Abstract
As the most predominant RNA epigenetic regulation in eukaryotic cells, N6-methyladenosine (m6A) plays a critical role in human tumorigenesis and cancer progression. However, the biological function and molecular mechanism of m6A regulation in naso-pharyngeal carcinoma (NPC) remain elusive. Here, we showed that Wilms’ tumor 1-associating protein (WTAP) expression was apparently upregulated in NPC, and increased WTAP was associated with poor prognosis. WTAP upregulated in NPC was fine-tuned by KAT3A-mediated H3K27 acetylation. Functionally, WTAP was required for the growth and metastasis of NPC. Mechanistically, lncRNA DIAPH1-AS1 was identified as a bona fide m6A target of WTAP. WTAP-mediated m6A modification of DIAPH1-AS1 enhanced its stability relying on the m6A reader IGF2BP2-dependent pathway. Furthermore, DIAPH1-AS1 acted as a molecular adaptor that promoted MTDH-LASP1 complex formation and upregulated LASP1 expression, ultimately facilitating NPC growth and metastasis. Thus, WTAP-mediated DIAPH1-AS1 m6A methylation is required for NPC tumorigenesis and metastasis.
Subject terms: Cancer genetics, Metastasis, Epigenetics
Introduction
Nasopharyngeal carcinoma (NPC) is a metastasis-prone primary nasopharynx malignancy that is endemic in southern China, where the undifferentiated type occurs most frequently [1]. Unfortunately, because of the high invasion and early metastasis characteristics of NPC, patients diagnosed with NPC frequently present with advanced disease at first diagnosis [2]. Despite progress in diagnosis and systemic treatment based on radiotherapy, the prognosis of patients with NPC remains unsatisfactory [3]. Consequently, there is an urgent need to determine the mechanism underlying NPC growth and metastasis, which would facilitate the development of specific therapeutic strategies against NPC. Accumulating evidence shows that epigenetic dysregulation, such as DNA methylation [4], non-coding RNA [5, 6], and super-enhancers [7], contributes substantially to the occurrence and progression of NPC. However, these studies predominantly focused on epigenetic alterations at the transcriptional level, while the post-transcriptional modifications in NPC have not been explored further.
Post-transcriptional modifications of eukaryotic RNAs, mainly including N6-methyladenosine (m6A), N1-methyladenosine (m1A), and 5-methylcytosine (m5C), have been proven to be fine-tuning chemical structural features of basic RNAs, with critical roles in regulating transcript stability, translation, alternative splicing (AS), subcellular localization, and phase separation [8]. Among them, m6A is the most prevalent in higher eukaryote mRNA and non-coding RNA. In mammals, it chiefly occurs on the adenine of an RRACH (R corresponds to G or A; H corresponds to A, C, or U) sequence, which is catalyzed by a large methyltransferase complex containing methyltransferase-like 3/14 (METTL3/14), Wilms tumor 1-associated protein (WTAP), and Vir-like m6A methyltransferase associated (VIRMA). In contrast, two m6A demethylases, AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated (FTO), play central roles in selectively removing the methyl code from their target RNAs. Subsequently, the fate of m6A-modified RNAs depends on the reader proteins that typically interpret and mediate the functional outcome of dynamic m6A deposition, which ultimately regulates gene expression and cell fate [9–11].
Recently, the dysregulation of m6A modification has been associated with diverse biological processes, including cancer development. For instance, ALKBH5 promotes the tumorigenicity of glioblastoma stem-like cells by decreasing the m6A modification on FOXM1 (forkhead box M1) nascent transcripts and thus maintaining its expression [12]. METTL3 was proven to accelerate cellular glycolysis and promote the progression of gastric cancer (GC) by upregulating the m6A modification on HDGF (heparin binding growth factor) mRNA and enhancing its RNA stability through the insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3)-dependent pathway [13]. Intriguingly, METTL14 exerts its tumor-inhibitory function by downregulating an oncofetal long noncoding RNA (lncRNA) XIST in a METTL14-YTH N6-methyladenosine RNA binding protein 2 (YTHDF2)-mediated, m6A-dependent manner [14]. Our previous study found the low m6A deposition on the ZNF750 transcripts, which is likely correlated with the instability and low expression of ZNF750 in NPC [15]. These studies shed light on the function and prognostic value of m6A modulators that alter a substantial subset of crucial genes that participate in the intricate process of tumorigenesis and cancer development. However, the precise function and the underlying regulatory mechanisms of m6A modulators in NPC remain elusive.
In the present study, we aimed to investigate WTAP expression and function in NPC. The results showed that WTAP was noticeably upregulated in NPC and promoted NPC cell proliferation, migration, and invasion in vitro and in vivo. Mechanistically, WTAP-mediated m6A modification of the lncRNA DIAPH1-AS1, which was interpreted by m6A reader insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2). This positively modulated DIAPH1-AS1 expression, facilitated the Metadherin (MTDH)-LIM and SH3 protein 1 (LASP1) interaction, maintained LASP1 expression, and eventually promoted NPC growth and metastasis. These results suggest that WTAP is a novel prognostic and therapeutic target in NPC.
Materials and methods
Clinical specimens
This study was approved by the Institutional Review Board of Sun Yat-Sen University Cancer Center (GZR2020-220). Written informed consent was obtained from the patients before the study began. All human NPC paraffin-embedded biopsy tissues were obtained from patients with detailed clinical characteristics and long-term follow-up data from January 2004 to December 2013.
Cell culture
All the cell lines were authenticated and were kindly provided by Dr. M. Zeng (Sun Yat-Sen University Cancer Center). Human NPC cells were maintained in Roswell Park Memorial Institute (RPMI)−1640 medium (Invitrogen, Carlsbad, CA, USA) or Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen). The human immortalized nasopharyngeal epithelial cell lines NP69 and N2Tert were cultured in keratinocyte serum-free medium (KSFM, Invitrogen) supplemented with bovine pituitary extract (BD Biosciences, San Diego, CA, USA).
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was isolated using TRIzol (Invitrogen), and the cDNA was synthesized using HiScript III RT SuperMix (Vazyme, Nanjing, China). Quantitative real-time PCR (qPCR) assays were carried out using a CFX96 Touch real-time system (Bio-Rad, Hercules, CA, USA) with SYBR Green qPCR SuperMix-UDG reagents (Invitrogen). The threshold cycle number (CQ) was analyzed in triplicate for each sample. The CQ values for the target genes were normalized against that of GAPDH (glyceraldehyde-3-phosphate dehydrogenase). The primer sequences used for qRT-PCR are shown in Supplementary Table S1.
Western blotting analysis
Cells were lysed in Radioimmunoprecipitation assay (RIPA) buffer (Millipore, Billerica, MA, USA) containing protease and phosphatase inhibitors (Thermo Fisher Scientific, Waltham, MA, USA) to extract the total proteins. Western blotting was performed as described in our previous study [16]. The antibodies used in this study were: Anti-WTAP (1:1000, Abcam, Cambridge, MA, USA), anti-KAT3A (1:1000, Abcam), anti-IG2BP2 (1:2000, Abcam), anti-MTDH (1:1000, Abcam), anti-HA-tag (1:5000, Abcam), anti-LASP1 (1:2000, Abcam) and anti-α-tubulin (1:5000, Abcam).
Cell transfection and lentiviral infection
For RNA interference, the synthesized duplex RNA interference (RNAi) oligonucleotides targeting human KAT3A, WTAP, DIAPH1-AS1, IGF2BP2, MTDH, and LASP1 mRNA sequences were purchased from RiboBio (Guangzhou, China). A scrambled duplex RNA oligonucleotide was used as an RNA negative control. The sequences are listed in Supplementary Table S2. For gene overexpression, cDNAs comprising the open reading frames of human WTAP (EX-Z3234-M06), IGF2BP2 (EX-H1849-M02), MTDH (EX-I2253-M06), LASP1 (EX-M0555-M06) with sequences encoding an N-terminal HA-tag were purchased from GeneCopoeia (Guangzhou, China). The full-length, truncated variants and m6A methylated site mutations (wild type, A-T, A-G, A-Del) of lncRNA DIAPH1-AS1 were synthesized by Genscript (Nanjing, China) and cloned into the pcDNA3.1 (−) or pmir-GLO vectors to generate overexpression or dual-luciferase vectors. The empty vector was used as the control. Cells were transfected with the siRNA using Lipofectamine RNAiMAX (Invitrogen) or with plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturers’ instructions.
For short hairpin RNA (shRNA)-mediated WTAP stable knockdown, lentiviruses expressing shRNA clones for WTAP and a scrambled shRNA control were designed and then produced by GeneCopoeia (Supplementary Table S3). SUNE-1 and HONE-1 cells were plated at 50% confluence on six-well plates and infected with the virus at certain concentrations accordingly to the manufactures’ instructions. Target cells were selected using 1 μg/ml puromycin (Thermo Fisher Scientific, Waltham, USA) for 7 days.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed using a Pierce Magnetic ChIP Kit (Thermo Fisher Scientific). Briefly, cells were fixed using 1% formaldehyde and quenched with 125 mM glycine. Nuclei were harvested and sonicated to generate fragments of ~200 bp. For ChIP, 50 μl of sonicated chromatin was immunoprecipitated overnight with 5 μg of anti-H3K27ac (CST, Boston, MA, USA) or KAT3A (Abcam) antibodies. Beads with bound immunocomplexes were washed, eluted, and DNA was then isolated to perform qPCR. Input was prepared with 10% of the sonicated chromatin. The primers are listed in Supplementary Table S4.
Transwell migration and invasion assay
For Transwell invasion and migration assays, 5 × 104 (migration assay) or 1 × 105 (invasion assay) cells were seeded into the upper chambers in FBS-free media with or without Matrigel (BD Biosciences, San Jose, CA, USA), respectively. Medium containing 10% FBS was added to the lower chamber. After a 24-h of incubation, the cells located on the lower surface of upper chambers were fixed, stained with hematoxylin, and observed under an inverted microscope.
Cell proliferation and colony formation assays
For cell proliferation assay, 1 × 103 cells/well were seeded in a 96-well plate, and cell viability was detected every 24 h for 5 days using a Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. For colony formation assay, cells were seeded at a density of 400 cells/well in 6-well plates and cultured for approximately 10 days until colonies were detectable. The colonies were then fixed with methanol, stained with hematoxylin, and analyzed.
