Abstract
Streptococcus milleri NMSCC 061 was screened for antimicrobial substances and shown to produce a bacteriolytic cell wall hydrolase, termed millericin B. The enzyme was purified to homogeneity by a four-step purification procedure that consisted of ammonium sulfate precipitation followed by gel filtration, ultrafiltration, and ion-exchange chromatography. The yield following ion-exchange chromatography was 6.4%, with a greater-than-2,000-fold increase in specific activity. The molecular weight of the enzyme was 28,924 as determined by electrospray mass spectrometry. The amino acid sequences of both the N terminus of the enzyme (NH2 SENDFSLAMVSN) and an internal fragment which was generated by cyanogen bromide cleavage (NH2 SIQTNAPWGL) were determined by automated Edman degradation. Millericin B displayed a broad spectrum of activity against gram-positive bacteria but was not active against Bacillus subtilis W23 or Escherichia coli ATCC 486 or against the producer strain itself. N-Dinitrophenyl derivatization and hydrazine hydrolysis of free amino and free carboxyl groups liberated from peptidoglycan digested with millericin B followed by thin-layer chromatography showed millericin B to be an endopeptidase with multiple activities. It cleaves the stem peptide at the N terminus of glutamic acid as well as the N terminus of the last residue in the interpeptide cross-link of susceptible strains.
Most eubacteria are mechanically stabilized by the shape-determining peptidoglycan, which consists of polysaccharide chains that are connected by short peptides. Expansion of the bacterial cell wall during growth and splitting of the septum prior to cell separation require enzymes that cleave some of the existing covalent bonds within the peptidoglycan sacculus. These enzymes are collectively known as peptidoglycan hydrolases (24). Some have been characterized as N-acetylmuramidases, N-acetylglucosaminidases, N-acetylmuramyl-l-alanine amidases, endopeptidases, and transglycosidases (10). Most peptidoglycan hydrolases have been characterized as bacteriolytic enzymes by in vitro studies (29). Lysostaphin, which is active against Staphylococcus aureus, is one such enzyme. It has been suggested that lysostaphin may function catabolically to release nutrients from other staphylococci in the environment, with the producer cell being protected by a specific immunity protein (12, 18). The lytic effect of lysostaphin results from a direct attack on the integrity of the staphylococcal cell wall, specifically bringing about cleavage of the pentaglycine cross-link between the stem peptides of individual peptidoglycan subunits (4).
Zoocin A, a bacteriolytic cell wall hydrolase produced by Streptococcus zooepidemicus 4881, like lysostaphin, cleaves bonds within the peptide moiety of peptidoglycan (26). Zoocin A has been shown to inhibit the growth of all Streptococcus pyogenes strains and all S. zooepidemicus strains other than 4881 itself (14). Both enzymes, lysostaphin and zoocin A, possess a catalytic domain in the N-terminal portion and a substrate recognition domain in the C-terminal region. The catalytic domains also have considerable sequence similarity (25).
The oral streptococcus Streptococcus milleri NMSCC 061 inhibits the growth of a variety of bacterial species. In this article, we report the purification and mode of action of millericin B, produced by S. milleri NMSCC 061. We show that the mode of action is lytic, that lysis occurs as a direct result of the interaction of millericin B with the cell wall, and that millericin B cleaves the peptide moiety of susceptible peptidoglycan.
MATERIALS AND METHODS
Bacterial strains.
Stock cultures were stored in tryptone soy broth (TSB) (Oxoid) at −70°C as a 30% glycerol suspension. Working cultures were maintained on blood agar plates and subcultured every 2 weeks at 37°C.
Purification of millericin B.
