Abstract
Despite the rapid and broad implementation of CRISPR-Cas9-based technologies, convenient tools to modulate dose, timing, and precision remain limited. Building on methods using synthetic peptide nucleic acids (PNAs) to bind RNA with unusually high affinity, we describe guide RNA (gRNA) spacer-targeted, or ‘antispacer’, PNAs as a tool to modulate Cas9 binding and activity in cells in a sequence-specific manner. We demonstrate that PNAs rapidly and efficiently target complexed gRNA spacer sequences at low doses and without design restriction for sequence-selective Cas9 inhibition. We further show that short PAM-proximal antispacer PNAs achieve potent cleavage inhibition (over 2000-fold reduction) and that PAM-distal PNAs modify gRNA affinity to promote on-target specificity. Finally, we apply antispacer PNAs for temporal regulation of two dCas9-fusion systems. These results present a novel rational approach to nucleoprotein engineering and describe a rapidly implementable antisense platform for CRISPR-Cas9 modulation to improve spatiotemporal versatility and safety across applications.
INTRODUCTION
Off-target cleavage events are well-characterized consequences of bacteria-derived nucleases and threaten their safe application to human therapy (1,2). Additionally, as Cas9-based biotechnologies continue to rapidly develop across sectors (3,4), there is a need for modulating tools to further realize the potential of these systems and conveniently manipulate dose, timing and accuracy (5). Preferably such tools would feature user-friendly design and application, facile delivery, scalable synthesis, and sequence specificity for application to multiplexed experiments.
As a countermeasure to deleterious events, and to improve control over Cas9-derived systems, previous studies explored the application of anti-CRISPR (Acr) proteins and small molecules to directly bind and inhibit Cas9 and related CRISPR family proteins (6–8). However, anti-CRISPR systems feature notable limitations including toxicity and potential immunogenicity related to peptide expression, generalized inhibition mechanisms, modest potency, and restriction to a single Cas ortholog or CRISPR subtype (9,10). Nucleic acid-based inhibitors such as DNA aptamers and chemically modified gRNA-binding nucleic acids have also been explored with some success (11,12). In both cases, however, sugar-phosphate-backboned nucleic acids were only able to appreciably bind the generalized PAM-recognition components of gRNAs rather than the full guide spacer sequence, and thus lack true sequence-specificity. As an alternative, we sought to explore the use of peptide nucleic acid (PNA) molecules as universal sequence-selective antisense modifiers of CRISPR-Cas9 and RNA-guided nucleases.
PNAs are synthetic chimeric oligonucleotides modified to feature a neutrally charged polyamide backbone (Figure 1A) (13,14). Due to minimized repulsive forces between polymer backbones these molecules bind complementary DNA and RNA sequences with remarkably high affinity and specificity (15). For these reasons, PNAs have been widely employed as antisense or antigene molecules for applications such as gene targeting, microRNA modulation, and mRNA-binding protein identification (16–19). Meanwhile, CRISPR-Cas9 systems are known to target double-stranded DNA (dsDNA) sequences in an interaction that is dependent on protospacer adjacent motif (PAM) sequence recognition followed by base-pairing between a target DNA strand and a complementary spacer sequence coded by an internalized gRNA (R-loop formation, Figure 1B) (20,21). We hypothesized that PNAs, with high-affinity sequence-specific binding to RNA, may efficiently hybridize to gRNA spacer sequences to control the ability of Cas9 to interact with DNA sequences (Figure 1B). With an engineered hydrophobic protein-like backbone, we hypothesized that PNAs may be especially amenable to stable nucleoprotein complexation and accommodation within the Cas9 binding channel.
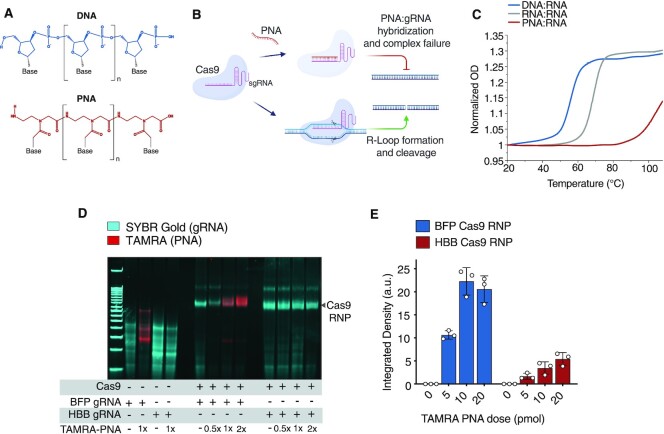
Figure 1.
Peptide nucleic acids efficiently hybridize gRNA spacer sequences in vitro. (A) Structures of DNA and PNA oligomers. (B) Schematic depicting antisense PNA hybridization to a Cas9 sgRNA spacer sequence. (C) Melting curves for DNA:RNA (blue), RNA:RNA (gray) and PNA:RNA (red) 20-bp duplexes. Data are shown as normalized OD (260nm) measurements across temperatures. (D) Electrophoretic mobility shift assay (EMSA) assay demonstrating antispacer PNA binding to sequence-matched sgRNA and Cas9 RNP on polyacrylamide gel (Supplementary Figure S1). Cas9 RNP band is labeled. (E) Antispacer PNA binding to Cas9 RNP loaded with BFP sgRNA (blue) and HBB sgRNA (red), described as TAMRA band integrated density from analysis of gels from three independent experiments. For (E), bars represent mean ± s.d. from n= 3 independent experiments, mean differences and P-values listed in Supplementary Table S1.
Here, we characterize PNAs as antisense modifiers of CRISPR-Cas9 with powerful inhibitory and modulatory properties. We demonstrate antispacer PNAs as a facile and design restriction-free platform and describe advancements in inhibitory potency and on-target specificity improvement while highlighting robust sequence selectivity. This study outlines a novel and accessible tool to expand the versatility and improve the safety of Cas9 applications and describes a novel approach for the regulation of nucleoproteins via chimeric synthetic antisense nucleic acid molecules.
MATERIALS AND METHODS
PNA synthesis and purification
Automated PNA synthesis
PNA oligomers were synthesized using a Biotage Initiator + Alstra microwave peptide synthesizer. The desired sequence was assembled automatically on 10% l-lysine loaded Rink Amide ChemMatrix resin (Sigma Aldrich, 727768-5G) using standard Fmoc chemistry. All Fmoc-aeg-PNA monomers were purchased from PNA BIO INC, Thousand Oaks. The oligomers were cleaved from the resin at room temperature using a trifluoroacetic acid (TFA): water: triisopropylsilane: (38:1:1) cocktail solution (60 min × 1). The crude PNAs were precipitated with cold ether, purified and characterized by reverse phase–high performance liquid chromatography (RP-HPLC) (5–95% ACN/water/0.1% TFA gradient) and MALDI-TOF spectroscopy (MALDI-TOF-MS Shimadzu AXIMA Confidence), respectively.
Manual PNA synthesis
TAMRA dye-labeled PNA oligomers were synthesized manually on 10% l-lysine loaded solid support (MBHA (4-methylbenzhydrylamine) resin, Peptides International, RMB-1045-PI) using standard Boc chemistry procedures. All Boc-aeg-PNA monomers were purchased from ASM Research Chemicals GmbH (Hannover, Germany). Kaiser tests were performed to ensure complete deprotection and coupling during each cycle. The oligomers were cleaved from the resin using a m-cresol: thioanisole: trifluoromethanesulfonic acid (TFMSA): trifluoroacetic acid (TFA) (1:1:2:6) cocktail solution (30 min × 2). The resulting mixtures were combined and the crude PNAs were precipitated with cold ether, purified, and characterized by reverse phase–high performance liquid chromatography (RP-HPLC) (5–95% ACN/water/0.1% TFA gradient) and MALDI-TOF spectroscopy (MALDI-TOF-MS Shimadzu AXIMA Confidence), respectively.
