Abstract
DDX58 encodes RIG-I, a cytosolic RNA sensor that ensures immune surveillance of nonself RNAs. Individuals with RIG-IE510V and RIG-IQ517H mutations have increased susceptibility to Singleton-Merten syndrome (SMS) defects, resulting in tissue-specific (mild) and classic (severe) phenotypes. The coupling between RNA recognition and conformational changes is central to RIG-I RNA proofreading, but the molecular determinants leading to dissociated disease phenotypes remain unknown. Herein, we employed hydrogen/deuterium exchange mass spectrometry (HDX-MS) and single molecule magnetic tweezers (MT) to precisely examine how subtle conformational changes in the helicase insertion domain (HEL2i) promote impaired ATPase and erroneous RNA proofreading activities. We showed that the mutations cause a loosened latch-gate engagement in apo RIG-I, which in turn gradually dampens its self RNA (Cap2 moiety:m7G cap and N1-2-2′-O-methylation RNA) proofreading ability, leading to increased immunopathy. These results reveal HEL2i as a unique checkpoint directing two specialized functions, i.e. stabilizing the CARD2-HEL2i interface and gating the helicase from incoming self RNAs; thus, these findings add new insights into the role of HEL2i in the control of antiviral innate immunity and autoimmunity diseases.
INTRODUCTION
DDX58 encodes RIG-I, an interferon receptor that ensures immune surveillance of exogenous RNAs (1). DDX58 gain-of-function pathogenic variants can drive elevated type I interferon production, leading to disturbances in immune function (2). For instance, Singleton-Merten syndrome (SMS) is a rare immunogenic disorder with variable clinical phenotypes, including psoriasis, acro-osteolysis, and glaucoma, and severe cases are observed with dental dysplasia and tendon rupture (3–5). RIG-I consists of N-terminal tandem caspase activation and recruitment domains (CARD1-2 or CARDs), a central helicase domain (HEL) containing two RecA-like domains (HEL1-2) and an insertion domain HEL2i, and a carboxyl terminal domain (CTD) (6–8). HEL2i is uniquely associated with duplex RNA-activated ATPases (DRAs), which are phylogenetically classified as a subgroup within helicase superfamily 2 (9,10). Dysfunctions of DDX58 variants (c.1529A > T [p.E510V], c.803G > T [p.C268F], c1118A > C [p.E373A] and c1551G > C [p.Q517H]) have been reported to be correlated with the increased clinical severities of type I interferonopathies, thereby contributing to the occurrence of tissue-specific, atypical and classic SMS defects (3–5). Atypical SMS RIG-I variants carry C268F and E373A mutations belonging to conserved SF2 helicase motifs I and II, respectively, forming a category of ATPase-deficient RIG-I variants (11–13). The molecular mechanisms of impaired RNA discrimination by atypical SMS RIG-Is have recently been characterized (2,14). SMS RIG-I variants carrying the E510V and Q517H mutations are located on the same HEL2i α-helix, and the former is spatially more distal than the latter to the CARD2 latch motif, which forms an intra-molecular interface with HEL2i in apo RIG-I (15,16). Therefore, these HEL2i mutations are distant from the ATPase cleft, suggesting an alternative mechanism not correlated with dysregulated ATP binding and hydrolysis function. However, how this genotype-phenotype gradient of DDX58 variants selectively determines the clinical severities of SMS remains enigmatic.
RIG-I in the resting state adopts an autoinhibited state, in which CARD2-HEL2i mimics a latch-gate interface that suppresses CARDs signaling (16–18). Upon RNA recognition, the HEL domain induces conformational changes that liberate the signaling CARDs (6,15,16). ATP binding impacts the RNA-dependent ATPase function model of RIG-I and facilitates HEL engagement and CARDs exposure (15). In addition, K63-polyUb chains serve as endogenous agonists for bridging RIG-I CARDs into a tetrameric conformation, which engages the MAVSCARD tetramer for the nucleation of MAVS fibers, a signaling cascade that culminates in interferon and cytokine production (1,8,19–21). RIG-I prefers to recognize short, blunt-ended, and double-stranded RNAs with a 5′-triphosphate moiety (5′ppp) or a 5′-diphosphate moiety (5′pp), which are considered pathogen-associated molecular pattern (PAMP) RNA features frequently present in viral RNA genomes or replication intermediates (22–24). Because of a variety of RNA editing enzymes, mRNAs of humans and other higher eukaryotes possess a 7-methylguanosine (m7G) cap at the 5′ end with either the first nucleotide ribose (N1-2′-O-M, Cap1 moiety) or the first two nucleotide riboses modified with 2′-O-methylation (N1-2-2′-O-M, Cap2 moiety) (25–27). RIG-I discriminates against N1-2′-O-M RNA due to a mechanism involving the N1-2′-O-M modification that specifically protects RIG-I CTD residue H830 from interacting with N1-2′-OH. This protection results in a weakened RNA recognition event by RIG-I HEL-CTD and a blunt CARDs opening event (15,28). Previous studies have revealed the existence of self RNAs that can be recognized by RIG-I in cellulo and in vivo (14,28–30). Understanding the mechanisms whereby RIG-IQ517H and RIG-IE510V erroneously recognize self RNA and the onset of different degrees of autoimmune conditions is of critical importance for therapeutic intervention.
In this study, we utilized amide hydrogen/deuterium exchange coupled with mass spectrometry (HDX-MS) and magnetic tweezers (MT) to dissect the molecular behaviors of the genotype-phenotype gradient of DDX58 variants, which encode RIG-IE510V and RIG-IQ517H, and investigate how they promote aberrant interferon production associated with mild and severe SMS phenotypes. HDX-MS has proven to be a robust biophysical approach to study receptor conformational equilibrium and dynamic changes in the context of a binding event (31). MT have been found highly effective for dissecting protein conformational states, dynamics of their transitions, and intermolecular interactions at the single-molecule level with high spatiotemporal resolution (32–35). In summary, our study uncovers new mechanistic insights into the dysregulated RIG-I proofreading of self RNAs. We show that this consequence results from a pair of HEL2i mutations at amino acid positions 510 and 517 that partially blunt the CARD2 interface in their apo forms. These RIG-I variants have functionally gained the ability to recognize self RNAs and activate themselves. Our results suggest the HEL2i helix as a novel target to design therapies against autoimmune diseases.
MATERIALS AND METHODS
Cloning, protein expression and purification
Human WT full-length RIG-I was cloned into pET-SUMO and pUNO vectors. Full-length RIG-I mutants E510V, Q517H, E373A and C268F were constructed in pET-SUMO and pUNO vectors by site-directed mutagenesis of full-length RIG-I. All constructs were confirmed by sequencing. RIG-I (WT and 4mut) was expressed in Escherichia coli strain Rosetta (DE3) (TransGen) as soluble proteins. The soluble fraction of protein was purified from the cell lysate using a HisTrap™ HP column (GE Healthcare). The protein was then digested with ULP1 protease to remove the 6xHis–SUMO, and RIG-I was further purified with a HiTrap™ Heparin HP column (GE Healthcare). Finally, purified protein was passed through a HiLoad™ 16/600 Superdex™ 200 (GE Healthcare) size exclusion column against 50 mM HEPES, pH 8.0, 150 mM NaCl, 2 mM DTT and 5% glycerol at 4°C, snap frozen in liquid nitrogen, and stored at –80°C.
K63–polyUb chain synthesis and production
Lys63-linked ubiquitin chains (K63-polyUbn) were generated from a reaction containing 1 mM ubiquitin, 1 μM mE1, 10 μM UbcH13 and 10 μM Uev1A in buffer (10 mM ATP, 20 mM PC, 1.2 U/ml IPP, 1.2 U/ml CPK, 50 mM Tris pH 7.5, 10 mM MgCl2, 2 mM DTT). The K63-polyUbn synthesis reaction was performed at 37°C for 3–6 h. Synthesized polyUbn chains were purified as described previously (21). Briefly, ubiquitin chains were diluted 5-fold into 50 mM ammonium acetate, pH 4.5, and 0.1 M NaCl and separated over a 0.1–1.0 M NaCl gradient in 50 mM ammonium acetate, pH 4.5, using a HiTrapTM Heparin HP column (GE Healthcare). Fractions were applied to a Superdex™ 200 Increase 10/300 GL column equilibrated with buffer (50 mM Tris, pH 7.5, 2 mM DTT) for final purification to homogeneity.
RNA preparation
All RNAs used in the study were chemically synthesized, purified by high-performance liquid chromatography, and analyzed by mass spectrometry (MS; Trilink and BioSynthesis RNA synthesis service). The lyophilized RNA was resuspended in 20 mM potassium phosphate buffer, pH 7.0.
