Abstract
Complete understanding of a biological system requires quantitation of metabolic fluxes that reflect its dynamic state. Various analytical chemistry tools, enzyme-based probes, and microscopy enable flux measurement. However, any method alone falls short of comprehensive flux quantitation. Here we show that integrating these techniques results in a systems-level quantitative map of absolute metabolic fluxes that constitute an indispensable dimension of characterizing phenotypes. Stable isotopes, mass spectrometry, and NMR spectroscopy reveal relative pathway fluxes. Biochemical probes reveal the physical rate of environmental changes. FRET- and SRS-based microscopy reveal targeted metabolite and chemical bond formation. These techniques are complementary and can be computationally integrated to reveal actionable information on metabolism. Integrative metabolic flux analysis using various quantitative techniques advances biotechnology and medicine.
Introduction
Metabolism is a dynamic network of biochemical reactions. Metabolites are the intermediates of metabolism, and their concentrations are informative because they are involved in biological processes ranging from gene regulation to signaling to biosynthesis. Metabolite levels, however, do not convey the full story of the state of biological systems. Metabolic fluxes (i.e., rates of biochemical activities) offer more actionable information because they reflect metabolism in motion [1].
Measurement of metabolic fluxes remains challenging because fluxes are intangible quantities that must be derived from directly measurable quantities such as concentrations. To infer fluxes in biological systems, researchers have used time-course or end-point measurement of extracellular metabolites in culture media. This approach reveals nutrient uptake and product secretion rates, but fails to inform intracellular and in vivo metabolic fluxes. To overcome this shortfall, isotope tracers are fed or infused into cells and animals. As isotope-labeled substrates traverse metabolism, the tracers leave metabolites with isotope labeling patterns that imply relative metabolic fluxes (e.g., relative pathway usage). Although these flux quantification approaches provide deeper insight into biological systems than metabolite levels, relative intracellular fluxes only partially depict dynamic metabolic states.
Absolute metabolic fluxes offer richer information about biological systems because they are physical quantities that connect biology to kinetic and thermodynamic principles. Absolute flux quantitation, however, often requires integration of multiple intracellular and extracellular measurements (Figure 1). Integrative analysis involves thoughtful design of experiments (e.g., choice of isotope tracers and sampling time points), generation of complementary data (e.g., transport fluxes and internal flux ratios) using various analytical tools, and computational frameworks that connect disparate data mathematically [1–3]. The resulting absolute flux quantifications provide a systems-level, coherent, and dynamic view of metabolism [4,5].
Figure 1. The scope of various quantitative tools for quantitation of metabolite levels and fluxes.
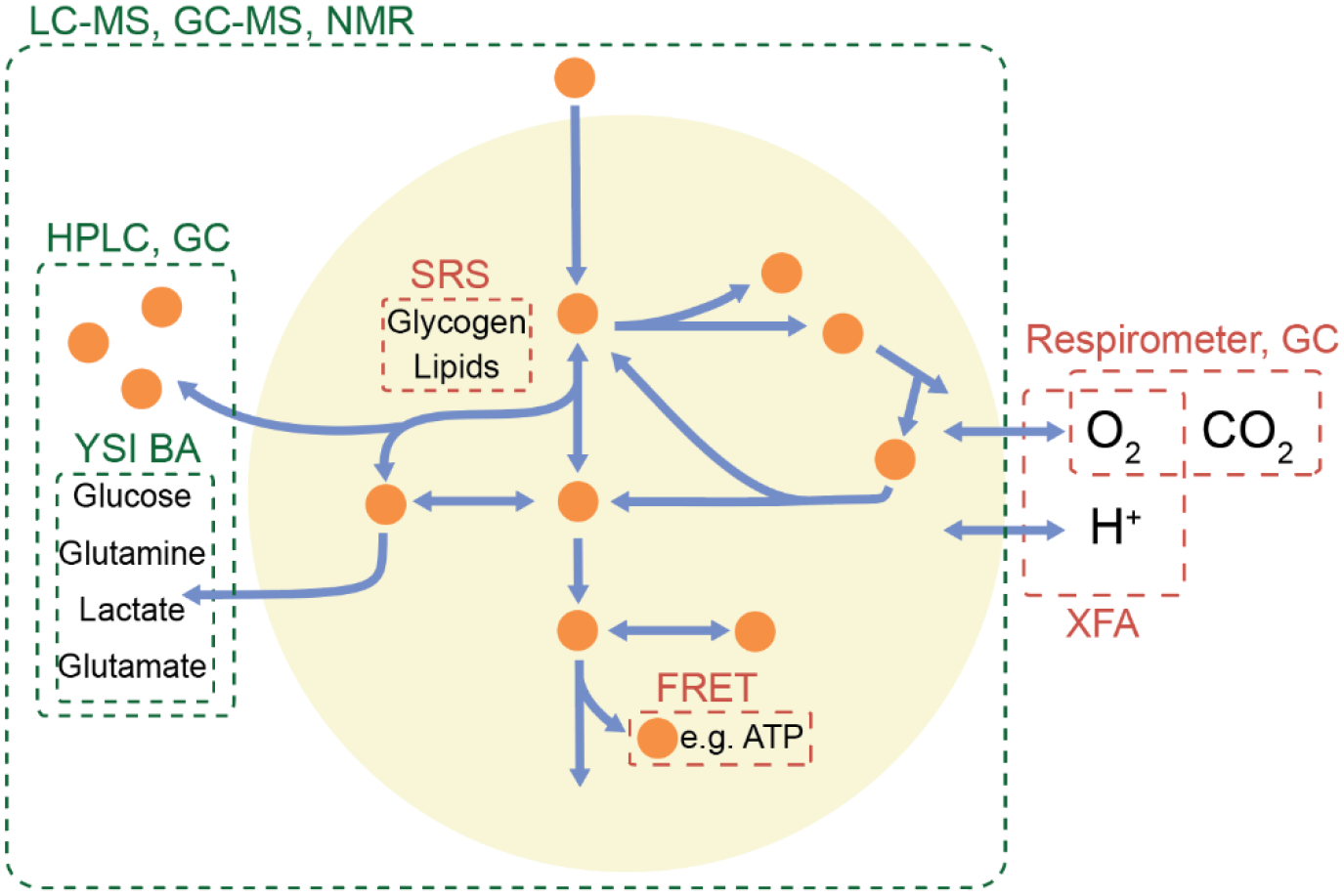
Both intracellular and extracellular metabolite measurements are needed to calculate absolute metabolic fluxes. Different tools can be used to measure compounds in different parts of metabolism. Tools shown in green measure concentrations, which can be converted to fluxes using time-course measurements and stable isotope tracers, while tools in red can directly measure rates. These tools can then be integrated to determine the fluxes across multiple metabolic pathways. LC-MS, liquid chromatography-mass spectrometry; GC-MS, gas chromatography-mass spectrometry; NMR, nuclear magnetic resonance; HPLC, high performance-liquid chromatography; GC, gas chromatography; YSI BA, Yellow Springs Instruments biochemical analyzer; SRS, stimulated Raman scattering; FRET, fluorescence resonance energy transfer; and XFA, Seahorse extracellular flux analyzer.