All PNA stock solutions (automated and manual) were prepared using nanopure water, and the concentrations were determined using a Thermo Scientific™ NanoDrop™ OneC microvolume spectrophotometer. The following extinction coefficients were used: 13 700 M–1cm–1 (A), 6600 M–1 cm–1 (C), 11 700 M–1 cm–1 (G), and 8600 M–1 cm–1 (T).
20mer PNAs were all synthesized with three l-lysine (K) residues on either C- or N- terminus to facilitate solubility. 10mer PNAs were synthesized with a single L-lysine (K) residue on either terminus. HPLC instrument set up consisted of: Waters 2998 Photodiode Array Detector, Waters 2545 Quaternary Gradient Module, Waters 2707 Autosampler.
In some cases, purified, synthesized, and characterized PNAs were acquired directly from PNA BIO Inc. Refer to Table S2 for all PNA sequences and sources used in this study.
UV spectroscopy thermal melting analysis
All samples were prepared by mixing a stoichiometric amount of each strand (1 μM) in 10 mM sodium phosphate buffer at pH 7.4 (Supplementary Table S4). The samples were pre-annealed at 95°C for 5 min and gradually cooled to room temperature prior to the melting experiment. UV melting experiments were performed using a Shimadzu UV-3600 Plus UV–Vis-NIR spectrophotometer equipped with a thermoelectrically controlled multicell holder. UV melting spectra were recorded after every 1°C temperature change by monitoring the absorbance at 260 nm from 20°C to 110°C to 20°C, with a heating/cooling ramp rate of 1.0°C/min. The heating and cooling curves were overlapped to confirm reversible denaturation. All spectra were plotted on Origin 2020 and smoothed using a 20-point adjacent averaging algorithm. The first derivative plots of the melting curves were generated to determine the melting temperature for each duplex.
EMSA assay and analysis
10 pmol of purified sgRNA (IDT, Alt-R CRISPR-Cas9 sgRNA) alone, or 10 pmol of sgRNA and 10 pmol of purified spCas9 nuclease (Sigma-Aldrich, CAS9PROT-250UG), were incubated at room temperature for 10 min in a 20 μl reaction in 1 × 3.1 Buffer (B7203S, NEB) to form RNP. Specified dose of TAMRA dye labelled PNA was diluted into 1 μl of water, added to wells, and incubated in a thermocycler for 30 min at 37°C. A BIORAD 5% TBE polyacrylamide gel was pre-run for 30 min at 10 mA. 5 μl of final binding reactions were loaded onto gels with BlueJuice loading buffer (Invitrogen) and run at 10 mA. Gels were first imaged using BIORAD GelDoc XRS+ with a Green Epi (BIORAD #170-8284) 605/50 filter to acquire TAMRA signal and then incubated in 1× SYBR Gold (Invitrogen) in 1× TBE buffer for 5 min before imaging by standard UV transillumination (Raw images in Supplementary Figure S1). For band density analysis, three independent experiments were loaded onto a single 26-well gel and run according to the specifications above. Integrated densities for TAMRA-Cas9 co-localized bands were determined across wells using ImageJ analyses, and background signal from each no PNA (0 pmol) control was subtracted for each experiment.
Cell culture
K562 cells (CCL-243, ATCC) were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS, Life Technologies). HEK293 cells (CRL-1573, ATCC) were maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS, Life Technologies). U2OS cells (HTB-96, ATCC) were maintained in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS, Life Technologies). All cell lines were tested and confirmed to be free of mycoplasma infection.
Cell lines
dCas9-VPR, CRISPRa
Stable K562-hEF1α-dCas9-VPR line was established by lentiviral transduction using Edit-R purified lentiviral particles (Horizon Discovery) and blasticidin selection (5 μg/ml, A1113903, Gibco). dCas9-VPR integration was confirmed by western blot with CRISPR-Cas9 antibody (7A9-3A3, Novus). dCas9-VPR expressing line was further transduced with Edit-R CRISPRa human POU5F1 sgRNA purified lentiviral particles (VSGH11902, Horizon Discovery, Supplementary Table S3) and puromycin selection (2 μg/ml, Gibco).
dCas9-p300 acetyltransferase
Stable K562-pLV-dCas9-p300 line was established from lentivirus generated from pLV-dCas9-p300-P2A-PuroR plasmid (Addgene: #83889) and Invitrogen Virapower packaging plasmids in 293FT cells (Invitrogen, R70007) and puromycin selection (2 μg/ml, Gibco). Lines were further transduced with lentivirus similarly generated with pLV-U6-UbC-eGFP-P2A-Bsr plasmids (Addgene: #83925) containing MYOD promoter targeted sgRNA sequences with blasticidin selection (5 μg/ml, Gibco, A1113903, Supplementary Table S3).
K562-BFP
BFP-expressing K562 reporter lines were acquired from the laboratory of Jacob Corn, and feature a lentiviral inserted BFP gene under puromycin selection (Addgene: #111092) (22).
Cas9 RNP formulation and nucleofections with PNA
For Cas9 RNP assembly, 50 pmol of sgRNA (IDT, Alt-R CRISPR-Cas9 sgRNA) and 45 pmol of SpCas9 (CP02, PNA BIO) were incubated at room temperature for 10 min in a 5 μl reaction in 1 × 3.1 Buffer (B7203S, NEB). For nucleofections, 1 × 106 cells, 5 μl Cas9 RNP, and 1 μl of Alt-R Cas9 electroporation enhancer (IDT) were resuspended in 100 μl of Lonza cell line solution (V4XC-2024/V4XC-1024, Lonza) and nucleofected using a Lonza 4D-Nucleofector X unit. Cas9 RNP treated cells were seeded in 2 ml of complete media and incubated for 96 h prior to harvesting for analysis.
For PNA pre-treatments, 1 × 106 cells and 1 μl of PNA diluted to desired concentration in water were similarly nucleofected using Lonza cell line kits and 4D-Nucleofector. PNA nucleofected cells were resuspended in 1.25 ml of complete media in 1.5 ml tubes and incubated at 37°C for 2 h before Cas9 RNP nucleofection as described above. After Cas9 RNP nucleofection cells were seeded in 2 ml of complete media and incubated for 96 h prior to harvesting and analysis.
In time-course experiments in Figure 2C, K562 samples were nucleofected with 5 μl RNP reactions with the addition of 100 pmol of ssODN donor in 1 μl of water. RNP nucleofected samples were incubated at 37°C in complete media before being spun down and nucleofected with 250 pmol of PNA at specified time points. After PNA delivery cells were seeded in 2 ml of complete media and incubated for 96 h prior to harvesting and analysis.