The purification of millericin B involved a four-step procedure which included ammonium sulfate precipitation, gel filtration, ultrafiltration, and ion-exchange chromatography. Batch cultures (2 × 2.5 liters) of S. milleri NMSCC 061 were grown in TSB at 30°C for 24 h. The cells were removed from the culture by centrifugation at 8,000 × g for 10 min at 4°C. Millericin B was recovered from the supernatant by the addition of ammonium sulfate to 75% saturation, precipitation on ice for 6 h, and collection of the precipitate by centrifugation at 16,000 × g for 20 min. The pellet was dissolved in 30 ml of 1 M NaCl–20 mM phosphate buffer (pH 7.0). This preparation was applied to a Sephadex G-75 column (2.5 by 56 cm; Pharmacia), equilibrated, and run at 1 ml/min in 1 M NaCl–20 mM phosphate buffer (pH 7.0). A sequence of 8-ml fractions was collected and assayed for millericin B activity. Active fractions were pooled and concentrated (30×) to a volume of 10 ml in an ultrafiltration chamber (Amicon) through a YM 10 membrane (Millipore) with a 10,000 molecular weight cutoff limit. The sample volume was adjusted to 300 ml with 20 mM phosphate buffer (pH 7.0) and refiltered. This was repeated several times to remove the NaCl from the sample. The sample was then applied to an SP-Sepharose HP column (1.6 by 10 cm; Pharmacia). The column was preequilibrated with 20 mM phosphate buffer (pH 7.0). A gradient of 0 to 90% buffer B (1.5 M NaCl in 20 mM phosphate, pH 7.0) applied over a period of 50 min was used to elute millericin B initially. The active fractions were pooled, rechromatographed, and separated under near-isocratic conditions (1.5 M NaCl) until a single active peak was obtained. The gradient for the second elution was from 80 to 85% buffer B over the same time as for the initial separation. This peak was collected as one fraction, lyophilized, and stored at 4°C until it was required. The purity of millericin B was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the method of Laemmli (15) with 12% discontinuous gels and a Mini-Protean II electrophoresis system (Bio-Rad, Richmond, Calif.). Analysis by mass spectrometry was conducted with a model API III quadruple mass spectrometer equipped with an IonSpray source (Sciex, Thornhill, Canada) at the Center for Mass Spectroscopy, University of Stellenbosch, South Africa. The mass spectrometry analysis indicated that millericin B was pure. All further analysis was done with the purified substance.
Purified millericin B was subjected to cyanogen bromide (CNBr) cleavage to generate internal fragments. The sample was solubilized in 50 μl of 70% formic acid. A crystal of CNBr was added, and the reaction tube was flushed with nitrogen upon closing. After 12 h of incubation at room temperature in the dark, the formic acid was evaporated under a vacuum. The digest was solubilized in 6 M guanidine-HCl–0.1 M Tris, pH 8.5, and 0.1 M dithiothreitol and resolved by reverse-phase high-performance liquid chromatography (HPLC). The N-terminal amino acid sequence was obtained with a model 491 Procise automated sequencer (Perkin-Elmer Applied BioSystems).
Preparation of cell walls.
Crude cell walls from Micrococcus luteus ATCC 46898 were prepared from overnight cultures grown in TSB at 30°C. The cells were collected after centrifugation at 8,000 × g for 10 min and resuspended in 20 mM phosphate buffer (pH 7.0). This suspension was incubated for 10 min in a boiling-water bath and centrifuged at 16,000 × g for 15 min (4°C). The pellet was washed twice with 20 mM phosphate buffer (pH 7.0) and resuspended in a minimal volume of the same buffer. This suspension was incorporated into agar plates and used for activity assays. Purified trypsinized cell walls were prepared from M. luteus and S. aureus NMSCC 133 as previously described (21). Teichoic acids were extracted by hydrolysis with 70% HF for 3 h at 0°C (16). HF was neutralized by addition of 6 N KOH, and the cell walls were washed 10 times with distilled water. Cell walls tended to aggregate and had to be resuspended by thorough ultrasonification (Virsonic 60; Virtis Company Inc., Gardiner, N.Y.) after each washing. A suspension of the cell walls in a polypropylene tube was sonicated for 5 min at 60 W output to homogeneity or until aggregation was no longer observed. Glass beads were added to the suspension to facilitate resuspension. They were assayed for purity by hydrolysis and reverse-phase HPLC of amino acids and amino sugars after derivatization with o-phthaldialdehyde (20).
Activity assay.
Agar plates containing crude cell wall extracts from M. luteus were used to assay for millericin B activity. An aliquot of 20 μl was placed in each prepared well, incubated at 30°C overnight, and checked for zones of lysis. The activity of millericin B was estimated by using twofold dilutions of the test preparations. The inhibitory concentration (defined as activity units [AU] per milliliter) of millericin B was given as the reciprocal of the highest twofold dilution to give a distinct zone of clearing. The activity spectrum for millericin B against several gram-positive bacteria (S. aureus NMSCC 133, Staphylococcus simulans 22, M. luteus ATCC 46898, Bacillus subtilis W23, Lactococcus lactis ATCC 8756, Listeria monocytogenes ATCC 2457, and Streptococcus agalactiae ATCC 2305) was determined by using the spot-on-lawn assay (13).
