Abstract
Among the mechanisms of suppression that T regulatory (Treg) cells exert to control the immune responses, the secretion of small extracellular vesicles (sEV) has been recently proposed as a novel contact‐independent immunomodulatory mechanism. Previous studies have demonstrated that Treg cells produce sEV, including exosomes, able to modulate the effector function of CD4+ T cells, and antigen presenting cells (APCs) such as dendritic cells (DCs) through the transfer of microRNA, cytokines, the production of adenosine, among others. Previously, we have demonstrated that Neuropilin‐1 (Nrp1) is required for Tregs‐mediated immunosuppression mainly by impacting on the phenotype and function of effector CD4+ T cells. Here, we show that Foxp3+ Treg cells secrete sEV, which bear Nrp1 in their membrane. These sEV modulate effector CD4+ T cell phenotype and proliferation in vitro in a Nrp1‐dependent manner. Proteomic analysis indicated that sEV obtained from wild type (wt) and Nrp1KO Treg cells differed in proteins related to immune tolerance, finding less representation of CD73 and Granzyme B in sEV obtained from Nrp1KO Treg cells. Likewise, we show that Nrp1 is required in Treg cell‐derived sEV for inducing skin transplantation tolerance, since a reduction in graft survival and an increase on M1/M2 ratio were found in animals treated with Nrp1KO Treg cell‐derived sEV. Altogether, this study describes for the first time that Treg cells secrete sEV containing Nrp1 and that this protein, among others, is necessary to promote transplantation tolerance in vivo via sEV local administration.
Keywords: extracellular vesicles, tolerance, treg cells, transplantation, neuropilin‐1
1. INTRODUCTION
Transplant rejection is still one major barrier to achieve long‐term allogeneic transplantation tolerance, despite established immunosuppressive drug treatment and improvements in surgical techniques, mainly due to chronic allograft rejection and toxicity associated with immunosuppression sustained for several years post‐transplantation (Safinia et al., 2015). Among several alternatives developed, the transfusion of T regulatory (Treg) cells as cell‐based therapy has been reported in several clinical studies, with additional ongoing trials pending for reporting (reviewed in detail elsewhere) (Romano et al., 2017). Treg cells are essential to maintain self‐tolerance in homeostasis and proper immune response completion, considering Treg cell‐related deficiency as the cause of autoimmune diseases and uncontrolled, undesired immune responses (Ohkura et al., 2013). These cells inhibit the effector function and proliferation of several leukocyte populations through a myriad of mechanisms (reviewed in depth by Josefowicz et al. (2012)). Treg cells are characterized by the expression of Foxp3, CD25, CD73 and Neuropilin‐1 (Nrp1), and the production of IL‐10 and IL‐35 (Wing et al., 2019).
Nrp1 is a membrane protein highly expressed in both murines and humans. Foxp3+ Treg cells (Battaglia et al., 2008; Gao et al., 2016; Weiss et al., 2012; Yadav et al., 2012). Nrp1 contains a large extracellular domain with affinity for a variety of ligands, such as semaphorins, several growth factors and even other membrane‐bound Nrp1 molecules (Campos‐Mora et al., 2013; Parker et al., 2012). Although initially a privileging function for Treg cells was attributed in the context of the immune synapse (Sarris et al., 2008), later reports described a cell‐intrinsic requirement of Nrp1 expression on Treg cells to maintain phenotypic stability and suppressor function (Overacre & Vignali, 2016). Treg cells with high Nrp1 expression have consistently shown increased suppressive function in the tumour microenvironment (Delgoffe et al., 2013; Overacre‐Delgoffe et al., 2017), sepsis (Gao et al., 2016), allogeneic materno‐foetal tolerance (Roy et al., 2017) and transplantation (Campos‐Mora et al., 2015; Yuan et al., 2013). In parallel, Nrp1 deficiency or its blockade impacts Treg cell phenotypic stability, immuno‐regulatory gene expression and suppressive function (Overacre‐Delgoffe et al., 2017). In line with these antecedents, other studies have related Nrp1 expression on T cells with immunosuppression, since mice with Nrp1 deletion on CD4+ T cells exhibited severe experimental autoimmune encephalomyelitis (EAE) (Roy et al., 2017; Solomon et al., 2011), and enhanced anti‐tumour immune response (Hansen et al., 2012). Furthermore, our recent work showed that the expression of Nrp1 is necessary on Treg cells to control IL‐10 secretion, to inhibit CD4+ T cell proliferation and pro‐inflammatory cytokine production, and to favour skin transplantation tolerance via suppressor T cells differentiation (Campos‐Mora et al., 2019).
As mentioned above, Treg cells use different mechanisms to exert immune suppression, and one of these processes, poorly studied in this T cell population, corresponds to the production of small extracellular vesicles (sEV) (Agarwal et al., 2014; Li et al., 2016). sEV comprises a heterogeneous subset of vesicles derived from endosomal compartments or plasma membrane, which shares features such as protein composition, round shape and a diameter between 30 and 200 nm (Jeppesen et al., 2019; Théry et al., 2018). sEV play an important role in intercellular communication, acting as “information packages” carrying proteins, nucleic acids, and other molecules from one cell to another, exerting an effect in recipient cells (Mathieu et al., 2019; Valadi et al., 2007). On T cells, TCR‐mediated activation has shown to increase the release of sEV, and it has been reported that in vitro activated murine Treg cells secrete more sEV in comparison with conventional T or B cells (Wen et al., 2017). Furthermore, a blockade in the ability of Treg cells to release exosomes drastically decreases its suppressor activity (Wen et al., 2017), and Treg cell‐derived sEV have shown intrinsic suppressive functions, inhibiting the proliferation and secretion of IL‐2 and IFN‐γ from activated T cells in vitro (Azimi et al., 2018; Smyth et al., 2013; Yu et al., 2013). Other studies have shown that Treg cell‐derived exosomes drive the differentiation of in vitro activated CD4+ T cells into IL‐10‐expressing Tr1‐like cells (Aiello et al., 2017) and favour the conversion of DCs to an anti‐inflammatory phenotype with increased production of IL‐10 and decreased production of IL‐6 in response to LPS treatment (Tung et al., 2018). Furthermore, Treg cell‐associated immune‐modulatory proteins such as CD73, CTLA‐4 and CD25 have been found in Treg cell‐derived sEV, but among these, only CD73 seems to contribute to the suppressive activity of these vesicles (Li et al., 2016; Smyth et al., 2013). Recently, it was reported that Treg cells secrete sEV containing the immunosuppressive cytokine IL‐35, which promotes T and B cells exhaustion (Sullivan et al., 2020). Other proteins, such as iNOS, were reported to be present on sEV from murine in vitro induced‐Treg cells, showing a negative effect on T cells by blocking their proliferation and inducing apoptosis (Aiello et al., 2017). in vivo, Treg cell‐derived sEV have shown to prolong allograft tolerance in mice and rat models (Aiello et al., 2017; Yu et al., 2013). Moreover, the Treg cell‐derived sEV effects on target cells are probably due to their complex molecular content, including proteins. For example, vesicle membrane proteins could initiate signalling pathways in target cells and several additional protein components could contribute to the functional change of target cells (Bebelman et al., 2020; Kim et al., 2020).
Considering that the presence of Nrp1 has been reported in exosomes derived from human (Lazar et al., 2015; Stamer et al., 2011) and mouse cells (Hassani & Olivier, 2013), we propose that Treg cell‐derived sEV contain Nrp1 in their vesicular membrane, and that Nrp1 participates in the function of these vesicles. Thus, Nrp1 could contribute to Treg cell‐mediated immune modulation in a contact‐independent manner.
In this work, we demonstrate that Foxp3+ Treg cells produce sEV containing Nrp1, and that wt Foxp3+ Treg cell‐derived sEV, and not those obtained from Nrp1KO Foxp3+ Treg cells, inhibit conventional T (convT) cells proliferation and modulate their gene expression in vitro. Interestingly, local administration of sEV acquired from Nrp1KO Foxp3+ Treg cells were unable to facilitate skin transplantation tolerance, which was correlated with less presence of M2 in the grafts and higher frequency of Th17 cells in dLN. In addition, proteomic analysis indicated that Foxp3+ Treg cell‐derived sEV contain several proteins linked to immune regulation, such as CD73, CD25, CD82, CD43 and lactate dehydrogenase type C (LDHC), which were less represented in sEV obtained from Nrp1KO Treg cells. Altogether, our study points out that Nrp1 contributes importantly to immune tolerance via sEV release from Treg cells.
2. MATERIALS AND METHODS
2.1. Mice
Female and male mice of 6–8 weeks old were used. Foxp3Cre‐YFP 92, Nrp1flox/flox 93 and RAG‐KO mice were purchased from The Jackson Laboratory (Maine, USA). BALB/c (H‐2d), C57Bl/6 (H‐2b), F1 (C57BL/6 × BALB/c, H‐2b/d) and Foxp3 GFP (CD45.1+ or Ly5.2) mice were maintained under pathogen‐free conditions at the animal facility located at Facultad de Medicina, of the Universidad de los Andes. All procedures were carried out according to the bioethics committee guidelines from Universidad de los Andes, and National Commission of Science and Technology (CONICYT).
2.2. sEV preparation and isolation
sEV‐free media was prepared as previously described (Théry et al., 2006). In brief, RPMI media was supplemented with penicillin/streptomycin (Corning, NY, USA), HEPES (Gibco, MD, USA), β‐mercaptoethanol (Sigma, MO, USA) and 20% FBS (Gibco). This medium was subjected to ultracentrifugation (at least 16 h) at 100,000 × g followed by filtration using 0.22 μm filter (Millipore, MA, USA). Then, the sEV‐depleted medium was diluted with no FBS‐supplemented RPMI until 10% FBS final concentration was reached. After antibody staining, splenic CD3+CD4+YFP‐ (CD4+Foxp3‐ T cells) and CD3+CD4+YFP+ (CD4+Foxp3+ T cells) were FACS‐sorted as CD4+ T conventional cells (convT) and Foxp3+ Treg cells. These cells were cultured at 1 × 106/ml in sEV‐free supplemented RPMI media and activated with plate‐bound anti‐CD3 (5 μg/mL, clone 145.2C11, Biolegend) and anti‐CD28 (2 μg/ml, clone 37.51, Biolegend) for 72 h. Conditioned media (CM) was collected and exosome‐enriched sEV were obtained as described previously (Théry et al., 2006). In short, cells plus cell debris were removed from CM by serial centrifugation at 300 × g for 5 min, 2000 × g for 20 min and 10,000 × g for 30 min. After filtration using 0.22 μm filter (Millipore), sEV were recovered after ultracentrifugation at 100,000 × g for 90 min at 4°C, followed by washing with PBS 1× and another step of ultracentrifugation. sEV were resuspended in ∼100 μl of PBS 1×, and both sEV and CM aliquots were stored at ‐80°C for further analysis.
2.3. Western blot
Western blots were performed as previously described (Lobb et al., 2015; Théry et al., 2006). In brief, sEV and cell samples were lysed in RIPA buffer (10 mM Tris‐HCl pH 6.8, 140 mM NaCl, 1 mM EDTA, 1% Triton X‐100, 0.1% sodium deoxycolate, 0.1% SDS) on ice for 20 min. Lysates were vortexed and spinned at 17,200 × g for 20 min, and lysate supernatants were kept at ‐80°C until further use. The protein concentration was determined using Pierce BCA Protein Assay Kit (Life Technologies, US). 5× reducing sample buffer (60 mM Tris‐HCl pH 6.8, 25% glycerol, 2% SDS, 5% β‐mercaptoethanol and 0.04% bromophenol blue) was added to lysates at proper proportion, samples were heated at 95°C for 10 min and cooled to room temperature before being resolved by 12% SDS‐polyacrylamide gel electrophoresis. Proteins were resolved under fully denaturing and reducing conditions, transferred to nitrocellulose membranes, blocked in 4% non‐fat powdered milk in PBS‐T (0.1% Tween‐20) and probed with antibodies. The following antibodies were used for Immunoblot: mouse anti‐Alix (clone 1A12; dilution 1:250), ‐Calnexin (clone AF18, dilution 1:250), ‐TSG101 (clone C‐2; dilution 1:250), ‐α‐Tubulin (clone B‐7, dilution 1:1000), all from Santa Cruz Biotechnologies (USA), anti‐CD73 (clone 1D7, dilution 1:500) from ThermoScientific (USA) and anti‐β‐actin (clone 8H10D10, dilution 1:10,000) from Cell Signaling Technologies (USA). When indicated, we performed stripping of the membranes using 62.5 mM Tris‐HCl (pH 6.8), 2% SDS, 0.7% β‐mercaptoethanol for 15 min at RT, and re‐probed using polyclonal anti‐Nrp1 (dilution 1:1000) from R&D Systems (USA). The following secondary antibodies were used: Goat Anti‐Mouse IgG (H+L) (dilution 1:8000; Jackson ImmunoResearch Laboratories, PA), and Rabbit anti‐Goat IgG (H+L) (dilution 1:3000; ThermoFischer Scientific, IL, USA). Protein bands were detected using Pierce ECL Western Blotting Substrate (ThermoFisher, USA) in a Mini HD9 equipment (Uvitec Ltd., Cambridge, UK).
