Abstract
The enzymatic degradation of amino acids in cheese is believed to generate aroma compounds and therefore to be essential for flavor development. Cystathionine β-lyase (CBL) can convert cystathionine to homocysteine but is also able to catalyze an α,γ elimination. With methionine as a substrate, it produces volatile sulfur compounds which are important for flavor formation in Gouda cheese. The metC gene, which encodes CBL, was cloned from the Lactococcus lactis model strain MG1363 and from strain B78, isolated from a cheese starter culture and known to have a high capacity to produce volatile compounds. The metC gene was found to be cotranscribed with a downstream cysK gene, which encodes a putative cysteine synthase. The MetC proteins of both strains were overproduced in strain MG1363 with the NICE (nisin-controlled expression) system, resulting in a >25-fold increase in cystathionine lyase activity. A disruption of the metC gene was achieved in strain MG1363. Determination of enzymatic activities in the overproducing and knockout strains revealed that MetC is essential for the degradation of cystathionine but that at least one lyase other than CBL contributes to methionine degradation via α,γ elimination to form volatile aroma compounds.
Lactic acid bacteria (LAB) are gram-positive bacteria widely used in a variety of dairy fermentation processes. They contribute to flavor formation and texture development in dairy products. Flavor development in cheese starts with the degradation of caseins by the activity of both rennet and the proteases and peptidases from LAB (44). The hydrolysis of caseins yields small peptides and free amino acids. These amino acids, specifically, the aromatic, branched-chain, and sulfur-containing residues, act as precursors for the formation of volatile aroma compounds (19). While the proteolytic and the peptidolytic systems of LAB have been extensively studied (20, 35), the reactions that follow proteolysis, leading to typical cheese flavor formation, are hardly known. The enzymatic and/or chemical conversions of amino acids are assumed to be essential for the formation of specific aroma compounds such as esters, thiols, aldehydes, and ketones (22, 28). The enzymatic degradation of amino acids may take place via different pathways involving several enzymes, for instance, deaminases, decarboxylases, or transaminases (22, 27, 33).
The enzyme cystathionine β-lyase (CBL) is able to degrade sulfur-containing amino acids and has been purified from Lactococcus lactis subsp. cremoris B78 (1). It is a tetrameric protein with identical subunits of approximately 35 to 40 kDa. CBL is a pyridoxal-5′-phosphate (PLP)-dependent enzyme, and its physiological function is the catalysis of the penultimate step in microbial and plant methionine biosynthesis, i.e., an α,β-elimination reaction from cystathionine to produce homocysteine, pyruvate, and ammonia. Subsequently, the homocysteine is methylated to form methionine (26). Although most of the L. lactis strains are auxotrophic for methionine, there are indications that a biosynthetic route for this amino acid exists and that it may be interrupted (9). CBL purified from L. lactis B78 is also able to degrade cystathionine via an α,γ elimination, resulting in the formation of cysteine, α-ketogluatarate, and ammonia. Furthermore, it is able to degrade other sulfur-containing amino acids via an α,γ-elimination reaction, which with methionine results in the production of methanethiol (1). This sulfur compound is a precursor of important flavor compounds in cheese, such as dimethyl disulfide (DMDS) and dimethyl trisulfide (DMTS) (37). Other Lactococcus and Lactobacillus enzymes involved in the conversion of amino acids to volatile compounds have recently been purified and characterized, i.e., aminotransferases and a cystathionine γ-lyase (5, 23, 40, 46). These enzymes are also able to convert methionine to volatile compounds.
In this paper we report the cloning, transcriptional analysis, and effects of disruption of the genes encoding CBLs from two different strains of L. lactis subsp. cremoris: strain B78, a strain isolated from a mixed-strain mesophilic starter used for the production of Gouda cheese, and MG1363, a plasmid-free model strain. Moreover, CBL was overproduced with the NICE (nisin-controlled expression) system, which uses the food-grade antimicrobial peptide nisin as the inducer (14, 31).
MATERIALS AND METHODS
Bacterial strains and media.