Renaturing SDS-PAGE (Zymograms).
SDS-polyacrylamide gels (12% [wt/vol] acrylamide) containing 0.2% (wt/vol) lyophilized M. luteus cell walls as a substrate were used for the detection of lytic activity. Upon completion of electrophoresis, the gels were soaked for 45 min in 250 ml of distilled water at room temperature with gentle agitation. The gels were then transferred to 250 ml of renaturation buffer (25 mM Tris-HCl [pH 7.5] containing 0.1% Triton X-100 and 10 mM MgCl2) and incubated with gentle rotation at 37°C for 16 h. The bands with lytic activity were observed as clear in the opaque gel. To enhance detection of the lytic bands, the gels were stained for 3 h in 0.1% methylene blue in 0.01% KOH and destained in distilled water. Samples were mixed 1:1 (vol/vol) with sample buffer at final concentrations of 1% SDS, 10% glycerol, 1 mM EDTA, 0.0025% (wt/vol) bromophenol blue, 5% β-mercaptoethanol, and 60 mM Tris-HCl (pH 6.5). The samples were heated for 10 min at 100°C and applied to the gel. Molecular weight standards (Promega) were run on the same gel, removed, and stained separately.
Appearance of N-acetylamino sugars and free amino groups in soluble fragments during lysis of bacterial cell walls.
Purified M. luteus cell walls (0.3 mg) were digested, at 30°C, with purified millericin PB (<1 μg/ml) in 20 mM phosphate buffer, pH 7.01. M. luteus cell walls digested with lysozyme under the same conditions were used as a control. Reaction mixtures were centrifuged, and the supernatant was dried under vacuum and redissolved in 1% K2B7O4. The appearance of reduced sugars was assayed by the Morgan-Elson test as previously described (10). The appearance of free amino groups was determined with Sanger reagent as previously described (12).
Determination of the N-terminal amino acids liberated.
Purified M. luteus, S. aureus, and S. milleri NMSCC 051 (nonproducer) cell walls (0.3 mg) were digested overnight with millericin B as described above. Undigested cell walls were harvested at 16,000 × g for 10 min, and the supernatant was lyophilized. Lyophilized samples were resuspended in 100 μl of 1% (wt/vol) K2B7O4, 10 μl of fluorodinitrobenzene reagent was added, and the mixture was incubated at 60°C for 30 min. After acidification with concentrated HCl (50 μl) the N-dinitrophenyl (DNP) derivatives of free amino acids were extracted three times with ether (100 μl). The residual ether was evaporated at 60°C. The samples were then hydrolyzed for 6 h at 95°C. The DNP derivatives of the N-terminal amino acids were extracted three times with ether, and residual ether was evaporated, dried under vacuum, and redissolved in 0.05 M NH3 and chromatographed on thin-layer plates of silica gel G (Merck). Free C termini of amino acids were detected by DNP-hydrazine derivatization followed by thin-layer chromatography. The plates were first developed with solvent A (n-butanol–1% ammonia [wt/vol], 1:1; upper phase) at room temperature. After being dried under a stream of cold air, the plates were developed with solvent B (chloroform-methanol-acetic acid, 85:14:1; single phase) at 2°C.
Bactericidal action of millericin B on M. luteus ATCC 46898 and S. aureus NMSCC 133 cells.
Suspensions of log-phase cells of M. luteus and S. aureus were centrifuged, washed twice, and resuspended in 20 mM phosphate buffer (pH 7.0) to an optical density at 600 nm (OD600) of 0.5. Aliquots of suitable dilutions taken at different time intervals were plated on TSB agar plates, and the number of CFU was recorded after 18 h of growth.
RESULTS
Purification of millericin B.