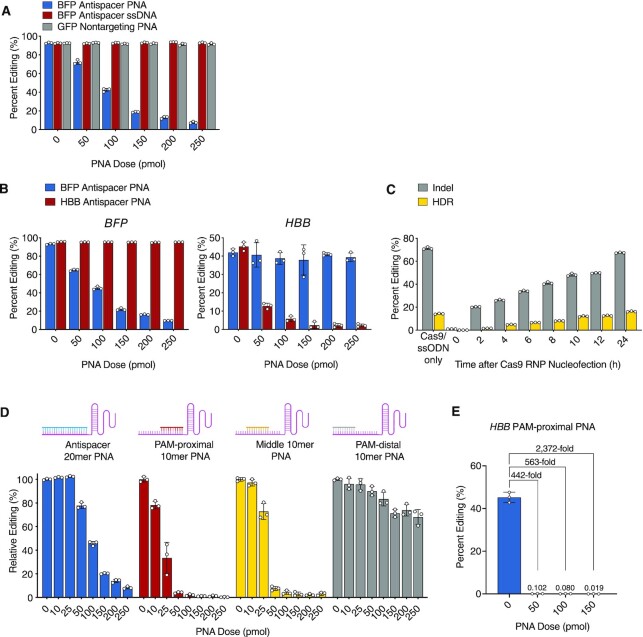
Figure 2.
Antispacer PNAs for sequence-specific Cas9 cleavage inhibition in human cells. (A) Percent BFP editing in K562-BFP cells pre-treated with BFP antispacer PNA (blue), BFP antisense ssDNA (red), or non-targeting GFP PNA (gray) followed by BFP-targeted Cas9 RNP treatment. (B) Percent BFP (left) and HBB (right) editing in K562-BFP cells pre-treated with BFP antispacer PNA (blue) or HBB antispacer PNA (red) followed by Cas9 RNP treatment. (C) Percent BFP indel (gray) and HDR (yellow) editing in K562-BFP cells treated with BFP antispacer PNA at specified timepoints relative to Cas9 RNP/donor ssODN nucleofection at time zero. (D) Relative percent BFP editing in K562-BFP cells pre-treated with 20mer full antispacer PNA (blue), 10mer PAM-proximal antispacer PNA (red), 10mer middle antispacer PNA (yellow), or 10mer PAM-distal antispacer PNA (gray) followed by BFP-targeted Cas9 RNP, as compared to untreated (0 pmol) condition. (E) Percent HBB editing in K562 cells pre-treated with 10mer PAM-proximal PNA followed by HBB-targeted Cas9 RNP. Mean fold changes in percent editing relative to untreated (0 pmol) condition are labelled. For (A–E), bars represent mean ± s.d. from n= 3 independent experiments, mean differences and P-values listed in Supplementary Table S1.
For pre-annealed PAM-distal PNA experiments, 50 pmol of sgRNA and 50 pmol of PAM-distal 10mer PNAs were incubated in a thermocycler at 37°C for 30 min in a 5 μl reaction in 1 × 3.1 Buffer (B7203S, NEB) to allow annealing. PNA annealing to sgRNAs was confirmed by gel shift on 5% polyacrylamide gel. 45 pmol of SpCas9 (CP02, PNA BIO) was then added at room temperature and incubated for 10 min to allow RNP formation. Resulting reaction was delivered to 1 × 106 K562 cells by a single nucleofection and treated as described above.
For Figure 4A and B, for each independent experiment, 6 replicates of 1 × 106 K562 stable line cells and 1 μl of PNA diluted to 250 μM in water were nucleofected using Lonza SF cell line kit and 4D-Nucleofector. Cells were pooled into cultures and maintained in 10mL of media at logarithmic growth phase (0.5–1.5 × 106 cells/ml) throughout the experiment. For each time-point 0.5 × 106 cells were removed from the culture for expression analysis (described below in RNA extraction and RT-PCR) and replaced with fresh complete media.
Figure 4.
Antispacer PNAs modulate activity of dCas9-fusion systems. (A) POU5F1 expression over time in K562-dCas9-VPR/POU5F1-sgRNA cells treated with 250 pmol of non-targeting BFP antispacer PNA (blue) and POU5F1 antispacer PNA (red). Relative mRNA was measured by RT-qPCR and presented as fold-change relative to non-transduced parental K562 line. Models were fit using a smoothing spline, λ = 0.5. Shaded regions represent 95% CI. (B) MYOD1 expression over time in K562-dCas9-p300/MYOD1-sgRNA cells treated with non-targeting BFP antispacer PNA (blue) and MYOD1 antispacer PNA (gray). Relative mRNA was measured by RT-qPCR and presented as fold-change relative to non-transduced parental K562 line. Models were fit using a smoothing spline, λ = 0.5. Shaded regions represent 95% CI. (C) H3K27ac percent occupancy (calculated by percent input method with 2% input) at selected timepoints relative to PNA nucleofection (250 pmol dose). Parent K562 (no dCas9-p300, yellow) and stable K562-dCas9-p300 lines prior to nucleofection are labelled. (A), points represent average measurements for three technical replicates from two independent experiments (n = 2), mean differences and P-values for each timepoint listed in Supplementary Table S1. (B) points represent average measurements for three technical replicates from three independent experiments (n = 3), mean differences and P-values for each timepoint listed in Supplementary Table S1. (C) Bars represent mean ± s.e.m. from average measurements for three technical replicates from n= 3 independent experiments, mean differences and P-values listed in Supplementary Table S1.
gRNA sequences and sources used in this study are listed in Supplementary Table S3 and PNA sequences are listed in Supplementary Table S2. ssODN in Supplementary Table S4.
Flow cytometry
K562 cells were spun down and resuspended in fresh complete media at 300 000 cells/ml 48 h prior to analysis. Samples for flow cytometry were fixed in complete media with 1% formaldehyde for 15 min before spinning down, resuspending in 300 μl PBS, and filtering through a cell strainer capped tube. Samples were analyzed for BFP and GFP fluorescence using a Cytoflex LX instrument (Beckman Coulter) using PacificBlue and FITC lasers, respectively, and quantification and gating was done using FloJo v10 software (Supplementary Figure S2). Background fluorescent drop out was determined by measurement of mock nucleofected lines in quadruplicate and subtracted from experimental values to determine reported editing frequency.
Targeted amplicon sequencing
Genomic DNA was purified from 0.5 × 106 cells using Promega ReliaPrep gDNA Tissue Miniprep System (A2052) and eluted in 50 μl of water. Library prep used 100ng of gDNA input with Ampliseq for Illumina Library Plus kit (20019101, Illumina) and used a custom primer pool designed to amplify a designed panel of genomic targets of interest (Supplementary Table S5). Libraries were indexed, pooled, and loaded onto a Mid-output (300 cycles, Illumina, FC-420–1004) cartridge for paired-end sequencing on an Illumina Miniseq instrument. FASTQ reads were subjected to quality analysis (Basespace FASTQC) and analyzed for indel frequency using Cas-Analyzer assessment tool (parameters: comparison range (R) – both ends of full amplicon sequence, Minimum frequency (n) = 1). gRNAs used in this study are listed in Supplementary Table S3, amplicon genomic coordinates listed in Supplementary Table S5.
In the case of EMX1 ON and OFF target amplicons, both regions were amplified from 200 ng of purified gDNA by Phusion High-Fidelity Polymerase (NEB, M0530S) and pooled and purified using QIAGEN PCR purification kit (28104, Qiagen). Library prep used 200 ng of purified amplicon input with Illumina DNA Prep kit (20025519, Illumina). EMX1 libraries were similarly indexed, pooled, sequenced, and analysed as above. Primers and genomic coordinates are listed in Supplementary Figures S4 and S5.
