Abstract
Innate immune sensor IFN-induced protein 16 (IFI16) exhibits anti-inflammatory effects via IFNβ and IFN-stimulated gene (ISG)15 induction in cancer cells. Epigallocatechin gallate (EGCG) is a potent natural DNA methyltransferase inhibitor (DNMTi). Previous studies revealed that conventional DNMTis, such as 5-azacytidine (5-aza-dc), induce IFI16 expression and EGCG decreases DNMT mRNA expression and global methylation (5mC) level via promoter demethylation of tumor suppressor genes in cancer cell lines. To the best of our knowledge, however, EGCG-mediated IFI16 promoter methylation status has been overlooked. Here, initial screening was performed to determine IFI16 expression and its correlation with DNMTs in cancer cell lines from various databases. Following treatment of breast cancer cell lines with 5-aza-dc, vitamin C and EGCG, expression levels of IFI16 and its downstream transcription targets IFNβ1 and ISG15 were assessed using RT-qPCR, and the 5mC level was assessed using ELISA. In silico molecular docking simulation was performed for all DNMTs to predict the mode of ligands binding with proteins. Finally, promoter methylation level in IFI16 gene was assessed following EGCG treatment. EGCG treatment induced IFI16 expression, interacted with certain amino acids residues in DNMT proteins and decreased 5mC level and promoter methylation of IFI16. The present results may provide a basis for targeting IFI16 expression as a therapeutic option in breast cancer cell lines.
Keywords: DNA methyltransferase, 5-methyl cytosine, epigallocatechin gallate, promoter methylation, IFN-induced protein 16
Introduction
IFN-induced protein 16 (IFI16) is a potent innate immune response evoker that amplifies DNA damage-mediated innate immune response in inflammatory disease (1–3). It also contributes to cell senescence (3). However, previous studies revealed the IFI16, which belongs to the pyrin and HIN domain family, serves a key role as an innate immune sensor and sensitizes foreign or intracellular double-stranded (ds)DNA (4,5). Although IFI16 predominantly sensitizes cytosolic DNA, another study showed that it can also translocate into nucleus to detect dsDNA (5,6).
The alternate splicing of mRNA yields three structural isoforms when IFI16 is transcribed: A, B and C (7,8). IFI16 gene comprises 200-amino acid repeats known as HIN-200 domain, which is classified as HIN-200-A or HIN-200-B. Both A and B domains are separated by the serine-threonine-proline-rich spacer region (9–11). Moreover, the basic region of 1–159 residues binds dsDNA at the N-terminus of the HIN-200-A subunit as well as the N-terminus of the HIN-200-A subunit to form a peptide sequence of amino acids with PYRIN domain that mediates the signal for nuclear localization of IFI16 from cytosol to nucleus (11).
IFI16 exerts its key innate immune responses in diverse manners. For example, upon recognition of cytosolic dsDNA by cyclic GMP-AMP synthase, it activates the downstream STING/TANK-binding kinase 1/IFN regulatory factor 3 pathway to induce transcription of IFNβ (12). Moreover, IFI16 binds with dsDNA and induces IFNβ transcription, which, in turn, induces IFN-stimulated gene 15 (ISG15) transcription via the common STING axis (13). Another study showed that IFI16 induces IFNβ expression while knockdown of IFI16 abrogates the IFNβ response (14). Transcriptional cross-talk between IFI16 and IFNβ has been proposed in a previous study (15). In addition to acting as a dsDNA sensor, IFI16 also serves as an ISG (16–18).
Previous studies have reported downregulated expression of IFI16 in cancer cells. For example, a study on the cytoplasm of prostate cancer cell lines demonstrated that IFI16 gene was not expressed or expressed in variant forms; the study also showed that functional overexpression of IFI16 halted colony formation (19). Another study showed that in certain cancer cells, loss of IFI16 expression facilitates cell survival even in the presence of low glucose (20). Moreover, IFI16 acts as a tumor suppressor gene, inhibits proliferation of hepatocellular carcinoma and triggers apoptosis (21). However, controversial findings have shown an oncogenic role of IFI16 in certain types of cancer cell, such as oral squamous and renal cell carcinoma and pancreatic adenocarcinoma (22–24).
Natural compounds, such as like vitamin C and green tea extract polyphenol Epigallocatechin gallate (EGCG), exert multiple effects on lipid antioxidation (25) and transcriptional machinery to modulate gene expression by targeting signaling pathways or transcription factors or epigenetically modulating gene expression (26). Moreover, EGCG has been shown to block DNA methyltransferase (DNMT) activity and reactivates methylation-mediated silenced genes, such as p16, in cancer cell lines (27). Additionally, EGCG and vitamin C increase DNA demethylation by inhibiting DNMT1/DNMT3b and modulating ten-eleven translocation enzymes, respectively (28). EGCG exhibits modulatory effects on innate and adaptive immune response stimuli in murine and human models (29,30). In addition, vitamin C also showed innate immune response booster through viral mimicry (pseudo-infection) in breast, colon, leukemia and hepatocellular cancer cell lines (31).
To the best of our knowledge, few studies have been conducted on epigenetic drug-mediated IFI16 expression. For example, IFI16 is acetylated in herpes simplex virus-1 (HSV-1) (32). Acetylation and deacetylation regulate nuclear and cytoplasmic localization of IFI16 (32). Another study showed that histone deacetylase inhibitor trichostatin A and CGK1026 induce IFI16 gene expression in prostate cancer cells (33). In addition, at a single-cell level, low dose treatment with DNA demethylating agents induces IFI16 gene expression (34). On the other hand, EGCG is a promoter demethylation-mediated epigenetic drug that induces expression of tumor suppressor genes in colon cancer cells (35). To the best of our knowledge, no previous study has investigated whether EGCG induces IFI16 expression by decreasing methylation of the IFI16 promoter. Therefore, the present study aimed to investigate this hypothesis.
Materials and methods
Preliminary screening of mRNA correlation, expression and immune responsiveness of targeted protein
IFI16 and DNMTs gene (Database of Genotypes and Phenotypes accession no. phs000424.v8.p2) expression was screened out using The Cancer Genome Atlas (TCGA) datasets through the UALCAN web server (ualcan.path.uab.edu/). The correlation between IFI16 and DNMTs (accession no. ENSG00000163565.17) were screened from the web server and validated by data retrieved from xenabrowser.net/. The immune responsiveness study of IFI16 was assessed by TIMER2.0 web server (timer.cistrome.org/). Finally, cell lines were screened from the Human Protein Atlas database where the targeted protein IFI16 gene expression was absent or downregulated. Based on the mRNA expression, SHSY5Y (neuroblastoma), T47D (ductal breast carcinoma), HepG2 (hepatocellular carcinoma), MCF-7 (breast cancer cell) and HeLa (cervical cancer cell) cell lines were identified.