Metabolic flux quantitation is ever-increasing in demand as biological research questions become increasingly complex. To meet this demand, development of more sophisticated and accessible techniques amenable to integrative flux analysis remains an active area of research. Recent advances in understanding human disease and augmenting metabolic engineering are attributable to our ability to quantitate metabolic fluxes. Here we highlight various analytical tools and techniques for the quantitation of absolute metabolic fluxes and their applications in biotechnology and medicine.
Integrative framework for quantitation of absolute metabolic fluxes
Absolute metabolic fluxes describe phenotypes with a glimpse of underlying mechanisms. Various metabolite and flux measurements from different analytical techniques can be mathematically combined into a coherent set of absolute fluxes using computational tools (Figure 2). For example, nutrient uptake and respiration rates using enzyme-based biosensors and extracellular flux analyzers provide transport fluxes in physical units. Isotope labeling data from mass spectrometry reveal the relative intracellular flux distributions, which can take on physical units by scaling to the measured transport fluxes. These data work in concert to piece together a more holistic quantitation of metabolism in action.
Figure 2. Integrating measurements from multiple techniques enables quantitative and comprehensive description of metabolism.
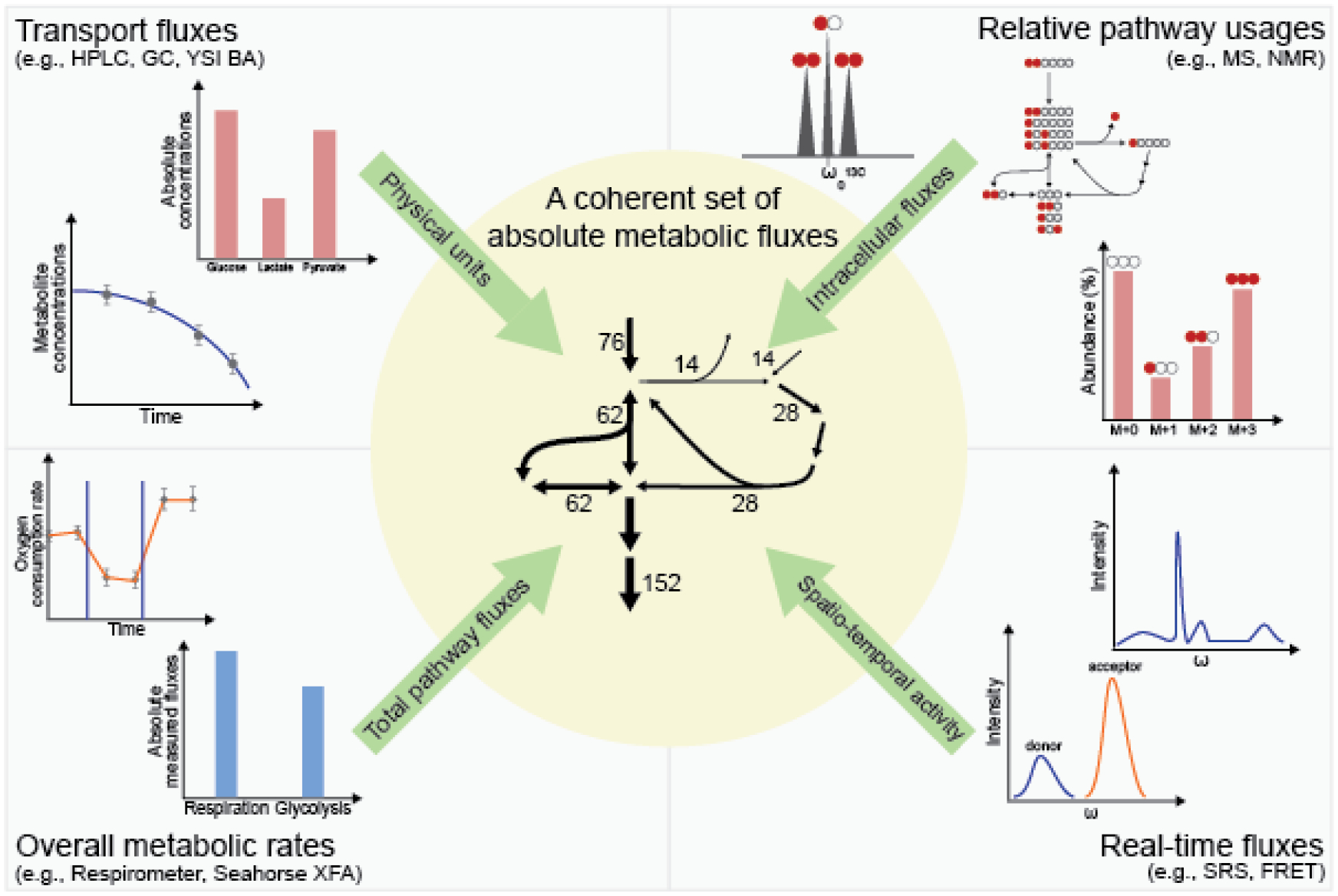
Different analytical techniques offer distinct metabolomic or fluxomic information. Transport fluxes provide physical units and can be measured by time-course measurement of extracellular metabolite concentrations using HPLC, GC, or enzyme-based biosensors (upper left). Isotope tracing techniques using MS or NMR give internal flux ratios that inform relative pathway usage (upper right). Tools such as the Seahorse XFA senses changes in environmental conditions like dissolved O2 and pH to compute overall metabolic rates (lower left). Techniques like FRET and SRS can characterize metabolic activity in real-time in live cells (lower right). Computational tools integrate these information to produce metabolic fluxes on a systems level (center).