HDX-MS
Peptide identification
Peptides were identified using tandem MS (MS/MS) with a Fusion Orbitrap mass spectrometer (Thermo Fisher). Product ion spectra were acquired in data-dependent mode with the top eight most abundant ions selected for the product ion analysis per scan event. The MS/MS data files were submitted to Proteome Discover 2.4 (Thermo Fisher) for high-confidence peptide identification.
HDX-MS analysis
Five micromolar purified RIG-I or RIG-I mutants was incubated with or without RNA (3p10l or Cap2-10l, molar ratio 1:1.2; K63-polyUbn (molar ratio 1:1) in buffer (50 mM HEPES, pH 7.5; 50 mM NaCl; 5% glycerol; 4 mM MgCl2; 2 mM DTT) for 30 min before the HDX reactions at 4°C. Four microliters of protein/protein complex with ligand/peptide was diluted into 16 μl D2O in exchange buffer (50 mM HEPES, pH 7.5; 50 mM NaCl; 2 mM DTT) and incubated for various HDX time points (i.e. 0, 10, 60, 300, 900 and 3600 s) at 4°C and quenched by mixing with 20 μl of ice-cold 3 M GHCl containing 1% trifluoroacetic acid. Each quenched sample was immediately injected into the LEAP Pal 3.0 HDX platform. Upon injection, samples were passed through an immobilized pepsin column (2 mm × 2 cm) at 120 μl min−1, and the digested peptides were captured on a C18 PepMap300 trap column (Thermo Fisher) and desalted. Peptides were separated across a 2.1 mm × 5 cm C18 separating column (1.9 μm Hypersil Gold, Thermo Fisher) with a linear gradient of 4–40% CH3CN and 0.3% formic acid over 6 min. Sample handling, protein digestion and peptide separation were conducted at 4°C. Mass spectrometric data were acquired using a Fusion Orbitrap mass spectrometer (Thermo Fisher) with a measured resolving power of 65 000 at m/z 400. HDX analyses were performed in triplicate, with single preparations of each protein ligand complex. The intensity weighted mean m/z centroid value of each peptide envelope was calculated and subsequently converted into a percentage of deuterium incorporation. Statistical significance for the differential HDX data was determined by an unpaired t test for each time point, a procedure that is integrated into the HDX Workbench software (36). Corrections for back exchange were made on the basis of an estimated 70% deuterium recovery and accounting for the known 80% deuterium content of the deuterium exchange buffer.
Data rendering
The HDX data from all overlapping peptides were consolidated to individual amino acid values using a residue averaging approach. Briefly, for each residue, the deuterium incorporation values and peptide lengths from all overlapping peptides were assembled. A weighting function was applied in which shorter peptides were weighted more heavily and longer peptides were weighted less. The weighted deuterium incorporation values were then averaged to produce a single value for each amino acid. The initial two residues of each peptide as well as prolines were omitted from the calculations.
EX1 kinetics analysis and curve fitting
The isotopic distributions of the MS peak (101–114, +3) were specifically analyzed by HX express2 software (37,38). Exponential 3P was used as the prediction model: a + b.exp(c.time (min)), where a is the asymptote, b is the scale, and c is the growth rate. The model was used to fit a curve to %D (response) and time (regressor) for each compound state. The inverse prediction was used to solve for the half-life (t1/2) for each compound state. Analysis of means was performed to identify the compound states with half-life (t1/2) values that were significantly different from the average half-life (t1/2) in a given comparison group.
Single-molecule pulling experiments in MT
Details of the application of single-molecule MT to profile the conformations of proteins have been recently published by our group (32,33) and other groups (34,39–41). Briefly, the prepared experimental chamber was modified with 0.5% (v/v) glutaraldehyde solution (Cat. no. G105905, Aladdin, China) for 45 min at room temperature (RT, 24 ± 2°C) and then incubated with 50 μg/ml Spy catcher and polystyrene bead solution (Cat. no. 17145-5, Polyscience, USA) for at least 4 h at RT or overnight at 4°C. After blocking with 1% BSA (Cat. no. 100-10SB, Lee Biosolutions, USA) PBS solution for at least 2 h at RT, the diluted RIG-I or RIG-IQ517H or truncated protein was flowed into the chamber for a 20-min incubation. Extra protein was eliminated by washing twice, and then BSA-blocked magnetic beads (Cat. no. 65801D, M270, Invitrogen, USA) were flowed into the chamber for a 20-min incubation to form linked protein tethers. In this study, RIG-I, RIG-IQ517H or truncated protein was genetically assembled with biotinylated AviTag and SpyTag on its N and C terminals, respectively, which allowed the protein to be captured in MT through specific interactions (Figure 2A, E and I).
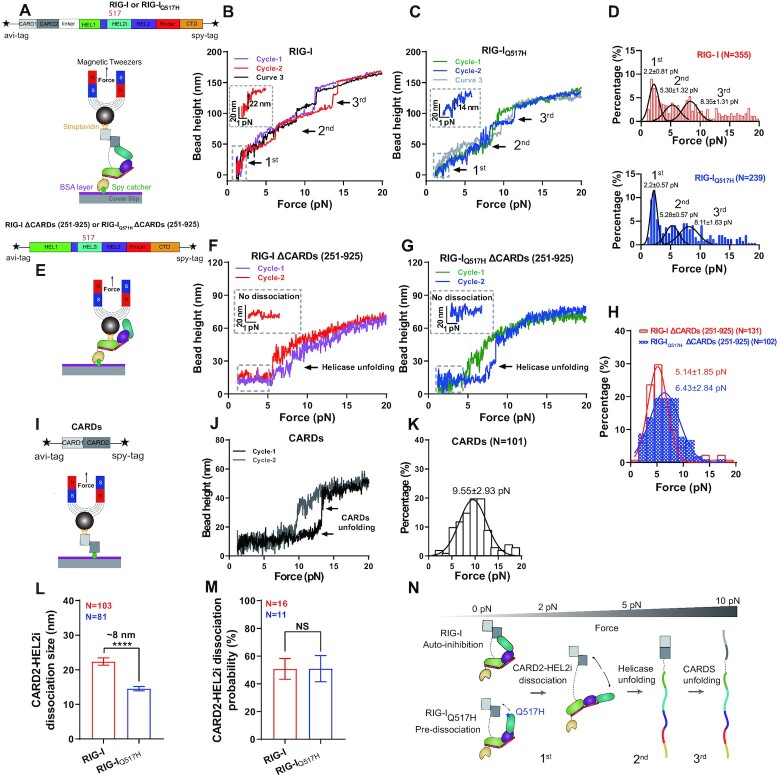
Figure 2.
Profiling the conformations of RIG-I and RIG-IQ517H by single-molecule magnetic tweezers (MT). (A) Schematic of RIG-I domains, tag engineering, and conformational profiling in MT. (B, C) Representative conformational curves of RIG-I (B) and RIG-IQ517H (C). Three main conformational changes are marked with arrows, wherein the first is magnified in a dashed box labeled with step size. (D) Force histogram for conformational changes of RIG-I (red) and RIG-IQ517H (blue). The three peaks obtained by Gaussian fitting are responses to the three main conformational changes in (B) and (C). (E) Schematic of the RIG-I helicase, tag engineering, and conformational profiling in MT. (F, G) Representative conformational curves of RIG-I ΔCARDs (F) and RIG-IQ517H ΔCARDs (G). Conformational changes are marked with arrows, wherein the 0–5 pN area is magnified in a dashed box. (H) Force histogram for conformational changes of RIG-I ΔCARDs (red) and RIG-IQ517H ΔCARDs (blue). The force peak obtained by Gaussian fitting is a response to the conformational change in (F) and (G). (I) Schematic of the RIG-I CARDs domain, tag engineering, and conformational profiling in MT. (J) Representative conformational curves of CARDs; conformational changes are marked with arrows. (K) Force histogram for conformational changes of CARDs. The force peak obtained by Gaussian fitting is a response to the conformational change in (J). (L) The size of CARD2-HEL2i dissociation of RIG-I and RIG-IQ517H. Data were derived from <3.75 pN, wherein the first conformational change occurred. (M) The dissociation probability of CARD2-HEL2i of RIG-I and RIG-IQ517H; results are represented from a single tether that was repeatedly stretched for at least 6 cycles. (N) Conformational changing profiles of RIG-I and RIG-IQ517H, wherein CARD2-HEL2i dissociation, helicase unfolding and CARDs unfolding occurred sequentially upon force. Data are presented as the mean ± SD in (D), (H) and (K) and as the mean ± SEM in (L) and (M); statistics indicate the significance of differences between compared sample states determined by Student's t test: ****P ≤ 0.0001; NS denotes nonsignificant in (L) and (M).
With the homemade MT, single-molecule pulling experiments were conducted in buffer containing 145 mM NaCl, 10 mM HEPES, 2 mM MgCl2 and 1% BSA at RT. The force loading rate was 1 pN/s, and the resting time between cycles was 50 s. The sampling rate of EMCCD was 200 Hz. The measured data were smoothed by a 0.1-s window for smoothly presenting force-bead height curves. A sudden increase in bead height (indicated by arrows, Figure 2B, C, F, G and J) was recognized as a conformational change of the protein.
