Abstract
Regulatory T cell (Treg) adoptive cell therapy (ACT) represents an emerging strategy for restoring immune tolerance in autoimmune diseases. Tregs are commonly purified using a CD4+CD25+CD127lo/- gating strategy, which yields a mixed population: 1) cells expressing the transcription factors, FOXP3 and Helios, that canonically define lineage stable thymic Tregs and 2) unstable FOXP3+Helios- Tregs. Our prior work identified the autoimmune disease risk-associated locus and costimulatory molecule, CD226, as being highly expressed not only on effector T cells but also, interferon-γ (IFN-γ) producing peripheral Tregs (pTreg). Thus, we sought to determine whether isolating Tregs with a CD4+CD25+CD226- strategy yields a population with increased purity and suppressive capacity relative to CD4+CD25+CD127lo/- cells. After 14d of culture, expanded CD4+CD25+CD226- cells displayed a decreased proportion of pTregs relative to CD4+CD25+CD127lo/- cells, as measured by FOXP3+Helios- expression and the epigenetic signature at the FOXP3 Treg-specific demethylated region (TSDR). Furthermore, CD226- Tregs exhibited decreased production of the effector cytokines, IFN-γ, TNF, and IL-17A, along with increased expression of the immunoregulatory cytokine, TGF-β1. Lastly, CD226- Tregs demonstrated increased in vitro suppressive capacity as compared to their CD127lo/- counterparts. These data suggest that the exclusion of CD226-expressing cells during Treg sorting yields a population with increased purity, lineage stability, and suppressive capabilities, which may benefit Treg ACT for the treatment of autoimmune diseases.
Keywords: CD226, Treg, lineage stability, suppressive function, autoimmune disease, adoptive cell therapy
Introduction
Human regulatory T cells (Tregs) possess the unique capacity to suppress innate and adaptive immune subsets throughout the body using a variety of mechanisms, including consumption of growth factors, degradation of inflammatory substrates, expression of negative regulators of costimulation, secretion of immunoregulatory cytokines, and trogocytosis (1–3). Impairment of Treg suppression in vivo leads to the proliferation of autoreactive T cells, which has been associated with the development of autoimmune diseases, such as type 1 diabetes (T1D) and systemic lupus erythematosus (SLE) (4, 5). Therefore, Tregs represent a critical target or even deliverable component of immunotherapies seeking to inhibit the pathogenesis of autoimmune diseases (6, 7).
Early proof-of-principle studies in the non-obese diabetic (NOD) mouse provide evidence that adoptive transfer of Tregs can reverse autoimmune diabetes (8–10). Translating this concept to patients with or at risk for T1D requires the isolation and subsequent ex vivo expansion of Tregs for adoptive cell therapy (ACT), due to the rarity of Tregs in both peripheral and umbilical cord blood (6, 11–14). Polyclonal autologous Treg-ACT was shown to be safe yet ineffective at preserving insulin production in individuals with recent-onset T1D (6), potentially due to limited Treg persistence in vivo. In a recent phase I clinical trial, low dose IL-2 bolstered polyclonal Treg engraftment in patients with T1D but also, imparted undesirable off-target expansion of cytotoxic cell subsets, such as activated natural killer (NK), mucosal associated invariant T (MAIT), and CD8+ T cells (15). Hence, there is a clear need to optimize Treg ACT, including through isolation of a Treg population that maintains lineage stability and suppressive functionality following ex vivo expansion.
Early Treg enrichment strategies relied on the observation that Tregs constitutively express the IL-2 receptor alpha chain (IL-2Rα/CD25), conferring a high affinity for the T cell growth factor, IL-2 (16). However, observations of activation-induced upregulation of CD25 on CD4+ conventional T cells (Tconv) (17, 18) supported the need for additional markers for effective Treg isolation (19). Current Treg isolation methods involve Fluorescence-Activated Cell Sorting (FACS) of CD4+CD25hi T cells with low to no expression of the IL-7 receptor, CD127 (20). However, CD127 can be downregulated by Tconv in response to signaling by IL-7 and other common γ-chain cytokines (20). Moreover, in instances of lymphopenia, increased serum levels of IL-7 are known to decrease CD127 expression on Tconv, significantly complicating efforts to isolate tolerogenic Tregs for ACT in patients with autoimmune diseases (21–23).
The CD127lo/- Treg isolation strategy yields a heterogeneous population containing both lineage stable FOXP3+Helios+ Tregs as well as FOXP3+Helios- Tregs, which are susceptible to phenotypic instability upon activation (24, 25). While subject to debate (26), the FOXP3+Helios+ transcription factor combination is generally accepted as identifying the thymically-derived Treg subset (tTregs) while FOXP3+Helios- designates the peripherally-induced Treg fraction (pTregs) (25, 27). Compared to tTregs, pTregs exhibit increased production of inflammatory cytokines, including IFNγ, as well as methylation at the conserved non-coding sequence 2 (CNS-2), referred to as the Treg-specific demethylation region (TSDR) (28). As a result, the CD4+CD25hiCD127lo/- population is predisposed to an outgrowth of less suppressive Tregs and increased expression of inflammatory/effector molecules during expansion, all of which may negatively impact ACT therapeutic efficacy (29). Furthermore, an increased proportion of IFNγ-secreting FOXP3+Helios- Tregs in patients with T1D versus healthy controls suggests that detrimental Treg plasticity may be augmented in settings of inflammation or autoimmunity (28).
Previous work in our laboratory characterizing the phenotype of IFNγ-secreting FOXP3+Helios- Tregs revealed high expression of the costimulatory molecule CD226 (30). CD226 is an activating costimulatory receptor associated with the initiation of Th1 and Th17 immune responses (31, 32). Following its ligation with CD112 or CD155 on antigen-presenting cells (APCs), CD226 becomes activated via phosphorylation of its immunoreceptor tyrosine-based activation motif (ITAM) (33), augmenting downstream Ras/MAPK signaling, which is known to result in increased secretion of the pro-inflammatory cytokines IFN-γ and IL-17A (31). In our studies, CD226 expression correlated positively with CD127 and negatively with FOXP3 expression; moreover, freshly isolated CD226lo Tregs exhibited increased demethylation at the TSDR as compared to CD226+ Tregs, suggesting high CD226 expression might be associated with an effector phenotype (30).
In addition to contributing to decreased regulatory function, CD226 has been identified to contain a potential gain-of-function risk variant contributing to a propensity for multiple autoimmune diseases including T1D, SLE, rheumatoid arthritis (RA), and multiple sclerosis (MS) (32, 34–36). We previously reported that knockout (KO) of Cd226 in NOD mice resulted in reduced severity of insulitis and diabetes incidence (37), and Wang et al. similarly observed that Cd226 KO reduced disease severity in an experimental autoimmune encephalomyelitis (EAE) mouse model of MS, further highlighting the role of CD226 in autoimmune disease pathogenesis (38).
To identify an improved surrogate surface marker for lineage stable Tregs, we performed extensive ex vivo analyses to evaluate the therapeutic potential of CD4+CD25+CD226- sorted T cells as compared to the conventional CD4+CD25+CD127lo/- strategy. Specifically, we hypothesized that this marker profile would allow for isolation and expansion of increased proportions of FOXP3+Helios+ Tregs, minimizing contamination of IFNγ-producing FOXP3+Helios- Tregs, to yield a more stable and functionally suppressive population
Materials and Methods
Human Subjects
Fresh peripheral blood mononuclear cell (PBMC) samples were isolated from human leukapheresis-enriched blood of healthy donors (median age: 22 years, range 18-39 years, N=20, 45% female) purchased from LifeSouth Community Blood Centers (Gainesville, FL).
CD4+ T Cell Enrichment From Human PBMC Samples
Before Treg isolation, CD4+ T cells were enriched by negative selection using a CD4+ T cell enrichment RosetteSep™ cocktail (StemCell Technologies, Vancouver, BC, Canada) according to the manufacturer’s instructions while autologous PBMCs required for suppression assays were isolated from unenriched peripheral blood. CD4+ T cell-enriched and unenriched components were diluted 1:1 with PBS and overlayed onto Ficoll-Paque Plus medium (Thermo Fisher, Waltham, MA, USA) for density gradient centrifugation (1200 x g, 20 min). PBMCs were suspended in Ammonium-Chloride-Potassium (ACK) Lysis Buffer (Gibco, Waltham, MA, USA), washed, and resuspended in PBS, according to the manufacturer’s instructions. Quantification of cell viability was accomplished by staining with Acridine Orange/Propidium Iodide (AO/PI) before reading on an Auto2000 Cellometer (Nexcelom Biosciences, Lawrence, MA, USA).
FACS Isolation of Treg Subsets
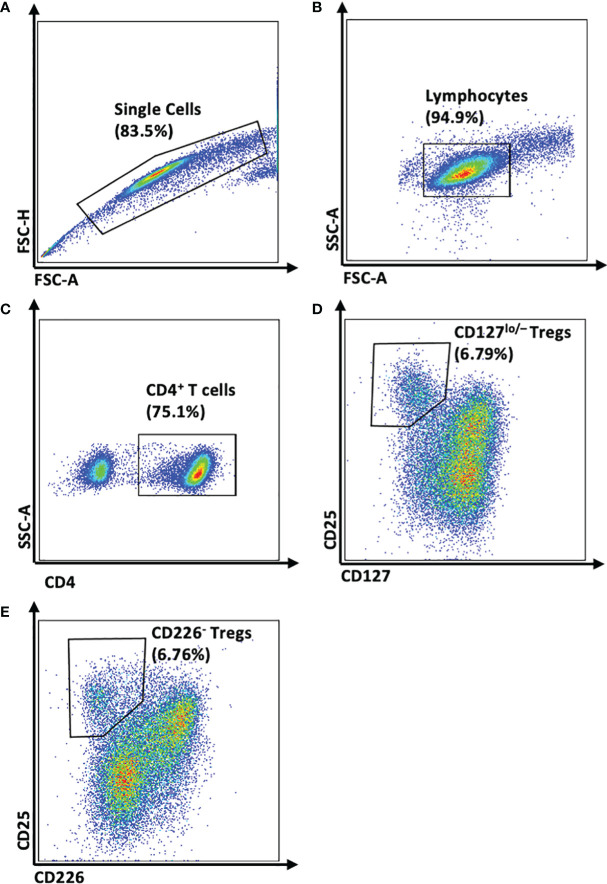
CD4+ T cell-enriched PBMCs were split and stained with: 1) CD4-BV510, CD25-APC, and CD127-PE or 2) CD4-BV510, CD25-APC, and CD226-PE-Cy7 (clone and manufacturer information provided in Table 1 ). Matched sets of CD4+CD25+CD127lo/- and CD4+CD25+CD226- Tregs were isolated ( Figure 1 ) using a FACSAria™ III Cell Sorter (Beckton Dickinson, Franklin Lakes, NJ, USA; CD4+CD25+CD127lo/- median sort purity: 96.1%, range: 85.8-99.9%, N=6; CD4+CD25+CD226- median sort purity: 97.8%, range: 88.7-99.9%, N=6).