Strains and plasmids used in this study are listed in Table 1. L. lactis cells were routinely grown at 30°C, unless stated otherwise, in media based on M17 medium (Merck, Darmstadt, Germany) supplemented with 0.5% (wt/vol) glucose (GM17). When appropriate, the media contained chloramphenicol (10 μg ml−1 for Escherichia coli or 7 μg ml−1 for L. lactis), tetracycline (5 μg ml−1), ampicillin (50 μg ml−1), or erythromycin (3 μg ml−1).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid(s) | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli MC1061 | 7 | |
| L. lactis | ||
| MG1363 | Plasmid free | 24 |
| NIZO B78 | Starter strain | NIZO collection |
| NZ9000 | MG1363 pepN::nisRK | 31 |
| MG1363ΔmetC | MG1363 derivative, ΔmetC | This study |
| Plasmids | ||
| pUC18, pUC19 | Ampr | 45 |
| pNZ8133 | Ampr, 4.8-kb pUC18 derivative carrying a 2.2-kb HindIII-PstI fragment from L. lactis MG1363 | This study |
| pNZ8134 | Ampr, 6.8-kb pUC19 derivative carrying a 4.2-kb EcoRI fragment from L. lactis MG1363 | This study |
| pNZ8135 | Ampr, 3.8-kb pUC19 derivative carrying a 1.2-kb BamHI-SalI fragment of the metC gene from L. lactis B78 | This study |
| pNZ8037 | Cmr, 3.4-kb, carrying the nisA promoter and MCS | 14 |
| pNZ8136 | Cmr, 4.5-kb pNZ8048 derivative carrying the MG1363 metC gene | This study |
| pNZ8137 | Cmr, 4.5-kb pNZ8048 derivative carrying the B78 metC gene | This study |
| pG+host8 | Tetr, 4.3-kb pWV01 thermosensitive replicon | 34 |
| pNZ8138 | Tetr, 4.9-kb pG+host8 derivative carrying a 665-bp internal fragment of the MG1363 metC gene | This study |
| pNZ9530 | Eryr, 7.0-kb pIL252 derivative containing nisR and nisK genes | 29 |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Eryr, erythromycin resistant; Tetr, tetracycline resistant; MCS, multiple cloning sites.
DNA isolation and manipulation.
E. coli MC1061 was used as an intermediate host for cloning and was handled by standard techniques (39). Isolation of E. coli plasmid DNA and standard recombinant techniques were performed as described by Sambrook et al. (39). Large-scale isolation of E. coli plasmid for nucleotide sequence analysis was performed with the Jet Star (Genomed GmbH, Bad Deynhausen, Germany) system, according to the instructions of the manufacturer. Isolation and transformation of L. lactis plasmid DNA were performed as described previously (15).
Construction of plasmids.
A degenerate oligonucleotide probe, 5′-GTNATHCAYGGNGGNATHTC-3′ (where H is A, C, or T; Y is C or T; and N is A, C, G, or T), derived from the NH2-terminal amino acid sequence of the CBL protein from L. lactis B78 (1) was designed for cloning of the MG1363 metC gene. A 2.2-kb HindIII-PstI chromosomal DNA fragment containing the 5′ region of the metC gene, which hybridized with this oligonucleotide in Southern hybridization, was cloned in pUC18, resulting in plasmid pNZ8133 (Table 1). The 3′ region of the metC gene was cloned in pUC19 as a 4.2-kb EcoRI chromosomal fragment that hybridized with an oligonucleotide with the sequence 5′-GCGGTTCTTTCGCTATTTTC-3′, based on the sequence of the 5′ part of the gene, generating plasmid pNZ8134.
The B78 metC gene was cloned by a PCR with two primers based on the sequence of the MG1363 metC gene: 5′-GCTTGGATCCATAAAAAAAGTTAATTCTGA-3′ and 5′-CTTCGTCGACTTCAACAGGACCAATGTGAG-3′, which contain a BamHI and SalI site, respectively (underlined). A 1.2-kb PCR fragment was obtained and cloned as a BamHI-SalI fragment in pUC19, resulting in plasmid pNZ8135. The nucleotide sequences of three clones derived from separate PCRs were analyzed.
Plasmid pNZ8037 (Table 1) was used for the construction of translational fusions of metC to the nisA promoter (PnisA). The MG1363 metC gene was cloned in this vector in two steps. The 5′ part of the gene was cloned in pNZ8037 digested with NcoI and PstI as a 265-bp RcaI-PstI PCR fragment. The RcaI and PstI sites were introduced with the primers 5′GTCCTCATGACAAGTATAAAAACTAAAG-3′, which contains two nucleotide substitutions (boldface) to generate an RcaI site (underlined) at the start codon of metC, and 5′-TGATCTCCTGCAGAAAATAGCG-3′, which contains a PstI site (underlined). The 3′ part of the gene was cloned as a PstI-HindIII fragment from pNZ8134, resulting in pNZ8136. A similar strategy was followed for the construction of pNZ8137, which carries B78's metC translationally coupled to PnisA, with the primers 5′-GGCCTCATGACAAGTTTAAAAAC-3′, which contains one nucleotide substitution (boldface) to generate an RcaI site (underlined), and 5′-TGATCTCCTGCAGAAAATAAAG-3′, which contains a PstI site (underlined). This PCR product was cloned in the vector pNZ8037, which had been digested with PstI and NcoI. The 3′ part of the gene was cloned as an EcoRI-HindIII fragment generated from pNZ8135, resulting in pNZ8137.