2.4. Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) was performed using a NanoSight NS300 (Malvern Instruments, UK), equipped with a 532 nm laser and a 565 nm LP filter. PBS 1× was used to dilute the starting samples. Three videos of 60 s were recorded for each sample, with temperature monitored along the readings. Videos recorded for each sample were analysed with the NTA Software in order to determine size and concentration of particles from samples of resuspended sEV. The Nanosight instrument was calibrated using 100‐nm diameter polystyrene latex beads and 100‐nm diameter 532‐nm green fluorescence standards (all from Malvern Panalytical, UK).
2.5. In vitro modulation assay
Splenic CD4+CD25‐ T cells were magnetically isolated (MACS, Miltenyi Biotec, Germany) and cultured in 96‐well plates for 3 days, previously coated with 5 μg/ml anti‐CD3 (clone 145‐2C11, Biolegend) and 2 μg/ml anti‐CD28 (clone 37.51, Biolegend) in complete RPMI media. Cells received 5 μg/ml of Tregs‐derived sEV at 0 and 24 h of culture. After 48 h cells were harvested and RNA was isolated using the RNeasy Kit (Qiagen, Germany), following manufacturer's instructions. cDNA libraries were prepared with iScript cDNA synthesis kit (BioRad, USA). Then, mRNA expression levels of Foxp3, IL‐10, IFN‐γ and IL‐17A were measured in Mx3000P qPCR system (Agilent Technologies, CA) using SsoFast EvaGreen qPCR Supermix (BioRad) as fluorescent detector. Nucleotide sequences of used primers are shown in Table 1. For analysis, relative expression was determined using the ΔΔCt method, using 18s as housekeeping control.
TABLE 1.
List of primers used in this study for qRT‐PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Foxp3 | CTG CTG GCA AAT GGA GTC TG | CCA GAG ACT GCA CCA CTT CT |
| Il‐10 | TGG GTT GCC AAG CCT TAT | AGA AAT CGA TGA CAG CGC CTC |
| Ifnγ | GCC AAG TTT GAG GTC AAC AAC C | ATC TCT TCC CCA CCC CGA AT |
| Il‐17a | AGCA GCGA TCAT CCCT CAAA | CTTC ATTG CGGT GGAG AGTCC |
| 18s | GCC CGA AGC GTT TAC TTT GA | TTG CGC CGG TCC AAG AAT TT |
2.6. In vitro suppression assay
Splenic CD4+CD25–Nrp1–Foxp3GFP– cells were sorted from Foxp3 GFPCD45.1+ mice as responder convT cells and labelled with 5 μM CellTrace Violet (CTV, ThermoFisher Scientific, MA, USA). Responder cells (5 × 104) were polyclonally activated with plate‐bound 5 μg/ml anti‐CD3 (clone 145‐2c11) and 2 μg/ml anti‐CD28 (clone 37.51, both from Biolegend) alone or co‐cultured with wt Foxp3YFP/Cre+ Treg cells (at 1:1 Treg:convT ratio), or treated with 5 μg/ml of sEV derived from wt Treg, Nrp1KO Treg or convT cells, in round‐bottom, 96‐well plates in 200 μl sEV‐free supplemented RPMI for 72 h. Responder T cell proliferation was analysed by tracking CTV dilution by flow cytometry using FACSCanto II (BD Biosciences). Suppression was calculated as previously described (Mcmurchy & Levings, 2012). In short, it uses the formula % Suppression = (1 – DITreg/DITresp) × 100% (where DITreg stands for the division index of responder cells with Treg cells, and DITresp stands for the division index of responder cells activated without treatment).
2.7. Skin transplantation and sEV treatment
Spleen and lymph node CD4+ T cells were pre‐enriched using the mouse CD4+ T Cell Isolation Kit (MACS, Miltenyi Biotec, CA, USA). CD4+Foxp3GFP–Nrp1– cells were sort‐purified as convT cells from Foxp3 GFPCD45.1+ reporter mice, and CD4+Foxp3Cre/YFP+ were FACS‐sorted as Tregs from Foxp3 Cre/YFPCD45.2+ (wt). Then, convT cells (1.5 × 105) were intravenously injected alone or co‐transferred with Treg cells (5 × 104) into RAG‐KO mice, one day before transplant surgery. Some animals in separated groups received 100 μl PBS 1× (vehicle) or a solution of 5 μg of sEV derived from wt or Nrp1KO Tregs, by subcutaneous injection surrounding the grafting area. Tail skin (∼1 cm2) from C57BL/6 (syngeneic) or F1 (allogeneic) donors was transplanted onto the dorsal area of RAG‐KO recipient mice, as described previously (Beilke & Gill, 2007). Survival of skin allografts was evaluated twice per week and grafts were considered rejected when 80% of the initial graft had disappeared or become necrotic.
2.8. Preparation of tissue cell suspensions
Fifteen days post‐transplant mice were euthanized, graft draining lymph nodes (dLNs) and skin grafts were harvested for flow cytometry analysis. dLNs were mechanically disaggregated in PBS 1× containing 5% FBS, and single cell suspension was obtained by filtering in a 40 μm cell strainer (BD Falcon) and maintained at 4°C. Skin‐infiltrating leukocytes were obtained as previously described (Galvez‐Cancino et al., 2019). Briefly, skin grafts excised from transplanted animals were cut in small fragments and digested in 1 ml of non‐supplemented RPMI medium containing 5 mg/ml collagenase D and 5 μg/ml DNase I for 30 min at 37°C (both from Roche). Then, digested skin pieces were mechanically disaggregated in a 40 μm cell strainer (BD Falcon) and single cell suspensions were obtained. After a washing step with supplemented RPMI medium, cells were digested a second time with 1 ml of supplemented RPMI medium containing 25 μg/ml DNase I for 5 min on ice. Finally, cells were stimulated for cytokine expression analysis, or directly stained for surface marker expression with antibodies as described below.
2.9. Flow cytometry
After cell viability staining with ZombieDye NIR (Biolegend, CA, USA), cell suspensions were stained in PBS 1 × 5% heat‐inactivated FBS (Gibco, USA) with the following antibodies: CD3e (clone 145‐2c11), CD4 (clone RM4‐5), CD11b (clone M1/70), CD11c (clone N418), CD25 (clone PC61), CD45.1 (clone A20), CD45.2 (clone 104), CD206 (clone C068C2), I‐A/I‐E (clone M5/114.15.2), Ly‐6C (clone HK1.4), Ly‐6G (clone 1A8), NK1.1 (clone PK136), Nrp1 (clone 3E12), all from Biolegend, CA, USA. Surface staining was performed for 30 min at 4°C. Intracellular staining was performed with anti‐Foxp3 (clone FJK‐16s) and the Fixation/Permeabilization Staining Kit (all from eBioscience, CA, USA), following manufacturer's instruction. For cytokine expression analysis, cells were activated with 50 ng/ml PMA (Sigma) and 1 μg/ml Ionomycin (Sigma) in RPMI containing 10% FBS and Brefeldin‐A (eBioscience, CA) for 4 h. Cells were washed, stained for surface markers, and treated with the Fixation/Permeabilization Staining Kit (all from eBioscience, CA, USA), following manufacturer's instructions. Intracellular cytokine staining was performed with the following antibodies: IFN‐γ (clone XMG1.2), IL‐17A (clone TC11‐18H10.1, both from Biolegend). Cells were analysed on FACSCanto II (BD Biosciences, USA) or FACS‐sorted in a FACSAria II (BD Biosciences, USA). Data analysis was performed on FlowJo software (CA, USA).
2.10. Mass spectrometric analysis of sEV content
sEV proteins were separated using polyacrylamide gradient gel electrophoresis. Each lane was divided into 6 sections to perform in‐gel digestion according to Kolodziej et al., 2016.
Liquid chromatography followed by tandem‐mass spectrometry (MS/MS) of the sample fractions was performed on a hybrid dual‐pressure linear ion trap/orbitrap mass spectrometer (LTQ Orbitrap Velos Pro, Thermo Scientific) equipped with an EASY‐nLC Ultra HPLC (Thermo Scientific). Peptide samples were dissolved in 10 μl of 2% acetonitrile/0.1% trifluoric acid and fractionated on a 75‐μm i.d., 25‐cm PepMap C18‐column, packed with 2 μm resin (Dionex, Germany). Separation was achieved by applying a gradient of 2% to 35% acetonitrile in 0.1% formic acid over 150 min at a flow rate of 300 nl/min. An Orbitrap full MS scan was followed by up to 20 LTQ MS/MS runs using collision‐induced dissociation (CID) fragmentation of the most abundantly detected peptide ions. The essential MS settings were as follows: full MS (resolution, 60,000; mass to charge ratio range, 400–2000); MS/MS (Linear Trap; minimum signal threshold, 500; isolation width, 2 Da; dynamic exclusion time setting, 30 s; and singly charged ions were excluded from the selection). Normalized collision energy was set to 35%, and activation time was set to 10 ms. Raw data processing, protein identification and result comparison were performed by PEAKS Studio 8.0 (Bioinformatics Solutions Inc., Canada). The false discovery rate was set to <1%.
2.11. Bioinformatics analysis
The differentially expressed proteins of each sEV sample were analysed by Venn diagrams using VENNY 2.1. Gene ontology analysis, Functional Annotation, Gene Functional Classification and Gene ID Conversion were performed using GeneOntology Consortium (Ashburner et al., 2000; The Gene Ontology, 2019; Thomas et al., 2003) and PANTHER version 11 (Mi et al., 2017; Thomas et al., 2003). Heatmaps of protein expression data were perform using HeatMapper (Babicki et al., 2016), which calculates the sample‐to‐sample distances using the Euclidean distance measurement method between the samples and clustering method of average linkage.
2.12. Statistical analysis
Statistical analysis was performed using Student's t‐test, two‐way ANOVA and log‐rank test using the software GraphPad Prism (CA, USA). Differences with p‐values less than 0.05 were considered significant. * < 0.05; ** < 0.01; *** < 0.001; ns, not significant.
3. RESULTS
3.1. Foxp3+ Treg cells produce Nrp1+ sEV
In previous reports, our group and others reported that Nrp1 deficiency in Foxp3+ Treg cells impaired their capacity to exert in vitro, contact‐independent immune suppression, using a 0.4 μm pore diameter transwell system (Campos‐Mora et al., 2019; Delgoffe et al., 2013). This observation suggested that Nrp1 might be involved in a Treg cell‐mediated suppressive mechanism that it is not fully described yet: the production of sEV with immunosuppressive properties (Li et al., 2016). To unveil this potential mechanism, we first isolated sEV from polyclonally activated Foxp3+ Treg cells obtained from Foxp3 YFP/Cre (wild‐type control, wt), Foxp3 YFP/Cre Nrp1 +/flox (heterozygous, het), and Foxp3 YFP/Cre Nrp1 flox/flox (Nrp1KO) animals. These sEV were isolated following the ultracentrifugation approach (Théry et al., 2006), enriching for 100,000 g pellet (which was used throughout the study), and quantified using Nano Tracking Analysis (NTA)‐based technique and characterized using Western Blot (Buzás et al., 2017; Théry et al., 2006) (the reader should note that there may be a possibility that non‐sEV associated factors may contribute to the observed effects described below). We found that the mean particle size of all isolated sEV was similar (154.2 ± 43.4 for wt Treg cells sEV; 159.7 ± 25.7 for het Treg cells sEV; and 139.9 ± 67.7 for Nrp1KO Treg cells sEV), regardless of the genotype of Treg cells used as source (Figure 1a). When we consider the cell number and the empiric dilution for NTA measurements, we found that all three Foxp3+ Treg cells genotypes are capable of secreting similar amounts of sEV (∼ 2.5 × 108 sEV/106 cultured Treg cells) (Figure 1b). In addition, we found that Treg cells tend to secrete higher amounts of sEV compared to convT cells (∼1.0 × 108 sEV/106 cultured convT), although this result is not statistically significant (Figure 1b) (Okoye et al., 2014). We also characterized Treg cell‐sEV studying the expression of canonical markers associated with exosomes‐like vesicles. Immunoblot analysis performed to total protein extracts of Treg cell‐derived sEV, probing for the presence of the proteins Alix and Tsg‐101 (specific markers for vesicles of endocytic origin) (Lötvall et al., 2014; Witwer et al., 2013), and the absence of calnexin (a marker of vesicles of endoplasmic reticulum origin), confirmed that Treg cell‐derived vesicles obtained from wt and Nrp1KO genotypes were enriched in sEV (Lötvall et al., 2014) (Figure 1c). Some components of cellular cytoskeleton (such as Tubulin subunits) have been reported as present in exosomes from leukocytes and non‐leukocyte cells (Buschow et al., 2010; Mathivanan et al., 2010) however, we could not detect the presence of α‐Tubulin in sEV samples isolated from Treg cells, independently of their expression of Nrp1 (Figure 1c). Together, these results strongly suggest that indeed these nanometric vesicles isolated from in vitro cultured Foxp3+ Treg cells correspond to exosome‐enriched sEV and, importantly, the deficiency of Nrp1 in Foxp3+ Treg cells does not affect sEV production and/or secretion.