Discrimination of viral and self RNAs by RIG-I or RIG-IQ517H in MT
A single RIG-I or RIG-IQ517H protein that displayed CARD2-HEL2i dissociation upon force was determined in MT, and then the force was permanently maintained at a very low level (∼1.3 pN), which not only ensured the associated state of CARD2-HEL2i most of the time but also enabled us to track the length of the RIG-I or RIG-IQ517H protein in real time. After the RIG-I or RIG-IQ517H protein was verified, HEPES buffer was flowed slowly into the chamber, and the length of the RIG-I or RIG-IQ517H protein was tracked in real time for more than 5 min to collect the control data. Finally, 5 μM 3p10l or Cap2-10l RNA was slowly flowed into the chamber, and the protein length was tracked in real time for ∼10 min to collect experimental data. The protein length difference between adding HEPES buffer and adding RNA was measured and treated as the RNA-driven conformational change of RIG-I or RIG-IQ517H, which represented the ability of RIG-I or RIG-IQ517H to discriminate viral and self RNAs.
Experiments were conducted at RT in buffer containing 145 mM NaCl, 10 mM HEPES, 2 mM MgCl2 and 1% BSA. At least three replicate experiments were conducted in each group (Figure 4H). The sampling rate of EMCCD was 200 Hz. The measured data were smoothed by a 1-s window for smoothly presenting time-relative length curves.
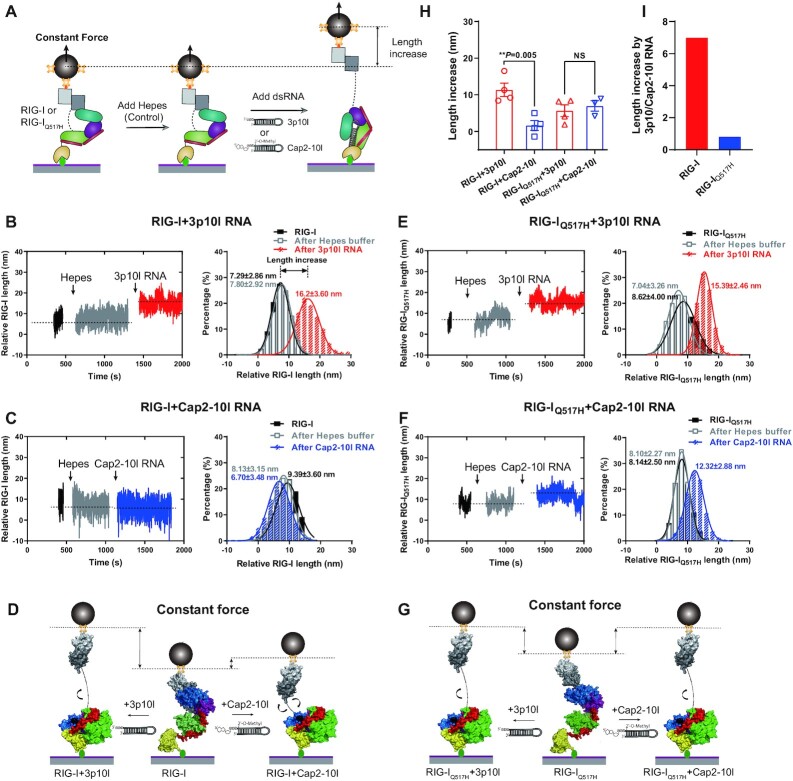
Figure 4.
RIG-IQ517H extends its conformation unbiasedly upon RNA proofreading. (A) Schematic revealing the RIG-I and RIG-IQ517H discrimination of viral and self RNAs in MT. When RIG-I and RIG-IQ517H were captured with a low force (∼1.3 pN) in MT, HEPES buffer flowed in as the control, and then the length increase of RIG-I or RIG-IQ517H driven by RNAs was measured. (B, C) Representative data of the length increase of RIG-I driven by 3p10l RNA (B) and RIG-I driven by Cap2-10l RNA (C), wherein the relative RIG-I length under different conditions was quantified by Gaussian fitting. (D) Schematic of the length increase of RIG-I driven by 3p10l RNA or Cap2-10l RNA. (E, F) Representative data of the length increase of RIG-IQ517H driven by 3p10l RNA (E) and RIG-IQ517H driven by Cap2-10l RNA (F), wherein the relative RIG-IQ517H length under different conditions was quantified by Gaussian fitting. (G) Schematic of the length increase of RIG-IQ517H driven by 3p10l RNA or Cap2-10l RNA. (H) Length increases in RIG-I and RIG-IQ517H driven by 3p10l RNA or Cap2-10l RNA. Each dot is derived from repeated experiments. (I) Length increases driven by 3p10l RNA and Cap2-10l RNA. Data are presented as the mean ± SD in (B), (C), (E) and (F) and as the mean ± SEM in (H); statistics indicate significance of differences between compared sample states determined by Student's t test: **P ≤ 0.01; NS represents nonsignificant in (H).
BIOMOL green ATPase assay
Purified WT RIG-I or RIG-I mutants (0.3 μM) were incubated with different concentrations of RNA (3p10l or Cap2-10l; 0 μM, 0.0006 μM, 0.003 μM, 0.015 μM, 0.03 μM, 0.06 μM, 0.12 μM, 0.24 μM, and 0.48 μM) in a total of 50 μl ATPase buffer (50 mM HEPES, pH 7.5, 50 mM NaCl, 4 mM MgCl2, 2 mM DTT) for 30 min. ATP (Sigma) was added to the reaction mixture to a final concentration of 2 mM, and the mixtures were incubated at room temperature for 15 min. Free phosphate concentration was determined using BIOMOL Green reagent (Enzo Life Sciences) in a microplate format, and absorbance was measured at OD630 nm. The significance of differences between groups was evaluated by unpaired Student's t test.
Interferon dual reporter assays
HEK293T cells were plated in 12-well plates in Dulbecco's modified Eagle medium (HyClone) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin. At 50% density, cells were cotransfected with pUNO plasmids encoding RIG-I or mutants (0.2 μg), IFN-β promoter-driven firefly luciferase reporter plasmid (1 μg) and a constitutively expressed Renilla luciferase reporter plasmid (pRL-TK, 50 ng) at a 1:2 ratio of DNA:Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The medium was changed 6 h post-transfection. After 12 h, cells were additionally transfected with the respective RNA (poly(I:C) or 3p10l or Cap2-10l, 0.5 μg) using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Cells were lysed 12 h post-stimulation, and IFN-β promoter activity was measured using the Dual Luciferase Reporter Assay System (Promega) and a PerkinElmer EnVision Multilabel Reader. Firefly luciferase activity was normalized against Renilla luciferase activity. Error bars represent the standard deviations of three independent experiments.
IRF3 phosphorylation assay
Cells were washed with cold PBS and lysed by SDS loading buffer. Cell lysates were separated on 10% SDS–PAGE gels and transferred to PVDF membranes. Membranes were incubated with p-IRF3 antibody (Abcam, ab76493) or IRF3 antibody (Abcam, ab68481) following blocking in 5% nonfat milk. Protein bands were visualized by chemiluminescence according to the manufacturer's instructions (Kindle Biosciences).
Statistical analysis
Unpaired Student's t test was used to evaluate differences between two experimental groups in all cases. Significant differences are denoted by *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, or ****P ≤ 0.0001. N.S. represents nonsignificant. In general, three biological replicates (n) for each condition were performed.