Analyses of cell cycle and cell apoptosis
Cell cycle and apoptosis analyses were performed using the Cell Cycle Detection Kit (KeyGEN BioTECH, Jiangsu, China) and Annexin V-FITC/PI Cell Apoptosis Detection Kit (KeyGEN BioTECH) according to the manufacturer’s directions. For cell cycle analysis, the NPC cells were collected, washed with 1× PBS, and suspended at a concentration of 1 × 106/ml. Then, cells were fixed with 70% ice-cold ethanol overnight and treated with RNase. Finally, cells were stained with Propidium Iodide (PI) for 30 min at 4 °C and submitted for cell cycle analysis on Flow Cytometry (Ex = 488 nm).
For cell apoptosis analysis, 5 × 105 NPC cells were suspended in 500 μl 1× Binding Buffer and stained with 1 μl Annexin V-FITC and 5 μl PI for 5 min at room temperature. Then, cells were submitted to cell apoptosis analysis on Flow Cytometry (Ex = 488 nm; Em = 530 nm). The cell cycle and apoptosis phenotypes were analyzed using FlowJoTM software (v9).
Quantification of global m6A levels
Total RNA was isolated from NPC cells with or without WTAP silenced using TRIzol, treated with DNase I (NEB, Ipswich, USA), and the total polyadenylated RNA (poly(A) + mRNA) was isolated using GenElute™ mRNA Miniprep Kit (Sigma-Aldrich, MO, USA) by incubating total RNA with Oligo(dT) polystyrene beads for 10 min at room temperature. After washing to remove contaminants, the poly(A) + mRNA is eluted and sent for conducting m6A level quantification. Equal amounts of total poly(A) + mRNA (200 ng) were coated on assay wells. Capture antibody solution and detection antibody solution were added respectively to assay wells with appropriate dilution concentration. The m6A levels were colorimetrically measured by reading the absorbance at OD450 nm, and then calculations were performed based on the standard curve.
m6A dot blots
m6A dot blots were conducted as described in other studies [17]. Briefly, total RNA was isolated from NPC cells using TRIzol reagent. After fragmentation using RNA Fragmentation Module (NEB), equal amounts of serial-diluted RNA were adopted for conducting m6A immunoprecipitation (MeRIP) using Magna MeRIPTM m6A kit (Millipore) according to the manuscript instructions. The eluted RNA was added into a nitrocellulose membrane (Boster-Bio, Wuhan, China) fixed with 96-wells Bio-Dot® Module (Bio-Rad) and then cross-linked with UV crosslinker. After blocking and incubating with the anti-m6A antibody (1:500; Synaptic Systems, Goettingen, Germany), the membrane was incubated with secondary antibodies and then detected using chemiluminescence with the ChemiDocTM MP Imaging System (Bio-Rad).
m6A microarray analysis
m6A microarray analysis was conducted by Aksomics Company (Shanghai, China) according to previous studies [18, 19]. Briefly, total RNAs were extracted from cells and incubated with m6A antibody for immunoprecipitation (IP). The modified RNAs (IP) and unmodified RNAs (Sup) were labeled with Cy5 and Cy3, respectively. Cy5 labeled cRNAs (IP) were fragmented and hybridized to a human m6A epitranscriptomic microarray (Arraystar, Rockville, USA) that contained 44,122 mRNA and 12,496 lncRNA degenerate probes. The hybridized arrays were scanned using an Agilent Scanner G2505C. Differentially m6A-methylated RNAs were identified by filtering with the P-value < 0.05 and log2(|Fold Changes (FC)|) > 0.5.
MeRIP-qPCR
MeRIP was conducted to examine m6A modification of genes using the Magna MeRIPTM m6A kit (Millipore) according to the manufacturer’s instructions. Briefly, 10 μg aliquot of anti-m6A antibody (Synaptic Systems) was conjugated to ChIP grade protein A/G magnetic beads overnight at 4°C. Three hundred microgram aliquot of fragmented total RNA was then incubated with the antibody in IP buffer with protease inhibitor and RNase inhibitor. The m6A modified RNA was then eluted with elution buffer, purified through Phenol/Chloroform/Isoamyl alcohol (25:24:1, Millipore) extraction, and then analyzed via quantitative RT-PCR assays. The primer sequences are shown in Supplementary Table S1.
Dual-luciferase reporter assay
Luciferase activity was measured by the Dual-luciferase Reporter Assay System (Promega, Madison, USA) according to the manuscript’s instructions. Briefly, NPC cells were co-transfected with each Firefly luciferase reporter construct in combination with the Renilla luciferase construct, and both luciferase activities in cell extracts at 48 h after transfection were measured using a Steady-Glo® Luciferase Assay System (Promega). The Firefly luciferase activity was normalized to the Renilla luciferase activity that reflects expression efficiency.
RNA pulldown
The biotin-labeled RNA pulldown probes were synthesized by Sangon Biotech (Shanghai, China), and the sequences are shown in Supplementary Table S5. The biotin-coupled RNA complex was pulled down by incubating cell lysates with streptavidin-coated magnetic beads (Invitrogen) under the manufacture’s instruction [20]. The bound proteins were eluted and sent for LC/MS analysis (Fitgene Biotech, Guangzhou, China) or western blot analysis. The MS results were derived by the Fitgene Biotechnology Company. The scores reported in the table were derived from ions scores as a non-probabilistic basis for ranking protein hits. The ions score is −10×Log(P), where P is the probability that the observed match is a random event. Individual ions scores > 23 indicate identity or extensive homology (P < 0.05).
RNA immunoprecipitation (RIP)
RIP was conducted using Magna RIPTM Kit (Millipore) following the manufacturer’s protocol. Protein A/G magnet beads coated with 10 μg of specific antibody or normal IgG were incubated with cell lysates at 4 °C overnight. The co-precipitated RNAs were then isolated by elution buffer, purified by Phenol/Chloroform/Isoamyl alcohol (25:24:1), and subsequently subjected for quantitative RT-PCR analysis.
Fluorescence in-situ hydration (FISH) and immunofluorescence (IF)
FISH and IF double staining were performed to detect the co-localization and interaction of DIAPH1-AS1 and IGF2BP2 or MTDH. Alexa Fluor 555-labeled DIAHPH1-AS1 FISH probes were designed and synthesized by RiboBio. Briefly, cells were seeded on glass slides and left overnight. After fixation and permeabilization, cells were incubated with DIAHPH1-AS1 FISH probes, followed by anti-IGF2BP2 (Abcam; 1:100) or anti-MTDH antibodies (Abcam; 1:100). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma). Images were acquired using a fluorescence microscope (Olympus FV1000, Tokyo, Japan) and Z-Stack was performed using a laser confocal microscope (Zeiss LSM-880 Fast AiryScan, Germany).
Immunoprecipitation (IP)
IP assay was performed using PierceTM Co-Immunoprecipitation Kit (Thermo Fisher) following the manufacturer’s instructions. Cells were lysed with IP buffer and sent for centrifugation. The supernatant was incubated with anti-MTDH antibodies (Abcam), anti-LASP1 antibodies or anti-hemagglutinin (HA) antibodies (Abcam) for one hour. Immunocomplexes were recovered and washed four times in IP buffer, and then sent for LC/MS analysis (Fitgene) or western blot analysis.
Animal experiments
Establishment and analysis of BALB/c nude mice subcutaneous xenograft and metastasis models were performed as we described before [4]. About in vivo tumorigenesis model, 1 × 106 indicated SUNE-1 cells were subcutaneously injected into the flanks of nude mice. The tumor volumes were measured every 4 days. On day 32 after injection, the mice were sacrificed and the subcutaneous tumors were excised and weighed. For the inguinal lymph node metastasis model, 2 × 105 indicated SUNE-1 cells were injected into the footpads of nude mice. Following a 6-week growth, the mice were sacrificed, with their footpad tumors and inguinal lymph nodes detached. All tumors and inguinal lymph nodes were paraffin-embedded and cut into 5 μm tissue sections for subsequent analysis. All animal experiments were approved by the Institutional Animal Care and Use Committee, Sun Yat-sen University Cancer Center (L102012019110C).
In-situ hybridization (ISH) staining
The ISH probes specific for human lncRNA DIAPH1-AS1 were obtained for Sangon Biotech (Shanghai, China) and the sequence was list in Supplementary Table S6. ISH assay was performed using the ISH Kit (Boster-Bio) according to the manufacturer’s instructions. Briefly, after deparaffinization, rehydration, and digestion, the samples were incubated with digoxin-labeled ISH probes and corresponding antibodies. The sections were stained using streptavidin horseradish peroxidase and visualized with 3,3′-diaminobenzidine (DAB).
Immunohistochemistry (IHC) staining
For IHC staining, the specimens were incubated with anti-WTAP antibodies (1:200, Abcam) at 4 °C overnight, followed by incubation of biotin-labeled corresponding secondary antibody. The avidin biotinylated peroxidase complex methods were used to determine the relative expression of the target protein to visualize the bound antibodies. For the IHC staining analysis, a semi-quantified scoring criterion called immunoreactivity-scoring (IRS) system was used as previous instructions [21], in which the staining intensity was designated as either not existent (0), weak (1), moderate (2), or strong (3), and the proportion of cell staining was scored as either no cells stained (0), less than 25% of cells stained (1), 25–50% of cells stained (2), 51–75% of cells stained (3) or more than 75% of cells stained (4). The IRS was calculated by the multiplication of these two variables. For statistical analysis, cases were grouped as either low WTAP expression (IRS 0–6) or high WTAP expression (IRS 7–12).
Statistical analyses
Statistical analyses were carried out using GraphPad Prism (version 8.0; GraphPad Inc., La Jolla, CA, USA), IBM SPSS (version 23.0; IBM Corp., Armonk, NY, USA), and R (version 3.6) software for Windows. Data were obtained from at least three independent experiments and statistical significance was analyzed using a two-tailed unpaired Student’s t-test between two groups and by one-way analysis of variance (ANOVA) followed by a Bonferroni test for multiple comparisons. A P-value of <0.05 was considered statistically significant.