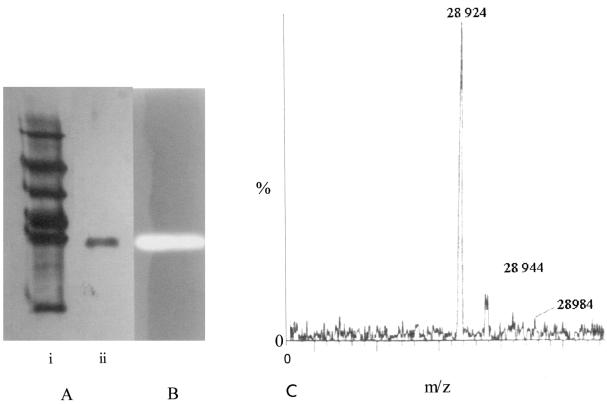
The purification stages of millericin are shown in Table 1. Millericin B is secreted into the supernatant of a growing culture up to a level of approximately 40 AU/ml. Optimal recovery of activity was during the mid-log phase of growth. The first step of purification involved ammonium sulfate precipitation. Activity was recovered from the 70% ammonium sulfate fraction at approximately 7.68 × 104 AU. Considerable loss of activity was noticed during the gel filtration step (results not shown). The addition of 1 M NaCl to the running buffer increased the recovery of millericin activity by 80-fold, from 26.8 to 2,146.4 AU/mg. After the gel filtration step, the sample was diluted in 20 mM phosphate and reconcentrated by ultrafiltration by passing it several times through a membrane with a cutoff value of 10 kDa. This step served to both desalt the sample and concentrate the active protein. Repetitive ultrafiltration did not result in any significant loss of millericin activity. After ion-exchange chromatography, millericin did not elute as a single peak but rather as part of a broad peak. The active fractions were pooled, desalted by ultrafiltration, and reseparated with a near-isocratic gradient within the elution range of millericin (80 to 85% B). This resulted in the elution of millericin B as a single peak. The purity of this peak was assessed by SDS-PAGE (Fig. 1) and found to contain a single band with an estimated molecular mass of 28 kDa. Activity was linked to this band by using renaturation SDS-PAGE with gels containing purified M. luteus cell walls (Fig. 1).
TABLE 1.
Stepwise purification of the bacteriolytic enzyme millericin from the culture supernatant of S. milleri NMSCC 061
| Purification step | Protein (mg) | Inhibitory activity (AU) | Sp act (AU/mg) | Yield (%) | Times purified |
|---|---|---|---|---|---|
| Supernatant | 105,000 | 2.0 × 105 | 1.9 | 100 | 1 |
| Ammonium sulfate | 2,862 | 7.68 × 104 | 26.8 | 38 | 14.1 |
| Sephadex G-75 | 8.5 | 1.6 × 104 | 2,146.4 | 8 | 1,129.6 |
| Sepharose HP | 4 | 1.28 × 104 | 4,292.8 | 6.4 | 2,259.3 |
FIG. 1.
(A) Polyacrylamide (12%) gel electrophoresis of purified millericin. Lanes: i, mid-range molecular weight standards (Promega); ii, purified millericin after ion-exchange chromatography. (B) Gel slice of a 12% acrylamide zymogram containing 0.1% M. luteus cell walls stained with 0.1% methylene blue in 0.01% KOH following renaturation of millericin PB in 0.1% Triton X-100 and 10 mM MgCl2. (C) Electrospray mass spectrometry of millericin B after ion-exchange chromatography.
Inhibitory activity of millericin B.
The range of inhibitory activity of millericin B against gram-positive bacteria was determined by an agar plate assay. Millericin B was added to wells in agar plates containing crude cell walls of the indicator cells. Activity was detected against several gram-positive bacteria tested. These included L. monocytogenes, S. agalactiae, L. lactis, and a nonproducing S. milleri strain, NMSCC 051. In contrast, B. subtilis was resistant to the action of millericin B. Escherichia coli, the only gram-negative bacterium tested, was not susceptible to millericin activity even when 5 mg of millericin B/ml was used. The diameter of the lytic zones produced in M. luteus growth was double that when other indicator cell walls were used.
N-terminal amino acid sequencing.
The amino-terminal amino acid sequence of purified millericin B was determined by automated Edman degradation. The first 12 amino acid residues were as follows: NH2-Ser-Glu-Asn-Asp-Phe-Ser-Leu-Ala-Met-Val-Ser-Asp. CNBr cleavage allowed us to obtain the sequence of the first 10 residues of an internal fragment by automated Edman degradation. The sequence obtained was as follows: Ser-Ile-Gln-Thr-Asn-Ala-Pro-Trp-Gly-Leu.
Effect of millericin B on the viability of M. luteus ATCC 46898 cultures.