Integrative flux quantitation requires mathematical models of metabolic networks and computational tools. Flux balance analysis (FBA) explores the feasible flux space defined by imposing mass balance and steady state constraints on metabolite levels and finds the set of fluxes that optimizes an imposed cellular objective, such as maximizing biomass production. Measurement of transport fluxes, thermodynamics, and gene expression data are commonly used to constrain the feasible flux space [6]. Metabolic flux analysis (MFA) finds the set of fluxes that best recapitulate the observed growth rate, transport fluxes, and isotope labeling patterns across metabolites resulting from stable isotope tracers [5]. Isotopically non-stationary (INST) MFA combines absolute metabolite concentrations with kinetic isotope labeling information for absolute fluxes [7]. Kinetic flux profiling (KFP) is an example of INST MFA that converts the time development of metabolite isotope labeling and absolute concentration into flux in a linear pathway [8]. These isotope-based flux quantitation frameworks impose mass balances on all isotopologues of each metabolite and assume metabolic pseudo-steady state. When this assumption is deemed inappropriate, dynamic metabolic flux analysis (DMFA) enables quantitation of changing fluxes by kinetic and data-driven modeling [9,10]. These computational tools provide mathematical frameworks for integrative analysis and continue to improve [11].
Further incorporation of proteomics and transcriptomics into these flux quantitation frameworks deepens our understanding of cellular physiology [12]. Low or high expression levels of enzymes intrinsically set lower or upper bounds for fluxes. Thus, proteomics and transcriptomics can be integrated into constraint-based modeling to improve flux predictions [13,14]. The development of new techniques in biological imaging and targeted ionization for mass spectrometry has the potential to provide spatiotemporal insights into metabolic activity [15–17]. Analytical tools that yield complementary datasets facilitate integrative analysis and reveal an indispensable dimension of metabolic fluxes in characterizing phenotypes.
Analytical toolset for quantifying metabolic fluxes
Metabolic fluxes can be inferred by the changing levels of extracellular metabolites and the isotopic labeling patterns of intracellular and circulating metabolites. Some analytical tools, such as Seahorse Extracellular Flux Analyzer (XFA) and respirometer, report fluxes directly by converting raw measurements into rates in the background. Various analytical techniques have respective strengths and weaknesses and thus excel in different aspects of metabolite and flux measurement (Figure 3a). No single analytical technique is a one-stop solution to comprehensive flux quantitation (Figure 3b).
Figure 3. Analytical toolbox for absolute metabolic flux quantification.
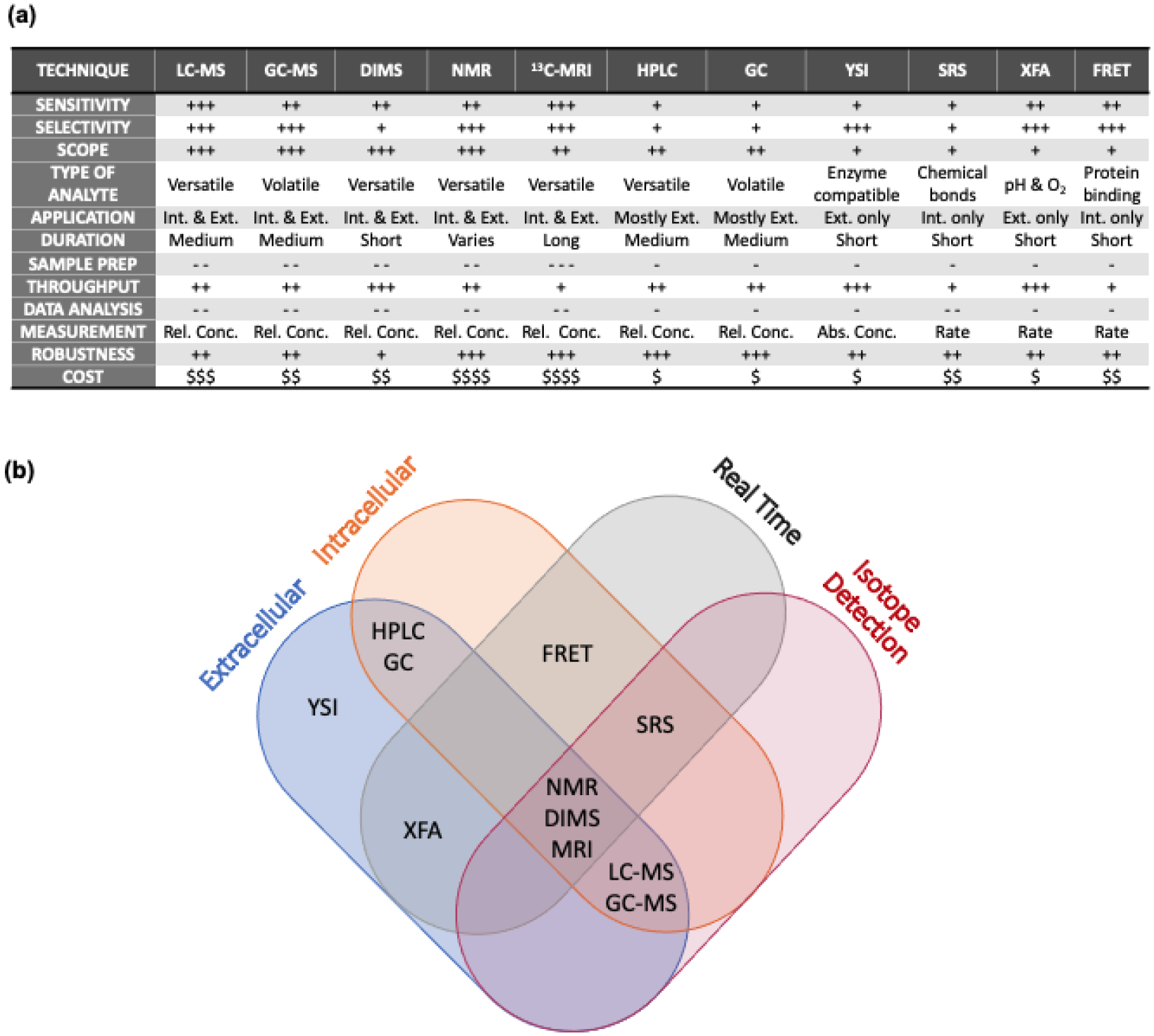
Each tool provides different information that can be integrated to calculate absolute metabolic fluxes and determine a full picture of metabolism. (a) Table comparing the strengths and weaknesses of 11 commonly used and emerging analytical tools. The scope relates to the breadth of analytes that can be measured, which in turn reflects the number of fluxes that can be quantified. (b) Venn diagram showing the types of information provided by each tool. DIMS, direct-injection mass spectrometry; and 13C MRI, hyperpolarized 13C magnetic resonance imaging.
Liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), and nuclear magnetic resonance spectroscopy (NMR) are widely used for comprehensive metabolite and flux quantitation. LC-MS and GC-MS are integral techniques capable of measuring analytes based on their physicochemical properties [18]. NMR is a nondestructive method that identifies and quantifies analytes based on the resonance frequencies characteristic of functional groups [19]. These properties enable reliable measurements of intracellular metabolites and, when enriched with isotopic tracers, their labeling patterns. Steady-state isotope labeling data reveals relative metabolic fluxes because flux distributions determine the fate of atoms. However, the measurements using these techniques must be converted to physical units by comparing to standards or well-characterized analytes of E. coli, yeast, or a mammalian cell line [20]. LC-MS, GC-MS, and NMR are excellent omics tools but require supplementation with external flux measurements to confer absolute flux quantitation in physical units.
Complementary tools allow absolute quantitation of metabolic fluxes. Enzyme-based amperometric biosensors (e.g., YSI Biochemistry Analyzer) use electrochemical probes and immobilized enzymes to rapidly measures absolute levels of select metabolites with minimal sample preparation. Even without MS, HPLC (e.g., with ultraviolet or refractive index detectors) and GC (e.g., with flame ionization detector) excel at quantifying many metabolites based on known concentrations and retention times of standards. By quantitating metabolite levels over time, these tools generate uptake and secretion fluxes [21].
Other tools may measure metabolic fluxes in real-time. The Seahorse Extracellular Flux Analyzer (XFA) measures pH and dissolved oxygen in culture media to assess cells’ extracellular acidification (ECAR) and oxygen consumption rate (OCR), which correspond to the rates of glycolysis and mitochondrial respiration [22]. Hyperpolarized (HP) 13C magnetic resonance imaging (MRI) has facilitated in vivo studies of metabolic flux. MRI detects the 1H NMR signal of endogenous water. Adding a hyperpolarized 13C probe enhances signal contrast to map metabolic activity based on enzymatic conversion of an injected 13C tracer. HP 13C MRI results in real-time absolute fluxes of select pathways in living organisms with a 10,000-fold increase in signal-to-noise ratio compared to conventional MRI [23,24].
Spatial resolution of metabolic activity can be accomplished by mass spectrometry. Matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a crystallized matrix of molecules to ionize biological analytes with a laser beam selectively targeting different locations. For higher spatial resolution, image-based nanoscale secondary ion mass spectrometry (nanoSIMS) is a promising technique for analyzing metabolic activity at the single-cell level. NanoSIMS is an ion microprobe technique that scans a cell surface with a primary ion beam, resulting in secondary electron and secondary ion images of different elemental and isotopic compositions. Its spatial resolution can reach the sub-micron range in the regime of single-cell metabolism [25].
Förster resonance energy transfer (FRET) and stimulated Raman scattering (SRS) microscopy allow measurement of intracellular fluxes at both cellular and subcellular resolution. FRET measures the rate of energy transfer between coupled fluorophores caused by conformational changes of proteins in response to protein-metabolite interactions, enabling flux measurements of single enzymatic steps [15]. SRS quantifies intracellular metabolites based on the vibrational characteristics of chemical bonds [16]. SRS microscopy is an emerging tool for studying metabolic fluxes using stable isotopes with limited biochemical perturbation and does not rely on auxiliary molecular labeling. Active development of SRS techniques to quantitate fluxes has led to real-time analysis of glucose metabolism, fatty acid synthesis, and compartmental metabolism [26–28]. Integration of these analytical techniques facilitates absolute flux quantitation across various metabolic pathways.
Absolute fluxes as a new dimension of phenotypes
This section highlights recent works utilizing integrative metabolic flux analysis in various fields (Figure 4).
Figure 4. Integrative metabolic flux analysis provides actionable information to advance biotechnology and medicine.
Quantitation of metabolic fluxes in cell culture systems, animal models, and clinical studies drives efficient bio-derived product synthesis and advances our understanding of health and disease. Created with BioRender.com
-. Metabolic engineering and biotechnology
Fluxomics has empowered metabolic engineering and biotechnology by revealing limiting factors in microbial metabolism and bioprocesses. Using 13C-MFA, Yao et al. found that production of acetol in E. coli is limited by NADPH supply [29]. Knowledge of metabolic flux distributions allows identification of competing metabolic pathways that divert fluxes from the productive path. Using INST-MFA analysis, Cheah et al. quantified correlations between multiple routes for pyruvate metabolism and overall aldehyde production in S. elongatus. Knocking down the pathways that are negatively correlated to aldehyde production resulted in a 50% increase in productivity [30]. These techniques also lead to rational design of culture strategies. Measurements of external metabolites in Chinese Hamster Ovary (CHO) cell cultures have served as constraints on transport fluxes in FBA to quantitatively simulate different feeding strategies in silico [31]. The upshot was a markedly improved CHO cell culture strategy for the production of biopharmaceuticals. In another bioproduct synthesis study, using 13C isotope tracing and flux analysis, Cheng et al. enhanced the production of hyaluronic acid, a high-value polymerizable bioproduct, in C. glutamicum [32].