FIGURE 1.
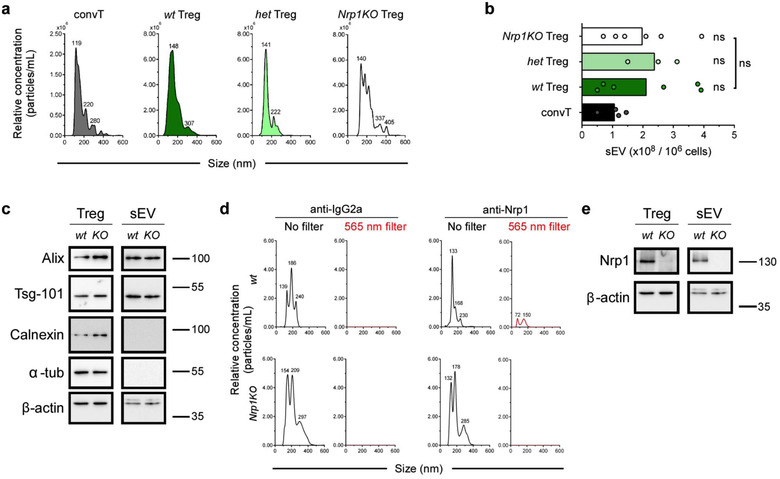
Nrp1 deletion on Foxp3+ Treg cells does not alter sEV secretion. Conventional T cells (convT) and Foxp3+ Treg cells from wt, het and Nrp1KO mice were sort‐purified and activated in sEV‐depleted cRPMI media with plate‐bound αCD3/αCD28 for 72 h. Conditioned media (CM) was harvested and sEV were obtained by ultracentrifugation. (a) Characterization of sEV size and relative concentration by nanoparticle tracking analysis (NTA) from convT (dark grey), and wt (dark green), het (light green) or Nrp1KO (white) Treg cells sEV. Numbers at peaks correspond to mean size reported by NanoSight. (b) Accumulated data of sEV production of aforementioned T cells populations. (c) Immunoblot analysis for sEV markers in the total lysate from Treg (Treg) cells or sEV from by wt and Nrp1KO animals. Numbers on the right side of the panel represent molecular size (kDa). (d) sEV from wt or Nrp1KO Treg cells were stained with PE‐coupled IgG2a (isotype, left) or anti‐Nrp1 antibodies (right), and then analyzed to determine size and relative concentration using no filter (black line) or 565 nm fluorescence filter (red line). Numbers at peaks correspond to mean size reported by NanoSight. (e) Immunoblot analysis of total lysates depicting Nrp1 expression in Treg cells and Treg‐derived sEV lysates after performing stripping of the membranes shown in (c). Numbers on the right side of the panel represent molecular size (kDa). For (b), bars represent mean, and each circle represents one sEV purification. Representative data from 3 (Het) to 6 (wt and Nrp1KO) sEV purifications. One‐way ANOVA (Tukey's multiple comparisons test), ns = not significant
Next, we analysed the presence of Nrp1 protein in sEV from wt and Nrp1KO Treg cells using two approaches: first, we stained the sEV with Phycoerithrin (PE)‐coupled anti‐Nrp1 antibody and measured PE fluorescence emission signal by NTA (Carnell‐Morris et al., 2017). Following this strategy, we only found PE signal in sEV obtained from wt Treg cells and not from those isolated from Nrp1KO Treg cells (Figure 1d). This observation was confirmed by immunoblot, where Nrp1 was found in both wt Treg cell lysates and wt Treg cell‐derived sEV lysates (Figure 1e), but was not detected in lysates from either Nrp1KO Treg cells or Nrp1KO Treg cell‐derived sEV (Figure 1e). Lastly, sEV shape and size were confirmed by SEM, finding semi‐rounded and ∼150 nm average size sEV, irrespective of the Treg cell genotype (data not shown). These results suggest that wt Treg cells secrete sEV “loaded” with Nrp1 molecules in its surface.
3.2. Nrp1 is required for Treg cell‐mediated suppression of convT via sEV
The expression of Nrp1 in T cells has long been associated with immune tolerance in several immune settings (Campos‐Mora et al., 2019; Campos‐Mora et al., 2015; Solomon et al., 2011), but the mechanisms by which Nrp1 expression is involved in controlling the immune response is not fully known. Recently, we reported that Nrp1 plays a role in modulating convT cells phenotype and function in vivo in a Treg cell‐dependent manner, as ex vivo alloreactive T cells displayed suppressive function when co‐transferred with Nrp1+ Treg cells but not when transferred with Nrp1KO Treg cells, suggesting that Nrp1 may play a role in generating CD4+ T cells with suppressive properties in the periphery (Campos‐Mora et al., 2019). Hence, the presence of Nrp1 in Treg cell‐derived sEV might be relevant for the immunosuppressive capability of these nanovesicles.
Therefore, we proceeded to study the in vitro suppressive function of wt and Nrp1KO‐Treg cell‐derived sEV on the proliferation of freshly isolated, CellTraceTMViolet‐stained CD4+CD25‐ convT cells polyclonally activated. We used 50,000 freshly isolated wt Treg cells as control for suppression, and convT cells‐derived sEV as biologically active sEV control. As previously described (Wahlgren et al., 2012), we found that 1 μg of convT cells‐derived sEV boosted convT cells proliferation compared with vehicle control (∼91% vs. 83% of proliferation, respectively) (Figure 2a). On the contrary, we observed that the treatment with 1 μg of wt Treg cell‐derived sEV (equivalent to 200,000 Treg cells) decreased convT cell proliferation and their overall division index (Figure 2a‐c). Conversely, Nrp1KO Treg cell‐derived sEV could not inhibit convT proliferation and division index as their wt counterpart (∼81% vs. ∼74% of convT cells in proliferation; and ∼0.9 vs. ∼0.75 for division indexes, respectively) (Figure 2a‐c). After calculating the mean suppression percentage, we confirmed that the suppressive function of Nrp1KO Treg cell‐derived sEV was lower than wt Treg cell‐derived sEV (∼25% vs. ∼1%, respectively) (Figure 2c). Altogether, these results suggest that the presence of Nrp1 in Treg cell‐derived sEV is required for optimal inhibition of convT cell proliferation in vitro. Next, we sought to evaluate whether Treg cell‐derived sEV modulate convT cells phenotype in vitro and to elucidate a potential involvement of Nrp1 in this process. For this, CD4+CD25– T cells were sorted from C57BL/6 spleens and polyclonally activated in the presence of PBS 1X (vehicle) or 5 μg/ml sEV obtained from wt or Nrp1KO Treg cells. After 48 h, CD4+ T cells were harvested and RNA was isolated to test for the expression of Foxp3, Il10, Ifng and Il‐17a transcripts. As observed, the addition of wt Treg cell‐derived sEV induced the expression of Foxp3, Il10 and Ifng mRNA, although the differences observed were not statistically significant, except for Foxp3 (Figure 2d). No change for Il‐17a mRNA was obtained. Furthermore, sEV isolated from Nrp1KO Treg cells did not induce changes in the expression of Foxp3 or Ifng mRNA, although a modest up‐regulation on Il‐17a mRNA levels was obtained with Nrp1KO Treg cell‐derived sEV.
FIGURE 2.
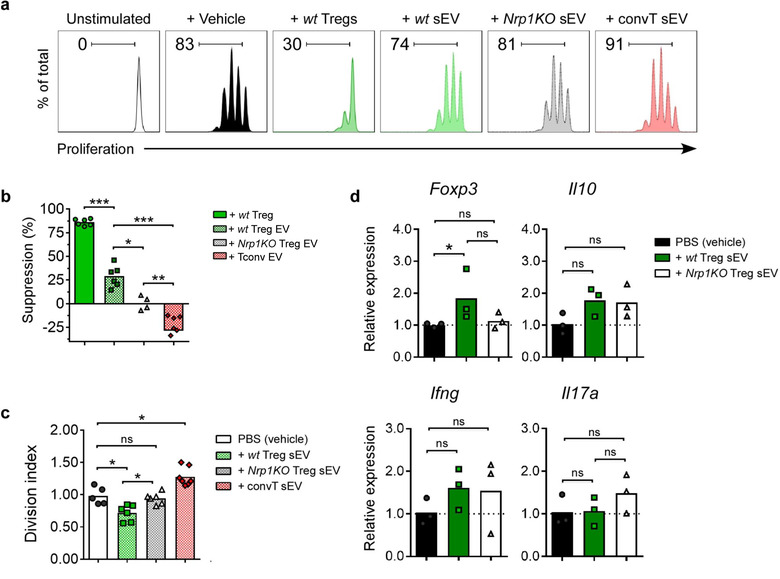
sEV from Nrp1+ Treg cells modulate convT cells' phenotype in vitro. Conventional T (convT) cells were stained with CTV and activated in vitro with plate‐bound anti‐CD3 (5 μg/ml)/anti‐CD28 (2 μg/ml) in sEV‐free media, alone or in the presence of wt Treg cells (suppression control), treated with PBS 1X (vehicle) or 5 μg/ml sEV from wt or Nrp1KO Treg cells, or convT‐derived sEV, for 72 h. (a) Representative histograms show proliferation of unstimulated convT (white), or activated convT treated with vehicle (black), or co‐cultured with wt Treg cells (dark green), or treated with sEV derived from wt (green dotted) or Nrp1KO Treg cells (black dotted) or convT cells (red dotted). Numbers at top‐left indicate frequency of proliferating convT cells. (b) Bar graph shows suppression (%) of convT cell proliferation, calculated as described in Material and Methods. (c) Bar graph shows Division Index of convT cells under the aforementioned conditions. (d) Cells were harvested at 48 h postsEV treatment, and gene expression for Foxp3, Il‐10, Ifng and Il‐17a was measured by qRT‐PCR on convT cells treated with vehicle (black) or with wt (dark green) or Nrp1KO Treg‐derived sEV. For b‐d, bars represent mean, and each point represents one independent experiment. One‐way ANOVA (Tukey's multiple comparisons test), * p < 0.05; ** p < 0.01; *** p < 0.001; ns = not significant. n = 3 independent experiments
3.3. Nrp1+ Treg cell‐derived sEV favor transplant tolerance by accumulating intragraft M2
The immunomodulatory role of murine Treg cell‐derived sEV has been evaluated in several studies as reviewed by Rojas et al. (2020). Moreover, it has been recently reported that human Treg cell‐derived sEVs can inhibit the proliferation and modify cytokine production of effector T cells protecting against human skin allograft damage (Tung et al., 2018). To elucidate the relevance of Nrp1 in Treg cell‐derived sEV, we tested wt and Nrp1KO Treg cell‐derived sEV capacity of inducing in vivo immune tolerance to skin allografts. Briefly, sEV was obtained from wt or Nrp1KO Treg cells and injected subcutaneously in the area surrounding the transplant bed (back of the mouse) of RAG‐KO mice previously receiving convT cells. Skin transplanted RAG‐KO mice without adoptively transferred convT cells were used as acceptance controls (Figure 3). The next day, mice were allografted and transplant survival was monitored three times per week. As depicted in Figure 3c‐d, RAG‐KO animals receiving convT cells reject the skin grafts, showing ∼10% of transplant survival, while the administration of wt Treg cell‐derived sEV prevents rejection, raising transplant survival to ∼57% (Figure 3b‐d). Conversely, the treatment with Nrp1KO Treg cell‐derived sEV did not protect against transplant rejection, obtaining only ∼28% of graft survival.
FIGURE 3.