Table 1.
Antibodies used for flow cytometry.
| Target | Clone | Fluorochrome | Vendor | Concentration | RRID |
|---|---|---|---|---|---|
| CD4 | SK3 | BV510 | BD Biosciences | 0.05 µg/mL | AB_2744424 |
| CD8 | RPA-T8 | PE-CF594 | BD Biosciences | 0.10 µg/mL | AB_11154052 |
| CD25 | BC96 | APC | BioLegend | 0.50 µg/mL | AB_314280 |
| CD25 | BC96 | BV605 | BioLegend | 0.50 µg/mL | AB_11218989 |
| CD39 | eBioA1 | APC | eBioscience | 0.50 µg/mL | AB_1963578 |
| CD40L | 24-31 | APC-Cy7 | BioLegend | 0.50 µg/mL | AB_2076096 |
| CD45RA | HI100 | BV605 | BioLegend | 0.10 µg/mL | AB_2563814 |
| CD73 | AD2 | PE | BD Pharmingen | 0.50 µg/mL | AB_393561 |
| CD127 | A019D5 | PE | BioLegend | 0.20 µg/mL | AB_1937251 |
| CD197 (CCR7) | 2-L1-A | APC-R700 | BD Biosciences | 0.10 µg/mL | AB_2869856 |
| CD226 | 11A8 | PE-Cy7 | BioLegend | 0.40 µg/mL | AB_2616645 |
| CLTA-4 | L3D10 | PE-Cy7 | BioLegend | 0.50 µg/mL | AB_2563098 |
| FOXP3 | 206D | Alexa Fluor 488 | BioLegend | 0.50 µg/mL | AB_430883 |
| FOXP3 | 259D | Alexa Fluor 488 | BioLegend | 0.50 µg/mL | AB_430887 |
| GITR | 621 | PE-Cy5 | BioLegend | 0.50 µg/mL | AB_2240646 |
| Helios | 22F6 | Pacific Blue | BioLegend | 0.25µg/mL | AB_10690535 |
| IL-10 | JES3-9D7 | BV421 | BioLegend | 0.08 µg/mL | AB_2632952 |
| IL-17A | BL168 | BV605 | BioLegend | 0.12 µg/mL | AB_2563887 |
| IFN-γ | 4S.B3 | BV570 | BioLegend | 0.10 µg/mL | AB_2563880 |
| PD-1 | EH12.2H7 | Alexa Fluor 647 | BioLegend | 0.50 µg/mL | AB_940471 |
| TGF-β1 | TW4-2F8 | Alexa Fluor 647 | BioLegend | 0.40 µg/mL | AB_2721298 |
| TGF-β1 | FNLAP | PerCP-eFluor 710 | eBioscience | 0.50 µg/mL | AB_2573900 |
| TNF | Mab11 | BV650 | BioLegend | 0.20 µg/mL | AB_2561355 |
Figure 1.
Gating Strategy for FACS Isolation of Paired CD4+CD25+CD127lo/- and CD4+CD25+CD226- Tregs. Representative flow plots demonstrate the method by which CD127lo/- or CD226- Tregs were isolated from CD4+ T-cell enriched PBMC using a BD FACSAriaIII Cell Sorter. (A) Singlet gating was performed using forward scatter area (FSC-A) versus forward scatter height (FSC-H). (B) Lymphocytes were gated on FSC-A and side scatter area (SSC-A). (C) From the CD4+ T cell fraction, (D) CD25+CD127lo/- Tregs and (E) CD25+CD226- Tregs were isolated.
Treg Expansion
Following FACS isolation, cells were expanded for 14 days ex vivo (13). In brief, Tregs were cultured in complete RPMI media (cRPMI; RPMI 1640 media Phenol Red w/o L-Glutamine (Lonza, Basel, CH-BS, Switzerland), 5mM HEPES (Gibco, Waltham, MA, USA), 5 mM MEM Non-Essential Amino Acids (NEAAs; Gibco), 2mM Glutamax (Gibco), 50 µg/mL penicillin (Gibco), 50 µg/mL streptomycin (Gibco), 20 mM sodium pyruvate (Gibco), 50 mM 2-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), 20 mM sodium hydroxide (Sigma-Aldrich) and 10% FBS (Genesee Scientific, El Cajon, CA, USA)) with Teceleukin recombinant human IL-2 (rhIL-2; Roche, Basel, CH-BS, Switzerland) at 300 IU/mL, with media and rhIL-2 being replaced every 3-4 days. Tregs were stimulated using MACS® GMP ExpAct Treg Beads (Miltenyi Biotec, Bergisch Gladbach, NW, Germany) at a 4:1 bead:cell ratio. Beads were replaced at day seven, and cells were expanded through day 14.
Analysis of TSDR Epigenetic Signature
Demethylation of the FOXP3-TSDR, or conserved non-coding sequence (CNS2), represents a robust epigenetic indicator of tTreg purity (39). We quantified demethylation within the FOXP3-TSDR by real-time polymerase chain reaction (RT-PCR) as previously described (30), with the following modifications. DNA extraction was conducted using the DNeasy® Blood & Tissue Kit (QIAGEN, Hilden, NW, Germany) as described by the manufacturer’s protocol. Following extraction, DNA was quantified using the Qubit™ Double-Stranded DNA (dsDNA) High Sensitivity (HS) Assay Kit (Invitrogen, Waltham, MA, USA) on the Qubit™ Fluorometer system (Invitrogen). Bisulfite conversion of DNA was conducted using the EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA). RT-PCR was performed using a StepOne™ system (Applied Biosystems, Waltham, MA, USA).
Flow Cytometric Analysis of Treg Phenotype
To assess the phenotype and purity of Tregs before and following 14 days of expansion, 1 x 105 CD4+CD25+CD226- and CD4+CD25+CD127lo/- Tregs were stained with Live/Dead™ Near-IR viability dye (Thermo Fisher) for 10 minutes at 4°C before washing with stain buffer (PBS + 2% FBS + 0.05% NaN3 w/v). Cells were then stained with an extracellular antibody cocktail, consisting of CD4-BV510, CD25-APC, CD45RA-BV605, CD127-PE, CD197-APC-R700, and CD226-PE-Cy7 for 30 minutes at 4°C (antibody clone and concentration are provided in Table 1 ). Cells were fixed and permeabilized using the eBioScience™ FOXP3 Transcription Factor Staining Buffer Set (Invitrogen) according to the manufacturer’s instructions, then stained with an intracellular transcription factor antibody cocktail, consisting of FOXP3-Alexa Fluor 488 and Helios-Pacific Blue ( Table 1 ). Data were collected on an Aurora 3L (16V-14B-8R) spectral flow cytometer (Cytek, Fremont, CA, USA), and analysis was conducted using FlowJo™ version 10.6.1 Software (BD Life Sciences, Ashland, OR, USA). Tregs were classified as CD4+CD25+FOXP3+, CD4+CD25+FOXP3+Helios+, and CD4+CD25+FOXP3+Helios- with phenotype established based on CD45RA and CD197 (CCR7) expression as follows: CD45RA+CCR7+ naïve, CD45RA-CCR7+ central memory (TCM), CD45RA-CCR7- effector memory (TEM), and CD45RA+CCR7- effector memory re-expressing CD45RA (TEMRA) cells. The detailed gating strategy is shown in Figure S1 . Protein expression levels were reported as stain indices [SI = geometric mean fluorescence intensity (gMFI) of the stained sample/gMFI of the applicable fluorescence minus one (FMO) control].
Flow Cytometric Analysis of Intracellular Cytokine Production
Following 14 days of ex vivo expansion as described above, MACS® GMP ExpAct Treg Beads were removed, then CD4+CD25+CD127lo/- and CD4+CD25+CD226- sorted Tregs were immediately assessed for intracellular cytokine expression. Cells were either stimulated with PMA (10 µg/mL; Thermo Fisher) and ionomycin (500 nM; Thermo Fisher) or unstimulated for four hours in the presence of GolgiStop (0.66 µL/mL; BioLegend, San Diego, CA, USA). Stimulated cells underwent staining for viability and extracellular markers, including CD4-BV510, CD25-APC, CD127-PE, CD226-PE-Cy7, and TGF-β1-PerCP-eFluor 710 ( Table 1 ), and were subsequently permeabilized as described above. Following permeabilization, cells were stained with the FOXP3-AF488 and Helios-Pacific Blue cocktail, as well as an intracellular cytokine cocktail consisting of IL-10-BV421, IL-17A-BV605, IFN-γ-BV570, TGF-β1-Alexa Fluor 647, and TNF-BV650 ( Table 1 ). Fold change of cytokine expression levels were assessed by dividing the gMFI of the stained, stimulated sample by the gMFI of the applicable stained, unstimulated control. Differences between fold change of cytokine expression are reported as Z-scores, [Z = (Mean fold change for Treg subset – Mean fold change for all Tregs assessed)/standard deviation of the sample].