For the construction of a knockout strain, a 665-bp internal fragment of the metC gene was generated by PCR with the oligonucleotides 5′-GTCCTCTAGAGGACTTGGACAACCTAAAG-3′, which contains an XbaI site (underlined), and 5′-GTCCCCCGGGCATTTGCCGAATGAG-3′, which contains an SmaI site (underlined), and cloned as an XbaI-SmaI fragment in the vector pG+host8, which contains a thermosensitive replicon (34). The integrity of all constructs containing PCR fragments was checked by DNA sequencing.
Nucleotide sequence analysis.
Automatic double-stranded DNA sequence analysis was performed on both strands with an ALFred DNA sequencer (Pharmacia Biotech). Sequencing reactions were accomplished with an AutoRead sequencing kit, initiated by using Cy5-labelled universal and reverse primers and continued with synthetic primers in combination with Cy5-13-dATP according to the instructions of the manufacturer (Pharmacia Biotech). Sequence data were assembled and analyzed with the PC/GENE program (version 6.70; IntelliGenetics). The sequences were compared with sequences in the SwissProt library (May 1999 release) by using TFASTA.
RNA isolation, Northern blotting, and primer extension analysis.
Total RNA was isolated from exponentially growing MG1363 and B78 cultures by the Macaloid method described by Kuipers et al. (30). The samples obtained from the induced cultures were treated with DNase I. For Northern blot analysis, RNA was separated on a 1% formaldehyde agarose gel, blotted, and hybridized as described previously (43). The probes used for hybridization were radiolabelled with [α-32P]dATP by nick translation. The blots were washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C, and autoradiographs were exposed for 20 h (wild-type strains) or 1 h (overproduction strains).
Primer extension was performed by annealing 20 ng of oligonucleotides to 25 μg of RNA as described by Kuipers et al. (30). A synthetic 18-mer oligonucleotide, 5′-CGGAACAGATACTGCTCC-3′, complementary to the 5′ sequence of metC gene, was used as a primer.
MetC overproduction.
For overexpression of the metC genes, an overnight culture of L. lactis NZ9000 harboring either pNZ8136 or pNZ8137 was transferred (1%) to fresh medium (GM17) and grown until an optical density at 600 nm of 0.5 was reached. The cells were induced with 0 or 2.5 ng of nisin ml−1 and grown for another 2 h (31). The cells were then harvested and the cell extracts (CEs) were prepared with a French press. These extracts were used for the enzymatic assays.
Enzymatic assays.
Cystathione lyase activity was monitored with either cystathionine or methionine as a substrate. Enzyme activity towards cystathionine was measured by determination of free-thiol group formation with DTNB (5,5′-dithiobis 2-nitrobenzoic acid) as described by Uren (42). PLP was added to the reaction mixture at a final concentration of 20 μM. Enzyme activity towards methionine was assayed by determination of volatile sulfur compounds. Methionine was used at a final concentration of 10 mM in 5 ml of the reaction mix, which contained 50 mM KPi buffer (pH 6.8) and 20 μM PLP. After the addition of the CEs, the reaction proceeded at 30°C for 5 h. The reaction was stopped by the addition of 5 mM hydroxylamine. The products formed were determined by dynamic headspace gas chromatographic analysis as described previously (1).
Protein concentrations were determined by a protein assay (Bio-Rad) based on the method of Bradford (4), with bovine serum albumin as the standard.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this study were submitted to GenBank under the accession no. AF131880 and AF131881.
RESULTS
Sequence analysis of the metC gene of L. lactis MG1363 and B78.
CBL has been purified from L. lactis B78, and its NH2-terminal amino acid sequence has been determined (1). The gene encoding this protein, designated metC, was cloned from two strains of L. lactis, MG1363 and B78, with a degenerate probe based on the NH2-terminal sequence, as was described in Materials and Methods. Sequence analysis of plasmids pNZ8133 and pNZ8134 (metC from MG1363) and pNZ8135 (metC from B78) revealed the presence of an open reading frame of 1,140 bp (Fig. 1). Both metC genes were preceded by a reasonable consensus ribosome binding site for L. lactis (AGAAAG) (8) and encoded a putative protein of 380 amino acids which show a perfect sequence identity to the NH2-terminal part of the L. lactis B78 CBL. The calculated molecular mass was 41 kDa, which is in agreement with the reported apparent molecular mass of 35 to 40 kDa for the B78 CBL (1).
FIG. 1.
Physical and genetic map of the MG1363 metCcysK operon. For EcoRI, HindIII, and PstI, only sites relevant for subcloning are indicated. Pr, promoter.
The lactococcal MetC sequences showed high homology to each other (84.0% identity at the DNA level and 94.3% at the protein level) and showed clear sequence homologies with CBLs from E. coli (43.2% identity), Haemophilus influenzae (48.7% identity), and Bacillus subtilis (52.8% identity); with cystathionine γ-synthase from E. coli (44.8% identity) and Helicobacter pylori (65.0% identity); and with cystathionine γ-lyase from B. subtilis (63.2% identity), all of which belong to the family of PLP-dependent enzymes (2, 21, 32, 41). A multiple-sequence alignment of the MetC sequence from L. lactis and the homologous PLP proteins revealed that 53 amino acid residues are strictly conserved and that the N-terminal part shows the highest variability (Fig. 2). All residues important for catalysis, as reported by Clausen et al. (10), are conserved in the lactococcal MetC proteins.