Results
WTAP is upregulated and correlates with poor prognosis in NPC
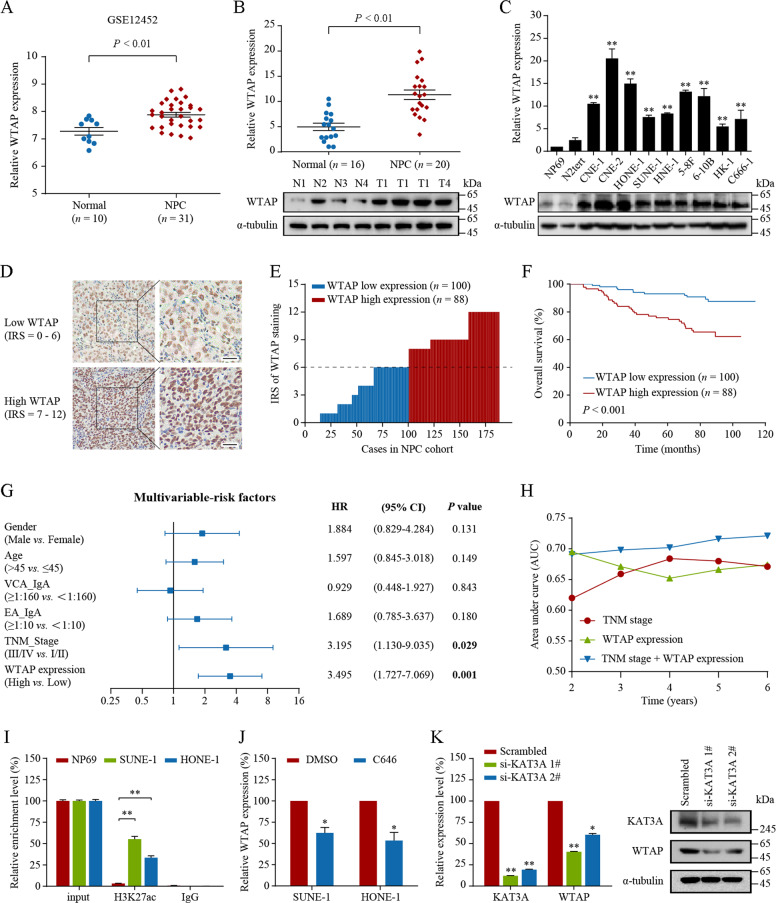
To investigate the potential role of m6A mediators in NPC, we first compared the mRNA levels of several key m6A regulators based on GEO database GSE12452 containing 31 NPC and 10 non-tumor nasopharyngeal epithelial tissues. WTAP was the most significantly differentially expressed m6A regulator and identified to be upregulated in NPC (Fig. 1A, P < 0.01). For verification, we detected the mRNA level of WTAP using quantitative real-time reverse transcription PCR (qRT-PCR) and found WTAP was remarkably higher in cancerous (n = 20) compared with those in samples (n = 16) (Fig. 1B, upper panel). Moreover, WTAP protein levels were also obviously increased in NPC compared to normal tissues (Fig. 1B, lower panel). Consistently, the WTAP mRNA and protein levels in nine NPC cell lines were significantly upregulated relative with those in the immortalized normal nasopharyngeal epithelial cell lines (NP69 and N2Tert, Fig. 1C).
Fig. 1. WTAP is upregulated in NPC and serves as a prognostic factor for NPC patients.
A The relative expression of WTAP in NPC (n = 31) and normal tissues (n = 10) based on data from the GEO database (GSE12452). B The relative expression of WTAP in NPC and normal nasopharyngeal epithelial tissues. C The relative expression of WTAP in NPC cell lines and two immortalized nasopharynx epidermal cells (NP69 and N2tert). WTAP was detected by qRT-PCR (upper panel) or western blotting (lower panel), and normalized to GAPDH or α-tubulin. D–H The prognostic value of WTAP in NPC was assessed by IHC staining in the NPC cohort (n = 188). The expression level of WTAP was measured using the WTAP immunoactivity score (IRS) system. D Two examples of the assigned IRS result are provided, with designations of 0–6 as WTAP low expression (n = 100) and 7–12 as WTAP high expression (n = 88). Scale bar: 25 μm. E The distribution of WTAP IRS in the above-mentioned NPC cohort (n = 188). F Kaplan–Meier overall survival (OS) estimate of WTAP expression in patients with NPC from our cohort (n = 188). G Multivariable Cox analysis of clinical prognostic parameters for OS of patients with NPC (n = 188). The hazard ratio (HR), 95% confidence interval (95% CI), and P-value are listed. H The time-dependent receiver operating characteristic (ROC) analysis showing the clinical risk score (TNM stage), the WTAP risk score, and the combined clinical and WTAP risk scores in the NPC cohort. I The enrichment of H3K27ac at the WTAP promoter was evaluated via ChIP-qPCR assays in NP69 and two NPC cell lines (SUNE-1 and HONE-1). J WTAP expression was measured by qRT-PCR in SUNE-1 and HONE-1 cells treated with C646 (10 μM) or DMSO for 48 h. K Quantitative RT-PCR (left panel) and western blotting (right panel) were used to measure KAT3A and WTAP expression in HONE-1 cells with KAT3A silencing. Data are presented as the mean ± SEM or mean ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.
We subsequently analyzed the clinical significance of WTAP in a cohort of NPC samples (n = 188) using immunohistochemistry analysis. Patients were divided into two groups (WTAP low and high expression, Fig. 1D‒E). WTAP high expression was significantly correlated with clinical outcomes of NPC patients (Supplementary Table S7), including locoregional failure (P = 0.036), distant metastasis (P = 0.027), and death (P < 0.001). Kaplan–Meier analysis revealed that high WTAP expression was associated significantly with poor overall survival (Fig. 1F, P < 0.001). Multivariable Cox regression analysis revealed that WTAP expression and TNM stage were independent prognostic factors for patients with NPC (Fig. 1G, both P < 0.05). To further evaluate the prognostic value of WTAP expression, we constructed an integrated prognostic model in combination with the TNM stage and WTAP expression. In this model, high-risk patients were defined as high WTAP expression with advanced NPC (n = 69, death events = 26), and low-risk patients were defined as low WTAP expression and/or early-stage NPC (n = 119, death events = 15). We then conducted time-dependent receiver operating characteristics (ROC) curve analysis, in which the TNM stage and WTAP expression were used separately or in combination. As shown in Fig. 1H, the integrated model had a higher accuracy for prognostic evaluation than either TNM stage or WTAP expression alone. These results support that WTAP is upregulated in NPC and might serve as a potential prognostic biomarker for patients with NPC.
KAT3A-mediated H3K27 acetylation transcriptionally activates WTAP in NPC
To explore the potential mechanism of WTAP upregulation in NPC, we analyzed WTAP promoter using the UCSC Genome Browser (http://generic.ucsc.edu/). As shown in Supplementary Fig. S1A, acetylation of H3K27 (H3K27ac) signals were enriched in the WTAP promoter region, indicating that WTAP might be regulated at the transcriptional level via histone acetylation. Using chromatin immunoprecipitation-quantitative real-time PCR (ChIP-qPCR) assays, we identified a gain of H3K27ac at WTAP promoter in NPC cell lines (SUNE-1 and HONE-1) compared with that in the control cell line NP69 (Fig. 1I, P < 0.01; Supplementary Fig. S1B). Furthermore, treatment with the histone acetyltransferase (HAT) inhibitor C646 markedly decreased the WTAP expression (Fig. 1J, P < 0.05; Supplementary Fig. S1C). Since CREB-binding protein (CREBBP, also known as KAT3A) is necessary for histone acetylation, we performed ChIP assays to confirm the enrichment of KAT3A at the WTAP promoter (Supplementary Fig. S1D, P < 0.01). Additionally, silencing KAT3A conspicuously reduced the enrichment of H3K27ac at the WTAP promoter (Supplementary Fig. S1E, P < 0.05), thereby decreasing WTAP expression (Fig. 1K, all P < 0.05). These data strongly support that WTAP frequently elevated in NPC, and this elevation is partially caused by KAT3A-mediated histone acetylation activation of its promoter.
Since the apparent epigenetic modification caused by Epstein-Barr virus (EBV) infection is inseparable from the malignant transformation process of nasopharyngeal epithelial cells [22, 23], we further investigated the contribution of DNA methylation to the dysregulation of WTAP expression in NPC. We first analyzed the DNA methylation profile from previous studies (GSE52068 and GSE62366) [24]. However, inconsistent results were found between two studies, indicating that the DNA methylation level of WTAP could not be definitively determined (Supplementary Fig. S1F, G). To explore this further, we treated cells with DNA demethylation regent 5-Aza-2′-deoxycytidine (DAC) and found that the DAC treatment was nearly unable to boost the expression of WTAP in the immortalized nasopharyngeal epithelial cell line N2Tert (Supplementary Fig. S1H, fold change = 1.16). Furthermore, pan-cancer multivariable survival analysis using an online DNA methylation analysis tool MethSurv (https://biit.cs.ut.ee/methsurv/) revealed that DNA methylation rate of WTAP was not a specific risk factor for all cancer types (Supplementary Fig. S1I, all not significant). Collectively, these results indicated that the DNA hypomethylation of the WTAP promoter was not the main contributor for the WTAP upregulation in NPC. We also explored whether the EB virus infection has influence on the upregulated H3K27ac level of WTAP in NPC cells. ChIP-qPCR assays showed that EBV-positive (C666-1) and EBV-negative (CNE-1 and CNE-2) NPC cells shared similar H3K27ac enrichment level in the WTAP promoter region (Supplementary Fig. S1J). These results indicated that EBV infection may not the main reason for the upregulated H3K27ac modification level of WTAP in NPC.
