Abstract
In medium supplemented with chondroitin sulfate, Flavobacterium heparinum synthesizes and exports two chondroitinases, chondroitinase AC (chondroitin AC lyase; EC 4.2.2.5) and chondroitinase B (chondroitin B lyase; no EC number), into its periplasmic space. Chondroitinase AC preferentially depolymerizes chondroitin sulfates A and C, whereas chondroitinase B degrades only dermatan sulfate (chondroitin sulfate B). The genes coding for both enzymes were isolated from F. heparinum and designated cslA (chondroitinase AC) and cslB (chondroitinase B). They were found to be separated by 5.5 kb on the chromosome of F. heparinum, transcribed in the same orientation, but not linked to any of the heparinase genes. In addition, the synthesis of both enzymes appeared to be coregulated. The cslA and cslB DNA sequences revealed open reading frames of 2,103 and 1,521 bp coding for peptides of 700 and 506 amino acid residues, respectively. Chondroitinase AC has a signal sequence of 22 residues, while chondroitinase B is composed of 25 residues. The mature forms of chondroitinases AC and B are comprised of 678 and 481 amino acid residues and have calculated molecular masses of 77,169 and 53,563 Da, respectively. Truncated cslA and cslB genes have been used to produce active, mature chondroitinases in the cytoplasm of Escherichia coli. Partially purified recombinant chondroitinases AC and B exhibit specific activities similar to those of chondroitinases AC and B from F. heparinum.
Glycosaminoglycans (GAGs) are a group of linear heterogeneous polysaccharides made up of repeating disaccharide units of glucosamine or galactosamine and uronic acid (3). They are divided into four main groups: chondroitin sulfates, including dermatan sulfate (chondroitin sulfate B); heparin and heparan sulfate; hyaluronic acid; and keratan sulfate (in which the uronic acid is replaced by galactose). As the carbohydrate components of proteoglycans, they are thought to be involved in the regulation of various cellular processes such as adhesion, differentiation, migration, and proliferation (3, 15, 16). Chondroitin sulfates are the most common forms of GAGs found in proteoglycans.
Flavobacterium heparinum (also classified as Cytophaga heparina [2], Sphingobacterium heparinum [41], and Pedobacter heparinus [39]), a gram-negative, nonpathogenic soil bacterium isolated by Payza and Korn (29), synthesizes five GAG lyases: three heparinases and two chondroitinases. All of these enzymes have been purified and characterized as heparinase I (HepI) (23, 44), HepII, and HepIII (23), all of which degrade either heparin, heparan sulfate, or both; chondroitinase AC (ChnAC), a 75-kDa enzyme which degrades both chondroitin sulfates A and C; and ChnB, a dermatan sulfate-degrading 55-kDa enzyme (8, 26, 43).
Chondroitin lyase activities have also been reported in other bacteria. Proteus vulgaris synthesizes two chondroitinase ABC lyases, each with a different mode of action (10, 43). Chondroitin lyases I and II were identified in Bacteroides thetaiotaomicron (22). Chondroitin lyase activity was also reported in Arthrobacter aurescens (12, 13), Porphyromonas gingivalis W50 (37), Streptococcus intermedius (36), and several other bacteria, as reviewed by Linhardt et al. (21). The genes coding for chondroitin sulfate ABC endolyase and chondroitin lyase II from P. vulgaris and B. thetaiotaomicron, respectively, have been cloned and expressed in Escherichia coli with enzymatic activity (1, 9, 35). However, none of these chondroitin lyases harbor the unique substrate specificity of ChnB from F. heparinum, which can only degrade dermatan sulfate.
At the molecular level, very little is known about the regulation of GAG lyase expression in F. heparinum. Maximal expression levels of heparinases are obtained when heparin is used as the sole carbon source (6, 7), while optimal expression of chondroitinases is reached when chondroitin sulfates are used (25, 43). Sulfate repression and glucose effects have been observed in the regulation of heparinase expression (7). In addition, GAG lyase enzymes allow F. heparinum to use heparin, heparin sulfate, and the chondroitins as sole sources of carbon, nitrogen, and energy. GAG molecules are catabolized to smaller polysaccharides, mainly disaccharides, with 4,5-unsaturated uronic acid residues at their nonreducing ends (3). It is interesting that all five GAG lyases are located in the periplasmic space of F. heparinum (8, 46). Knowledge of the mechanism of enzyme induction, the enzymes involved in their translocation across the cytoplasmic membrane, or the elements that control their gene expression in F. heparinum is limited.
Until recently, little was known about the structure of the chondroitinases from F. heparinum. Both chondroitinases have been crystallized (4, 19), and their structures have been resolved (J. Fethiere, B. Eggimann, and M. Cygler, submitted for publication; W. Huang, A. Matte, Y. Li, Y. S. Kim, R. J. Linhardt, H. Su, and M. Cygler, submitted for publication). It was shown that the two chondroitinases differ significantly in terms of their crystal forms and three-dimensional structures (4, 19; Fethiere et al. and Huang et al., submitted for publication).
F. heparinum, unlike most bacteria, is capable of posttranslational glycosylation. Four GAG lyases (HepI, HepII, ChnAC, and ChnB) are glycosylated (18). Thus far, documentation of glycosylation in bacteria has been limited to structural or secreted proteins (11). Structural analysis of the carbohydrate moieties of these GAG lyases using nuclear magnetic resonance and mass spectroscopy show them to be O linked to either serine or threonine with the consensus sequence Asp-Ser or Asp-Thr (14, 17). This is consistent with the O glycosylation consensus sequence of glycoproteins synthesized by F. meningosepticum (30).
In this paper, we report the isolation and characterization of the genes encoding the chondroitin lyase enzymes of F. heparinum. We also describe the relationship of these genes within the F. heparinum chromosome and the expression of active chondroitin lyases in E. coli.
MATERIALS AND METHODS
Bacterial strains, phages, and plasmids.