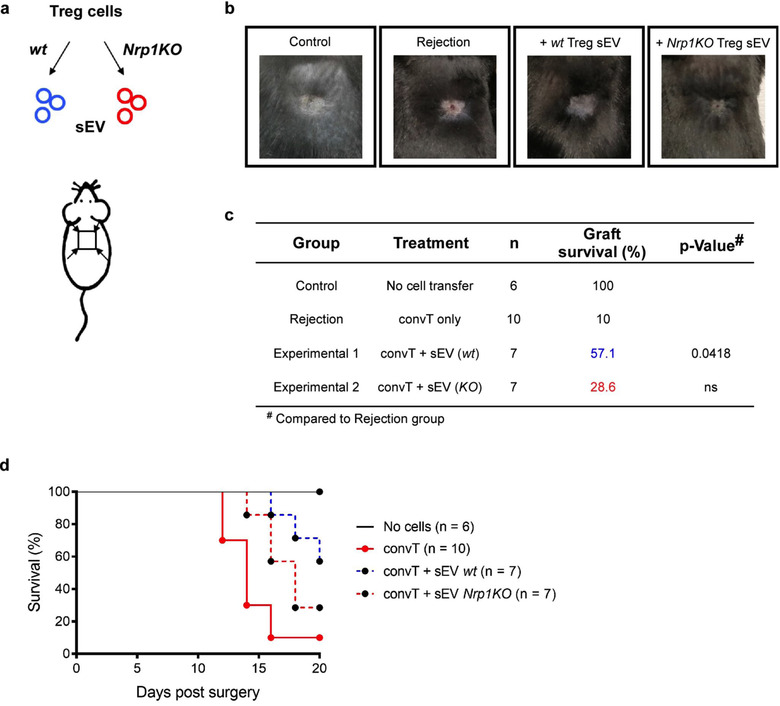
Nrp1KO Treg cell‐derived sEV fails to induce skin graft tolerance. RAG‐KO mice received 1.5 × 105 convT cells by intravenous injection, and one dose of PBS 1× (vehicle) or sEV derived from wt or Nrp1KO Treg cells by subcutaneous injection. Next day, all mice were transplanted with allogeneic skin grafts, and 20 days post‐transplant graft survival was evaluated. (a) Representative scheme depicts sEV isolation from wt (blue) or Nrp1KO (red) Foxp3+ Tregs, and the location of sEV administration before skin transplantation. (b) Representative photographs showing the outcome of graft survival at day 20 after surgeries in mice with no cell transfer (control group), mice receiving only convT cells (No treatment, rejection group) or mice receiving convT cells and sEV obtained from wt or Nrp1KO Treg cells. (c) Table showing experimental groups, treatment, number of animals per group (n), graft survival (%) and p‐value at day 20 postsurgery. For c, N‐1 Chi‐squared test. n = 3 independent experiments. (d) Survival plot showing the percentage of skin graft survival, including the number of animals per group
During the progression of the immune response against the allograft, several immune populations are recruited to the transplanted tissue by released inflammatory signals, among others (Béland et al., 2015). It has been described that immune cells from the innate compartment, such as monocytes/macrophages and NK cells, also play a pivotal role in the final outcome of the response, determining allograft tolerance or rejection (Benichou et al., 2012; Ordikhani et al., 2020). Accordingly, we studied cell infiltration into the skin grafts and dLN in rejecting animals and those treated with wt or Nrp1KO Treg cells‐derived sEV, in order to elucidate the effect of Treg cells sEV treatment on allograft leukocyte infiltration and the role of Nrp1 in this phenomenon. After following enzymatic digestion of the skin grafts, we analysed different cell populations residing in the tissue. We evaluated the presence of CD4+ T cells, Type‐1 Macrophages or M1 (CD11b+MHC‐II+/highCD206‐), Type‐2 Macrophages or M2 (CD11b+MHC‐II+/‐CD206+), NK cells (CD11b+NK1.1+), conventional Dendritic Cells or cDC (CD11c+MHC‐II+) and Neutrophils (CD11c‐CD11b+Ly6G+) (Crane et al., 2014; Rose et al., 2012; Rusinkevich et al., 2019), Figure S1 and S2. As depicted, we found a robust population of M1 cells, which did not vary among the three experimental groups (∼30%; Figure 4a,b). On the contrary, the frequencies of M2 cells displayed variations: whereas ∼5% of M2 cells were found in the rejecting and ∼8% in wt Treg cell‐derived sEV conditions, animals treated with Nrp1KO Treg cells‐derived sEV reached a significant reduction to ∼2% (Figure 4a and c). Consequently, the M1/M2 ratio was higher in the Nrp1KO Treg cell‐derived sEV‐treated group compared to animals receiving wt Treg cell‐derived sEV, Figure 4d. In contrast, when we looked at these populations in transplant‐dLN, no differences were found, Figure 4h‐j. Furthermore, the frequencies of cDC, and NK cells in the skin grafts were unchanged among all groups tested, whereas the population of neutrophils was reduced in wt Treg cell‐derived sEV treated animals versus rejection controls (from ∼18% to 8%, respectively). In dLN, NK cells and Neutrophils remained constant in all groups, but the frequency of cDC was decreased in Nrp1KO Treg cell‐derived sEV‐treated group compared to rejecting animals (from ∼10% to 6%, respectively), Figure S3. These results suggest that Nrp1‐bearing sEV may be involved in M2 cells accumulation in the skin transplants. To complement this data, we studied the frequencies of CD4+ T cell populations in skin grafts and transplant‐dLN, focusing in Th1 (CD4+IFNγ+), Th17 (CD4+IL‐17+) and Foxp3+ Treg cells. In this case, we did not find differences in Th1, Th17 and Treg cell populations between wt and Nrp1KO Treg cell‐derived sEV treated groups, either in the skin grafts (Figure 4e‐g) or transplant‐dLN (Figure 4k‐m).
FIGURE 4.
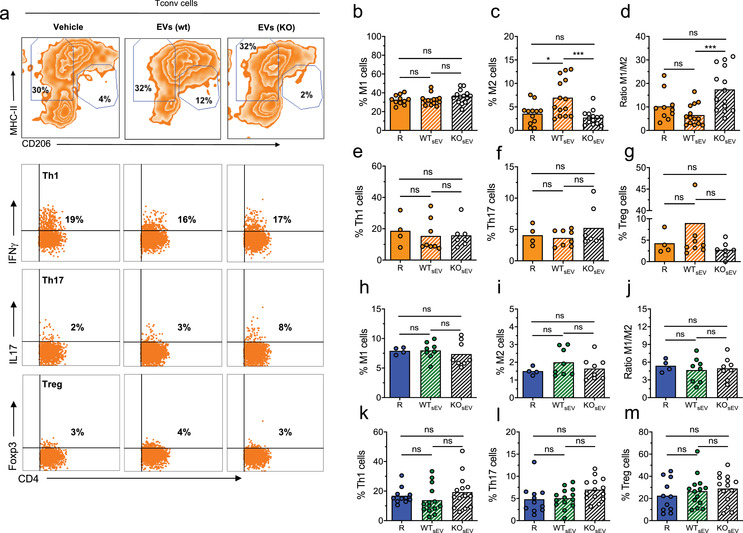
Nrp1+ Treg‐derived sEV induces transplant tolerance by modulating intragraft M1/M2 ratio. Graft‐draining lymph nodes and skin grafts were harvested from transplanted animals, and single cell suspensions were obtained for analysis. From skin preparations, CD45+CD11b+ macrophages were gated, and expression of MHC‐II and CD206 we used to define M1‐type (MHC‐II+/high CD206‐) and M2‐type (MHC‐IIlow CD206+/high) by flow cytometry. (a) Representative contour plots show the frequency (%) of graft‐infiltrating M1‐type, M2‐type macrophages, IFN‐γ+ Th1, IL‐17A+ Th17 and Foxp3+ Treg cells from animals treated with vehicle (rejection group) or treated with wt Treg‐ or Nrp1KO Treg cells‐derived sEV. Below, bar graphs represent (b) the frequency of M1‐type macrophages; (c) the frequency of M2‐type macrophages; (d) the M1/M2 ratio; (e) the frequency of Th1 cells; (f) the frequency of Th17 cells; and (g) the frequency of Foxp3+ Treg cells detected in skin‐infiltrating lymphocyte samples from mice of untreated group (R, dark orange), treated with sEV derived from wt Treg (dashed orange) or Nrp1KO Treg cells (dashed white). From graft‐draining lymph node (dLN) preparations, bar graphs show the frequency of (h) the frequency of M1‐type macrophages; (i) the frequency of M2‐type macrophages; (j) the M1/M2 ratio; (k) the frequency of Th1 cells; (l) the frequency of Th17 cells; and (m) the frequency of Foxp3+ Treg cells present in graft dLN of rejecting mice (R, blue), and those treated with sEV obtained from wt (dashed green) or Nrp1KO (dashed white). For (b)‐(m), One‐way ANOVA (Tukey's multiple comparison test). * p < 0.05; ** p < 0.01; *** p < 0.001; ns = not significant. n = 3 independent experiments, with n = 3–8 mice group, per experiment
3.4. Deletion of Nrp1 on Foxp3+ Treg cells affects the cargo composition of sEV
After evaluating the immune‐suppressive capacity of wt versus Nrp1KO Treg cell‐derived sEV, an extensive proteomic analysis was performed to assess whether the deficiency of Nrp1 on Treg cells affects the cargo composition of their sEV in comparison to wt control. The composition of sEV obtained from wt and Nrp1KO Treg cells was determined using label‐free liquid chromatography‐ tandem mass spectrometry (LC‐MS/MS)‐based proteomics.
The proteomes of two biological replicates for each sample (wt or Nrp1KO derived sEV) were analysed and compared, obtaining a total of 172 unique proteins identified across the combined samples. Venn diagram analysis showed that 84 of the identified sEV proteins were present in both types of sEV, while 51 proteins were found exclusively on wt and 37 exclusively on Nrp1KO Treg cell‐derived sEV (Figure 5a).
FIGURE 5.
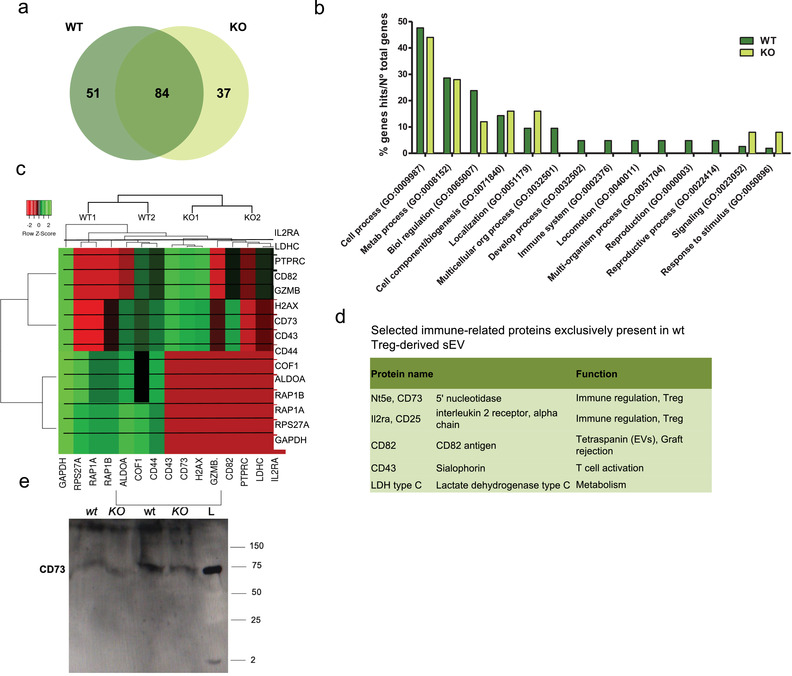
Analysis of sEV proteome reveals that Nrp1+ Treg cell‐derived sEV are enriched in proteins with immunosuppressive potential. sEV derived from wt or Nrp1KO Treg cells were isolated and proteomic analysis was performed as described in Material and Methods. (a) The lists of proteins expressed in the different sEV samples were compared using Venn diagram (VENNY 2.1). 172 proteins were analyzed in total, of these 51 were expressed only in wt Treg cells, and 37 in Nrp1KO Treg cell‐derived sEV. (b) GO‐biological processes (GO‐BP) analysis showed GO: 0002376 “'”immune response'’ as exclusively enriched in wt Treg cell‐derived sEV (p < 0.05). (c) Heatmap of differentially expressed proteins from wt and Nrp1KO Treg cell‐derived sEV samples (p < 0.05). The differential expression of the different proteins was analyzed using HeatMapper. Log2 signal intensity values for any single protein were resized to Row Z‐Score scale (from − 2, the lowest expression to + 2, the highest expression for a single protein). (d) Table including five more relevant proteins with immunosuppressive potential. We also included the synonymous of protein name and a brief description of the biological function associated to the immune response. (e) Western blot analysis for evaluating the presence of CD73 on Treg cell lysates obtained from wt or Nrp1KO mice (KO) and sEV enriched from in vitro cultured Treg cells isolated from wt or Nrp1KO animals (KO). All conditions were normalized to load 30 μg of total protein. L: ladder
To elucidate the biological functions of the proteins differentially expressed in wt and Nrp1KO Treg cell‐derived sEV, we performed Gene Ontology (GO) enrichment analysis comparing the 51 proteins exclusively expressed on wt versus the 37 proteins exclusively found on Nrp1KO Treg‐cell‐derived sEVs using Panther GenOntology Consortium. Among the GO Biological Process (GO‐BP) categories that were significantly involved (adjusted p value < 0.05), we found a higher enrichment in the categories of “Cell Process,” “Metabolic Process” and “Biological Regulation” in samples derived from sEV from wt Treg cells over Nrp1KO Treg cell‐derived sEV (Figure 5b). Interestingly, we also found that categories like “Immune system Process,” “Locomotion” and “Reproductive Process” are exclusively enriched in samples derived from wt Treg cell‐derived sEV.