RESULTS
RIG-I HEL2i mutations destabilize the CARD2-HEL2i engagement and lead to increased solvent exposure
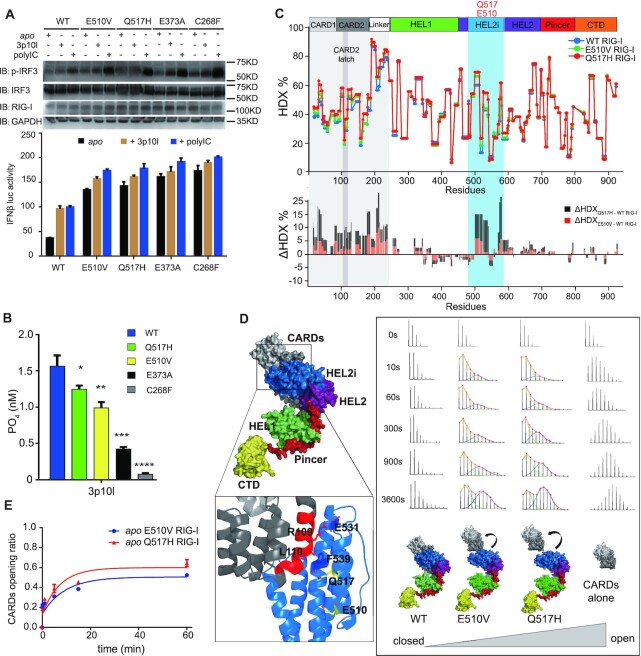
We first conducted the IFN-β reporter assay to examine the roles of RIG-IQ517H and RIG-IE510V in inducing downstream signaling pathways and compared these pathways with those of WT RIG-I, RIG-IE373A and RIG-IC268F. In the absence of RNA stimuli, the expression of all SMS RIG-Is stimulated a more than twofold increase in IFN-β signal response compared to the signal observed with WT RIG-I (Figure 1A). In the presence of 3p10l and polyIC treatment, all RIG-Is activated IRF3 phosphorylation and the IFN-β signaling pathway in HEK293T cells (Figure 1A). In contrast to RIG-IE373A and RIG-IC268F, which had impaired ATPase hydrolysis function, RIG-IE510V and RIG-IQ517H exhibited efficient RNA-dependent ATPase activity similar to the RNA-dependent ATPase activity of WT RIG-I (Figure 1B). These observations suggest that compared with ATPase-deficient SMS RIG-Is, RIG-IE510V and RIG-IQ517H possess an alternative mechanism to induce autoimmune disorder. Sequence alignment indicates that residue E510 is strictly conserved in vertebrate species (Supplementary Figure S1). Q517 is conserved except in mouse and rabbit RIG-Is wherein residue H is present at the equivalent amino acid position (Supplementary Figure S1). The CARD2 latch residues, however, are poorly conserved (Supplementary Figure S1). Next, comparative HDX-MS analysis between apo proteins (RIG-I versus RIG-IE510V vs. RIG-IQ517H) was performed to study protein dynamics in solutions and monitor large structural rearrangements (Figure 1C and Supplementary Figure S2). Single amide consolidated ΔHDX analysis of RIG-IQ517H vs. RIG-I revealed that apo RIG-IQ517H appears to exhibit increased solvent exchange activity (10% < ΔHDX < 25%, P < 0.001) at regions constituting CARDs, Linker and HEL2i gate domains (Figure 1C and Supplementary Figure S2). Analysis of ΔHDX values of RIG-IE510V versus RIG-I revealed that RIG-IE510V also showed increased solvent exposure in the aforementioned regions of RIG-IE510V, although to lesser extents than RIG-IQ517H (Figure 1C and Supplementary Figure S2). The presence of a RIG-I RNA agonist to RIG-I increased solvent exchange (ΔHDX %) in CARDs and HEL2i regions, resulting in the dissociation of CARDs and HEL2i (15,16) (Supplementary Figure S3). The observation of increased solvent exposure associated with RIG-I variants led us to examine the local dynamics of the CARD2 latch peptide (103–114), which governs CARDs partial unfolding events in solutions (15,16). Intriguingly, this latch motif exhibited an EX1 kinetic regime that displays bimodal isotope clusters associated with RIG-IE510V and RIG-IQ517H but not RIG-I or CARDs (1-228) (Figure 1D). The EX1 kinetics appears if the unfolding rate is much slower than the intrinsic exchange rate of the amide hydrogens within the unfolding region (42). It results in a bimodal distribution signature wherein the lower mass envelope corresponds to molecules that have not yet exchanged (not yet unfolded) and the higher mass envelope corresponds to molecules that have undergone exchange (molecules that have unfolded) (16,43–45). And the increase in the intensity of the higher mass isotopic envelope over time (at higher m/z value) could indicate the rate of unfolding or opening during the unfolding event. We determined the CARDs structural transition rate from the lower MS envelope to the higher MS envelope by fitting two Gaussian peaks to the bimodal isotope cluster and dividing the entire peak area by the area of higher MS envelope or the isotope cluster area of the unfolded state (U/(U + F)) (44,45). An exponential 3P with the prediction model: a + b ×exp(c.time(min)) could further accommodate the ratio of unfolded state in each data point and calculate the half-life (t1/2) of CARDs partial unfolding. The presence of E510V and Q517H mutations allowed increased CARDs partial unfolding events with half-lives (t1/2) of 5.08 and 6.46 min, respectively (Figure 1E). Similar as CARDs region, the linker region (191–239), which is intrinsically disordered, displayed increased solvent exchange when associated with RIG-I variants (Figure 1C and Supplementary Figure S2). These observations suggest that the HEL2i mutations destabilize the CARD2-HEL2i interface, promoting partial opening of the closed, autoinhibited conformation of RIG-I, which in turn causes some residues normally buried at the CARD-Hel2i interface and the linker residues to become more exposed and undergo increased solvent exchange (46). And the degree of solvent exposure at CARDs and linker region is higher when associated with apo RIG-IQ517H, as compared with apo RIG-IE510V.
Figure 1.
RIG-I HEL2i mutations destabilize the CARD2-HEL2i engagement and lead to increased solvent exposure. (A) Examination of IRF3 phosphorylation and IFN-β reporter assays of SMS RIG-I variants. The indicated SMS RIG-I constructs were expressed in the presence and absence of RNA treatment to examine their contributions to the induction of IFN-β responses. Data are presented as the mean values ± SEM; n = 3 independent experiments; statistics indicate the significance of differences between WT and the respective mutant determined by Student's t test: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. N.S. denotes nonsignificant. (B) ATP hydrolysis assay of SMS RIG-I variants bound with 3p10l RNA. (Data are presented as the mean values ± SEM, n = 3 independent experiments; statistics indicate significance of differences between compared sample states determined by Student's t test: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. N.S. denotes nonsignificant.). (C) Single amide consolidated HDX-MS profiling of apo RIG-I, RIG-IE510V and RIG-IQ517H. The y- and x-axes illustrate the D% and domain arrangement of RIG-I, respectively. In the lower panel, a positive value on the y-axis (ΔHDX %) represents deprotection (solvent exposure) against deuterium exchange in the corresponding region depicted along the x-axis of apo RIG-IE510V and apo RIG-IQ517H, as compared with apo RIG-I. (D) An apo RIG-I model is shown on the left panel, and the CARD2-HEL2i interface is highlighted, wherein the CARD2 latch peptide (103–114) is colored red. The analysis of the EX1 kinetics is shown on the right panel. It displays the MS spectra of CARD2 latch peptide (103–114) derived from the indicated complexes at the indicated on-exchange time points. The abundance of each mass population (high and low) is determined by fitting a bimodal Gauss-function to the isotopic envelope by HX express2 software. (E) CARDs partial opening rate calculated from EX1 analysis. Half-life (t1/2) of respective partial unfolding event is determined by fitting an exponential 3P with the prediction model: a + b ×exp(c.time(min)), where a is the asymptote, b is the scale and c is the growth rate, is used to fit a curve to %D (response) and time (regressor).
RIG-IQ517H adopts a more extended conformation than RIG-I
Given the large variations in HDX dynamics between RIG-I and RIG-IQ517H, we utilized single-molecule MT to profile their conformations and free energy landscape. We anchored a single RIG-I or RIG-IQ517H protein in MT with an N-terminal AviTag to the magnetic bead and a C-terminal SpyTag to the coverslip surface (Figure 2A). Upon force increment, a single RIG-I or RIG-IQ517H protein sequentially displayed a sudden increase in bead height (SBHI) resulting from either RIG-I’s intermolecular interactions or specific domain unfolding (33) (Figure 2B and C). Three main regions corresponding to 0–4, 4–7 and 7–12 pN hierarchically occurred and were classified as first, second and third SBHI events, respectively (Figure 2B–D). We zoomed in to quantify the displacements of the first SBHI event, ∼22 and ∼14 nm for RIG-I and RIG-IQ517H, respectively (Figure 2B and C). The forces (2.2 ± 0.81 versus 2.2 ± 0.57 pN) that induced the first SBHI event were not significantly different between the proteins (Figure 2D).