Flow Cytometric Analysis of Treg Activation Markers
Following 14 days of ex vivo expansion, CD4+CD25+CD127lo/- and CD4+CD25+CD226- sorted Tregs were labeled with CellTrace™ Violet (CTV; Thermo Fisher) as recommended by the manufacturer, then cultured with no PBMCs or stimulation (0 hour condition) or with autologous PBMCs at a 1:1 ratio in the presence of soluble anti-CD3 (8 µg/mL, Clone OKT3, BioLegend, RRID: AB_11150592) and soluble anti-CD28 (4 µg/mL, Clone CD28.2, Thermo Fisher, RRID: AB_468926) for 24 or 48 hours. Cells were stained for viability with Live/Dead™ Blue viability dye (Thermo Fisher) and underwent surface staining for CD4-BV510, CD25-BV605, PD-1-AF647, CD39-APC, CD73-PE, CTLA-4-PE-Cy7, GITR-PE-Cy5, and CD40L-APC-Cy7 ( Table 1 ). The cells were subsequently permeabilized as described above and stained with FOXP3-AF488 and Helios-Pacific Blue before flow cytometric assessment on a Cytek Aurora 5L (16UV-16V-14B-10YG-8R) spectral flow cytometer and analyzed in FlowJo version 10.6.1 Software.
Treg Suppression Assays
Post-expansion CD4+CD25+CD226- Tregs and CD4+CD25+CD127lo/- Tregs were collected on day 14 and immediately labeled with cell proliferation dye (CPD-eFluor 670; Biolegend), while autologous PBMCs were labeled with CTV as recommended by the manufacturers’ protocol. Tregs were co-cultured with PBMCs (Treg : PBMC ratios of 1:1, 1:2, 1:4, 1:8, 1:16, 1:32) in the presence of soluble anti-CD3 (8 µg/mL, Clone OKT3) and soluble anti-CD28 (4 µg/mL, Clone CD28.2) in triplicate for 96 hours. Replicates were pooled, subjected to surface staining for CD4-BV510 and CD8-PE-CF594 ( Table 1 ), assessed using a Cytek Aurora 5L (16UV-16V-14B-10YG-8R) spectral flow cytometer, and analyzed in FlowJo version 10.6.1 Software. Percent suppression of responder cells was established by the division index (DI) method using proliferation modeling (40).
Statistical Analysis
Generation of figures and statistical analysis were conducted using GraphPad Prism version 9.2.0 (GraphPad Software, San Diego, CA, USA). Data were analyzed by two-way ANOVA with Bonferroni’s post hoc test for multiple testing correction unless otherwise stated. Area under the curve (AUC) values were compared using paired t-tests (41). The p-value ≤ 0.05 was considered significant.
Results
CD25 Expression is Elevated on CD4+CD25+CD226- Tregs
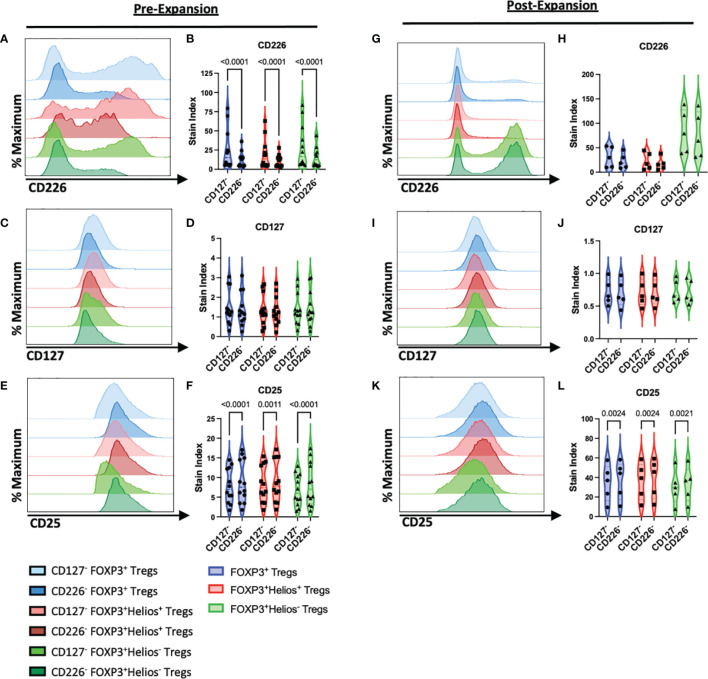
To characterize the efficacy of the sorting strategies in isolating CD4+CD25+CD226- versus CD4+CD25+CD127lo/- Tregs ( Figure 1 ), we examined surface expression levels of CD226, CD127, and CD25 on CD4+CD25+FOXP3+ total Tregs, FOXP3+Helios+ Tregs, as well as FOXP3+Helios- Tregs by flow cytometry ( Figure S1 ), both prior to and following ex vivo expansion (42). As expected, CD226 expression was significantly lower on CD4+CD25+CD226- sorted cells, including total FOXP3+ Tregs (0.49-fold, p<0.0001) as well as FOXP3+Helios+ (0.55-fold, p<0.0001) and FOXP3+Helios- subsets (0.60-fold, p<0.0001; Figures 2A, B ). Prior to expansion, CD4+CD25+CD226- and CD4+CD25+CD127lo/- sorted Tregs displayed comparably low CD127 expression ( Figures 2C, D ). Yet, the CD4+CD25+CD226- isolation strategy yielded a significantly higher CD25 gMFI on Tregs (1.13-fold, p<0.0001), including both the FOXP3+Helios+ (1.08-fold, p=0.0011) and FOXP3+Helios- subsets (1.22-fold, p<0.0001; Figures 2E, F ). As a result of expansion, CD4+CD25+CD226- sorted Tregs re-expressed CD226 at similar levels to CD4+CD25+CD127lo/- Tregs following ex vivo expansion, with the most dramatic upregulation of CD226 occurring in the FOXP3+Helios- fraction ( Figures 2G, H ). However, CD127 levels remained comparably low across all Treg subsets ( Figures 2I, J ), and CD25 levels remained augmented on CD4+CD25+CD226- versus CD4+CD25+CD127lo/- sorted cells, including total Tregs (1.08-fold, p=0.0024), FOXP3+Helios+ Tregs (1.08-fold, p=0.0024) and FOXP3+Helios- Tregs (1.10-fold, p=0.0021; Figures 2K, L ).
Figure 2.
CD4+CD25+CD226- Tregs Exhibit Increased CD25 Expression. Representative histograms show expression of cell surface markers on CD127lo/- (lighter blue) and CD226- CD4+CD25+FOXP3+ Tregs (darker blue), CD127lo/- (lighter red) and CD226- CD4+CD25+FOXP3+Helios+ Tregs (darker red), and CD127lo/- (lighter green) and CD226- CD4+CD25+FOXP3+Helios- Tregs (darker green) with violin plots showing stain index (SI) fold change from FMO controls, (A–F) prior to expansion (n =12 biological with n = 2 technical replicates) and (G–L) following 14 days of ex vivo expansion (n = 5 biological with n = 2 technical replicates). (A, B, G, H) CD226, (C, D, I, J) CD127, (E, F, K, L) CD25. Significant P-values are reported on the figure for two-way ANOVA with Bonferroni correction for multiple comparisons of Treg isolation conditions from matched subjects.
CD4+CD25+CD226- Tregs Maintain Higher Purity and Lineage Stability
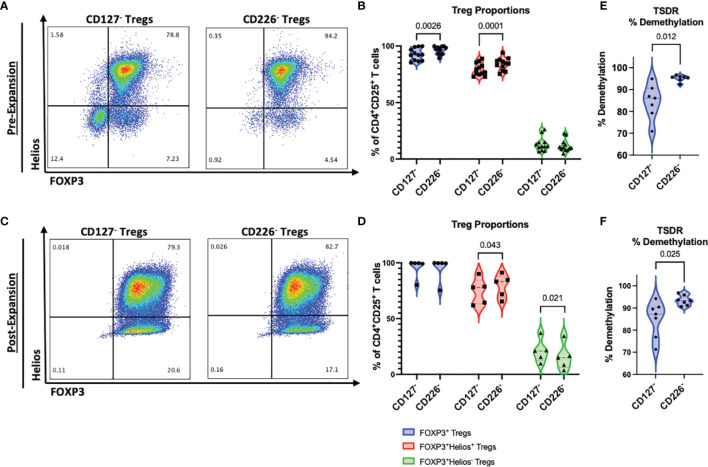
We previously identified CD4+CD25+CD127lo/-CD226+ Tregs as a subset with a higher proportion of IFNγ-producing pTregs, as compared to CD4+CD25+CD127lo/-CD226- Tregs (30). To evaluate the potential of using a CD4+CD25+CD226- sort for isolation of more lineage stable Tregs, as compared to the typical CD4+CD25+CD127lo/- strategy, we examined the expression of the canonical Treg lineage-defining transcription factors, FOXP3 and Helios, using flow cytometry ( Figure S1 ). We identified significantly increased percentages of FOXP3+ Tregs prior to expansion (+3.60%, p=0.0026), including an increased proportion of FOXP3+Helios+ Tregs (+4.70%, p=0.0001) within the CD4+CD25+CD226- sorted population, compared to CD4+CD25+CD127lo/- ( Figures 3A, B ). Importantly, these differences were not related to variations in donor sex ( Figure S2A ) or age ( Figure S2B ); though, we did identify a significantly non-zero slope (p=0.023) suggesting that the isolation of CD127lo/- FOXP3+Helios+ Tregs increased with age for our data set ( Figure S2B ). Following a 14-day expansion period, significant increases in the FOXP3+Helios+ Treg (+3.57%, p=0.043) and decreases in the FOXP3+Helios- Treg (-4.43%, p=0.021) subpopulations were observed from CD4+CD25+CD226- versus CD4+CD25+CD127lo/- sorted cells, despite comparable frequencies of total Tregs ( Figures 3C, D ). These data suggest that isolation of CD4+CD25+CD226- Tregs may yield a more stable Treg population after ex vivo expansion, without compromising post-expansion yield ( Figure S3 ).