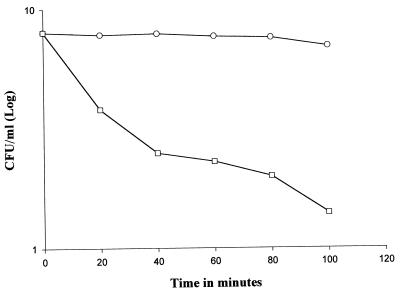
The addition of millericin B to stationary-phase M. luteus cultures resulted in an immediate decrease in the viable count (Fig. 2) compared with a culture that received milliQue (MQ) water. The viability of M. luteus decreased by 50% within the first 20 min of incubation. After 100 min of incubation, no viable cells were recovered.
FIG. 2.
Effect of millericin B on the growth of M. luteus ATCC 46898. Symbols: ○, control (addition of MQ water); □, test (addition of millericin).
Mode of action.
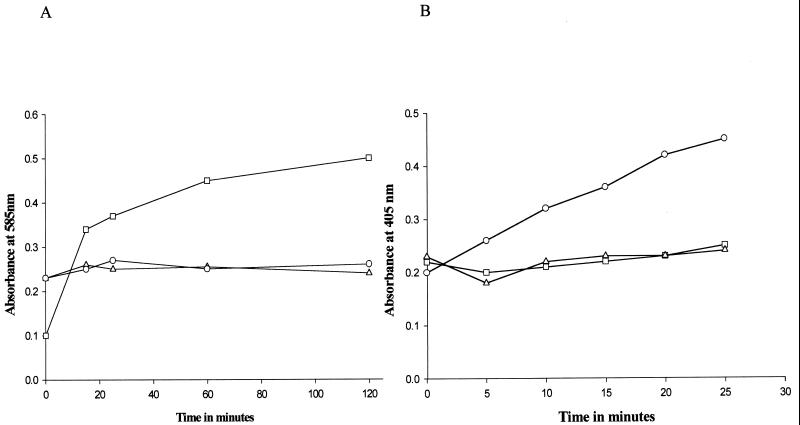
Millericin B was tested for the hydrolysis of the glycan bond between the N-acetylmuramic acid and N-acetylglucosamine repeating subunits of peptidoglycan by the Morgan-Elson test (10). The liberation of free reducing sugars was monitored photometrically. An increase in the liberation of reducing sugars was detected in the presence of lysozyme when M. luteus peptidoglycan was used as a substrate (Fig. 3A). There was no significant increase in the liberation of reducing sugars in the presence of millericin B. Substrate in the presence of 20 mM phosphate buffer was used as a control. In all cases, the OD585 reported is the mean value of triplicate experiments performed.
FIG. 3.
(A) Liberation of free reducing sugars from the cell walls of M. luteus ATCC 46898 after digestion with a cell wall-degrading enzyme, analyzed by the Morgan-Elson test (12). Symbols: □, cell walls digested with lysozyme; ○, cell walls digested with millericin B; ▵, cell wall in the presence of 20 mM phosphate buffer (pH 7.0). (B). Liberation of free amino groups from the digestion of M. luteus ATCC 46898 cell walls with millericin B by N-dinitrophenol (DNP) derivatization (12). Symbols: ○, cell walls digested with millericin B; □, cell walls digested with lysozyme; ▵, cell walls in the presence of 20 mM phosphate buffer (pH 7.0).
The detection of endopeptidase activity was done by a colorimetric assay with N-dinitrophenyl derivatization (10). M. luteus peptidoglycan digested with millericin B showed an increase in the liberation of free amino groups (Fig. 3B). Controls contained substrate but no enzyme and showed no significant change in absorbance, demonstrating that the rise in absorbance in the test was due to the activity of millericin B. As a negative control, M. luteus peptidoglycan was digested with lysozyme and tested for the liberation of free amino groups. There was no significant increase in OD405, indicating that no free amino groups were being liberated.
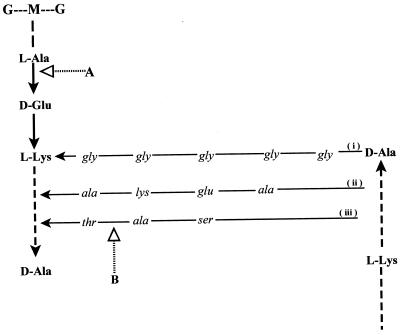
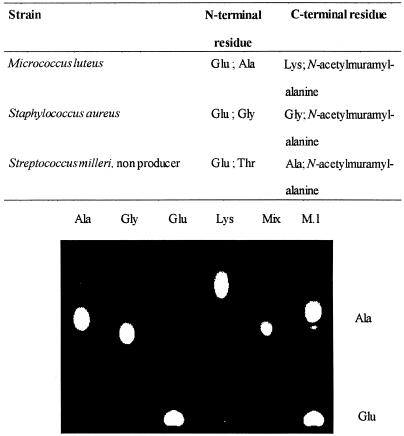
Thin-layer chromatography showed that a possible cleavage site for millericin B activity in M. luteus peptidoglycan was the glutamic acid of the stem peptide and the N terminus of the alanine within the interpeptide bridge (Fig. 4). Similar results were obtained for S. aureus and an S. milleri strain. In contrast to these results, no cleavage was shown against the S. milleri NMSCC 061 producer strain.