Because RIG-I adopts an autoinhibited state by CARD2-HEL2i engagement, we hypothesized that the 1st SBHI results from the dissociation of CARD2 from HEL2i and that the ∼8 nm difference in the SBHI displacement derives from the different initial conformations between apo RIG-I and apo RIG-IQ517H. To test these hypotheses, we produced truncated proteins RIG-IΔCARDs and RIG-IQ517HΔCARDs, which lack the signaling CARD domain (Figure 2E). As expected, when tethered with RIG-IΔCARDs and RIG-IQ517HΔCARDs, single-molecule MT failed to detect the first SBHI event observed at 2 pN with the full-length receptors (Figure 2F and G). In contrast, the second conformational change corresponding to 4–7 pN observed with the full-length receptors remained (Figure 2H) in these truncated proteins. We therefore conclude that the observed second conformational change derives mainly from the unfolding of the helicase domain in RIG-I and RIG-IQ517H. To verify the remaining element in RIG-I, we tethered the CARDs domain in MT (Figure 2I) and, as expected, found the third conformational change corresponding to 7–12 pN, which depicted the unfolding of the CARDs domain in RIG-I (Figure 2J and K). Moreover, we found that the size of CARD2-HEL2i dissociation in RIG-IQ517H was ∼8 nm shorter than that in RIG-I, although the dissociation probability showed no significant difference between the two proteins (Figure 2L and M). It is also important to note that although these mechanically unfolding events observed in the MT experiments might not be physiologically relevant, these data indicate that RIG-IQ517H adopts a ‘predissociated’ state, in which RIG-I ‘pre-extends’ an ∼8 nm conformational change and saves ∼4.29 kBT Gibbs free energy on the way to fully release CARDs (47) (assuming an equilibrium state at the peak force). We dissected the conformations and conformational changes of both RIG-I and RIG-IQ517H, and MT data showed that RIG-I helicase domain unfolds before CARDs domain (Figure 2N). The observed difference in initial conformation between the two proteins revealed by MT data was consistent with the large variations in HDX dynamics that were observed between apo RIG-IQ517H and apo RIG-I.
Impaired RNA proofreading capabilities associated with RIG-IQ517H and RIG-IE510V
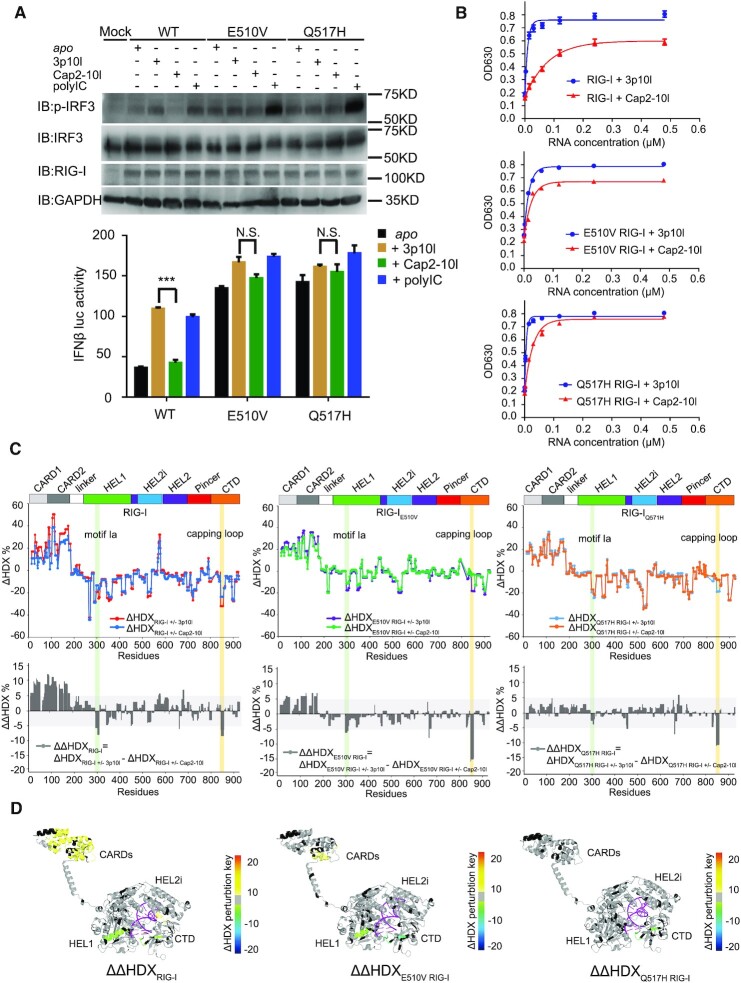
To understand the dysregulated mechanism of RNA surveillance by RIG-IQ517H and RIG-IE510V, we introduced two synthetic RNA ligands—3p10l and Cap2-10l—to represent viral and self RNA, respectively. Two 10 base pair (bp) hairpin duplex RNAs (5′-GAAUAUAAUAGUGAUAUUAUAUUC-3′) with the same nucleotide backbone were either synthesized with a 5′ triphosphate moiety (5′ppp, 3p10l RNA that mimics viral RNA) or a triphosphate moiety capped with m7G and modified with 2′-O-methylation at the first two nucleotide ribose guanines (N1-2-2′-O-M, Cap2-10l RNA that mimics self RNA) (15). In the cell-based reporter assay, 3p10l and polyIC, but not Cap2-10l, stimulated a more than twofold increase in IFN-β response and stronger IRF3 phosphorylation immnoblots than those observed in the untreated state (Figure 3A). However, this phenomenon of RNA discrimination against Cap2-10l RNA was not observed with RIG-IQ517H or RIG-IE510V in the presence or absence of 3p10l, Cap2-10l and polyIC treatment, suggesting that RIG-IQ517H and RIG-IE510V constitutively signal in HEK293T cells (Figure 3A). Next, we conducted RNA-dependent ATPase activity assays to decipher the molecular profiles of impaired RNA discrimination by RIG-IQ517H and RIG-IE510V. A distinguished ATPase activity profile was observed with KM values of 9 ± 1 nM for the RIG-I & 3p10l complex and 61 ± 6 nM for the RIG-I & Cap2-10l complex (Figure 3B and Table 1). In particular, RIG-IE510V displayed KM values of 13 ± 2 and 28 ± 3 nM for the 3p10l and Cap2-10l complexes, respectively, whereas RIG-IQ517H displayed KM values of 7 ± 1 and 21 ± 2 nM for the 3p10l and Cap2-10l complexes, respectively (Figure 3B and Table 1). It is important to note that although the KM value of Cap2-10l is relatively high, comparative analysis of ATPase (maximum stimulation values) showed that Cap2-10l was capable to maximally stimulate 67.9 +/- 0.7% of that observed for 3p10l, which further reinforced Cap2-10l as a RIG-I partial agonist or a self-like ligand (Figure 3B and Table 1). However, this RNA discrimination, as examined by ATPase maximum stimulation activity, gradually became indistinguishable between the RIG-IE510V (84.6 ± 2.3%) and RIG-IQ517H (98.7 ± 2.4%) profiles (Figure 3B and Table 1). These kinetic data suggest that compared with RIG-I, RIG-IE510V and RIG-IQ517H gradually lose the ability to exclude self RNA.
Figure 3.
Impaired RNA proofreading capabilities associated with RIG-IQ517H and RIG-IE510V. (A) Examination of IRF3 phosphorylation and IFN-β reporter assays of RIG-IE510V and RIG-IQ517H upon respective RNA treatment. Data are presented as the mean values ± SEM; n = 3 independent experiments; statistics indicate significance of differences between WT and the respective mutant determined by Student's t test: *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001. N.S. denotes nonsignificant. (B) RNA-dependent ATPase activity profiles of the indicated protein complexes. (C) Single amide consolidated HDX-MS profiling of RIG-I, RIG-IE510V and RIG-IQ517H upon RNA discrimination between 3p10l and Cap2-10l. In the uppor panel, the y- and x-axes illustrate the HDX perturbations (ΔHDX%) of respective RNA binding event and domain arrangement of RIG-I or its HEL2i variants, respectively. A positive value or a negative value in the y axis (ΔHDX%) represents protection or deprotection against deuterium exchange in the corresponding region of the receptor depicted in the x axis when a RNA binding event (induced by 3p10l or Cap2-10l) takes place. In the lower panel, it displays the comparative HDX analysis (ΔΔHDX%) between 3p10l- and Cap2-10l-mediated HDX perturbations on RIG-I, RIG-IE510V and RIG-IQ517H. ΔΔHDX% values (ΔΔHDX = ΔHDXRIG-I(WT/mut) ± 3p10l - ΔHDXRIG-I(WT/mut) ± Cap2-10l) could indicate the conformational perturbations of RIG-I or its variants upon RNA discrimination. (D) Differential single amino acid consolidation HDX data are mapped onto the full-length RNA-bound RIG-I structure model in the ribbon, as shown by the representation of ΔΔHDX profiles of RIG-I, RIG-IE510V and RIG-IQ517H (Supplementary Figure S5, columns (xiii, xiv and xv), and Figure S8). Percentages of deuterium differences are color-coded according to the HDX key (below). Black, regions that have no sequence coverage and include proline residues that have no amide hydrogen exchange activity; gray, no statistically significant changes between compared states; purple, duplex RNA ligand.
Table 1.