Figure 3.
CD226- Tregs maintain higher purity than conventionally sorted CD127lo/- Tregs. Tregs from each FACS method were examined for FOXP3 and Helios expression (A, B) at day 0 following isolation (n = 12 biological with n = 2 technical replicates) and (C, D) at day 14 following ex vivo expansion (n = 5 biological with n = 2 technical replicates). (A, C) Representative flow plots pre-gated on live CD4+CD25+ cells show percentages of Treg subsets for CD127lo/- sorted Tregs and CD226- sorted Tregs. (B, D) Percentages of CD4+CD25+FOXP3+ Tregs (blue), CD4+CD25+FOXP3+Helios+ Tregs (red), and CD4+CD25+FOXP3+Helios- Tregs (green) per FACS isolation method. P-values are reported on the figure for two-way ANOVA with Bonferroni post-hoc correction for multiple comparisons of Treg isolation conditions from matched subjects. (E, F) Percent demethylation at the TSDR of CD127lo/- and CD226- sorted Treg cultures, (E) pre-expansion and (F) post-expansion. Significant P-values are reported on the figure for paired T-tests, n = 7 biological with n = 2 technical replicates.
tTregs display a distinct epigenetic profile, including the selective demethylation of the FOXP3-TSDR region (39). We therefore evaluated levels of TSDR methylation by RT-PCR. This analysis showed increased levels of TSDR demethylation in the CD4+CD25+CD226- Treg population both before (+10.69%, p=0.012) and following expansion (+8.46%, p=0.025) compared to Tregs isolated by the CD4+CD25+CD127lo/- marker profile ( Figures 3E, F ). These data corroborate our flow cytometry results identifying a higher purity of lineage stable Tregs in CD4+CD25+CD226-sorted cells ( Figures 3A–D ). Together, these results demonstrate high purity and lineage stability of CD4+CD25+CD226- Tregs throughout ex vivo expansion.
CD4+CD25+CD226- Tregs Display a More Naïve Phenotype
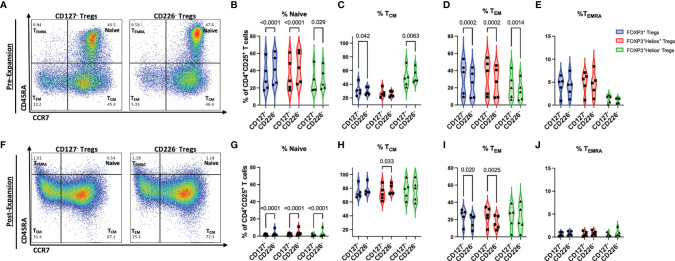
We next sought to assess the extent of differentiation in CD4+CD25+CD226- and CD4+CD25+CD127lo/- Tregs, pre- and post-expansion. Before expansion, CD4+CD25+CD226- Tregs were found to contain significantly more naïve total Tregs (+8.92%, p<0.0001), FOXP3+Helios+ Tregs (+8.58%, p<0.0001), and FOXP3+Helios- Tregs (+2.95%, p=0.029), as well as fewer TCM total Tregs (-1.78%, p=0.042), yet TCM FOXP3+Helios- Treg frequencies were increased versus CD4+CD25+CD127lo/- Tregs (+2.54%, p=0.0063, Figures 4A–C ). Pre-expansion CD4+CD25+CD226- Tregs also comprised fewer TEM total Tregs (-6.97%, p=0.0002), FOXP3+Helios+ Tregs (-6.75%, p=0.0002), FOXP3+Helios- Tregs (-5.21%, p=0.0014), with no significant differences in TEMRA compared to CD4+CD25+CD127lo/- Tregs ( Figures 4D, E ). Additionally, CD4+CD25+CD226- cells represented a significantly greater proportion (1.89-fold, p=0.019) and absolute cell count (p=0.037) within the CD4+CD25+CD127lo/- sorted Treg population, as compared to CD4+CD25+CD127lo/-CD45RA+ cells ( Figure S4 ), suggesting that CD226- enrichment for naïve and TCM cells does not compromise post-sort yield as drastically as a four-marker CD4+CD25+CD127lo/-CD45RA+ naïve Treg isolation strategy (43, 44).
Figure 4.
CD226- Tregs display a more immunoregulatory phenotype than CD127lo/- Tregs. Memory differentiation of Tregs from each FACS method was assessed by CD45RA and CD197 (CCR7) expression (A–E) at day 0 following isolation and (F–J) at day 14 following ex vivo expansion (n = 5 biological with n = 2 technical replicates). (A, F) Representative flow plots pre-gated on live CD4+CD25+ cells show percentages for each T cell memory subset for CD127lo/- sorted Tregs and CD226- sorted Tregs. Relative proportions of (B, G) CD45RA+CCR7+ naïve, (C, H) CD45RA-CCR7+ TCM, (D, I) CD45RA-CCR7- TEM, and (E, J) CD45RA+CCR7- TEMRA subsets from CD127lo/- sorted Treg and CD226- sorted Treg cultures after gating on CD4+CD25+ FOXP3+ Tregs (blue), CD4+CD25+FOXP3+Helios+ Tregs (red), and CD4+CD25+FOXP3+Helios- Tregs (green). P-values are reported on the figure for two-way ANOVA with Bonferroni post-hoc correction for multiple comparisons of Treg isolation conditions from matched subjects.
Post-expansion, higher percentages of remaining naïve Tregs in the total Treg (+1.47%, p<0.0001), FOXP3+Helios+ (+1.79%, p<0.0001), and FOXP3+Helios- (+1.58%, p<0.0001) subpopulations, as well as higher percentages of TCM-differentiated FOXP3+Helios+ Tregs (+4.96%, p=0.033) were observed in the CD4+CD25+CD226- sorted Tregs as compared to CD4+CD25+CD127lo/- sorted cells ( Figures 4F–H ). Additionally, CD4+CD25+CD226- Treg expansion yielded significantly lower percentages of TEM-differentiated total Tregs (-5.63%, p=0.020) and FOXP3+Helios+ Tregs (-8.05%, p=0.0025), with no differences in TEMRA-differentiated Tregs observed post-expansion ( Figures 4I, J ). Collectively, these data support the use of CD4+CD25+CD226- for the isolation of naïve Tregs which differentiate more readily into a TCM as opposed to TEM phenotype after expansion. This finding has potential implications for Treg ACT in settings of autoimmunity and transplantation: specifically, the preferential outgrowth of TCM from CD226- Tregs may lead to better engraftment efficiency and localization to secondary lymphoid organs where autoimmune priming and graft versus host disease (GvHD) are initiated (45).
CD4+CD25+CD226- Tregs Display a More Immunoregulatory Cytokine Profile
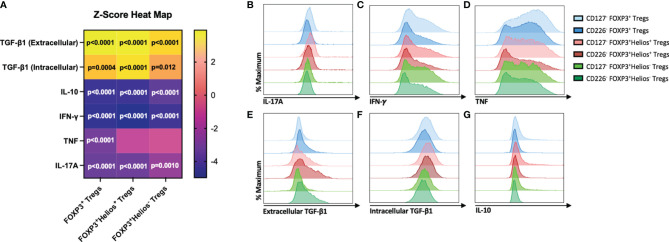
The production of pro-inflammatory cytokines is a hallmark of Treg instability, which may contribute to a loss of immune tolerance in autoimmune disorders (30, 46). Therefore, we sought to determine whether CD4+CD25+CD226- Tregs possess a more immunoregulatory cytokine profile than CD4+CD25+CD127lo/- Tregs. Flow cytometric assessment of cytokine production in unstimulated cells revealed no differences in cytokine production at rest ( Figure S5 ), but following PMA/Ionomycin stimulation, we observed decreased pro-inflammatory IL-17A expression by CD4+CD25+CD226- sorted Tregs within the total Treg (0.92-fold, p<0.0001), FOXP3+Helios+ (0.92-fold, p<0.0001), and FOXP3+Helios- (0.94-fold, p=0.0010) populations ( Figures 5A, B ). Additionally, significant decreases were observed in IFN-γ expression by CD4+CD25+CD226- sorted total Tregs (0.86-fold, p<0.0001), FOXP3+Helios+ (0.85-fold, p<0.0001), and FOXP3+Helios- (0.87-fold, p<0.0001) as well as TNF expression by CD4+CD25+CD226- sorted total Tregs (0.80-fold, p<0.0001), both of which are pro-inflammatory cytokines associated with a Th1 effector profile ( Figures 5A, C, D ) (47, 48). We found significantly increased expression of both extracellular and intracellular TGF-β1 by CD4+CD25+CD226- sorted total Tregs (1.87-fold, p<0.0001; 1.15-fold, p=0.0004, respectively), FOXP3+Helios+ Tregs (1.55-fold, p<0.0001; 1.16-fold, p<0.0001), and FOXP3+Helios- Tregs (1.56-fold, p<0.0001; 1.10-fold, p=0.012) ( Figures 5A, E, F ). Given that TGF-β1 is critical for inhibiting Th1 differentiation (49, 50), these findings corroborate the observations of decreased pro-inflammatory cytokine expression by CD4+CD25+CD226- Tregs. Interestingly, expression of the anti-inflammatory cytokine IL-10 was significantly decreased in CD4+CD25+CD226- sorted total Tregs (0.83-fold, p<0.0001), FOXP3+Helios+ Tregs (0.83-fold, p<0.0001), and FOXP3+Helios- Tregs (0.85-fold, p<0.0001) ( Figures 5A, G ). IL-10, however, is commonly produced by Tr1-like T cells, which differentiate from conventional CD4+ T cells and may only transiently express FOXP3 (2), providing a potential mechanism for its decreased production. This observation is consistent with CD4+CD25+CD226- T cell sorting resulting in a decreased frequency of effector subsets. Overall, these data suggest that CD4+CD25+CD226- Tregs maintain a more immunoregulatory cytokine profile than the CD4+CD25+CD127lo/- counterpart.