WTAP facilitates NPC cell proliferation and metastasis in vitro and in vivo
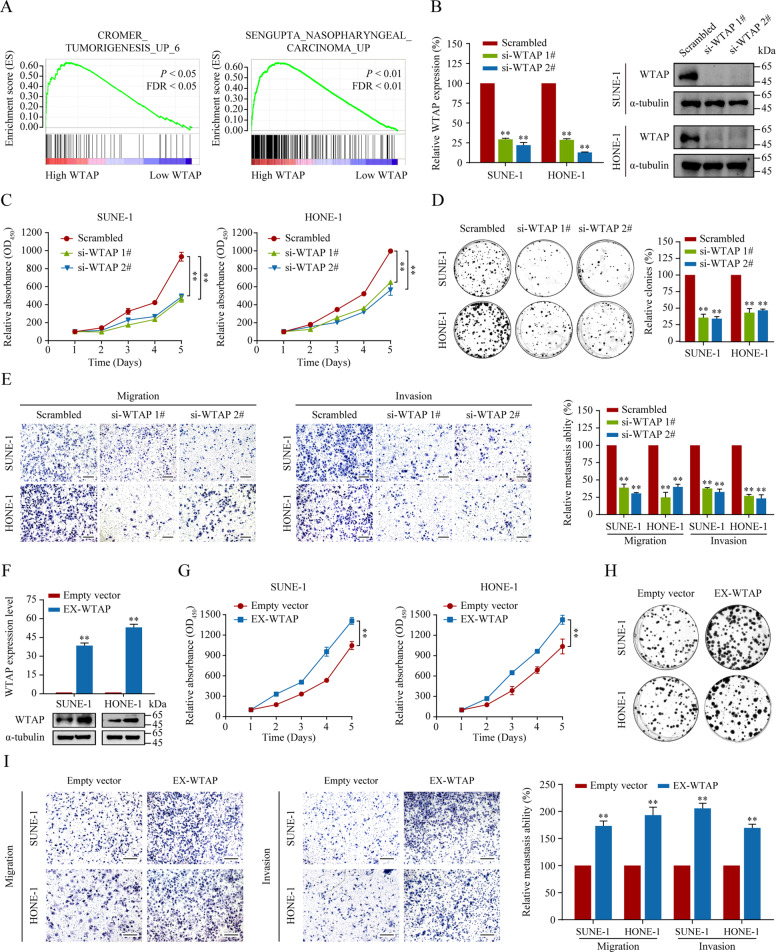
To gain an insight into the function of WTAP, we implemented gene set enrichment analysis (GSEA) based on NPC microarray data in the GEO database (GSE12452). The GESA results revealed that upregulated WTAP expression was closely associated with NPC development (Fig. 2A, P < 0.05). Specifically, we found that multiple oncogenic-related gene sets correlated positively with high WTAP expression (Supplementary Fig. S2A‒C, all P < 0.05), suggesting a crucial regulatory role of WTAP in NPC progression.
Fig. 2. WTAP facilitates NPC cell proliferation and metastasis in vitro.
A GSEA of data from the GEO database (GSE12452) revealed that WTAP expression correlated positively with tumorigenesis and NPC carcinogenesis. B siRNA-mediated interference was used to knockdown WTAP in SUNE-1 and HONE-1 cells. Knockdown efficiency was determined using qRT-PCR (left panel) and western blotting (right panel) as indicated. C, D Cell proliferation was analyzed using CCK-8 (C) and colony formation (D) assays in SUNE-1 and HONE-1 cells with or without WTAP silencing. E Representative image (left panel) and statistical analysis (right panel) of Transwell migration and invasion assays in SUNE-1 and HONE-1 cells with or without WTAP knockdown. Scale bar: 200 μm. F Overexpress efficiency of WTAP was assessed using qRT-PCR (upper panel) and western blotting (lower panel). G, H Cell proliferation was analyzed using CCK-8 (G) and colony formation (H) assays in SUNE-1 and HONE-1 cells with or without WTAP overexpression. I Representative image (left panel) and statistical analysis (right panel) of Transwell migration and invasion assay in SUNE-1 and HONE-1 cells with WTAP overexpression. Scale bar: 200 μm. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.
To confirm these, we characterized the oncogenic phenotypes in NPC cells (SUNE-1 and HONE-1) with WTAP silencing (Fig. 2B, all P < 0.01). Cell Counting Kit-8 (CCK-8) and colony formation assays revealed that knockdown of WTAP apparently impaired the proliferation ability of NPC cells (Fig. 2C, D, all P < 0.01), which were confirmed by counting the NPC cells directly (Supplementary Fig. S3A, B). Additionally, to explain the reason for the deceleration of cell proliferation rates upon WTAP silencing, we analyzed the cell cycle and apoptosis phenotypes in NPC. Fluorescence-activated cell sorting (FACS) analysis illustrated that restrained WTAP expression induced cell cycle arrest in the G1 phase (Supplementary Fig. S3C, D) and stimulated cell apoptosis compared with that in the controls (Supplementary Fig. S3E, F), indicating that knockdown of WTAP inhibited NPC cell proliferation by prolonging the G1 phase and inducing cellular apoptosis.
Then, Transwell assays showed that silencing WTAP markedly inhibited SUNE-1 and HONE-1 cells migration and invasion (Fig. 2E, all P < 0.01). We further determined whether upregulation of WTAP could promote NPC cell progression. We upregulated WTAP expression by infecting NPC cells with control lentivirus or lentivirus that overexpressed WTAP (Fig. 2F, P < 0.01). As expected, functional studies showed that cell proliferation as well as migratory and invasive capacities were increased upon exogenous overexpression of WTAP (Fig. 2G‒I, all P < 0.01).
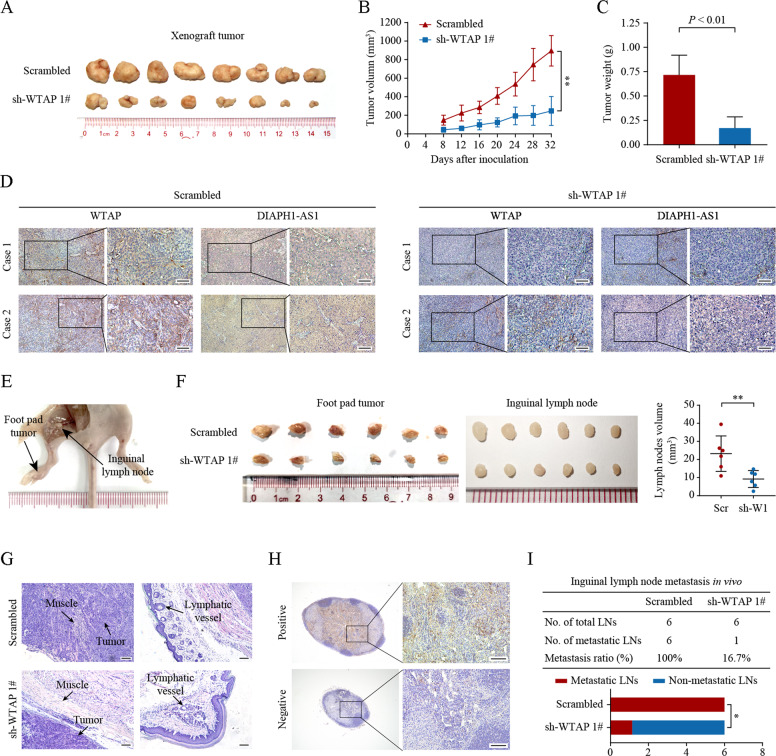
To further evaluate the effect of WTAP on NPC tumor growth and metastasis in vivo, we established xenograft growth and inguinal lymph node metastasis models. Notably, xenografts formed by SUNE-1 cells bearing sh-WTAP exhibited a significantly slower growth rate and lower tumor weight compared to scrambled controls (Fig. 3A‒D). Furthermore, the volumes of primary footpad tumors and metastatic inguinal lymph nodes in the WTAP knockdown group were apparently smaller than those in the control group (Fig. 3E‒F). Silencing WTAP caused a less aggressive phenotype towards the skin and lymphatic vessels of the primary tumors (Fig. 3G). Besides, the metastasis ratio of inguinal lymph nodes was significantly lower in WTAP silencing group (Fig. 3H, I). Taken together, these findings suggest that WTAP might exert its oncogenic role in NPC by promoting cellular proliferation, migration, and invasion.
Fig. 3. Knockdown of WTAP disturbs the proliferation and invasion of NPC in vivo.
A–D Xenograft tumors are formed by SUNE-1-shWTAP or -shNC cells in nude mice. A Representative image of the xenograft tumors. B, C Quantitative analysis of the tumor volumes (B) and tumor weights (C) of the xenografts. Tumor volume was compared at indicated time points and the tumor weight was measured at the endpoint. D Xenograft tumor sections stained for WTAP and DIAPH1-AS1 were examined by IHC and ISH. Scale bar: 50 μm. E–I SUNE-1 cells stably expressing sh-WTAP or a scrambled control shRNA were transplanted into the footpad of nude mice to construct an inguinal lymph node metastasis model. E Representative image of the inguinal lymph node metastasis model. F Representative image of the footpad tumors (left) and inguinal lymph nodes (right). G Representative images of microscopic primary footpad tumors stained with H&E. The overall mouse muscle tissues were disrupted and surrounded by tumor tissues in the negative control group, while the extent of muscle destruction in the WTAP-silenced group was markedly less (left). Compared with the control group, the number of invaded lymphatic vessels apparently decreased in the WTAP-silenced group (right). Scale bar: 100 μm. H Representative images of IHC staining with pan-cytokeratin in inguinal lymph nodes. The staining of keratin in lymph nodes is negative under normal circumstances, but positive when lymph node metastasis of NPC occurs. Scale bar: 100 μm. I Inguinal lymph node metastatic ratios. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01.
WTAP stabilizes lncRNA DIAPH-AS1 in an m6A-dependent manner
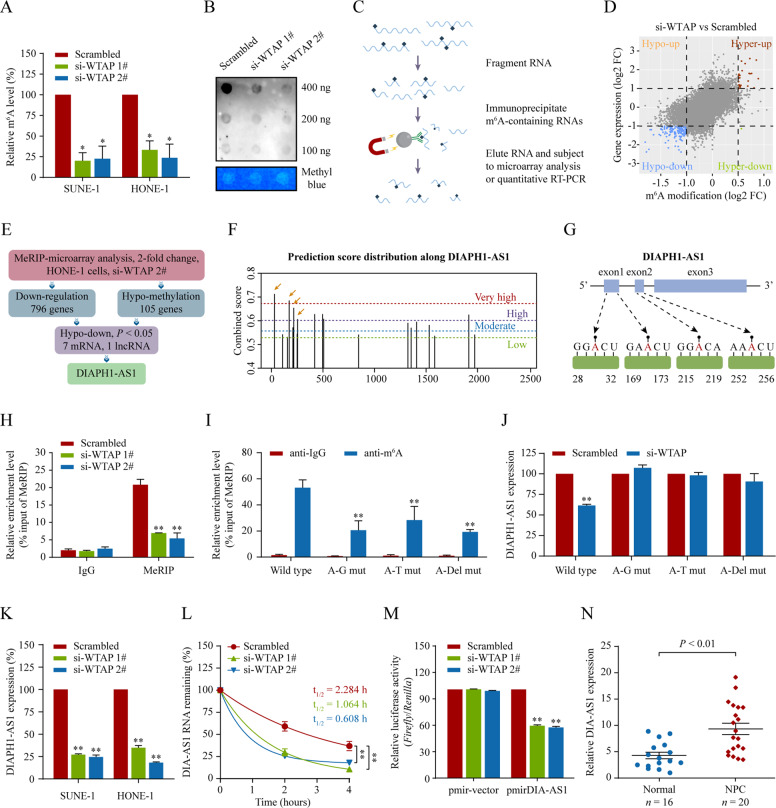
As a key N(6)-methyltransferase, WTAP exerts its biological function by modulating the m6A modification of mRNA and non-coding RNAs (ncRNAs) and consequently regulating its biogenesis, decay, and transport [25, 26]. To explore the underlying mechanisms by which WTAP drives NPC malignant phenotype, we first performed m6A quantification assays and found a reduced global m6A levels in NPC cells upon WTAP silencing (Fig. 4A, all P < 0.05), which was confirmed by the m6A dot blot assays (Fig. 4B). To identify potential RNA targets of WTAP, we carried out transcriptome-wide m6A immunoprecipitation (MeRIP) followed by microarray analysis in HONE-1 cells with or without WTAP silencing (Fig. 4C). We found a global hypomethylation of m6A in the transcription level upon WTAP knockdown (Fig. 4D). Using the PANTHER classification analysis tool (http://www.pantherdb.org) [27], we found a significant enrichment of m6A-modified transcripts in several tumor malignant phenotypes (Supplementary Fig. S4). As GSEA results indicated that WTAP might participate in ncRNA processing (Supplementary Fig. S2D‒F, all P < 0.01), we were motivated to explore whether WTAP affects cancer biology by regulating ncRNAs.