As analytical and computational techniques for absolute flux quantitation become more standardized and accessible, efforts to increase their throughput are on the rise. Automated robotic platforms that can rapidly quantify fluxes en masse have been used to demonstrate the robustness of central carbon pathways across 180 E. coli strains [33]. Metabolic fluxes through synthesis pathways in cell-free systems can also be analyzed in high-throughput with the use of self-assembled monolayers for MALDI-based mass spectrometry. O’Kane et al. developed this technique to optimize the cell-free synthesis of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA), a key precursor to isoprenoids [34]. High-throughput approaches are in active development and accelerate the design-build-test-learn cycle in synthetic biology. Automated biofoundries that adopt high-throughput synthetic biology approaches facilitate the discovery and robust production of a wide array of bio-inspired materials monomers (e.g., mandelic and hydroxymandelic acid) [35]. Integrating quantitative flux analysis into biofoundries will boost the synthesis of target products via pathway and culture optimization. For example, with knowledge of carbon fluxes, enhancement of erythromycin synthesis was achieved through redirection of carbons towards biosynthetic precursors [36]. Biofoundries increasingly combine quantitative metabolic flux data with rapid strain engineering to synergistically produce next-generation monomers of growing complexity and value [37].
Fluxomics has expanding applicability in biotechnology. Glycolytic and fatty acid fluxes have recently been used as a parameter to assess human skeletal muscle models built from 3D-printed tissue biomaterials [38]. Measuring fluxes in cells from bio-printed scaffolds allows researchers to better design biomaterials that can rigorously model human tissues or be safely incorporated into humans. Fluxomics is growing as a tool for enhancing bioproduct synthesis and tissue engineering as well as for emerging research avenues across biotechnology.
-. Nutrition, health, and medicine
Quantitative knowledge of metabolic fluxes enhances our understanding of health and disease. The study of many nutrition related diseases such as type 2 diabetes, obesity, and nonalcoholic fatty liver disease (NAFLD) benefit from metabolic flux analysis. For instance, experiments feeding [6,6-2H2]glucose into mice with a fat and sugar rich diet showed that NAFLD progresses to nonalcoholic steatohepatitis (NASH) through upregulation of hepatic tricarboxylic acid (TCA) flux and anaplerosis [39]. A ketogenic diet, in comparison, decreased hepatic mitochondrial flux and alleviated NAFLD in obese human subjects [40]. In a double-blind randomized human trial, long term consumption of beverages sweetened with fructose and sucrose, but not glucose, altered lipid metabolism to increase liver fat, a hallmark of developing NAFLD [41]. These repercussions may be curbed by controlling fructose intake to be slower than intestinal clearance capacity [42]. Increased use of quantitative flux analysis in vivo has revealed new insights on whole-body metabolism and nutrition.
Advancements in fluxomics techniques have shed light on the dynamic nature of metabolism. LC-MS-based quantitation of fluxes in mice showed that, although some specialized tissues rapidly catabolize circulating glucose to produce circulating lactate, glucose is mainly stored as glycogen that is later broken down for necessary glycolytic intermediates [43]. When liver-based gluconeogenesis is disrupted by knockout of cytosolic phosphoenolpyruvate carboxykinase in mice, glucose homeostasis is maintained by upregulating renal gluconeogenesis [44]. In hepatocellular carcinoma and NAFLD, nonfunctional glycine N-methyltransferase is the leading cause for disturbed hepatic gluconeogenesis and diverted gluconeogenic precursors to fuel lipogenic polyamine and transsulfuration pathways [45]. Another study demonstrated that glucose is the primary nutrient source to the TCA cycle in physiological contexts across different animal strains and tissues [46]. In hepatic gluconeogenesis, the TCA cycle is maintained via anaplerosis by mitochondrial pyruvate carboxylase (PC) activity. Loss of hepatic PC function, as in the case of hypoglycemia, resulted in upshifted ketogenesis to maintain beta-oxidation and TCA cycling [47]. Another cardiac study that traced [U-13C]glucose revealed that pressure and volume overload increases pyruvate and lactate flux in the heart [48]. Arnould et al. found that the loss of cellular prion protein (PrPC) in neurodegenerative diseases reduces glycolytic flux relative to fatty acid degradation and induces oxidative stress [49]. These examples highlight the role of quantitative flux analysis in advancing our understanding of complex biological systems.
Metabolic flux quantitation provides new insights into therapeutic target discovery and understanding the mechanism of action of drugs. Metformin, a hypoglycemic drug, lowers endogenous glucose production by modulating the redox state and suppressing hepatic gluconeogenesis [50]. In rats treated with metformin, differential inhibition of metabolic fluxes from different gluconeogenic substrates lactate and glycerol depended on cytosolic NADH levels. In another study, metformin lowered the age-related inflammation in CD4+ T-cells ex vivo and promoted compensatory oxidative phosphorylation over glycolysis [51]. Dichloroacetate (DCA) is another widely used drug that activates pyruvate dehydrogenase and promotes apoptosis by inhibiting pyruvate dehydrogenase kinase. Increased TCA cycle flux upon DCA treatment led to metabolic reprogramming in tumor cells and activated glutamine anaplerosis [52]. Subsequently, a multidrug treatment combining DCA with a glutaminase inhibitor improved inhibition of tumor growth [52]. In addition, DCA treatment decreased glycolytic flux [53] that is compensated by increased pentose phosphate pathway flux and serine synthesis flux in a leukemia cell line, thus offering an additional chemotherapeutic target for multidrug treatments [54].
Imaging and microscopy augment metabolic flux quantitation in higher eukaryotes by adding spatial and real-time information. Using HP 13C MRI to measure the production of hyperpolarized 13C-lactate upon hyperpolarized 13C-pyruvate injection, visualization of glycolytic flux in human metastatic prostate cancer captured therapeutic response [55]. In mice, HP 13C MRI was used to assess neuroinflammation by measuring pyruvate-to-lactate conversion [56]. Microscopy-based flux quantitation enables cellular and subcellular resolution of metabolic activity. A FRET-based method for quantitation of intracellular glycolytic metabolites was developed to measure glycolytic flux in motion in individual endothelial cells [57]. This study showed how cell motility was a result of glycolysis-driven cytoskeleton assembly. RNA-based sensors are another solution to metabolic heterogeneity found in multicellular systems. To measure glycolytic flux in single cells, Ortega et al. developed an RNA-based metabolite sensor for measuring intracellular fructose-1,6-bisphosphate whose concentration correlates with glycolytic flux [58]. These emerging techniques unveil the truly dynamic nature of metabolism and pave new ways to deconstruct metabolic heterogeneity. Therefore, full integration of imaging and microscopy with metabolic flux quantitation is a promising future research direction.