Figure 5.
CD226- Tregs exhibit a less inflammatory cytokine profile than CD127lo/- Tregs. Cytokine production by 14-day ex vivo expanded CD127lo/- sorted Treg and CD226- sorted Treg cultures was examined by flow cytometry following a four-hour PMA/Ionomycin stimulation in the presence of a protein transport inhibitor. (A) Heat map shows the mean fold change of protein expression (z-score) of cytokines (rows) by CD226- FOXP3+, FOXP3+Helios+, and FOXP3+Helios- Treg populations (columns) compared to CD127lo/- Treg populations. n = 9 biological with n = 2 technical replicates. Significant P-values are reported on the figure for two-way ANOVA with Bonferroni’s multiple comparisons between Treg isolation conditions of matched subjects. Representative histograms show expression of (B) IL-17A, (C) IFN-γ, (D) TNF, (E) Extracellular TGF-β1, (F) Intracellular TGF-β1, and (G) IL-10 for CD127lo/- (lighter blue) and CD226- CD4+CD25+FOXP3+ Tregs (darker blue), CD127lo/- (lighter red) and CD226- CD4+CD25+FOXP3+Helios+ Tregs (darker red), and CD127lo/- (lighter green) and CD226- CD4+CD25+FOXP3+Helios- Tregs (darker green).
CD4+CD25+CD226- Tregs Present Increased Surface Expression of PD-1 and CD39 Following Activation
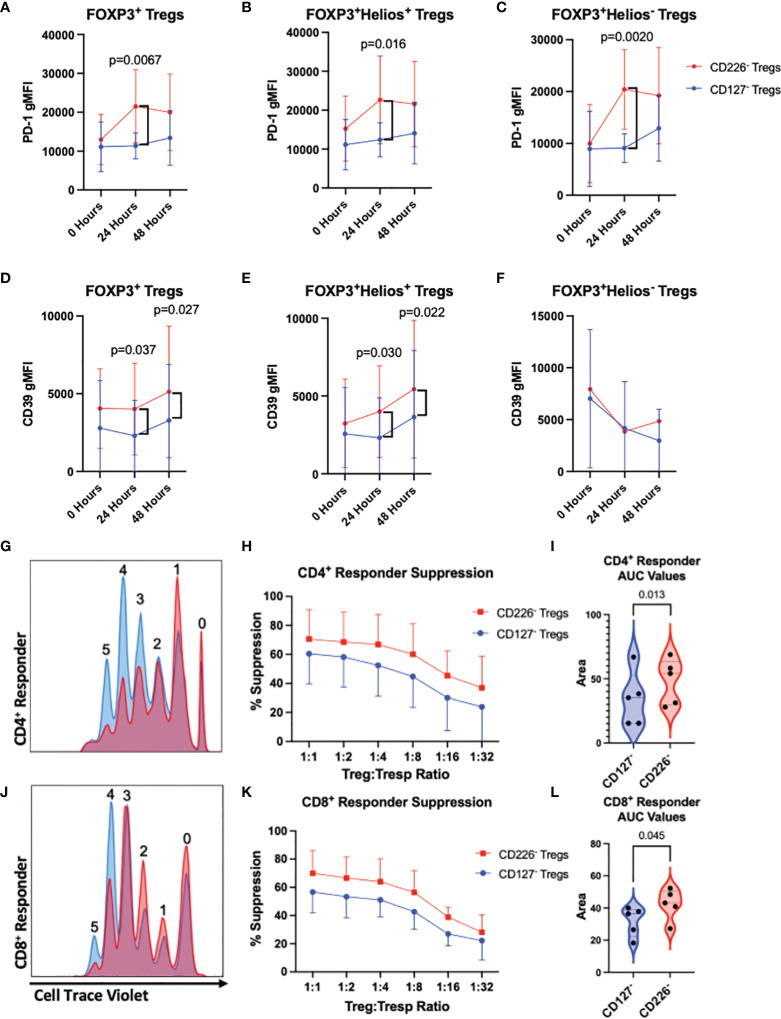
To identify whether CD4+CD25+CD226- sorted Tregs demonstrate differential expression of proteins associated with Treg-mediated suppression, we evaluated PD-1, CD39, CD73, CD40L, GITR, and CTLA-4 by flow cytometry on Tregs expanded from CD226- and CD127lo/- preparations, prior to and following activation by autologous PBMCs. After 24 hours of stimulation, CD226- sorted Tregs displayed increased expression of PD-1 on both total FOXP3+ Tregs (1.89-fold, p=0.0067) as well as within the FOXP3+Helios+ Treg (1.82-fold, p=0.016) and FOXP3+Helios- Treg subsets (2.24-fold, p=0.0020) as compared to CD127lo/- sorted counterparts; however, there were no significant differences observed at baseline or after 48 hours of stimulation ( Figures 6A–C ). Furthermore, CD226- Tregs exhibited significantly increased expression of CD39 at 24 and 48 hours in both total Tregs (1.75-fold, p=0.037; 1.56-fold, p=0.027, respectively) and the FOXP3+Helios+ subset (1.73-fold, p=0.030; 1.47-fold, p=0.022, respectively) compared to CD127lo/- Tregs ( Figures 6D–F ). Compared to CD127lo/- sorted Tregs, CD226- sorted Tregs did not exhibit any significant differences in surface expression of CD73, CD40L, GITR, or CTLA-4, including after stimulation ( Figure S6 ). Taken together, these data suggest that CD226- Tregs may exhibit enhanced suppression of responder T cells via the PD-1/PD-L1 and CD39/CD73 ectonucleotidase pathways (51, 52).
Figure 6.
CD226- Tregs demonstrate an increased suppressive phenotype and ex vivo suppressive capabilities as compared to CD127lo/- Tregs. (A–C) PD-1 and (D–F) CD39 expression was assessed by flow cytometry on total FOXP3+ Tregs (A, D), FOXP3+Helios+ Tregs (B, E), and FOXP3+Helios- Tregs (C, F) from 14-day ex vivo expanded CD127lo/- sorted Tregs versus CD226- Tregs following co-culture with autologous PBMCs in the presence of soluble α-CD3 and α-CD28 for 0, 24, or 48 hours. n = 5 biological replicates. Significant P-values reported on the figure for two-way ANOVA with Bonferroni’s multiple comparison between Treg isolation conditions of matched subjects. CD127lo/- sorted Treg (blue) and CD226- sorted Treg cultures (red) were by expanded for 14-days ex vivo, then labeled with Cell Proliferation Dye eFluor670 and co-cultured in decreasing two-fold dilutions with CellTrace Violet-labeled autologous responder PBMCs in the presence of soluble α-CD3 and α-CD28 antibodies for four days before flow cytometric assessment of (G–I) CD4+ and (J–L) CD8+ T cell proliferation. (G, J) Representative dye dilution plots demonstrating suppression of responders. Percent suppression of (H) CD4+ and (K) CD8+ responders was quantified by the division index method, and comparisons between FACS conditions were made using (I, L) area under the curve (AUC) values for each percent suppression curve (40, 41). Data reflects n = 5 biological with n = 3 technical replicates. Significant P-values are reported on the figure for paired T-tests comparing Treg isolation conditions from matched subjects.
CD4+CD25+CD226- Tregs Demonstrate Increased Ex Vivo Suppressive Capabilities
During expansion, Treg cultures can be prone to lineage instability as well as outgrowth of Tconv contaminants, ultimately impacting therapeutic potential by reducing suppressive capabilities (53). Given that CD4+CD25+CD226- sorted Tregs exhibited a greater frequency of lineage stable FOXP3+Helios+ Tregs than CD4+CD25+CD127lo/- Tregs, corroborated by epigenetic, differentiation and cytokine profile data, we sought to understand how sorting method might impact suppressive capacity. To accomplish this, we conducted dual-color ex vivo suppression assays using serial dilutions of expanded Tregs (sorted via CD4+CD25+CD127lo/- or CD4+CD25+CD226-) with autologous whole PBMC responder cells. CD226- sorted Tregs exhibited significantly increased suppression of both CD4+ and CD8+ effector T cell subsets, as demonstrated by decreased division indices for CTV labeled responder cells, as compared to those observed in CD127lo/- sorted Treg cocultures ( Figures 6G–L ). Importantly, we did not observe dilution of CPD in either Treg population, suggesting that differences in suppression were not due to further expansion of CD226- sorted Tregs. These data suggest that isolation of CD4+CD25+CD226- Tregs yields a more suppressive Treg population that may improve ACT efficacy compared to conventional CD4+CD25+CD127lo/- Tregs.
Discussion
The requirement for long-term stability following engraftment in Treg-ACT necessitates the use of a combination of surface markers that function as a surrogate for the tTreg lineage-defining transcription factors. In this study, we compared the commonly employed Treg FACS isolation method using the CD4+CD25+CD127lo/- marker profile to an alternative CD4+CD25+CD226- approach. Importantly, sorted cells were assessed both pre- and post-expansion with the latter rested for 7 days prior to phenotypic, epigenetic, and functional characterization in order to mitigate transient upregulation of Helios in pTreg and contaminating Tconv (54) as a potential confounding factor. The resulting data demonstrate that isolation of CD4+CD25+CD226- Tregs yields a greater frequency of lineage stable FOXP3+Helios+ Tregs, a reduced percentage of FOXP3+Helios- Tregs, and increased demethylation at the FOXP3 TSDR compared to CD4+CD25+CD127lo/- Tregs. Among total Treg, FOXP3+Helios+ Treg and FOXP3+Helios- Treg subsets, increased expression of CD25 was observed on CD4+CD25+CD226- sorted cells without compromising the low CD127 expression typically achieved using the CD4+CD25+CD127lo/- strategy. Together, this suggests that CD4+CD25+CD226- Tregs may have a higher avidity for IL-2, which is putatively reported to result in downstream pSTAT5-signaling to reinforce lineage stability, fitness, and function (55–57). While the increase in purity we observed is modest relative to CD4+CD25+CD127lo/- Tregs, we expect that these small improvements in initial purity may have a significant biological impact on the long-term survival and stability of a transferred population in ACT applications (58).