RNA extraction and RT-qPCR
For RNA extractions 0.5 × 106 K562 cells were spun down, washed once with PBS, homogenized using QIAshredder columns (QIAGEN 79656) and processed using QIAGEN RNeasy mini kits (74106) before eluting in 50 μl of water. Resulting RNA was used as input for high-capacity cDNA Reverse Transcription Kit (Applied Biosystems) and resulting reaction was diluted 1:5 with water. RT-qPCR reactions were conducted in triplicate for each sample using a StepOnePLUS Real-Time PCR System (Applied Biosystems) and Taqman Fast Advanced Master Mix (Applied Biosystems) with TaqMan gene specific probes (Supplementary Table S4). Expression relative to the untreated K562 parent line was recorded as fold-change and calculated from averaged CT values using the ΔΔCT method.
ChIP-qPCR
K562 cells (6 × 106 cells) were diluted to 0.4 × 106 cells/ml and cross-linked with a final concentration of 1% formaldehyde (Sigma-Aldrich, 252549) for each IP. Chromatin was prepared and sheared according to manufacturer protocol using SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling, #9003) and QSONICA Q800R3 sonicator for nuclear lysis. For each condition, 10 μg of chromatin was incubated with 3 μg of H3K27ac antibody (abcam ab4729) rotating overnight at 4°C. Chromatin was incubated with 30 μl of protein G magnetic beads (Cell Signaling, #70024) and washed, eluted, reverse cross-linked, and purified according to manufacturer protocol. ChIP-qPCR reactions were conducted in technical triplicate for 2% input and IP samples using a StepOnePLUS Real-Time PCR System (Applied Biosystems), SimpleChIP Universal qPCR Mastermix (Cell Signaling, #88989), and MYOD1 target specific primers (Supplementary Table S4). Percent occupancy values were calculated by percent input method from 2% input samples for each replicate.
Cell viability assays
1 × 106 K562 cells were nucleofected with specified doses of Cas9 RNP, PNA, or ssDNA using Lonza 4D-nucleofector. Cells were then serially diluted and seeded at 1,000 cells/well into 96-well plates and incubated for 72 h. Finally, cell viability was measured in-plate using CellTiter-Glo Luminescent Cell Viability Assay (G7570, Promega) with a Synergy H1 Multi-Mode Microplate Reader (Biotek) and graphed as percent viability relative to mock (0 pmol) conditions for each experiment.
Statistics
Graphing and statistical analysis were performed for each data set using Prism 9 (v9.3.0) software unless otherwise mentioned. All relevant differences and P-values from this study are summarized in Supplementary Table S1. For Figures 1E, 2A–E, 3B and Supplementary Figure S4C–F, comparisons relative to control untreated conditions were calculated using Dunnett's multiple comparisons test after confirming significant two-way ANOVA interactions. For Figure 3D, E and Supplementary Figures S4G, H and S5A–D, an unpaired Student's t-test was used to compare untreated and PNA-treated conditions. For comparisons between conditions in Figures 1E, 3F and 4C Tukey's multiple comparisons test was used after confirming significant two-way ANOVA interactions. For Figure 3C and Supplementary Figure S4A,B, a simple linear regression analysis was preformed and used to determine best-fit line and 95% CI. For Figure 4A, B, Šídák's multiple comparisons test was used to compare mean relative mRNA expression for each timepoint. Figures 4A, B, graphs and models were generated using JMP Pro (v15.0.0) software to fit a smoothing spline, λ = 0.5 and 95% CI.
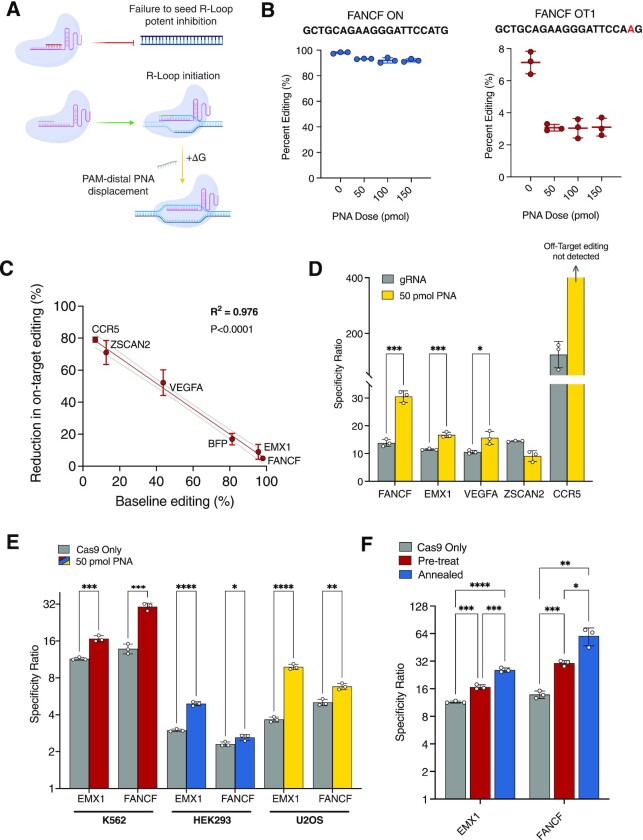
Figure 3.
Short PAM-distal PNAs modulate gRNA affinity to improve on-target specificity. (A) Schematic depicting Cas9 RNP-bound PAM-proximal antispacer PNA (red) for enzyme inhibition (top) and PAM-distal antispacer PNA (gray) for increasing the energetic demand for R-loop formation by requiring PNA displacement (bottom). (B) Percent FANCF editing at on-target (blue) and off-target (OT1, red) sites in K562 cells pre-treated with 10mer PAM-distal PNA followed by FANCF-targeted Cas9 RNP. gRNA spacer sequence is highlighted above graphs in bold with target mismatches highlighted in red. (C) Graph plotting percent reduction in on-target editing for PAM-distal 10mer PNA 50 pmol conditions as a function of mean baseline percent editing in untreated (0 pmol) condition in K562 cells. Best-fit line (red) was determined by simple linear regression model (Y = –0.7961X + 83.67) and dashed lines represent 95% CI. Calculated R2 value (0.976) and corresponding gene targets are labelled on plot. (D) Cas9 target specificity ratios for untreated (gray) and PAM-distal PNA pre-treated (50 pmol, yellow) K562 cells. CCR5 specificity cannot be represented as a discrete value because PAM-distal PNA treated off-target events were not detectable (specificity ratio → ∞). (E) EMX1 and FANCF Cas9 target specificity ratios for untreated (gray) and PAM-distal PNA pre-treated K562 (red), HEK293 (blue) and U2OS (yellow) cell lines. (F) EMX1 and FANCF Cas9 specificity ratios in K562 cells. Conditions represent Cas9 only/no PNA treatment (gray), PAM-distal PNA pre-treatment followed by Cas9 RNP (pre-treat, 50 pmol, red), and PAM-distal PNA pre-annealed to Cas9 RNP complex and delivered as a single treatment (annealed, 50 pmol, blue). For (B, C), points represent mean ± s.d. from n= 3 independent experiments, P-values listed in Supplementary Table S1. For (D–F) bars represent mean ± s.d. from n= 3 independent experiments, mean differences and P-values listed in Supplementary Table S1.