The bacterial strains, plasmids, and λ phages used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Genotype and relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli XL-1 Blue | Stratagene | |
| F. heparinum | ATCC 13125 | |
| λ phages | ||
| λ-DASH II | Stratagene | |
| λIB7 | λ-DASH II phage containing cslA | This study |
| λIB8 | λ-DASH II phage containing cslB | This study |
| Plasmids | ||
| pBluescript KS+ | Stratagene | |
| pIB11 | 400-bp amplicon of cslA generated with primers OAC-2 and OAC-4 | This study |
| pIB12 | 400-bp amplicon of cslA generated with primers OAC-1 and OAC-3 | This study |
| pIB13 | 800-bp amplicon of cslA generated with primers OAC-4 and OAC-3 | This study |
| pIB14 | 300-bp amplicon of cslB generated with primers OB-1 and OB-2 | This study |
| pIB15 | pBluescript KS+ vector with 6-kb BamHI DNA fragment from λIB7 | This study |
| pIB16 | pBluescript KS+ vector with 5-kb HindIII DNA fragment from λIB8 | This study |
| pIBX1 | E. coli expression vector | 40 |
| pIBX7 | pIBX1 vector with cslA | This study |
| pIBX8 | pIBX1 vector with cslB | This study |
Growth and induction conditions.
The minimal medium and growth conditions used for F. heparinum were described by Zimmermann et al. (45). In the study of cslA and cslB gene expression in F. heparinum (see Table 4), growth was at 23°C for 24 h in minimal medium supplemented with (i) 0.2% (wt/vol) glucose, (ii) 0.2% (wt/vol) glucose and 1% (wt/vol) chondroitin sulfate A (Sigma Chemical Co., St. Louis, Mo.), or (iii) 1% (wt/vol) chondroitin sulfate A. Cells were then subcultured in the same medium for an additional 8 h of growth at 23°C.
TABLE 4.
Synthesis of ChnAC, ChnB, and HepI in F. heparinum grown in media containing various carbon sourcesa
| Enzyme | Avg activity (mIU ml−1A600 unit−1)b ± SE when grown in:
|
|||
|---|---|---|---|---|
| Gluc | Glu + CS-Ad | CS-Ae | Ratiof | |
| HepI | <5.0 | 20.8 ± 2.6 | 88.1 ± 3.3 | 4.2 |
| ChnAC | <5.0 | 30.8 ± 0.9 | 79.5 ± 8.3 | 2.6 |
| ChnB | <5.0 | 10.5 ± 1.5 | 29.4 ± 1.2 | 2.8 |
Values are based on three individual experiments.
One unit of heparin-, chondroitin sulfate A-, or dermatan sulfate-degrading activity is defined as the amount of enzyme that will liberate 1 μmol of product per min at 30°C and pH 8.
Glu, glucose-only medium.
Glu + CS-A, glucose medium with 1% chondroitin sulfate A.
CS-A, 1% chondroitin sulfate A medium.
Ratio of activity obtained in chondroitin sulfate A medium compared to medium containing glucose with chondroitin sulfate A.
E. coli was grown in Luria broth at the temperatures indicated below. λ phage was grown and amplified as recommended by the supplier (Stratagene, La Jolla, Calif.). Ampicillin was provided at a concentration of 200 μg/ml. Expression of the recombinant chondroitinases was achieved by growing E. coli XL-1 Blue harboring either plasmid pIBX7 (ChnAC) or pIBX8 (ChnB) at 37°C overnight in Luria broth containing ampicillin. Cells were subcultured in the same medium and grown to an A600 of approximately 0.5. Cultures were induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Boehringer Mannheim, Montreal, Quebec, Canada) and were incubated at 25, 30, and 37°C for 5, 3.5, and 2 h, respectively.
Purification of chondroitinases.
The chondroitinases were purified from F. heparinum as previously described (8). Recombinant chondroitinases were purified as previously described (8) but with the following modifications. Homogenates of E. coli cultures synthesizing the recombinant proteins were diluted with 10 mM sodium phosphate buffer (pH 7). The homogenate containing recombinant ChnAC (rChnAC) was applied to a CM-Sepharose Fast Flow column (16 mm [inside diameter] by 100 mm; Pharmacia, Mississauga, Ontario, Canada) and eluted at a flow rate of 6 ml/min with a linear gradient of 0 to 1 M sodium acetate in 10 mM sodium phosphate buffer (pH 7). rChnAC eluted from the column at about 400 to 500 mM sodium acetate. The homogenate containing rChnB was applied to a Cellufine Sulfate affinity chromatography column (16 mm [inside diameter] by 25 mm; Amicon Inc., Oakville, Ontario, Canada) and eluted at a flow rate of 2.5 ml/min with a linear gradient of 0 to 400 mM NaCl in 10 mM sodium phosphate buffer (pH 7). rChnB eluted from the column at about 100 to 200 mM NaCl.
Enzyme assays.
Enzyme assays were performed as previously described (42) but with the following modifications. Pepstatin and phenylmethylsulfonyl fluoride were omitted from the phosphate-buffered saline (PBS). Fractions were analyzed for chondroitin sulfate A-, chondroitin sulfate C-, and dermatan sulfate-degrading activities in reaction buffers composed of either chondroitin sulfate A (Sigma Chemical Co.), chondroitin sulfate C (Sigma Chemical Co.), or dermatan sulfate (Celsus Laboratories Inc., Cincinnati, Ohio) at 500 μg/ml in 50 mM Tris, pH 8. For HepI expression (see Table 4), heparin-degrading activity was measured as described previously (44).
Clostripain digestion and peptide sequence analysis.
The enzymes were subjected to digestion using clostripain (EC 3.4.22.8; Sigma Chemical Co.). Preactivated clostripain was added to either chondroitinase at 1 to 2% (wt/wt) in a buffer comprised of 25 mM sodium phosphate, 0.2 mM calcium acetate, and 2.5 mM dithiothreitol (Sigma Chemical Co.) at pH 7.5 ± 0.1 and incubated for 2 to 3 h at 37°C. The reaction mixtures were applied to a Brownlee C8 reverse-phase high-pressure liquid chromatography column (2.1 mm [inside diameter] by 3.0 cm; Applied BioSystems, Mississauga, Ontario, Canada), and the individual peptide fragments were eluted at a flow rate of 0.2 ml/min with a linear gradient of 0 to 60% acetonitrile in 0.1% trifluoroacetic acid. Peptide fragments recovered from the reaction mixtures were subjected to amino acid sequence analysis using a 470A gas-phase protein sequencer (Applied Biosystems, Foster City, Calif.) at the Biotechnology Research Institute in Montreal, Quebec, Canada.
Molecular biology techniques.