Cell culture and treatment
MCF-7 cell line was procured from American Type Culture Collection and sub-cultured in DMEM supplemented with 10% FBS (both UFC Biotech) and 1% penicillin and incubated at 37°C in a 5% CO2 incubator. Upon 60–80% confluence, cells were trypsinized, seeded (3,000 cells/well) in 6-well plates and incubated at 37°C overnight to ensure that cells were healthy without any contamination. Based on previous studies, half maximal inhibitory concentration doses for EGCG were added to MCF-7 and T47D cell lines at 40 and 20 µM for 48 h, respectively and incubated at 37°C (36,37). At the same time, 5-azacytadine (5-aza-dc) and vitamin C were added at 60 and 240 µM, respectively, and incubated at 37°C for 48 h.
cDNA synthesis and quantitative (q)PCR
Total RNA was extracted using PureLink™ RNA Mini kit (cat. no. 1944999; Thermo Fisher Scientific, Inc.). A total of 100 ng/µl RNA from untreated and treated MCF-7 and T47D cell lines was transcribed into cDNA using High Capacity cDNA Synthesis kit (cat. no. 00656567; Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. IFI16, IFNβ, ISG15 primer sequences were used as previously described (31,38,39) (Table I). DNMTs primers were designed using the UCSC genome browser (genome.ucsc.edu/; Table I). Reverse transcription (RT)-qPCR was performed using PowerUp SYBR Green Master Mix (cat. no. 1805029; Applied Biosystems; Thermo Fisher Scientific, Inc.). Thermocycling conditions were as follows: 50°C for 2 min, 95°C for 2 min, 95°C for 15 sec and 60°C for 1 min. RT-qPCR was performed using ABI 7300 Prism. The numbers of transcripts were normalized to RPLP0 (forward Primer; 5′ATGTGGGCTTTGTGTTCACC3′ and Reverse Primer; 5′TCCAGTCTTGATCAGCTGCA3′) and calculated via the 2−ΔΔCq method (40).
Table I.
Reverse transcription-quantitative PCR primers.
| Gene | Forward primer, 5′-3′ | Reverse primer, 5′-3′ |
|---|---|---|
| IFI16 | CTCGGAGAGCTCGGACAG | TACCTATGACGACGCTGCTG |
| ISG15 | GCCTCAGCTCTGACACC | CGAACTCATCTTTGCCAGTACA |
| IFNB1 | TCTGGCACAACAGGTAGTAGGC | GAGAAGCACAACAGGAGAGCAA |
| DNMT1 | CAGCAACGGGCAGATGTTTC | CGGAGGGTGCTTTGTAGATG |
| DNMT3a | CTACGCACCACCTCCACCAG | CAATGTTCCGGCACTTCTGC |
| DNMT3b | GAGTCCATTGCTGTTGGAACCG | ATGTCCCTTTGTCGCCAACCT |
| Methylated IFI16 | TTCGAGTAGTTGGGATTATAGGC | TAATACAAAATTAACTAAACGCGAT |
| Unmethylated IFI16 | TTTTTTGAGTAGTTGGGATTATAGGT | AAATAATACAAAATTAACTAAACACAAT |
IFI, IFN-induced protein; ISG, IFN-stimulated gene; DNMT, DNA methyltransferase.
Extraction of genomic DNA
Genomic DNA was extracted from untreated and treated MCF-7 and T47D cell lines using a DNAbler kit (havensci.com/; cat. no. DE95050). A total of 200 µl digestion buffer was added per sample followed by 20 µl Proteinase K and RNase A. The sample was vortexed and centrifuged at 1,000 × g for 10 min in 25°C followed by 5 min incubation in a heat block (55-60°C). A total of 200 µl lysis buffer was added, vortexed and centrifuged at 10,000 × g for 5 min in 25°C. Then, 99% ethanol was added, followed by short vortex and centrifugation at 10,000 × g for 15 sec in 25°C The ethanol-lysis buffer content was transferred into the nuclease-free spin column and centrifuged at 10,000 × g for 1 min in 25°C. Then, 500 µl wash buffer was added according to the manufacturer's instructions and, 50 µl elution buffer was added to the spin column and centrifuged at 8,000 × g for 2 min at 25°C to elute genomic DNA.
Determination of global methylation (5mC) levels
MethylFlash™ 5mC ELISA Easy kit (cat. no. P-1030, EpiGentek) was used to assess 5mC level in MCF-7 and T47D cell lines in untreated and treated conditions. According to manufacturer's protocol, binding solution was added to 200 ng genomic DNA/sample in 8 wells followed by addition of 5 mC antibody and developer and stop solution. Finally, optical density was measured at 450 nm using BioTek ELISA microplate reader.
Protein, ligand retrieval and molecular docking simulation
The crystallographic protein structure of DNMT1, DNMT3a and DNMT3b (ID nos. 4wxx, 6brr, 6kdl) was retrieved from the Protein Database Bank web database (rcsb.org/). The 2D structure of 5-aza-dc (PubChem ID 9444), vitamin C (PubChem ID 54670067), EGCG PubChem ID 65064), S-adenosyl methionine (SAM) (PubChem ID 34755) and S-adenosyl homocysteine (SAH) (PubChem ID 439155) were retrieved from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) database as sdf format. The 5-aza-dc was selected as a positive control for DNMT proteins. The proteins were preprocessed using Biovia Discovery Studio visualizer (3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/visualization/) version (16.1.0) (41). For molecular docking, PyRX software version 0.8 (https://pyrx.sourceforge.io/) (42) was used via integrated Open Bable to optimize ligands. Autodock wizards were used to create maximum grid box dimensions. After running docking via Run Autodock in Vina wizard, the ligand in a complex with the protein was visualized using Biovia Discovery Studio visualizer version (16.1.0) (41).
Bisulfite modification and methylation-specific qPCR
Using EpiJET DNA Bisulfite Conversion kit (cat. no. 00596381; Thermo Fisher Scientific, Inc.), bisulfite conversion of genomic DNA was performed according to the manufacturer's protocol. The bisulfite-modified DNA template was used for qPCR using kit (Hi-Tech Green 2X qPCR Universal Mix, cat. no. GQM-M-001-10, Molequle-On). Whole gene sequences were selected for IFI16 and submitted to Metprimer online software (urogene.org/methprimer/) v1.1 beta for methylated and unmethylated primer design (Table I). During primer design, CpG islands were selected. Bisulfite-converted DNA was used for qPCR using Methylation specific PCR primers and thermocycling conditions as follows: 50°C for 2 min, 95°C for 2 min, 95°C for 15 sec and 60°C for 1 min. For gel electrophoresis, 1.7% (w/v) agarose gel was prepared in 1X Tris acetate EDTA with ethidium bromide staining. qPCR product was mixed with 6X DNA gel loading dye for separation via electrophoresis. Amplified products were visualized using GelDoc Biorad imaging system.