RESULTS
In vitro characterization of antispacer PNAs
For antispacer PNAs to impart an appreciable effect on Cas9 activity the affinity of a PNA for a given gRNA spacer sequence must exceed that of the DNA target strand. We began by comparing the thermal stabilities of a 20mer RNA oligonucleotide (consisting of a spacer sequence designed to target the blue fluorescent protein (BFP) gene) bound to either a complementary target DNA sequence from the BFP gene (length 20nt), a complementary RNA oligomer (length 20nt), or a complementary ‘antispacer’ PNA (length 20 bases). We applied UV spectroscopy thermal analyses to generate melting curves for DNA:RNA, RNA:RNA and PNA:RNA duplexes. We found that PNAs demonstrated a far higher affinity for the RNA spacer sequence (Tm > 100°C) as compared to target sequence DNA or a complementary RNA oligonucleotide (Tm = 56°C and 69°C, respectively, Figure 1C). Following this observation, we next designed an electrophoretic mobility shift assay (EMSA) with purified Cas9 single guide RNA (sgRNA) and Cas9 ribonucleoprotein (RNP) with or without a TAMRA dye-labeled BFP antispacer PNA to determine the extent to which PNAs bind gRNA spacer sequences within RNP complexes in vitro. We found that PNA-TAMRA signals strongly co-localized with both sequence-matched sgRNA and RNP bands (BFP target), but not sgRNA or RNP containing a heterologous spacer sequence (HBB target) (Figure 1D, Supplementary Figure S1). These results indicate that PNAs stably bind complementary gRNA spacer sequences as part of the Cas9 RNP complex. Quantitative analysis of TAMRA-Cas9 RNP gel bands revealed a dose-dependent increase in PNA binding between 5 pmol (0.5×) and 10 pmol (1×) conditions (2.09-fold increase, P < 0.0001, Figure 1E). However, 20 pmol (2×) doses revealed no significant additional binding compared to 10 pmol (P > 0.6), suggesting that the Cas9 RNP may be efficiently bound at near 1:1 PNA:sgRNA ratios (Figure 1E). Thus, in vitro, PNAs efficiently and tightly hybridize both free and Cas9-complexed gRNA spacer sequences at low stoichiometric ratios.
Antispacer PNAs achieve sequence-selective Cas9 cleavage inhibition in human cells
We next sought to determine if antispacer PNAs prevent Cas9 RNP-mediated cleavage and editing of a BFP reporter gene in human cells. For these experiments, PNAs were nucleofected into cells 2 h prior to Cas9 RNP transfection, thus requiring PNAs to hybridize spacer targets in the intracellular environment. We synthesized a BFP-targeted 20mer antispacer PNA (designed to bind the gRNA targeting the BFP gene) and compared its effect to a nontargeting PNA (corresponding to a sequence in the GFP gene, length 20 bases) and to an antispacer single-stranded DNA oligonucleotide (ssDNA, length 20nt, with the same sequence as the BFP-targeted PNA) in Cas9 RNP-nucleofected K562-BFP cells. Flow cytometry measurements of BFP fluorescent drop out revealed a robust dose-dependent inhibitory effect of antispacer PNA on Cas9-mediated BFP gene disruption and indel formation at picomole doses (P < 0.0001, Figure 2A, Supplementary Figure S2), whereas there was no effect of the antispacer DNA nor of the heterologous GFP PNA (P > 0.3 across conditions). To further establish the sequence-specific effect of PNAs on Cas9 activity, we compared the inhibitory effects of two 20mer antispacer PNAs: one designed to bind to the gRNA matching a target sequence in the BFP gene and the other to a gRNA targeting the human beta-globin gene (HBB). We tested both PNAs against Cas9 RNPs loaded with sgRNAs targeting the respective BFP or HBB sites in the K562-BFP cell line. Editing was quantified either by flow cytometry for BFP or by high-throughput amplicon sequencing in the case of HBB. We found that only sequence-matched PNAs inhibited gene editing at their respective loci (P < 0.0001 across conditions) with no detectable reduction in editing across doses for unmatched PNAs (P > 0.48 across conditions, Figure 2B), showing the specificity of action for antispacer PNAs in human cells.
Next, we considered the possibility that PNAs might act, in part, by binding to target genomic DNA rather than, or in addition to, binding to the gRNA. To investigate this possibility, we tested 20mer PNAs complementary to both strands of the BFP target DNA sequence, including the PAM and flanking 20nt regions and measured their effects on editing. We found that only PNAs with complementarity to the PAM-proximal gRNA spacer sequence showed any inhibitory activity (P < 0.0001, Supplementary Figure S3). PNAs with complementarity to DNA sequences but not the gRNA were ineffective (P > 0.34 across conditions, Supplementary Figure S3). We conclude that PNAs inhibit Cas9 primarily via hybridization to gRNA spacer sequences rather than to the target DNA.
We next investigated whether PNAs could be introduced into cells at varying times after Cas9 RNP transfection to prevent further editing activity. For these experiments, we not only assayed for BFP knockout by indel formation (BFP–, GFP–) but also for homology directed repair (HDR)/template-mediated editing of BFP to GFP when Cas9 RNP is combined with a single-stranded oligodeoxynucleotide (ssODN) donor instructing a single codon change (BFP–, GFP+, Supplementary Figure S2A and B). We chose to assay for both HDR and NHEJ repair outcomes to confirm the scaled effects of Cas9 inhibition for both repair pathway outcomes. ssODN templates were designed to avoid complementarity with the PNA to prevent hybridization events. The antispacer PNA demonstrated the ability to rapidly and proportionally block both indel formation (NHEJ) and template-mediated editing (HDR) across time-points up to 24 h, at which time most gene modification had occurred, and the overall editing levels were close to those seen in non-PNA treated cells (Cas9 RNP/ssODN only, Figure 2C). Together, these studies demonstrate that antispacer PNAs act in human cells in a sequence-specific fashion to robustly prevent on-target Cas9-mediated cleavage and editing. These results show that this effect can be dose and time-adjusted to tune overall levels of activity.
Short PAM-proximal antispacer PNAs demonstrate superiorly potent Cas9 cleavage inhibition
Next, to evaluate the impact of PNA length and binding position on Cas9 inhibition, we synthesized smaller 10mer antispacer PNAs to scan across the BFP spacer sequence and determined their effects on editing. In addition to the fully complementary 20mer antispacer PNA, 10mer PNAs were synthesized to bind across the PAM-proximal, middle, and PAM-distal spacer sequences of a BFP-targeted gRNA (Figure 2D). We found that a 10mer PNA targeted to the PAM-proximal spacer demonstrated highly effective Cas9 inhibitory activity in cells, with a 24.5-fold reduction in editing noted at 1:1 PNA:sgRNA molar ratios (50 pmol) and a 342.6-fold reduction at 5:1 ratios (250 pmol, Figure 2D). A separate amplicon sequencing analysis of a 10mer PAM-proximal PNA targeted to an HBB gRNA in K562 cells also demonstrated excellent inhibition (Figure 2E). For HBB target editing we measured a 443-fold reduction at 1:1 PNA:sgRNA ratios (50 pmol, 45.2% versus 0.102% editing) and near complete abrogation of editing by 150 pmol doses (6 total indels detected across 28 104 analyzed amplicons over three replicates, mean 2372-fold reduction, Figure 2E). Middle antispacer PNAs also demonstrated potent inhibition (32.4-fold reduction at 250 pmol), though slightly weaker than PAM-proximal targeting (Figure 2E). We hypothesize that improved inhibitory activity of 10mer PNAs over the full 20mer antispacer PNA may be attributed to size and the ability of the molecule to enter the Cas9 binding channel. The PAM-distal antispacer PNA, however, demonstrated an attenuated ability to block BFP editing across doses, achieving only 10% reduction at 50 pmol doses (versus 96.0% reduction for PAM-proximal PNA) and 32% reduction at 250 pmol doses (versus 99.7% for PAM-proximal). These data emphasize the necessity of the PAM-proximal spacer sequence for Cas9 targeting and inhibitory activity and are consistent with previous studies on the mechanisms and kinetics of Cas9 R-loop formation (20,23). Importantly, at very low stoichiometric doses (1:1), PAM-proximal PNAs demonstrated superior inhibition activity in cells (at least 10-fold improvement) as compared to published values for purified inhibitors such as Acrs, small molecules, DNA-based aptamers, and small modified nucleic acid inhibitors, indicating the high potency of appropriately designed PNAs for Cas9 inhibition (7,11–12).