Conclusion
Metabolic fluxes offer uniquely valuable information about biological systems because they quantify pathway activities and provide regulatory insights. Knowledge of both metabolite concentrations and fluxes would bring us closer to the full picture of dynamic metabolism. Recent advances in analytical techniques and computational tools have rendered flux quantitation more accessible than ever before. The easier access to metabolic flux information has aided the advancement of metabolic engineering and therapeutic development as well as our understanding of metabolic regulation in health and disease. Nonetheless, comprehensive absolute metabolic flux quantitation remains a specialized method, requiring case-by-case consideration of experimental design and integration of multiple analytical and computational techniques. In the future, technological advances may streamline absolute flux quantitation at the subcellular as well as the systems level, and in real time. Broad adoption of fluxes (i.e., utilizing rates as well as levels) as a means of phenotypic characterization, discovery, and engineering would accelerate and revolutionize biotechnology and medicine.
Highlights.
Integrating analytical techniques leads to absolute metabolic flux quantitation.
Emerging techniques enable single-cell and real-time flux quantitation.
Metabolic fluxes offer insights into metabolic control.
Application of robust metabolic control contributes to biotechnology and medicine.
A promising future direction is spatiotemporal resolution of metabolic fluxes.
Acknowledgements
The authors would like to thank the members of the Park lab and the UCLA Metabolomics Center for helpful discussion. This work was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number R35GM143127 and the BioPACIFIC Materials Innovation Platform of the National Science Foundation under Award Number DMR-1933487. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the National Science Foundation.
References
• Of special interest
•• Of outstanding interest
- 1.Dai Z, Locasale JW: Understanding metabolism with flux analysis: From theory to application. Metabolic engineering 2017, 43:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen DK, Young JD: Tracing metabolic flux through time and space with isotope labeling experiments. Current opinion in biotechnology 2020, 64:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long CP, Antoniewicz MR: High-resolution 13C metabolic flux analysis. Nature Protocols 2019, 14:2856–2877. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Wondisford FE, Song C, Zhang T, Su X: Metabolic Flux Analysis—Linking Isotope Labeling and Metabolic Fluxes. Metabolites 2020, Vol 10, Page 447 2020, 10:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniewicz MR: A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications. Metabolic Engineering 2021, 63:2–12. [DOI] [PubMed] [Google Scholar]
- 6.Orth JD, Thiele I, Palsson BO: What is flux balance analysis? Nature Biotechnology 2010 28:3 2010, 28:245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheah YE, Young JD: Isotopically nonstationary metabolic flux analysis (INST-MFA): putting theory into practice. Current Opinion in Biotechnology 2018, 54:80–87. [DOI] [PubMed] [Google Scholar]
- 8.Yuan J, Bennett BD, Rabinowitz JD, Scarino J, Munger J, Shenk T: Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nature Protocols 2008 3:8 2008, 3:1328–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniewicz MR: Dynamic metabolic flux analysis — tools for probing transient states of metabolic networks. Current Opinion in Biotechnology 2013, 24:973–978. [DOI] [PubMed] [Google Scholar]
- 10.Feng X, Xu Y, Chen Y, Tang YJ: Integrating Flux Balance Analysis into Kinetic Models to Decipher the Dynamic Metabolism of Shewanella oneidensis MR-1. PLOS Computational Biology 2012, 8:e1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rahim M, Ragavan M, Deja S, Merritt ME, Burgess SC, Young JD: INCA 2.0: A tool for integrated, dynamic modeling of NMR- and MS-based isotopomer measurements and rigorous metabolic flux analysis. Metabolic Engineering 2022, 69:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santiago-Rodriguez TM, Hollister EB: Multi ‘omic data integration: A review of concepts, considerations, and approaches. Seminars in Perinatology 2021, 45:151456. [DOI] [PubMed] [Google Scholar]
- 13.Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V, et al. : Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nature Protocols 2019 14:3 2019, 14:639–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gopalakrishnan S, Maranas CD: 13C metabolic flux analysis at a genome-scale. Metabolic Engineering 2015, 32:12–22. [DOI] [PubMed] [Google Scholar]
- 15.Soleja N, Manzoor O, Nandal P, Mohsin Mohd: FRET-based nanosensors for monitoring and quantification of alcohols in living cells. Organic & Biomolecular Chemistry 2019, 17:2413–2422. [DOI] [PubMed] [Google Scholar]
- 16.Hu F, Shi L, Min W: Biological imaging of chemical bonds by stimulated Raman scattering microscopy. Nature Methods 2019 16:9 2019, 16:830–842. [DOI] [PubMed] [Google Scholar]
- 17.Nuñez J, Renslow R, CliffIII JB, Anderton CR: NanoSIMS for biological applications: Current practices and analyses. Biointerphases 2017, 13:03B301. [DOI] [PubMed] [Google Scholar]
- 18.Mairinger T, Sanderson J, Hann S: GC–QTOFMS with a low-energy electron ionization source for advancing isotopologue analysis in 13 C-based metabolic flux analysis. Analytical and Bioanalytical Chemistry 2019, 411:1495–1502. [DOI] [PubMed] [Google Scholar]
- 19.Emwas A-H, Roy R, McKay RT, Tenori L, Saccenti E, Gowda GAN, Raftery D, Alahmari F, Jaremko L, Jaremko M, et al. : NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JO, Rubin SA, Xu Y-F, Amador-Noguez D, Fan J, Shlomi T, Rabinowitz JD: Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nature Chemical Biology 2016 12:7 2016, 12:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. •.Moiz B, Garcia J, Basehore S, Sun A, Li A, Padmanabhan S, Albus K, Jang C, Sriram G, Clyne AM: 13C Metabolic Flux Analysis Indicates Endothelial Cells Attenuate Metabolic Perturbations by Modulating TCA Activity. Metabolites 2021, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Authors use 13C Metabolic Flux Analysis with measurements from LC-MS and YSI to study the effects of metabolic inhibitors of polyol, pentose phosphate and hexosamine biosynthetic pathways on endothelial metabolism in human umbilical vein endothelial cells. Fidarestat, DHEA and Azaserine were found to increase TCA activity.