Statistical analysis
Unpaired t-test was used to compare two groups; >2 groups were compared using one-way ANOVA followed by post hoc Tukey's post hoc test. Data were analyzed using GraphPad Prism software (version 9; GraphPad Software, Inc.). P≤0.05 was considered to indicate a statistically significant difference. The data are presented as the mean ± SD of one independent experiment performed in triplicate.
Results
IFI16 is differentially expressed in different tumor cell lines, inversely correlated with differential DNMT mRNA expression and correlated with immune responsiveness
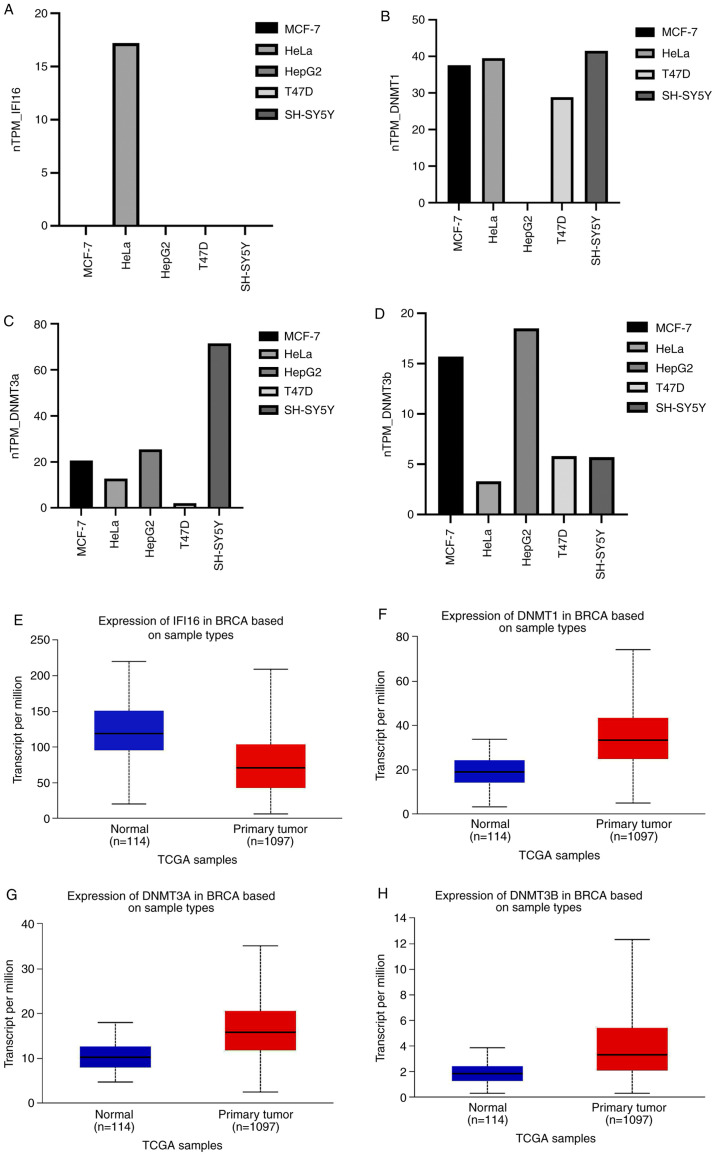
IFI16 expression was screened in various tumor and normal cell lines. IFI16 expression was not found in MCF-7, T4D, SHSY5, HepG2 cell lines but was observed in HeLa (Fig. 1A). DNMT mRNA expression was inversely associated with IFI16 mRNA expression in most cell lines, except HeLa (Fig. 1A-D). Moreover, TGCA omics data showed that the breast cancer (BRCA) has a significantly lower expression of IFI16 (Fig. 1E) but higher expression of DNMT1 (Fig. 1F), DNMT3 (Fig. 1G) and DNMT3B (Fig. 1H). An inverse correlation between expression of IFI16 and DNMTs was observed in the breast cancer cell line however this was not statistically significant (Fig. S1A). The immune responsiveness study of IFI16 showed a moderate correlation with different immune infiltrating cells such as CD8+/4+ T cells, macrophages, dendritic cells and neutrophils (Fig. S1B). Hence, the screening results indicated IFI16 had impact on immune infiltrating cells.
Figure 1.
Differential expression of IFI16 and DNMTs in cancer cell lines and immune response of IFI16. (A) Different cancer cell lines showing IFI16 gene expression. (B) Different cancer cell lines showing (B) DNMT1, (C) DNMT3a and (D) DNMT3b gene expression. (E) TCGA samples data showing IFI16 gene expression (P=1.8×10−12) in breast cancer cell line. (F) TCGA samples data showing DNMT1 gene expression (P<10−12) in breast cancer cell line. (G) TCGA samples data showing DNMT3A gene expression (P=1.6×10−12) in breast cancer cell line. and (H) TCGA samples data showing DNMT3B gene expression (P=1.6×10−12) in breast cancer cell line. IFI, IFN-induced protein; DNMT, DNA methyltransferase; TPM, transcripts per million; TCGA, The Cancer Genome Atlas.
DNMTi analog EGCG induces IFI16 and IFN-associated gene expression in breast cancer cell lines
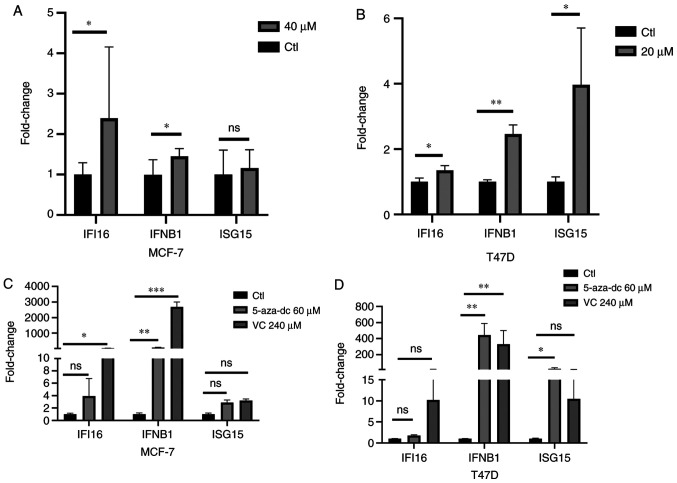
The extracted RNAs from EGCG-treated cell lines were used to investigate IFI16 expression. The results showed that 40 µM EGCG induced expression of IFI16 in the MCF-7 cell line and 20 µM EGCG treatment induced expression in the T47D cell line (Fig. 2A and B). As IFI16 acts as an upstream target for transcribing IFNβ1 and ISG15 (43), expression of IFNβ1 and ISG15 was assessed in both cell lines. In MCF-7 cells, 40 µM treatment induced IFNβ1 expression but did not affect ISG15 gene expression (Fig. 2A). By contrast, 20 µM EGCG induced IFNβ1 and ISG15 gene expression in the T47D cell line (Fig. 2B). These data suggested that EGCG treatment induced expression of IFI16 and its downstream targeted genes expression.