K M and ATPase activity profiles
| WT | E510V | Q517H | |
|---|---|---|---|
| KM (3p10l-RNA) | 9 ± 1 nM | 13 ± 2 nM | 7 ± 1 nM |
| KM (Cap2-10l-RNA) | 61 ± 6 nM | 28 ± 3 nM | 21 ± 2 nM |
| Maximum ATPase activity (Normalized OD630) | |||
| 3p10l-RNA | 0.632 ± 0.020 | 0.551 ± 0.011 | 0.602 ± 0.018 |
| Cap2-10l-RNA | 0.429 ± 0.018 | 0.470 ± 0.006 | 0.594 ± 0.009 |
| Cap2-10l/3p10l-RNA | 67.9 ± 0.7% | 84.6 ± 2.3% | 98.7 ± 2.4% |
Compararative HDX-MS studies were further employed to explore the impaired RNA proofreading capabilities associated with RIG-IQ517H and RIG-IE510V when compared to RIG-I, as they could undergo conformational changes to different extents during RNA discrimination between 3p10l and Cap2-10l. We firstly analyzed ΔHDXRIG-I(WT/mut) ± 3p10l and ΔHDXRIG-I(WT/mut) ± Cap2-10l to dissect the comformational changes during RNA discrimination mediated by individual RIG-I receptor (Figure 3C, Supplementary Figure S4–S7 and Table 2). Compared with Cap2-10l binding, 3p10l binding induced a greater magnitude of protection against solvent exchange in regions constituting the CTD capping loop (843–856), HEL1 motif Ia (296–310) and motif Ic (343–351) and HEL2i gate motif (522–539) (Figure 3C, Supplementary Figure S4, S5 and Table 2). A further comparison analysis of the ΔHDX data revealed that a larger surface area of RIG-I CARDs allowed increased solvent exchange activities in the presence of 3p10l when compared with the presence of Cap2-10l (Figure 3C, Supplementary Figure S4, S5 and Table 2). These observations suggest that RIG-I could discriminate against Cap2-10l and undergo dissociated activation profile upon RNA proofreading. We further introduced ΔΔHDX analysis to measure the degree of comformational perturabations (ΔΔHDX = ΔHDXRIG-I(WT/mut) ± 3p10l – ΔHDXRIG-I(WT/mut) ± Cap2-10l) (Figure 3C, Supplementary Figure S4–S7 and Table 2). The conformational perturbations in ΔΔHDX (%) display the relative differential HDX between RIG-I(WT/mut) & 3p10l and RIG-I(WT/mut) & Cap2-10l complexes, it measures the degree of conformational changes of receptor when the Cap2-10l mediated binding event is subtracted from that observed with 3p10l. As a result, ΔΔHDX (%) could be used to quantify the receptor's ability to discriminate between 3p10l and Cap2-10l. Single amino acid consolidation view of ΔΔHDX values was mapped to a RNA bound RIG-I structural model for visualization (Figure 3D, Supplementary Figure S4 and Table 2). ΔΔHDX analysis of RIG-IE510V and RIG-IQ517H detected a similar conformational perturbation profle in the CTD capping loop (843–856) as compared with that of RIG-I, suggesting that the CTD capping loop derived from individual RIG-I receptor still possesses the potency to discriminate against Cap2-10l (Figure 3C, D, Supplementary Figure S4–S7 and Table 2). The largest degree of conformational perturbation (a 5–15% increase in ΔΔHDX, P < 0.001) upon RNA discriminaton was detected in the CARDs region associated with RIG-I. However, examinations of the ΔΔHDX profiles of CARDs region of RIG-IE510V and RIG-IQ517H revealed a reduced (∼6% increase in ΔΔHDX) and an almost completely neutralized ΔΔHDX perturbation profile (0 < ΔΔHDX < 5%, P < 0.001), respectively (Figure 3C, D, Supplementary Figure S4–S7 and Table 2). In the HEL1 motif Ia region, a gradual decrease with respect to the degree of conformational perturbations (ΔΔHDX %) was also observed in RIG-I, RIG-IE510V and RIG-IQ517H profiles (Figure 3C, D, Supplementary Figure S4–S7 and Table 2). These results indicate that although CTD preserves the ability to discriminate between 5′ppp and N1-2-2′-O-M moieties, the HEL2i and HEL1 play a major role in surveilling the RNA backbone by RIG-IE510V and RIG-IQ517H. The HEL2i mutations impair the receptor's capability to discriminate against a self-like RNA Cap2-10l.
Table 2.
HDX data of RIG-I, RIG-IE510V, and RIG-IQ517H upon RNA proofreading of 3p10l and Cap2-10l. Differential ΔHDX and ΔΔHDX views of the indicated protein or protein complex upon binding to 3p10l or Cap2-10l. The values listed under each HDX experiment are the averaged differences in the percentage of deuterium incorporation of the corresponding peptide derived from two different states across all exchange time points (i.e. 0, 10, 60, 300 and 900 s). In the comparison of two samples, the perturbation %D is determined by calculating the difference between the samples. HDX Workbench colors each peptide according to the smooth color gradient HDX perturbation key (D%) shown in the figure. Differences in %D between -5% and 5% are considered nonsignificant and are colored gray according to the HDX perturbation key. A negative value represents decreased deuterium incorporation or stabilization, while a positive value represents increased deuterium incorporation or destabilization in the corresponding region of the receptor when a binding event takes place. Blank regions represent undetected peptides for the corresponding experiment
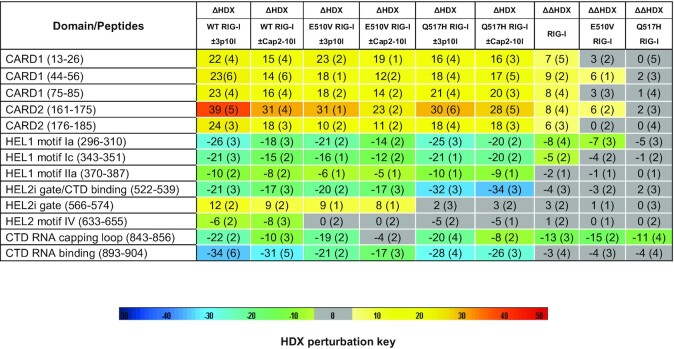
|
To test whether these observed differences in CARD dynamics could influence receptor binding to K63-polyUb chains and CARDs oligomerization, we performed sequential HDX studies to probe the RNA-protein complex in the presence and absence of K63-linked polyUb chains (Supplementary Figures S4–S7). HDX analysis of complexes of RIG-I and RIG-IE510V bound with Cap2-10l and 3p10l RNA revealed differential interactions of K63-Ub9 with the respective CARDs surface (Supplementary Figures S4–S6). However, the presence of K63-Ub9 in the RIG-IQ517H-RNA complexes resulted in a similar threshold of protection against deuterium exchange in the CARDs region (Supplementary Figures S4 and S7). These data further reveal the disrupted effects of the gated mechanism of RIG-IQ517H at the distal CARDs signaling domain upon RNA discrimination.
RIG-IQ517H extends its conformation stochastically upon RNA proofreading
We further utilized MT experiment to quantify the length change of RIG-IQ517H upon RNA binding in real time. First, we verified the CARD2-HEL2i dissociation event of RIG-I or RIG-IQ517H with MT and then kept the loading force constant at ∼1.3 pN to avoid disrupting the CARD2-HEL2i association. Second, to test how RNA stimulation changes the RIG-I conformations, we sequentially flowed HEPES buffer (as a control) and RNAs (3p10l or Cap2-10l) into the MT chamber to be incubated with the RIG-I protein and simultaneously tracked the relative length changes of RIG-I or RIG-IQ517H in real time (Figure 4A). The addition of HEPES buffer not significantly altered the RIG-I length in all tested states (Figure 4B and C). The presence of 3p10l RNA with RIG-I increased the length of RIG-I by ∼10 nm, whereas RIG-I molecules displayed an ∼3 nm change upon the addition of Cap2-10l RNA (Figure 4B–D). Unlike the findings with RIG-I, 3p10l RNA and Cap2-10l RNA induced a similar length extension (4–7 nm) in RIG-IQ517H (Figure 4E–G). These results indicate that RIG-IQ517H enables to extend its conformation stochastically upon 3p10l and Cap2-10l binding, which further suggests that RIG-IQ517H loses the discrimination against Cap2-10l RNA.
DISCUSSION
The coupling between RNA recognition and conformational changes is central to RIG-I RNA proofreading behaviors, but the molecular determinants of abnormal conformational transitions are difficult to assess experimentally. This study demonstrates the abilities of HDX-MS and MT and provides a quantitative method for studying HEL2i-mediated RNA gating as the critical first step in the discrimination of self and nonself RNAs.
