S rDNA gene (a), amplified 356 bp and the virulence genes (b,c), amplified 396 and 180 bp for toxA and fliC genes, respectively. Lane (L): 100 bp Ladder ‘’Marker’’, Lane): 1–5), the examined samples, Lane Pos: Positive control, Lane Neg: Negative control.
Figure 2 shows the XRD analysis of silver nanoparticles before and after drug Virokill loading. As shown, characteristic peaks for silver and its oxides AgO and Ag2O appeared in the diffractogram. The peaks at 38, 56.1 and 64.4 could be attributed to the diffraction planes of (111), (142) and (220) of silver respectively50,51. Peaks at 27.4 and 31.9 corresponds to (110) and (111) planes of Ag2O, respectively49. While peaks at 30.9, 33.3 and 45 corresponds to (111), (202) and (132) planes of AgO, respectively52,53.
Figure 3 shows the FTIR spectra of the prepared silver nanoparticles. The broad band around 3400 cm−1 can be assigned to the stretching of O–H bond in adsorbed water molecules. The low intensity peak at 600 cm−1 originates from the Ag–O bond54. After drug Virokoll loading, the characteristic peaks form S–O stretching vibration for SO42− and HSO5− in the per-oxy-mono-sulfate appeared at approximately 1440 cm−1 and 1116 cm−1, respectively55.
Referring to Transmission electron microscopy (TEM) image (Fig. 4a of silver nanoparticles it shows that the particles have a spherical like shape with diameters ranging between 36–60 nm. After the Virokill loading, the TEM image (Fig. 4b) shows that the Virokill completely encapsulates the silver nanoparticles Ag NPs within its matrix.
Results illustrated in Table 4 showed that the sensitivity of P. aeruginosa was significantly high to Virokill/AgNPs (200/1000) mg/L after 24 h followed by Ag/NPs (200/1000) mg/l after 24 h at P < 0.001 (70.0 and 55.0%, respectively). On the other hand, it showed a significant high degree of resistance to Peracetic acid 0.25% and 0.5% after 30 min and 1 h of exposure (90.0, 75.0% and 80.0, 65.0%, respectively). Generally, the pathogen showed a variable degree of resistance to most of the used disinfectants at different contact times and concentrations, yet this resistance decreases with the increase of both concentration and exposure time of this disinfectant. Despite the common use of these disinfectants in dairy farms from awhile, and even so these results might be shocking but not actually surprising which might be referred due to the insufficient cleaning of the farm and removal of organic matter and/or the misuse of disinfectants (following the label instructions)56,57. Eventually that have led to increase resistance of food borne pathogens to biocides58 with the probability of cross transfer of genes that responsible for their resistance to both disinfectants and antibiotics in the environment this became of great interest to researchers everywhere to overcome and/or to prevent this from happening so saving money and protecting humans' life59–61. The significant high degree of bacterial resistance recorded in this study might be attributed to the ability of these pathogens to form biofilm and owning virulence genes which have been proved during the study, also might be due the modest hygienic practices followed in the farm under the study, where presence of organic matter suggests the establishment of biofilm formation on the corresponding surfaces40 and improper use of disinfectants according to the manufacturers' instructions58. On the contrary to our results Gibson et al.62 proved that the use of acids such peracetic acid significantly (P < 0.05) affected the viability of S. aureus and P. aeruginosa and it was not necessary to use another disinfectant for cleaning. Stewart et al.63 reported that chloramines (chlorosulfates as Virokill) were able to penetrate biofilm 6–8 times sooner than free chlorine however these bacteria were highly resistant to both agents. Ag/NPs were proven to damage the bacterial cell membranes and finally their death64. Moreover, silver nanoparticles are known for their lethal effect to different bacterial pathogens65 with special reference to the shape and size of nanoparticles65 that was detected by TEM in this study. Since Virokill showed better results than per-acetic acid it was loaded on Ag/NPs to improve its penetration into the bacterial cells particularly in biofilm and Ag/NPs acts in synergism together with the disinfectant to improve its biocidal efficiency66.
Table 4.