Isolation of chromosomal DNA, cloning, and DNA manipulation techniques were performed as described by Sambrook et al. (32). T4 DNA ligase and restriction endonucleases were purchased from New England Biolabs (Mississauga, Ontario, Canada). DNA fragments destined for ligation were first separated by agarose gel electrophoresis, and the DNA was extracted from the gel with a Geneclean I kit (Bio 101, Inc., La Jolla, Calif.). DNA of bacterial transformants was isolated for restriction analysis using the RPM kit (Bio 101).
DNA amplification.
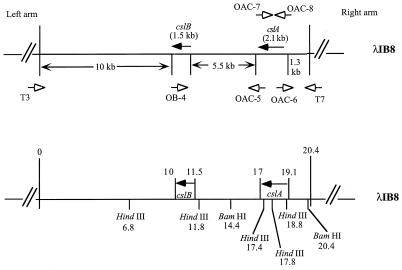
Oligonucleotide primer synthesis, preparation of genomic F. heparinum DNA for use as a template, and analysis of PCR-generated products were all performed as previously described (40). PCR amplification was performed as described by Mullis et al. (27) with the modifications described previously (40). For the gene mapping experiment (see Fig. 5), intact λIB8 was used and the PCR products were generated by using a Taq extender kit (Stratagene) as recommended by the manufacturer.
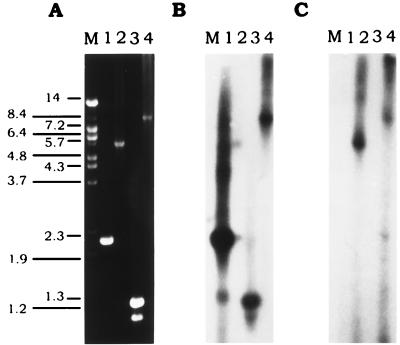
FIG. 5.
Southern blots mapping the proximity and orientation of the cslA and cslB genes on the F. heparinum genome. (A) Ethidium bromide-stained agarose gel containing PCR products. (B) Autoradiograph of a Southern blot probed with a BamHI fragment from pIBX7 containing the cslA ORF. (C) Autoradiograph of a Southern blot probed with a BamHI-XmnI fragment from pIBX8 containing the cslB ORF. Lanes: M, DNA molecular size markers (sizes [in kilobases] are indicated on the left); 1, 1 μg of the PCR product of cslA obtained with primers OAC-7 and OAC-8; 2, 1 μg of the PCR product, comprised of small portions of cslA and cslB and the intergenic region between the two genes, obtained with primers OAC-5 and OB-4; 3, 1 μg of the PCR product, containing a small portion of cslA and the region 5′ of cslA, obtained with primers OAC-6 and T7; 4, 1 μg of the PCR product, composed of cslB, cslA, the intergenic region between the two genes, and the region 5′ of cslA, obtained with primers T7 and OB-4. The DNA sequence of T3 is 5′-AATTAACCCTCACTAAAGGG-3′, and that of T7 is 5′-GTAATACGACTCACTATAGGGC-3′. DNA sequences of the other primers are listed in Table 2. In all cases, λIB8 was used as the template.
DNA hybridization.
DNA for Southern blotting (38) was digested with the appropriate restriction endonuclease, separated by gel electrophoresis, and transferred to a nylon membrane (Hybond-N; Amersham, Oakville, Ontario, Canada). Plaque and dot blots, labelling of probes with [α-32P]dATP, hybridizations, and washes were performed as previously described (40).
F. heparinum chromosomal DNA library screening.
A λ-Dash II chromosomal F. heparinum library was screened for the cslA and cslB genes as previously described (40).
Cloning of the cslA region.
Degenerate oligonucleotide primers were designed by using the clostripain peptides. Primer pairs OAC-4 plus OAC-2 and OAC-1 plus OAC-3 both generated 400-bp PCR fragments, while primer pair OAC-4 plus OAC-3 generated an 800-bp PCR fragment (Table 2). These PCR fragments were cloned into the vector pBluescript KS+ to form plasmids pIB11, pIB12, and pIB13. The 800-bp insertion from pIB13 was used as a probe to screen the F. heparinum λ-Dash II genomic library. Phage λIB7, containing a 6-kb BamHI fragment, hybridized strongly to this insertion. This 6-kb fragment was isolated and cloned into pBluescript KS+ to produce plasmid pIB15.
TABLE 2.
Primer sequences and their amino acid coordinates in ChnAC and ChnB
| Enzyme and primer | Primer sequencea | Amino acidsb |
|---|---|---|
| chnAC | ||
| OAC-1 | 5′ ggatcc GAR TAY TAY AAY ATH ATG CCN GT 3′ | 396–403 |
| OAC-2 | 5′ ggatcc ACN GGC ATD ATR TTR TAR TAY TC 3′ | 403–396 |
| OAC-3 | 5′ ggatcc TCN GGR AAR TAR TAN CCD ATN GCR TCR TG 3′ | 531–522 |
| OAC-4 | 5′ ggatcc TAY ATG GAY TTY AAY GTN GAR GG 3′ | 280–287 |
| OAC-5 | 5′ GTC CTT GCC AAT ACC AAC CAG 3′ | 602–608 |
| OAC-6 | 5′ CTG CGG AGT GGC AAT TTC 3′ | 135–130 |
| OAC-7 | 5′ TTT CAG TTC AAC CGT TGC ACC 3′ | 700–694 |
| OAC-8 | 5′ ggatcc ATG CAG CAG ACC GGT ACT GCA GAA 3′ | 23–29 |
| OAC-9 | 5′ ggatcc CCT AGA TTA CTA CCA TCA AAA 3′ | 3′ of cslA |
| chnB | ||
| OB-1 | 5′ ggatcc CAR ATY GCC GAY GGN ACN TAT AAA GA 3′ | 48–56 |
| OB-2 | 5′ ggatcc GGC NSK ATT GGC TTC RTC AAA 3′ | 144–138 |
| OB-4 | 5′ ATC TTT ATA AGT CCC ATC GGC 3′ | 56–50 |
| OB-5 | 5′ ggatcc ATG CAG GTT GTT GCT TCA AAT GAA ACT 3′ | 26–33 |
| OB-6 | 5′ ggaatcaattc ACC GGG ATG ATC 3′ | 3′ of cslB |
Lowercase nucleotides represent the BamHI and XmnI restriction sites incorporated at the 5′ ends of the primers for use in cloning. The nucleotide codes used are those recommended by the International Union of Pure and Applied Chemistry-IUB Biochemical Nomenclature Commission.
Amino acid coordinates are those of the precursor proteins.
Cloning of the cslB region.