Next, we performed a heatmap graph of variance using HeatMapper. Each sample is shown in the heatmap according to their protein expression represented with a band intensity (arbitrary units) conveyed as single values for each protein (Figure 5c). We observed that some proteins derived from wt Treg cell‐derived sEV are exclusively overexpressed with respect to those isolated from Nrp1KO Treg cells, particularly, proteins related to immune modulation such as IL2RA, CD73, CD82, CD43 and LDHC (Figure 5d). Preliminary western blot experiment suggests that at least CD73 is less abundant in sEV isolated from Nrp1KO Treg cells compared to wt Treg cells (Figure 5e). A complete list of proteins identified in sEV isolated from wt and Nrp1KO Treg cells are shown in Table 2 and Table 3, respectively (Supplementary Material).
TABLE 2.
List of proteins exclusively found in sEV isolated from wt Treg cells
| Gene name | Uniprot ID |
|---|---|
| RE1‐silencing transcription factor | Q8VIG1 |
| Phosphoglycerate kinase 1 | P09411 |
| Sodium/potassium‐transporting ATPase subunit beta‐3 | P97370 |
| Myb‐binding protein 1A | Q7TPV4 |
| Anaphase‐promoting complex subunit 1 | P53995 |
| 5′‐nucleotidase | Q61503 |
| Membrane‐associated guanylate kinase, WW and PDZ domain‐containing protein 1 | Q6RHR9 |
| Actin, cytoplasmic 1 | P60710 |
| Peroxiredoxin‐2 | Q61171 |
| Poly(rC)‐binding protein 1 | P60335 |
| GRIP and coiled‐coil domain‐containing protein 2 | Q8CHG3 |
| Heterochromatin protein 1‐binding protein 3 | Q3TEA8 |
| Alpha‐2‐HS‐glycoprotein | P29699 |
| Protein ITPRID2 | Q922B9 |
| Neutral amino acid transporter B(0) | P51912 |
| 60S ribosomal protein L24 | Q8BP67 |
| Perilipin‐1 | Q8CGN5 |
| Histone H2AX | P27661 |
| 60S ribosomal protein L27a | P14115 |
| Receptor‐type tyrosine‐protein phosphatase C | P06800 |
| Leukosialin | P15702 |
| ATP synthase subunit alpha, mitochondrial | Q03265 |
| Lysosomal acid glucosylceramidase | P17439 |
| Vitrin | Q8VHI5 |
| Serine/arginine repetitive matrix protein 2 | Q8BTI8 |
| Histone H1t | Q07133 |
| Keratin, type I cytoskeletal 12 | Q64291 |
| L‐lactate dehydrogenase C chain | P00342 |
| Heat shock protein HSP 90‐alpha | P07901 |
| Sodium/potassium‐transporting ATPase subunit alpha‐2 | Q6PIE5 |
| RNA‐binding protein FUS | P56959 |
| Tumour necrosis factor receptor superfamily member 18 | O35714 |
| 60S ribosomal protein L15 | Q9CZM2 |
| Peripherin | P15331 |
| Alpha‐1‐antitrypsin 1–4 | Q00897 |
| Basigin | P18572 |
| T‐complex protein 1 subunit theta | P42932 |
| 60S ribosomal protein L10a | P53026 |
| Testis‐expressed protein 2 | Q6ZPJ0 |
| Adhesion G‐protein coupled receptor V1 | Q8VHN7 |
| Dihydropyrimidinase‐related protein 2 | O08553 |
| Elongation factor 2 | P58252 |
| Ubiquitin carboxyl‐terminal hydrolase BAP1 | Q99PU7 |
| Splicing factor, proline‐ and glutamine‐rich | Q8VIJ6 |
| Elongation factor 1‐alpha 2 | P62631 |
| 60S ribosomal protein L27 | P61358 |
| EF‐hand and coiled‐coil domain‐containing protein 1 | Q9JJF6 |
| Alpha‐1‐antitrypsin 1–3 | Q00896 |
| Interleukin‐2 receptor subunit alpha | P01590 |
| Ubiquitin‐associated and SH3 domain‐containing protein A | Q3V3E1 |
| CD82 antigen | P40237 |
TABLE 3.
List of proteins exclusively found in sEV isolated from Nrp1KO Treg cells
| Gene name | Uniprot ID |
|---|---|
| Ras‐related protein Rap‐1A | P62835 |
| Keratin, type II cytoskeletal 7 | Q9DCV7 |
| Obscurin | A2AAJ9 |
| Glyceraldehyde‐3‐phosphate dehydrogenase, testis‐specific | Q64467 |
| Monocarboxylate transporter 1 | P53986 |
| Arf‐GAP with SH3 domain, ANK repeat and PH domain‐containing protein 2 | Q7SIG6 |
| A‐kinase anchor protein 13 | E9Q394 |
| Eukaryotic initiation factor 4A‐I | P60843 |
| Keratin, type I cytoskeletal 40 | Q6IFX3 |
| 60S ribosomal protein L10‐like | P86048 |
| Keratin, type I cuticular Ha2 | Q62168 |
| Keratin, type II cuticular Hb1 | Q9ERE2 |
| Annexin A2 | P07356 |
| Keratin, type II cuticular 87 | Q6IMF0 |
| Keratin, type I cuticular Ha3‐I | Q8K0Y2 |
| Hydrocephalus‐inducing protein | Q80W93 |
| Glial fibrillary acidic protein | P03995 |
| Tubulin alpha‐1A chain | P68369 |
| Keratin, type I cuticular Ha6 | B1AQ75 |
| Nestin | Q6P5H2 |
| Ubiquitin‐40S ribosomal protein S27a | P62983 |
| 60S ribosomal protein L29 | P47915 |
| Collagen alpha‐1(XVI) chain | Q8BLX7 |
| Mitogen‐activated protein kinase kinase kinase 12 | Q60700 |
| Triosephosphate isomerase | P17751 |
| Segment polarity protein dishevelled homolog DVL‐2 | Q60838 |
| Gem‐associated protein 5 | Q8BX17 |
| Sperm‐associated antigen 1 | Q80ZX8 |
| Keratin, type I cuticular Ha1 | Q61765 |
| Nuclease‐sensitive element‐binding protein 1 | P62960 |
| 60S ribosomal protein L10 | Q6ZWV3 |
| Myeloperoxidase | P11247 |
| Ras‐related protein Rap‐1b | Q99JI6 |
| Dystonin | Q91ZU6 |
| Keratin, type II cuticular Hb4 | Q99M73 |
| Keratin, type I cuticular Ha5 | Q497I4 |
| Protocadherin Fat 3 | Q8BNA6 |
4. DISCUSSION
Treg cells secrete sEV as part of their immune suppression mechanisms (Li et al., 2016; Okoye et al., 2014; Smyth et al., 2013), and some proteins highly expressed in Treg cells (CD73, CD25, CTLA‐4) were reported to be present in sEV derived from these cells (Agarwal et al., 2014). Among them, it was found that CD73 contributes to the immunomodulatory function of these sEV (Smyth et al., 2013). However, it has not been described yet whether other markers expressed in Treg cells are present in their sEV (such as PD‐1, GITR, or Nrp1). The Nrp1 protein has only been reported in sEV derived from macrophages and tumour cells, both human and mouse (Demory Beckler et al., 2013; Hassani & Olivier, 2013; Lazar et al., 2015; Stamer et al., 2011). To date, there are no reports involving Nrp1 in the biogenesis, cargo content or secretion of sEV of any cell type; thus, the possibility that the production of sEV by Treg cells could be altered by Nrp1 deficiency was addressed. In this work we demonstrated for the first time that Nrp1 is present in sEV secreted by wt Treg cells, and that Nrp1 deficiency in Treg cells does not impair the release (or the quantity) of sEV released by in vitro activated murine Treg cells. In addition, when we analysed protein extracts from these sEV fractions, we did not observe changes in the presence of proteins classically associated with sEV, such as Tsg‐101 and Alix, which are part of the endosomal sorting complex required for transport (ESCRT) complexes, (Latifkar et al., 2019) in sEV derived from Nrp1KO Treg cells. Therefore, these results suggest that Nrp1 may not participate in the biogenesis of Treg cell‐derived sEV activated in vitro, although further research should be performed to confirm this observation.
Moreover, this study showed that only wt Treg cell‐derived sEV exerted suppressive function in vitro, decreasing the proliferation and cell division (division index) of polyclonally‐activated convT cells On average, treatment with Nrp1KO Treg cell‐derived sEV did not inhibit convT proliferation, suggesting that the presence of Nrp1 on Foxp3+ Treg cell‐derived sEV (or their cargo) is involved in the suppressive function of sEV, either directly or indirectly. The contribution of Nrp1 to the function of sEV from Treg cells could be intrinsic to the Treg cells themselves, due to the role of Nrp1 in maintaining phenotypic stability of these cells (Overacre & Vignali, 2016).
On the other hand, the proteomic cargo of sEV has an evident importance and remains as the focus of significant interest, because it can provide valuable information about their intracellular origin, their potential “homing” (cell targeting) or the sEV cellular effect once received/incorporated on target cells (Schey et al., 2015; Vagner et al., 2019). As mentioned above, some highly expressed Treg cells proteins such as CD73, CD25 and CTLA‐4 have been also found in wt Treg cell‐derived sEV. Our results from the proteomic analysis showed that CD73 was over‐represented in wt Treg cell‐derived sEV, as previously described by Smyth and collaborators (Smyth et al., 2013), which could lead to the increased ability of the of wt Treg cell‐derived sEV to suppress the proliferation of activated T lymphocytes. We also found increased expression of CD25 or IL2RA in wt Treg cell‐derived sEV, a molecule that is also associated with increased immunosuppressive potential. The role of CD73 and CD25 on Treg cell's function, and the implications of their down‐regulation, have been reported (Smyth et al., 2013). CD82 is a tetraspanin associated with lipid rafts in the cell membrane, that it has been described as an exosome‐enriched marker, and it can elicit inhibitory effects on cell migration (Huang et al., 2020; Yeung et al., 2018). Among immune cells, CD82 is expressed by DCs and T cells, where it reduces cell motility through regulation of cytoskeleton proteins, such as RhoA, and this tetraspanin also promotes actin polymerization (Delaguillaumie et al., 2004; Delaguillaumie et al., 2002). Therefore, it is possible that Nrp1+ Treg cell‐derived sEV, containing cytoskeleton‐modulating proteins, could participate in the modulation of immune cell migration to the inflammation site and the infiltration into the allograft.
LDHC is an isoenzyme of LDH preferentially expressed in testis and associated with glucose and ATP in mammalian germ cells (Gupta, 2012), and its presence in sEV from different cell types has been reported (Anand et al., 2018; Bruno et al., 2009; Gangoda et al., 2017). Furthermore, the activation marker CD43 is coded by the gen Spn (Fulton et al., 2010), and it has been reported that it collaborates with P‐selectin glycoprotein ligand‐1 to mediate T cell migration to inflamed skin (Matsumoto et al., 2007). Activated Treg cells express CD43 on the surface membrane during viral infection (Fulton et al., 2010; Joedicke et al., 2014). Also, it has been shown that CD43 serves as a marker for a sub‐population of skin‐homing Treg cells expressing Il10 and Gzmb that inhibit immune responses in a murine model of contact hypersensitivity (Ikebuchi et al., 2016). CD43 deficiency decreases Treg cell frequency and worsens overall mortality in a murine model of sepsis through a mechanism that is still not fully described (Fay et al., 2018). Considering that CD43 expression on Treg cells could be important for Treg cells' beneficial effect during sepsis, it is reasonable to suggest that the presence of CD43 on Treg cell‐derived sEV could participate in the control of immune dysregulation and mortality.
We think that convT cells (and other inflammatory cells) receiving sEV with less abundance of these immune regulation‐related proteins could explain the less effective suppressive activity of sEV purified from Nrp1KO Treg cells, as observed in vitro (convT cell proliferation) and in vivo (transplant experiments).
There are few reports describing that the treatment with Foxp3+ Treg cell‐derived sEV extended allograft survival. Using a rat kidney transplantation model, a study found that intravenous injection of sEV derived from Treg cells could delay transplant rejection and inhibit the proliferation of allo‐reactive T cells (Yu et al., 2013). Using the same model, another study showed that Treg cell‐derived sEV increase allograft survival by decreasing pro‐inflammatory cytokine levels in the serum of treated animals (Aiello et al., 2017). Hence, we set out to analyse the participation of Nrp1 in the tolerogenic function of sEV derived from Treg cells in vivo, using a murine skin transplantation model. Our results indicated that only Nrp1+ Treg cell‐derived sEV were able to promote allograft survival, whereas the Nrp1KO sEV counterpart was unable to prevent rejection, proving that Nrp1 expression could also be relevant for the function of sEV derived from Foxp3+ Treg cells. The content of these sEV could play a key role in inducing transplant tolerance. In this context, previous reports showed that DCs transduced to express the immunosuppressive cytokines IL‐10 and TGF‐β, were capable of secreting sEV containing these cytokines, and these sEV were capable of inhibiting inflammation in models of rheumatoid arthritis and experimental colitis (Cai et al., 2012; Kim et al., 2005). A recently published report showed for the first time that sEV released by murine Treg cells possess IL‐35 associated at sEV surface, and these Treg cell‐derived sEV not only induced T‐cell and B‐cell anergy, but also promote tolerance to heart transplants through infectious tolerance (Sullivan et al., 2020). Previous results by our group showed that Nrp1KO Treg cells secreted decreased levels of IL‐10 in vivo during skin transplant rejection (Campos‐Mora et al., 2019). If cytokines like IL‐10 were contained in Treg cell‐derived sEV, it is possible that decreased suppressive function of sEV derived from Nrp1KO Treg cells may also be partially attributed to lower IL‐10 content.