The bactericidal efficacy of tested disinfectants, silver nanoparticles and its loaded forms against P. aeruginosa traits.
| Tested disinfectant/bacteria (no.) | Conc (mg/L) | Sensitivity pattern (%) of P. aeruginosa isolates at different exposure times | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 30 min | 1 h | 24 h | ||||||||
| S | I | R | S | I | R | S | I | R | ||
| Peracteic acid® | 0.25% | 10.0 | 0.0 | 90.0 | 15.0 | 5 | 80 | 30.0 | 20.0 | 50.0 |
| 0.5% | 20.0 | 5 | 75.0 | 25.0 | 10 | 65 | 35.0 | 25.0 | 40.0 | |
| Virokill® | 0.25 | 15.0 | 10 | 75.0 | 20.0 | 15 | 65 | 25.0 | 20.0 | 55.0 |
| 0.5 | 20.0 | 10 | 70.0 | 25.0 | 20 | 55 | 30.0 | 30.0 | 40.0 | |
| 1.0% | 35.0 | 0.0 | 65.0 | 30.0 | 10 | 60 | 45.0 | 10.0 | 45.0 | |
| AgNPs | 150 | 15.0 | 10 | 75.0 | 15.0 | 10 | 75 | 20.0 | 15.0 | 65.0 |
| 200 | 20.0 | 15 | 65.0 | 30.0 | 10 | 60 | 55.0 | 15.0 | 30.0 | |
| Virokill/AgNPs | 150 | 25.0 | 5 | 70.0 | 30.0 | 20 | 50 | 40.0 | 20.0 | 40.0 |
| 200 | 30.0 | 10 | 60.0 | 35.0 | 25.0 | 40.0 | 70.0 | 20.0 | 10.0 | |
| P value | 0.001 | 0.001 | 0.001 | |||||||
S Sensitive, I Intermediate, R Resistant.
Conclusion
From the current study it was concluded that P. aeruginosa prevailed in dairy farm environment and the more there is a lack of hygiene the more their ability to form biofilm will increase and eventually become more virulent and resistant to both disinfectants and antibiotics. The increased resistance to antimicrobial agents possesses a risk to humans' and animals' health through the infection of dairy animals' and contamination of dairy products. Therefore, we recommend to design and activate a routine cleaning and disinfection program that initially clean and remove organic matter which protects these pathogens and hinder the efficacy of the used disinfectant starting from the animal house and passing through the entire system of dairy production. Using Virokill/AgNPs composite (200/1000) mg/L that was proven its efficacy and allowing it to act on the pathogen for 24 h to maintain clean and healthy dairy environment, increase the milk quality and improve the dairy industry.
Author contributions
S.A.A.A.A. and M.B.E.D.M., both authors sheared the conception and design of the study, collection, analysis, interpretation of data, drafting the manuscript and grammar revision while, R.M. made the characterization and interpretation of the nanoparticles.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Erskine RJ, et al. Pseudomonas mastitis: Difficulties in detection and elimination from contaminated wash water systems. J. Am. Vet. Med. Assoc. 1987;7:811–815. [PubMed] [Google Scholar]
- 2.Blanc DS, et al. Frequency and molecular diversity of P. aeruginosa upon admission and during hospitalization: A prospective epidemiologic study. Clin. Microbiol. Infect. 1998;4:242–247. doi: 10.1111/j.1469-0691.1998.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 3.Weizhong C, et al. CRISPR/Cas9-based genome editing in P. aeruginosa and cytidine deaminase-mediated base editing in pseudomonas species. I Sci. 2018;31(6):222–231. doi: 10.1016/j.isci.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazuki H, et al. Characterization of P. aeruginosa isolates from dogs and cats in Japan: Current status of antimicrobial resistance and prevailing resistance mechanisms. Microbiol. Immunol. 2012;56:123–127. doi: 10.1111/j.1348-0421.2011.00416.x. [DOI] [PubMed] [Google Scholar]
- 5.Sela S, et al. Phenotypic and genotypiccharacterization of P. aeruginosa strains isolated from mastitis outbreaks in dairy herds. J. Dairy Res. 2010;4(4):425–429. doi: 10.1017/S0022029907002610. [DOI] [PubMed] [Google Scholar]
- 6.Ohnishi M, et al. Antimicrobial susceptibilities and bacteriological characteristics of bovine P. aeruginosa and Serratia marcescens isolates from mastitis. Vet. Microbiol. 2011;154:202–207. doi: 10.1016/j.vetmic.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Simões M, Simões LC, Vieira MJ. A review of current and emergent biofilm control strategies. LWT Food Sci. Technol. 2010;43:573–583. doi: 10.1016/j.lwt.2009.12.008. [DOI] [Google Scholar]
- 8.The KH, et al. Thermo-resistant enzyme-producing bacteria isolated from the internal surfaces of raw milk tankers. Int. Dairy J. 2011;21:742–747. doi: 10.1016/j.idairyj.2011.04.013. [DOI] [Google Scholar]
- 9.Aysel U, Özgür C, Belma A. Characterization of Pseudomonas spp. from seawater of the southwest coast of Turkey. J. Biol. Environ. Sci. 2012;6(16):15–23. [Google Scholar]
- 10.World Health Organization (WHO). Media Centre. News Release. WHO publishes list of bacteria for which new antibiotics are urgently needed?http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/. Accessed March (2017).