Partial guessmer and degenerate oligonucleotide primers were designed with BamHI restriction sites incorporated at their 5′ ends. Primer pair OB-1 plus OB-2 (Table 2) generated a 300-bp PCR fragment which was cloned into pBluescript KS+ to produce plasmid pIB14. This insertion was used to screen the F. heparinum λ-Dash II genomic library. Phage λIB8 was found to hybridize strongly with the pIB14 insertion and contained a 5-kb HindIII fragment, which was cloned into pBluescript KS+ to produce plasmid pIB16.
DNA sequence analysis.
DNA sequences were determined by the dideoxy-chain termination method of Sanger et al. (33).
Construction of plasmids pIXB7 and pIXB8.
Inserts were obtained by DNA amplification. Oligonucleotides OAC-8 and OB-5 (Table 2), specific to cslA and cslB, respectively, were designed to contain BamHI restriction sites at their 5′ ends, followed by an ATG start site inserted immediately preceding the codon for the first amino acid of the mature protein (Q23 in the case of ChnAC; Q26 for ChnB). The other oligonucleotides, OAC-9 and OB-6 (Table 2), corresponding to cslA and cslB, respectively, were designed to hybridize downstream of the stop codon, followed by either a BamHI (OAC-9) or an XmnI (OB-6) restriction site at the 3′ end. Amplicons of the expected sizes, about 2.1 and 1.5 kb for cslA and cslB, respectively, were inserted into the corresponding restriction sites of the expression vector pIBX1 (40). DNA sequence analysis was carried out to ensure that no errors were introduced during amplification.
Antibody production.
Polyclonal antibodies against ChnAC and ChnB were raised in rabbits. A standard immunization procedure was employed, and ChnAC, ChnB, and HepI affinity columns were prepared as described by Su et al. (40). Anti-ChnAC or anti-ChnB antibodies were purified from sera preabsorbed on separate HepI columns to eliminate any cross-reactivity with other GAG lyases from F. heparinum. The unbound fractions were collected and purified over the respective ChnAC or ChnB antigen column. Bound antibodies (anti-ChnAC or anti-ChnB) were eluted in 0.1 M glycine (pH 2.0).
Protein and Western blot analysis.
Protein concentrations were determined by the Bradford dye-binding procedure using the Bio-Rad protein assay (Bio-Rad Laboratories); bovine serum albumin was the standard used. Protein electrophoresis was performed on crude E. coli extracts by using precast, discontinuous sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel electrophoresis (PAGE) gels (Bio-Rad Laboratories), while 10% polyacrylamide gels were used for the purified chondroitinases. Reducing gels were run in which 5% 2-mercaptoethanol (Sigma Chemical Co.) was added to the sample loading buffer. Quantities loaded per lane were equivalent to an optical density at 600 nm 0.15 U of or approximately 2 μg for the crude and purified samples, respectively. Proteins were visualized with 0.1% Coomassie brilliant blue R-250 (Bio-Rad Laboratories). Western blot analysis was performed as previously described (40), except that the primary antibodies used were rabbit anti-ChnAC antibodies diluted 1:50,000 or rabbit anti-ChnB antibodies diluted 1:25,000.
Protein stability assay.
Both recombinant E. coli and wild-type F. heparinum purified chondroitinases were subjected to heat inactivation as follows. The enzymes were diluted in PBS (10 mM sodium phosphate buffer [pH 7], 150 mM NaCl) to an initial total protein concentration of 50 μg/ml. Samples were incubated at 37°C for 6 h and then diluted in PBS containing bovine serum albumin at 1 mg/ml and assayed for the appropriate activities.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the cslA and cslB sequences are U27583 and U27584, respectively.
RESULTS
Isolation of the F. heparinum gene for ChnAC, cslA.
The amino acid sequences of three clostripain-digested ChnAC peptides were used as templates to design degenerate oligonucleotides (Table 2). F. heparinum-specific amplicons were obtained and cloned. DNA sequence analysis indicated that plasmid pIB13, which contained the 800-bp amplicon, consisted of the two 400-bp amplicons found in plasmids pIB11 and pIB12. The deduced amino acid sequence corresponded to cslA-derived peptide sequences (data not shown). Using the insertion from pIB13 as a probe, a 6-kb BamHI fragment was isolated from a λ-Dash II F. heparinum chromosomal DNA library, cloned, and subjected to DNA sequence analysis.
A region of approximately 3.2 kb was sequenced, and a single continuous open reading frame (ORF) of 2,103 bp, encoding 700 amino acid residues, was deduced. The N-terminal peptide sequence of mature ChnAC, as reported by Gu et al. (8), lacks amino acids 1 to 22, implying a signal peptide which is 22 residues long. The mature ChnAC protein consisted of 678 amino acid residues with a calculated molecular mass of 77,169 Da. The G+C content of the cslA gene, 44.2 mol%, is similar to that reported for F. heparinum chromosomal DNA (43 mol%) (39).
Isolation of the F. heparinum gene for ChnB, cslB.
Degenerate primers designed from clostripain-generated ChnB peptides (Table 2) were used to create a 300-bp PCR amplicon from F. heparinum chromosomal DNA which was cloned to form plasmid pIB14. DNA sequence analysis of this 300-bp amplicon confirmed that a portion of cslB had been cloned. A 5-kb HindIII fragment, isolated from a λ-Dash II F. heparinum chromosomal DNA library by using the insert from pIB14 as a probe, was cloned to create plasmid pIB16 and subjected to DNA sequence analysis.
A single 1,521-bp ORF coding for 506 amino acid residues was identified after sequencing of a portion of the insertion. The mature protein's N-terminal sequence, as reported by Gu et al. (8), mapped to the deduced peptide sequence of the cslB gene. The putative signal peptide is 25 amino acids long. Mature chnB is composed of 481 amino acid residues with a calculated molecular mass of 53,563 Da. The G+C content of this gene is 43.6 mol%.
Expression of cslA and cslB in E. coli.
Due to the fact that the signal sequence from F. heparinum was not functional when the gene for HepI was expressed in E. coli (34), both ORFs were amplified by PCR and modified to include an ATG initiator codon immediately preceding the N-terminal residue of the mature protein. The DNA fragments were then cloned into expression vector pIBX1. After induction at various temperatures, the E. coli strains which contained plasmids pIBX7 and pIBX8 were analyzed for recombinant protein expression.