Compared with RIG-I, MDA5 gain-of-function variants are associated with a wider spectrum of autoimmune diseases in humans, such as systemic lupus erythematosus (SLE), lupus-like nephritis, Aicardi-Goutières syndrome(AGS), dermatomyositis, rheumatoid arthritis, and type 1 diabetes (48–51). These MDA5 gain-of-function mutations are associated with HEL1-HEL2 that impair the receptor's ATPase activity (48–51). Similarly, SMS RIG-I variants (E373A and C268F) are linked to impaired ATPase activity(15). However, there is no reported HEL2i mutation associated with MDA5 that could lead to these autoimmune diseases. This could be due to the fact that MDA5 lacks HEL2i-CARD2 interface and adopts an open CARDs conformation in solutions (21). As a result, the HEL2i domain of MDA5 plays a distinctive role in recognizing self RNA and placing CARDs activation as compared with RIG-I. In addition, sequence alignment of RLR HEL2i region further reveals that residue E510 is well-conserved, which contrasts with the observation that residue Q517 renders little conservation among RLRs (Supplementary Figure S1). It therefore suggests that the loosened gating mechanism of Q517H variant is relatively specific in inducing hyperactivation of RIG-I in humans. Intrigulingly, it is also important to note that the HEL2i domain is uniquely associated with specialized RNA-activated ATPases (DRAs) that include RNA sensors, RIG-I-like receptors, the ribonuclease Dicer, and DRH-1 and DRH-3 involved in the siRNA pathway of Caenorhabditis elegans (9,10,52). Besides its role involved in RNA backbone scanning and binding, the HEL2i domain of Dicer and LGP2 plays a regulatory role in interacting with other cytosolic proteins and their signaling pathways (10,53,54).
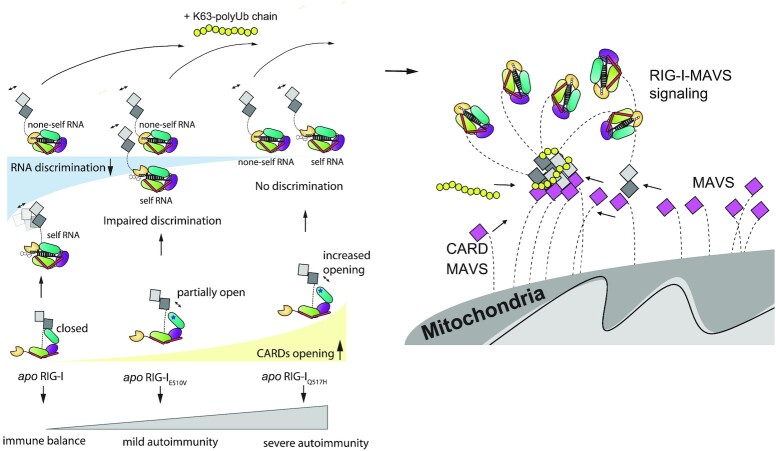
In summary, we show that RIG-I HEL2i domain governs the immune balance and hemostasis of interferon signaling pathways. Our work provides a mechanistic understanding of how the genotype-phenotype gradient of DDX58 variants contributes to different pathogeneses of mild and classic SMS syndromes (Figure 5). Our work also reveals the critical role of HEL2i in coordinating RNA sensing and CARDs signaling, identifying HEL2i as a novel target for RIG-I-based therapeutics to treat autoimmune diseases. In future studies, it is important to explore the general molecular link of how RLR mutations associate with specific clinical symptoms and their severities.
Figure 5.
Cartoon representations of RIG-I autoimmune signaling events. In the resting state, RIG-I adopts an autoinhibited conformation via CARD2-HEL2i interaction. RIG-I can recognize nonself RNA and discriminate against self RNA, resulting in distinguished CARDs opening events. Unlike WT RIG-I, apo RIG-IE510V and apo RIG-IQ517H are coupled with a reduced gradient of CARD2-HEL2i engagement. RIG-IE510V can distinguish self from nonself RNA via its HEL-CTD module and activate CARDs differently but to lesser extents than RIG-I. RIG-IQ517H, however, fails to distinguish self from nonself RNA and results in fast CARDs unfolding events at similar unfolding rates. The subsequent K63-polyUb9 can bind to activated CARDs and trigger CARDs oligomerizations prior to MAVS activation.
DATA AVAILABILITY
HDX-MS data has been deposited in the PRIDE database (Project accession: PXD031831).
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank Dr Tingjun Hou and Dr Xueping Hu for assistance on the computational analysis and discussions for the project.
Author contributions: J.Z. initiated the project and participated in project design and analysis with W.C., D.L. and D.H.L.; W.C. and P.Y.F. designed, performed and wrote the MT experiment portion of the manuscript. Y.X.L., B.S. and W.J.S. prepared all protein samples in this study; J.Z. set up the HDX-MS platform at SIMM; Y.X.L. and J.Z. designed and performed the HDX-MS analysis and analyzed the HDX-MS data; Y.X.L. and B.S. performed the luciferase assays and IRF3 activation assays; Y.X.L., B.S. and W.J.S. performed the ATPase assays; and C.L. and D.H.L. provided insightful discussions. Y.X.L., B.S., P.Y.F., W.C. and J.Z. prepared the figures; and P.Y.F., W.C., D.H.L. and J.Z. wrote the manuscript and accommodated comments from all authors.
Contributor Information
Yixuan Lei, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, Zhejiang, China; The Drug Research Center of Immunological Diseases, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Panyu Fei, Department of Cell Biology and Department of Cardiology of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China; School of Mechanical Engineering, Zhejiang University, Hangzhou 310027, Zhejiang, China.
Bin Song, The Drug Research Center of Immunological Diseases, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Wenjia Shi, The Drug Research Center of Immunological Diseases, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; School of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing 210023, Jiangsu, China.
Cheng Luo, Department of Cell Biology and Department of Cardiology of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China; The Chemical Biology Center, Drug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; School of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, UCAS, Hangzhou 310024, Zhejiang, China.
Dahai Luo, Lee Kong Chian School of Medicine, NTU Institute of Structural Biology, School of Biological Sciences, Nanyang Technological University, 636921, Singapore.
Dan Li, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, Zhejiang, China.
Wei Chen, Department of Cell Biology and Department of Cardiology of the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China; Liangzhu Laboratory, Zhejiang University Medical Center, 1369 West Wenyi Road, Hangzhou 311121, China; Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, the MOE Frontier Science Center for Brain Science & Brain-Machine Integration, State Key Laboratory for Modern Optical Instrumentation Key Laboratory for Biomedical Engineering of the Ministry of Education, College of Biomedical Engineering and Instrument Science, Zhejiang University, Hangzhou 310058, Zhejiang, China.
Jie Zheng, The Drug Research Center of Immunological Diseases, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; School of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing 210023, Jiangsu, China; School of Pharmaceutical Science and Technology, Hangzhou Institute for Advanced Study, UCAS, Hangzhou 310024, Zhejiang, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Shanghai Municipal Science and Technology Major Project; National Natural Science Foundation of China [81971538]; Shanghai PuJiang Talent program [PJ20190001389 to J.Z.]; National Natural Science Foundation of China [31971237 to W.C.]; Natural Science Foundation of Zhejiang Province [LZ19H300001 to D.L.]; Singapore Ministry of Health's National Medical Research Council under its Open Fund Individual Research Grant (OF-IRG) [NMRC/OFIRG/0075/2018 to D.H.L.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rehwinkel J., Gack M.U.. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020; 20:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng J., Chang M.R., Stites R.E., Wang Y., Bruning J.B., Pascal B.D., Novick S.J., Garcia-Ordonez R.D., Stayrook K.R., Chalmers M.J.et al.. HDX reveals the conformational dynamics of DNA sequence specific VDR co-activator interactions. Nat. Commun. 2017; 8:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jang M.A., Kim E.K., Now H., Nguyen N.T.H., Kim W.J., Yoo J.Y., Lee J., Jeong Y.M., Kim C.H., Kim O.H.et al.. Mutations in DDX58, which encodes RIG-I, cause atypical singleton-merten syndrome. Am. J. Hum. Genet. 2015; 96:266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferreira C.R., Crow Y.J., Gahl W.A., Gardner P.J., Goldbach-Mansky R., Hur S., de Jesus A.A., Nehrebecky M., Park J.W., Briggs T.A.. DDX58 and classic singleton-merten syndrome. J. Clin. Immunol. 2019; 39:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prasov L., Bohnsack B.L., El Husny A.S., Tsoi L.C., Guan B., Kahlenberg J.M., Almeida E., Wang H., Cowen E.W., De Jesus A.A.et al.. DDX58(RIG-I)-related disease is associated with tissue-specific interferon pathway activation. J. Med. Genet. 2021; 59:294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo D., Ding S.C., Vela A., Kohlway A., Lindenbach B.D., Pyle A.M.. Structural insights into RNA recognition by RIG-I. Cell. 2011; 147:409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang F., Ramanathan A., Miller M.T., Tang G.Q., Gale M., Patel S.S., Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011; 479:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peisley A., Wu B., Xu H., Chen Z.J., Hur S.. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014; 509:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo D., Kohlway A., Pyle A.M.. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol. 2013; 10:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohlway A., Luo D., Rawling D.C., Ding S.C., Pyle A.M.. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013; 14:772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beyer D.C., Ghoneim M.K., Spies M.. Structure and mechanisms of SF2 DNA helicases. Adv. Exp. Med. Biol. 2013; 767:47–73. [DOI] [PubMed] [Google Scholar]