- 11.Moradali MF, Ghods S, Rehm BH. Lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017;7(39):1–29. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haiko J, Westerlund-Wikström B. The role of the bacterial flagellum in adhesion and virulence. Biology. 2013;2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm. FEMS Microbiol. Rev. 2013;37(6):849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Wrafy F, Brzozowska E, Górska S, Gamian A. Pathogenic factors of P. aeruginosa—the role of biofilm in pathogenicity and as a target for phage therapy. Postepy. Hig. Med. Dosw. 2017;71:78–91. doi: 10.5604/01.3001.0010.3792. [DOI] [PubMed] [Google Scholar]
- 15.Marchand S, et al. Biofilm formation in milk production and processing environments; Influence on milk quality and safety. Compr. Rev. Food Sci. Food Saf. 2012;11:133–147. doi: 10.1111/j.1541-4337.2011.00183.x. [DOI] [Google Scholar]
- 16.Abdallah M, et al. Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 2014;196:453–472. doi: 10.1007/s00203-014-0983-1. [DOI] [PubMed] [Google Scholar]
- 17.Drenkard E. Antimicrobial resistance of P. aeruginosa biofilms. Microbes Infect. 2003;5:1213–1219. doi: 10.1016/j.micinf.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Elmoslemany AM, et al. The association between bulk tank milk analysis for raw milk quality and on-farm management practices. Prev. Vet. Med. 2010;95(1–2):32–40. doi: 10.1016/j.prevetmed.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Malek AA, Matthew J, Deborah W, Hogan A. Absence of membrane phosphatidylcholine does not affect virulence and stress tolerance phenotypes in the opportunistic pathogen P. aeruginosa. PLoS ONE. 2012;7(2):e30829. doi: 10.1371/journal.pone.0030829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dua K, Shukla SD, Tekade RK, Hansbro PM. Whether a novel drug delivery system can overcome the problem of biofilms in respiratory diseases. Drug Deliv. Transl. Res. 2017;7(1):179–187. doi: 10.1007/s13346-016-0349-0. [DOI] [PubMed] [Google Scholar]
- 21.Haji SH. Detection of biofilm formation in p aeruginosa isolates from clinical specimens. Zanco J. Pure Appl. Science. 2018;30(4):83–89. [Google Scholar]
- 22.Deshmukh SP, Patil SM, Mullani SB, Delekar SD. Silver nanoparticles as an effective disinfectant: A review. Mater. Sc. Eng. C. 2019;97:954–965. doi: 10.1016/j.msec.2018.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, Silver S. Silver as a biocide: Will resistance become a problem? Nat. Biotechnol. 1998;16(10):e888. doi: 10.1038/nbt1098-888. [DOI] [PubMed] [Google Scholar]
- 24.Munoz MA, Zadoks RN. Short communication: Patterns of fecal shedding of Klebsiella by dairy cows. J. Dairy Sc. 2007;90:1220–1224. doi: 10.3168/jds.S0022-0302(07)71610-7. [DOI] [PubMed] [Google Scholar]
- 25.Kerig NR, Holt JG. Bergey's Manual of Systemic Bacteriology. 8Th. Williams and Willkins; 1984. [Google Scholar]
- 26.Holtz, J.G. et al. Bergey’s manual of determinative Bacteriology. In: L Spilker T, Coenye T, Vandamme P, LiPuma JJ (ed). PCR-Based Assay for Differentiation of P. aeruginosa from Other Pseudomonas Species Recovered from Cystic Fibrosis Patients. 9th Ed.. Journal of clinical microbiology, pp 2074–2079 (2000). [DOI] [PMC free article] [PubMed]
- 27.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of P. aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J. Clin. Microbiol. 2004;42:2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matar GM, et al. Transcription levels of P. aeruginosa exotoxin a gene and severity of symptoms in patients with otitis externa. Curr. Microbiol. 2002;45:350–354. doi: 10.1007/s00284-002-3703-z. [DOI] [PubMed] [Google Scholar]
- 29.Ghadaksaz A, Fooladi AAA, Hosseini HH, Amin M. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J. Appl. Biomed. 2015;13(1):61–68. doi: 10.1016/j.jab.2014.05.002. [DOI] [Google Scholar]