FIG. 2.
Alignment of the amino acid sequences of the metC genes from L. lactis MG1363 (CBL-LLMG1363) and L. lactis B78 CBL-LLB78 with those of other members of the PLP-dependent protein family. Sequences were deduced from the following GenBank accession numbers: P56069 (Helicobacter pylori cystathionine γ-synthase [CGS-HPYL]), U93874 (B. subtilis cystathionine γ-lyase [CGL-BSUB]), Z99110 (B. subtilis CBL), P00935 (E. coli [ECOL] cystathionine γ-synthase), P06721 (E. coli CBL), and P44527 (Haemophilus influenzae [HIN] CBL). Conserved residues are indicated in white letters on a black background and with an asterisk underneath the sequences. The boxed region corresponds to a PLP binding site.
Downstream of the metC gene of L. lactis MG1363, we found a second open reading frame in the same orientation (Fig. 1), which encoded a putative 318-amino-acid protein that showed high sequence identity with cysteine synthases from B. subtilis (44.4% identity) (3), Mycobacterium tuberculosis (42.9% identity) (12), E. coli (40.4% identity) (6), and Salmonella enterica serovar Typhimurium (39.1% identity) (6). A putative ribosome binding site (AGGAA) was located 6 bp upstream of the start codon, but possible −35 and −10 promoter regions could not be identified upstream of the start codon.
Transcriptional analysis of the MG1363 metCcysK operon.
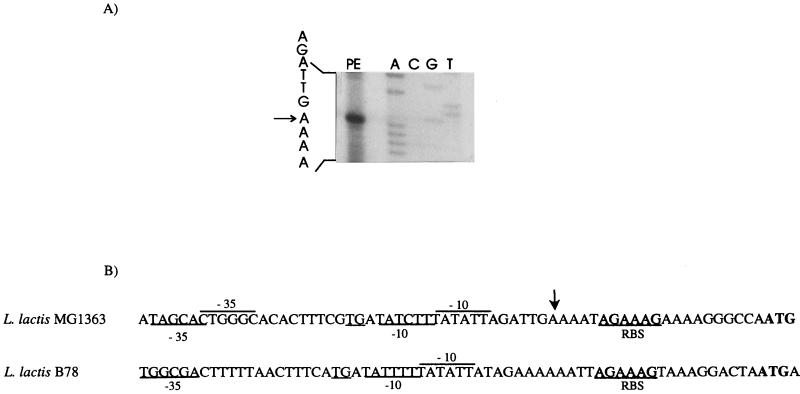
Primer extension experiments were performed on total RNA isolated from L. lactis MG1363 to identify the transcription initiation site. The transcription start site was found at the adenine residue located 20 nucleotides upstream of the translational start site (Fig. 3A). It is not preceded by clear consensus −35 and −10 sequences (Fig. 3B) (16). Downstream of the stop codon of the cysK gene of MG1363 we found a hairpin structure (ΔG = −15 kcal mol−1) which might function as a transcription termination site. The region upstream of the B78 metC start codon differed slightly from that of MG1363 (Fig. 3B).
FIG. 3.
(A) Transcription initiation site of the metC gene, determined by primer extension (PE, primer extension product). The DNA sequence ladder from L. lactis MG1363 was obtained with the same primer and is shown for size comparison (lanes A, C, G, and T). The arrow indicates the deduced transcription initiation site. (B) Comparison of the promoter regions from L. lactis MG1363 and B78. The predicted −35 and −10 regions and ribosome binding site (RBS) are indicated.
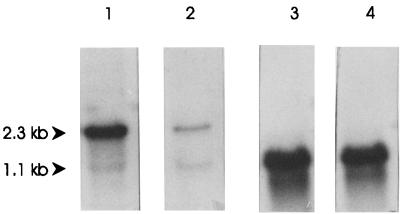
Northern hybridization experiments were carried out with RNAs from L. lactis MG1363 and B78. Hybridization with a metC-specific probe revealed a dominant 2.3-kb transcript that corresponds to a bicistronic metCcysK transcript and a less abundant 1.1-kb metC transcript in strain MG1363. In contrast to what occurred with MG1363, the intensities of both transcripts were more equal in strain B78, and the transcription level for B78 metCcysK was lower than for MG1363 metCcysK (Fig. 4). These results indicate that the metC and cysK genes form an operon in L. lactis.
FIG. 4.
Northern blot hybridization of RNAs isolated from L. lactis MG1363 (lane 1), L. lactis B78 (lane 2), and L. lactis NZ9000 harboring plasmid pNZ8136 (lane 3) or pNZ8137 (lane 4) induced with nisin. The membrane was hybridized with an internal gene fragment of MG1363 metC (lanes 1 and 3) or B78 metC (lanes 2 and 4). The predicted sizes of the different transcripts are indicated by arrowheads.