- 13. Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008; 37:317–336. [DOI] [PubMed] [Google Scholar]
- 14. Lassig C., Matheisl S., Sparrer K.M., de Oliveira Mann C.C., Moldt M., Patel J.R., Goldeck M., Hartmann G., Garcia-Sastre A., Hornung V.et al.. Correction: ATP hydrolysis by the viral RNA sensor RIG-I prevents unintentional recognition of self-RNA. Elife. 2016; 5:e14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng J., Wang C., Chang M.R., Devarkar S.C., Schweibenz B., Crynen G.C., Garcia-Ordonez R.D., Pascal B.D., Novick S.J., Patel S.S.et al.. HDX-MS reveals dysregulated checkpoints that compromise discrimination against self RNA during RIG-I mediated autoimmunity. Nat. Commun. 2018; 9:5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng J., Yong H.Y., Panutdaporn N., Liu C., Tang K., Luo D.. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2015; 43:1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramanathan A., Devarkar S.C., Jiang F., Miller M.T., Khan A.G., Marcotrigiano J., Patel S.S.. The autoinhibitory CARD2-Hel2i interface of RIG-I governs RNA selection. Nucleic. Acids. Res. 2016; 44:896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kowalinski E., Lunardi T., McCarthy A.A., Louber J., Brunel J., Grigorov B., Gerlier D., Cusack S.. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011; 147:423–435. [DOI] [PubMed] [Google Scholar]
- 19. Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z.J.. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010; 141:315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang X., Kinch L.N., Brautigam C.A., Chen X., Du F., Grishin N.V., Chen Z.J.. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012; 36:959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song B., Chen Y., Liu X., Yuan F., Tan E.Y.J., Lei Y., Song N., Han Y., Pascal B.D., Griffin P.R.et al.. Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains. Immunity. 2021; 54:2218–2230. [DOI] [PubMed] [Google Scholar]
- 22. Schlee M., Roth A., Hornung V., Hagmann C.A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G.et al.. Recognition of 5 ' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009; 31:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T.. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004; 5:730–737. [DOI] [PubMed] [Google Scholar]
- 24. Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J.et al.. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014; 514:372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramanathan A., Robb G.B., Chan S.H.. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016; 44:7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F.G., Khan A.G., Patel S.S., Marcotrigiano J.. Structural basis for m7G recognition and 2 '-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Furuichi Y., Shatkin A.J.. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000; 55:135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S.et al.. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-Methylated self RNA. Immunity. 2015; 43:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H.X., Liu Z.X., Sun Y.P., Zhu J., Lu S.Y., Liu X.S., Huang Q.H., Xie Y.Y., Zhu H.B., Dang S.Y.et al.. Rig-I regulates NF-kappaB activity through binding to Nf-kappab1 3′-UTR mRNA. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6459–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malathi K., Dong B., Gale M. Jr, Silverman R.H. Small self-RNA generated by RNase l amplifies antiviral innate immunity. Nature. 2007; 448:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng J., Strutzenberg T., Pascal B.D., Griffin P.R.. Protein dynamics and conformational changes explored by hydrogen/deuterium exchange mass spectrometry. Curr. Opin. Struct. Biol. 2019; 58:305–313. [DOI] [PubMed] [Google Scholar]
- 32. Hu W., Zhang Y., Fei P., Zhang T., Yao D., Gao Y., Liu J., Chen H., Lu Q., Mudianto T.et al.. Mechanical activation of spike fosters SARS-CoV-2 viral infection. Cell Res. 2021; 31:1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu P., Zhang T., Liu B., Fei P., Cui L., Qin R., Zhu H., Yao D., Martinez R.J., Hu W.et al.. Mechano-regulation of Peptide-MHC class i conformations determines TCR antigen recognition. Mol. Cell. 2019; 73:1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Le S.M., Yu M., Yan J.. Direct single-molecule quantification reveals unexpectedly high mechanical stability of vinculin-talin/alpha-catenin linkages. Sci. Adv. 2019; 5:eaav2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu M., Le S.M., Ammon Y.C., Goult B.T., Akhmanova A., Yan J.. Force-Dependent regulation of talin-kank1 complex at focal adhesions. Nano Lett. 2019; 19:5982–5990. [DOI] [PubMed] [Google Scholar]
- 36. Pascal B.D., Willis S., Lauer J.L., Landgraf R.R., West G.M., Marciano D., Novick S., Goswami D., Chalmers M.J., Griffin P.R.. HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass. Spectrom. 2012; 23:1512–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weis D.D., Wales T.E., Engen J.R., Hotchko M., Ten Eyck L.F.. Identification and characterization of EX1 kinetics in H/D exchange mass spectrometry by peak width analysis. J. Am. Soc. Mass Spectr. 2006; 17:1498–1509. [DOI] [PubMed] [Google Scholar]
- 38. Guttman M., Weis D.D., Engen J.R., Lee K.K.. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J. Am. Soc. Mass Spectr. 2013; 24:1906–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yao M., Goult B.T., Klapholz B., Hu X., Toseland C.P., Guo Y., Cong P., Sheetz M.P., Yan J.. The mechanical response of talin. Nat. Commun. 2016; 7:11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo Z.L., Hong H.Y., Yuan G.H., Qian H., Li B., Cao Y., Wang W., Wu C.X., Chen H.. Hidden intermediate state and second pathway determining folding and unfolding dynamics of GB1 protein at low forces. Phys. Rev. Lett. 2020; 125:198101. [DOI] [PubMed] [Google Scholar]
- 41. del Rio A., Perez-Jimenez R., Liu R.C., Roca-Cusachs P., Fernandez J.M., Sheetz M.P.. Stretching single talin rod molecules activates vinculin binding. Science. 2009; 323:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arrington C.B., Robertson A.D.. Correlated motions in native proteins from MS analysis of NH exchange: evidence for a manifold of unfolding reactions in ovomucoid third domain. J. Mol. Biol. 2000; 300:221–232. [DOI] [PubMed] [Google Scholar]
- 43. Fang J., Engen J.R., Beuning P.J.. Escherichia coli processivity clamp beta from DNA polymerase III is dynamic in solution. Biochemistry. 2011; 50:5958–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang B., Stjepanovic G., Shen Q., Martin A., Hurley J.H.. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 2015; 22:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trelle M.B., Madsen J.B., Andreasen P.A., Jorgensen T.J.. Local transient unfolding of native state PAI-1 associated with serpin metastability. Angew. Chem. 2014; 53:9751–9754. [DOI] [PubMed] [Google Scholar]
- 46. Schweibenz B.D., Devarkar S.C., Solotchi M., Craig C., Zheng J., Pascal B.D., Gokhale S., Xie P., Griffin P.R., Patel S.S.. The intrinsically disordered CARDs-Helicase linker in RIG-I is a molecular gate for RNA proofreading. EMBO J. 2022; e109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bustamante C., Chemla Y.R., Forde N.R., Izhaky D.. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004; 73:705–748. [DOI] [PubMed] [Google Scholar]
- 48. Martinez A., Varade J., Lamas J.R., Fernandez-Arquero M., Jover J.A., de la Concha E.G., Fernandez-Gutierrez B., Urcelay E.. Association of the IFIH1-GCA-KCNH7 chromosomal region with rheumatoid arthritis. Ann. Rheum. Dis. 2008; 67:137–138. [DOI] [PubMed] [Google Scholar]
- 49. Molineros J.E., Maiti A.K., Sun C., Looger L.L., Han S.Z., Kim-Howard X., Glenn S., Adler A., Kelly J.A., Niewold T.B.et al.. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLoS Genet. 2013; 9:e1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nejentsev S., Walker N., Riches D., Egholm M., Todd J.A.. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009; 324:387–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rice G.I., Del Toro Duany Y., Jenkinson E.M., Forte G.M., Anderson B.H., Ariaudo G., Bader-Meunier B., Baildam E.M., Battini R., Beresford M.W.et al.. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type i interferon signaling. Nat. Genet. 2014; 46:503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li K., Zheng J., Wirawan M., Trinh N.M., Fedorova O., Griffin P.R., Pyle A.M., Luo D.. Insights into the structure and RNA-binding specificity of caenorhabditis elegans Dicer-related helicase 3 (DRH-3). Nucleic Acids Res. 2021; 49:9978–9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lenoir J.J., Parisien J.P., Horvath C.M.. Immune regulator LGP2 targets Ubc13/UBE2N to mediate widespread interference with K63 polyubiquitination and NF-kappaB activation. Cell Rep. 2021; 37:110175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilson R.C., Tambe A., Kidwell M.A., Noland C.L., Schneider C.P., Doudna J.A.. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell. 2015; 57:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HDX-MS data has been deposited in the PRIDE database (Project accession: PXD031831).