- 30.Niveditha S, et al. The isolation of the biofilm formation of uro-pathogens in the patients with catheter associated urinary tract infections (UTIs) J. Clin. Diagn. Res. 2012;6(9):1478–1482. doi: 10.7860/JCDR/2012/4367.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sileikaite A, et al. Analysis of silver nanoparticles produced by chemical reduction of silver salt solution. Mater. Sci. 2006;12:287–291. [Google Scholar]
- 32.Meyer B, Cookson B. Does microbial resistance or adaptation to biocides create a hazard in infection prevention and control? J. Hosp. Infect. 2010;76:200–205. doi: 10.1016/j.jhin.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, et al. Antimicrobial nanomaterial for water disinfection and microbial control: Potential applications and implications. Water Res. 2008;42:4591–4602. doi: 10.1016/j.watres.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 34.Fagundes CM, et al. Presence of Pseudomonas spp. related to different phases of the milking process with different hygienic managements and in refrigerated milk. Rural. 2006;36:568–572. doi: 10.1590/S0103-84782006000200032. [DOI] [Google Scholar]
- 35.Silby MW, et al. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011;35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee S, et al. Detection and characterization of pathogenic P. aeruginosa from bovine subclinical mastitis in West Bengal, India. Vet. World. 2017;10(7):738–742. doi: 10.14202/vetworld.2017.738-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banda R, Nduko J, Matofari J. Bacterial biofilm formation in milking equipment in Lilongwe, Malawi. J. Food Qual. Haz. Cont. 2020;7:142–148. [Google Scholar]
- 38.Ma L, et al. Analysis of P. aeruginosa conditional psl variants reveal roles for the psl polysaccharide in adhesion and maintaining biofilm structure post-attachment. J. Bacteriol. 2006;188:8213–2821. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidal A, et al. Microbiological diagnosis and antimicrobial sensitivity profiles in diseased free-living raptors. Avn. Path. 2017;46:442–450. doi: 10.1080/03079457.2017.1304529. [DOI] [PubMed] [Google Scholar]
- 40.Castron H, et al. Genomic epidemiology and phenotyping reveal on-farm persistence and cold adaptation of raw milk outbreak-associated Yersinia pseudotuberculosis. Front. Microbiol. 2019;10:1049–1060. doi: 10.3389/fmicb.2019.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cloete TE. Resistance mechanisms of bacteria to antimicrobial compounds. Intern. Biodeter. Biodegrad. 2003;51:277–282. doi: 10.1016/S0964-8305(03)00042-8. [DOI] [Google Scholar]
- 42.Davies D. biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 43.Bjarnsholt T, et al. P. aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Ped. Pul. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 44.Milivojevic D, et al. Biofilm-forming strains ability and infection potential of P aeruginosa strains isolated from animals and humans. Path. Dis. 2018;76(4):1–14. doi: 10.1093/femspd/fty041. [DOI] [PubMed] [Google Scholar]
- 45.Ngo TP, Bui HL, Luong HN, Bui TV. Identification of biofilm formation bacterial strains isolatedfromraw milk in BenTre Province Vietnam. Chem. Eng. Trans. 2021;87:607–612. [Google Scholar]
- 46.Campodónico V, et al. Evaluation of flagella and flagellin of P. aeruginosa as vaccines. Infect. Immun. 2010;78:746–755. doi: 10.1128/IAI.00806-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013;37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passador L, et al. Expression of P. aeruginosa virulence genes require cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 49.Wedekind JE, et al. Refined crystallographic structure of P. aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 2001;314:823–837. doi: 10.1006/jmbi.2001.5195. [DOI] [PubMed] [Google Scholar]