Overproduction of MetC in L. lactis.
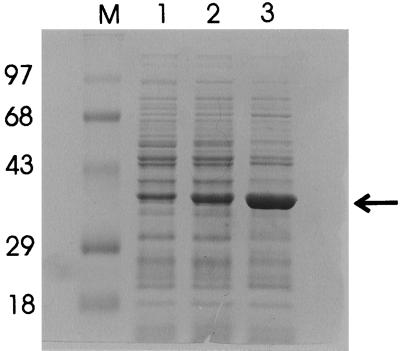
The metC gene was overexpressed with the NICE system (14, 31). The genes from both strains were cloned under the control of the nisA promoter by a PCR strategy, resulting in plasmids pNZ8136 and pNZ8137. These plasmids were introduced in L. lactis NZ9000 (31), which contains the nisRK signal transduction genes. Cultures of cells harboring the expression plasmids or plasmid pNZ8037 were induced with nisin. CFEs from induced and uninduced cultures were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, which showed the clear overproduction of a 39-kDa protein in induced cultures (Fig. 5). The induced cultures of L. lactis harboring pNZ8137 (metC from B78) showed a higher amount of MetC than that of the strain harboring pNZ8136 (metC from MG1363). In order to establish whether these differences in production levels corresponded with the transcriptional levels, RNAs were isolated from nisin-induced cultures of both strains, and Northern blot analysis revealed comparable levels of messenger RNA. The metC transcripts from the induced plasmid were more abundant than the chromosomal metCcysK transcript (Fig. 4).
FIG. 5.
Coomassie blue-stained gel after sodium dodecyl sulfate-polyacrylamide gel electrophoresis of extracts of L. lactis NZ9000 harboring pNZ8136, which contains MG1363 metC (lanes 1 [uninduced] and 2 [induced], or harboring pNZ8137, which contains B78 metC (lane 3 [induced]). The cultures were induced with 2.5 ng of nisin per ml. Lane M, molecular weight markers. Molecular weights (in thousands) are noted at the left. The location of the overproduced proteins is indicated (arrow).
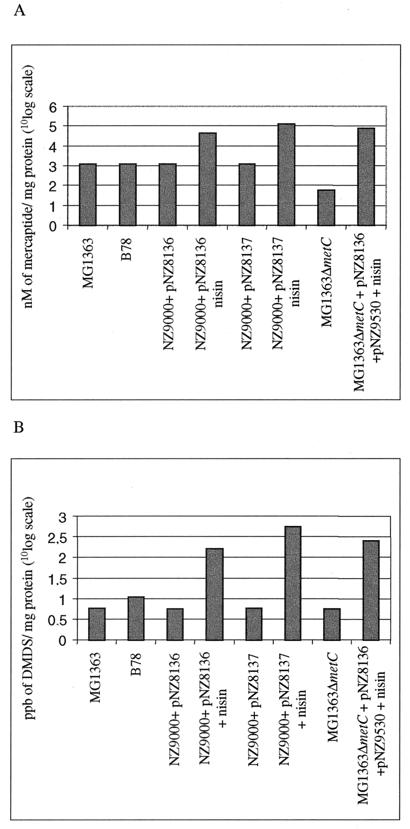
To analyze if the increase in the amount of MetC was related to an increment in CL enzyme activity, several assays were carried out with different substrates. Enzyme activity towards cystathionine was assayed by determining the formation of free-thiol groups (Fig. 6A). Methionine was also evaluated as a substrate by determining the formation of the volatile compound, DMDS, by headspace gas chromatography (Fig. 6B). Both wild-type strains (MG1363 and B78) showed similar activities towards cystathionine, but B78 revealed a higher production of volatile compounds (DMDS) from methionine than that of MG1363 (Fig. 6B). Overproduction of MG1363 MetC resulted in an increase in the enzyme activity of more than 25 times, while overproduction of B78 MetC resulted in a 100-fold increase in enzyme activity compared to that of the wild-type MG1363 strain.
FIG. 6.
Cystathionine lyase activity in CEs of L. lactis strains towards cystathionine (A), expressed as namolar concentrations of mercaptide formed from conversion of free thiols by DTNB, or methionine (B), expressed as parts per billion of DMDS formed. Nisin (2.5 ng/ml) was added where indicated. The values on the y axes are given in a 10log-unit scale. Values are the means of five independent measurements. Standard errors were less than 10% for each value indicated.
Disruption of metC.