Fig. 4. WTAP-mediated m6A methylation stabilizes lncRNA DIAPH1-AS1.
A, B Total RNA m6A contents in WTAP-knockdown HONE-1 and SUNE-1 cells. Total RNAs were submitted to global m6A levels quantification analysis (A) and dot blot assays (B). C Schematic outline of the MeRIP followed by microarray and qPCR analysis. D MeRIP followed by microarray analysis to identify WTAP-mediated RNA methylation. Distribution of genes with significantly altered m6A levels and expression levels in HONE-1 cells with WTAP knockdown. E The hypomethylated lncRNA DIAPH1-AS1 was identified as downregulated expression in HONE-1 cells with WTAP knockdown. F The enriched and specific m6A peak distribution of DIAPH1-AS1 transcripts predicted by SRAMP. The yellow arrows indicate the m6A enrichment peaks in 1‒281nt of DIAPH1-AS1 transcripts. G Diagram showing the position of m6A motifs with a high combined score within DIAPH1-AS1 transcripts. H m6A enrichment in DIAPH1-AS1 transcripts (1–281 nt) in control and WTAP-silencing cells using MeRIP-qPCR. I MeRIP-qPCR assays to analyze the m6A-modification levels of DIAPH1-AS1 in HONE-1 cells transfected with DIAPH1-AS1 wild type and its mutants expression. A–G mut, adenine residues substituted by guanine; A–T mut, adenine residues substituted by thymine; A–Del mut, adenine residues deleted. J Quantitative RT-PCR for detecting DIAPH1-AS1 expression in HONE-1 cells transfected with DIAPH1-AS1 wild type or mutant constructs together with WTAP siRNAs or its scramble. K Quantitative RT-PCR analysis of DIAPH1-AS1 expression in SUNE-1 and HONE-1 cells with WTAP knockdown. L DIAPH1-AS1 RNA stability in control and WTAP-silenced cells. Quantitative RT-PCR of DIAPH1-AS1 expression at various time points after actinomycin D (10 μg/mL) treatment. M Relative luciferase activity in HONE-1 cells co-transfected with luciferase reporter pmirGLO-DIAPH1-AS1 or its vector pmirGLO (luciferase reporter) and WTAP siRNAs or its scramble control. Data appear as the relative ratio of Firefly to Renilla luciferase activities. N Relative expression of DIAPH1-AS1 in NPC (n = 20) and normal nasopharyngeal epithelial tissues (n = 16) measured by qRT-PCR. Data are presented as the means ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.
Among the hypomethylated lncRNAs in WTAP-silenced cells, DIAPH1-AS1 expression was markedly downregulated (|log2FC|> 1, P < 0.05) accompanied by drastically decreased m6A modification (Fig. 4E). Thus, we assumed that WTAP might regulate DIAPH1-AS1 expression in an m6A-dependent manner. For verification, we first predicted the m6A sites along the DIAPH1-AS1 sequence using the SRAMP prediction tool (http://www.cuilab.cn/sramp) [28]. There were strong m6A peak enrichment and four m6A sites (RRACH motifs) with a high confidence threshold predicted at 1–281 nt (exon 1/2 of 3) of DIAPH1-AS1 (Fig. 4F‒G). Consistently, MeRIP-qPCR confirmed that WTAP knockdown drastically decreased the m6A levels of DIAPH1-AS1 (Fig. 4H). To investigate whether m6A modification is dependent on these m6A residues, we constructed wild-type and mutant DIAPH1-AS1 vectors, in which the adenine residues in predicted m6A motifs in DIAPH1-AS1 were substituted by guanine (A–G mut), thymine (A–T mut), or deleted (Del-mut), respectively. As expected, MeRIP-qPCR showed that the m6A levels on DIAPH1-AS1 mutant transcripts were apparently decreased relative to the wild-type transcript, suggesting that the identified m6A motifs within 1–281 nt on DIAPH1-AS1 were predominantly responsible for its m6A modification (Fig. 4I).
Furthermore, we investigated whether m6A modification on DIAPH1-AS1 affected its expression. As expected, silencing WTAP significantly decreased DIAPH1-AS1 expression, while the constructs containing the mutated m6A residues had no effect on DIAPH1-AS1 expression (Fig. 4J‒K). We then analyzed the half-life of DIAPH1-AS1 in WTAP-silenced HONE-1 cells using an actinomycin D chase experiment and a one-phase exponential decay model with 18 S rRNA normalization. The half-life of DIAPH1-AS1 was approximately 2.28 h in control cells, whereas silencing WTAP increased DIAPH1-AS1 turnover by about 25–50% (0.61‒1.06 h) (Fig. 4L). In line with these results, dual-luciferase reporter assays revealed that knockdown of WTAP substantially inhibited luciferase activity of DIAPH1-AS1 reporter construct but not empty vector (Fig. 4M, all P < 0.01). Correspondingly, immunohistochemistry and in-situ hybridization staining showed a significant decrease of DIAPH1-AS1 expression in tumors of the WTAP knockdown group (Fig. 3D). These results suggest that WTAP stabilizes DIAPH1-AS1 by promoting its m6A modification. The role of DIAPH1-AS1 in NPC remains largely unknown. Quantitative RT-PCR showed that DIAPH1-AS1 was upregulated in NPC (n = 20) compared with normal tissues (n = 16) (Fig. 4N, P < 0.01). In vitro and in vivo functional studies revealed that silencing DIAPH1-AS1 inhibited NPC cell proliferation, migration, and invasion (Supplementary Fig. S5A–J).
In summary, WTAP maintains DIAPH1-AS1 stability via an m6A-dependent mechanism, and their similar loss-of-function phenotypes support the notion that WTAP targets lncRNA DIAPH1-AS1 to control NPC growth and metastasis.
IGF2BP2 recognizes WTAP-mediated DIAPH1-AS1 m6A modification
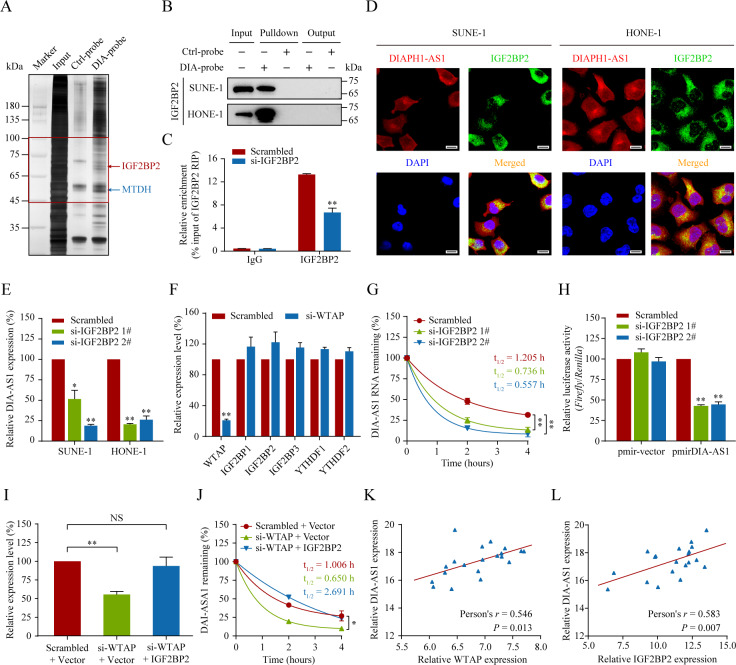
To identify the m6A reader that interprets the m6A modification of DIAPH1-AS1 and mediates its biological processing, we synthesized biotin-labeled DIAPH1-AS1 probes and implemented an RNA pulldown assay followed by mass spectrometry (Supplementary Table S8). Among DIAPH1-AS1 pulldown proteins, IGF2BP2 attracted our interest and confirmed by western blotting (Fig. 5A‒B). IGF2BP2 belongs to the IGF2BP family, in which a distinct group of m6A readers bind to m6A-modified transcripts and regulate its decay [29]. To determine whether IGF2BP2 was a specific m6A reader of DIAPH1-AS1, we performed RNA immunoprecipitation (RIP) assays and found that DIAPH1-AS1 was markedly enriched using anti-IGF2BP2 antibodies, which was undermined when IGF2BP2 silenced (Fig. 5C, P < 0.01). Fluorescent in situ hybridization (FISH) accompanied by IF revealed that DIAPH1-AS1 co-localized with IGF2BP2 in the cytoplasm (Fig. 5D and Supplementary Fig. S6A‒F).
Fig. 5. IGF2BP2 interprets WTAP-mediated DIAPH1-AS1 m6A modification.