Figure 2.
Differential expression of IFI16 and IFN-associated genes following EGCG and DNMTi treatment. IFI16 and IFN-associated gene expression in EGCG-treated (A) MCF-7 and (B) T47D cell lines. IFI16 and IFN-associated gene expression in DNMTi-treated (C) MCF-7 and (D) T47D cell lines. The data are presented as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. IFI, IFN-induced protein; DNMTi, DNA methyltransferase inhibitor; EGCG, Epigallocatechin gallate; Ctl, control; ISG, IFN-stimulated gene; 5-aza-dc, 5-Azacytadine; VC, vitamin C; ns, not significant.
Gene expression levels in both cell lines were also assessed following treatment with conventional DNMTis, such as 5-aza-dc and vitamin C. IFI16, IFNβ1 and ISG15 gene expression in MCF-7 cell line was induced following treatment with 60 µM 5-aza-dc or 240 µM vitamin C (Fig. 2C) although the slight induction of ISG15 gene expression results were not statistically significant. Similar results were also observed in the T47D cell line in which IFI16 gene expression was induced following treatment with 5-aza-dc and vitamin C but this was not statistically significant (Fig. 2D). Moreover, both treatments significantly induced the IFNβ1 gene expression in T47D cell line, while ISG15 gene expression was significantly induced with 5-aza-dc treatment (Fig. 2D). Hence, DNMTi or vitamin C may induce the IFI16 gene and its downstream targeted genes IFNβ1 and ISG15.
DNMTi analog EGCG decreases DNMT expression in breast cancer cell lines
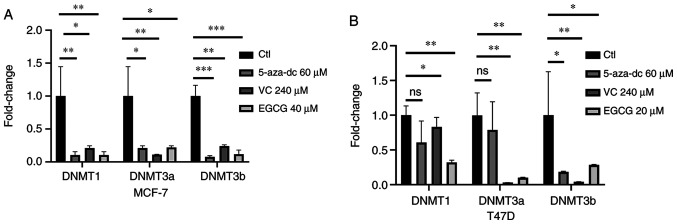
5-aza-dc and vitamin C are potent DNMTis that modify the epigenome (31,44). Hence, DNMT gene expression was assessed following treatment with EGCG, 5-aza-dc or vitamin C in MCF-7 and T47D cell lines. DNMT1, DNMT3a and DNMT3b gene expression levels were notably decreased following treatment with 60 µM 5-aza-dc or 240 µM vitamin C in both cell lines (Fig. 3A and B). Additionally, 40 and 20 µM EGCG treatment decreased expression of all DNMTs in both MCF-7 and T47D cell lines (Fig. 3A and B). These data indicated that DNMTi, vitamin C and EGCG may affect DNMT gene expression.
Figure 3.
Differential expression of DNMTs following EGCG and DNMTi treatment. DNMT expression in EGCG- or DNMTi-treated (A) MCF-7 and (B) T47D cell lines. The data are presented as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. DNMTi, DNA methyltransferase inhibitor; EGCG, Epigallocatechin gallate; Ctl, control; 5-aza-dc, 5-Azacytadine; VC, vitamin C; ns, not significant.
Receptor grid box generation and molecular docking simulation: In silico interaction of EGCG with DNMT
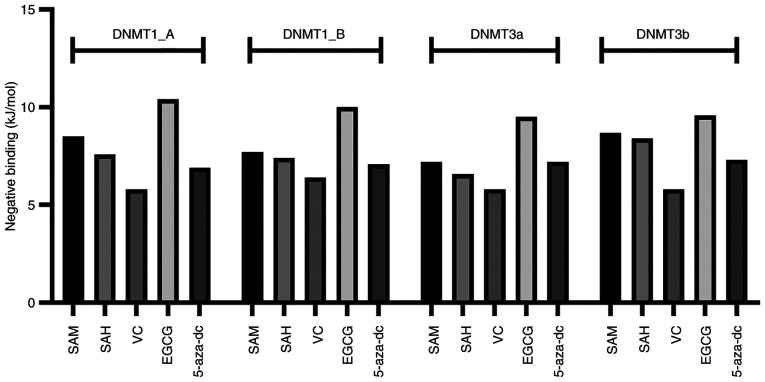
Molecular docking-based computational techniques have been considered as a mechanistic tool (45) for in silico drug design. For DNMT1_A, grid box comprised X=205.34, Y=88.76 and Z=145.45 points and spaced dimension was centered on DNMT1_A protein at X=−23.06, Y=47.81 and Z=−23.78. The binding energy of SAM, SAH, vitamin C, EGCG and 5-aza-dc with DNMT1_A was −8.5, −7.6, −5.8, −10.4 and −6.9 kJ/mol, respectively (Fig. 4). For DNMT1_B, grid box comprised X=120.97, Y=89.06 and Z=95.42 points and spaced dimension was centered on the DNMT1_B protein at X=17.63, Y=21.36 and Z=−51.18. The binding energy of SAM, SAH, vitamin C, EGCG and 5-aza-dc with DNMT1_B was −7.7, −7.4, −6.4, −10.0 and −7.1 kJ/mol, respectively (Fig. 4). For DNMT3a, grid box comprised of X=83.85, Y=87.88 and Z=60.33 points and spaced dimension was centered on X=−20.31, Y=−56.03 and Z=20.19. The binding energy of SAM, SAH, vitamin C, EGCG and 5-aza-dc with DNMT3a was −7.2, −6.6, −5.8, −9.5 and −7.2 kJ/mol, respectively (Fig. 4). For DNMT3b, the grid box comprised X=83.85, Y=87.88 and Z=60.33 points and spaced dimension was centered on the DNMT3b protein at X=−20.31, Y=−9.01 and Z=−3.88. The binding energy of SAM, SAH, vitamin C, EGCG and 5-aza-dc with DNMT3b was −8.7, −8.4, −5.8, −9.6 and −7.3 kJ/mol, respectively (Fig. 4).
Figure 4.
In silico docking simulation of different DNMTs with SAM, SAH, 5-aza-dc, VC and EGCG. Docking score as binding energy of all compounds with DNMT1_A chain, DNMT1_B chain, DNMT3a and DNMT3b. DNMT, DNA methyltransferase; 5-aza-dc, 5-Azacytadine; VC, vitamin C; EGCG, Epigallocatechin gallate; SAM, S-adenosyl methionine; SAH, S-adenosyl homocysteine.