For the construction of a metC knockout strain, an internal fragment of the metC gene of MG1363 was amplified by PCR and cloned in pG+host8, which has a thermosensitive replicon (34). Single-crossover mutants that were able to grow at 37°C under antibiotic selection were obtained as described by Maguin et al. (34), and plasmid integration was confirmed by Southern blot and PCR analyses. One colony was selected for further characterization and designated MG1363ΔmetC. The enzymatic activity in L. lactis MG1363ΔmetC was assayed with the different substrates and compared to that of MG1363 (Fig. 6). When cystathionine was used as the substrate for this knockout strain, a dramatic decrease in activity was observed (Fig. 6A). In contrast, no differences were observed when methionine was the substrate of the reaction (Fig. 6B), indicating that other lyases might be more prominent for this conversion.
Complementation of the metC knockout strain.
To ensure that the loss of the MetC activity in the MG1363ΔmetC strain was caused by the disruption of metC exclusively and not due to the expected polar effect on cysK expression, the strain was complemented with the MG1363 metC gene. In order to enable the use of the NICE system for the expression of the metC gene, plasmid pNZ9530 (29) was cotransformed with pNZ8136 into the knockout strain. The resulting strain had CL activity comparable to that of MG1363ΔmetC, but upon induction with nisin, we observed an overexpression of metC, resulting in an overproduction of CL activity (Fig. 6). These results show that CL activity is encoded mainly by the metC gene. The higher activity of the complemented knockout strain relative to that of the MG1363 MetC-overproducing strain is probably caused by the difference in concentrations of NisR and NisK, which originate from plasmid- and chromosome-located nisR and nisK genes, respectively.
DISCUSSION
Genes encoding CBLs have been cloned and characterized from several microorganisms, like E. coli, Salmonella serovar Typhimurium, Bordetella avium, B. subtilis, and H. influenzae (2, 21, 25, 32, 36). This enzyme catalyzes the penultimate step in microbial methionine biosynthesis. A CBL from L. lactis B78 that can also convert methionine into volatile products and therefore is likely to be involved in flavor formation during dairy fermentations like cheese ripening has been purified (1). We have cloned and characterized the metC genes encoding CBLs from two different L. lactis strains, MG1363, a plasmid-free model strain, and B78, a natural isolate from a mesophilic cheese starter culture.
The lactococal MetC proteins are highly similar and show high resemblances with other PLP-dependent enzymes, such as cystathionine γ-lyases and cystathionine γ-synthases. The characteristic residues of PLP proteins, determined for E. coli (10), are strictly conserved in the two MetC proteins (Fig. 2). They include the residues involved in cofactor binding (Tyr45, Arg47, Gly75, and Thr195), residues important for catalysis (Tyr99, Glu142, Asp171, Asn172, and Lys196), and the general α-carboxylate binding group (Arg358) (10, 11).
The metC gene is located in an operon with the cysK gene. In Salmonella serovar Typhimurium the metC gene is also cotranscribed with another gene, homologous to the yghB gene from E. coli (36). In E. coli, B. subtilis, or H. influenzae, the metC gene occurs as a single transcriptional unit (2, 21, 32, 38).
The cysK gene product of L. lactis is homologous to cysteine synthases, which catalyze the formation of cysteine from O-acetyl-l-serine and sulfide, the final step of cysteine biosynthesis. Additionally, both the lactococcal CBL and cystathionine γ-lyase are able to form cysteine from cystathionine (1, 5).
In E. coli, expression of the metC gene is controlled by the concentration of methionine in the medium (38). Recently, Dias and Weimer (17) observed that the CL activity in L. lactis is regulated by the amount of methionine and cysteine in the medium. We show that metC and cysK form one operon, which may imply that metCcysK gene expression is regulated by methionine and cysteine concentrations and that this can be considered a combined autoregulation of the methionine and cysteine biosynthesis routes. Future experiments need to verify whether MetC and CysK activities are regulated and, if so, whether regulation takes place at the transcriptional or posttranscriptional level.
Overproduction of enzymes that are involved in the development of flavor compounds in cheese is an attractive feature for the dairy industry. The overproduction of MetC proteins with the NICE system resulted in a 25- to 100-fold increase in the activities of the enzyme towards different substrates, as is shown in Fig. 6. Interestingly, both the enzyme activity and the amount of protein were higher for the strain overproducing B78 MetC than for that overproducing MG1363 MetC (Fig. 5 and 6). Northern blot analysis revealed no differences at the transcriptional level between both overproducing strains (Fig. 4). In spite of the high degree of similarities between the MG1363 and B78 metC genes, a comparison of the codon usages revealed that there were significantly more preferred codons for L. lactis (9), and fewer nonpreferred codons, in the B78 metC sequence than in that of MG1363 (data not shown), which might account for a better translation efficiency and higher production level of MetC from L. lactis B78. A better translation efficiency of the B78 metC gene does not result in higher CBL activity in the wild-type strain than that in MG1363 (Fig. 6). This observation might be explained by differences in transcription levels for both genes, as a higher mRNA level was detected for the MG1363 metC gene in the wild-type situation (Fig. 4). This discrepancy in gene expression is likely to be caused by the differences in the promoter regions (Fig. 3B). The higher transcription level of the metC gene in MG1363 and the better translation efficiency of the B78 metC transcript probably result in comparable enzyme activities in both strains.