Short PAM-distal antispacer PNAs modulate gRNA target affinity to improve on-target specificity
We next sought to explore whether antispacer PNAs could be leveraged to improve the on-target specificity of Cas9 RNP activity, defined here as the ratio of on-target to off-target editing rates. Previous work demonstrated that manipulating 5′ PAM-distal regions of sgRNAs by truncation or with engineered RNA hairpins improves specificity in Cas9 systems, likely by selectively preventing lower affinity off-target R-loop formation (23–25). Further, strategic gRNA spacer sequence base modifications have also been used to affect thermodynamic barriers and favorably modulate specificity (26). We hypothesized that generating PAM-distal PNA:gRNA duplexes with 10mer PNAs in cells may similarly interrupt R-loop formation preferentially at off-target binding sites, thereby increasing specificity (Figure 3A). To test this, PAM-distal antispacer 10mer PNAs were designed for gRNAs targeting five selected genomic sites in the FANCF, EMX1, ZSCAN2, CCR5 and VEGFA genes, each with known off-target activity in the genome. We pre-treated K562 cells with nucleofected PAM-distal 10mer PNAs followed by Cas9 RNP 2 h later and quantified on-target and respective off-target editing rates using high-throughput amplicon sequencing. In the case of the clinically relevant target FANCF, a 10mer PAM-distal PNA reduced overall off-target editing by 57.1% at a 1:1 PNA:sgRNA ratio (50 pmol, 7.13% versus 3.07%) while on-target editing was reduced by only 4.9% (50 pmol, 98.0% versus 93.2%, Figure 3B). This 11.6-fold difference in percent editing reduction supports the hypothesis that gRNAs bound by PAM-distal PNAs incur an energetic penalty affecting successful R-loop formation (net ΔG increase, Figure 3A). PAM-distal PNAs cause overall thermodynamic stability to decrease and are demonstrated to favor high-affinity on-target cleavage as opposed to lower energy off-target cleavage. As further evidence, we found that lower affinity gRNAs were more susceptible to on-target inhibition by 10mer PAM-distal PNAs. Percent reduction in on-target editing strongly correlated with in silico predicted on-target affinity scores (27) (R2 = 0.853, P < 0.0001, Supplementary Figure S4A) as well as measured mean baseline on-target editing in K562 cells (R2 = 0.976, P < 0.0001, Figure 3C). Thus, we demonstrate that percent editing reduction for PAM-distal PNA-bound gRNAs predictably decreases as a function of both predicted and empiric measures of affinity.
In agreement with our models, off-target editing decreased by a larger margin as compared to on-target editing for four out of five loci, and sites with the largest differences in percent on-target versus off-target editing demonstrated the largest increases in specificity (Figure 3D, Supplementary Figure S4B–F). In the case of ZSCAN2, both on-target and off-target editing decreased by a large margin (80.3% and 66.3%, respectively), and we did not observe an overall increase in specificity (Figure 3D, Supplementary Figure S4D). For CCR5, on-target editing decreased by 79.1% while off-target editing in the CCR2 gene was rendered undetectable (Figure 3D, Supplementary Figure S4E). Based on these relationships, we suspect further fine-tuning of PNA length and binding strength may overcome high on-target reduction in lower affinity targets, such as ZSCAN2 and CCR5, while maintaining large reductions in off-target editing.
Next, we explored the ability of PAM-distal PNAs to improve on-target specificity in additional cell lines. We selected gRNAs with the highest off-target activity from our analysis, FANCF and EMX1, and pre-treated K562, HEK293, and U2OS cell lines with PAM-distal PNAs prior to Cas9 RNP nucleofection. As expected, we observed significantly increased specificity ratios for both targets across all three cell lines (P < 0.05 across conditions, Figure 3E). Notably, K562, HEK293 and U2OS lines demonstrated specificity improvements across widely variable baseline off-target editing activity for both EMX1 (8.33%, 32.7% and 26.8%, respectively, Supplementary Figures S4F, S5A and B) and FANCF (7.13%, 28.4% and 19.8%, respectively, Figure 3B and Supplementary Figure S5C and D).
Finally, we tested pre-annealing a PAM-distal 10mer PNA to FANCF and EMX1 sgRNAs prior to Cas9 RNP formation for delivery as a single complex. We found that both pre-annealed FANCF and EMX1 Cas9 RNP–PNA complexes showed further improved specificity over pre-treatment approaches (Figure 3F, Supplementary Figure S4G and H). We hypothesize this further improvement in specificity is due to more efficient PNA:RNA duplex formation in vitro. Importantly, this result indicates the feasibility of delivering PNA-bound ribonucleoprotein as a single functional complex.
Together, these data demonstrate that antispacer PNAs can be rationally designed to impart two distinct modulation effects on Cas9 activity: superior sequence-specific inhibition via PAM-proximal spacer targeting, and affinity manipulation and specificity enhancement via PAM-distal binding.
Antispacer PNAs modulate spatiotemporal activity of dCas9 fusion systems in a sequence-specific manner
Beyond use as a nuclease for gene knockout and editing, nuclease-deficient variants of Cas9 (dCas9) are widely used to create fusion systems for sequence-specific localization of effectors. Precise control over these systems via sequence-specific dCas9 binding modulation has the potential to further expand the versatility of applications and provide improved control for biological investigations. Given the robust effects of antispacer PNAs on Cas9-mediated cleavage, we sought to test the extent to which PNAs can modulate the functional activity of two well-established dCas9 fusion effectors: a CRISPRa (dCas9-VPR) transcriptional activator and a histone acetyltransferase-fused epigenome editor (dCas9-p300). We applied PNAs to a stably integrated CRISPRa system in K562 cells targeting a POU5F1 promoter to constitutively upregulate the gene, and measured gene expression over time following PNA transfection (Figure 4A). After the introduction of 250 pmol of a 20mer POU5F1-targeted antispacer PNA, POU5F1 expression rapidly dropped to the level of the parent line expression by 48 h (Figure 4A). By 120 h (day 5) expression began to return to its upregulated baseline (Figure 4A). This effect was specific to the sequence-matched antispacer PNA, as a nontargeting (BFP) PNA control did not suppress CRISPRa activity at the POU5F1 locus, which remained upregulated and highly expressed (Figure 4A). Next, we used PNAs to modulate the appearance and disappearance of epigenetic marks using a fused acetyltransferase (dCas9-p300) epigenetic editing system (28). At baseline this system produced increased H3K27ac histone modification near the promoter region of the MYOD1 gene and demonstrated robust gene upregulation (Figure 4B and C). MYOD1 gRNA-targeted PNAs were able to drive down epigenetic-mediated gene upregulation by 24 h (Figure 4B and C). By 168 h (7 days) expression returned to the fully upregulated state (Figure 4B). Corroborating these observations, we saw a significant decrease in H3K27ac occupancy at the MYOD1 promoter region by 24 and 48 h after PNA treatment followed by a subsequent increase at 240 h as measured by ChIP-qPCR (Figure 4C). Collectively, these data demonstrate that antispacer PNAs can temporally manipulate stable dCas9 fusion systems and influence downstream biology. Results from two independent systems suggest that PNA blocking happens rapidly and in a sequence-specific manner following a single PNA dose, with full effects noted by 24–48 h and persistence for at least an additional 2–3 days after that.