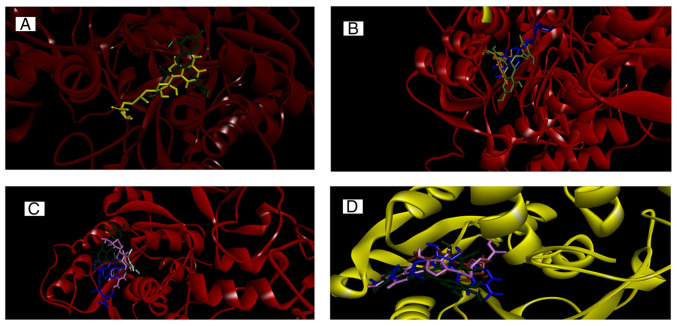
In silico molecular docking simulation predicted that SAM and EGCG compounds bind in similar positions. SAM was bound at Glu:1168, Phe:1145, Glu:1266, Arg:1310, Thr:1526, Thr:1528 and Asn:1578 residue (Fig. S2), whereas EGCG bound at Met:1169, Pro:1225, Glu:1168, Glu:1266 and Arg:1310 residue (Fig. S2). Though more conventional H bond interactions were found in the case of SAM, other molecular bonding interactions, such as π-anion and π-alkyl, in addition to four conventional H bonds, were also observed in the EGCG-DNMT1_A complex (Fig. S2). Furthermore, 5-aza-dc was found to interact with DNMT1_A by interacting with Glu:562, Glu:566, Asp:565, Ser:570 and Gln:687 residue with 3 conventional H bonds (Fig. S2). Vitamin C bound at Gln:369, Cys:409, Ser:436 and Glu:494 residue with 4 conventional H bonds (Fig. S2). SAH bound at Glu:562, ASP:565, Glu:566, Pro:574 and Arg:690 with conventional H bond and alkyl interactions (Fig. S2). The predicted site of SAM and EGCG interaction is presented in Fig. 5A.
Figure 5.
In silico interaction of EGCG, SAM, SAH, 5-aza-dc and vitamin C with DNMTs. (A) Predicted binding sites for SAM (yellow) and EGCG (green) in DNMT1_A chain. (B) Predicted binding sites for 5-aza-dc (orange), SAM (blue) and EGCG (green) in DNMT1_B chain. (C) Predicted binding sites for SAM (blue), SAH (pink), EGCG (green) and vitamin C (white) in DNMT3a. (D) Predicted binding sites for SAM (blue), SAH (pink), EGCG (green) and 5-aza-dc (orange) in DNMT3b. DNMT, DNA methyltransferase; 5-aza-dc, 5-Azacytadine; VC, vitamin C; EGCG, Epigallocatechin gallate; SAM, S-adenosyl methionine; SAH, S-adenosyl homocysteine.
For DNMT1_B, 5-aza-dc interacted with Phe:1145, Ser:1146, Leu:1151 and Asn:1578 with four conventional H bonds and three C-H bonds. SAM interacted with Phe:1145, Glu:1168, Glu:1266, Arg:1310, Gln:1536, Arg:1574 and Asn:1578 with eight conventional H bonds. EGCG interacted with DNMT1_B at Ser:1146, Glu:1168, Pro:1224/1225, Glu:1266 and Ala:1579 with four conventional H bonds, and one each of π-anion, π-alkyl and C-H bonds (Fig. S3). SAH and vitamin C interacted with DNMT1_B at Glu:562, Asp:565, Ser:570, Asp:571, Glu:572, Gln:594, Arg:595 Val:658 and Met:1232, Arg:1276, Ser:1277, Val:1344 residues, respectively. Nine conventional H and one alkyl bond were found in the case of SAH, while vitamin C exhibited four conventional H bonds (Fig. S3). The predicted site of 5-aza-dc, SAM and EGCG interaction is presented in Fig. 5B.
Docking study of DNMT3a showed that SAM, SAH, EGCG and vitamin C interacted with DNMT3a as follows: SAM, Gly:707, Cys:710, Arg:792, Arg:891 with four conventional H bonds; SAH, Phe:640, Pro:709, Glu:765, Arg:891 with four conventional H bonds and one each π-sulfur and alkyl bond; EGCG, Phe:640, Gly:642, Val:665, Ser:663, Gly:685, Asp:686, Gly:707, Pro:709, Asn:711, Arg:891 with five H, one C-H, one π-σ, one π-alkyl, one π-cation, two π-π T-shaped bond and vitamin C, Phe:640, Pro:709, Glu:765 and Arg:891 (Fig. S4). 5-aza-dc interacted with Phe:640, Gly:707, Glu:765, Arg:891, Trp:893 residues with five H and one π-σ bond (Fig. S4). The predicted site of SAM SAH, EGCG and vitamin C interaction is presented in Fig. 5C.
Docking study of DNMT3b showed that 5-aza-dc, SAM, SAH and EGCG interacted with DNMT3b as follows: 5-aza-dc, Asp:582, Ser:610, Arg:832, Trp:834 with two H and one π-σ bond; SAM, Glu:605, Val:606/628, Ser:649, Glu:697 residues with eight H bonds and one attractive charge interaction; SAH, Phe:581, Gly:583, Thr:586, Glu:605, Cys:607, Asp:627, Val:628, Arg:832, Trp:834 with seven H and one each C-H and alkyl bond and EGCG, Phe:581, Ser:610, Val:606/628, Pro:650, Cys:651, Arg:832 with four H and one each π-σ, π-sulfur, π-cation, π-alkyl and π-π T-shaped interactions (Fig. S5). DNMT3b and vitamin C interacted with Asp:582, Ile:584, Thr:586, Gly:648, Arg:832 with five H and one π-donor H bond (Fig. S5). The predicted site of SAM SAH, 5-aza-dc and EGCG interaction is presented in Fig. 5D.
EGCG, but not vitamin C, decreases 5mC level
As EGCG is a potent DNMTi (46), its effect on 5mC level in both cell lines. In both cell lines, 60 µM 5-aza-dc decreased 5mC level but this was only significant in MCF-7 cells (Fig. 6A and B). Additionally, 40 µM EGCG significantly decreased 5mC level in MCF-7 cell line (Fig. 6A). Furthermore, 20 µM EGCG induced a greater 5mC decrease in the T47D cell line (Fig. 6B). Vitamin C treatment significantly increased 5mc level in both cell lines (Fig. 6A and B). These data suggested that EGCG decreased 5mC level in breast cancer cell lines.
Figure 6.
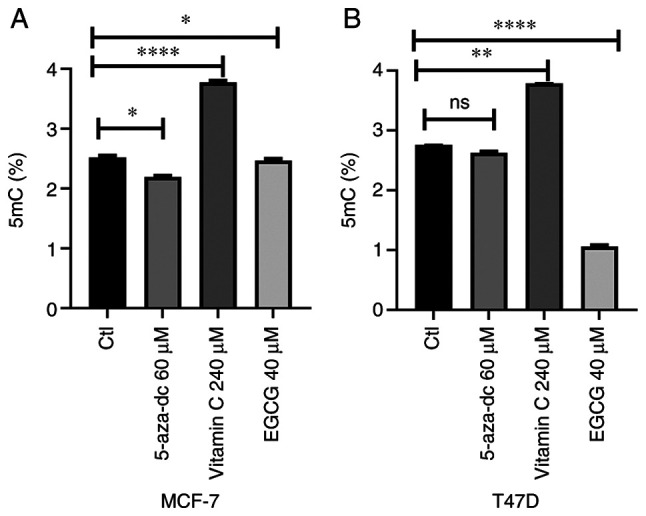
5mC level following treatment with DNA methyltransferase inhibitors. 5mC following 5-aza-dc, vitamin C and EGCG treatment in (A) MCF-7 and (B) T47D cell lines. The data are presented as the mean ± SD. *P<0.05, **P<0.01 and ****P<0.0001. 5mC, global methylation; EGCG, Epigallocatechin gallate; ns, not significant; Ctl, control.