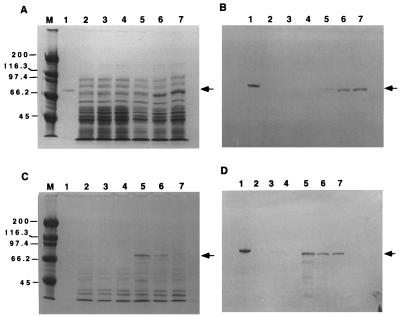
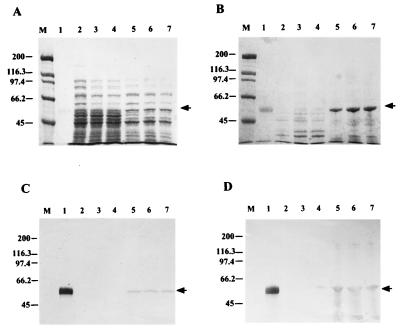
Protein electrophoresis indicated that both proteins were expressed in both soluble and insoluble forms at all induction temperatures. Intense bands of 70 and 55 kDa, corresponding to ChnAC and ChnB, respectively, were seen in E. coli fractions harboring plasmids pIBX7 (ChnAC) (Fig. 1A and C) and pIBX8 (ChnB) (Fig. 2A and B) but not in fractions with the expression plasmid pIBX1 (Fig. 1A and C and 2A and B). Western blot analysis of duplicate gels using antibodies specific for ChnAC (Fig. 1B and D) or ChnB (Fig. 2C and D) confirmed only those bands expressed by pIBX7 or pIBX8 as being ChnAC or ChnB. In the case of either recombinant protein, they were expressed mostly in the insoluble form at elevated induction temperatures whereas they were more predominant in the soluble form at lower temperatures.
FIG. 1.
SDS-PAGE and Western blot analysis of recombinant ChnAC in E. coli induced at various temperatures. Both soluble (A and B) and insoluble (C and D) fractions (0.15 OD600 unit) were loaded onto SDS–7.5% PAGE reducing gels. Panels: A and C, Coomassie blue R-250-stained gels; B and D, Western blots of duplicates of gels A and C transferred to nitrocellulose membrane filters. Lanes: 1, wild-type F. heparinum ChnAC (1 μg) (positive control); 2 to 4, pIBX1 samples (negative control) induced at 37, 30, and 25°C, respectively; 5 to 7, pIBX7 samples induced at 37, 30, and 25°C, respectively; M, molecular size markers. An arrow indicates the position of ChnAC. The value on the left are molecular masses in kilodaltons.
FIG. 2.
SDS-PAGE and Western blot analysis of ChnB in E. coli induced at various temperatures. Both soluble (A and C) and insoluble (B and D) fractions (0.15 OD600 unit) were loaded onto SDS–7.5% PAGE reducing gels. Panels: A and B, Coomassie blue R-250-stained gels; C and D, Western blots of duplicates of gels A and B transferred to nitrocellulose membrane filters. Lanes: 1, wild-type F. heparinum ChnB (1 μg) (positive control); 2 to 4, pIBX1 samples (negative control) induced at 25, 30, and 37°C, respectively; 5 to 7, pIBX8 samples induced at 25, 30, and 37°C, respectively; M, molecular size markers. An arrow indicates the position of rchnB. The values to the left are molecular masses in kilodaltons.
Enzyme activity analysis of both recombinant proteins showed them to be biologically active (Table 3). However, activity was only detected in the soluble cell extracts and not in the insoluble forms (data not shown). The level of enzyme activity was inversely correlated to the induction temperature, and the highest expression levels were reached at 25°C. For rChnAC, the ratio of chondroitin sulfate A-degrading activity to chondroitin sulfate C-degrading activity was approximately 1.5:1, which was similar to that observed for the native F. heparinum enzyme (8).
TABLE 3.
Chondroitin sulfate A-, chondroitin sulfate C-, and dermatan sulfate-degrading activities in soluble extracts from E. coli strains containing expression plasmidsa
| Temp (°C) and substrate | Avg activity (mIU ml−1A600 unit−1) ± SEb
|
||
|---|---|---|---|
| pIBX1 | pIBX7 | pIBX8 | |
| 37 | |||
| CS-Ac | <3.0 | 17.6 ± 3.3 | NDf |
| CS-Cd | <3.0 | 11.7 ± 1.6 | ND |
| DSe | <3.0 | ND | 13.7 ± 2.2 |
| 30 | |||
| CS-A | <3.0 | 36.7 ± 0.6 | ND |
| CS-C | <3.0 | 26.0 ± 0.5 | ND |
| DS | <3.0 | ND | 21.7 ± 2.4 |
| 25 | |||
| CS-A | <3.0 | 46.9 ± 3.0 | ND |
| CS-C | <3.0 | 31.3 ± 1.4 | ND |
| DS | <3.0 | ND | 62.5 ± 2.4 |
Values are based on three individual experiments.
One unit of chondroitin sulfate A-, chondroitin sulfate C-, or dermatan sulfate-degrading activity is defined as the amount of enzyme that will liberate 1 μmol of product per min at 30°C and pH 8.
CS-A, chondroitin sulfate A.
CS-C, chondroitin sulfate C.
DS, dermatan sulfate.
ND, not determined.
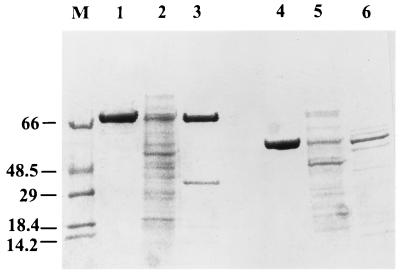
Purified rChnAC and rChnB were analyzed by protein electrophoresis, activity measurement, and stability assay. The sizes of rChnAC and rChnB were similar to those of ChnAC and ChnB from F. heparinum, respectively (Fig. 3, lanes 3 and 1 for ChnAC and lanes 6 and 4 for ChnB). The recombinant proteins also had specific activities similar to those of their wild-type counterparts (data not shown). However, rChnAC and ChnAC behaved differently when subjected to heat inactivation. Approximately 50% of rChnAC activity remained after its exposure to 37°C for 6 h, while ChnAC from F. heparinum retained full activity after similar treatment. rChnB and ChnB were also subjected to heat inactivation, and neither lost any activity.
FIG. 3.