- 22.Perry CGR, Kane DA, Lanza IR, Neufer PD: Methods for Assessing Mitochondrial Function in Diabetes. Diabetes 2013, 62:1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardenkjaer-Larsen JH, Leach AM, Clarke N, Urbahn J, Anderson D, Skloss TW: Dynamic nuclear polarization polarizer for sterile use intent. NMR in Biomedicine 2011, 24:927–932. [DOI] [PubMed] [Google Scholar]
- 24.Granlund KL, Tee SS, Vargas HA, Lyashchenko SK, Reznik E, Fine S, Laudone V, Eastham JA, Touijer KA, Reuter VE, et al. : Hyperpolarized MRI of Human Prostate Cancer Reveals Increased Lactate with Tumor Grade Driven by Monocarboxylate Transporter 1. Cell Metabolism 2020, 31:105–114.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chadwick GL, Otero FJ, Gralnick JA, Bond DR, Orphan VJ: NanoSIMS imaging reveals metabolic stratification within current-producing biofilms. Proceedings of the National Academy of Sciences of the United States of America 2019, 116:20716–20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du J, Su Y, Qian C, Yuan D, Miao K, Lee D, Ng AHC, Wijker RS, Ribas A, Levine RD, et al. : Raman-guided subcellular pharmaco-metabolomics for metastatic melanoma cells. Nature Communications 2020 11:1 2020, 11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shou J, Oda R, Hu F, Karasawa K, Nuriya M, Yasui M, Shiramizu B, Min W, Ozeki Y: Super-multiplex imaging of cellular dynamics and heterogeneity by integrated stimulated Raman and fluorescence microscopy. iScience 2021, 24:102832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. ••.Zhang L, Shi L, Shen Y, Miao Y, Wei M, Qian N, Liu Y, Min W: Spectral tracing of deuterium for imaging glucose metabolism. Nature Biomedical Engineering 2019 3:5 2019, 3:402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study developed a microscopy technique that used stimulated Raman-scattering imaging of deuterium to trace glucose anabolic utilization in tumor, brain, intestine, and liver mouse tissues.
- 29. ••.Yao R, Li J, Feng L, Zhang X, Hu H: 13C metabolic flux analysis-guided metabolic engineering of Escherichia coli for improved acetol production from glycerol. Biotechnology for Biofuels 2019, 12:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. ••.Cheah YE, Xu Y, Sacco SA, Babele PK, Zheng AO, Johnson CH, Young JD: Systematic identification and elimination of flux bottlenecks in the aldehyde production pathway of Synechococcus elongatus PCC 7942. Metabolic Engineering 2020, 60:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cheah et. al rationally engineered 50% higher aldehyde production in S. elongatus using GC-MS based MFA data and a design-build-test-learn approach to strain improvement.
- 31.Huang Z, Xu J, Yongky A, Morris CS, Polanco AL, Reily M, Borys MC, Li ZJ, Yoon S: CHO cell productivity improvement by genome-scale modeling and pathway analysis: Application to feed supplements. Biochemical Engineering Journal 2020, 160:107638. [Google Scholar]
- 32.Cheng F, Yu H, Stephanopoulos G: Engineering Corynebacterium glutamicum for high-titer biosynthesis of hyaluronic acid. Metabolic Engineering 2019, 55:276–289. [DOI] [PubMed] [Google Scholar]
- 33.Bergès C, Cahoreau E, Millard P, Enjalbert B, Dinclaux M, Heuillet M, Kulyk H, Gales L, Butin N, Chazalviel M, et al. : Exploring the Glucose Fluxotype of the E. coli y-ome Using High-Resolution Fluxomics. Metabolites 2021, Vol 11, Page 271 2021, 11:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Kane PT, Dudley QM, McMillan AK, Jewett MC, Mrksich M: High-throughput mapping of CoA metabolites by SAMDI-MS to optimize the cell-free biosynthesis of HMG-CoA. Science Advances 2019, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson CJ, Carbonell P, Jervis AJ, Yan C, Hollywood KA, Dunstan MS, Currin A, Swainston N, Spiess R, Taylor S, et al. : Rapid prototyping of microbial production strains for the biomanufacture of potential materials monomers. Metabolic Engineering 2020, 60:168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You D, Wang M-M, Yin B-C, Ye B-C: Precursor Supply for Erythromycin Biosynthesis: Engineering of Propionate Assimilation Pathway Based on Propionylation Modification. ACS Synthetic Biology 2019, doi: 10.1021/ACSSYNBIO.8B00396. [DOI] [PubMed] [Google Scholar]
- 37.Lu Z, Zhang X, Dai J, Wang Y, He W: Engineering of leucine-responsive regulatory protein improves spiramycin and bitespiramycin biosynthesis. Microbial Cell Factories 2019 18:1 2019, 18:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khodabukus A, Madden L, Prabhu NK, Koves TR, Jackman CP, Muoio DM, Bursac N: Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle. Biomaterials 2019, 198:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasenour CM, Kennedy AJ, Bednarski T, Trenary IA, Eudy BJ, da Silva RP, Boyd KL, Young JD: Vitamin E does not prevent Western diet-induced NASH progression and increases metabolic flux dysregulation in mice. Journal of Lipid Research 2020, 61:707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luukkonen PK, Dufour S, Lyu K, Zhang XM, Hakkarainen A, Lehtimäki TE, Cline GW, Petersen KF, Shulman GI, Yki-Järvinen H: Effect of a ketogenic diet on hepatic steatosis and hepatic mitochondrial metabolism in nonalcoholic fatty liver disease. Proceedings of the National Academy of Sciences of the United States of America 2020, 117:7347–7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geidl-Flueck B, Hochuli M, Németh Á, Eberl A, Derron N, Köfeler HC, Tappy L, Berneis K, Spinas GA, Gerber PA: Fructose- and sucrose- but not glucose-sweetened beverages promote hepatic de novo lipogenesis: A randomized controlled trial. Journal of Hepatology 2021, 75:46–54. [DOI] [PubMed] [Google Scholar]
- 42.Jang C, Wada S, Yang S, Gosis B, Zeng X, Zhang Z, Shen Y, Lee G, Arany Z, Rabinowitz JD: The small intestine shields the liver from fructose-induced steatosis. Nature Metabolism 2020, 2:586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.TeSlaa T, Bartman CR, Jankowski CSR, Zhang Z, Xu X, Xing X, Wang L, Lu W, Hui S, Rabinowitz JD: The Source of Glycolytic Intermediates in Mammalian Tissues. Cell Metabolism 2021, 33:367–378.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. •.Rahim M, Hasenour CM, Bednarski TK, Hughey CC, Wasserman DH, Young JD: Multitissue 2H/13C flux analysis reveals reciprocal upregulation of renal gluconeogenesis in hepatic PEPCK-C–knockout mice. JCI Insight 2021, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors established an isotopic flux modeling approach that can quantify hepatic and renal TCA activity and gluconeogenic and anaplerotic fluxes in parallel. This article uncovered the shared role of liver and kidney in maintaining glucose homeostasis via regulation of gluconeogenic flux through PEPCK-C.