A, B Identification of the DIAPH1-AS1-protein complexes pulled down by DIAPH1-AS1 junction probes with proteins extracted from HONE-1 cells, followed by silver staining (A) and western blotting analysis (B). The specific bands (45–100 kDa, indicated in the red box) were cut off and subjected to mass spectrometry (MS) analysis. C IGF2BP2 was immunoprecipitated and RIP-qPCR was used to assess the association of DIAPH1-AS1 with IGF2BP2. D FISH and IF double staining in SUNE-1 and HONE-1 cells showing the co-localization of DIAPH1-AS1 (Cy3; Red) and IGF2BP2 (Green) in the cytoplasm; Nuclei are stained blue (DAPI). Scale bar: 20 μm. E Quantitative RT-PCR analysis of DIAPH1-AS1 expression in SUNE-1 and HONE-1 cells with or without WTAP silencing. F Quantitative RT-PCR analysis of mRNA levels of m6A readers (IGF2BP1, IGF2BP2, IGF2BP3, YTHDF1 and YTHDF2) in HONE-1 cells with or without WTAP silencing. G DIAPH1-AS1 RNA stability in control and IGF2BP2-silenced cells. Quantitative RT-PCR of DIAPH1-AS1 at the indicated time points after treatment with actinomycin D (10 μg/mL). H. Relative luciferase activity in HONE-1 cells co-transfected with luciferase reporter pmirGLO or pmirGLO-DIAPH1-AS1 and IGF2BP2 siRNAs or the control. I Quantitative RT-PCR analysis of DIAPH1-AS1 expression in HONE-1 cells co-transfected with si-WTAP and the IGF2BP2 overexpression vector or the corresponding control. J DIAPH1-AS1 RNA stability in HONE-1 cells co-transfected with si-WTAP and the IGF2BP2 overexpression vector or the corresponding control. Quantitative RT-PCR of DIAPH1-AS1 at the indicated time points after treatment with actinomycin D (10 μg/mL). K, L Pearson correlation analysis of DIAPH1-AS1 and WTAP mRNA (K), as well as DIAPH1-AS1 and IGF2BP2 mRNA (L) in 20 NPC tissues, determined using qRT-PCR. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.
IGF2BP2 was reported to be an RNA stabilizer for m6A-modified RNA; therefore, we investigated whether IGF2BP2 enhances DIAPH1-AS1 stability via an m6A-dependent manner. Consistent with our hypothesis, DIAPH1-AS1 expression was down-regulated significantly after IGF2BP2 inhibition (Fig. 5E, all P < 0.01), whereas no association was found between the WTAP and IGF2BP2 expression (Fig. 5F and Supplementary Fig. S7A, B). More importantly, DIAPH1-AS1 degraded more quickly in IGF2BP2-silencied cells (Fig. 5G, all P < 0.01). And silencing IGF2BP2 obviously reduced the luciferase activity of DIAPH1-AS1 reporter construct (Fig. 5H, all P < 0.01). To further validate that the WTAP-mediated m6A modification of DIAPH1-AS1 may maintain its IGF2BP2-dependent stability, we overexpressed IGF2BP2 in HONE-1 cells with WTAP silencing. As shown in Fig. 5I, J, the loss of WTAP drastically reduced DIAPH1-AS1 expression and destroyed its RNA stabilization, which could be reversed by overexpression of IGF2BP2. We also observed that both the expressions of WTAP and IGF2BP2 were positively correlated with DIAPHA1-AS1 expression (Fig. 5K, L and Supplementary Fig. S7 C, D, all P < 0.05). Our data indicate that the m6A reader IGF2BP2 interprets WTAP-mediated lncRNA DIAPH1-AS1 m6A modification and maintains its stability by preventing its degradation.
DIAPH1-AS1 acts as an adapter to form MTDH-LASP1 complex
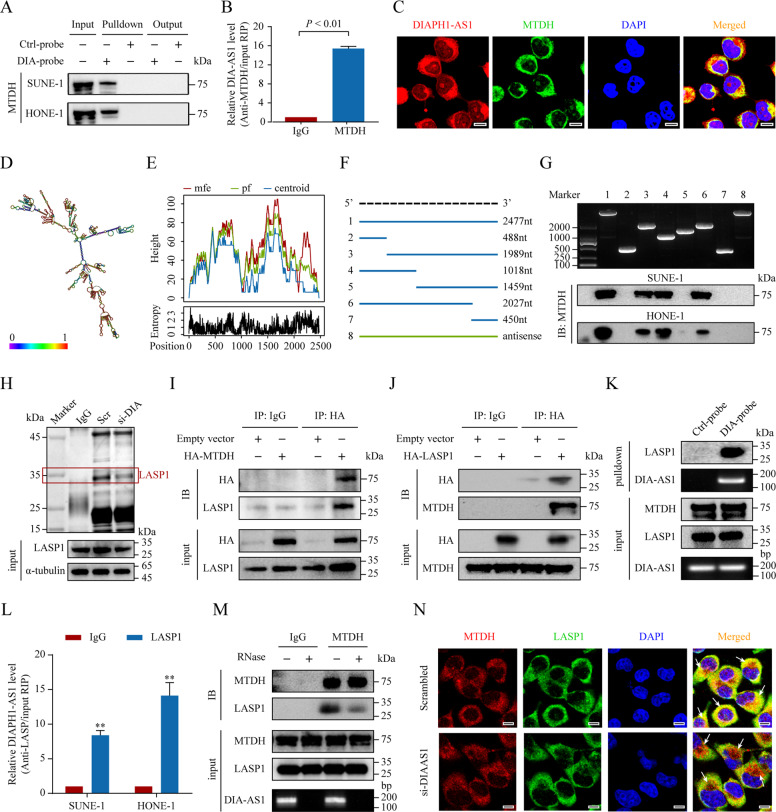
Certain lncRNAs substantially exert their functions by interacting with diverse proteins, such as signaling mediators or transcription factors [30, 31]. To explore the mechanism by which DIAPH-AS1 regulates NPC progression, we re-analyzed the above mass spectrometry results and identified MTDH as the top hit (Fig. 5A). Western blotting validated MTDH as the interacting protein of DIAPH1-AS1 (Fig. 6A). RIP assays showed that DIAPH1-AS1 could specifically bind to MTDH (Fig. 6B), and they mainly co-localized in the cytoplasm (Fig. 6C and Supplementary Fig. S8A‒F). To determine the specific region through which DIAPH1-AS1 interacts with MTDH, we first predicted the secondary structure and minimum free energy (MFE) structure of DIAPH1-AS1 using the RNAfold WebServer (http://rna.tbi.univie.ac.at/) (Fig. 6D–E). Accordingly, RNA pulldown assays using different segments of DIAPH1-AS1 demonstrated that nucleotides 489–1018 of DIAPH1-AS1 could directly bind to MTDH (Fig. 6F–G).
Fig. 6. DIAPH1-AS1 acts as an adapter to form MTDH-LASP1 complex.
A RNA pulldown assay showing the direct interaction of DIAPH1-AS1 with the MTDH protein. B Association of endogenous MTDH and DIAPH1-AS1 was detected using an RIP-qPCR assay. HONE-1 cell lysates were immunoprecipitated with the anti-MTDH antibodies. C FISH and IF double staining showing the subcellular co-localization of DIAPH1-AS1 and MTDH. Scale bar: 20 μm. D, E The secondary structure (D) and minimum free energy (MFE) structure (E) of DIAPH1-AS1 was analyzed using the online tool RNAfold WebServer. F, G Deletion mapping of the MTDH-binding domain in DIAPH1-AS1. F Diagrams of full-length DIAPH1-AS1 and its deletion fragments. G Top, the in vitro–transcribed full-length DIAPH1-AS1 and deletion fragments with the correct sizes are indicated. Bottom, western blotting analysis for MTDH pulled down by different DIAPH1-AS1 fragments. H An immunoprecipitation assay was performed to detect the interaction between MTDH and LASP1 in HONE-1 cells with or without DIAPH1-AS1 silencing. Isolated proteins were resolved by SDS-PAGE followed by silver staining. Differential bands were cut for mass spectrometry. I, J HONE-1 cells transfected with HA-tagged MTDH vectors (I), HA-tagged LASP1 vectors (J) or control vectors were lysed for co-IP assays using anti-HA antibodies or normal rabbit IgG. K The interaction between DIAPH1-AS1 and LASP1 was confirmed using RNA pulldown assays and western blotting analysis. L The association of endogenous LASP1 and DIAPH1-AS1 was detected using an RIP-qPCR assay. M Immunoblotting to detect of exogenous MTDH and LASP1 immunoprecipitated by anti-MTDH antibodies with or without RNase If treatment in HONE-1 cells. N The interaction between MTDH and LASP1 in HONE-1 cells, with or without DIAPH1-AS1 silencing was assessed using immunofluorescence double staining. Scale bar: 20 μm. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.
MTDH can promote tumor progression by forming interconnected protein complexes [32, 33]. LncRNAs are known to exert their oncogenic functions by enhancing protein-protein interaction [34]. We speculated that DIAPH1-AS1 acted as an adapter to form MTDH-protein complexes. Thus, we performed immunoprecipitation-mass spectrometry analysis in HONE-1 cells after DIAPH1-AS1 knockdown. LASP1 was pulled down by MTDH antibody, which was weakened by DIAPH1-AS1 knockdown (Fig. 6H and Supplementary Table S9). Co-IP assays using HA-MTDH and HA-LASP1 overexpressing vectors validated the endogenous MTDH-LASP1 protein complex (Fig. 6I, J). Moreover, the interaction of DIAPH1-AS1 and LASP1 was confirmed by biotin-RNA pulldown (Figs. 5A and 6K), as well as by an RNA immunoprecipitation assay (Fig. 6L). Next, we tested whether the interaction between MTDH and LASP1 was dependent on the existence of DIAPH1-AS1. Cell lysates were treated with either ribonuclease If (RNase If) or RNase inhibitor (RNaseIn), followed by co-IP assays with anti-MTDH antibodies. The results showed that LASP1 could be co-immunoprecipitated by MTDH in the RNaseIn-treated samples, whereas treatment with RNase If abolished this complex formation (Fig. 6M). Meanwhile, DIAPH1-AS1 knockdown reduced the co-localization of MTDH and LASP1 in the cytoplasm (Fig. 6N). Overall, our data indicate that the formation of the MTDH-LASP1 complex in NPC requires their specific interaction with DIAPH1-AS1.