SDS-PAGE analysis of purified chondroitinases from E. coli and F. heparinum stained with Coomassie blue R-250. Approximately 2 μg of each sample was loaded onto SDS–10% PAGE reducing gels. Lanes: 1 to 3, ChnAC; 4 to 6, ChnB; 1 and 4, from F. heparinum; 2 and 5, E. coli homogenates; 3, CM-Sepharose-purified rChnAC; 6, Cellufine Sulfate affinity-purified rchnB; M, molecular weight markers. The values to the left are molecular masses in kilodaltons.
Southern blot analysis.
To investigate the relationship among the GAG lyase genes in F. heparinum, a Southern blot analysis of its chromosome was carried out in which cslA, cslB, and the heparinase genes hepA (34), hepB (40), and hepC (40) were used to probe PCR products corresponding to the 5′ and 3′ regions of cslA and cslB, as well as digests of λIB7 and λIB8. The cslA probe cross-hybridized to the amplicon corresponding to the region 5′ of the cslB gene and also to a fragment from λIB8 (data not shown). It was therefore concluded that cslA was 5′ of cslB and that phage λIB8 contained both genes. No cross-hybridization signals were observed with the heparinase genes (data not shown), suggesting that they are not proximally linked to the chondroitinase genes.
To determine the exact distance between the two genes and the orientation of the two ORFs with respect to one another, an experiment using PCR methodology and Southern blot analysis was performed. Oligonucleotides OAC-6 and OAC-5 were designed to correspond to the 5′ and 3′ ends of the cslA gene, respectively, and were used in a PCR in conjunction with oligonucleotide OB-4, corresponding to the 5′ end of the cslB gene (Fig. 4). An amplicon was obtained only with primer pair OAC-5 plus OB-4 (Fig. 5A, lane 2). These results indicated that the cslA gene was 5′ of the cslB gene and both genes were in the same orientation. The size of the PCR product, 5.5 kb, indicated the distance between the two genes. A PCR product of a similar size was also amplified by using F. heparinum chromosomal DNA as the template (data not shown). The identity of these PCR products (Fig. 5A) was further confirmed by Southern hybridization experiments in which the cslA and cslB genes were used as probes (Fig. 5B and C).
FIG. 4.
Schematic diagram of λIB8 containing both cslA and cslB. The directions of the ORFs of the two genes are indicated by labeled arrows with solid heads. The approximate sizes of the genomic regions on both sides of the genes are indicated between the arrows. The arrows with open heads indicate the locations of the PCR primers and the directions of synthesis from the primers. T3 and T7 indicate the locations of the primers derived from the T3 and T7 promoter regions in the lambda arms. OB-4 indicates the location of the primer derived from the DNA sequence at the end of the ORF of cslB. OAC-5, OAC-6, OAC-7, and OAC-8 indicate the locations of primers derived from 3′ and 5′ DNA sequences at the ends of the ORF of cslA. A partial restriction map is also provided, and positions are indicated in kilobases.
Induction of cslA and cslB in F. heparinum.
An experiment was conducted to analyze the synthesis of both chondroitinases when F. heparinum is grown in various media (Table 4). No ChnAC or ChnB activity was detected when the bacteria were grown in glucose-only medium, as was the case for HepI. They were expressed to detectable levels with the addition of the inducer chondroitin sulfate A to the medium. The highest level was reached when glucose was eliminated from the medium. A 2.5-fold increase in the expression levels of both chondroitinase enzymes was observed when the bacteria were grown in chondroitin sulfate A-only medium, compared to a 4-fold increase in HepI expression under similar conditions. The data suggest that the cslA and cslB genes were coregulated.
DISCUSSION
We have reported here the isolation and DNA sequence analysis of the cslA and cslB genes coding for ChnAC and ChnB, respectively. They were also expressed in E. coli with biological activity. This is the first report describing the cloning and expression of these enzymes from F. heparinum.
The calculated molecular masses of mature ChnAC and ChnB, derived from the DNA sequences, are 77,169 and 53,563 Da, respectively. Two carbohydrate moieties were identified in ChnAC and determined to be 1,194 and 1,080 Da. Only one was found in ChnB, and it had a molecular mass of 1,180 Da (17). Therefore, the total molecular mass of ChnAC is estimated to be 79,443 Da and that of ChnB is estimated to be 54,743 Da. These values closely reflect those of 79,557 and 54,779 Da as determined by mass spectroscopic analysis (17) for ChnAC and ChnB, respectively.
Signal sequences were observed in the coding regions of both genes, and their existence is consistent with their translocation to the periplasmic space of F. heparinum, as demonstrated by Linhardt's group and our laboratory (8). The leader peptides are typical of prokaryotic signal sequence regions (28). Moreover, the N termini of the mature proteins are also characteristic of exported bacterial proteins in that they lack a net positive charge (42). Glutamine is the +1 amino acid at the signal peptidase processing site in both chondroitinases and also is present in the heparinases from F. heparinum (40). This suggests that a similar translocation mechanism is involved.
A BLASTX search (www.ncbi.nlm.nih.gov/BLAST; version 2.0.6, released 16 September 1998) for both the cslA gene and its deduced peptide sequence was conducted. No significant homology with any previously published DNA sequences was found. However, a search using the deduced peptide sequence of the cslA gene revealed a high degree of homology between ChnAC and a putative secreted lyase from Streptomyces coelicolor (31) and a group of hyaluronate lyases from group B streptococci (20). In addition, homology was noted to the chondroitin sulfate ABC endolyase from P. vulgaris and chondroitin lyase II from B. thetaiotaomicron in the C-terminal region of ChnAC, a region that has been suggested to be the active site of chondroitin sulfate ABC endolyase (35). Finding these sequence similarities was not surprising because all of these enzymes are also polysaccharide lyases. Therefore, ChnAC may have arisen from the same ancestor as these lyases and may have a similar three-dimensional structure and reaction mechanism.
A BLASTX search conducted for the cslB gene and its deduced amino acid sequence showed no significant homology to any other previously published DNA sequence. However, the deduced amino acid sequence revealed significant homology to a polysaccharide lyase, alginate lyase from Pseudomonas sp. (24), in the N-terminal region of both proteins. The ChnB protein might share structural similarities and an evolutionary relationship with the alginate lyase.
F. heparinum ChnAC and ChnB shared no homology at either the DNA or the peptide level. This dissimilarity is consistent with data from Cygler's laboratory (Fethiere et al. and Huang et al., submitted), in which the determined crystal structures of the two chondroitinases were very different. Detailed structure and function studies will be performed by using the sequence data reported herein.