DISCUSSION
In summary, we designed, synthesized, and characterized a class of synthetic high-affinity gRNA-binding PNA molecules capable of rationally manipulating Cas9 interactions with dsDNA targets. Using PNAs with PAM-proximal homology we show potent Cas9 inhibition in human cells by targeting the most consequential interactions driving R-loop formation (23). We found that this approach results in superior inhibitory potency at low stoichiometric ratios with the important added benefit of sequence specificity. We postulate, in contrast to other approaches to Cas9 inhibition, that PNAs may be applicable as sequence-selective regulators of multiplexed Cas9 systems such as large-scale screens.
In this work, we also characterize the affinity-dependent effects of PAM-distal targeted antispacer PNAs and demonstrate that this approach can be leveraged to predictably improve Cas9 specificity. Because off-target sites, by definition, occur at lower affinity sites as compared to on-target loci, we predict specificity to increase for most Cas9 targets. We note in our study, however, that lower affinity gRNAs were less amenable to specificity improvement using PAM-distal 10mer PNAs. Importantly, PAM-distal PNAs can be further adjusted in length and chemically modified via sidechain substitution at the gamma(γ) position in the polyamide backbone to fine-tune binding strength for improved effect (29,30). For example, for low-affinity targets such as CCR5 and ZSCAN2, shorter 8mer or 6mer PAM-distal PNAs may result in less on-target inhibition effects and more optimally adjust specificity. Conversely, for high-affinity targets such as FANCF or EMX1, increasing PNA affinity can further improve specificity by further reducing off-target effects and maintaining high on-target editing. In addition to increasing PNA length beyond 10 bases, gamma(γ) position modification with chemical sidechains such as a hydroxymethly group (serine) or polyethlene glycol (mini-PEG) can improve PNA helical organization and increase binding strength (29,31). Notably, previous work from Verona et al. has shown that one can computationally predict the effects of length and γ-modifications on PNA helical preorganization to fine-tune PNA affinity for complementary RNA targets (30). Coupled with models predicting antispacer PNA influence on gRNA target affinity, as we describe in our study (Figure 3C and Supplementary Figure S4A and B), these tools have the potential to predict optimized PNA formulations for individual gRNA targets. Incorporating predicted gRNA affinity, off-target rates, and PNA binding strength, future in silico methods may identify targets most suitable for off-target mitigation using PNAs. Future studies informed by the phenomena we describe here may further optimize Cas9 specificity across targets to reduce genotoxicity and improve safety for use in human therapeutics.
While PNAs offer clear advantages as a synthetic nuclease-resistant technology with powerful nucleic acid binding properties, their application poses some challenges (32). Firstly, PNAs are not as commercially accessible as conventional nucleic acids, and historically PNA synthesis required laborious manual chemical synthesis and relevant expertise. However, with the availability of suitable monomers, methods as outlined in this study are available to automate PNA synthesis using an in-lab benchtop peptide synthesizer and HPLC system (see Materials and Methods for both automated and manual synthesis protocols (33)). Using this approach, we were able to design, synthesize, and purify PNAs ready for delivery within 72 h and with minimal hands-on time. Additionally, with a hydrophobic polyamide backbone, PNAs may demonstrate poor aqueous solubility and aggregation relative to classic nucleic acids, especially sequences with high purine content. Notably, this effect is minimized for shorter PNAs (<30 bases), such as those used in this study, and can be further offset with the inclusion of hydrophilic N- and C-terminal l-lysine residues. Toxicity, potentially related to sequence, may also present possible challenges for application. Viability studies in nucleofected human K562 cells using a variety of PNA sequences and lengths used in this study demonstrated minimal cellular toxicity across doses (Supplementary Figure S6). Prior studies have similarly noted low toxicity across a range of PNA doses in other cell types including U251 and HeLa lines and in primary human CD34+ cells (34–36). These findings are consistent with previous studies in vivo in mice that found little to no toxicity across multiple delivery methods for systemic treatments of PNA formulations (31,37–39).
Finally, in contrast to sequence-restricted or protein-coded approaches, we report an accessible and rapidly implementable platform for CRISPR-Cas9 modulation that features facile PNA oligomer design and application. This technology provides new methods for enhanced control over Cas9-derived experimental systems and may offer a tool to improve the safety of human therapeutic applications. Further, because antispacer PNAs target gRNA sequences, they are theoretically applicable to all current and future Cas9 and dCas9-based systems, and likely other RNA-guided RNP systems. Beyond CRISPR-Cas9, the principles applied in this study can inform future approaches to manipulate the character and activity of nucleoproteins via stable antisense binding and modification with PNAs.
DATA AVAILABILITY
Raw sequencing data for experiments from this study are available at Bioproject/SRA (BioProject ID PRJNA750752).
Cas-Analyzer is an instant assessment tool for high-throughput sequencing data for genome edited cells (http://www.rgenome.net/cas-analyzer/#!) and was used for analysis of gene editing outcomes from Illumina Miniseq sequencing data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Hegan and A. Dhawan for their assistance. We thank J. Corn and C. Richardson for providing K562-BFP cells used in this study. We also thank S. Oyaghire, S. Yeon, and H. Li for their assistance with experiments. We thank Yale Flow Cytometry for their assistance with flow cytometry analysis. We thank the Keck Oligo Synthesis Resource at Yale for their assistance with primer synthesis services. Illustrations generated using BioRender.com software.
Contributor Information
Nicholas G Economos, Department of Therapeutic Radiology, Yale School of Medicine, New Haven, CT 06520 USA; Department of Genetics, Yale School of Medicine, New Haven, CT 06520 USA.
Elias Quijano, Department of Therapeutic Radiology, Yale School of Medicine, New Haven, CT 06520 USA; Department of Genetics, Yale School of Medicine, New Haven, CT 06520 USA.
Kelly E W Carufe, Department of Therapeutic Radiology, Yale School of Medicine, New Haven, CT 06520 USA; Department of Genetics, Yale School of Medicine, New Haven, CT 06520 USA.
J Dinithi R Perera, Department of Therapeutic Radiology, Yale School of Medicine, New Haven, CT 06520 USA.
Peter M Glazer, Department of Therapeutic Radiology, Yale School of Medicine, New Haven, CT 06520 USA; Department of Genetics, Yale School of Medicine, New Haven, CT 06520 USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01HL139756, R35CA197574, U01AI145965 to P.M.G.]; N.G.E. was supported by National Research Service Award fellowship [F30HL149185] from NIH NHLBI; N.G.E. and E.Q. were supported by MSTP training grant [T32GM007205]. Funding for open access charge: NIH [U01AI145965].
Conflict of interest statement. N.G.E., E.Q. and P.M.G. are inventors on US patent application no. 63/197,879 submitted by and assigned to Yale University, which covers compositions and methods for CRISPR-Cas9 modification using PNAs.