In our model, treatment with these Treg cell‐derived sEV consisted of a single dose of 5 μg by subcutaneous administration at the dorsal region of the animals, flanking the skin area where grafting would be performed the next day. In this way, wt Treg cell‐derived sEV effectively promoted transplant survival. Transferred convT cells or endogenous APCs recruited to the allograft site are likely to capture these sEV (Agarwal et al., 2014; Li et al., 2016; Rojas et al., 2020). Therefore, we analysed the frequency of several innate immune populations infiltrating the skin grafts, and we found that the treatment with wt Treg cell‐derived sEV increased the proportion of M2 cells. Macrophages are critical mediators of immune responses against transplants, and M1 cells have shown to infiltrate and contribute to graft loss through inflammatory mechanisms (Liu et al., 2012). Conversely, CD206‐expressing M2 cells (also known as “alternative‐activated macrophages”) have anti‐inflammatory capacity promoting the resolution of inflammation, facilitate wound healing and angiogenesis, and therefore, they may promote allograft damage repair (Li et al., 2019). In a murine corneal allograft model, it was shown that M2 macrophages were present in animals that did not reject the transplants (Oh et al., 2013). However, other studies have reported that M2 cells might contribute to chronic rejection by promoting fibrosis and vasculopathies both in mice models and in graft biopsies from patients (Nadeau et al., 1995; Toki et al., 2014; Wu et al., 2016; Yang et al., 2003), suggesting that further research is needed to determine the participation of M2 cells in allograft rejection. Nevertheless, recent evidence also suggests that macrophages can participate in transplantation tolerance, as the case of regulatory macrophages (Mregs), a distinct macrophage subset with suppressive functions mediated by an iNOS‐dependent mechanism (Riquelme et al., 2013). Mregs are capable to inhibit CD8+ T cell immunity, promote Treg cell expansion and induce graft tolerance (Conde et al., 2015; Hutchinson et al., 2011). Some studies have shown that Mregs/Treg cell and M2/Treg cell cross‐talk is crucial to the maintenance of immune tolerance (Ikebuchi et al., 2016), due to the capacity of M2 cells to induce Treg cells (Savage et al., 2008), or due to the acquisition of an immunomodulatory phenotype on M2 linked to the presence of Treg cell‐derived IL‐10 and TGF‐β (Okeke & Uzonna, 2019). Considering this, it is remarkable that allograft rejection was associated to M1/M2 ratio increase in animals treated with a single dose of Nrp1KO Treg cell‐derived sEV, suggesting that the presence of Nrp1 on these vesicles could be important for proper intercellular communication and crosstalk between M2 and Treg cells.
Lastly, we found that Nrp1 was present in wt Treg cell‐derived sEV samples by western blot assays and fluorescence detection using NTA; however, in the proteomic analysis we were not able to detect Nrp1 in Treg cell‐derived sEV samples. Missing peptides are common in MS‐based proteomic data. In fact, it is common to have 20%–40% of all attempted intensity measures missing. This can happen in several ways: (i) the peptide is present in low abundances, and in some samples the peak intensities are not high enough to be detected or for the corresponding ions to be selected for MS/MS fragmentation, (ii) competition for charge in the ionization process, by which some ion species are liable to be dominated by others, and (iii) peptides whose chemical or physical structure cause them to get trapped in the LC column, among others.
Taken together, this study reveals that Nrp1 expression participates in the function of Treg cell‐derived sEV, and that its depletion significantly dampens sEV immunosuppressive function, which proposes a novel role for Nrp1 in the immunosuppressive mechanism of Treg cells to control the immune response and to promote tolerance.
AUTHOR CONTRIBUTIONS
Mauricio Campos‐Mora: Data curation; Formal analysis; Methodology; Writing – original draft. Javiera De Solminihac: Data curation. Carolina Rojas: Data curation; Formal analysis. Cristina Padilla: Data curation. Rodrigo Pacheco: Resources. Thilo Kaehne: Data curation; Formal analysis; Methodology. Úrsula Wyneken: Formal analysis; Methodology. Karina Pino‐Lagos: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Supervision; Validation; Writing – review & editing.
CONFLICT OF INTEREST
All authors declare no conflict of interest.
Supporting information
Supplementary Figure 1. Gating strategy for identification of NK cells, cDC, Neutrophils and Macrophages (Type‐1 and ‐2). The image depicts the gating strategy to analyze NK cells, cDC, Neutrophils and Macrophages from skin grafts. The transplants were removed and processed as indicated in the Material and method section. To obtain the frequencies of the mentioned cell populations, we draw a gate for leukocyte size followed by a gate on CD45+ cells. From this one, we identified NK cells as those expressing CD11b and NK1.1 markers. Neutrophils were discriminated as those cells expressing CD11b and Ly6G but lacking CD11c. Conventional DC were selected as CD11c+MHC‐II+/high cells, and macrophages as CD11b+CD206‐MHC‐II+/high for M1 and CD11b+CD206+/high MHC‐IIlow for M2.
Supplementary Figure 2. Gating strategy for T cell subsets analysis. This figure shows the gating strategy to identify T cell population from skin graft‐draining lymph nodes. In this case, the gating included a first region based on size for leukocytes, followed by CD45 expression. Then, we show plots depicting the expression of CD4 and Foxp3 (for Treg cells), CD4 and IFN‐γ (for Th1 cells), and CD4 with IL‐17 (for Th17 cells).
Supplementary Figure 3. Frequencies of cDC, NK cells and Neutrophils, in addition to CD4+ T cells expressing CD25 and Nrp1 are depicted in plots corresponding to samples obtained from skin grafts (top set, orange code) and draining lymph nodes (bottom set, blue and green code). Data from rejecting mice (R), and those treated with sEV obtained from wt (WTsEV) or Nrp1KO (KOsEV) are shown. One‐way ANOVA (Tukey's multiple comparison test). * p < 0,05; ** p < 0,01; *** p < 0,001; ns = not significant. n = 3 independent experiments, with n = 3‐8 mice group, per experiment.
ACKNOWLEDGEMENTS
We are grateful to Dr. Óscar Cerda (Facultad de Medicina, Universidad de Chile) for scientific advice and to Mrs. María Alejandra Espinoza (Centro Ciencia & Vida, Santiago, Chile) for technical assistance. This work was supported by FONDECYT Regular Grant 1210654 (K.P‐L.). M.C.‐M., C.R. and C.P. are fellows of the National Scholarship CONICYT numbers 21150907, 2110841 and 21210187 for graduate studies, respectively.
Campos‐Mora, M. , DeSolminihac, J. , Rojas, C. , Padilla, C. , Kurte, M. , Pacheco, R. , Kaehne, T. , Wyneken, Ú. , & Pino‐Lagos, K. (2022). Neuropilin‐1 is present on Foxp3+ T regulatory cell‐derived small extracellular vesicles and mediates immunity against skin transplantation. Journal of Extracellular Vesicles, 11, e12237. 10.1002/jev2.12237
REFERENCES
- Agarwal, A. , Fanelli, G. , Letizia, M. , Tung, S. L. , Boardman, D. , Lechler, R. , Lombardi, G. , & Smyth, L. A. (2014). Regulatory T cell‐derived exosomes: Possible therapeutic and diagnostic tools in transplantation. Frontiers in Immunology, 5, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello, S. , Rocchetta, F. , Longaretti, L. , Faravelli, S. , Todeschini, M. , Cassis, L. , Pezzuto, F. , Tomasoni, S. , Azzollini, N. , Mister, M. , Mele, C. , Conti, S. , Breno, M. , Remuzzi, G. , Noris, M. , & Benigni, A. (2017). Extracellular vesicles derived from T regulatory cells suppress T cell proliferation and prolong allograft survival. Science Reports, 7, 11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, S. , Foot, N. , Ang, C. ‐. S. , Gembus, K. M. , Keerthikumar, S. , Adda, C. G. , Mathivanan, S. , & Kumar, S. (2018). Arrestin‐Domain Containing Protein 1 (Arrdc1) regulates the protein cargo and release of extracellular vesicles. Proteomics, 18, 1800266. [DOI] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C. A. , Blake, J. A. , Botstein, D. , Butler, H. , Cherry, J. M. , Davis, A. P. , Dolinski, K. , Dwight, S. S. , Eppig, J. T. , Harris, M. A. , Hill, D. P. , Issel‐Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J. C. , Richardson, J. E. , Ringwald, M. , Rubin, G. M. , & Sherlock, G. (2000). Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics, 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi, M. , Ghabaee, M. , Moghadasi, A. N. , Noorbakhsh, F. , & Izad, M. (2018). Immunomodulatory function of Treg‐derived exosomes is impaired in patients with relapsing‐remitting multiple sclerosis. Immunologic Research, 66, 513–520. [DOI] [PubMed] [Google Scholar]
- Babicki, S. , Arndt, D. , Marcu, A. , Liang, Y. , Grant, J. R. , Maciejewski, A. , & Wishart, D. S. (2016). Heatmapper: Web‐enabled heat mapping for all. Nucleic Acids Research, 44, W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia, A. , Buzzonetti, A. , Monego, G. , Peri, L. , Ferrandina, G. , Fanfani, F. , Scambia, G. , & Fattorossi, A. (2008). Neuropilin‐1 expression identifies a subset of regulatory T cells in human lymph nodes that is modulated by preoperative chemoradiation therapy in cervical cancer. Immunology, 123, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebelman, M. P. , Crudden, C. , Pegtel, D. M. , & Smit, M. J. (2020). The convergence of extracellular vesicle and GPCR biology. Trends in Pharmacological Sciences, 41, 627–640. [DOI] [PubMed] [Google Scholar]
- Beilke, J. N. , & Gill, R. G. (2007). Frontiers in nephrology: The varied faces of natural killer cells in transplantation–Contributions to both allograft immunity and tolerance. Journal of the American Society of Nephrology: JASN, 18, 2262–2267. [DOI] [PubMed] [Google Scholar]
- Béland, S. , Désy, O. , Vallin, P. , Basoni, C. , & De Serres, S. A. (2015). Innate immunity in solid organ transplantation: An update and therapeutic opportunities. Expert review of clinical immunology, 11, 377–389. [DOI] [PubMed] [Google Scholar]
- Benichou, G. , Tonsho, M. , Tocco, G. , Nadazdin, O. , & Madsen, J. C. (2012). Innate immunity and resistance to tolerogenesis in allotransplantation. Frontiers in Immunology, 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, S. , Grange, C. , Deregibus, M. C. , Calogero, R. A. , Saviozzi, S. , Collino, F. , Morando, L. , Busca, A. , Falda, M. , Bussolati, B. , Tetta, C. , & Camussi, G. (2009). Mesenchymal stem cell‐derived microvesicles protect against acute tubular injury. Journal of the American Society of Nephrology: JASN, 20, 1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschow, S. I. , Balkom, B. W. M. , Aalberts, M. , Heck, A. J. R. , Wauben, M. , & Stoorvogel, W. (2010). MHC class II‐associated proteins in B‐cell exosomes and potential functional implications for exosome biogenesis. Immunology and Cell Biology, 88, 851–856. [DOI] [PubMed] [Google Scholar]
- Buzás, E. I. , Gardiner, C. , Lee, C. , & Smith, Z. J. (2017). Single particle analysis: Methods for detection of platelet extracellular vesicles in suspension (excluding flow cytometry). Platelets, 28, 249–255. [DOI] [PubMed] [Google Scholar]
- Cai, Z. , Zhang, W. , Yang, F. , Yu, L. , Yu, Z. , Pan, J. , Wang, L. , Cao, X. , & Wang, J. (2012). Immunosuppressive exosomes from TGF‐β1 gene‐modified dendritic cells attenuate Th17‐mediated inflammatory autoimmune disease by inducing regulatory T cells. Cell Research, 22, 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos‐Mora, M. , Contreras‐Kallens, P. , Gálvez‐Jirón, F. , Rojas, M. , Rojas, C. , Refisch, A. , Cerda, O. , & Pino‐Lagos, K. (2019). CD4+Foxp3+T regulatory cells promote transplantation tolerance by modulating effector CD4+ T cells in a neuropilin‐1‐dependent manner. Frontiers in Immunology, 10, 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos‐Mora, M. , Morales, R. A. , Gajardo, T. , Catalán, D. , & Pino‐Lagos, K. (2013). Neuropilin‐1 in transplantation tolerance. Frontiers in Immunology, 4, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos‐Mora, M. , Morales, R. A. , Pérez, F. , Gajardo, T. , Campos, J. , Catalan, D. , Aguillón, J. C. , & Pino‐Lagos, K. (2015). Neuropilin‐1+ regulatory T cells promote skin allograft survival and modulate effector CD4+ T cells phenotypic signature. Immunology and Cell Biology, 93, 113–119. [DOI] [PubMed] [Google Scholar]