FIG. 4.
Fragments of the primary structure, stem peptide, and interpeptide cross bridge of peptidoglycans found in S. aureus (i), M. luteus (ii), and S. milleri (iii). A and B, cleavage sites of millericin B as determined by DNP derivatization and DNP-hydrazine derivatization.
DISCUSSION
The purification of peptidoglycan hydrolases usually involves extracting the enzymes from intact cells by using a high concentration of salts and affinity chromatography with resins with covalently linked peptidoglycan (3). However, S. milleri NMSCC 061 secretes millericin B directly into the supernatant. This simplified the purification because a single precipitation step with ammonium sulfate enabled us to harvest 38% of the total amount of millericin present in the supernatant. During the early stages of purification, millericin B activity could be masked by the presence of cellular proteins and medium components. The possibility of its binding to the carbohydrate moiety of the gel matrix may explain the lack of activity in the eluting fractions. The addition of NaCl possibly reduces these interactions. Reverse-phase HPLC of the active fractions obtained after gel filtration was unsuccessful. The use of an SP-Sepharose ion-exchange column linked to an HPLC system gave more satisfactory results. The elution of millericin B from this column was done under very-high-salt conditions. We were able to purify millericin B to homogeneity and detect its activity on an SDS-PAGE renaturation gel. Comparison of the first 10 amino acids as well as the sequence obtained from the internal fragment to existing protein sequences in the databases (SWISSPROT, BLAST, and NCBI) revealed no significant sequence similarity to any known proteins.
The antimicrobial activity of millericin B occurs by cleaving bonds in the peptide moiety of the peptidoglycan of susceptible organisms. Lysostaphin-treated S. aureus cells show a rapid reduction in both viable count and OD (22). M. luteus cultures treated with millericin B showed simultaneous loss of viability, suggesting that as with lysostaphin, lysis occurred as a direct result of millericin B activity against the cell wall.
Peptidoglycan is insoluble as a result of the extensive cross-linking of the subunits. Hydrolysis of sufficient cross-links will bring about solubilization of the cell wall and consequent lysis (27). Several methods exist to detect the enzymatic nature of the hydrolysis reaction, most of which measure the increase in OD of a particular component released via a colorimetric assay (10). The application of these assays in the present study enabled the activity of millericin B to be measured. Millericin B was characterized as an endopeptidase, cleaving bonds within the peptide moiety of peptidoglycan. Several strains were sensitive to millericin B, and all had the same stem peptide sequence (l-Ala, d-Glu, l-Lys, d-Ala) but different cross-links in their peptidoglycans. In contrast, millericin B-treated B. subtilis and E. coli cells showed no loss in viability, even after 5 mg of enzyme/ml was added and the incubation periods were prolonged. This could indicate that resistance is due to the absence of a specific cleavage site because the cross-links in both these organisms are of the A1γ type and they both have diaminopimelic acid substituted for lysine in their stem peptides (23). Modification of peptidoglycan can also affect sensitivity to peptidoglycan hydrolases. B. cereus peptidoglycan is resistant to lysozyme because of the unacetylated amino groups on the majority of its glucosamine residues, and it can be converted to a lysozyme-sensitive form by acetylation with acetic anhydride (2, 11). Conversely, the lysozyme resistance of the peptidoglycans of other organisms is due to O-acetylation of aminosugars, and these peptidoglycans can be made lysozyme sensitive by de-O-acetylation with a dilute base (5, 9, 19). Accessory cell wall polymers, such as teichoic acids or lipoteichoic acids, can also affect the susceptibility of bacteria to a number of peptidoglycan hydrolases (1, 3, 6, 8). The millericin B producer strain, S. milleri NMSCC 061, did not show any susceptibility to millericin B. For most bacteriocins, including lysostaphin, the producer strain is protected from the effects of its own bacteriocin through the action of a specific immunity factor (7, 12, 28). The lysostaphin resistance gene (epr) encodes a protein that specifies the modification of peptidoglycan cross bridges in S. simulans 22 and S. aureus (7). The millericin B producer strain may also utilize a similar mechanism to protect itself from millericin B.