Disruption of metC resulted in a notable decrease in the CL activity towards cystathionine (Fig. 6A). This result clearly indicates that the metC gene encodes a protein which catalyzes the formation of homocysteine from cystathionine, as has been reported before (18, 38). Although most of the L. lactis strains are auxotrophic for methionine, the biosynthetic route for this amino acid seems to exist in L. lactis, as some autotrophic strains are known (9). In many L. lactis strains, the methionine synthesis pathway is probably interrupted due to the adaptation of L. lactis for growth in a rich medium like milk (9, 13).
As has been reported by several authors, the enzymes involved in methionine biosynthesis are also important for methionine degradation in food fermentation (1, 17). Although the main catabolic reaction of CBL is the conversion of cystathionine to homocysteine, the CBL purified from L. lactis B78 is also able to catalyze the conversion of methionine to methanethiol, which can subsequently be oxidized to DMDS and DMTS (1). We show that extracts of L. lactis cells overproducing MetC yield larger amounts of DMDS formed from methionine than the wild type (Fig. 6B). However, no significant differences in the DMDSs formed were found between the knockout strain and the wild-type strain. This implies that enzymes other than CBL, like cystathionine γ-lyase (5), play a more prominent role in the conversion of methionine. Moreover, in addition to conversion by the CBL or cystathionine γ-lyase activity, methionine can be converted to volatile flavor compounds by aminotransferase activity (23, 46).
In conclusion, we have cloned, disrupted, overexpressed, and transcriptionally characterized the metC gene from two L. lactis strains. Although CBL is not essential for the conversion of methionine to volatile products, its overproduction results in an increase in flavor compound formation. Future studies will focus on the role of MetC in dairy product flavor formation and regulatory factors acting upon it.
ACKNOWLEDGMENTS
We thank Roland Siezen and Michiel Kleerebezem for critically reading the manuscript.
M.F. was supported by a scholarship of the Ministerio de Educacion y Cultura of Spain. Part of this work was supported by the EU research grant FAIR CT97-3173.
REFERENCES
- 1.Alting A C, Engels W J M, van Schalkwijk S, Exterkate F A. Purification and characterization of cystathionine β-lyase from Lactococcus lactis subsp. cremoris B78 and its possible role in flavor development in cheese. Appl Environ Microbiol. 1995;61:4037–4042. doi: 10.1128/aem.61.11.4037-4042.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belfaiza J, Parsot C, Martel A, de la Tour C B, Margarita D, Cohen G N, Saint-Girons I. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci USA. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A, Sorokin A, Ehrlich S D. Mapping of the 150 kb spoIIIC-pheA region of the Bacillus subtilis chromosome using long accurate PCR and three yeast artificial chromosomes. Microbiology. 1996;142:3017–3020. doi: 10.1099/13500872-142-11-3017. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bruinenberg P G, de Roo G, Limsowtin G K Y. Purification and characterization of cystathionine γ-lyase from Lactococcus lactis subsp. cremoris SK11: possible role in flavor compound formation during cheese maturation. Appl Environ Microbiol. 1997;63:561–566. doi: 10.1128/aem.63.2.561-566.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne C R, Monroe R S, Ward K A, Kredich N M. DNA sequences of the cysK regions of Salmonella typhimurium and Escherichia coli and linkage of the cysK regions to ptsH. J Bacteriol. 1988;170:3150–3157. doi: 10.1128/jb.170.7.3150-3157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 8.Chiaruttini C, Milet M. Gene organization, primary structure and RNA processing analysis of a ribosomal RNA operon in Lactococcus lactis. J Mol Biol. 1993;230:57–76. doi: 10.1006/jmbi.1993.1126. [DOI] [PubMed] [Google Scholar]
- 9.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–38. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 10.Clausen T, Huber R, Laber B, Pohlenz H D, Messerschmidt A. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine β-lyase from Escherichia coli at 1.83 Å. J Mol Biol. 1996;262:202–224. doi: 10.1006/jmbi.1996.0508. [DOI] [PubMed] [Google Scholar]
- 11.Clausen T, Laber B, Messerschmidt A. Mode of action of cystathionine β-lyase. J Biol Chem. 1997;378:321–326. [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;11:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Delorme C, Godon J J, Ehrlich S D, Renault P. Gene activation in Lactococcus lactis: histidine biosynthesis. J Bacteriol. 1993;175:4391–4399. doi: 10.1128/jb.175.14.4391-4399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vos W M, Vos P, Dehaard H, Boerrigter I. Cloning and expression of the Lactococcus lactis ssp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 16.de Vos W M, Simons G. Gene cloning and expression systems in lactococci. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, United Kingdom: Chapman & Hall; 1994. pp. 52–105. [Google Scholar]