DIAPH1-AS1 confers NPC progression via MTDH-LASP1 complex
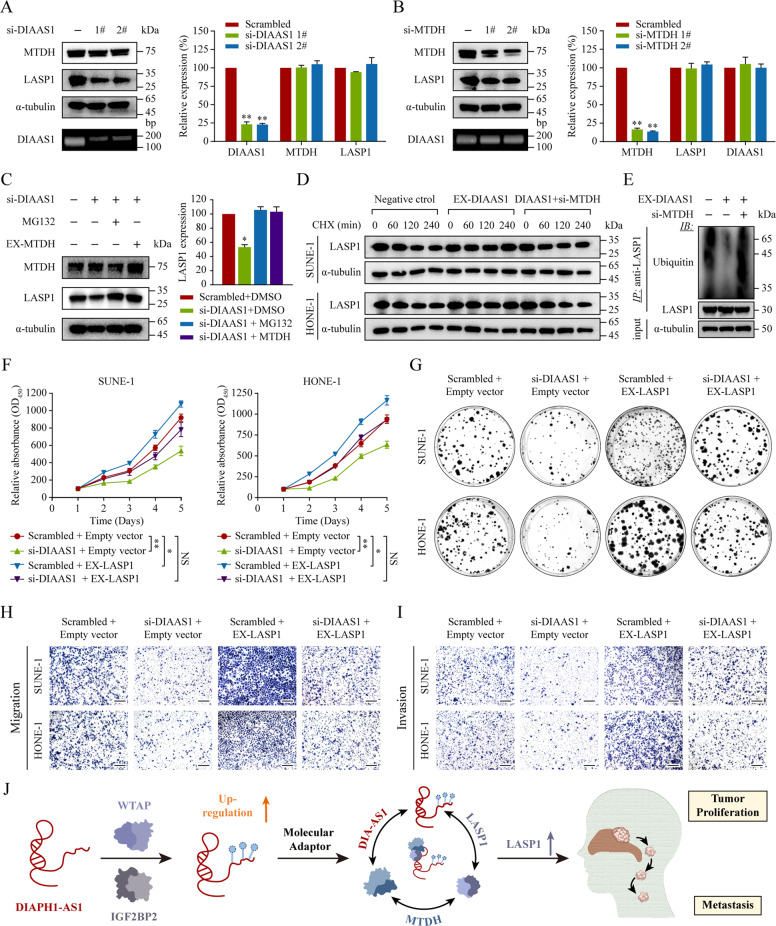
To better understand how DIAPH1-AS1 exerts its function through the MTDH-LASP1 complex, we initially assessed MTDH and LASP1 expression and found silencing DIAPH1-AS1 had no impact on their mRNA, while the LASP1 protein was dramatically decreased (Fig. 7A). Since MTDH can stabilize its binding protein to promote tumor progression [32, 35], we evaluated its influence on LASP1 expression. The results showed that knockdown of MTDH decreased LAPS1 protein, while its mRNA remained constant (Fig. 7B), which indicated that LASP1 was regulated at the post-transcriptional level. Therefore, we silenced DIAPH1-AS1 and treated the cells with MG132 (proteasome inhibitor) or MTDH-overexpressing vector. As shown in Fig. 7C, the downregulation of LASP1 caused by DIAPH1-AS1 knockdown could be rescued by MG132 treatment or MTDH overexpression. We further exposed cells to the protein synthesis inhibitor cycloheximide (CHX) and found that overexpression of DIAPH1-AS1 significantly prolonged the half-life of the LASP1 protein, while downregulation of MTDH repressed this effect (Fig. 7D). Finally, overexpression of DIAPH1-AS1 resulted in decreased ubiquitination of LASP1, which could be rescued by MTDH silencing (Fig. 7E). Jointly, these results demonstrate that DIAPH1-AS1 can stabilize LASP1 via its mediated MTDH-LASP1 interaction.
Fig. 7. DIAPH1-AS1 confers NPC progression via MTDH-LASP1 complex.
A, B The expression levels of MTDH, LASP1, and DIAPH1-AS1 in NPC cells with or without DIAPH1-AS1 (A) or MTDH (B) silencing were detected using western blotting (left) and qRT-PCR (right). C Western blotting analysis of MTDH and LASP1 levels in HONE-1 cells with or without DIAPH1-AS1 silencing, and treated with MG132 (10 mmol/l) for 24 h or transfected with MTDH overexpression vectors. D SUNE-1 and HONE-1 cells co-transfected with DIAPH1-AS1 overexpressing vector and si-MTDH or the corresponding negative control, were treated with 50 µg/ml cycloheximide (CHX) for the indicated time points and harvested for western blotting analysis to assess the protein stability and half-life of LASP1. E The immunoprecipitation (IP) experiment with anti-LASP1 antibodies, followed by western blotting with anti-ubiquitin antibodies to detect ubiquitinated LASP1 in HONE-1 cells co-transfected as indicated. F–I The effect of the ectopic expression of LASP1 to rescue the si-DIAPH1-AS1-mediated downregulation of NPC cell proliferation and metastasis. SUNE-1 and HONE-1 cells were co-transfected with si-DIAPH1-AS1 and LASP1 overexpressing vectors or the corresponding negative control. Then CCK-8 assays (F) and colony formation assays (G) for cell proliferation and Transwell assays for cell invasion (H, I) were performed. Scale bar: 200 μm. J A graphic illustration of WTAP promoting tumor growth and metastasis of NPC by modulating lncRNA DIAPH1-AS1 m6A modification and disrupting its RNA decay. Data are presented as the mean ± SD. *P < 0.05, **P < 0.01. The experiments were repeated at least three times independently.
To learn whether DIAPH1-AS1 exerts tumor-promoting functions by modulating LASP1 expression, we conducted in vitro functional rescue experiments by overexpressing LASP1 in cells with or without DIAPH1-AS1 knockdown. CCK-8 and colony formation assays showed that overexpression of LASP1 reversed the suppressive effect of DIAPH1-AS1 knockdown on the proliferation ability of NPC cells (Fig. 7F, G and Supplementary Fig. S9A). Similar results were obtained by transwell migration and invasion assays (Fig. 7H, I and Supplementary Fig. S9B‒C). Collectively, these data demonstrate that lncRNA DIAPH1-AS1 stabilizes LASP1, and therefore promotes NPC growth and metastasis.
Discussion
The occurrence and development of cancers is essentially a disease of gene control, which is affected by both genetic abberations and epigenetic modifications. Dysregulation of epigenetic modification, including DNA methylation, histone variants and modifications, nucleosome positioning, and RNA modifications [36], has been implicated in various types of cancers. Among the RNA modifications, m6A is the most prevalent one in human mRNA and non-coding RNA [37]. In recent years, RNA m6A modification has emerged as a critical regulator for the fate of RNA, such as RNA splicing, mRNA translocation, and degradation. Previous studies demonstrated that dysregulation of m6A RNA modification correlated closely with a variety of vital bioprocesses, including cancer development [38–40]. As a reversible epigenetic modification, m6A is driven by m6A methyltransferases (METTL3, METTL14, WTAP, and VRIMA, termed “writers”), removed by m6A demethylases (FTO and ALKBH5, termed “erasers”), and recognized by m6A binding proteins (IGF2BPs, YTHDCs, and YTHDFs, termed “readers”). Among these m6A modulators, WTAP has been studied extensively in multiple tumors. Previous studies have reported that WTAP functions as an oncogenic m6A modulator in hepatocellular carcinoma [25], pancreatic cancer [41], osteosarcoma [42], renal cell carcinoma [43], head and neck squamous cell carcinoma [44], and B-cell lymphoma [45]. However, the biological function of m6A modulators is highly dependent on the cellular context. The role of m6A modification, as well as the expression imbalance of m6A modulators, in NPC remains elusive. In the present study, we showed that WTAP was upregulated in NPC and promoted NPC growth and metastasis via its m6A catalytic activity. The upregulation of WTAP in NPC was fine-tuned by histone acetylation revealing the interconnected regulatory network between chromatin modification and RNA epigenetics. Moreover, survival analysis indicated that WTAP was a potential marker for NPC prognosis. However, it requires further investigation and validation before preclinical application.
The epigenetic modification caused by EBV infection is inseparable from the malignant transformation process of nasopharyngeal epithelial cells, especially for NPC that originate in endemic region [22, 23]. Therefore, it is reasonable to speculate EBV infection might play a role in aberrantly upregulation of WTAP in NPC. However, we did not observe significant difference in H3K27ac modification level of WTAP between EBV-negative and EBV-positive NPC cells. More importantly, integration analysis of DNA methylation in NPC from GEO dataset, DNA demethylation treatment of nasopharyngeal cell lines and survival analysis collectively indicated that DNA methylation of WTAP is not a crucial factor for WTAP upregulation in NPC. These findings suggested WTAP regulation is independent of EBV infection and its associated epigenetic alteration. The underlying mechanisms triggering the dysregulation of WTAP in NPC remain to be further investigated in the future.
Recent studies have revealed that lncRNAs could be marked by m6A-methylation for subsequent RNA stability, RNA-miRNA interaction, and RNA-protein interaction [46–48]. As significant master regulators in human cells, lncRNAs were demonstrated to participate critically in multiple stages of carcinogenesis and tumor progression [49, 50]. Recent studies discovered that lncRNAs exert positive or negative regulatory functions in NPC. For instance, an oncogenic lncRNA, PVT1, serves as a molecular scaffold to activate lysine acetyltransferase 2A (KAT2A) and stabilizes hypoxia inducible factor 1 alpha (HIF-1α) in NPC [51]. Our previous study identified lncRNA as a crucial driver of NPC progression and chemoresistance by interacting with ACLY protein and regulating cellular acetyl-CoA metabolism [52]. These clues not only emphasized the vital role of m6A modification in the fate of functional RNAs, but also reminded us to explore whether the m6A modification could affect cancer biology via modulating m6A modification of non-coding RNAs. To this end, we employed MeRIP assays following microarray analysis to compare the gene expression and m6A modification profiles in NPC cells with or without WTAP silencing. DIAPH1-AS1 was one of the lncRNAs that showed differentially expression and m6A modification in the microarray analysis. To a large extent, the fate of m6A-modified RNAs depends on the recognition by different kinds of m6A readers [53]. As one of the distinct families of m6A readers, IGF2BPs was proved to facilitate the stability and storage of their target RNAs in a m6A-dependent manner [29]. This implies that a specific m6A reader should also be identified to provide a deeper insight into the role of WTAP-mediated m6A modification in NPC. Consequently, we showed that WTAP-mediated m6A modification stabilized DIAPH1-AS1 by enhancing its recognition by IGF2BP2.