Analysis of the cell wall digests of susceptible peptidoglycan showed two possible cleavage sites (Fig. 4). The first site is at the alpha amino group of glutamic acid (Fig. 5) This cleavage site was present on all types of peptidoglycan tested. The second site appears to be located within the interpeptide bridge. Cleavage occurred at the alpha amino group of the C-terminal residue in the interpeptide bridge. This phenomenon of multiple enzymatic activities has also recently been linked to the murein hydrolase of the staphylococcal phage φ11 (17). Deletion mutant analysis showed that the enzyme had both d-alanyl-glycyl endopeptidase and N-acetylmuramyl-l-alanyl amidase activities. The phage φ11 enzyme consists of three domains, an endopeptidase domain, an amidase domain, and a cell wall-targeting domain. In contrast, lysostaphin only cuts at the alpha amino group of a glycine residue, indicating that lysostaphin only cuts within the interpeptide bridge and not the stem peptide of S. aureus peptidoglycan (22).
FIG. 5.
Amino acid residues detected after digestion of peptidoglycan from three bacterial strains, DNP derivatization was used to detect free N termini and DNP-hydrazine derivatization was used for the C termini. A representative thin-layer chromatograph of cell walls from M. luteus appears as an example. Mix, mixture of standard amino acids that occur in the cell wall of M. luteus; M.l., M. luteus.
ACKNOWLEDGMENTS
This work was partially supported by grants from the Foundation for Research and Development of South Africa and the BONFOR program of the Medical Faculty, University of Bonn.
We also thank M. van der Merwe of the Biochemistry Department, University of Stellenbosch, for the mass spectrometry analysis and R. Chauhan of the Molecular Biology Unit, University of Natal, for the amino acid sequencing analysis.
REFERENCES
- 1.Amako K, Umeda A, Murata K. Arrangement of peptidoglycan in the cell wall of Staphylococcus spp. J Bacteriol. 1982;150:844–850. doi: 10.1128/jb.150.2.844-850.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki Y, Nakatani T, Wakayama K, Ito E. Occurrence of N-nonsubstituted glucosamine residues in peptidoglycan of lysozyme resistant cell walls of Bacillus cereus. J Biol Chem. 1972;247:6312–6322. [PubMed] [Google Scholar]
- 3.Bierbaum G, Sahl H-G. Autolytic system of Staphylococcus simulans 22: influence of cationic peptides on activity of N-acetylmuramoyl-l-alanine amidase. J Bacteriol. 1987;169:5452–5458. doi: 10.1128/jb.169.12.5452-5458.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browder H P, Zygmunt W A, Young J R, Tavorina P A. Lysostaphin: enzymatic mode of action. Biochem Biophys Res Commun. 1965;19:383–389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- 5.Clarke A J. Extent of peptidoglycan O-acetylation in the tribe Proteae. J Bacteriol. 1993;175:4550–4553. doi: 10.1128/jb.175.14.4550-4553.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleveland R F, Holtje J-V, Wicken A J, Tomasz A, Daneo-Moore L, Shockman G D. Inhibition of bacterial wall lysins by lipoteichoic acids and related compounds. Biochem Biophys Res Commun. 1975;67:1128–1135. doi: 10.1016/0006-291x(75)90791-3. [DOI] [PubMed] [Google Scholar]
- 7.DeHart H P, Heath H E, Heath L S, LeBlanc P A, Sloan G L. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Environ Microbiol. 1995;61:1475–1479. doi: 10.1128/aem.61.4.1475-1479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer W, Rösel P, Koch H U. Effect of alanine ester substitution and other structural features of lipoteichoic acids on their inhibitory activity against autolysins of Staphylococcus aureus. J Bacteriol. 1981;146:467–475. doi: 10.1128/jb.146.2.467-475.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghuysen J-M, Strominger J L. Structure of the cell wall of Staphylococcus aureus, strain Copenhagen. II. Separation and structure of disaccharides. Biochemistry. 1963;2:1119–1125. doi: 10.1021/bi00905a036. [DOI] [PubMed] [Google Scholar]