- Carnell‐Morris, P. , Tannetta, D. , Siupa, A. , Hole, P. , & Dragovic, R. (2017). Analysis of extracellular vesicles using fluorescence nanoparticle tracking analysis. Methods in Molecular Biology, 1660, 153–173. [DOI] [PubMed] [Google Scholar]
- Conde, P. , Rodriguez, M. , Van D Touw, W. , Jimenez, A. , Burns, M. , Miller, J. , Brahmachary, M. , Chen, H.‐M. , Boros, P. , Rausell‐Palamos, F. , Yun, T. J. , Riquelme, P. , Rastrojo, A. , Aguado, B. , Stein‐Streilein, J. , Tanaka, M. , Zhou, L. , Zhang, J. , Lowary, T. L. , … Ochando, J. (2015). DC‐SIGN(+) macrophages control the induction of transplantation tolerance. Immunity, 42, 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, M. J. , Daley, J. M. , Van Houtte, O. , Brancato, S. K. , Henry, W. L. , & Albina, J. E. (2014). The monocyte to macrophage transition in the murine sterile wound. Plos One, 9, e86660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaguillaumie, A. , Harriague, J. , Kohanna, S. , Bismuth, G. , Rubinstein, E. , Seigneuret, M. , & Conjeaud, H. (2004). Tetraspanin CD82 controls the association of cholesterol‐dependent microdomains with the actin cytoskeleton in T lymphocytes: Relevance to co‐stimulation. Journal of Cell Science, 117, 5269–5282. [DOI] [PubMed] [Google Scholar]
- Delaguillaumie, A. , Lagaudrière‐Gesbert, C. , Popoff, M. R. , & Conjeaud, H. (2002). Rho GTPases link cytoskeletal rearrangements and activation processes induced via the tetraspanin CD82 in T lymphocytes. Journal of Cell Science, 115, 433–443. [DOI] [PubMed] [Google Scholar]
- Delgoffe, G. M. , Woo, S.‐R. , Turnis, M. E. , Gravano, D. M. , Guy, C. , Overacre, A. E. , Bettini, M. L. , Vogel, P. , Finkelstein, D. , Bonnevier, J. , Workman, C. J. , & Vignali, D. A. A. (2013). Stability and function of regulatory T cells is maintained by a neuropilin‐1‐semaphorin‐4a axis. Nature, 501, 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demory Beckler, M. , Higginbotham, J. N. , Franklin, J. L. , Ham, A. J. , Halvey, P. J. , Imasuen, I. E. , Whitwell, C. , Li, M. , Liebler, D. C. , & Coffey, R. J. (2013). Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Molecular & Cellular Proteomics: MCP, 12, 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, K. T. , Chihade, D. B. , Chen, C.‐W. , Klingensmith, N. J. , Lyons, J. D. , Ramonell, K. , Liang, Z. , Coopersmith, C. M. , & Ford, M. L. (2018). Increased mortality in CD43‐deficient mice during sepsis. Plos One, 13, e0202656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton, R. B. , Meyerholz, D. K. , & Varga, S. M. (2010). Foxp3+ CD4 regulatory T cells limit pulmonary immunopathology by modulating the CD8 T cell response during respiratory syncytial virus infection. Journal of Immunology, 185, 2382–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez‐Cancino, F. , Lopez, E. , & Lladser, A. (2019). Analysis of tissue‐resident immune cells from mouse skin and lungs by flow cytometry. Methods in Molecular Biology, 1913, 217–222. [DOI] [PubMed] [Google Scholar]
- Gangoda, L. , Liem, M. , Ang, C. ‐. S. , Keerthikumar, S. , Adda, C. G. , Parker, B. S. , & Mathivanan, S. (2017). Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics, 1, 1600370. [DOI] [PubMed] [Google Scholar]
- Gao, Y. L. , Chai, Y. F. , Qi, A. L. , Yao, Y. , Liu, Y. C. , Dong, N. , Wang, L. J. , & Yao, Y. M. (2016). Neuropilin‐1highCD4⁺CD25⁺ regulatory T cells exhibit primary negative immunoregulation in sepsis. Mediators of Inflammation, 2016, 7132158. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gupta, G. S. (2012). LDH‐C4: A target with therapeutic potential for cancer and contraception. Molecular and Cellular Biochemistry, 371, 115–127. [DOI] [PubMed] [Google Scholar]
- Hansen, W. , Hutzler, M. , Abel, S. , Alter, C. , Stockmann, C. , Kliche, S. , Albert, J. , Sparwasser, T. , Sakaguchi, S. , Westendorf, A. M. , Schadendorf, D. , Buer, J. , & Helfrich, I. (2012). Neuropilin 1 deficiency on CD4+Foxp3+ regulatory T cells impairs mouse melanoma growth. Journal of Experimental Medicine, 209, 2001–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani, K. , & Olivier, M. (2013). Immunomodulatory impact of leishmania‐induced macrophage exosomes: A comparative proteomic and functional analysis. PLoS Neglected Tropical Diseases, 7, e2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Hays, F. A. , Tomasek, J. J. , Benyajati, S. , & Zhang, X. A. (2020). Tetraspanin CD82 interaction with cholesterol promotes extracellular vesicle‐mediated release of ezrin to inhibit tumour cell movement. Journal of Extracellular Vesicles, 9, 1692417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, J. A. , Riquelme, P. , Sawitzki, B. , Tomiuk, S. , Miqueu, P. , Zuhayra, M. , Oberg, H. H. , Pascher, A. , Lützen, U. , Janßen, U. , Broichhausen, C. , Renders, L. , Thaiss, F. , Scheuermann, E. , Henze, E. , Volk, H.‐D. , Chatenoud, L. , Lechler, R. I. , Wood, K. J. , … Fändrich, F. (2011). Cutting Edge: Immunological consequences and trafficking of human regulatory macrophages administered to renal transplant recipients. Journal of Immunology, 187, 2072–2078. [DOI] [PubMed] [Google Scholar]
- Ikebuchi, R. , Teraguchi, S. , Vandenbon, A. , Honda, T. , Shand, F. H. W. , Nakanishi, Y. , Watanabe, T. , & Tomura, M. (2016). A rare subset of skin‐tropic regulatory T cells expressing Il10/Gzmb inhibits the cutaneous immune response. Science Reports, 6, 35002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177, 428–445.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joedicke, J. J. , Dietze, K. K. , Zelinskyy, G. , & Dittmer, U. (2014). The phenotype and activation status of regulatory T cells during Friend retrovirus infection. Virologica Sinica, 29, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz, S. Z. , Lu, L.‐F. , & Rudensky, A. Y. (2012). Regulatory T cells: Mechanisms of differentiation and function . Annual Review of Immunology, 30, 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. , Lee, S. , Shin, E. , Seong, K. M. , Jin, Y. W. , Youn, H. , & Youn, B. (2020). The emerging roles of exosomes as EMT regulators in cancer. Cells, 9(4), 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.‐H. , Lechman, E. R. , Bianco, N. , Menon, R. , Keravala, A. , Nash, J. , Mi, Z. , Watkins, S. C. , Gambotto, A. , & Robbins, P. D. (2005). Exosomes derived from IL‐10‐treated dendritic cells can suppress inflammation and collagen‐induced arthritis. Journal of Immunology, 174, 6440–6448. [DOI] [PubMed] [Google Scholar]
- Kolodziej, A. et al., (2013). High Resolution Quantitative Synaptic Proteome Profiling of Mouse Brain Regions After Auditory Discrimination Learning. J Vis Exp, 118. [DOI] [PMC free article] [PubMed]
- Kolodziej, A. et al., (2016). High Resolution Quantitative Synaptic Proteome Profiling of Mouse Brain Regions After Auditory Discrimination Learning, (in eng), J Vis Exp, no. 118, Dec 15 2016. [DOI] [PMC free article] [PubMed]
- Latifkar, A. , Hur, Y. H. , Sanchez, J. C. , Cerione, R. A. , & Antonyak, M. A. (2019). New insights into extracellular vesicle biogenesis and function. Journal of Cell Science, 132(13), jcs222406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, I. , Clement, E. , Ducoux‐Petit, M. , Denat, L. , Soldan, V. , Dauvillier, S. , Balor, S. , Burlet‐Schiltz, O. , Larue, L. , Muller, C. , & Nieto, L. (2015). Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell & Melanoma Research, 28, 464–475. [DOI] [PubMed] [Google Scholar]
- Li, J. , Li, C. , Zhuang, Q. , Peng, B. , Zhu, Y. , Ye, Q. , & Ming, Y. (2019). The evolving roles of macrophages in organ transplantation. Journal of Immunology Research, 2019, 5763430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Liu, C. , Yu, Z. , & Wu, M. (2016). New insights into regulatory T cells: Exosome‐ and non‐coding RNA‐mediated regulation of homeostasis and resident Treg cells. Frontiers in Immunology, 7, 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Xiao, X. , Demirci, G. , Madsen, J. , & Li, X. C. (2012). Innate NK cells and macrophages recognize and reject allogeneic nonself in vivo via different mechanisms. Journal of Immunology, 188, 2703–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb, R. J. , Becker, M. , Wen Wen, S. , Wong, C. S. F. , Wiegmans, A. P. , Leimgruber, A. , & Möller, A. (2015). Optimized exosome isolation protocol for cell culture supernatant and human plasma. Journal of Extracellular Vesicles, 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall, J. , Hill, A. F. , Hochberg, F. , Buzás, E. I. , Di Vizio, D. , Gardiner, C. , Gho, Y. S. , Kurochkin, I. V. , Mathivanan, S. , Quesenberry, P. , Sahoo, S. , Tahara, H. , Wauben, M. H. , Witwer, K. W. , & Théry, C. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for extracellular vesicles. Journal of Extracellular Vesicles, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu, M. , Martin‐Jaular, L. , Lavieu, G. , & Théry, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell‐to‐cell communication. Nature Cell Biology, 21, 9–17. [DOI] [PubMed] [Google Scholar]
- Mathivanan, S. , Lim, J. W. , Tauro, B. J. , Ji, H. , Moritz, R. L. , & Simpson, R. J. (2010). Proteomics analysis of A33 immunoaffinity‐purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue‐specific protein signature. Molecular & Cellular Proteomics: MCP, 9, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, M. , Shigeta, A. , Furukawa, Y. , Tanaka, T. , Miyasaka, M. , & Hirata, T. (2007). CD43 collaborates with P‐selectin glycoprotein ligand‐1 to mediate E‐selectin‐dependent T cell migration into inflamed skin. Journal of Immunology, 178, 2499–2506. [DOI] [PubMed] [Google Scholar]
- Mcmurchy, A. N. , & Levings, M. K. (2012). Suppression assays with human T regulatory cells: A technical guide. European Journal of Immunology, 42, 27–34. [DOI] [PubMed] [Google Scholar]
- Mi, H. , Huang, X. , Muruganujan, A. , Tang, H. , Mills, C. , Kang, D. , & Thomas, P. D. (2017). PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Research, 45, D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau, K. C. , Azuma, H. , & Tilney, N. L. (1995). Sequential cytokine dynamics in chronic rejection of rat renal allografts: Roles for cytokines RANTES and MCP‐1. Proceedings of the National Academy of Sciences of the United States of America, 92, 8729–8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J. Y. , Lee, H. J. , Ko, A. Y. , Ko, J. H. , Kim, M. K. , & Wee, W. R. (2013). Analysis of macrophage phenotype in rejected corneal allografts. Investigative Ophthalmology & Visual Science, 54, 7779–7784. [DOI] [PubMed] [Google Scholar]
- Ohkura, N. , Kitagawa, Y. , & Sakaguchi, S. (2013). Development and maintenance of regulatory T cells. Immunity, 38, 414–423. [DOI] [PubMed] [Google Scholar]