It is important to note that the data herein were derived from PBMC samples from healthy subjects (i.e., the general population). We speculate that the differences observed between CD4+CD25+CD127- and CD4+CD25+CD226- Tregs may be more prominent in autoimmune subjects, particularly during periods of acute inflammation where IL-2R signaling defects have been observed (59–61). This concept is critical when considering the transfer of islet antigen-specific Tregs created using genetically-modified T cell receptors (TCR) or chimeric antigen receptors (CAR) that could potentially become directly pathogenic toward islets and/or β-cells in situations of Treg instability (62–64).
To further characterize CD4+CD25+CD226- Tregs, we assessed T cell memory differentiation markers and found increased proportions of naïve Tregs both before and after expansion, along with reduced proportions of effector memory Tregs post-expansion as compared to the traditional CD4+CD25+CD127lo/- strategy. These results suggest that CD4+CD25+CD226- may not only serve as a better set of markers to identify lineage stable Tregs but also, to avoid effector contaminants. This notion is supported by prior work by Hoffmann and colleagues who initially demonstrated that CD45RA+ naïve Tregs displayed increased stability upon expansion as compared to CD45RA- memory Tregs (65), a finding that we have consistently replicated from both umbilical cord and adult peripheral blood samples (13, 28, 66). Previous studies have identified the marker profile CD4+CD25+CD127lo/-CD45RA+ as selecting for predominately naïve tTregs; however, this isolation method yields a much smaller population than required for many ACT applications (43, 44). While our three-marker sorting strategy enriches for naïve Tregs, it also captures CD4+CD25+CD226- memory Tregs, resulting in a greater FACS yield compared to a CD4+CD25+CD127lo/-CD45RA+ strategy. Hence, the CD4+CD25+CD226- sorting strategy strikes a practical balance between the desire to enrich for naïve Tregs versus CD4+CD25+CD127lo/- isolation but also, maximize cell yield as compared to CD4+CD25+CD127lo/-CD45RA+ isolation. Similarly, while TIGIT has been identified as a marker of lineage stable tTregs, we previously reported that TIGIT+ cells had a limited expansion capacity and therefore, would not produce enough Tregs for ACT (30, 31). Importantly, we observed a significantly lower proportion of TEM-differentiated Tregs and a higher proportion of TCM-differentiated Tregs expanded ex vivo from the CD226- Treg population. This observation suggests that CD4+CD25+CD226- Tregs may not only be longer-lived, but potentially, localize more readily in secondary lymphoid organs (17). This remains a critical issue for Treg-ACT applications in the context of T1D, as Tregs must be able to migrate to sites of inflammation and priming, specifically to the pancreatic draining lymph nodes where recent studies have revealed the presence of a stem-cell like CD8+ T cell progenitor population that significantly contributes to pancreatic β-cell destruction in the NOD model of T1D (67).
During our assessment of the therapeutic potential of this CD4+CD25+CD226- Treg subset, we found decreased expression of the pro-inflammatory cytokines IL-17A, TNF, and IFN-γ, associated with Th17 and Th1 responses, in comparison to CD4+CD25+CD127lo/- Tregs following PMA/Ionomycin stimulation. This finding is especially important in the context of autoimmunity, as Th1 and Th17 effectors have been associated with several autoimmune diseases, including T1D and MS, suggesting that CD4+CD25+CD226- Treg isolation may potentially deplete these pathogenic Tregs and present reduced risk of pro-inflammatory ex-Treg outgrowth compared to CD4+CD25+CD127lo/- Treg isolation (68–70). Beyond this, PMA/Ionomycin stimulated CD226- sorted Tregs had increased intracellular and surface expression of the immunoregulatory molecule, TGF-β1 (71). Following TCR activation, CD226- Tregs had increased expression of the immunoregulatory checkpoint molecules, PD-1 and CD39, which are associated with programmed cell death and ATP hydrolysis, respectively (52, 72). Inhibition or dysregulation of these checkpoint regulators has been associated with the development of autoreactivity (73, 74). Finally, given our observation that CD226- sorted Tregs had higher CD25 expression post-expansion, it is possible that the augmented suppressive capacity observed could, at least in part, be related to increased competition for IL-2. Altogether, our data support the notion that differences in IL-2 consumption, cytokine production, and contact-dependent mechanisms may all contribute toward the increased level of suppression observed with CD226- versus CD127lo/- sorted Tregs.
Tregs are emerging as a powerful therapeutic modality in a broad array of autoimmune settings (75, 76). While our study supports CD226- Tregs as a robust population to yield stable FOXP3+Helios+ Tregs, we note the limitation that our studies were conducted using general population control samples. Further research is needed to determine if CD226- Tregs will provide increased purity and stability in patients with active autoimmune disease. Indeed, there are a number of outstanding questions regarding the TCR repertoire, homing receptors, and in vivo trafficking of CD226- Tregs relative to CD127lo/- Tregs (77).
These findings also raise a number of considerations regarding ACT with CD226- Tregs and therapeutic targeting of the CD226 pathway in situations of autoimmunity. On one hand, our data related to CD226 being highly expressed on effector T cells supports additional studies targeting this pathway in vivo to block destructive autoimmunity. However, this approach should be taken with some caution, as we note that CD226 is also highly expressed by IL-10-secreting Tr1-like T cells (78). Thus, any immunotherapy seeking to inhibit CD226 on Tregs would likely need to be carefully dosed to increase Treg stability without compromising CD226-mediated Tr-1 like T cell function. Furthermore, these results support the continued investigation of the precise mechanisms by which reduced CD226 expression allows for increased Treg lineage stability and suppressive capacity. We note that additional studies are currently underway in our laboratory to assess the impact of CD226 on Tregs using both targeted biologics, along with global and conditional knockout approaches in animal models (37), as well as through gene targeting approaches in human Tregs. In summary, our findings present a novel method to generate a highly stable and suppressive Treg subset for use in ACT by initially eliminating Tregs expressing the costimulatory molecule, CD226.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
MB: writing – original draft, writing – review and editing, formal analysis, visualization, validation, investigation, project administration, and supervision. LP: investigation, writing – review and editing, and funding acquisition. SH, JA, LS, KN, and EC: investigation and writing – review and editing. HS, CF, and AP: writing - review and editing. MS: writing - review and editing and funding acquisition. TB: conceptualization, writing – review and editing, funding acquisition, project administration, and supervision. TB is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to the article and approved the submitted version.
Funding
Funding was provided by the National Institutes of Health through the support of grants to LDP (T32 DK108736) and to TMB (R01 DK106191, HIRN UG3/UH3 DK122638, P01 AI042288). Additional programmatic support was provided by Diabetes Research Connection to MS (DRC Project 45) and programmatic support by Leona M. and Harry B. The Helmsley Charitable Trust.