REFERENCES
- 1. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O.et al.. DNA targeting specificity of RNA-guided cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang X.-H., Tee L.Y., Wang X.-G., Huang Q.-S., Yang S.-H.. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. - Nucleic Acids. 2015; 4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018; 9:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doudna J.A., Charpentier E.. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. [DOI] [PubMed] [Google Scholar]
- 5. Shivram H., Cress B.F., Knott G.J., Doudna J.A.. Controlling and enhancing CRISPR systems. Nat. Chem. Biol. 2021; 17:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shin J., Jiang F., Liu J.-J., Bray N.L., Rauch B.J., Baik S.H., Nogales E., Bondy-Denomy J., Corn J.E., Doudna J.A.. Disabling cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017; 3:e1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B.et al.. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019; 177:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marino N.D., Pinilla-Redondo R., Csörgő B., Bondy-Denomy J.. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods. 2020; 17:471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marino N.D., Zhang J.Y., Borges A.L., Sousa A.A., Leon L.M., Rauch B.J., Walton R.T., Berry J.D., Joung J.K., Kleinstiver B.P.et al.. Discovery of widespread type i and type v CRISPR-Cas inhibitors. Science. 2018; 362:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meeske A.J., Jia N., Cassel A.K., Kozlova A., Liao J., Wiedmann M., Patel D.J., Marraffini L.A.. A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science. 2020; 369:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barkau C.L., O’Reilly D., Rohilla K.J., Damha M.J., Gagnon K.T.. Rationally designed Anti-CRISPR nucleic acid inhibitors of CRISPR-Cas9. Nucleic Acid Ther. 2019; 29:136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao J., Inomata R., Kato Y., Miyagishi M.. Development of aptamer-based inhibitors for CRISPR/Cas system. Nucleic Acids Res. 2020; 49:1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egholm M., Buchardt O., Christensen L., Behrens C., Freier S.M., Driver D.A., Berg R.H., Kim S.K., Norden B., Nielsen P.E.. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993; 365:566. [DOI] [PubMed] [Google Scholar]
- 14. Nielsen P.E., Egholm M., Buchardt O.. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjugate Chem. 1994; 5:3–7. [DOI] [PubMed] [Google Scholar]
- 15. Pellestor F., Paulasova P.. The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur. J. Hum. Genet. 2004; 12:694. [DOI] [PubMed] [Google Scholar]
- 16. Zeng F., Peritz T., Kannanayakal T.J., Kilk K., Eiríksdóttir E., Langel U., Eberwine J.. A protocol for PAIR: PNA-assisted identification of RNA binding proteins in living cells. Nat. Protoc. 2006; 1:920–927. [DOI] [PubMed] [Google Scholar]
- 17. Fabbri E., Manicardi A., Tedeschi T., Sforza S., Bianchi N., Brognara E., Finotti A., Breveglieri G., Borgatti M., Corradini R.et al.. Modulation of the biological activity of microRNA-210 with peptide nucleic acids (PNAs). ChemMedChem. 2011; 6:2192–2202. [DOI] [PubMed] [Google Scholar]
- 18. Peter E.N. Gene targeting and expression modulation by peptide nucleic acids (PNA). Curr. Pharm. Des. 2010; 16:3118–3123. [DOI] [PubMed] [Google Scholar]
- 19. Economos N.G., Oyaghire S., Quijano E., Ricciardi A.S., Saltzman W.M., Glazer P.M.. Peptide nucleic acids and gene editing: perspectives on structure and repair. Molecules. 2020; 25:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anders C., Niewoehner O., Duerst A., Jinek M.. Structural basis of PAM-dependent target DNA recognition by the cas9 endonuclease. Nature. 2014; 513:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 23. Farasat I., Salis H.M.. A biophysical model of CRISPR/Cas9 activity for rational design of genome editing and gene regulation. PLoS Comput. Biol. 2016; 12:e1004724–e1004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kocak D.D., Josephs E.A., Bhandarkar V., Adkar S.S., Kwon J.B., Gersbach C.A.. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019; 37:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ryan D.E., Taussig D., Steinfeld I., Phadnis S.M., Lunstad B.D., Singh M., Vuong X., Okochi K.D., McCaffrey R., Olesiak M.et al.. Improving CRISPR–Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2017; 46:792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R.et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016; 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015; 33:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dragulescu-Andrasi A., Rapireddy S., Frezza B.M., Gayathri C., Gil R.R., Ly D.H.. A simple γ-backbone modification preorganizes peptide nucleic acid into a helical structure. J. Am. Chem. Soc. 2006; 128:10258–10267. [DOI] [PubMed] [Google Scholar]
- 30. Verona M.D., Verdolino V., Palazzesi F., Corradini R.. Focus on PNA flexibility and RNA binding using molecular dynamics and metadynamics. Sci. Rep. 2017; 7:42799–42799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bahal R., Ali McNeer N., Quijano E., Liu Y., Sulkowski P., Turchick A., Lu Y.-C., Bhunia D.C., Manna A., Greiner D.L.et al.. In vivo correction of anaemia in β-thalassemic mice by γPNA-mediated gene editing with nanoparticle delivery. Nat. Commun. 2016; 7:13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta A., Mishra A., Puri N.. Peptide nucleic acids: advanced tools for biomedical applications. J. Biotechnol. 2017; 259:148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Braasch D.A., Nulf C.J., Corey D.R.. Synthesis and purification of peptide nucleic acids. Curr. Protoc. Nucleic Acid Chem. 2002; Chapter 4:Unit 4.11. [DOI] [PubMed] [Google Scholar]
- 34. Turner J.J., Ivanova G.D., Verbeure B., Williams D., Arzumanov A.A., Abes S., Lebleu B., Gait M.J.. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans -activation in cells. Nucleic Acids Res. 2005; 33:6837–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gasparello J., Manicardi A., Casnati A., Corradini R., Gambari R., Finotti A., Sansone F.. Efficient cell penetration and delivery of peptide nucleic acids by an argininocalix[4]arene. Sci. Rep. 2019; 9:3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McNeer N.A., Chin J.Y., Schleifman E.B., Fields R.J., Glazer P.M., Saltzman W.M.. Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol. Ther. 2011; 19:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaubey B., Tripathi S., Pandey V.N.. Single acute-dose and repeat-doses toxicity of anti-HIV-1 PNA TAR-penetratin conjugate after intraperitoneal administration to mice. Oligonucleotides. 2008; 18:9–20. [DOI] [PubMed] [Google Scholar]
- 38. McNeer N.A., Schleifman E.B., Cuthbert A., Brehm M., Jackson A., Cheng C., Anandalingam K., Kumar P., Shultz L.D., Greiner D.L.et al.. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 2013; 20:658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schleifman E.B., McNeer N.A., Jackson A., Yamtich J., Brehm M.A., Shultz L.D., Greiner D.L., Kumar P., Saltzman W.M., Glazer P.M.. Site-specific genome editing in PBMCs with PLGA Nanoparticle-delivered PNAs confers HIV-1 resistance in humanized mice. Mol. Ther. - Nucleic Acids. 2013; 2:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for experiments from this study are available at Bioproject/SRA (BioProject ID PRJNA750752).
Cas-Analyzer is an instant assessment tool for high-throughput sequencing data for genome edited cells (http://www.rgenome.net/cas-analyzer/#!) and was used for analysis of gene editing outcomes from Illumina Miniseq sequencing data.