EGCG induces IFI16 gene expression by decreasing DNA methylation
Since EGCG decreased 5mC level in both cell lines, promoter methylation status was assessed in IFI16 promoter. EGCG treatment decreased methylation signaling in the promoter of the IFI16 gene in MCF-7 cell line (Fig. 7A). The decrease in methylation signaling in IFI16 was greater in the T47D cell line than the MCF-7 cell line (Fig. 7B). The present data suggesting that EGCG decreases methylation level in the IFI16 gene promoter.
Figure 7.
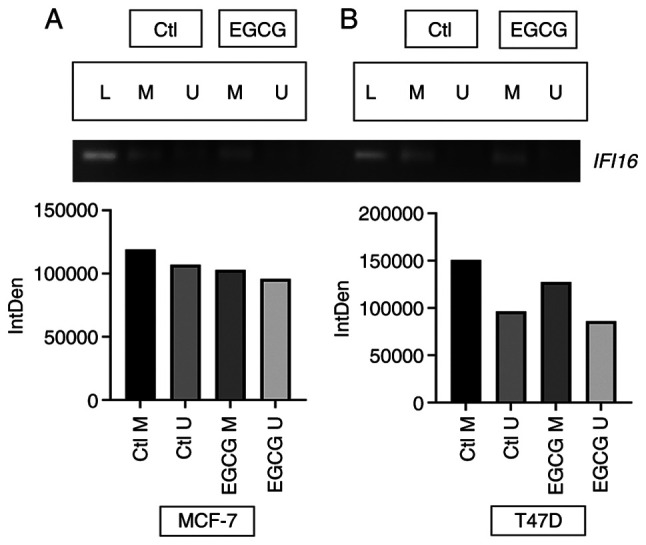
EGCG treatment demethylates IFI16 promoter. IFI16 gene promoter demethylation in (A) MCF-7 and (B) T47D cell lines following EGCG treatment. L, ladder; IntDen, Integrated density; Ctl, control; M, methylated; U, unmethylated; IFI, IFN-induced protein; EGCG, Epigallocatechin gallate.
Discussion
Nucleic acid sensors, such as IFI16 DNA sensor, generate an innate immune response in tumor cells (47). IFI16 senses ds nucleic acid ligands and relays molecular signaling to induce inflammatory innate immune response via IFN stimulation. Previous epigenetic studies of these sensors in various cancer cell lines have shown that DNMTi and vitamin C induce their expression (31,38,48). EGCG-mediated epi-transcriptomic based regulation of IFI16 sensor is still not well studied. Hence, the aim of the present study was to investigate this in breast cancer lines.
IFI16 gene expression was screened in cancer cell lines, including MCF-7, T47D, SHSY5, HepG2 and HeLa, from the Human Protein Atlas. The present study aimed to investigate epigenetic mechanisms, such as DNA methylation-mediated epigenetic transcriptional regulation of IFI16 gene; therefore, DNMT mRNA expression was assessed in cancer cell lines. The TCGA dataset for IFI16 and DNMT genes was validated in breast cancer cell line from online data sources (UALCAN and UCSC Xena browser). The present study found a negative association between IFI16 and DNMT gene expression. Hence, MCF-7 and T47D cell lines were treated with DNMTis, like EGCG, 5-aza-dc and vitamin C and expression of IFI16 and DNMTs was assessed. EGCG treatment induced IFI16 gene expression in both cell lines and this was greater in the MCF-7 cell line. As IFI16 stimulates IFN and ISG response, IFNβ1 and ISG15 expression was assessed; EGCG upregulated IFNβ1 gene expression in both cell lines and this was greatest in MCF-7 cell line. To the best of our knowledge, the effect of EGCG on expression of these genes has not been reported previously. Expression of these genes was measured following treatment with 60 µM 5-aza-dc or 240 µM vitamin C, which resulted in significant upregulation of mRNA expression for these genes. Previously, it has been shown that IFI16 and ISG15 gene expression is upregulated following treatment with vitamin C and 5-aza-dc alone or combination in HCT116, SNU398 and HL60 cancer cell lines (31). Additionally, 5-aza-dc treatment induces IFI16, ISG15 and IFNβ1 expression in ovarian cancer cell lines (48).
In addition, 5-aza-dc, vitamin C and EGCG treatment decreased DNMT1, DNMT3a and DNMT3b mRNA expression. A previous study showed that 20 µM EGCG and 5µM 5-aza-dc treatment decrease DNMTs mRNA expression and DNMT activity and reactivates tumor suppressor genes by decreasing promoter methylation levels in various breast cancer lines (49).
As 5-aza-dc, vitamin C and EGCG decreased DNMTs mRNA expression, 5mC levels were assessed post-treatment. In both MCF-7 and T47D cell lines, 5-aza-dc slightly decreased 5mC% level. Moreover, EGCG decreased 5mC levels in MCF-7 cells and a significant decrease in 5mC level in T47D cells was observed. A previous study on skin cancer cells revealed EGCG mediated declination of 5mC level and suggested that EGCG-mediated methylation is slow and might be more effective as a treatment when used for long periods (>72 h) (50). In the present study, no effect of vitamin C on the 5mC level was observed in any cell line. Binding of vitamin C outside of the binding pocket for DNMT1 and DNMT3b might be the reasons or any other factors that might be involved in the regulation of vitamin C-mediated 5mC regulation. A previous study showed the effect of vitamin C on methylation is reversible (51). DNMT3A activity regulation is governed by 5mC-binding protein MeCP2 and histone 3 tail modification (52). Ubiquitin-like With PHD and Ring Finger Domains 1 can regulate DNMT1 (53). Vitamin C may not functionally affect all these factors.
In silico molecular docking simulation showed that EGCG exhibited the greatest binding energy with DNMT proteins. 5-aza-dc showed the second highest binding energy with DNMTs. SAM and SAH were used as a control for docking simulation. The lowest binding energy was observed with vitamin C for all DNMTs. Additionally, docking interaction and predicted binding sites for all compounds showed that SAM and EGCG bound in a similar position across all DNMTs. Binding pocket was not specified during docking simulation to avoid bias docking; docking simulation demonstrated that compounds bind with certain amino acid residues in the binding pocket site. Similar amino acid residues interactions were also found in several previous molecular docking simulations of EGCG and DNMT (27,54,55).