Conflict of Interest
Author HS was employed in ROSALIND, Inc. Author CF was employed in NanoString Technologies, Inc. Authors TB, HS, and CF share intellectual property related to the use of CD226- Tregs for the treatment of autoimmune diseases.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the blood donors who participated in these studies. We thank members of the Brusko Laboratory at the University of Florida Diabetes Institute for discussions and technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.873560/full#supplementary-material
References
- 1. Tang Q, Bluestone JA. The Foxp3+ Regulatory T Cell: A Jack of All Trades, Master of Regulation. Nat Immunol (2008) 9(3):239–44. doi: 10.1038/ni1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roncarolo MG, Gregori S, Bacchetta R, Battaglia M, Gagliani N. The Biology of T Regulatory Type 1 Cells and Their Therapeutic Application in Immune-Mediated Diseases. Immunity (2018) 49(6):1004–19. doi: 10.1016/j.immuni.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 3. Akkaya B, Oya Y, Akkaya M, Al Souz J, Holstein AH, Kamenyeva O, et al. Regulatory T Cells Mediate Specific Suppression by Depleting Peptide-MHC Class II From Dendritic Cells. Nat Immunol (2019) 20(2):218–31. doi: 10.1038/s41590-018-0280-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hull CM, Peakman M, Tree TIM. Regulatory T Cell Dysfunction in Type 1 Diabetes: What's Broken and How can We Fix It? Diabetologia (2017) 60(10):1839–50. doi: 10.1007/s00125-017-4377-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buckner JH. Mechanisms of Impaired Regulation by CD4(+)CD25(+)FOXP3(+) Regulatory T Cells in Human Autoimmune Diseases. Nat Rev Immunol (2010) 10(12):849–59. doi: 10.1038/nri2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bluestone JA, Buckner JH, Fitch M, Gitelman SE, Gupta S, Hellerstein MK, et al. Type 1 Diabetes Immunotherapy Using Polyclonal Regulatory T Cells. Sci Transl Med (2015) 7(315):315ra189. doi: 10.1126/scitranslmed.aad4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dall'Era M, Pauli ML, Remedios K, Taravati K, Sandova PM, Putnam AL, et al. Adoptive Treg Cell Therapy in a Patient With Systemic Lupus Erythematosus. Arthritis Rheumatol (2019) 71(3):431–40. doi: 10.1002/art.40737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tarbell KV, Yamazaki S, Olson K, Toy P, Steinman RM. CD25+ CD4+ T Cells, Expanded With Dendritic Cells Presenting a Single Autoantigenic Peptide, Suppress Autoimmune Diabetes. J Exp Med (2004) 199(11):1467–77. doi: 10.1084/jem.20040180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tonkin DR, He J, Barbour G, Haskins K. Regulatory T Cells Prevent Transfer of Type 1 Diabetes in NOD Mice Only When Their Antigen is Present In Vivo. J Immunol (2008) 181(7):4516–22. doi: 10.4049/jimmunol.181.7.4516 [DOI] [PubMed] [Google Scholar]
- 10. Tang Q, Henriksen KJ, Bi M, Finger EB, Szot G, Ye J, et al. In Vitro-Expanded Antigen-Specific Regulatory T Cells Suppress Autoimmune Diabetes. J Exp Med (2004) 199(11):1455–65. doi: 10.1084/jem.20040139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marek-Trzonkowska N, Myśliwec M, Siebert J, Trzonkowski P. Clinical Application of Regulatory T Cells in Type 1 Diabetes. Pediatr Diabetes (2013) 14(5):322–32. doi: 10.1111/pedi.12029 [DOI] [PubMed] [Google Scholar]
- 12. Brusko TM, Koya RC, Zhu S, Lee MR, Putnam AL, McClymont SA, et al. Human Antigen-Specific Regulatory T Cells Generated by T Cell Receptor Gene Transfer. PloS One (2010) 5(7):e11726. doi: 10.1371/journal.pone.0011726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Seay HR, Putnam AL, Cserny J, Posgai AL, Rosenau EH, Wingard JR, et al. Expansion of Human Tregs From Cryopreserved Umbilical Cord Blood for GMP-Compliant Autologous Adoptive Cell Transfer Therapy. Mol Ther Methods Clin Dev (2017) 4:178–91. doi: 10.1016/j.omtm.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Putnam AL, Brusko TM, Lee MR, Liu W, Szot GL, Ghosh T, et al. Expansion of Human Regulatory T-Cells From Patients With Type 1 Diabetes. Diabetes (2009) 58(3):652–62. doi: 10.2337/db08-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rouxel O, Da Silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, et al. Cytotoxic and Regulatory Roles of Mucosal-Associated Invariant T Cells in Type 1 Diabetes. Nat Immunol (2017) 18(12):1321–31. doi: 10.1038/ni.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 Regulates FOXP3 Expression in Human CD4+CD25+ Regulatory T Cells Through a STAT-Dependent Mechanism and Induces the Expansion of These Cells In Vivo . Blood (2006) 108(5):1571–9. doi: 10.1182/blood-2006-02-004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenblum MD, Way SS, Abbas AK. Regulatory T Cell Memory. Nat Rev Immunol (2016) 16(2):90–101. doi: 10.1038/nri.2015.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T Cell Lineage Specification by the Forkhead Transcription Factor Foxp3. Immunity (2005) 22(3):329–41. doi: 10.1016/j.immuni.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 19. Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, et al. CD127 Expression Inversely Correlates With FoxP3 and Suppressive Function of Human CD4+ T Reg Cells. J Exp Med (2006) 203(7):1701–11. doi: 10.1084/jem.20060772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ukena SN, Höpting M, Velaga S, Ivanyi P, Grosse J, Baron U, et al. Isolation Strategies of Regulatory T Cells for Clinical Trials: Phenotype, Function, Stability, and Expansion Capacity. Exp Hematol (2011) 39(12):1152–60. doi: 10.1016/j.exphem.2011.08.010 [DOI] [PubMed] [Google Scholar]
- 21. Ash S, Yarkoni S, Askenasy N. Lymphopenia is Detrimental to Therapeutic Approaches to Type 1 Diabetes Using Regulatory T Cells. Immunol Res (2014) 58(1):101–5. doi: 10.1007/s12026-013-8476-x [DOI] [PubMed] [Google Scholar]
- 22. Puronen CE, Thompson WL, Imamichi H, Beq S, Hodge JN, Rehm C, et al. Decreased Interleukin 7 Responsiveness of T Lymphocytes in Patients With Idiopathic CD4 Lymphopenia. J Infect Dis (2012) 205(9):1382–90. doi: 10.1093/infdis/jis219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schulze-Koops H. Lymphopenia and Autoimmune Diseases. Arthritis Res Ther (2004) 6(4):178–80. doi: 10.1186/ar1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shevyrev D, Tereshchenko V. Treg Heterogeneity, Function, and Homeostasis. Front Immunol (2019) 10:3100. doi: 10.3389/fimmu.2019.03100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lam AJ, Uday P, Gillies JK, Levings MK. Helios Is a Marker, Not a Driver, of Human Treg Stability. Eur J Immunol (2022) 52(1):75–84. doi: 10.1002/eji.202149318 [DOI] [PubMed] [Google Scholar]
- 26. Elkord E. Helios Should Not Be Cited as a Marker of Human Thymus-Derived Tregs. Commentary: Helios(+) and Helios(-) Cells Coexist Within the Natural FOXP3(+) T Regulatory Cell Subset in Humans. Front Immunol (2016) 7:276. doi: 10.3389/fimmu.2016.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yates K, Bi K, Haining WN, Cantor H, Kim HJ. Comparative Transcriptome Analysis Reveals Distinct Genetic Modules Associated With Helios Expression in Intratumoral Regulatory T Cells. Proc Natl Acad Sci USA (2018) 115(9):2162–7. doi: 10.1073/pnas.1720447115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McClymont SA, Putnam AL, Lee MR, Esensten JH, Liu W, Hulme MA, et al. Plasticity of Human Regulatory T Cells in Healthy Subjects and Patients With Type 1 Diabetes. J Immunol (2011) 186(7):3918–26. doi: 10.4049/jimmunol.1003099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giganti G, Atif M, Mohseni Y, Mastronicola D, Grageda N, Povoleri GA, et al. Treg Cell Therapy: How Cell Heterogeneity can Make the Difference. Eur J Immunol (2021) 51(1):39–55. doi: 10.1002/eji.201948131 [DOI] [PubMed] [Google Scholar]
- 30. Fuhrman CA, Yeh WI, Seay HR, Saikumar Lakshmi P, Chopra G, Zhang L, et al. Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226. J Immunol (2015) 195(1):145–55. doi: 10.4049/jimmunol.1402381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lozano E, Joller N, Cao Y, Kuchroo VK, Hafler DA. The CD226/CD155 Interaction Regulates the Proinflammatory (Th1/Th17)/Anti-Inflammatory (Th2) Balance in Humans. J Immunol (2013) 191(7):3673–80. doi: 10.4049/jimmunol.1300945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shapiro MR, Thirawatananond P, Peters L, Sharp RC, Ogundare S, Posgai AL, et al. De-Coding Genetic Risk Variants in Type 1 Diabetes. Immunol Cell Biol (2021) 99(5):496–508. doi: 10.1111/imcb.12438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, et al. (CD155) and Nectin-2 (PRR-2/Cd112). Int Immunol (2004) 16(4):533–8. doi: 10.1093/intimm/dxh059 [DOI] [PubMed] [Google Scholar]
- 34. Qiu ZX, Zhang K, Qiu XS, Zhou M, Li WM. CD226 Gly307Ser Association With Multiple Autoimmune Diseases: A Meta-Analysis. Hum Immunol (2013) 74(2):249–55. doi: 10.1016/j.humimm.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 35. Mattana TC, Santos AS, Fukui RT, Mainardi-Novo DT, Costa VS, Santos RF, et al. CD226 Rs763361 Is Associated With the Susceptibility to Type 1 Diabetes and Greater Frequency of GAD65 Autoantibody in a Brazilian Cohort. Mediators Inflamm (2014) 2014:694948. doi: 10.1155/2014/694948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. (IMSGC) IMSGC . The Expanding Genetic Overlap Between Multiple Sclerosis and Type I Diabetes. Genes Immun (2009) 10(1):11–4. doi: 10.1038/gene.2008.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shapiro MR, Yeh WI, Longfield JR, Gallagher J, Infante CM, Wellford S, et al. CD226 Deletion Reduces Type 1 Diabetes in the NOD Mouse by Impairing Thymocyte Development and Peripheral T Cell Activation. Front Immunol (2020) 11:2180. doi: 10.3389/fimmu.2020.02180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang N, Yi H, Fang L, Jin J, Ma Q, Shen Y, et al. CD226 Attenuates Treg Proliferation via Akt and Erk Signaling in an EAE Model. Front Immunol (2020) 11:1883. doi: 10.3389/fimmu.2020.01883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huehn J, Polansky JK, Hamann A. Epigenetic Control of FOXP3 Expression: The Key to a Stable Regulatory T-Cell Lineage? Nat Rev Immunol (2009) 9(2):83–9. doi: 10.1038/nri2474 [DOI] [PubMed] [Google Scholar]