- 17.Dias B, Weimer B. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl Environ Microbiol. 1998;64:3320–3326. doi: 10.1128/aem.64.9.3320-3326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwivedi C M, Ragin R C, Uren J R. Cloning, purification and characterization of β-cystathionase from Escherichia coli. Biochemistry. 1982;21:3064–3069. doi: 10.1021/bi00256a005. [DOI] [PubMed] [Google Scholar]
- 19.Engels W J M, Visser S. Development of cheese flavour from peptides and amino acids by cell-free extracts of Lactococcus lactis subsp. cremoris B78 in a model system. Neth Milk Dairy J. 1996;50:3–17. [Google Scholar]
- 20.Exterkate F A, Alting A C. The role of starter peptidases in the initial proteolytic events leading to amino acids in Gouda cheese. Int Dairy J. 1995;5:15–28. [Google Scholar]
- 21.Fleischmann R D, Adams M D, White O, Clayton R A, Kierkness E F, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 22.Gao S, Oh D H, Broadbent J R, Johnson M E, Weimer B C, Steele J L. Aromatic amino acid catabolism by lactococci. Lait. 1997;77:371–381. [Google Scholar]
- 23.Gao S, Steele J L. Purification and characterization of oligomeric species of an aromatic amino acid aminotransferase from Lactococcus lactis subsp. lactis S3. J Food Biochem. 1998;22:197–211. [Google Scholar]
- 24.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentry-Weeks C R, Keith J M, Thompson J. Toxicity of Bordetella avium beta-cystathionase toward MC3T3-E1 osteogenic cells. J Biol Chem. 1993;268:7298–7314. [PubMed] [Google Scholar]
- 26.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1988. p. 46. [Google Scholar]
- 27.Hemme D, Bouillanne C, Métro F, Desmazeaud M J. Microbial catabolism of amino acids during cheese ripening. Sci Aliment. 1982;2:113–123. [Google Scholar]
- 28.Imhof R, Bosset J O. Relationships between micro-organisms and formation of aroma compounds in fermented dairy products. Z Lebensm-Unters-Forsch. 1994;198:267–276. [Google Scholar]
- 29.Kleerebezem M, Beerthuyzen M M, Vaughan E E, de Vos W M, Kuipers O P. Controlled expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl Environ Microbiol. 1997;63:4581–4584. doi: 10.1128/aem.63.11.4581-4584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 31.Kuipers O P, de Ruyter P G G A, Kleerebezem M, de Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 32.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 33.Law B A. Proteolysis in relation to normal and accelerated cheese ripening. In: Fox P F, editor. Cheese: chemistry, physics and microbiology. Vol. 1. London, England: Elsevier Applied Science; 1987. pp. 365–392. [Google Scholar]
- 34.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mierau I, Kunji E R, Venema G, Kok J. Casein and peptide degradation in lactic acid bacteria. Biotechnol Genet Eng Rev. 1997;14:279–301. doi: 10.1080/02648725.1997.10647945. [DOI] [PubMed] [Google Scholar]
- 36.Park Y M, Stauffer G V. DNA sequence of the metC gene and its flanking regions from Salmonella typhimurium LT12 and homology with the corresponding sequence of Escherichia coli. Mol Gen Genet. 1989;216:164–169. doi: 10.1007/BF00332246. [DOI] [PubMed] [Google Scholar]
- 37.Parliment T H, Kolor M G, Rizzo D J. Volatile components of Limburger cheese. J Agric Food Chem. 1982;30:1006–1008. [Google Scholar]
- 38.Saint-Girons I, Parsot C, Zakin M M, Bârzu O, Cohen G N. Methionine biosynthesis in enterobacteriaceae: biochemical, regulatory, and evolutionary aspects. Crit Rev Biochem. 1988;23(Suppl. 1):1–42. doi: 10.3109/10409238809083374. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook H, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Smacchi E, Gobbetti M. Purification and characterization of cystathionine γ-lyase from Lactobacillus fermentum DT41. FEMS Microbiol Lett. 1998;166:197–202. doi: 10.1111/j.1574-6968.1998.tb13890.x. [DOI] [PubMed] [Google Scholar]
- 41.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 42.Uren J R. Cystathionine β-lyase from Escherichia coli. Methods Enzymol. 1987;143:483–486. doi: 10.1016/0076-6879(87)43086-3. [DOI] [PubMed] [Google Scholar]
- 43.van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 44.Visser F W M. Contribution of enzymes from rennet, starter bacteria and milk to proteolysis and flavour development in cheese. 3. Protein breakdown: analysis of the soluble nitrogen and amino acid nitrogen fractions. Neth Milk Dairy J. 1997;31:210–239. [Google Scholar]
- 45.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Yvon M, Thirouin S, Rijnen L, Fromentier D, Gripon J C. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl Environ Microbiol. 1997;63:414–419. doi: 10.1128/aem.63.2.414-419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]