- 50.Govarthanan M, et al. Biosynthesis and characterization of silver nanoparticles using panchakavya, an Indian traditional farming formulating agent. Int. J. Nanomed. 2014;2:e1593. doi: 10.2147/IJN.S58932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang H, Ren Y, Wang T, Wang C. Preparation and antibacterial activities of Ag/Ag + /Ag 3+ nanoparticle composites made by pomegranate (Punica granatum ) rind extract. Results Phys. 2016;6:299–304. doi: 10.1016/j.rinp.2016.05.012. [DOI] [Google Scholar]
- 52.Dhoondia ZH, Chakraborty H. Lactobacillus mediated synthesis of silver oxide nanoparticles. Nanomater. Nanotechnol. 2012;2:1–7. doi: 10.5772/55741. [DOI] [Google Scholar]
- 53.Gauri B, Vidya K, Sharada D, Shobha W. Synthesis and characterization of Ag/AgO nanoparticlesas alcohol sensor. Res. J. Chem. Env. 2016;2:1–5. [Google Scholar]
- 54.Sabzi M, Dezfuli MS. A study on the effect of compositing silver oxide nanoparticles by carbon on the electrochemical behavior and electronic properties of zinc silver oxide batteries. Int. J. Appl. Ceram. Technol. 2018;15:1446–14458. doi: 10.1111/ijac.13047. [DOI] [Google Scholar]
- 55.Widyaningtyas, A.L., Yulizar, Y. & Apriandanu, D.O.B.Ag2O nanoparticles fabrication by Vernonia amygdalina Del. leaf extract: synthesis, characterization, and its photocatalytic activities. IOP Conf. Ser. Mater. Sci. Eng. e12022 (2019).
- 56.Dong X, et al. Monodispersed CuFe2O4 nanoparticlesanchored on natural kaolinite as highly efficient peroxymonosulfate catalyst for bisphenol A degradation. Appl. Catal. B. Environ. 2019;253:206–217. doi: 10.1016/j.apcatb.2019.04.052. [DOI] [Google Scholar]
- 57.Olukemi OA, Funmilayo OA. The efficacy of the commonly used hospital disinfectants on P. aeruginosa. Int. Res. J. Microbiol. 2011;2(7):226–229. [Google Scholar]
- 58.Al-Jailawi MH, Ameen RS, Al-Jeboori MR. Effect of disinfectants on antibiotics susceptibility of P. aeruginosa. J. Appl. Biotechnol. 2013;1:54–63. doi: 10.5296/jab.v1i1.4038. [DOI] [Google Scholar]
- 59.Morente EO, et al. Biocidetolerance in bacteria. Int. J. Food Microbiol. 2013;162(1):13–25. doi: 10.1016/j.ijfoodmicro.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 60.Franci G, et al. Silver nanoparticles as potential antibacterial agents. Molecules. 2015;20:8856–8874. doi: 10.3390/molecules20058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinto Y, et al. Split brain: Divided perception but undivided consciousness. Brain. 2017;2:1–7. doi: 10.1093/brain/aww358. [DOI] [PubMed] [Google Scholar]
- 62.Gibson H, Taylor JH, Hall KE, Holah JT. Effectiveness of cleaning techniques used in the food industry in terms of the removal of bacterial biofilms. J. Appl. Microbiol. 1999;84:41–48. doi: 10.1046/j.1365-2672.1999.00790.x. [DOI] [PubMed] [Google Scholar]
- 63.Stewart PS, Rayner J, Roe F, Rees WM. Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J. Appl. Microbiol. 2001;91:525–532. doi: 10.1046/j.1365-2672.2001.01413.x. [DOI] [PubMed] [Google Scholar]
- 64.Periasamy S, et al. How Staphylococcus aureus biofilms develop their characteristic structure. PNAS. 2012;109(4):1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berton V, et al. Study of the interaction between silver nanoparticles and Salmonella as revealed by transmission electron microscopy. J. Prob. Health. 2015;3:e123. [Google Scholar]
- 66.Liu HL, Dai SA, Fu KY, Hsu SH. Antibacterial properties of silver nanoparticles in three different sizes and their nanocomposites with a new waterborne polyurethane. Int. J. Nanomedicine. 2010;5:1017–1028. doi: 10.2147/IJN.S14572. [DOI] [PMC free article] [PubMed] [Google Scholar]