- 40. Roederer M. Interpretation of Cellular Proliferation Data: Avoid the Panglossian. Cytometry A (2011) 79(2):95–101. doi: 10.1002/cyto.a.21010 [DOI] [PubMed] [Google Scholar]
- 41. Akimova T, Levine MH, Beier UH, Hancock WW. Standardization, Evaluation, and Area-Under-Curve Analysis of Human and Murine Treg Suppressive Function. Methods Mol Biol (2016) 1371:43–78. doi: 10.1007/978-1-4939-3139-2_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chauvin JM, Zarour HM. TIGIT in Cancer Immunotherapy. J Immunother Cancer (2020) 8(2):e000957. doi: 10.1136/jitc-2020-000957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arroyo Hornero R, Betts GJ, Sawitzki B, Vogt K, Harden PN, Wood KJ. CD45RA Distinguishes CD4+CD25+CD127-/Low TSDR Demethylated Regulatory T Cell Subpopulations With Differential Stability and Susceptibility to Tacrolimus-Mediated Inhibition of Suppression. Transplantation (2017) 101(2):302–9. doi: 10.1097/TP.0000000000001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lei H, Kuchenbecker L, Streitz M, Sawitzki B, Vogt K, Landwehr-Kenzel S, et al. Human CD45RA(-) FoxP3(hi) Memory-Type Regulatory T Cells Show Distinct TCR Repertoires With Conventional T Cells and Play an Important Role in Controlling Early Immune Activation. Am J Transplant (2015) 15(10):2625–35. doi: 10.1111/ajt.13315 [DOI] [PubMed] [Google Scholar]
- 45. Liu T, Zhang D, Zhang Y, Xu X, Zhou B, Fang L, et al. Blocking CD226 Promotes Allogeneic Transplant Immune Tolerance and Improves Skin Graft Survival by Increasing the Frequency of Regulatory T Cells in a Murine Model. Cell Physiol Biochem (2018) 45(6):2338–50. doi: 10.1159/000488182 [DOI] [PubMed] [Google Scholar]
- 46. Kannan AK, Su Z, Gauvin DM, Paulsboe SE, Duggan R, Lasko LM, et al. IL-23 Induces Regulatory T Cell Plasticity With Implications for Inflammatory Skin Diseases. Sci Rep (2019) 9(1):17675. doi: 10.1038/s41598-019-53240-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Almawi WY, Tamim H, Azar ST. Clinical Review 103: T Helper Type 1 and 2 Cytokines Mediate the Onset and Progression of Type I (Insulin-Dependent) Diabetes. J Clin Endocrinol Metab (1999) 84(5):1497–502. doi: 10.1210/jcem.84.5.5699 [DOI] [PubMed] [Google Scholar]
- 48. Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM, et al. Autoreactive T Cell Responses Show Proinflammatory Polarization in Diabetes But a Regulatory Phenotype in Health. J Clin Invest (2004) 113(3):451–63. doi: 10.1172/JCI19585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sad S, Mosmann TR. Single IL-2-Secreting Precursor CD4 T Cell can Develop Into Either Th1 or Th2 Cytokine Secretion Phenotype. J Immunol (1994) 153(8):3514–22. [PubMed] [Google Scholar]
- 50. Ishigame H, Zenewicz LA, Sanjabi S, Licona-Limón P, Nakayama M, Leonard WJ, et al. Excessive Th1 Responses Due to the Absence of TGF-β Signaling Cause Autoimmune Diabetes and Dysregulated Treg Cell Homeostasis. Proc Natl Acad Sci U S A (2013) 110(17):6961–6. doi: 10.1073/pnas.1304498110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gu J, Ni X, Pan X, Lu H, Lu Y, Zhao J, et al. Human CD39hi Regulatory T Cells Present Stronger Stability and Function Under Inflammatory Conditions. Cell Mol Immunol (2017) 14(6):521–8. doi: 10.1038/cmi.2016.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Francisco LM, Sage PT, Sharpe AH. The PD-1 Pathway in Tolerance and Autoimmunity. Immunol Rev (2010) 236:219–42. doi: 10.1111/j.1600-065X.2010.00923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Baeten P, Van Zeebroeck L, Kleinewietfeld M, Hellings N, Broux B. Improving the Efficacy of Regulatory T Cell Therapy. Clin Rev Allergy Immunol (2021) 62:363–81. doi: 10.1007/s12016-021-08866-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios Expression is a Marker of T Cell Activation and Proliferation. PloS One (2011) 6(8):e24226. doi: 10.1371/journal.pone.0024226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Toomer KH, Lui JB, Altman NH, Ban Y, Chen X, Malek TR. Essential and non-Overlapping IL-2rα-Dependent Processes for Thymic Development and Peripheral Homeostasis of Regulatory T Cells. Nat Commun (2019) 10(1):1037. doi: 10.1038/s41467-019-08960-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 Programs the Development and Function of CD4+CD25+ Regulatory T Cells. Nat Immunol (2003) 4(4):330–6. doi: 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- 57. Passerini L, Allan SE, Battaglia M, Di Nunzio S, Alstad AN, Levings MK, et al. STAT5-Signaling Cytokines Regulate the Expression of FOXP3 in CD4+CD25+ Regulatory T Cells and CD4+CD25- Effector T Cells. Int Immunol (2008) 20(3):421–31. doi: 10.1093/intimm/dxn002 [DOI] [PubMed] [Google Scholar]
- 58. Marshall GP, Cserny J, Perry DJ, Yeh WI, Seay HR, Elsayed AG, et al. Clinical Applications of Regulatory T Cells in Adoptive Cell Therapies. Cell Gene Ther Insights (2018) 4(1):405–29. doi: 10.18609/cgti.2018.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sobel ES, Brusko TM, Butfiloski EJ, Hou W, Li S, Cuda CM, et al. Defective Response of CD4(+) T Cells to Retinoic Acid and Tgfβ in Systemic Lupus Erythematosus. Arthritis Res Ther (2011) 13(3):R106. doi: 10.1186/ar3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brusko TM, Wasserfall CH, Clare-Salzler MJ, Schatz DA, Atkinson MA. Functional Defects and the Influence of Age on the Frequency of CD4+ CD25+ T-Cells in Type 1 Diabetes. Diabetes (2005) 54(5):1407–14. doi: 10.2337/diabetes.54.5.1407 [DOI] [PubMed] [Google Scholar]
- 61. Brusko TM, Wasserfall CH, Hulme MA, Cabrera R, Schatz D, Atkinson MA. Influence of Membrane CD25 Stability on T Lymphocyte Activity: Implications for Immunoregulation. PloS One (2009) 4(11):e7980. doi: 10.1371/journal.pone.0007980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB Costimulation Ameliorates T Cell Exhaustion Induced by Tonic Signaling of Chimeric Antigen Receptors. Nat Med (2015) 21(6):581–90. doi: 10.1038/nm.3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weber EW, Parker KR, Sotillo E, Lynn RC, Anbunathan H, Lattin J, et al. Transient Rest Restores Functionality in Exhausted CAR-T Cells Through Epigenetic Remodeling. Science (2021) 372(6537):eaba1786. doi: 10.1126/science.aba1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cabello-Kindelan C, Mackey S, Sands A, Rodriguez J, Vazquez C, Pugliese A, et al. Immunomodulation Followed by Antigen-Specific T. Diabetes (2020) 69(2):215–27. doi: 10.2337/db19-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hoffmann P, Eder R, Boeld TJ, Doser K, Piseshka B, Andreesen R, et al. Only the CD45RA+ Subpopulation of CD4+CD25high T Cells Gives Rise to Homogeneous Regulatory T-Cell Lines Upon In Vitro Expansion. Blood (2006) 108(13):4260–7. doi: 10.1182/blood-2006-06-027409 [DOI] [PubMed] [Google Scholar]
- 66. Motwani K, Peters LD, Vliegen WH, El-Sayed AG, Seay HR, Lopez MC, et al. Human Regulatory T Cells From Umbilical Cord Blood Display Increased Repertoire Diversity and Lineage Stability Relative to Adult Peripheral Blood. Front Immunol (2020) 11:611. doi: 10.3389/fimmu.2020.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gearty SV, Dündar F, Zumbo P, Espinosa-Carrasco G, Shakiba M, Sanchez-Rivera FJ, et al. An Autoimmune Stem-Like CD8 T Cell Population Drives Type 1 Diabetes. Nature (2022) 602(7895):156–61. doi: 10.1038/s41586-021-04248-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sawant DV, Vignali DA. Once a Treg, Always a Treg? Immunol Rev (2014) 259(1):173–91. doi: 10.1111/imr.12173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Y, Liu Y, Chu CQ. Th17 Cells in Type 1 Diabetes: Role in the Pathogenesis and Regulation by Gut Microbiome. Mediators Inflamm (2015) 2015:638470. doi: 10.1155/2015/638470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jung MK, Kwak JE, Shin EC. IL-17a-Producing Foxp3 + Regulatory T Cells and Human Diseases. Immune Netw (2017) 17(5):276–86. doi: 10.4110/in.2017.17.5.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bommireddy R, Doetschman T. TGFbeta1 and Treg Cells: Alliance for Tolerance. Trends Mol Med (2007) 13(11):492–501. doi: 10.1016/j.molmed.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, Giometto R, et al. Expression of Ectonucleotidase CD39 by Foxp3+ Treg Cells: Hydrolysis of Extracellular ATP and Immune Suppression. Blood (2007) 110(4):1225–32. doi: 10.1182/blood-2006-12-064527 [DOI] [PubMed] [Google Scholar]
- 73. Fletcher JM, Lonergan R, Costelloe L, Kinsella K, Moran B, O'Farrelly C, et al. CD39+Foxp3+ Regulatory T Cells Suppress Pathogenic Th17 Cells and are Impaired in Multiple Sclerosis. J Immunol (2009) 183(11):7602–10. doi: 10.4049/jimmunol.0901881 [DOI] [PubMed] [Google Scholar]
- 74. Granados HM, Draghi A, Tsurutani N, Wright K, Fernandez ML, Sylvester FA, et al. Programmed Cell Death-1, PD-1, Is Dysregulated in T Cells From Children With New Onset Type 1 Diabetes. PloS One (2017) 12(9):e0183887. doi: 10.1371/journal.pone.0183887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Romano M, Fanelli G, Albany CJ, Giganti G, Lombardi G. Past, Present, and Future of Regulatory T Cell Therapy in Transplantation and Autoimmunity. Front Immunol (2019) 10:43. doi: 10.3389/fimmu.2019.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brusko TM, Putnam AL, Bluestone JA. Human Regulatory T Cells: Role in Autoimmune Disease and Therapeutic Opportunities. Immunol Rev (2008) 223:371–90. doi: 10.1111/j.1600-065X.2008.00637.x [DOI] [PubMed] [Google Scholar]
- 77. Shevach EM. Foxp3+ T Regulatory Cells: Still Many Unanswered Questions—A Perspective After 20 Years of Study. Front Immunol (2018) 9:1048. doi: 10.3389/fimmu.2018.01048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gregori S, Roncarolo MG. Engineered T Regulatory Type 1 Cells for Clinical Application. Front Immunol (2018) 9:233. doi: 10.3389/fimmu.2018.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.