The role of the glycosylation of these chondroitinase enzymes from F. heparinum remains unclear. The unglycosylated forms of both enzymes remain active, as was shown in the gene expression studies of both the cslA and cslB genes in E. coli (Table 3). Similar results were observed when the hepA and hepB genes were expressed in E. coli (34, 40). However, whether or not the absence of this glycosylation is of consequence to the specific activity or proper folding of these enzymes is uncertain. In addition, it has been suggested that protein glycosylation, especially O glycosylation, is related to protein stability (5) and perhaps this is the function of this posttranslational modification in F. heparinum. It seems that the nonglycosylated form of rChnAC produced by E. coli was not as stable as the native, glycosylated form from F. heparinum. This was not the case for rChnB from E. coli. The glycosylated and nonglycosylated forms had similar stabilities when subjected to heat denaturation. However, the results from this experiment are inconclusive because the enzymes purified from E. coli were less pure, with different modifications at their N termini. Further biochemical studies of the nonglycosylated forms of these enzymes from F. heparinum are needed to provide more conclusive answers to this question. We are currently working on creating a genetic system for F. heparinum which will be used to address these questions.
Expression of the chondroitinase enzymes in E. coli will provide an efficient means for their production. The ChnB activity measured in the E. coli cell extracts was higher than that in F. heparinum, as shown in Tables 3 and 4, even when F. heparinum was grown in a medium containing chondroitin sulfate A as the sole carbon source. Expression of ChnB reached 62.5 mIU ml−1 A600 unit−1 in E. coli, compared to 29.4 mIU ml−1 A600 unit−1 in F. heparinum. Slightly lower levels of expression of the cslA gene were found in E. coli than in F. heparinum, i.e., activities of 46.9 and 79.5 mIU ml−1 A600 unit−1, respectively. However, the high biomass of E. coli fermentation and well-established techniques will offset these slightly lower specific activities observed in the expression of the cslA gene in E. coli. The findings of this study will provide an alternative source for the chondroitinase enzymes' production.
ACKNOWLEDGMENTS
We thank France Dumas from the Biotechnology Research Institute in Montreal, Quebec, Canada, for performing the amino acid sequence analysis of clostripain-digested peptides and Pamela Danagher for her efforts in producing the antibodies. We are indebted to Peter Lau (Biotechnology Research Institute in Montreal, Quebec, Canada) for his encouragement and critical review of the manuscript. We are grateful to Zhongqi Shao (IBEX Technologies Inc.) for his comments and helpful suggestions. Our thanks are also extended to Allan Matte (Biotechnology Research Institute in Montreal, Quebec, Canada) for his careful reading of the manuscript.
REFERENCES
- 1.Cheng Q, Yu M C, Reeves A R, Salyers A A. Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J Bacteriol. 1995;177:3721–3727. doi: 10.1128/jb.177.13.3721-3727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen P. Description and taxonomic status of Cytophaga heparina (Payza and Korn) comb. nov. (basionym: Flavobacterium heparinum Payza and Korn 1956) Int J Syst Bacteriol. 1980;30:473–475. [Google Scholar]
- 3.Ernst S, Langer R, Cooney C L, Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Crit Rev Biochem Mol Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 4.Fethiere J, Shilton B H, Li Y, Allaire M, Laliberte M, Eggimann B, Cygler M. Crystallization and preliminary analysis of chondroitinase AC from Flavobacterium heparinum. Acta Crystallogr. 1998;D54:279–280. doi: 10.1107/s0907444997009037. [DOI] [PubMed] [Google Scholar]
- 5.Furukawa K, Kobata A. Protein glycosylation. Curr Opin Biotechnol. 1992;3:554–559. doi: 10.1016/0958-1669(92)90085-w. [DOI] [PubMed] [Google Scholar]
- 6.Galliher P M, Cooney C L, Langer R, Linhardt R J. Heparinase production by Flavobacterium heparinum. Appl Environ Microbiol. 1981;41:360–365. doi: 10.1128/aem.41.2.360-365.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galliher P M, Linhardt R J, Conway L J, Langer R, Cooney C L. Regulation of heparinase synthesis in Flavobacterium heparinum. Eur J Appl Microbiol Biotechnol. 1982;15:252–257. [Google Scholar]
- 8.Gu K, Linhardt R J, Laliberté M, Gu K, Zimmermann J. Purification, characterization and specificity of chondroitin lyases and glycuronidase from Flavobacterium heparinum. Biochem J. 1995;312:569–577. doi: 10.1042/bj3120569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guthrie E P, Shoemaker N B, Salyers A A. Cloning and expression in Escherichia coli of a gene coding for a chondroitin lyase from Bacteroides thetaiotaomicron. J Bacteriol. 1985;164:510–515. doi: 10.1128/jb.164.2.510-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamai A, Hashimoto N, Mochizuki H, Kato F, Makiguchi Y, Horie K, Suzuki S. Two distinct chondroitin sulfate ABC lyases. J Biol Chem. 1997;272:9123–9130. doi: 10.1074/jbc.272.14.9123. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann J L, O'Gaora P, Gallagher A, Thole J E R, Young D B. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 1996;15:3547–3554. [PMC free article] [PubMed] [Google Scholar]
- 12.Hiyama K, Okada S. Amino acid composition and physiochemical characterization of chondroitinase from Arthrobacter aurescens. J Biochem. 1975;78:1183–1190. doi: 10.1093/oxfordjournals.jbchem.a131015. [DOI] [PubMed] [Google Scholar]
- 13.Hiyama K, Okada S. Action of chondroitinases. III. Ionic strength effects and kinetics in the action of chondroitinase AC. J Biochem. 1977;82:429–463. [PubMed] [Google Scholar]
- 14.Huang L, Van Halbeek H, Eggimann B, Zimmermann J. Structural characterization of the novel O-linked carbohydrate structure of Flavobacterium heparinum heparinase I. Glycobiology. 1995;5:712. [Google Scholar]