- 10.Ghuysen J-M, Tripper D J, Strominger J L. Enzymes that degrade cell walls. Methods Enzymol. 1966;8:685–699. [Google Scholar]
- 11.Hayashi H, Araki Y, Ito E. Occurrence of glucosamine residues with free amino groups in cell wall peptidoglycan from bacilli as a factor responsible for resistance to lysozyme. J Bacteriol. 1973;113:592–598. doi: 10.1128/jb.113.2.592-598.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heath H E, Heath L S, Nitterauer J D, Rose K E, Sloan G L. Plasmid-encoded lysostaphin endopeptidase resistance of Staphylococcus simulans biovar staphylolyticus. Biochem Biophys Res Commun. 1989;160:1106–1109. doi: 10.1016/s0006-291x(89)80117-2. [DOI] [PubMed] [Google Scholar]
- 13.Jack R W, Tagg J R. Factors affecting production of the group A streptococcus bacteriocin SA-FF22. J Med Microbiol. 1992;36:132–138. doi: 10.1099/00222615-36-2-132. [DOI] [PubMed] [Google Scholar]
- 14.James S M, Tagg J R. A search within the genera Streptococcus, Enterococcus, and Lactobacillus for the organisms inhibitory to mutans streptococci. Microb Ecol Health Dis. 1988;1:153–162. [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lipkin D, Philips B E, Abrell J W. The action of hydrogen fluoride on nucleotides and other esters of phosphorus (V) acids. J Org Chem. 1969;34:1539–1547. [Google Scholar]
- 17.Navarre W W, Ton-That H, Faull K F, Schneewind O. Multiple enzymatic activities of the murein hydrolase from staphylococcal phage φ11. J Biol Chem. 1999;274:15847–15856. doi: 10.1074/jbc.274.22.15847. [DOI] [PubMed] [Google Scholar]
- 18.Recsei P A, Gruss A D, Novick R P. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci USA. 1987;84:1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenthal R S, Blundell J K, Perkins H R. Strain-related differences in lysozyme sensitivity and extent of O-acetylation of gonococcal peptidoglycan. Infect Immun. 1982;37:826–829. doi: 10.1128/iai.37.2.826-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahl H-G, Grossgarten M, Widger W R, Cramer W A, Brandis H. Structural similarities of the staphylococcin-like peptide Pep-5 to the peptide antibiotic nisin. Antimicrob Agents Chemother. 1985;27:836–840. doi: 10.1128/aac.27.5.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahl H-G, Hahn C, Brandis H. Interaction of the staphylococcin-like peptide Pep-5 with cell walls and isolated cell wall components of gram-positive bacteria. Zentlbl Bakteriol Hyg A. 1985;260:197–205. doi: 10.1016/s0176-6724(85)80115-2. [DOI] [PubMed] [Google Scholar]
- 22.Schindler C A, Schuhardt V T. Lysostaphin: a new bacteriolytic agent from the staphylococcus. Proc Natl Acad Sci USA. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. 36:407–477. [DOI] [PMC free article] [PubMed]
- 24.Shockman G D, Holtje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 131–166. [Google Scholar]
- 25.Simmonds R S, Simpson W, Tagg J R. Cloning and sequence analysis of zoocin A, a Streptococcus zooepidemicus gene encoding a bacteriocin-like inhibitory substance having a domain structure similar to that of lysostaphin. Gene. 1997;189:255–261. doi: 10.1016/s0378-1119(96)00859-1. [DOI] [PubMed] [Google Scholar]
- 26.Simmonds R S, Pearson L, Kennedy R C, Tagg J R. Mode of action of a lysostaphin-like bacteriolytic agent produced by Streptococcus zooepidemicus 4881. Appl Environ Microbiol. 1996;62:4536–4541. doi: 10.1128/aem.62.12.4536-4541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strominger J L, Ghuysen J M. Mechanism of enzymatic bacteriolysis. Science. 1967;156:213–221. doi: 10.1126/science.156.3772.213. [DOI] [PubMed] [Google Scholar]
- 28.Tagg J R, Dajani A S, Wannamaker L W. Bacteriocins of gram positive bacteria. Microbiol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward J B, Williamson R. Bacterial autolysins: specificity and function. In: Nombela C, editor. Microbial cell wall synthesis and autolysins. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1984. pp. 159–166. [Google Scholar]