- Okeke, E. B. , & Uzonna, J. E. (2019). The pivotal role of regulatory T cells in the regulation of innate immune cells. Frontiers in Immunology, 10, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye, I. S. , Coomes, S. M. , Pelly, V. S. , Czieso, S. , Papayannopoulos, V. , Tolmachova, T. , Seabra, M. C. , & Wilson, M. S. (2014). MicroRNA‐containing T‐regulatory‐cell‐derived exosomes suppress pathogenic T helper 1 cells. Immunity, 41, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordikhani, F. , Pothula, V. , Sanchez‐Tarjuelo, R. , Jordan, S. , & Ochando, J. (2020). Macrophages in organ transplantation. Frontiers in Immunology, 11, 582939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overacre, A. E. , & Vignali, D. A. (2016). T(reg) stability: To be or not to be. Current Opinion in Immunology, 39, 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overacre‐Delgoffe, A. E. , Chikina, M. , Dadey, R. E. , Yano, H. , Brunazzi, E. A. , Shayan, G. , Horne, W. , Moskovitz, J. M. , Kolls, J. K. , Sander, C. , Shuai, Y. , Normolle, D. P. , Kirkwood, J. M. , Ferris, R. L. , Delgoffe, G. M. , Bruno, T. C. , Workman, C. J. , & Vignali, D. A. A. (2017). Interferon‐gamma drives Treg fragility to promote anti‐tumor immunity. Cell, 169, 1130–1141.e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, M. W. , Guo, H.‐F. , Li, X. , Linkugel, A. D. , & Vander Kooi, C. W. (2012). Function of members of the neuropilin family as essential pleiotropic cell surface receptors. Biochemistry, 51, 9437–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme, P. , Tomiuk, S. , Kammler, A. , Fändrich, F. , Schlitt, H. J. , Geissler, E. K. , & Hutchinson, J. A. (2013). IFN‐γ‐induced iNOS expression in mouse regulatory macrophages prolongs allograft survival in fully immunocompetent recipients. Molecular Therapy: The Journal of the American Society of Gene Therapy, 21, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, C. , Campos‐Mora, M. , Cárcamo, I. , Villalón, N. , Elhusseiny, A. , Contreras‐Kallens, P. , Refisch, A. , Gálvez‐Jirón, F. , Emparán, I. , Montoya‐Riveros, A. , Vernal, R. , & Pino‐Lagos, K. (2020). T regulatory cells‐derived extracellular vesicles and their contribution to the generation of immune tolerance. Journal of Leukocyte Biology, 108(3), 813–824. [DOI] [PubMed] [Google Scholar]
- Romano, M. , Tung, S. L. , Smyth, L. A. , & Lombardi, G. (2017). Treg therapy in transplantation: A general overview. Transplant International: Official Journal of the European Society for Organ Transplantation, 30, 745–753. [DOI] [PubMed] [Google Scholar]
- Rose, S. , Misharin, A. , & Perlman, H. (2012). A novel Ly6C/Ly6G‐based strategy to analyze the mouse splenic myeloid compartment. Cytometry. Part A: The Journal of the International Society for Analytical Cytology, 81, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Bag, A. K. , Singh, R. K. , Talmadge, J. E. , Batra, S. K. , & Datta, K. (2017). Multifaceted role of neuropilins in the immune system: Potential targets for immunotherapy. Frontiers in Immunology, 8, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinkevich, V. , Huang, Y. , Chen, Z.‐Y. , Qiang, W. , Wang, Y.‐G. , Shi, Y.‐F. , & Yang, H.‐T. (2019). Temporal dynamics of immune response following prolonged myocardial ischemia/reperfusion with and without cyclosporine A. Acta Pharmacologica Sinica, 40, 1168–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safinia, N. , Scotta, C. , Vaikunthanathan, T. , Lechler, R. I. , & Lombardi, G. (2015). Regulatory T cells: Serious contenders in the promise for immunological tolerance in transplantation. Frontiers in Immunology, 6, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris, M. , Andersen, K. G. , Randow, F. , Mayr, L. , & Betz, A. G. (2008). Neuropilin‐1 expression on regulatory T cells enhances their interactions with dendritic cells during antigen recognition. Immunity, 28, 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, N. D. L. , De Boer, T. , Walburg, K. V. , Joosten, S. A. , Van Meijgaarden, K. , Geluk, A. , & Ottenhoff, T. H. M. (2008). Human anti‐inflammatory macrophages induce Foxp3+ GITR+ CD25+ regulatory T cells, which suppress via membrane‐bound TGFbeta‐1. Journal of Immunology, 181, 2220–2226. [DOI] [PubMed] [Google Scholar]
- Schey, K. L. , Luther, J. M. , & Rose, K. L. (2015). Proteomics characterization of exosome cargo. Methods (San Diego, Calif.), 87, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, L. A. , Ratnasothy, K. , Tsang, J. Y. S. , Boardman, D. , Warley, A. , Lechler, R. , & Lombardi, G. (2013). CD73 expression on extracellular vesicles derived from CD4+ CD25+ Foxp3+ T cells contributes to their regulatory function. European Journal of Immunology, 43, 2430–2440. [DOI] [PubMed] [Google Scholar]
- Solomon, B. D. , Mueller, C. , Chae, W.‐J. , Alabanza, L. M. , & Bynoe, M. S. (2011). Neuropilin‐1 attenuates autoreactivity in experimental autoimmune encephalomyelitis. Proceedings of the National Academy of Sciences of the United States of America, 108, 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamer, W. D. , Hoffman, E. A. , Luther, J. M. , Hachey, D. L. , & Schey, K. L. (2011). Protein profile of exosomes from trabecular meshwork cells. Journal of Proteomics, 74, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J. A. , Tomita, Y. , Jankowska‐Gan, E. , Lema, D. A. , Arvedson, M. P. , Nair, A. , Bracamonte‐Baran, W. , Zhou, Y. , Meyer, K. K. , Zhong, W. , Sawant, D. V. , Szymczak‐Workman, A. L. , Zhang, Q. , Workman, C. J. , Hong, S. , Vignali, D. A. A. , & Burlingham, W. J. (2020). Treg‐cell‐derived IL‐35‐coated extracellular vesicles promote infectious tolerance. Cell Reports, 30, 1039–1051.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucleic Acids Research , (2019). The Gene Ontology Resource: 20 years and still GOing strong. 47(D1), D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Amigorena, S. , Raposo, G. , & Clayton, A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology, Chapter 3, Unit 3.22. [DOI] [PubMed]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, P. D. , Campbell, M. J. , Kejariwal, A. , Mi, H. , Karlak, B. , Daverman, R. , Diemer, K. , Muruganujan, A. , & Narechania, A. (2003). PANTHER: A library of protein families and subfamilies indexed by function. Genome Research, 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki, D. , Zhang, W. , Hor, K. L. M. , Liuwantara, D. , Alexander, S. I. , Yi, Z. , Sharma, R. , Chapman, J. R. , Nankivell, B. J. , Murphy, B. , & O'connell, P. J. (2014). The role of macrophages in the development of human renal allograft fibrosis in the first year after transplantation. American Journal of Transplantation: Official journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 14, 2126–2136. [DOI] [PubMed] [Google Scholar]
- Tung, S. L. , Boardman, D. A. , Sen, M. , Letizia, M. , Peng, Q. , Cianci, N. , Dioni, L. , Carlin, L. M. , Lechler, R. , Bollati, V. , Lombardi, G. , & Smyth, L. A. (2018). Regulatory T cell‐derived extracellular vesicles modify dendritic cell function. Science Reports, 8, 6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, T. , Chin, A. , Mariscal, J. , Bannykh, S. , Engman, D. M. , & Di Vizio, D. (2019). Protein composition reflects extracellular vesicle heterogeneity. Proteomics, 19, 1800167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi, H. , Ekström, K. , Bossios, A. , Sjöstrand, M. , Lee, J. J. , & Lötvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Wahlgren, J. , Karlson, T. D. L. , Glader, P. , Telemo, E. , & Valadi, H. (2012). Activated human T cells secrete exosomes that participate in IL‐2 mediated immune response signaling. Plos One, 7, e49723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, J. M. , Bilate, A. M. , Gobert, M. , Ding, Y. , Curotto De Lafaille, M. A. , Parkhurst, C. N. , Xiong, H. , Dolpady, J. , Frey, A. B. , Ruocco, M. G. , Yang, Y. , Floess, S. , Huehn, J. , Oh, S. , Li, M. O. , Niec, R. E. , Rudensky, A. Y. , Dustin, M. L. , Littman, D. R. , & Lafaille, J. J. (2012). Neuropilin 1 is expressed on thymus‐derived natural regulatory T cells, but not mucosa‐generated induced Foxp3+ T reg cells. Journal of Experimental Medicine, 209, 1723–1742. s1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, C. , Seeger, R. C. , Fabbri, M. , Wang, L. , Wayne, A. S. , & Jong, A. Y. (2017). Biological roles and potential applications of immune cell‐derived extracellular vesicles. Journal of Extracellular Vesicles, 6, 1400370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing, J. B. , Tanaka, A. , & Sakaguchi, S. (2019). Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity, 50, 302–316. [DOI] [PubMed] [Google Scholar]
- Witwer, K. W. , Buzás, E. I. , Bemis, L. T. , Bora, A. , Lässer, C. , Lötvall, J. , Nolte‐‘T Hoen, E. N. , Piper, M. G. , Sivaraman, S. , Skog, J. , Théry, C. , Wauben, M. H. , & Hochberg, F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 2, 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. , Zhao, Y. , Xiao, X. , Fan, Y. , Kloc, M. , Liu, W. , Ghobrial, R. M. , Lan, P. , He, X. , & Li, X. C. (2016). Graft‐infiltrating macrophages adopt an M2 phenotype and are inhibited by purinergic receptor P2×7 antagonist in chronic rejection. American Journal of Transplantation: Official journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 16, 2563–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, M. , Louvet, C. , Davini, D. , Gardner, J. M. , Martinez‐Llordella, M. , Bailey‐Bucktrout, S. , Anthony, B. A. , Sverdrup, F. M. , Head, R. , Kuster, D. J. , Ruminski, P. , Weiss, D. , Von Schack, D. , & Bluestone, J. A. (2012). Neuropilin‐1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. Journal of Experimental Medicine, 209, 1713–1722. s1711‐1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Reutzel‐Selke, A. , Steier, C. , Jurisch, A. , Tullius, S. G. , Sawitzki, B. , Kolls, J. , Volk, H. D. , & Ritter, T. (2003). Targeting of macrophage activity by adenovirus‐mediated intragraft overexpression of TNFRp55‐Ig, IL‐12p40, and vIL‐10 ameliorates adenovirus‐mediated chronic graft injury, whereas stimulation of macrophages by overexpression of IFN‐gamma accelerates chronic graft injury in a rat renal allograft model. Journal of the American Society of Nephrology: JASN, 14, 214–225. [DOI] [PubMed] [Google Scholar]
- Yeung, L. , Hickey, M. J. , & Wright, M. D. (2018). The many and varied roles of tetraspanins in immune cell recruitment and migration. Frontiers in Immunology, 9, 1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Huang, C. , Song, B. , Xiao, Y. , Fang, M. , Feng, J. , & Wang, P. (2013). CD4+CD25+ regulatory T cells‐derived exosomes prolonged kidney allograft survival in a rat model. Cellular Immunology, 285, 62–68. [DOI] [PubMed] [Google Scholar]
- Yuan, Q. , Hong, S. , Shi, B. , Kers, J. , Li, Z. , Pei, X. , Xu, L. , Wei, X. , & Cai, M. (2013). CD4(+)CD25(‐)Nrp1(+) T cells synergize with rapamycin to prevent murine cardiac allorejection in immunocompetent recipients. Plos One, 8, e61151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Gating strategy for identification of NK cells, cDC, Neutrophils and Macrophages (Type‐1 and ‐2). The image depicts the gating strategy to analyze NK cells, cDC, Neutrophils and Macrophages from skin grafts. The transplants were removed and processed as indicated in the Material and method section. To obtain the frequencies of the mentioned cell populations, we draw a gate for leukocyte size followed by a gate on CD45+ cells. From this one, we identified NK cells as those expressing CD11b and NK1.1 markers. Neutrophils were discriminated as those cells expressing CD11b and Ly6G but lacking CD11c. Conventional DC were selected as CD11c+MHC‐II+/high cells, and macrophages as CD11b+CD206‐MHC‐II+/high for M1 and CD11b+CD206+/high MHC‐IIlow for M2.
Supplementary Figure 2. Gating strategy for T cell subsets analysis. This figure shows the gating strategy to identify T cell population from skin graft‐draining lymph nodes. In this case, the gating included a first region based on size for leukocytes, followed by CD45 expression. Then, we show plots depicting the expression of CD4 and Foxp3 (for Treg cells), CD4 and IFN‐γ (for Th1 cells), and CD4 with IL‐17 (for Th17 cells).
Supplementary Figure 3. Frequencies of cDC, NK cells and Neutrophils, in addition to CD4+ T cells expressing CD25 and Nrp1 are depicted in plots corresponding to samples obtained from skin grafts (top set, orange code) and draining lymph nodes (bottom set, blue and green code). Data from rejecting mice (R), and those treated with sEV obtained from wt (WTsEV) or Nrp1KO (KOsEV) are shown. One‐way ANOVA (Tukey's multiple comparison test). * p < 0,05; ** p < 0,01; *** p < 0,001; ns = not significant. n = 3 independent experiments, with n = 3‐8 mice group, per experiment.