- 45.Hughey CC, Trefts E, Bracy DP, James FD, Donahue EP, Wasserman DH: Glycine N-methyltransferase deletion in mice diverts carbon flux from gluconeogenesis to pathways that utilize excess methionine cycle intermediates. Journal of Biological Chemistry 2018, 293:11944–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. •.Liu S, Dai Z, Cooper DE, Kirsch DG, Locasale JW: Quantitative Analysis of the Physiological Contributions of Glucose to the TCA Cycle. Cell Metabolism 2020, 32:619–628.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors showed that, TCA cycle flux is reduced and renal gluconeogenesis and ketogenesis activated when liver gluconeogenesis is suppressed in pyruvate carboxylase (PC) knockout mice model. Loss of PC led to increased oxidative stress and elevated inflammation in liver.
- 47. •.Cappel DA, Deja S, Duarte JAG, Kucejova B, Iñigo M, Fletcher JA, Fu X, Berglund ED, Liu T, Elmquist JK, et al. : Pyruvate-Carboxylase-Mediated Anaplerosis Promotes Antioxidant Capacity by Sustaining TCA Cycle and Redox Metabolism in Liver. Cell Metabolism 2019, 29:1291–1305.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This integrative study combined LC-MS, GC-MS, 1H NMR, and Seahorse EFA to validate the role of mitochondrial pyruvate carboxylase (PC) in gluconeogenesis. The authors found that PC is essential for maintain biosynthesis, oxidation, and urea cycle function.
- 48.Schnelle M, Chong M, Zoccarato A, Elkenani M, Sawyer GJ, Hasenfuss G, Ludwig C, Shah AM: In vivo [U-13 C]glucose labeling to assess heart metabolism in murine models of pressure and volume overload. Am J Physiol Heart Circ Physiol 2020, 319:422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. •.Arnould H, Baudouin V, Baudry A, Ribeiro LW, Ardila-Osorio H, Pietri M, Caradeuc C, Soultawi C, Williams D, Alvarez M, et al. : Loss of prion protein control of glucose metabolism promotes neurodegeneration in model of prion diseases. PLOS Pathogens 2021, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mass spectrometry, NMR and Seahorse measurements are integrated to elucidate the role of cellular prion protein (PrPC) in brain metabolism and how the loss of PrPC function contributes to neurodegenerative diseases. The study found that loss of PrPC reduces glycolytic flux, increases fatty acid degradation and increases oxidative stress.
- 50.Madiraju AK, Qiu Y, Perry RJ, Rahimi Y, Zhang XM, Zhang D, Camporez JPG, Cline GW, Butrico GM, Kemp BE, et al. : Metformin inhibits gluconeogenesis via a redox-dependent mechanism in vivo. Nature Medicine 2018, 24:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. •.Bharath LP, Agrawal M, McCambridge G, Nicholas DA, Hasturk H, Liu J, Jiang K, Liu R, Guo Z, Deeney J, et al. : Metformin Enhances Autophagy and Normalizes Mitochondrial Function to Alleviate Aging-Associated Inflammation. Cell Metabolism 2020, 32:44–55.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]; A relationship between metabolic fluxes and age-related inflammation were established using T cells isolated from human subjects. Metformin reversed inflammation in Th17 inflammation in T cells by promoting compensatory oxidative phosphorylation over glycolysis.
- 52.Schoonjans CA, Joudiou N, Brusa D, Corbet C, Feron O, Gallez B: Acidosis-induced metabolic reprogramming in tumor cells enhances the anti-proliferative activity of the PDK inhibitor dichloroacetate. Cancer Letters 2020, 470:18–28. [DOI] [PubMed] [Google Scholar]
- 53.Feuerecker B, Biechl P, Veltkamp C, Saur D, Eisenreich W: Metabolic response of pancreatic carcinoma cells under treatment with dichloroacetate. Metabolites 2021, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cha JW, Jin X, Jo S, An YJ, Park S: Metabolic mechanisms of a drug revealed by distortion-free13C tracer analysis. Chemical Science 2021, 12:4958–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen H-Y, Aggarwal R, Bok RA, Ohliger MA, Zhu Z, Lee P, Gordon JW, van Criekinge M, Carvajal L, Slater JB, et al. : Hyperpolarized 13C-pyruvate MRI detects real-time metabolic flux in prostate cancer metastases to bone and liver: a clinical feasibility study. Prostate Cancer and Prostatic Diseases 2019 23:2 2019, 23:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.le Page LM, Guglielmetti C, Najac CF, Tiret B, Chaumeil MM: Hyperpolarized 13C magnetic resonance spectroscopy detects toxin-induced neuroinflammation in mice. NMR in Biomedicine 2019, 32:e4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu D, Harrison DL, Szasz T, Yeh C-F, Shentu T-P, Meliton A, Huang R-T, Zhou Z, Mutlu GM, Huang J, et al. : Single-cell metabolic imaging reveals a SLC2A3-dependent glycolytic burst in motile endothelial cells. Nature Metabolism 2021 3:5 2021, 3:714–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. ••.Ortega AD, Takhaveev V, Vedelaar SR, Long Y, Mestre-Farràs N, Incarnato D, Ersoy F, Olsen LF, Mayer G, Heinemann M: A synthetic RNA-based biosensor for fructose-1,6-bisphosphate that reports glycolytic flux. Cell Chemical Biology 2021, 28. [DOI] [PubMed] [Google Scholar]; The authors developed an RNA-based sensor to capture glycolytic flux using fructose-1,6-bisphosphate (FBP) concentration as a proxy. This emerging single-cell flux quantitation technique underscores the metabolic heterogeneity found amongst cell populations and provides a foundation for future microscopy-based analyses.