DIAPH1-AS1, located on chromosome 5q31.3, is a novel lncRNA without any functional or biological research in NPC. Mechanistically, a substantial proportion of lncRNAs carry out their biological responses as molecular adapters that enhance protein-protein interactions. In such cases, the molecular functions (such as chromatin regulation, protein phosphorylation, and subcellular accumulation) of these binding proteins may be affected [31, 54]. In liver cancer, lncRNA HULC was identified as a modular scaffold that directly binds to two proteins, lactate dehydrogenase A (LDHA) and pyruvate kinase M2 (PKM2), to promote their phosphorylation levels, which consequently boosts cellular glycolysis [34]. Similarly, in gastric cancer, lncRNA GClnc1 modulates the interaction of the WD repeat domain 5 (WDR5)-KAT2A protein complex, which alters the state of histone modification, and ultimately transcriptionally activates its target genes [55]. In our hand, DIAPH1-AS1 was demonstrated to promote NPC growth and metastasis as a modular scaffold for MTDH-LASP1 complexes, in which it stabilizes the LASP1 protein by modulating its ubiquitin-dependent proteasome degradation through MTDH. MTDH was recently implicated in several cancer-related processes because of its recruitment of different signaling molecules through the peptide motifs over its entire sequence [56]. In breast cancer, MTDH interacts and stabilizes Staphylococcal nuclease and tudor domain containing 1 (SND1) protein under stress conditions during carcinogenesis [32]. Consistent with these findings, MTDH was also found to bind to LASP1 and elevate its abundance at the post-transcriptional level. Remarkably, the formation of the MTDH-LASP1 complex is partially dependent on the existence of a molecular scaffold, lncRNA DIAPH1-AS1. However, because of the complex and diverse mechanisms by which large ncRNAs carry out their regulatory functions, further studies are required to verify this model, emphasizing the need for deeper research on lncRNA-protein interaction.
Based on our results, we proposed a molecular mechanism model in which WTAP maintains lncRNA DIAPH1-AS1 stability in an m6A-dependent way, which facilitates the formation of the MTDH-LASP1 complex, protecting LASP1 from ubiquitin degradation, and ultimately promoting NPC growth and metastasis (Fig. 7J). This study highlights the evolving roles of WTAP-mediated m6A modification on lncRNAs in NPC, and identified WTAP as a potential biomarker and target for NPC.
Supplementary information
Acknowledgements
We thank Dr. M. Zeng from Sun Yat-Sen University Cancer Center for providing NPC and immortalized nasopharyngeal epithelial cell lines.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81902762, No. 82002220), the Health & Medical Collaborative Innovation Project of Guangzhou City, China (No. 201604020003, 201803040003), the Special Support Program of Sun Yat-sen University Cancer Center (16zxtzlc06), the Natural Science Foundation of Guangdong Province (No. 2019A1515012045), the Innovation Team Development Plan of the Ministry of Education (No. IRT_17R110), and the National Key R&D Program of China (2016YFC0902000). The datasets generated and/or analyzed during the current study are available at the Research Data Deposit public platform (www.researchdata.org.cn).
Author contributions
Conceptualization, Z.X.L., Y.S.; Data production, analysis and investigation, Z.X.L., Z.Q.Z., P.Y.Y., L.L., G.Q.Z., J.W.L., L.L.Z., F.P.C., Y.Q.L., C.F.W., F.L., and J.M.; Writing, Review and Edit, Z.Q.Z, Z.X.L., and Y.S.; Supervision, N.L., Y.S.; Funding acquisition, L.L. and Y.S. The author(s) read and approved the final manuscript.
Data availability
The source data that support the findings of this study are available to readers. MeRIP profiles are accessible at the GEO repository under accession number GSE160644.
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Institutional Review Board of Sun Yat-Sen University Cancer Center (GZR2020-220). All animal experiments conducted was compliant with Institutional Animal Care and Use Committee, Sun Yat-sen University Cancer Center (L102012019110C).
Footnotes
Edited by A Degterev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhi-Xuan Li, Zi-Qi Zheng, Pan-Yang Yang, Li Lin, Guan-Qun Zhou.
Contributor Information
Na Liu, Email: liun1@sysucc.org.cn.
Ying Sun, Email: sunying@sysucc.org.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00905-w.
References
- 1.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Chen L, Hu GQ, Zhang N, Zhu XD, Yang KY, et al. Gemcitabine and cisplatin induction chemotherapy in nasopharyngeal carcinoma. N Engl J Med. 2019;381:1124–35. doi: 10.1056/NEJMoa1905287. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Li WF, Chen NY, Zhang N, Hu GQ, Xie FY, et al. Induction chemotherapy plus concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: a phase 3, multicentre, randomised controlled trial. Lancet Oncol. 2016;17:1509–20. doi: 10.1016/S1470-2045(16)30410-7. [DOI] [PubMed] [Google Scholar]
- 4.Ren X, Yang X, Cheng B, Chen X, Zhang T, He Q, et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun. 2017;8:14053. doi: 10.1038/ncomms14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–41. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- 6.Wen X, Tang X, Li Y, Ren X, He Q, Yang X, et al. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. Int J Mol Sci. 2016;17:1956.
- 7.Yuan J, Jiang YY, Mayakonda A, Huang M, Ding LW, Lin H, et al. Super-enhancers promote transcriptional dysregulation in nasopharyngeal carcinoma. Cancer Res. 2017;77:6614–26. doi: 10.1158/0008-5472.CAN-17-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–8. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, You Y, Lu Z, Yang J, Li P, Liu L, et al. N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science. 2019;365:1171–6. doi: 10.1126/science.aax4468. [DOI] [PubMed] [Google Scholar]
- 11.Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA. 2017;23:1754–69. doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang PP, He QP, Lei Y, Li YQ, Wen X, Hong MZ, et al. m 6 A-mediated ZNF750 repression facilitates nasopharyngeal carcinoma progression. Cell Death Dis. 2018;9:1169. doi: 10.1038/s41419-018-1224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–26. doi: 10.1158/0008-5472.CAN-19-0799. [DOI] [PubMed] [Google Scholar]
- 17.Nagarajan A, Janostiak R, Wajapeyee N. Dot blot analysis for measuring global N(6)-methyladenosine modification of RNA. Methods Mol Biol. 2019;1870:263–71. doi: 10.1007/978-1-4939-8808-2_20. [DOI] [PubMed] [Google Scholar]
- 18.Yan G, Yuan Y, He M, Gong R, Lei H, Zhou H, et al. m(6)A methylation of precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol Ther Nucleic Acids. 2020;19:421–36. doi: 10.1016/j.omtn.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chokkalla AK, Mehta SL, Kim T, Chelluboina B, Kim J, Vemuganti R. Transient focal ischemia significantly alters the m(6)A epitranscriptomic tagging of RNAs in the brain. Stroke. 2019;50:2912–21. doi: 10.1161/STROKEAHA.119.026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weichert W, Denkert C, Schmidt M, Gekeler V, Wolf G, Köbel M, et al. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br J Cancer. 2004;90:815–21. doi: 10.1038/sj.bjc.6601610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birdwell C, Queen K, Kilgore P, Rollyson P, Trutschl M, Cvek U, et al. Genome-wide DNA methylation as an epigenetic consequence of Epstein-Barr virus infection of immortalized keratinocytes. J Virol. 2014;88:11442–58. doi: 10.1128/JVI.00972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chua M, Wee J, Hui E, Chan A. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–24. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 24.Jiang W, Liu N, Chen X, Sun Y, Li B, Ren X, et al. Genome-wide identification of a methylation gene panel as a prognostic biomarker in nasopharyngeal carcinoma. Mol Cancer Ther. 2015;14:2864–73. doi: 10.1158/1535-7163.MCT-15-0260. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14:703–21. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaikkonen MU, Adelman K. Emerging roles of non-coding RNA transcription. Trends Biochem. Sci. 2018;43:654–67. doi: 10.1016/j.tibs.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18:499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 32.Wan L, Lu X, Yuan S, Wei Y, Guo F, Shen M, et al. MTDH-SND1 interaction is crucial for expansion and activity of tumor-initiating cells in diverse oncogene- and carcinogen-induced mammary tumors. Cancer Cell. 2014;26:92–105. doi: 10.1016/j.ccr.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emdad L, Sarkar D, Su ZZ, Lee SG, Kang DC, Bruce JN, et al. Astrocyte elevated gene-1: recent insights into a novel gene involved in tumor progression, metastasis and neurodegeneration. Pharmacol Ther. 2007;114:155–70. doi: 10.1016/j.pharmthera.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Li Y, Yan S, Wang H, Shao X, Xiao M, et al. Interactome analysis reveals that lncRNA HULC promotes aerobic glycolysis through LDHA and PKM2. Nat Commun. 2020;11:3162. doi: 10.1038/s41467-020-16966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, He K, Yan S, Yang Y, Gao X, Zhang M, et al. Metadherin/Astrocyte elevated gene-1 positively regulates the stability and function of forkhead box M1 during tumorigenesis. Neuro-oncology. 2017;19:352–63. doi: 10.1093/neuonc/now229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–68. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 37.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 38.Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–88. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020;19:104. doi: 10.1186/s12943-020-01216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 2020;19:88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li BQ, Liang ZY, Seery S, Liu QF, You L, Zhang TP, et al. WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing Fak mRNA in pancreatic cancer. Cancer Lett. 2019;451:48–57. doi: 10.1016/j.canlet.2019.02.043. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Li Y, Zhi S, Ding Z, Wang W, Peng Y, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11:659. doi: 10.1038/s41419-020-02847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang J, Wang F, Cheng G, Si S, Sun X, Han J, et al. Wilms’ tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J Exp Clin Cancer Res. 2018;37:40. doi: 10.1186/s13046-018-0706-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao X, Cui L. Development and validation of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am J Cancer Res. 2019;9:2156–69. [PMC free article] [PubMed] [Google Scholar]
- 45.Kuai Y, Gong X, Ding L, Li F, Lei L, Gong Y, et al. Wilms’ tumor 1-associating protein plays an aggressive role in diffuse large B-cell lymphoma and forms a complex with BCL6 via Hsp90. Cell Commun Signal. 2018;16:50. doi: 10.1186/s12964-018-0258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–94. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. N6-methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906–20. doi: 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Chen W, Lian J, Zhang H, Yu B, Zhang M, et al. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell Death Differ. 2020;27:695–710. doi: 10.1038/s41418-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng ZQ, Li ZX, Guan JL, Liu X, Li JY, Chen Y, et al. Long noncoding RNA TINCR-mediated regulation of acetyl-CoA metabolism promotes nasopharyngeal carcinoma progression and chemoresistance. Cancer Res. 2020;80:5174–88. doi: 10.1158/0008-5472.CAN-19-3626. [DOI] [PubMed] [Google Scholar]
- 53.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–50. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 56.Guo F, Wan L, Zheng A, Stanevich V, Wei Y, Satyshur KA, et al. Structural insights into the tumor-promoting function of the MTDH-SND1 complex. Cell Rep. 2014;8:1704–13. doi: 10.1016/j.celrep.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data that support the findings of this study are available to readers. MeRIP profiles are accessible at the GEO repository under accession number GSE160644.