- 15.Jackson R L, Busch S J, Cardin A D. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 16.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 17.Laliberte M, Eggimann B, Zimmermann J J F, Huang L, van Halbeek H. Determination of the glycosylation sites of glycosaminoglycan lyases from Flavobacterium heparinum. Protein Sci. 1996;5(Suppl. 1):435s. [Google Scholar]
- 18.Lechner J. Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem. 1989;58:173–194. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Matte A, Su H, Cygler M. Crystallization and preliminary X-ray analysis of chondroitinase B from Flavobacterium heparinum. Acta Crystallogr. 1999;D55:1055–1057. doi: 10.1107/s0907444999002097. [DOI] [PubMed] [Google Scholar]
- 20.Lin B, Hollingshead S K, Coligan J E, Egan M L, Baker J R, Pritchard D G. Cloning and expression of the gene for group B streptococcal hyaluronate lyase. J Biol Chem. 1994;269:30113–30116. [PubMed] [Google Scholar]
- 21.Lindhardt R J, Galliher P M, Cooney C L. Polysaccharide lyases. Appl Biochem Biotechnol. 1986;12:135–177. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- 22.Linn S, Chan T, Lipeski L, Salyers A A. Isolation and characterization of two chondroitin lyases from Bacteroides thetaiotaomicron. J Bacteriol. 1983;156:859–866. doi: 10.1128/jb.156.2.859-866.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohse D L, Linhardt R J. Purification and characterization of heparin lyases from Flavobacterium heparinum. J Biol Chem. 1992;267:24347–24355. [PubMed] [Google Scholar]
- 24.Maki H, Mori A, Fujiyama K, Kinoshita S, Yoshida T. Cloning, sequence analysis and expression in Escherichia coli of a gene encoding an alginate lyase from Pseudomonas sp. OS-ALG-9. J Gen Microbiol. 1993;139:987–993. doi: 10.1099/00221287-139-5-987. [DOI] [PubMed] [Google Scholar]
- 25.Michelacci Y M, Dietrich C P. Isolation and partial characterization of an induced chondroitinase B from Flavobacterium heparinum. Biochem Biophys Res Commun. 1974;56:973–980. doi: 10.1016/s0006-291x(74)80284-6. [DOI] [PubMed] [Google Scholar]
- 26.Michelacci Y M, Dietrich C P. A comparative study between a chondroitinase B and a chondroitinase AC from Flavobacterium heparinum. Biochem J. 1975;151:121–129. doi: 10.1042/bj1510121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp Quant Biol. 1986;51:263–269. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 28.Palzkill T, Le Q, Wong A, Botstein D. Selection of functional peptide cleavage sites from a library of random sequences. J Bacteriol. 1994;176:563–568. doi: 10.1128/jb.176.3.563-568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payza A N, Korn E D. Bacterial degradation of heparin. Nature (London) 1956;177:88–89. doi: 10.1038/177088a0. [DOI] [PubMed] [Google Scholar]
- 30.Plummer T H, Tarentino A L, Hauer C R. Novel, specific O-glycosylation of secreted Flavobacterium meningosepticum proteins. Asp-Ser and Asp-Thr-Thr consensus sites. J Biol Chem. 1995;270:13192–13196. doi: 10.1074/jbc.270.22.13192. [DOI] [PubMed] [Google Scholar]
- 31.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasisekharan R, Bulmer M, Moremen K W, Cooney C L, Langer R. Cloning and expression of heparinase I gene from Flavobacterium heparinum. Proc Natl Acad Sci USA. 1993;90:3660–3664. doi: 10.1073/pnas.90.8.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato N, Shimada M, Nakajima H, Oda H, Kimura S. Cloning and expression in Escherichia coli of the gene encoding the Proteus vulgaris chondroitin ABC lyase. Appl Microbiol Biotechnol. 1994;41:39–46. doi: 10.1007/BF00166079. [DOI] [PubMed] [Google Scholar]
- 36.Shain H, Homer K A, Beighton D. Degradation and utilisation of chondroitin sulphate by Streptococcus intermedius. J Med Microbiol. 1996;44:372–380. doi: 10.1099/00222615-44-5-372. [DOI] [PubMed] [Google Scholar]
- 37.Smith A J, Greenman J, Embery G. Detection and possible biological role of chondroitinase and heparitinase enzymes produced by Porphyromonas gingivalis W50. J Periodontal Res. 1997;32:1–8. doi: 10.1111/j.1600-0765.1997.tb01376.x. [DOI] [PubMed] [Google Scholar]
- 38.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 39.Steyn P L, Segers P, Vancanneyt M, Sandra P, Kersters K, Joubert J J. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int J Syst Bacteriol. 1998;48:165–177. doi: 10.1099/00207713-48-1-165. [DOI] [PubMed] [Google Scholar]
- 40.Su H, Blain F, Musil R A, Zimmermann J J F, Gu K, Bennett D C. Isolation and expression in Escherichia coli of hepB and hepC, genes coding for the glycosaminoglycan-degrading enzymes heparinase II and heparinase III, respectively, from Flavobacterium heparinum. Appl Environ Microbiol. 1996;62:2723–2734. doi: 10.1128/aem.62.8.2723-2734.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi M, Yokota A. Proposals of Sphingobacterium faecium sp. nov., Sphingobacterium piscium sp. nov., Sphingobacterium heparinum comb. nov., Sphingobacterium thalpophilum comb. nov. and two genospecies of genus Sphingobacterium, and synonymy of Flavobacterium yabuuchiae and Sphingobacterium spiritivorum. J Gen Appl Microbiol. 1992;38:465–482. [Google Scholar]
- 42.Von Heijne G. Net N-C charge imbalance may be important for signal sequence function in bacteria. J Mol Biol. 1986;192:287–290. doi: 10.1016/0022-2836(86)90365-7. [DOI] [PubMed] [Google Scholar]
- 43.Yamagata T, Saito H, Habuchi O, Suzuki S. Purification and properties of bacterial chondroitinases and chondrosulfatases. J Biol Chem. 1968;243:1523–1535. [PubMed] [Google Scholar]
- 44.Yang V C, Linhardt R J, Bernstein H, Cooney C L, Langer R. Purification and characterization of heparinase from Flavobacterium heparinum. J Biol Chem. 1985;260:1849–1857. [PubMed] [Google Scholar]
- 45.Zimmermann J J, Langer R, Cooney C L. Specific plate assay for bacterial heparinase. Appl Environ Microbiol. 1990;56:3593–3594. doi: 10.1128/aem.56.11.3593-3594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimmermann J J, Oddie K, Langer R, Cooney C L. The release of heparinase from the periplasm space of Flavobacterium heparinum by three-step osmotic shock. Appl Biochem Biotechnol. 1991;30:137–148. doi: 10.1007/BF02921681. [DOI] [PubMed] [Google Scholar]