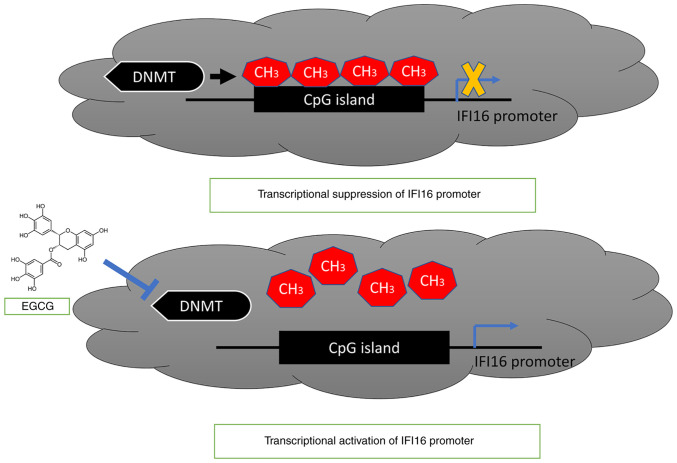
As EGCG mediated a decrease in 5mC level and induction of IFI16 gene expression, the effect of EGCG on status of IFI16 gene promoter was assessed. Following EGCG treatment, methylation signaling was decreased in both cell lines. MCF-7 cell lines showed low decrease in methylation signaling following EGCG treatment, whereas T47D showed a greater decrease in methylation signaling following EGCG treatment. This may be due to low decrease in 5mC level in EGCG-treated MCF-7 cell line and greater 5mC decrease in EGCG-treated T47D cell line. However, a recent study revealed that IFI16 is hypomethylated in glioblastoma (56). To the best of our knowledge, EGCG-mediated reactivation of the IFI16 gene by decreasing promoter methylation has not previously been reported. IFI16 gene promoter methylation is DNMT-mediated, whereas EGCG treatment can block the DNMTs activity and reduce the methylation of IFI16 gene promoter, thus activating transcription of IFI16 gene (Fig. 8).
Figure 8.
EGCG effect on IFI16 gene promoter. The proposed model showed IFI16 gene promoter is methylated by DNMT-mediated catalysis of methyl group incorporation. EGCG treatment can block the DNMT activity and in turn decrease methylation level in IFI16 gene promoter, leading to transcriptional activation of IFI16 gene. IFI, IFN-induced protein; EGCG, Epigallocatechin gallate; DNMT, DNA methyltransferase.
As a potent natural DNMTi, EGCG induces expression of innate immune sensor IFI16 by decreasing promoter methylation in breast cancer cells. The present findings provide a basis to investigate whether natural DNMTis (such as EGCG) exert beneficial effects by inducing immune response.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Deanship Scientific Research at King Abdulaziz University, Jeddah (grant no. G:257-130-1441).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MIK designed and conceptualized the study. SMN and WHA performed experiments, analyzed the data and wrote the manuscript. MIK and WHA confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Choubey D, Panchanathan R. IFI16, an amplifier of DNA-damage response: Role in cellular senescence and aging-associated inflammatory diseases. Ageing Res Rev. 2016;28:27–36. doi: 10.1016/j.arr.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Ouchi M, Ouchi T. Role of IFI16 in DNA damage and checkpoint. Front Biosci. 2008;13:236–239. doi: 10.2741/2673. [DOI] [PubMed] [Google Scholar]
- 3.Duan X, Ponomareva L, Veeranki S, Panchanathan R, Dickerson E, Choubey D. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol Cancer Res. 2011;9:589–602. doi: 10.1158/1541-7786.MCR-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stratmann SA, Morrone SR, van Oijen AM, Sohn J. The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex. ELife. 2015;4:e11721. doi: 10.7554/eLife.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choubey D, Deka R, Ho SM. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci. 2008;13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- 8.Johnstone RW, Trapani JA. Transcription and growth regulatory functions of the HIN-200 family of proteins. Mol Cell Biol. 1999;19:5833–5838. doi: 10.1128/MCB.19.9.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: More than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Choubey D, Duan X, Dickerson E, Ponomareva L, Panchanathan R, Shen H, Srivastava R. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: Role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30:371–380. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veeranki S, Choubey D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: Regulation of subcellular localization. Mol Immunol. 2012;49:567–571. doi: 10.1016/j.molimm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21:548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhat N, Fitzgerald KA. Recognition of cytosolic DNA by cGAS and other STING-dependent sensors. Eur J Immunol. 2014;44:634–640. doi: 10.1002/eji.201344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, Fitzgerald KA, Kurt-Jones EA. Interferon γ-inducible Protein (IFI) 16 transcriptionally regulates Type I interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem. 2014;289:23568. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopitar-Jerala N. The role of interferons in inflammation and inflammasome activation. Front Immunol. 2017;8:873. doi: 10.3389/fimmu.2017.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itsui Y, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, Koyama T, Takeda Y, Nakagawa M, Kakinuma S, Sekine Y. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J Viral Hepat. 2006;13:690–700. doi: 10.1111/j.1365-2893.2006.00732.x. [DOI] [PubMed] [Google Scholar]
- 17.Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xin H, Curry J, Johnstone RW, Nickoloff BJ, Choubey D. Role of IFI 16, a member of the interferon-inducible p200-protein family, in prostate epithelial cellular senescence. Oncogene. 2003;22:4831–4840. doi: 10.1038/sj.onc.1206754. [DOI] [PubMed] [Google Scholar]
- 20.Duan X, Ponomareva L, Veeranki S, Choubey D. IFI16 induction by glucose restriction in human fibroblasts contributes to autophagy through activation of the ATM/AMPK/p53 pathway. PLoS One. 6:e19532, 20122. doi: 10.1371/journal.pone.0019532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin W, Zhao Z, Ni Z, Zhao Y, Du W, Chen S. IFI16 restoration in hepatocellular carcinoma induces tumour inhibition via activation of p53 signals and inflammasome. Cell Prolif. 2017;50:e12392. doi: 10.1111/cpr.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo Y, Nagai K, Nakahata S, Saito Y, Ichikawa T, Suekane A, Taki T, Iwakawa R, Enari M, Taniwaki M, et al. Overexpression of the DNA sensor proteins, absent in melanoma 2 and interferon-inducible 16, contributes to tumorigenesis of oral squamous cell carcinoma with p53 inactivation. Cancer Sci. 2012;103:782–790. doi: 10.1111/j.1349-7006.2012.02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu B, Zheng X, Sun Z, Cao P, Zhang J, Wang W. IFI16 can be used as a biomarker for diagnosis of renal cell carcinoma and prediction of patient survival. Front Genet. 2012;12:599952. doi: 10.3389/fgene.2021.599952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JX, Cheng CS, Gao HF, Chen ZJ, Lv LL, Xu JY, Shen XH, Xie J, Zheng L. Overexpression of interferon-inducible Protein 16 promotes progression of human pancreatic adenocarcinoma through interleukin-1β-induced tumor-associated macrophage infiltration in the tumor microenvironment. Front Cell Dev Biol. 2021;9:640786. doi: 10.3389/fcell.2021.640786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Intra J, Kuo SM. Physiological levels of tea catechins increase cellular lipid antioxidant activity of vitamin C and vitamin E in human intestinal Caco-2 cells. Chem Biol Interact. 2007;169:91–99. doi: 10.1016/j.cbi.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cione E, La Torre C, Cannataro R, Caroleo MC, Plastina P, Gallelli L. Quercetin, epigallocatechin gallate, curcumin, and resveratrol: From dietary sources to human MicroRNA modulation. Molecules. 2019;25:63. doi: 10.3390/molecules25010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 28.Beetch M, Harandi-Zadeh S, Shen K, Lubecka K, Kitts DD, O'Hagan HM, Stefanska B. Dietary antioxidants remodel DNA methylation patterns in chronic disease. Br J Pharmacol. 2020;177:1382–1408. doi: 10.1111/bph.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo CL, Chen TS, Liou SY, Hsieh CC. Immunomodulatory effects of EGCG fraction of green tea extract in innate and adaptive immunity via T regulatory cells in murine model. Immunopharmacol Immunotoxicol. 2014;36:364–370. doi: 10.3109/08923973.2014.953637. [DOI] [PubMed] [Google Scholar]
- 30.Nance Cl, Mata M, McMullen A, McMullen A, Shearer WT. Regulation of innate immune recognition of viral infection by epigallocatechin gallate. J Allergy Clin Immunol. 2014;133:AB246. doi: 10.1016/j.jaci.2013.12.876. [DOI] [Google Scholar]
- 31.Liu M, Ohtani H, Zhou W, Ørskov AD, Charlet J, Zhang YW, Shen H, Baylin SB, Liang G, Grønbæk K, Jones PA. Vitamin C increases viral mimicry induced by 5-aza-2′-deoxycytidine. Proc Natl Acad Sci USA. 2016;113:10238–10244. doi: 10.1073/pnas.1612262113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alimirah F, Chen J, Davis FJ, Choubey D. IFI16 in human prostate cancer. Mol Cancer Res. 2007;5:251–259. doi: 10.1158/1541-7786.MCR-06-0269. [DOI] [PubMed] [Google Scholar]
- 34.Takeshima H, Yoda Y, Wakabayashi M, Hattori N, Yamashita S, Ushijima T. Low-dose DNA demethylating therapy induces reprogramming of diverse cancer-related pathways at the single-cell level. Clin Epigenetics. 2020;12:142. doi: 10.1186/s13148-020-00937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris J, Moseley VR, Cabang AB, Coleman K, Wei W, Garrett-Mayer E, Wargovich MJ. Reduction in promotor methylation utilizing EGCG (epigallocatechin-3-gallate) restores RXRα expression in human colon cancer cells. Oncotarget. 2016;7:35313–35326. doi: 10.18632/oncotarget.9204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CY, Han Z, Li X, Xie HH, Zhu SS. Mechanism of EGCG promoting apoptosis of MCF-7 cell line in human breast cancer. Oncol Lett. 2017;14:3623–3627. doi: 10.3892/ol.2017.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moradzadeh M, Hosseini A, Erfanian S, Rezaei H. Epigallocatechin-3-gallate promotes apoptosis in human breast cancer T47D cells through down-regulation of PI3K/AKT and Telomerase. Pharmacol Rep. 2017;69:924–928. doi: 10.1016/j.pharep.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rialdi A, Campisi L, Zhao N, Lagda AC, Pietzsch C, Ho JSY, Martinez-Gil L, Fenouil R, Chen X, Edwards M, et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016;352:aad7993. doi: 10.1126/science.aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Islam MR, Awal MA, Khames A, Abourehab MAS, Samad A, Hassan WMI, Alam R, Osman OI, Nur SM, Molla MHR, et al. Computational identification of druggable bioactive compounds from catharanthus roseus and avicennia marina against colorectal cancer by targeting thymidylate synthase. Mol. 2022;27:2089. doi: 10.3390/molecules27072089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikwu FA, Shallangwa GA, Mamza PA, Uzairu A. In silico studies of piperazine derivatives as potent anti-proliferative agents against PC-3 prostate cancer cell lines. Heliyon. 2020;6:e03273. doi: 10.1016/j.heliyon.2020.e03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zevini A, Olagnier D, Hiscott J. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017;38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nur SM, Rath S, Ahmad V, Ahmad A, Ateeq B, Khan MI. Nutritive vitamins as epidrugs. Crit Rev Food Sci Nutr. 2021;61:1–13. doi: 10.1080/10408398.2020.1712674. [DOI] [PubMed] [Google Scholar]
- 45.de Ruyck J, Brysbaert G, Blossey R, Lensink MF. Molecular docking as a popular tool in drug design, an in silico travel. Adv Appl Bioinform Chem. 2016;9:1–11. doi: 10.2147/AABC.S105289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Won JL, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005;68:1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 47.Okude H, Ori D, Kawai T. Signaling through nucleic acid sensors and their roles in inflammatory diseases. Front Immunol. 2021;11:625833. doi: 10.3389/fimmu.2020.625833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. Erratum: Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;152:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng J, Shi W, Guo H, Long W, Wang Y, Qi J, Liu J, Xu Y. The inhibitory effect of (−)-Epigallocatechin-3-Gallate on breast cancer progression via reducing SCUBE2 methylation and DNMT activity. Molecules. 2019;24:2899. doi: 10.3390/molecules24162899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p 16 INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajavelu A, Lungu C, Emperle M, Dukatz M, Bröhm A, Broche J, Hanelt I, Parsa E, Schiffers S, Karnik R, et al. Chromatin-dependent allosteric regulation of DNMT3A activity by MeCP2. Nucleic Acids Res. 2018;46:9044–9056. doi: 10.1093/nar/gky715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bostick M, Jong KK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 54.Khan MA, Hussain A, Sundaram MK, Alalami U, Gunasekera D, Ramesh L, Hamza A, Quraishi U. (−)-Epigallocatechin-3-gallate reverses the expression of various tumor-suppressor genes by inhibiting DNA methyltransferases and histone deacetylases in human cervical cancer cells. Oncol Rep. 2015;33:1976–1984. doi: 10.3892/or.2015.3802. [DOI] [PubMed] [Google Scholar]
- 55.Yiannakopoulou EC. Targeting DNA methylation with green tea catechins. Pharmacology. 2015;95:111–116. doi: 10.1159/000375503. [DOI] [PubMed] [Google Scholar]
- 56.Alivand MR, Najafi S, Esmaeili S, Rahmanpour D, Zhaleh H, Rahmati Y. Integrative analysis of DNA methylation and gene expression profiles to identify biomarkers of glioblastoma. Cancer Genet. 2021:258–259. 135–150. doi: 10.1016/j.cancergen.2021.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.